Abstract
BCL-2 proteins are known to engage each other to determine the fate of a cell after a death stimulus. However, their evolutionary conservation and the many other reported binding partners suggest an additional function not directly linked to apoptosis regulation. To identify such a function, we studied mice lacking the BH3-only protein BIM. BIM−/− cells had a higher mitochondrial oxygen consumption rate that was associated with higher mitochondrial complex IV activity. The consequences of increased oxygen consumption in BIM−/− mice were significantly lower body weights, reduced adiposity and lower hepatic lipid content. Consistent with reduced adiposity, BIM−/− mice had lower fasting blood glucose, improved insulin sensitivity and hepatic insulin signalling. Lipid oxidation was increased in BIM−/− mice, suggesting a mechanism for their metabolic phenotype. Our data suggest a role for BIM in regulating mitochondrial bioenergetics and metabolism and support the idea that regulation of metabolism and cell death are connected.
Type 2 diabetes is characterised by insulin resistance and reduced functional mass of pancreatic beta cells. In addition, the content and oxidative capacity of mitochondria is reduced in insulin responsive tissues such as skeletal muscle, liver, and white adipose tissue of subjects with insulin resistance and type 2 diabetes, and these changes are believed to contribute to diabetes pathogenesis.1
The inner mitochondrial membrane is the site of oxidative phosphorylation (OXPHOS) and energy metabolism. OXPHOS involves the transfer of electrons liberated during β-oxidation and the citric acid cycle across the protein complexes of the electron transport chain.2 This transfer of electrons, together with the pumping of protons from the mitochondrial matrix to the intermembrane space is required for generation of ATP and release of heat.2
In contrast, the outer mitochondrial membrane is important in apoptotic cell death signalling. In mammalian cells, the intrinsic apoptosis pathway, also known as the BCL-2 regulated or mitochondrial pathway, is activated by cellular stresses such as radiation exposure, DNA damage and growth factor withdrawal.3 Activation of the pro-apoptotic BH3-only BCL-2 family members, such as BIM or PUMA, inhibits the pro-survival BCL-2 family members, including BCL-2, BCL-XL, and MCL-1, allowing activation of the pro-apoptotic multi-BH domain effectors BAX and BAK,3, 4 leading to mitochondrial outer membrane permeabilisation with consequent release of cytochrome c into the cytoplasm, and activation of the caspase cascade that eventually causes cell death.3
BIM is a potent BH3-only factor that can bind with high affinity to all the pro-survival members of the BCL-2 family.3 It mediates apoptosis in a variety of cell types in response to cellular stresses, such as growth factor deprivation, ER stress, oxidative stress, and corticosteroid exposure.5, 6, 7, 8 Although BIM is expressed at very low levels in the absence of stress, it has been observed in the mitochondria of non-apoptotic cells, in association with the translocase of the outer membrane complex that imports proteins into the mitochondria.9, 10 This indicates that it may have a mitochondrial role, even in resting cells. Further, the protein sequence of BIM is conserved across species,11 and binding partners additional to pro-survival BCL-2 proteins have been described.9 We have therefore studied BIM-deficient mice to investigate whether BIM could have a physiological role in resting cells separate to its apoptotic function. BIM−/− mice had increased lipid oxidation, associated with increased activity of mitochondrial complex IV and an increased mitochondrial oxygen consumption rate (OCR) at a cellular level. As a consequence, BIM-deficient mice had lower fasting blood glucose, improved insulin sensitivity and reduced adiposity. Our results suggest that BIM has a role in mitochondrial function that is independent of apoptosis, and its deficiency results in altered whole-body metabolic homoeostasis.
Results
BIM-deficient cells have increased mitochondrial OCR and reduced mitochondrial membrane potential
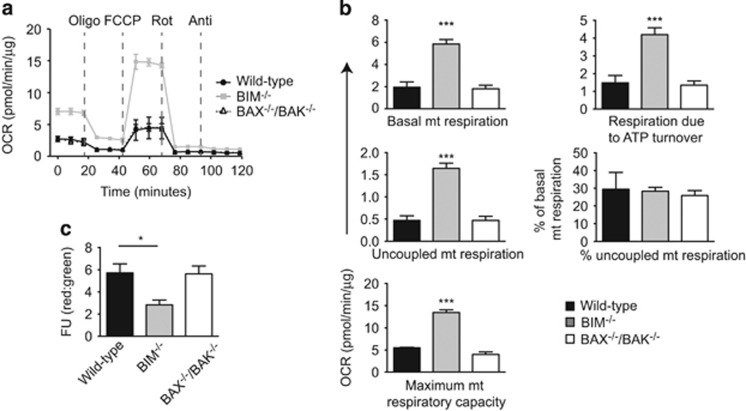
We investigated whether BIM is involved in regulating cellular mitochondrial oxidative metabolism. The mitochondrial OCR was measured in mouse embryonic fibroblasts (MEF) under serum-free conditions. BIM−/− MEFs had substantially higher basal OCR than wild-type controls (Figures 1a and b). Similar to wild-type cells, ~70% of basal OCR in BIM−/− MEFs was coupled to ATP synthesis suggesting that there was a proportional increase in uncoupled and ATP-coupled OCR in BIM−/− cells (Figures 1a and b). Strikingly, we noticed an increase of more than threefold in the maximal mitochondrial respiratory capacity of BIM−/− MEFs compared with wild-type controls (Figures 1a and b). Similar differences were seen in basal and maximal cellular respiratory capacity (Supplementary Figure 1A). The rate of oxygen consumption coupled to flux through complex-I and complex-III was determined after sequentially adding rotenone and antimycin A to the cells and was also higher in BIM−/− cells (Supplementary Figure 1A). The mitochondrial OCR of BAX−/−BAK−/− MEFs was not different from wild-type controls, suggesting that increased OCR in BIM−/− cells may be separate from its pro-apoptotic function (Figures 1a and b) These data indicate that in addition to its known role as a pro-apoptotic protein, BIM is also involved in regulating mitochondrial OXPHOS.
Figure 1.
BIM-deficient cells have increased mitochondrial oxygen consumption rate (OCR) and reduced mitochondrial potential. (a) Mitochondrial OCR was measured in wild-type, BIM−/− and BAX−/−BAK−/− MEFs using a Seahorse XF24 bioanalyzer, n=3 independent MEF isolations/genotype. After recording three baseline OCR measurements, oligomycin (Oligo), FCCP, rotenone (Rot) and antimycin A (Anti) were sequentially added to the cells and OCR measurements taken thrice after each treatment. (b) Quantification of parameters of mitochondrial respiration measured by the Seahorse bioanalyzer. n=3 independent experiments. ***P<0.001 BIM−/− versus wild-type MEF (one-way ANOVA). (c) Mitochondrial potential was measured in wild-type, BIM−/− and BAX−/−BAK−/− MEFs by JC-1 staining. n=3 independent experiments, *P<0.05 BIM−/− versus wild-type MEFs (one-way ANOVA). Data show mean±S.E.M.
Mitochondrial membrane potential, measured by JC-1 staining, was lower in BIM−/− MEFs than wild-type MEFs (Figure 1c), indicating changes in ionic equilibrium across the inner mitochondrial membrane. In contrast, the intensity of JC-1 staining in BAX−/−BAK−/− MEFs was similar to wild-type controls (Figure 1c). The reduction in mitochondrial potential could be a consequence of increased mitochondrial ATP turnover in BIM−/− MEFs. These data show that BIM has a fundamental role in regulating mitochondrial function, cellular bioenergetics, and energy expenditure.
Hepatocytes from BIM−/− mice had a similar amount of mitochondrial DNA compared with wild-type hepatocytes (Supplementary Figure 1B). BIM deficiency resulted in increased mean mitochondrial cross-sectional area in hepatocytes (Supplementary Figure 1C & D). This could be owing to altered mitochondrial fusion or fission.12, 13 BIM-deficient cells also had reduced number of mitochondria per cell and mitochondria were relatively more round in shape, which is likely to be due to their bigger size (Supplementary Figure 1C & D).
BIM-deficient hepatocytes have increased activity of complex IV
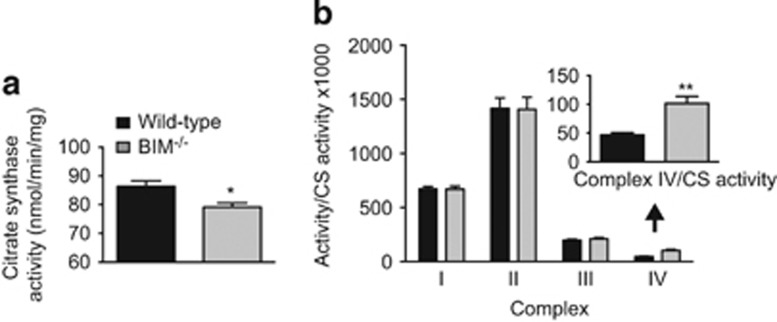
To understand how BIM could control OCR, activity of mitochondrial complexes I–IV was measured in liver mitochondrial preparations from control and BIM−/− mice. These assays assess the intrinsic capacity of mitochondrial complexes independent of cellular energetics, which was assessed in the experiments using MEFs. Citrate synthase activity (a marker for mitochondrial content) was slightly reduced in BIM−/− livers (Figure 2a). We observed an increase of about twofold in the activity of complex IV in BIM-deficient liver tissue, whereas no difference was observed in the activities of other complexes, indicating that the effects of BIM deficiency are specific to complex IV and not generalised to the entire OXPHOS machinery (Figure 2b).
Figure 2.
BIM deficiency increases activity of complex IV. (a,b) Activity of (a) citrate synthase and (b) mitochondrial electron transport chain complexes (I–IV) measured in liver samples of wild-type and BIM−/− mice and normalised to citrate synthase activity. Complex IV activity is enlarged. n=6 mice/genotype. Data show mean±S.E.M. *P<0.05, **P<0.01 BIM−/− versus wild-type liver tissue samples (Student’s t-test)
In blue native PAGE (BN-PAGE) analysis, a cocktail of antibodies against complexes I, II, III, and IV (anti-OXPHOS) showed no difference in the expression of individual complexes (Supplementary Figure 2). Further, no difference in the assembly of CI/CIII/CIV supercomplexes was observed between wild-type and BIM−/− liver tissue samples suggesting that loss of BIM does not increase complex IV activity by facilitating improved assembly of supercomplexes (Supplementary Figure 2).
Lipid oxidation and energy expenditure is greater in BIM-deficient mice than wild-type controls
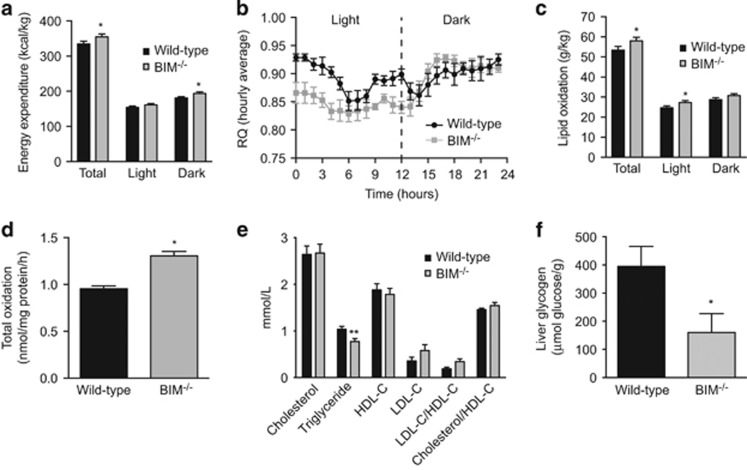
Our data in MEFs show that BIM regulates cellular energy expenditure that is supported by mitochondrial oxidative metabolism. Mitochondria are an important site of cellular macronutrient metabolism. This includes glucose oxidation via the TCA cycle. In addition, β-oxidation of lipids and the subsequent oxidation of the resultant acetyl-CoA, also through the TCA cycle, produces reducing equivalents for the generation of ATP molecules through OXPHOS.14, 15 To find out whether changes in mitochondrial OXPHOS owing to BIM deficiency alter whole-body energy expenditure and substrate metabolism, we studied mice in metabolic cages. Consistent with data obtained in MEFs, mice lacking BIM had increased energy expenditure (Figure 3a). These mice had a lower respiratory quotient under the basal conditions of light cycle (Figure 3b), which was associated with increased lipid utilisation by BIM−/− mice (Figure 3c). Carbohydrate oxidation was reduced in the light cycle in BIM−/− mice (Supplementary Figure 3A), indicating that BIM also regulates whole-body substrate utilisation. No significant differences between wild-type and BIM−/− mice were observed in physical activity (Supplementary Figure 3B).
Figure 3.
BIM-deficient mice have altered lipid and glucose metabolism. (a) Energy expenditure (kcal/kg), (b) mean respiratory quotient (RQ) measured hourly for 24 h (from 7am to 7am), and (c) lipid oxidation (g/kg), measured in metabolic cages for chow-fed male mice at 18 weeks of age, n=7–8 mice/genotype. *P<0.05 BIM−/− versus wild-type mice (Student’s t-test). Energy expenditure and lipid oxidation normalised to mouse weight (g/kg body mass). (d) Total palmitate oxidation measured in hepatocytes isolated from 20-week-old chow-fed male wild-type and BIM−/− mice. One representative experiment is shown. *P<0.05 BIM−/− versus wild-type mice calculated from fold change of pooled data from n=3 independent experiments (Student’s t-test). (e) Concentrations of triglyceride and cholesterol species in blood collected from 24-week-old fasted (6 h) chow-fed male mice. n=5-8 mice/genotype. **P<0.01 BIM−/− versus wild-type mice (Student’s t-test). (f) Liver glycogen content in samples harvested from 24-week-old male fasted (6 h) chow-fed mice. n=8-9 mice/genotype. *P<0.01 BIM−/− versus wild-type mice (Student’s t-test). Data show mean±S.E.M.
BIM-deficient mice have altered lipid and glucose metabolism in the liver
The liver is a key organ in regulating glucose and fat metabolism in the body. We examined metabolism of these micronutrients in the liver. Isolated hepatocytes lacking BIM had a greater capacity to oxidise palmitate than wild-type hepatocytes (Figure 3d). Further, BIM−/− mice had lower serum triglycerides, whereas cholesterol concentrations were similar between wild-type and BIM−/− mice (Figure 3e). We hypothesise that owing to increased energy expenditure, BIM−/− mice metabolise the glycogen stores relatively rapidly and this is followed by greater lipid metabolism in the resting state, resulting in reduced hepatic lipid and glycogen content. Indeed, fasting liver glycogen content was reduced in BIM−/− livers (Figure 3f). Although serum albumin was slightly reduced, deficiency of BIM did not affect concentrations of total proteins (Supplementary Figure 3C) and liver enzymes (not shown) in the serum, suggesting that synthetic liver function in BIM−/− mice is overall normal.
BIM deficiency is associated with reduced body weight and adiposity
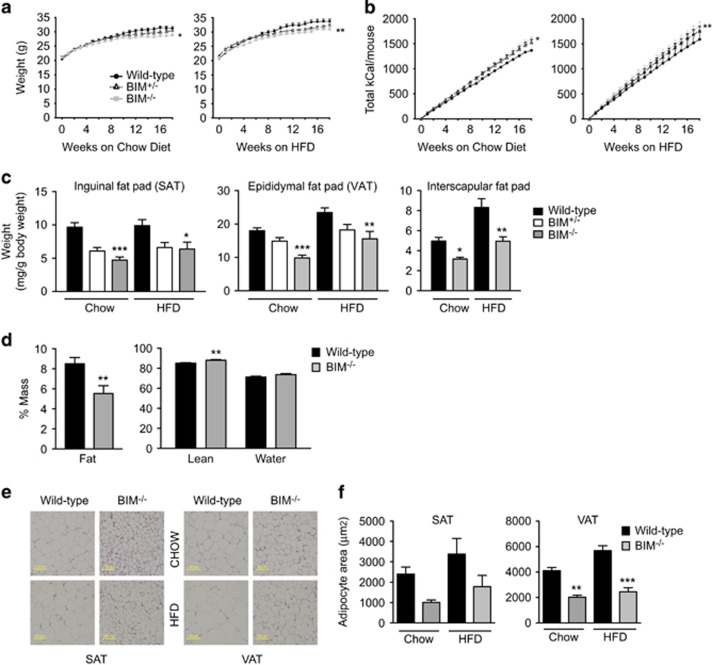
Given the increased energy expenditure and lipid oxidation, and favourable serum lipid profile in BIM-deficient mice, we next investigated total body weight and adiposity in mice on a chow diet and those challenged by a 45% high-fat diet (HFD) for a total of 18 weeks. Compared with wild-type mice on similar diet, mice lacking BIM gained less weight (Figure 4a). Consistent with increased energy expenditure, BIM−/− mice had slightly greater cumulative food intake than the wild-type controls, despite the reduced weight gain (Figure 4b). Heterozygous BIM+/− mice were intermediate between wild-type and knockout mice showing that effects of BIM deficiency are allele dosage dependent (Figures 4a and b). BIM deficiency did not affect length (Supplementary Figure 4A) or histological structure of the small intestine (Supplementary Figure 4B), suggesting that reduced weight in BIM−/− mice is not associated with altered intestinal morphology that could potentially affect nutrient absorption.
Figure 4.
BIM-deficient mice have reduced body weight and adiposity. (a,b) Body weights in g (a) and cumulative calorie intake (b) of wild-type, BIM+/− and BIM−/− male mice on chow (left) or high-fat diet (right) measured weekly from 6 weeks of age for 18 weeks. n=14–16 mice/genotype. *P<0.05, **P<0.01 BIM−/− versus wild-type mice on similar diet (two-way ANOVA). (c) Weight of inguinal fat pad (SAT), epididymal fat pad (VAT), and interscapular brown fat pad measured after 18 weeks (24 weeks of age) and normalised to total body weight (mg/g body weight). n=9–16 mice/genotype *P<0.05, **P<0.01, ***P<0.001 BIM−/− versus wild-type mice on similar diet (one-way ANOVA). (d) Body composition determined at 18 weeks of age in chow-fed mice by MRI scanning. % fat, lean, and water mass are shown. n=7–8 mice/genotype. **P<0.001 BIM−/− versus wild-type mice (Student’s t-test). (e) Formalin-fixed sections of inguinal fat pads (SAT; left) and epididymal fat pads (VAT; right) isolated from 24-week-old chow-fed or HFD-fed male wild-type mice or BIM−/− mice stained with H&E. Representative images of n=4 mice/genotype. Scale bar 100 μm. (f) Quantification of adipocyte cross-sectional area of SAT and VAT from chow-fed and HFD-fed mice, n=4 mice/genotype. **P<0.01, ***P<0.001 BIM−/− versus wild-type mice on similar diet (one-way ANOVA). Data show mean±S.E.M.
Consistent with lower body weight, BIM-deficient mice also had lower visceral (VAT) and subcutaneous (SAT) white fat pad mass and interscapular brown fat mass on chow and HFDs indicating a generalised decrease in adiposity in BIM−/− mice (Figure 4c). This was confirmed by MRI at 18 weeks of age, with reduced percentage fat mass and increased percentage lean body mass and unchanged percentage mass of water observed in BIM−/− compared with wild-type mice (Figure 4d). Adipocyte cell size was smaller in BIM-deficient mice than wild-type counterparts on both a chow diet and HFD (Figures 4e and f). Liver weights were similar across all the mouse groups (Supplementary Figure 4C).
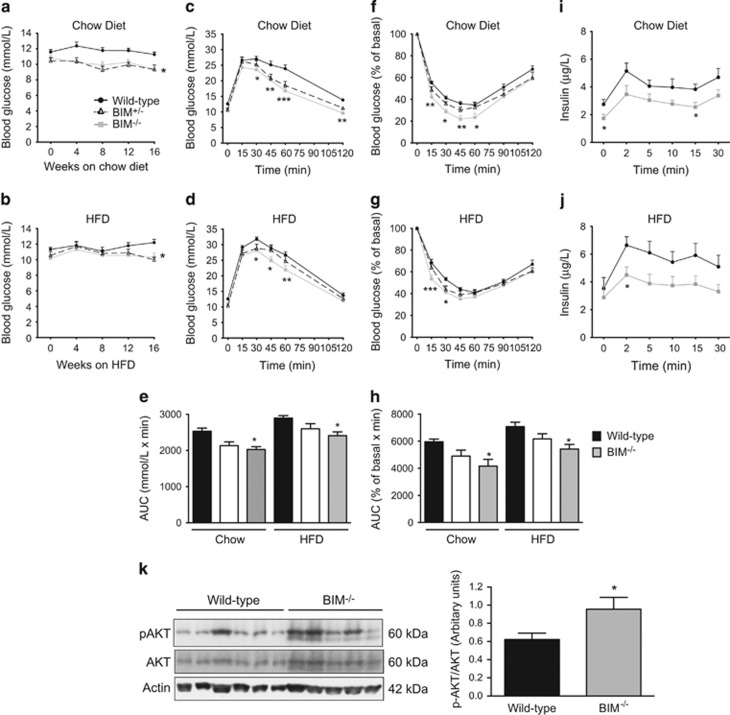
Deficiency of BIM improves glucose homoeostasis in mice
Increased energy expenditure and reduced body fat is commonly associated with improvements in insulin sensitivity and glucose homoeostasis.16 Compared with mice on a chow diet, high-fat feeding increased fasting blood glucose in wild-type mice (Figures 5a and b). BIM−/− and BIM+/− mice had lower fasting blood glucose concentrations when compared with chow or high-fat-fed wild-type mice (Figures 5a and b). No significant differences in non-fasting blood glucose were observed (Supplementary Figure 4D). Lower fasting blood glucose could be owing to enhanced release of insulin from islets or improved sensitivity to insulin in peripheral tissues.
Figure 5.
Improved glucose homoeostasis in BIM-deficient mice. (a,b) Blood glucose concentration (mmol/l) measured every four weeks after 7–8 h of fasting for mice on chow (a) and high-fat diet (b). n=14–16 mice/genotype. *P<0.05 BIM−/− versus wild-type mice on similar diet (two-way ANOVA). (c, d) IP-GTT performed on 20-week-old mice after 6 h of fasting by injecting 2 g/kg of glucose. Results for the chow-fed mice (c) and high-fat-fed mice (d) are shown. n=14–16 mice/genotype. (e) Area under the curve (AUC) for the IP-GTT data from chow and high-fat-fed mice. n=14–16 mice/genotype. For (c–e) *P<0.05, **P<0.01 and ***P<0.001 BIM−/− versus wild-type mice on similar diet (two-way ANOVA). (f, g) IP-ITT was performed on 18-week-old mice after 4 h of fasting by injecting 0.75 U/kg of insulin. Results are expressed as a percentage of basal blood glucose concentration and are shown for the chow-fed mice (f) and high-fat-fed mice (g). n=14–16 mice/genotype. (h) Area under the curve (AUC) for the IP-ITT data from chow and high-fat-fed mice. n=14–16 mice/genotype. For (f–h) *P<0.05, **P<0.01 and ***P<0.001 BIM−/− versus wild-type mice on similar diet (two-way ANOVA). (i,j) Plasma insulin concentrations for samples obtained during IV-GTT determined by ELISA. Results for the chow-fed mice (i) and high-fat-fed mice (j) are shown. n=9–11 mice/genotype. *P<0.05 (Student’s t-test). (k) Western blot of phosphorylated AKT (pAKT), total AKT, and actin (loading control) performed on liver lysates isolated from 20-week-old male mice after 6 h of fasting. Liver samples were isolated 10 min after intraperitoneal injection of insulin. n=5–6 mice/genotype. Quantification of western blots for pAKT normalised to total AKT. *P<0.05 BIM−/− versus wild-type mice on chow diet (Student’s t-test). Data show mean±S.E.M.
Islet function was analysed by intraperitoneal glucose tolerance testing in 20-week-old mice, after 14 weeks on either chow or high-fat diet. BIM−/− mice had improved glucose tolerance compared with wild-type mice (Figures 5c and e). High-fat feeding made the mice glucose intolerant, but BIM−/− mice still had improved glucose tolerance compared with BIM+/+ mice (Figures 5d and e). Glucose tolerance in heterozygous BIM+/− mice was intermediate between wild-type and BIM−/− mice (Figures 5c and e).
Insulin mediated glucose disposal (measured in an intraperitoneal insulin tolerance test) was significantly increased in mice lacking BIM compared with wild-type controls (Figures 5f and h). Although the high-fat diet impaired sensitivity to insulin in all mice, BIM−/− mice remained more sensitive to injected insulin than BIM+/+ mice (Figures 5g and h). Sensitivity of heterozygous BIM+/− mice to insulin was intermediate between wild-type and BIM−/− mice (Figures 5f and h).
Hyperinsulinaemia is a compensatory response by islets to high-fat-induced insulin resistance.17 Deficiency of BIM was associated with lower plasma insulin concentrations after intravenous glucose injection compared with wild-type controls under both dietary conditions (Figures 5i and j and Supplementary Figure 4E). These data suggest that the increased insulin sensitivity in BIM-deficient mice results in reduced insulin requirements than wild-type mice to maintain their basal glucose and to normalise hyperglycaemia induced by high-fat feeding. Consistent with this we observed no differences in islet size or fractional insulin or glucagon-positive areas (Supplementary Figure 4F&G). Both basal and glucose-stimulated insulin secretion were the same in wild-type and BIM-deficient islets (Supplementary Figure 4H). Overall, these data suggest that improved peripheral tissue insulin sensitivity, and not islet function, results in the metabolic phenotype of BIM−/− mice.
Phosphorylation of Akt was greater in the livers of insulin-treated BIM−/− mice, suggesting that improved insulin signalling contributes to better insulin sensitivity in these mice (Figure 5k). No difference was observed in AMP-activated protein kinase (AMPK) phosphorylation between BIM+/+ and BIM−/− livers suggesting that increased insulin signalling is not secondary to AMPK mediated metabolic changes (such as AMPK driven increased lipid metabolism) in BIM-deficient hepatocytes (Supplementary Figure 5A). We observed a similar trend of increased Akt phosphorylation in the skeletal muscle (gastrocnemius) of BIM−/− mice (Supplementary Figure 5B).
Age-related intra-cellular lipid droplet accumulation,18 visualised by histological analysis and oil red O staining, was reduced in hepatocytes of BIM-deficient mice compared with wild-type controls and this was observed in both chow-fed and high-fat-fed mice (Supplementary Figure 5C-E). Reduced hepatic lipid content may be the result of reduced lipogenesis, however, we did not observe any change in lipogenic or lipid uptake enzyme expression in liver or visceral fat (Supplementary Figure 5F). Ectopic lipid accumulation in the liver reduces hepatic insulin sensitivity.19 Therefore, reduced hepatocyte lipid accumulation in BIM-deficient mice could contribute to increased hepatic insulin sensitivity.
Discussion
In this study, we show that BIM regulates mitochondrial OXPHOS and its deficiency leads to increased mitochondrial oxygen consumption and activity of complex IV, the terminal enzyme in the electron transport chain. Mitochondria are the major site of cellular ATP synthesis and oxidation of lipids, so changes in mitochondrial OXPHOS are likely to affect whole-body macronutrient metabolism. In particular, liver and muscle are the main sites of lipid oxidation in the body, and most likely to be affected by increasing complex IV activity. Taking advantage of the fact that BIM-deficient mice survive to adulthood and do not show any significant pathology on a C57BL/6 genetic background at a young age, we found that altered bioenergetics observed with BIM deficiency led to increased use of lipids for resting energy metabolism resulting in reduced body fat, reduced glycogen stores and increased insulin sensitivity. Consistent with our data, feeding a high-fat diet to mice for 12 weeks induced insulin resistance and reduced the expression of complex IV in visceral fat.20 However, mice lacking the complex IV assembly factor Surf1 have reduced complex IV activity and this is associated with increased mitochondrial biogenesis, reduced adiposity, increased lipid oxidation and improved insulin sensitivity.21 This shows that the impact of alterations in complex IV activity on insulin sensitivity and substrate metabolism is dependent on the underlying mechanism and metabolic context. Our study shows that increased mitochondrial activity caused by BIM deficiency is a mechanism for improving insulin sensitivity.
One caveat of our work is the observation that BIM-deficient mice have perturbed immune homoeostasis. In BIM-deficient mice, autoreactive T cells escape deletion in the thymus. However, widespread autoimmunity is avoided because a subset of FoxP3+CD4+ Treg cells is preferentially increased in BIM−/− mice.22 This shift in the balance of immune subsets could contribute to the improved insulin sensitivity in BIM−/− mice. However, we did not observe any changes in immune cell populations in the VAT of BIM−/− mice (not shown). Also, it is likely that all tissues contribute to the metabolic phenotype in BIM−/− mice, and tissue-specific knockout mice will display a partial phenotype. Preliminary data in hepatocyte-specific knockouts show insulin sensitivity consistent with the whole-body data (ENG and HET unpublished data).
Our data suggest that BIM inhibits both ATP-coupled and uncoupled respiration and this is seen in non-apoptotic conditions in the absence of apoptotic stimuli known to activate BIM. Consistent with this, increased accumulation of mitochondrial reactive oxygen species (ROS) and reduced mitochondrial respiration were observed in neuroblastoma cells conditionally expressing BIM.23 BIM has tumour-suppressor properties24, 25 and it is possible that BIM-mediated inhibition of mitochondrial activity provides the initial trigger for tumour cell apoptosis by generating ROS in the mitochondria.23 Therefore, it is possible that metabolic effects of BIM could be connected with apoptosis, and elucidating this connection will be determined in future studies.
Changes in mitochondrial OXPHOS have been observed previously in vitro in cells with altered expression of the pro-survival factors MCL-1, BCL-2, and BCL-XL.13, 26, 27, 28 Overexpression of these pro-survival factors increased mitochondrial activity, and although the effects on OXPHOS are not exactly the same, they are consistent with our findings that deletion of BIM increased mitochondrial function and complex IV activity. The exact mechanism by which BIM regulates these effects remains unclear. It was recently reported that a variant of BCL-2 is associated with altered insulin sensitivity in a large genome wide association study.29 BCL-2 is also associated with type 2 diabetes30 and body fat distribution.31 It is currently unknown if BIM directly binds to a subunit of complex IV or if its deletion leads to increased availability of pro-survival factors in the mitochondrial matrix. The highly unstructured nature of BIM32 suggests that it may have a secondary role in the mitochondria through association with another protein, such as one of the pro-survival factors. Although the pro-apoptotic proteins BAD33 and BNip334 have demonstrated roles in mitochondrial function and liver energy metabolism, PUMA did not have a similar metabolic phenotype.35 Our new data showing a role of pro-apoptotic BIM in metabolism, together with previous data showing an effect of pro-survival members of the same family13, 26, 27, 28 suggest that the family of proteins is involved in both cell death and metabolism.
AMPK and peroxisome proliferator-activated receptor proteins are important regulators of lipid oxidation,14, 36 and have been implicated in protection from apoptosis of BIM-deficient neuronal cells, hepatocytes and beta cells.37, 38, 39 Further, activation of AMPK has been shown to increase mitochondrial content and activity of complexes III and IV.40 This raises the possibility that by inhibiting apoptosis, BIM deficiency could permit prolonged and sustained AMPK activation, resulting in increased oxidative metabolism of fatty acids. However, we did not observe evidence that BIM and AMPK mediate cross-talk between apoptosis and metabolism, and therefore we suggest that the increased lipid oxidation in BIM−/− mice is not likely to be AMPK mediated, but depends on downstream increase in mitochondrial activity.
Overall, we have shown that in addition to its known role as a potent inducer of apoptosis, BIM is involved in the regulation of mitochondrial oxygen consumption and complex IV activity and its loss results in increased lipid metabolism and reduced body fat. Future work will determine what domain(s) in BIM is important in regulating mitochondrial activity. Targeting the mitochondrial respiratory chain to oxidise fatty acids and improve insulin sensitivity is a novel treatment option for type 2 diabetes. For example, glimins, a new class of drug that act by increasing the mitochondrial capacity to oxidise fatty acids, are as effective as metformin in preliminary human studies.41 The lipid-lowering effects of metformin are dependent on AMPK target enzymes ACC1 and ACC2,42 and metformin treatment does not lead to substantial increases in mitochondrial OXPHOS.43 It is possible that targeting the mitochondrial role of BIM could be an option to amplify the lipid-lowering effects of metformin by increasing mitochondrial activity, providing of course that this does not impact on BIM’s critical role in regulation of cell death.
Materials and methods
Mice
Mice deficient in Bim generated on a pure C57BL/6 genetic background were kindly provided by Dr Philippe Bouillet (Walter and Eliza Hall Institute of Medical Research, Parkville, VIC, Australia).5 BIM−/− and BIM+/− mice were bred at St Vincent’s Institute. Wild-type C57BL/6 mice were purchased from Animal Resources Centre (Canning Vale, WA, Australia). All animals were housed at St Vincent’s Institute Bioresources Centre, and experiments were approved by the St Vincent’s Hospital Animal Ethics Committee.
High-fat diet and metabolic analysis
Six-week-old male BIM−/−, BIM+/− or BIM+/+ mice on C57BL/6 background were randomly placed on a chow diet (Specialty Feeds, Glenn Forest, Australia) or a HFD (45% kCal/fat, Specialty Feeds) for 18 weeks. Mice were maintained on a 12 h light/12 h dark cycle. Mice were weighed and their food intake was calculated weekly. Non-fasting blood glucose was measured every 2 weeks while fasting blood glucose was measured after 7–8 h of fasting at 0, 6, 10, 14, and 18 weeks using a glucometer (Accu Chek Performa, Roche, Basel, Switzerland). At 18 weeks, mice were killed and pancreas, liver, inguinal fat pad (SAT), epididymal fat pad (VAT), and interscapular fat pad (brown adipose tissue) were weighed, and collected for further analysis.
Mice lacking BIM and wild-type controls fed a chow diet were housed in metabolic cages (Columbus Instruments, Columbus, OH, USA) for 48 h at 18 weeks of age. After 24 h of acclimatisation, physical activity, energy expenditure and substrate utilisation were measured by indirect calorimetry.44 Body composition was determined by MRI scanning (EchoMRI, Houston, TX, USA).
Blood samples were obtained from fasted mice by cardiac puncture. Serum was isolated from the blood samples by centrifugation and the measurements for lipids, proteins and liver enzymes were performed on a clinical chemistry analyser (Beckman Coulter AU5800 Clinical Chemistry System, Brea, CA, USA).
Insulin and glucose tolerance tests
Mice were fasted for 4 h at 18 weeks of age before performing an intraperitoneal insulin tolerance test. Insulin (Actrapid, Novo Nordisk, Bagsværd, Denmark), at a dose of 0.75 U/kg, was injected intra-peritoneally and blood glucose was measured at 0, 15, 30, 45, 60, 90, and 120 min after tail bleeding.
At 20 weeks of age, mice were fasted for 6 h before an intraperitoneal glucose tolerance test. Glucose solution (2 g/kg, Baxter, Deerfield, IL, USA) was injected and blood glucose was measured at 0, 15, 30, 45, 60, and 120 min. An intravenous glucose tolerance (IV-GTT) test was performed on 24-week old mice after 6 h of fasting according to previously described methods.45 In brief, 1 g/kg glucose was injected and plasma samples were obtained at 0, 2, 5, 10, 15, and 30 min to measure insulin concentration by ELISA (Mercodia, Uppsala, Sweden).
Glucose-stimulated insulin secretion
Islets of Langerhans were isolated as described previously.46 Twenty islets of similar size were pre-incubated for 30 min in Krebs-Ringer bicarbonate (KRB) buffer containing 3 mM glucose and 0.1% BSA, after which they were incubated for 30 min with KRB buffer containing either 20 mM glucose or 3 mM glucose and 0.1% BSA. The secreted insulin was measured using a rat/mouse insulin ELISA kit (Mercodia).
Western blotting and blue native PAGE
Twenty-week-old male mice were fasted for 6 h and then injected intra-peritoneally with PBS or insulin (0.65 mU/g). Liver and muscle samples were collected in liquid nitrogen 10 min after insulin injection. Lysates were prepared and western blotting was performed according to previously described methods.47 Anti-phospho-S473-Akt, anti-Akt, anti phosphoT172-AMPK and anti-pan-alpha AMPK antibodies from Cell Signaling Technology (Danvers, MA, USA), and anti-BIM antibody from Alexis Biochemicals (Plymouth Meeting, PA, USA) were used. Blotting with anti-actin or anti-tubulin antibodies (Santa Cruz Biotechnology, Santa Cruz, CA, USA) were used for loading control.
Blue native polyacrylamide gel electrophoresis (BN-PAGE) of liver tissue samples was performed as described.48 Immunoblotting was performed using an antibody cocktail (Abcam, Cambridge, UK) against NDUFB8 (ab110242), CII-30kDa (ab14714), CIII-core protein 2 (ab14745), CIV subunit I (ab14705) and CV alpha subunit (ab14748).
Seahorse studies and JC-1 staining
Wild-type, BIM−/−, and BAX−/−BAK−/− MEFs, isolated from mouse lines made on a mixed B6/129 background and subsequently backcrossed onto B6 for more than six generations, were analysed on a Seahorse XF24 Flux Analyzer (Seahorse Bioscience, North Billerica, MA, USA) in serum-free conditions according to a previously described protocol.49 After plating 25 000 MEFs/well and recording three basal OCR measurements, readings were taken after sequentially injecting oligomycin (1 μM), FCCP (1 μM), rotenone (1 μM), and antimycin A (1 μM). Calculations of parameters of mitochondrial respiratory function49 included subtraction of non-mitochondrial respiration from all mitochondrial respiration parameters. The data were normalised to total protein content/well measured by BCA assay (Thermo Fisher Scientific, Waltham, MA, USA) according to manufacturer's instructions.
Mitochondrial potential was measured in wild-type, BIM−/−, and BAX−/−BAK−/− MEFs using JC-1 dye (10 μg/ml) according to manufacturer’s instructions. After three washes with PBS, red and green fluorescence was measured using an excitation wavelength of 488nm and emission wavelengths of 522 and 605 nm respectively, using a Flexstation II microplate reader (Molecular Devices, Sunnyvale, CA, USA). Activity of electron transport chain complexes was measured in liver tissue samples as previously described.50
Histology
For all the histological analysis, tissue was fixed in formalin, and sections (5 μm) were stained according to standard procedures. Guinea pig anti-insulin antibody (Dako, Glostrup, Denmark) was detected with Alexa Fluor555-conjugated goat anti-guinea pig IgG antibody (Life Technologies, Carlsbad, CA, USA). Mouse anti-glucagon antibody (Sigma-Aldrich, St. Louis, MO, USA) was detected with Alexa Fluor647-conjugated goat anti-mouse IgG antibody (Life Technologies). Nuclei were stained with DAPI reagent (Life Technologies). Images were analysed on a Nikon A1R-A1 confocal microscope (Nikon Corporation, Tokyo, Japan). The percentage of insulin and glucagon-positive staining per islet was analysed using ImageJ software (NIH, Bethesda, MD, USA).
Hepatic lipid content was analysed on frozen sections of BIM+/+ and BIM−/− liver by oil red O staining according to a previously published protocol.51 Slides were imaged using an Aperio digital slide scanner (Leica, Wetzlar, Germany) and oil red O-positive areas were quantified using the Aperio software.
Electron microscopy
Transmission electron microscopy was performed as described previously.52 For isolated hepatocytes, cells were fixed in 2% paraformaldehyde, 0.05% glutaraldehyde in 0.08 M Sorensens’s phosphate buffer then processed using standard procedures. Ultrathin sections ~70 nm thickness were cut on Ultracut-S ultramicrotome (Leica, Wein, Austria) and contrasted with uranyl acetate and lead citrate. Images were analysed by ImageJ software (NIH).
Real-time PCR
Total RNA was isolated using Nucleospin RNA kits (Macherey-Nagel, Düren, Germany) and reverse transcribed using a High Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA, USA) following the manufacturer’s protocol. Real-time PCR was performed using AmpliTaq Gold DNA Polymerase (Applied Biosystems) and a LightCycler 480 (Roche). TaqMan primers for Scd1, Srebf1, Pparg, Acaca, Fasn, Cd36 and Actin-b were purchased from Applied Biosystems. Data analysis was performed using the ΔΔCT method: relative mRNA expression levels were calculated by normalising to the signal for Actin-b mRNA in each sample and then comparison with wild-type samples.
Palmitate oxidation assay
Measurement of palmitate oxidation capacity of hepatocytes using [14C] palmitate has been described previously.42 The radioactivity of the culture medium as well as the acid soluble intermediates after chemical extraction of cellular lipids was measured. Total oxidation was then calculated as a function of both [14C] CO2 produced and incomplete oxidation products.
Liver glycogen quantification
Liver tissues were harvested from 24-week-old male mice after 6 h of fasting. Glycogen content was measured using a glucose oxidase reaction-based glycogen assay kit according to manufacturer’s directions (Sigma).
Statistical analysis
Statistical analysis was performed using GraphPad Prism Software (San Diego, CA, USA). All data shown as bar graphs are mean±S.E.M. Data were analysed by t-test, one-way or two-way ANOVA with Bonferroni’s or Tukey’s post test for comparison of multiple columns (as appropriate). A P-value of <0.05 was considered statistically significant and *P<0.05, **P<0.01 and ***P<0.001. Area under the curve was calculated by applying the trapezoidal method.
Acknowledgments
We thank Lorraine Elkerbout, Stacey Fynch, William Stanley, Jasmine McLeod, Lara Yachou-Wos, Sam Thorburn, Eva Orlowski (all St Vincent’s Institute), Adrienne Laskowski (Murdoch Childrens Research Institute), Philippe Bouillet, Andreas Strasser, Lorraine O’Reilly, Marco Herold, Margs Brennan, David Huang, David Segal (The Walter and Eliza Hall Institute), Sara Ellis, Jill Danne and Chad Johnson (Peter MacCallum Cancer Centre), Greg Steinberg (McMaster University, Canada) and Sof Andrikopoulos (University of Melbourne) for experimental advice, technical assistance, reagents and critical reading of the manuscript. This work was funded by a National Health and Medical Research Council of Australia (NHMRC) Project grant, a NHMRC and Juvenile Diabetes Research Foundation joint special programme grant, and fellowships from the NHMRC. This work received support from the Operational Infrastructure Support Scheme of the Government of Victoria.
Author contributions
JAW, SG, CT, ENG, AEF, TC, JG, EGP, DS, CV, and CS performed experiments, analysed data and revised the manuscript. JAW, SG, BK, TWHK, SLM, and HET designed the study and wrote the manuscript. MTR, DRT, and BEK contributed to conception, design and interpretation of this work, provided essential reagents and critically revised the manuscript.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by L Scorrano
The authors declare no conflict of interest.
Supplementary Material
References
- Szendroedi J, Phielix E, Roden M. The role of mitochondria in insulin resistance and type 2 diabetes mellitus. Nat Rev Endocrinol 2012; 8: 92–103. [DOI] [PubMed] [Google Scholar]
- Papa S, Martino PL, Capitanio G, Gaballo A, De Rasmo D, Signorile A et al. The oxidative phosphorylation system in mammalian mitochondria. Adv Exp Med Biol 2012; 942: 3–37. [DOI] [PubMed] [Google Scholar]
- Czabotar PE, Lessene G, Strasser A, Adams JM. Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat Rev Mol Cell Biol 2014; 15: 49–63. [DOI] [PubMed] [Google Scholar]
- Giam M, Huang DC, Bouillet P. BH3-only proteins and their roles in programmed cell death. Oncogene 2008; 27: S128–S136. [DOI] [PubMed] [Google Scholar]
- Bouillet P, Metcalf D, Huang DC, Tarlinton DM, Kay TW, Kontgen F et al. Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science 1999; 286: 1735–1738. [DOI] [PubMed] [Google Scholar]
- Ekoff M, Kaufmann T, Engstrom M, Motoyama N, Villunger A, Jonsson JI et al. The BH3-only protein Puma plays an essential role in cytokine deprivation induced apoptosis of mast cells. Blood 2007; 110: 3209–3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlacher M, Michalak EM, Kelly PN, Labi V, Niederegger H, Coultas L et al. BH3-only proteins Puma and Bim are rate-limiting for gamma-radiation- and glucocorticoid-induced apoptosis of lymphoid cells in vivo. Blood 2005; 106: 4131–4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthalakath H, O'Reilly LA, Gunn P, Lee L, Kelly PN, Huntington ND et al. ER stress triggers apoptosis by activating BH3-only protein Bim. Cell 2007; 129: 1337–1349. [DOI] [PubMed] [Google Scholar]
- Frank DO, Dengjel J, Wilfling F, Kozjak-Pavlovic V, Hacker G, Weber A. The pro-apoptotic BH3-only protein Bim interacts with components of the translocase of the outer mitochondrial membrane (TOM). PLoS One 2015; 10: e0123341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Swanson BJ, Wang M, Hildeman DA, Schaefer BC, Liu X et al. Constitutive association of the proapoptotic protein Bim with Bcl-2-related proteins on mitochondria in T cells. Proc Natl Acad Sci USA 2004; 101: 7681–7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jette CA, Flanagan AM, Ryan J, Pyati UJ, Carbonneau S, Stewart RA et al. BIM and other BCL-2 family proteins exhibit cross-species conservation of function between zebrafish and mammals. Cell Death Differ 2008; 15: 1063–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman SB, Chen Y-b QiB, McCaffery JM, Rucker EB, Goebbels S et al. Bcl-xL increases mitochondrial fission, fusion, and biomass in neurons. J Cell Biol 2009; 184: 707–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perciavalle RM, Stewart DP, Koss B, Lynch J, Milasta S, Bathina M et al. Anti-apoptotic MCL-1 localizes to the mitochondrial matrix and couples mitochondrial fusion to respiration. Nat Cell Biol 2012; 14: 575–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houten SM, Wanders RJ. A general introduction to the biochemistry of mitochondrial fatty acid beta-oxidation. J Inherit Metab Dis 2010; 33: 469–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hue L, Taegtmeyer H. The Randle cycle revisited: a new head for an old hat. Am J Physiol Endocrinol Metab 2009; 297: E578–E591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauner H. The impact of pharmacotherapy on weight management in type 2 diabetes. Int J Obes Relat Metab Disord 1999; 23: S12–S17. [DOI] [PubMed] [Google Scholar]
- Andrikopoulos S, Massa CM, Aston-Mourney K, Funkat A, Fam BC, Hull RL et al. Differential effect of inbred mouse strain (C57BL/6, DBA/2, 129T2) on insulin secretory function in response to a high fat diet. J Endocrinol 2005; 187: 45–53. [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhou D, Zhang F, Tu Y, Xia Y, Wang H et al. Liver Patt1 deficiency protects male mice from age-associated but not high-fat diet-induced hepatic steatosis. J Lipid Res 2012; 53: 358–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel VT, Shulman GI. Mechanisms for insulin resistance: common threads and missing links. Cell 2012; 148: 852–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins TD, Holden CR, Sansbury BE, Gibb AA, Shah J, Zafar N et al. Metabolic remodeling of white adipose tissue in obesity. Am J Physiol Endocrinol Metab 2014; 307: E262–E277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deepa SS, Pulliam D, Hill S, Shi Y, Walsh ME, Salmon A et al. Improved insulin sensitivity associated with reduced mitochondrial complex IV assembly and activity. FASEB J 2013; 27: 1371–1380. [DOI] [PubMed] [Google Scholar]
- Zhan Y, Zhang Y, Gray D, Carrington EM, Bouillet P, Ko HJ et al. Defects in the Bcl-2-regulated apoptotic pathway lead to preferential increase of CD25 low Foxp3+ anergic CD4+ T cells. J Immunol 2011; 187: 1566–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagenbuchner J, Kuznetsov A, Hermann M, Hausott B, Obexer P, Ausserlechner MJ. FOXO3-induced reactive oxygen species are regulated by BCL2L11 (Bim) and SESN3. J Cell Sci 2012; 125: 1191–1203. [DOI] [PubMed] [Google Scholar]
- Aichberger KJ, Gleixner KV, Mirkina I, Cerny-Reiterer S, Peter B, Ferenc V et al. Identification of proapoptotic Bim as a tumor suppressor in neoplastic mast cells: role of KIT D816V and effects of various targeted drugs. Blood 2009; 114: 5342–5351. [DOI] [PubMed] [Google Scholar]
- Egle A, Harris AW, Bouillet P, Cory S. Bim is a suppressor of Myc-induced mouse B cell leukemia. Proc Natl Acad Sci USA 2004; 101: 6164–6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alavian KN, Li H, Collis L, Bonanni L, Zeng L, Sacchetti S et al. Bcl-xL regulates metabolic efficiency of neurons through interaction with the mitochondrial F1FO ATP synthase. Nat Cell Biol 2011; 13: 1224–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YB, Aon MA, Hsu YT, Soane L, Teng X, McCaffery JM et al. Bcl-xL regulates mitochondrial energetics by stabilizing the inner membrane potential. J Cell Biol 2011; 195: 263–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZX, Pervaiz S. Involvement of cytochrome c oxidase subunits Va and Vb in the regulation of cancer cell metabolism by Bcl-2. Cell Death Differ 2010; 17: 408–420. [DOI] [PubMed] [Google Scholar]
- Walford GA, Gustafsson S, Rybin D, Stancakova A, Chen H, Liu CT et al. Genome-wide association study of the modified Stumvoll Insulin Sensitivity Index identifies BCL2 and FAM19A2 as novel insulin sensitivity loci. Diabetes 2016; 65: 3200–3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena R, Elbers CC, Guo Y, Peter I, Gaunt TR, Mega JL et al. Large-scale gene-centric meta-analysis across 39 studies identifies type 2 diabetes loci. Am J Hum Genet 2012; 90: 410–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shungin D, Winkler TW, Croteau-Chonka DC, Ferreira T, Locke AE, Magi R et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature 2015; 518: 187–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinds MG, Smits C, Fredericks-Short R, Risk JM, Bailey M, Huang DC et al. Bim, Bad and Bmf: intrinsically unstructured BH3-only proteins that undergo a localized conformational change upon binding to prosurvival Bcl-2 targets. Cell Death Differ 2007; 14: 128–136. [DOI] [PubMed] [Google Scholar]
- Gimenez-Cassina A, Garcia-Haro L, Choi CS, Osundiji MA, Lane EA, Huang H et al. Regulation of hepatic energy metabolism and gluconeogenesis by BAD. Cell Metab 2014; 19: 272–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick D, Zhang W, Beaton M, Marsboom G, Gruber M, Simon MC et al. BNip3 regulates mitochondrial function and lipid metabolism in the liver. Mol Cell Biol 2012; 32: 2570–2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litwak SA, Loh K, Stanley WJ, Pappas EG, Wali JA, Selck C et al. p53-upregulated-modulator-of-apoptosis (PUMA) deficiency affects food intake but does not impact on body weight or glucose homeostasis in diet-induced obesity. Sci Rep 2016; 6: 23802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill HM, Holloway GP, Steinberg GR. AMPK regulation of fatty acid metabolism and mitochondrial biogenesis: implications for obesity. Mol Cell Endocrinol 2013; 366: 135–151. [DOI] [PubMed] [Google Scholar]
- Concannon CG, Tuffy LP, Weisova P, Bonner HP, Davila D, Bonner C et al. AMP kinase-mediated activation of the BH3-only protein Bim couples energy depletion to stress-induced apoptosis. J Cell Biol 2010; 189: 83–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kefas BA, Cai Y, Ling Z, Heimberg H, Hue L, Pipeleers D et al. AMP-activated protein kinase can induce apoptosis of insulin-producing MIN6 cells through stimulation of c-Jun-N-terminal kinase. J Mol Endocrinol 2003; 30: 151–161. [DOI] [PubMed] [Google Scholar]
- Meisse D, Van de Casteele M, Beauloye C, Hainault I, Kefas BA, Rider MH et al. Sustained activation of AMP-activated protein kinase induces c-Jun N-terminal kinase activation and apoptosis in liver cells. FEBS Lett 2002; 526: 38–42. [DOI] [PubMed] [Google Scholar]
- Dugan LL, You YH, Ali SS, Diamond-Stanic M, Miyamoto S, DeCleves AE et al. AMPK dysregulation promotes diabetes-related reduction of superoxide and mitochondrial function. J Clin Invest 2013; 123: 4888–4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vial G, Chauvin MA, Bendridi N, Durand A, Meugnier E, Madec AM et al. Imeglimin normalizes glucose tolerance and insulin sensitivity and improves mitochondrial function in liver of a high-fat, high-sucrose diet mice model. Diabetes 2015; 64: 2254–2264. [DOI] [PubMed] [Google Scholar]
- Fullerton MD, Galic S, Marcinko K, Sikkema S, Pulinilkunnil T, Chen ZP et al. Single phosphorylation sites in Acc1 and Acc2 regulate lipid homeostasis and the insulin-sensitizing effects of metformin. Nat Med 2013; 19: 1649–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Montalvo A, Mercken EM, Mitchell SJ, Palacios HH, Mote PL, Scheibye-Knudsen M et al. Metformin improves healthspan and lifespan in mice. Nat Commun 2013; 4: 2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur V, Connor T, Sanigorski A, Martin SD, Bruce CR, Henstridge DC et al. Disruption of the class IIa HDAC Corepressor complex increases energy expenditure and lipid oxidation. Cell Rep 2016; 16: 2802–2810. [DOI] [PubMed] [Google Scholar]
- Kooptiwut S, Zraika S, Thorburn AW, Dunlop ME, Darwiche R, Kay TW et al. Comparison of insulin secretory function in two mouse models with different susceptibility to beta-cell failure. Endocrinology 2002; 143: 2085–2092. [DOI] [PubMed] [Google Scholar]
- McKenzie MD, Dudek NL, Mariana L, Chong MM, Trapani JA, Kay TW et al. Perforin and Fas induced by IFNgamma and TNFalpha mediate beta cell death by OT-I CTL. Int Immunol 2006; 18: 837–846. [DOI] [PubMed] [Google Scholar]
- Gurzov EN, Tran M, Fernandez-Rojo MA, Merry TL, Zhang X, Xu Y et al. Hepatic oxidative stress promotes insulin-STAT-5 signaling and obesity by inactivating protein tyrosine phosphatase N2. Cell Metab 2014; 20: 85–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittig I, Braun HP, Schagger H. Blue native PAGE. Nat Protoc 2006; 1: 418–428. [DOI] [PubMed] [Google Scholar]
- Martin SD, Morrison S, Konstantopoulos N, McGee SL. Mitochondrial dysfunction has divergent, cell type-dependent effects on insulin action. Mol Metab 2014; 3: 408–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier AE, Thorburn DR. Biochemical analyses of the electron transport chain complexes by spectrophotometry. Methods Mol Biol 2012; 837: 49–62. [DOI] [PubMed] [Google Scholar]
- Mehlem A, Hagberg CE, Muhl L, Eriksson U, Falkevall A. Imaging of neutral lipids by oil red O for analyzing the metabolic status in health and disease. Nat Protoc 2013; 8: 1149–1154. [DOI] [PubMed] [Google Scholar]
- Shackleton M, O'Reilly LA, Sutherland KD, Bath ML, Ellis S, Strasser A et al. Impaired lactation in mice expressing dominant-negative FADD in mammary epithelium. Dev Dyn 2009; 238: 1010–1016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.