Abstract
In 1984, we investigated the t(14;18) chromosomal translocations that frequently occur in patients with follicular lymphoma. We first identified a locus on chromosome 18 involved in these translocations with the chromosome 14 containing the immunoglobulin heavy chain locus. Within this region on chromosome 18, we then discovered a gene that we called BCL2, which was activated by the translocations. Since that time, many studies determined that BCL2 is one of the most important oncogenes involved in cancer by inhibiting apoptosis. In 2002, we studied 13q deletions in chronic lymphocytic leukemia (CLL) and found that the microRNA cluster miR-15a/miR-16-1 (miR-15/16) is deleted by 13q deletions. In 2005, we discovered that miR-15/16 function as tumor suppressors by directly targeting BCL2. Thus the loss of two negative regulators of BCL2 expression results in overexpression of BCL2. Very recently, a specific BCL2 inhibitor ABT-199 (Venetoclax) was developed and approved by FDA for CLL treatment. Thus it took 32 years from fundamental discovery of a critical oncogene to the development of a drug capable to cure CLL. In this review, we discuss the discovery, functions and clinical relevance of miR-15/16 and BCL2.
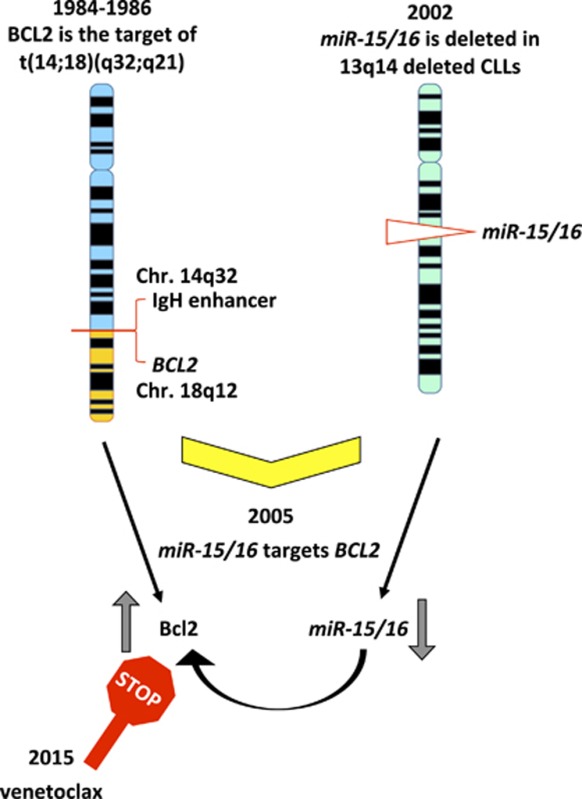
Graphical Abstract
Facts
BCL2 is a key oncogene involved in human cancer by inhibiting apoptosis.
miR-15/16 is a tumor-suppressor microRNA cluster deleted in most CLLs.
The miR-15/16 cluster targets BCL2.
Venetoclax is a specific BCL2 inhibitor that can cure CLL.
Open Questions
Are there other important targets of miR-15/16 in CLL?
Can miR-15/16 itself be of pharmaceutical use in CLL?
Can Venetoclax be used in combination with inhibitors of other miR-15/16 targets?
Discovery of BCL2
In 1984, we studied t(14;18) chromosomal translocations that almost invariably occurred in patients with follicular lymphoma, a malignant B-cell lymphoma. At first, we identified and cloned a locus on chromosome 18 involved in this translocation.1 We then cloned recombinant DNA probes spanning the breakpoint on chromosome 18 from cells derived by patients with follicular lymphoma carrying this translocation.1, 2 These probes could detect DNA rearrangements in most of the cases of follicular lymphoma, and almost all chromosome 18 rearrangements were clustered within a short DNA fragment.1, 2 Northern blotting analysis using previously obtained DNA probes detected several transcripts of approximately 6 kb in length in various tissues.1, 2 By screening cDNA libraries using breakpoint DNA probes and sequencing obtained clones, we determined the DNA sequence of the BCL2 gene and the protein sequence of the encoded BCL2 protein.3 Since that time, many studies contributed to determine that BCL2 is one of the most important oncogenes involved in cancer by inhibiting apoptosis4 and causes lymphoma development, particularly in context with c-MYC overexpression.5 The antiapoptotic function of BCL2 has been described and reviewed numerous times and newest advances in this field are described in other articles of this issue; thus in this review, we will focus on the regulation of BCL2 expression by microRNAs.
Chronic Lymphocytic Leukemia, 13q Deletions and miR-15/16
Chronic lymphocytic leukemia (CLL) is the most common human leukemia. CLL represents >30% of all lymphoid malignancies diagnosed each year in the United States and represents one-third of all leukemia cases.6 Interestingly, CLL is more common in men than in women. Very often, newly diagnosed CLL patients can live with mild disease and can survive without treatment for many years.6 Leukemic CLL cells can survive in tissue culture conditions for several weeks but usually do not proliferate and morphologically look similar to mature B-cells.6, 7 CLL is a heterogeneous disease and mostly defined as an expansion of a rare population of CD5-positive B-cells, as CD5 is commonly expressed on T-cells and not on mature B-cells.6, 7 There are several prognostic markers of CLL. For the most part, CLL patients showing high ZAP-70 (zeta-chain-associated protein kinase 70) expression and unmutated IgH VH have a clinically aggressive form of CLL and need treatment sooner than patients expressing low levels of ZAP-70 and mutated IgH VH, though this general rule has many exceptions.6, 7
CLL is a fascinating disease to study as almost all CLL cases (>80%) show genomic aberrations, and in most cases, these rearrangements occur at several chromosomal locations. The most common chromosomal abnormalities detectable in CLL cells include deletions at 13q (60%), 11q (18%), 17p (8%), and trisomy 12 (12–16%).8, 9 Recent studies demonstrated that trisomy 12 in aggressive CLL is associated with NOTCH1 (Notch homolog 1, translocation-associated (Drosophila)) activating mutations;10 17p deletions often involve inactivation of TP53 and miR-3676 (which targets TCL1, a critical oncogene in aggressive CLL);11, 12 and 11q deletions may involve the ATM (ataxia-telangiectasia mutated) gene.13 Generally, patients showing 11q and 17p deletions have more aggressive clinical course than patients with 13q deletions.9
In 2000–2002, we studied 13q14 deletions in CLL to determine which gene(s) are targets of this most common deletion in CLL. We believed that 13q14 had to contain a very important tumor-suppressor gene involved in the pathogenesis of CLL. Interestingly, in addition to CLL, the same region is often deleted in other malignancies. For example, almost 50% of mantle cell lymphomas (a rare type of B-cell non-Hodkin’s lymphoma) have genomic aberrations at 13q14,14 and the same region is frequently deleted in ~60% of prostate cancers.15 We and many other research groups extensively studied the 13q14 genomic region using positional cloning and sequencing approaches. We were able to sequence a region of >1 Mb within the 13q14-deleted region in our attempts to identify a specific target of 13q14 deletions.16, 17
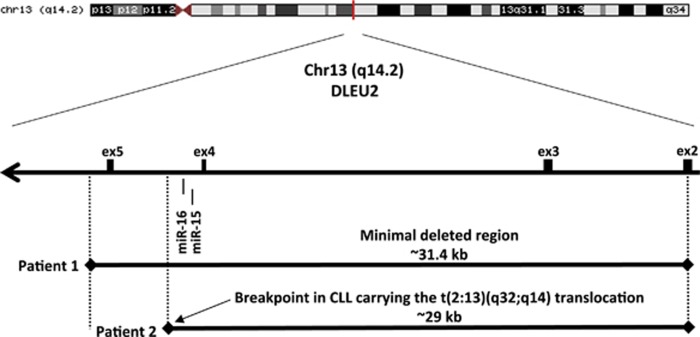
Despite the extensive amount of work including loss of heterozygosity (LOH) studies, sequencing and expression analysis of all protein-coding genes within and around the 13q14-deleted region, the affected gene(s) remained elusive, as no point mutations or loss of expression of any conventional genes were found.16, 17, 18, 19 In 2001–2002, we worked with several interesting CLL cases showing small deletions and translocations at 13q14. We generated several somatic cell hybrids to isolate the relevant chromosomes that included a rearranged 13q34 allele. One of these hybrids (derived from CLL cells of Patient 1) contained a small (~30 kb) deletion located between exons 2 and 5 of the DLEU2 gene (Figure 1). Another hybrid (derived from CLL cells of Patient 2) contained a translocation breakpoint within the same region.20, 21 Interestingly, DLEU2 itself was extensively studied previously, and no mutations in the coding region or downregulation of its expression were found.7, 20, 21 Thus by investigating these cases with a small deletion and a translocation, we came to the conclusion that the tumor suppressor must have been located within a 30 kb deletion. But all protein-coding genes were excluded. At that time, genomic databases were frequently updated, one of these updates contained microRNAs, and we finally found that a cluster of two microRNA genes, miR-15a and miR-16-1, was located at the translocation breakpoint (Figure 1).20 As miR-15/16 was the only gene in the region, we carried out a study to determine its expression in CLL. Northern blotting analysis of 60 13q-deleted specimens derived from patients with CLL revealed that expression of miR-15/16 was downregulated in approximately 70% of CLL cases compared with normal CD5+ B-cells.20 As none of the other genes in the region showed any involvement in CLL,16, 17, 21 and as the translocation breakpoint and the small deletion involved the miR-15/16 loss, we concluded that miR-15/16 is a target of 13q deletions in CLL.20 This was the first example that an alteration in the non-coding genome could be involved in cancer pathogenesis.20 MicroRNAs are a large class of non-coding RNAs involved in almost all cellular processes through the regulation of gene expression.22 MicroRNAs are first produced as long precursor molecules and then processed into hairpin structures of about 70–100 nt in length. These 70–100-nt stem-loop precursors are then processed into microRNAs.23 For the most part, mature microRNA molecules bind the 3′ UTRs of mRNAs through the conserved seed sequences located at the 5′-ends of the microRNAs by partial complementarity.23 This binding causes mRNA degradation and/or inhibits mRNA translation, resulting in decreased levels of gene expression.24
Figure 1.
miR-15/16 in 13q14-deleted region
The miR-15 and miR-16 Target BCL2
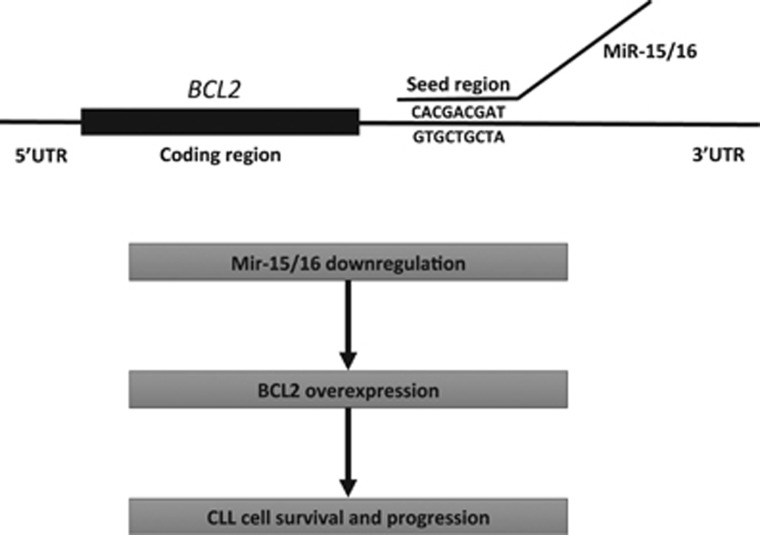
As we found that the miR-15/16 is lost in 13q14-deleted CLL cells, we hypothesized that these microRNAs might target important oncogenes responsible for B-cell transformation. Thus we searched available databases to identify oncogenes among the predicted miR-15/16 targets and focused on genes with possible role on CLL pathogenesis. Remarkably among top predicted targets in this search, we found BCL2, the gene we discovered in 1984, and found overexpressed in almost all CLL cases.25 By analyzing complementarity between miR-15/16 and the BCL2 mRNA, we found that the first 9 nucleotides, from the 5′-ends of both miR-15 and miR-16, are complementary to bases 3287–3279 in the 3′-end of the BCL2 cDNA.25 As mentioned above, BCL2 has a critical role in the malignant transformation of solid tissues as well as lymphoid cells. BCL2 protein mostly localizes in mitochondria, and it promotes survival and inhibits apoptosis by preventing the release of Cytochrome C from mitochondria into the cytoplasm.26, 27 Several B-cell malignancies (including follicular lymphomas and a fraction of diffuse B-cell lymphomas) carry a t(14;18)(q32;q21) chromosomal translocation. These translocations place immunoglobulin heavy-chain enhancers located at 14q32 near the BCL2 promoter resulting in upregulated BCL2 expression.1, 2 Interestingly, however, translocations involving 18q21 (where BCL2 is located) occur only in a very small minority of CLLs, whereas, on the other hand, overexpression of BCL2 is a hallmark of CLL.28, 29 Thus it seemed likely that BCL2 overexpression in CLL might be due to the deletion of miR-15/16 at 13q14. First, we studied the expression of miR-15, miR-16 and BCL2 in CLL and found that expression of both miR-15 and miR-16 was highest in the CLL samples with lowest BCL2 expression and was lowest in the CLL samples showing high BCL2 expression. We also determined that, in MEG-01 leukemic cells that do not express any miR-15/16, the exogenous expression of miR-15/16 resulted in BCL2 repression and induction of apoptosis. This was evidenced by cleavage of poly (ADP-ribose) polymerase and of pro-caspase 9.25 Interestingly, no apoptosis was detected when using mutated miR-15/16 (this mutation was found in two CLL patients, see below).
To demonstrate that miR-15/16 directly targeted BCL2, we carried out a series of luciferase assay experiments. We cloned a small fragment of the 3′ UTR of BCL2, which was predicted to interact with miR-15/16, into the 3′ UTR of the luciferase gene. Expression of miR-15/16 significantly decreased luciferase activity when the WT construct was used, whereas no difference was observed when a construct containing a mutated microRNA–mRNA interaction site was used.25 This showed that both microRNAs directly interact with and inhibit BCL2 expression (Figure 2).25 Overall, these results indicate that the loss of miR-15/16 is the main cause of BCL2 overexpression in CLL (Figure 2).
Figure 2.
miR-15/16 target BCL2 expression
miR-15/16: Mechanisms of Deregulation Beyond deletion
As our initial observation that miR-15/16 is deleted or downregulated in most of CLLs, additional studies indicated that miR-15/16 could also be downregulated by additional mechanisms. Allegra et al.30 showed that some CLLs have defective DROSHA processing that contributes to the loss of miR-15/16 expression. Sampath et al.31 and Allegra et al.30 demonstrated that histone deacetylases are overexpressed in CLL leading to the aberrant epigenetic silencing of miR-15/16 expression. Thus miR-15/16 can also be silenced by an epigenetic mechanism. As tumor suppressors are frequently mutated, and most of these mutations are loss-of-function mutations, it is very important to identify and characterize such mutations. Thus it is possible that mutations can also contribute to the loss of miR-15/16 in CLL. To identify such mutations, we carried out an additional study analyzing 75 CLL cases and 160 normal controls for mutations of several microRNAs, including miR-15/16.32 We found one germline mutation in the pre-miR-15/16 sequence in CLL samples derived from two CLL patients, and no mutations were found in the controls. This C-to-T mutation is located only seven base pairs after the end of the miR-16-1 precursor.32 We also analyzed this genomic region in all primates and determined that this region is strongly conserved, indicating that this mutation can have an important role in the processing of miR-16-1. This was confirmed by studying miR-16-1 expression levels in both patients by microchip analysis and Northern blotting. Both methods showed a significant decrease of expression of miR-16-1 in both patients.32 In addition, both patients had a monoallelic deletion at 13q14 while in their normal DNA the mutation was heterozygous. Thus in CLL cells from these patients one allele of miR-16-1 was deleted and the other one was mutated. Interestingly, the mother of one of these patients also had CLL and the sister of the same patient had breast cancer, suggesting an important role for this mutation or rare polymorphism.32
We also functionally tested this mutation and showed that it is a loss-of-function mutation, resulting in the decrease of miR-15/16 expression.32 All the data discussed above clearly showed that miR-15/16 is a target of 13q14 deletions and can also be mutated and downregulated by other mechanisms.
miR-15/16 Upstream and downstream Mechanisms
After these results, we investigated whether miR-15/16 functions as a tumor suppressor in vivo. We transiently transfected MEG-01 cells, which lack miR-15/16 expression, with miR-15/16 or empty vector and tested their tumorigenic activity upon inoculation in immunosuppressed mice. After 4 weeks, no tumors originated from cells transfected with miR-15/16, whereas, in sharp contrast, large tumors originated from cells transfected with the empty vector.33 Thus we concluded that miR-15 and miR-16 function as a tumor suppressor in MEG-01 leukemia cells. We also investigated the transcriptional and translational effects of miR-15/16 exogenous expression in MEG-01 cells. We found that miR-15/16 expression resulted in the induction of transcription of 265 genes and suppression of 3300 genes. Interestingly, among the repressed genes we found 85 predicted miR-15/16 targets. We also identified 27 proteins repressed by miR-15/16 using proteomics analysis (including BCL2). Most of these proteins were found to be involved in tumorigenesis, apoptosis or cell growth and eight of these proteins were predicted miR-15/16 targets.33
By analyzing the genomic region surrounding miR-15/16, we found several TP53-binding sites upstream of the cluster.34 To prove that p53 was bound to these sites, we carried out chromatin immunoprecipitation experiments. These analyses revealed that indeed p53 binds upstream of miR-15/16 cluster. Further experiments revealed that p53 transactivates the miR-15/16 cluster.34 Interestingly, the 3′ UTR of TP53 also contains binding sites for miR-15 and miR-16. We carried out a series of luciferase experiments and found that indeed these sites are targeted by miR-15/16. These results showed the interplay between miR-15/16 and p53: activation of p53 results in miR-15/16 overexpression, resulting in increased targeting of TP53 by miR-15/16, and decrease of p53 expression.34
Since our discovery that miR-15/16 targets BCL2, several reports described other important targets of miR-15/16. For example, miR-15/16 was found to inhibit Cyclin D1 in several malignancies, including bladder cancer and osteosarcoma.35, 36 MCL1 oncogene in CLL and the BMI1 oncogene in mantle call lymphoma were also validated as miR-15/16 targets.33, 37
Interestingly, miR-15/16 has tumor-suppressor functions not only in lymphoid malignancies. Reid et al.38 showed that miR-15/16 expression is inactivated in malignant pleural mesothelioma. Transfection of synthetic oligos mimicking miR-15/16 resulted in growth inhibition in mesothelioma cell lines and in reduction of tumor sizes in xenograft models.38
Mouse Models for miR-15/16
Supporting evidence for the tumor-suppressor activity of any gene is a tumor phenotype resulting upon its knockout in mouse models. To verify the function of miR-15/16, Klein et al.39 generated miR-15/16 knockout mouse models. Two knockout alleles were generated. One allele had the minimal deleted region (MDR) containing miR-15/16 and the Dleu2 gene; another allele had only miR-15/16 deleted.39 Both mouse models developed CD5+ B-cell proliferation and/or malignancies. At the age of 1 year, MDR mice had 50% of CD5 positive B-cells in their spleens similarly to miR-15/16 mice, which had a slightly lower number.39 At the age of 18 months, 26% of miR-15/16 mice and 42% of MDR mice developed B-cell proliferation or malignancies, while no WT controls developed any lymphoid malignancies.39 Generally, the MDR knockout phenotype was more severe than that of miR-15/16, suggesting that other sequences in the MDR locus may additionally contribute to the tumor-suppressor function of miR-15/16.39 Another interesting mouse model of CLL is the New Zealand Black (NZB) mouse strain.40 NZB is a naturally occurring mouse strain that late in life develops a CD5+ B-cell proliferation similar to CLL.40 Dr. Raveche’s laboratory carried out a genome-wide linkage study attempting to identify genomic loci responsible for the development of such CLL-like disease in NZB mice.41 They identified three genomic regions linked to the disease, which were located on chromosomes 14, 18, and 19. Intriguingly, the region on NZB chromosome 14 is homologous to the human 13q14 and contains mouse Dleu2 and miR-15/16.41 Sequencing of the mouse miR-15/16 region revealed a point mutation resembling a mutation previously found in miR-15/16 in human CLL samples.32, 41 Further investigation showed that this NZB mutation caused reduced the expression of miR-16 in these mice.41 These data confirmed the importance of miR-15/16 loss in CLL pathogenesis.
The tumor-suppressor function of miR-15a/16-1 cluster (referred above as miR-15/16) is well established as it is deleted, mutated and downregulated by other mechanisms in CLL and other cancers. The second member of miR-15/16 family (miR-15b/16-2 cluster) is located at 3q25.42 As miR-15b/16-2 is almost identical to miR-15a16-1, to determine whether miR-15b/16-2 could have any role in cancer, we generated miR-15b/16-2 knockout mice and studied their phenotype.42 Pathological examination of these knockout mice revealed enlarged spleens, twofolds to fourfolds compared with wild-type mice. The miR-15b/16-2 knockout mice developed CD5+ B-cell proliferation and lymphomas at the age of 15–18 months with a penetrance of 60%. FACS analysis revealed expanded CD5+ B-cell proliferation in the spleens of these mice. Interestingly, we observed only mild upregulation of BCL2 expression in these knockout mice, on the other hand we found more robust upregulation of several other predicted miR-15/16 targets (Ccnd1, Ccnd2, and Igf1r1) in spleen lymphocytes from these mice.42
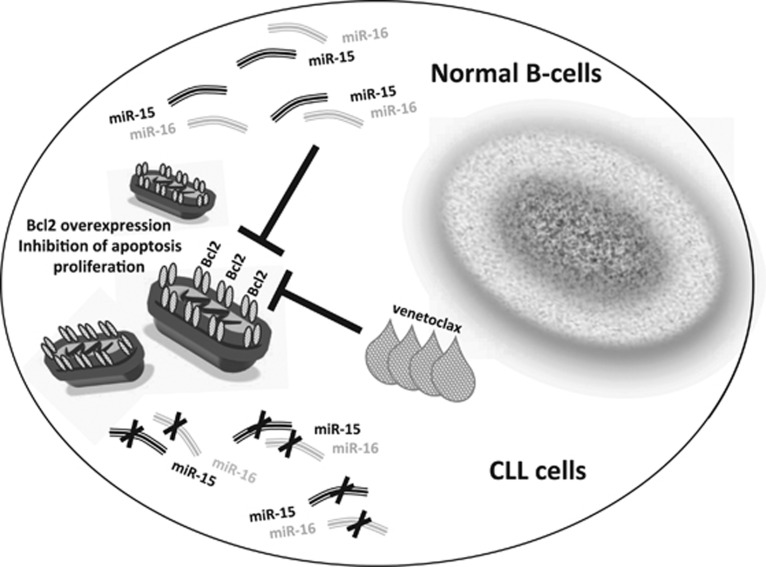
Venetoclax, a BCL2 Inhibitor in CLL
Generally, microRNAs are not used directly as drugs, mostly because they cannot be easily delivered in 100% of malignant cells. Thus, at present, miR-15/16 cannot be used as an anti-CLL drug directly. As we discussed above, BCL2 is overexpressed in almost all CLLs and could therefore be used as a target for development of drugs against CLL. There were several attempts in the past to develop BCL2 inhibitors. Most of them inhibited not only BCL2 but also other members of the BCL2 family, including BCLXL, making them toxic. More recently, Abbott was able to develop a drug targeting exclusively BCL2.43 The drug named ABT-199 (venetoclax) targets protein–protein interactions of BCL2. Venetoclax can induce complete remission in CLL patients, even in previously treated relapsed patients with 17p deletions, which are normally the most aggressive and difficult to treat.44 But even in these cases, venetoclax has a response rate of 80% and was recently approved by the FDA to treat these CLL cases (Figure 3).44 Venetoclax also showed promising results in treating different B-cell malignancies, including large B-cell lymphoma and follicular lymphoma.44
Figure 3.
Therapeutic implications of miR-15/16 targeting BCL2
Conclusions
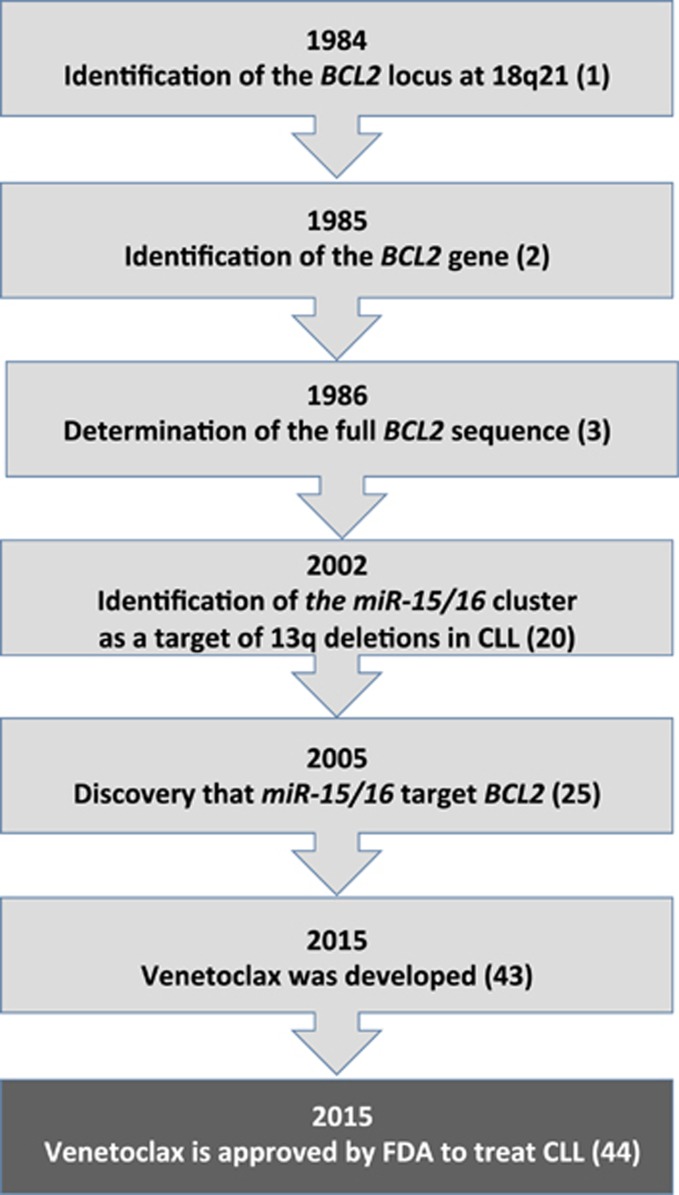
Since we first identified a locus on chromosome 18 involved in translocations in patients with follicular lymphoma and discovered a gene we named BCL2 that is activated by these translocations, it took >30 years to develop a drug inhibiting BCL2 and capable to cure CLL. Meanwhile, many studies showed the importance of BCL2 in many cancers, including other hematological malignancies. During this time, we identified the major regulators of BCL2 expression, miR-15 and miR-16, which are the first example of tumor-suppressor microRNAs, deleted or downregulated in >70% of CLLs. Thus, in the past 30 years, this research completed a full circle from the discovery of BCL2 and miR-15/16 to cancer treatment through a precision medicine approach (Figure 4).
Figure 4.
Major steps in the discovery of BCL2 and miR-15/16
Footnotes
Edited by F Pentimalli
The authors declare no conflict of interest.
References
- Tsujimoto Y, Finger LR, Yunis J, Nowell PC, Croce CM. Cloning of the chromosome breakpoint of neoplastic B cells with the t(14;18) chromosome translocation. Science 1984; 226: 1097–1099. [DOI] [PubMed] [Google Scholar]
- Tsujimoto Y, Cossman J, Jaffe E, Croce CM. Involvement of the bcl-2 gene in human follicular lymphoma. Science 1985; 228: 1440–1443. [DOI] [PubMed] [Google Scholar]
- Tsujimoto Y, Croce CM. Analysis of the structure, transcripts, and protein products of bcl-2, the gene involved in human follicular lymphoma. Proc Natl Acad Sci USA 1986; 83: 5214–5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaux DL, Cory S, Adams JM. Bcl-2 gene promotes haemopoietic cell survival and cooperates with c-myc to immortalize pre-B cells. Nature 1988; 335: 440–442. [DOI] [PubMed] [Google Scholar]
- Strasser A, Harris AW, Bath ML, Cory S. Novel primitive lymphoid tumours induced in transgenic mice by cooperation between myc and bcl-2. Nature 1990; 348: 331–333. [DOI] [PubMed] [Google Scholar]
- Sgambati M, Linet M, Devesa S. Chronic lymphocytic leukemia, epidemiological, familial, and genetic aspects. In: Cheson BD (ed). Chronic Lymphocytic Leukemias, Second Edition, Revised and Expanded,, Marcel Dekker, Inc: New York, USA, pp 33–622001. [Google Scholar]
- Bullrich F, Croce C. Molecular biology of chronic lymphocytic leukemia. In: Cheson BD (ed). Chronic Lymphocytic Leukemias, Second Edition, Revised and Expanded. Marcel Dekker, Inc: New York, USA, pp 9–322001. [Google Scholar]
- Edelmann J, Holzmann K, Miller F, Winkler D, Buhler A, Zenz T et al. High-resolution genomic profiling of chronic lymphocytic leukemia reveals new recurrent genomic alterations. Blood 2012; 120: 4783–4794. [DOI] [PubMed] [Google Scholar]
- Dohner H, Stilgenbauer S, Benner A, Leupolt E, Krober A, Bullinger L et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med 2000; 343: 1910–1916. [DOI] [PubMed] [Google Scholar]
- Balatti V, Bottoni A, Palamarchuk A, Alder H, Rassenti LZ, Kipps TJ et al. NOTCH1 mutations in CLL associated with trisomy 12. Blood 2012; 119: 329–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palamarchuk A, Yan PS, Zanesi N, Wang L, Rodrigues B, Murphy M et al. Tcl1 protein functions as an inhibitor of de novo DNA methylation in B-cell chronic lymphocytic leukemia (CLL). Proc Natl Acad Sci USA 2012; 109: 2555–2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balatti V, Rizzotto L, Miller C, Palamarchuk A, Fadda P, Pandolfo R et al. TCL1 targeting miR-3676 is codeleted with tumor protein p53 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA 2015; 112: 2169–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullrich F, Rasio D, Kitada S, Starostik P, Kipps T, Keating M et al. ATM mutations in B-cell chronic lymphocytic leukemia. Cancer Res 1999; 59: 24–27. [PubMed] [Google Scholar]
- Stilgenbauer S, Nickolenko J, Wilhelm J, Wolf S, Weitz S, Dohner K et al. Expressed sequences as candidates for a novel tumor suppressor gene at band 13q14 in B-cell chronic lymphocytic leukemia and mantle cell lymphoma. Oncogene 1998; 16: 1891–1897. [DOI] [PubMed] [Google Scholar]
- Dong JT, Boyd JC, Frierson HF Jr.. Loss of heterozygosity at 13q14 and 13q21 in high grade, high stage prostate cancer. Prostate 2001; 49: 166–171. [DOI] [PubMed] [Google Scholar]
- Bullrich F, Fujii H, Calin G, Mabuchi H, Negrini M, Pekarsky Y et al. Characterization of the 13q14 tumor suppressor locus in CLL: identification of ALT1, an alternative splice variant of the LEU2 gene. Cancer Res 2001; 61: 6640–6648. [PubMed] [Google Scholar]
- Migliazza A, Bosch F, Komatsu H, Cayanis E, Martinotti S, Toniato E et al. Nucleotide sequence, transcription map, and mutation analysis of the 13q14 chromosomal region deleted in B-cell chronic lymphocytic leukemia. Blood 2001; 97: 2098–2104. [DOI] [PubMed] [Google Scholar]
- Rondeau G, Moreau I, Bezieau S, Petit JL, Heilig R, Fernandez S et al. Comprehensive analysis of a large genomic sequence at the putative B-cell chronic lymphocytic leukaemia (B-CLL) tumour suppresser gene locus. Mutat Res 2001; 458: 55–70. [DOI] [PubMed] [Google Scholar]
- Mertens D, Wolf S, Schroeter P, Schaffner C, Dohner H, Stilgenbauer S et al. Down-regulation of candidate tumor suppressor genes within chromosome band 13q14.3 is independent of the DNA methylation pattern in B-cell chronic lymphocytic leukemia. Blood 2002; 99: 4116–4121. [DOI] [PubMed] [Google Scholar]
- Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA 2002; 99: 15524–15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekarsky Y, Calin GA, Aqeilan R. Chronic lymphocytic leukemia: molecular genetics and animal models. Curr Top Microbiol Immunol 2005; 294: 51–70. [DOI] [PubMed] [Google Scholar]
- Ambros V. The functions of animal microRNAs. Nature 2004; 431: 350–355. [DOI] [PubMed] [Google Scholar]
- Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell 2003; 113: 673–676. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116: 281–297. [DOI] [PubMed] [Google Scholar]
- Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA 2005; 102: 13944–13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Beato M, Sanchez-Aguilera A, Piris MA. Cell cycle deregulation in B-cell lymphomas. Blood 2003; 101: 1220–1235. [DOI] [PubMed] [Google Scholar]
- Grabow S, Waring P, Happo L, Cook M, Mason KD, Kelly PN et al. Pharmacological blockade of Bcl-2, Bcl-x(L) and Bcl-w by the BH3 mimetic ABT-737 has only minor impact on tumour development in p53-deficient mice. Cell Death Differ 2012; 19: 623–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada S, Andersen J, Akar S, Zapata JM, Takayama S, Krajewski S et al. Expression of apoptosis-regulating proteins in chronic lymphocytic leukemia: correlations with in vitro and in vivo chemoresponses. Blood 1998; 91: 3379–3389. [PubMed] [Google Scholar]
- Adachi M, Tefferi A, Greipp PR, Kipps TJ, Tsujimoto Y. Preferential linkage of Bcl-2 to immunoglobulin light chain gene in chronic lymphocytic leukemia. J Exp Med 1990; 171: 559–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allegra D, Bilan V, Garding A, Dohner H, Stilgenbauer S, Kuchenbauer F et al. Defective DROSHA processing contributes to downregulation of MiR-15/-16 in chronic lymphocytic leukemia. Leukemia 2014; 28: 98–107. [DOI] [PubMed] [Google Scholar]
- Sampath D, Liu C, Vasan K, Sulda M, Puduvalli VK, Wierda WG et al. Histone deacetylases mediate the silencing of miR-15a, miR-16, and miR-29b in chronic lymphocytic leukemia. Blood 2012; 119: 1162–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE et al. A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med 2005; 353: 1793–1801. [DOI] [PubMed] [Google Scholar]
- Calin GA, Cimmino A, Fabbri M, Ferracin M, Wojcik SE, Shimizu M et al. MiR-15a and miR-16-1 cluster functions in human leukemia. Proc Natl Acad Sci USA 2008; 105: 5166–5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbri M, Bottoni A, Shimizu M, Spizzo R, Nicoloso MS, Rossi S et alAssociation of a microRNA/TP53 feedback circuitry with pathogenesis and outcome of B-cell chronic lymphocytic leukemiaJAMA 2011; 305: 59–67. [DOI] [PMC free article] [PubMed]
- Cai CK, Zhao GY, Tian LY, Liu L, Yan K, Ma YL et al. miR-15a and miR-16-1 downregulate CCND1 and induce apoptosis and cell cycle arrest in osteosarcoma. Oncol Rep 2012; 28: 1764–1770. [DOI] [PubMed] [Google Scholar]
- Jiang QQ, Liu B, Yuan T. MicroRNA-16 inhibits bladder cancer proliferation by targeting Cyclin D1. Asian Pac J Cancer Prev 2013; 14: 4127–4130. [DOI] [PubMed] [Google Scholar]
- Teshima K, Nara M, Watanabe A, Ito M, Ikeda S, Hatano Y et al. Dysregulation of BMI1 and microRNA-16 collaborate to enhance an anti-apoptotic potential in the side population of refractory mantle cell lymphoma. Oncogene 2014; 33: 2191–2203. [DOI] [PubMed] [Google Scholar]
- Reid G, Pel ME, Kirschner MB, Cheng YY, Mugridge N, Weiss J et al. Restoring expression of miR-16: a novel approach to therapy for malignant pleural mesothelioma. Ann Oncol 2013; 24: 3128–3135. [DOI] [PubMed] [Google Scholar]
- Klein U, Lia M, Crespo M, Siegel R, Shen Q, Mo T et al. The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell 2010; 17: 28–40. [DOI] [PubMed] [Google Scholar]
- Raveche ES. Possible immunoregulatory role for CD5+B cells. Clin Immunol Immunopathol 1990; 56: 135–150. [DOI] [PubMed] [Google Scholar]
- Raveche ES, Salerno E, Scaglione BJ, Manohar V, Abbasi F, Lin YC et al. Abnormal microRNA-16 locus with synteny to human 13q14 linked to CLL in NZB mice. Blood 2007; 109: 5079–5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovat F, Fassan M, Gasparini P, Rizzotto L, Cascione L, Pizzi M et al. miR-15b/16-2 deletion promotes B-cell malignancies. Proc Natl Acad Sci USA 2015; 112: 11636–11641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souers AJ, Leverson JD, Boghaert ER, Ackler SL, Catron ND, Chen J et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat Med 2013; 19: 202–208. [DOI] [PubMed] [Google Scholar]
- Croce CM, Reed JC. Finally, an apoptosis-targeting therapeutic for cancer. Cancer Res 2016; 76: 5914–5920. [DOI] [PMC free article] [PubMed] [Google Scholar]