Abstract
Expression of the tumor suppressor gene NR4A3 is silenced in the blasts of acute myeloid leukemia (AML), irrespective of the karyotype. Although the transcriptional reactivation of NR4A3 is considered to have a broad-spectrum anti-leukemic effect, the therapeutic modalities targeting this gene have been hindered by our minimal understanding of the transcriptional mechanisms regulating its expression, particularly in human AML. Here we show the role of intragenic DNA hypermethylation in reducing the expression of NR4A3 in AML. Bisulfite sequencing analysis revealed that CpG sites at the intragenic region encompassing exon 3 of NR4A3, but not the promoter region, are hypermethylated in AML cell lines and primary AML cells. A DNA methyltransferase inhibitor restored the expression of NR4A3 following a reduction in DNA methylation levels at intragenic CpG sites. The in silico data revealed an enrichment of H3K4me1 and H2A.Z at exon 3 of NR4A3 in human non-malignant cells but that was excluded specifically in leukemia cells with CpG hypermethylation. This suggests that exon 3 represents a functional regulatory element involved in the transcriptional regulation of NR4A3. Our findings improve the current understanding of the mechanism underlying NR4A3 silencing and facilitate the development of NR4A3-targeted therapy.
Keywords: NR4A3, Tumor suppressor gene, Acute myeloid leukemia, DNA hypermethylation
1. Introduction
Nuclear family 4 subgroup A (NR4A) orphan nuclear receptors have no known natural ligands and consist of three mammalian members: 1) NR4A1 (Nur77), 2) NR4A2 (Nurr1), and 3) NR4A3 (Nor1). All three members share common structural properties in their carboxyl-terminal ligand and central DNA binding domains, while their amino-terminal domains are highly divergent [1–3]. All three receptors are widely expressed in different types of tissues and a variety of physiological signals can induce their expression, leading to the activation of NR4A target genes related to the cell cycle, apoptosis, inflammation, atherogenesis, metabolism, or DNA repair in a stimulus- and cell context-dependent manner [1,3,4]. In addition, the NR4A receptors are involved in tumorigenesis [2–5].
NR4A1 and NR4A3 are reportedly silenced in the blasts of patients with acute myeloid leukemia (AML) irrespective of the karyotype [6]. In line with this finding, Nr4a1−/−/Nr4a3−/− mice rapidly develop AML within one month after birth [6]. These results suggest that NR4A1 and NR4A3 function as tumor suppressor genes in myeloid malignancies and that NR4A receptors have a crucial role in the pathogenesis of AML [6]. Thus, unveiling the molecular mechanisms that regulate NR4A expression in AML would facilitate the development of novel therapies, including the transcriptional reactivation of the gene. However, the therapeutic modalities targeting NR4A receptors have been hindered by our minimal understanding of the mechanisms underlying reduced NR4A1 and NR4A3 expression, particularly in human AML cells.
Aberrant DNA methylation is a common mechanism in the pathogenesis of several types of cancer [7–13]. It is well-known that the expression of several tumor suppressor genes, such as p16 and MLH1, is repressed due to DNA hypermethylation at their promoter region [9]. Recently, tumorigenesis resulting from DNA hypermethylation at both promoter and transcribed regions has been reported [14]. In addition, several studies have shown both positive and negative correlations between intragenic DNA methylation and gene expression [14–17]. Due to the complex effects of abnormal DNA methylation on gene expression and tumorigenesis, it is essential to investigate the impact of aberrant DNA methylation on gene expression systematically at affected loci by individual tumor suppressors. Given that loss-of-function mutations in NR4A have not been reported in AML to date, we hypothesized that abnormal DNA methylation contributes to a reduction in NR4A expression in AML. In this study, we focused on NR4A3 and analyzed the DNA methylation status of NR4A3 in human AML cells. DNA hypermethylation at the promoter region of NR4A3 was not detected, while its intragenic DNA hypermethylation was associated with its reduced expression. Therefore, we propose the potential role of intragenic DNA hypermethylation in the transcriptional repression of NR4A3 in AML.
2. Materials and methods
2.1. Cells from human subjects and cell lines
We analyzed the bone marrow (BM) from AML patients and control subjects after obtaining written informed consent. Five BM cells from patients diagnosed with lymphoid neoplasia without BM invasion, idiopathic thrombocytopenic purpura, or Kikuchi’s disease were used as normal controls. The patient characteristics are shown in Table 1. Procedures were approved by the Human Investigation Review Committee at Chiba University Hospital. Mononuclear cells from BM samples were isolated on Ficoll-Paque PLUS (GE Healthcare, Pittsburgh, PA, USA). Specimens from control patients underwent CD34 positive selection by magnetic antibody-conjugated sorting (Miltenyi Biotech, Bergisch Gladbach, Germany). Primary AML cells were cultured in StemSpan serum-free expansion media (Stemcell Technologies, La Jolla, CA, USA) supplemented with 10 ng/mL recombinant human stem cell factor (SCF), Flt3 ligand (Flt3L), thrombopoietin (TPO), interleukin-3 (IL-3), and IL-6 (PeproTech, Rocky Hill, NJ, USA). All AML cell lines were maintained in RPMI 1640 supplemented with 10% fetal bovine serum (Thermo Scientific, Waltham, MS, USA) and 1% penicillin–streptomycin at 37 °C in a 5% CO2 atmosphere.
Table 1.
Patient Characteristics.
FAB, French–American–British classification; WBC, white blood cell; AML, acute myeloid leukemia; BM, bone marrow; F, female; M, male; DLBCL, diffuse large B-cell lymphoma; AITL, angioimmunoblastic T-cell lymphoma; ITP, idiopathic thrombocytopenic purpura.
| ID | Age | Sex | Disease | FAB | WBC (/μl) | Cytogenetics | Blasts in BM (%) |
|---|---|---|---|---|---|---|---|
| AML-1 | 59 | M | AML | M2 | 22000 | Normal | 88.0 |
| AML-2 | 39 | M | AML | M3 | 5300 | t(15;17) (q22;q12) | 77.0 |
| AML-3 | 64 | F | AML | M4 | 144100 | Normal | 82.2 |
| AML-4 | 58 | F | AML | M5a | 78500 | Normal | 80.8 |
| AML-5 | 72 | M | AML | M0 | 17500 | Complex karyotype | 91.6 |
| AML-6 | 39 | F | AML | M4 | 77800 | Complex karyotype | 83.8 |
| AML-7 | 50 | F | AML | M1 | 30700 | Normal | 77.2 |
| AML-8 | 41 | M | AML | M5a | 129700 | Normal | 88.2 |
| AML-9 | 69 | M | AML | M5a | 19000 | 46,XY,t(9;11)(p22;q23) | 92.2 |
| AML-10 | 58 | F | AML | M2 | 8100 | 45,X,-X,t(8;21)(q22;q22), del(9)(q) | 74.2 |
| Control-1 | 69 | M | DLBCL | – | – | – | – |
| Control-2 | 16 | F | AITL | – | – | – | – |
| Control-3 | 54 | F | ITP | – | – | – | – |
| Control-4 | 51 | M | DLBCL | – | – | – | – |
| Control-5 | 27 | M | Kikuchi’s disease | – | – | – | – |
2.2. Decitabine treatment
Human leukemia cell lines and primary AML cells were treated with different final concentrations of decitabine (DAC; Sigma-Aldrich, St Louis, MO, USA) or the same volume of phosphate buffered saline, replacing the medium and adding DAC or phosphate buffered saline every 24 h until harvested for RNA and DNA extraction.
2.3. Bisulfite sequencing
Genomic DNA was extracted following the procedures outlined in the Wizard Genomic DNA Purification Kit (Promega, San Luis Obispo, CA, USA) for cell lines and AllPrep DNA/RNA Mini Kit (Qiagen, Venlo, Netherlands) for human primary AML cells. The DNA was bisulfite-converted using MethylEasy Xceed Rapid DNA bisulfite modification kit (Human Genetic Signatures, North Ryde, NSW, Australia) according to the manufacture’s protocol. The modified DNA was amplified with bisulfite sequencing PCR (BSP). The BSP primers were designed by the online BiSearch software [18]. The sequences of the primers and information regarding the PCR settings are shown in Table 2. The BSP was carried out using EpiTaq HS (Takara Bio, Shiga, Japan). The PCR products were purified using NucleoSpin Gel and PCR clean-up (MACHEREY-NAGEL, Düren, Germany) and then cloned into the T-vector pMD20 (Takara Bio) for sequencing. For each amplicon, 8–12 individual clones were sequenced by Eurofins Genomics (Tokyo, Japan). The final sequence results were processed by online QUMA software [19].
Table 2.
The sequences of primers and conditions for bisulfite-sequencing PCR.
| Name | Analyzed regions relative to transcriptional start site of NR4A3 | No. of analyzed CpGs | Forward (5′ → 3′) | Reverse (5′ → 3′) | Annealing temperature (°C) | Cycle | Length(bp) |
|---|---|---|---|---|---|---|---|
| NR4A3-Region 1 | −551 to −24 | 51 | TTATGTAAGAGGAAAGTGTAGTT | CTCCATAAAATACCTAAAATAC | 54 | 48 | 528 |
| NR4A3-Region 2 | −37 to +485 | 47 | AGGTATTTTATGGAGAG | AAAAATCACAACTACTATCC | 54.4 | 40 | 522 |
| NR4A3-Region 3 | +6151 to +6535 | 15 | TTTGTTTTTGAGAGATTTTTTTT | AATAACTACTAATACTACTAC | 53.2 | 45 | 385 |
| NR4A3-Region 4 | +7116 to +7432 | 9 | GAGGGTTGTAAGGGTTTTTT | CAAATCCTTTTAAAACTCTAC | 57.7 | 40 | 317 |
2.4. In vitro colony-forming assay
Kasumi-1, THP-1 and MOLM-13 cells were seeded into a methyl-cellulose medium (Methocult M3234; Stemcell Technologies) with 10 ng/mL recombinant human SCF, Flt3L, TPO, IL-3, and IL-6. Colonies propagated in culture were counted on day 10.
2.5. Western blot analysis
Samples were separated by SDS-PAGE, transferred to a PVDF membrane, and detected by Western blotting using a mouse antibody against FLAG M2 (Sigma-Aldrich) and NR4A3 (Perseus Proteomics Inc, Tokyo, Japan).
2.6. Quantitative real-time polymerase chain reaction analysis
The total RNA was isolated using the RNeasy Mini Kit (Qiagen). The reverse transcription step was performed with the Prime-Script RT Reagent Kit (Takara Bio) according to the manufacturer’s instructions. The resulting cDNA samples were subjected to quantitative real-time PCR to measure the levels of NR4A3 mRNA using the TaqMan Assay-on-Demand kit with the StepOne Real-Time PCR System (Applied Biosystems, Norwalk, CT, USA). The Taq-Man probes were purchased from Applied Biosystems (NR4A3: Hs00545009 g1; GAPDH: Hs99999905 m1).
2.7. Retroviral vectors and virus production
Human NR4A3 cDNA tagged with a 3 × Flag was subcloned into a site upstream of an IRES-EGFP construct in pGCDNsam, a retroviral vector with a long terminal repeat (LTR) derived from a murine stem cell virus [20,21]. A recombinant vesicular stomatitis virus glycoprotein-pseudotyped high-titer retrovirus was generated by a 293GPG packaging cell line [22]. The virus in the supernatants of 293GPG cells was concentrated by centrifugation at 6000 × g for 16 h.
2.8. Transduction of AML cells
Kasumi-1, THP-1 and MOLM-13 cells overexpressing NR4A3 were generated by infection with the supernatants from transfected 293GPG cells in the presence of 5 μg/mL protamine sulfate for 72 h with subsequent sorting of GFP-positive cells using FACS Aria II (BD Biosciences, Bedford, MA, USA).
2.9. DNA methylation profiles and bioinformatics analysis
We used publically available DNA methylation profiles (all based on HumanMethylation450 K BeadChip) of AML patients from The Cancer Genome Atlas (TCGA) [23] and of healthy individuals (GSE40279, GSE35069) [24,25]. Beta values were converted into M-values, on which statistical analysis was performed. For Kaplan-Meier estimation of overall survival and relapse-free survival, we stratified samples by median DNA methylation levels and adjusted the results for multiple testing (log-rank test calculated in R/survival package).
2.10. Statistical analysis
The statistical significance of the data was determined with Student’s t-test using JMP 11 (SAS Institute Inc., Cary, NC, USA).
3. Results
3.1. Forced expression of NR4A3 by virus-mediated gene delivery suppresses the proliferation activity of human AML cells
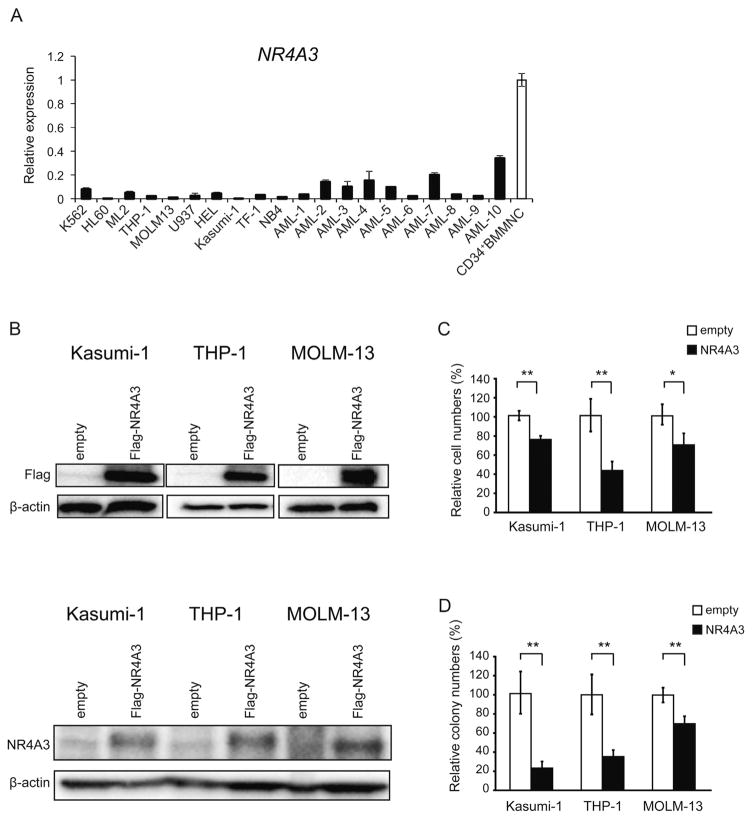
We first checked the expression levels of NR4A3 in 10 AML cell lines and 8 primary AML samples. The expression levels were drastically lower in all cell lines and primary AML samples compared to the controls (CD34+ BM mononuclear cells from control subjects) (Fig. 1A). To evaluate the tumor suppressive function of NR4A3 in human AML cells, we overexpressed NR4A3 in Kasumi-1, THP-1 and MOLM-13 cells using a virus-mediated gene delivery system. NR4A3 was successfully overexpressed in these cells (Fig. 1B), and led to the suppression of their growth as well as a significant reduction in the number of colonies generated in the methylcellulose (Fig. 1C and D). These results indicate that NR4A3 functions as a tumor suppressor gene in human AML as was reported for murine cells [6,26,27].
Fig. 1.
NR4A3 functions as a tumor suppressor gene in human acute myeloid leukemia (AML) cells. (A) Quantitative real-time PCR analysis of NR4A3 expression in AML cell lines, human primary AML samples and CD34+ bone marrow mononuclear cells (BMMNC) from control subjects (Control-3, Control-4 and Control-5). mRNA levels were normalized to GAPDH expression. The expression level in CD34+BMMNC is shown as the mean ± S.D. (n = 3). The expression level in AML cell lines and human primary AML samples relative to the mean level in CD34+BMMNC is represented as the mean ± S.D. for triplicate analyses. (B-D) Kasumi-1, THP-1 and MOLM-13 cells were transduced with either the control or NR4A3 retrovirus and were purified for the experiments by cell sorting using green fluorescent protein (GFP) as a marker 72 h after infection. (B) NR4A3 expressions 72 h after GFP sorting were detected by Western blotting using the anti-FLAG antibody (upper panel) and the anti-NR4A3 antibody (lower panel). (C) The growth of the AML cells transduced with NR4A3. The data are shown as the means ± S.D. for triplicate analyses. (D) Colony formations in methylcellulose were determined for AML cells transduced with NR4A3. Total colonies were scored on day 10. The percentage of colony numbers relative to the controls is shown as the means ± S.D. for triplicate analyses. *, P < 0.05; **, P < 0.01
3.2. Hypermethylated CpG sites are identified at the intragenic region encompassing exon 3 of NR4A3 in AML cell lines but not at its promoter region
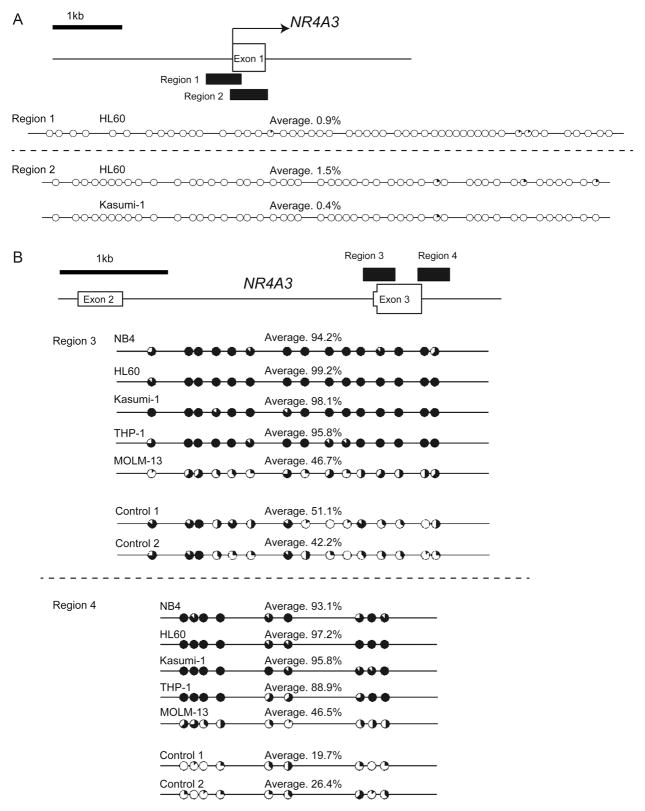
To test our hypothesis that DNA hypermethylation is involved in the mechanism of reduced NR4A3 expression, we assessed the DNA methylation status of NR4A3 by bisulfite sequencing analysis. We first investigated the DNA methylation status at the potential promoter sequences—Region 1 (528 bp) and Region 2 (623 bp)—spanning −551 to −24 and −38 to +584 relative to the transcription start site (TSS) of NR4A3, respectively (Fig. 2A). Unexpectedly, these regions were not hypermethylated in HL60 and Kasumi-1 cells (Fig. 2A). As intragenic CpG islands can also be preferentially methylated in cancer [28], we next focused on DNA methylation at the transcribed region. Intragenic DNA hyper-methylation affects the gene expression in both a positive and negative manner in cancer cells [14]. Notably, those associated with transcriptional repression have often been reported to be associated with H3K4me1 and H2A.Z enrichment peaks in human mammary epithelial cells (HMECs) [14]. This suggests that some transcribed regions with hypermethylated CpGs may represent functional regulatory elements, such as enhancers or alternative promoters in non-malignant cells [14]. Based on the results of this report, we assessed the distribution of H3K4 me1 and H2A.Z levels using the data from the Encyclopedia of DNA Elements (ENCODE; https://genome.ucsc.edu/ENCODE/, chromatin immunoprecipitation sequencing [ChIP-seq] data) to determine the region of the gene body to be used for DNA methylation analysis. The enrichment peaks of H3K4me1 and/or H2A.Z were observed in HMECs, human monocytes (CD14+ blood cells), and human B cells (CD20+ blood cells) at the region encompassing exon 3 of NR4A3 (Fig. S1). Thus, we assessed the DNA methylation profile in Region 3 (+6151 to +6535) and Region 4 (+7116 to +7432), including exon 3, which contains 15 and 9 CpG sites, respectively. Bisulfite sequencing analysis revealed that compared to the controls, the CpGs within these intragenic regions were highly methylated in NB4, HL60, Kasumi-1, and THP-1 cells and mildly methylated in MOLM-13 cells (Fig. 2B). Interestingly, H3K4me1 and H2A.Z enrichment peaks were not observed at exon 3 of NR4A3 in K562 cells from the ENCODE data, suggesting that these histone modifications were replaced by DNA methylation in AML cells (Fig. S1). Taken together, intragenic DNA hypermethylation of NR4A3 in AML cells was identified at the region encompassing exon 3, which potentially includes functional elements but not in the promoter region.
Fig. 2.
The hypermethylated region was identified at the region encompassing exon 3 of NR4A3 but not its promoter region.
(A–B) Targeted regions of bisulfite sequencing PCR are shown as black boxes. The methylation status of CpG sites at the potential promoter region (A) and at the region encompassing exon 3 (B) of NR4A3 was analyzed in AML cell lines and control samples (n = 2) using bisulfite sequencing. The percentages of methylation at each CpG site are indicated by black coloring within a pie chart. The percentages of methylation of the total CpG sites in the region of PCR products are also indicated as an average.
3.3. Decitabine restores the expression of NR4A3 and induces DNA demethylation at the region encompassing exon 3
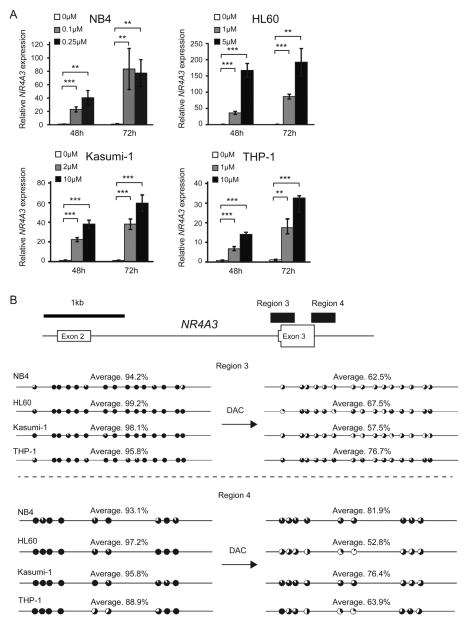
To evaluate the contribution of DNA hypermehylation at the region encompassing exon 3 of NR4A3 to its transcriptional repression, AML cell lines with heavily hypermethylated CpGs in this region were treated with the DNA methyltransferase inhibitor decitabine (DAC). DAC exposure restored the expression of NR4A3 in all AML cell lines in a dose- and time-dependent manner (Fig. 3A). Notably, the frequencies of methylated CpG sites in AML cells were reduced after DAC exposure (Fig. 3B). These results suggest that DNA hypermethylation of the region encompassing exon 3 of NR4A3 is associated with its reduced expression in AML cells.
Fig. 3.
Decitabine restored the silenced expression of NR4A3 accompanied by demethylation at the region encompassing exon 3 of NR4A3.
(A–B) AML cell lines, in which the region encompassing exon 3 is heavily methylated, were treated with different doses of decitabine (DAC) for 48 and 72 h. (A) mRNA levels of NR4A3 in the cells were evaluated by quantitative real-time PCR and normalized to GAPDH expression. Data are shown as the mean ± S.D. for triplicate analyses. (B) The methylation status of CpG sites at the region encompassing exon 3 of NR4A3 in AML cell lines was analyzed before and after DAC exposure using bisulfite sequencing. The percentages of methylation at each CpG site are indicated by black coloring in pie charts. The percentages of methylation of the total CpG sites in the region of PCR products are also indicated with an average. **, P < 0.01; ***, P < 0.001
3.4. DNA hypermethylation at the region encompassing exon 3 of NR4A3 is associated with its reduced expression in human primary AML samples
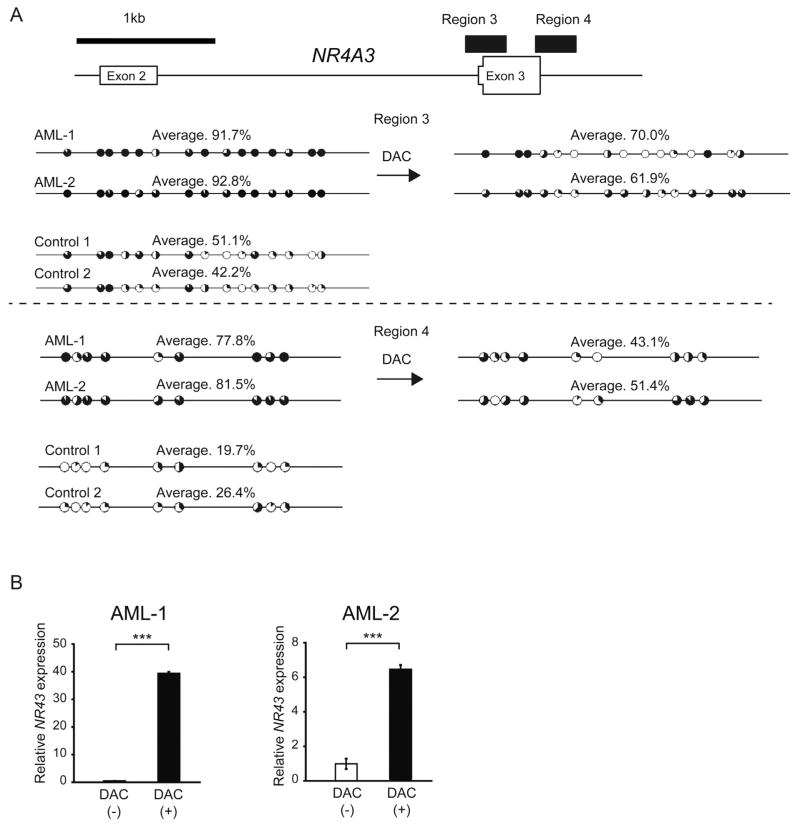
We next validated the results from AML cell lines in human primary AML samples. Hypermethylated intragenic DNA at the region encompassing exon 3 in AML samples was detected similarly to human AML cell lines (Figs. 4 A and Fig. S2A). Furthermore, DAC exposure restored the expression of NR4A3 and reduced the methylated levels of the region (Fig. 4B), although the change of its protein level could not be evaluated due to the paucity of our primary samples. Thus, DNA hypermethylation at the region encompassing exon 3 of NR4A3 is associated with its reduced expression in both AML cell lines and human primary AML samples.
Fig. 4.
DNA hypermethylation at the region encompassing exon 3 of NR4A3 is associated with its reduced expression in human primary AML samples.
(A) Two human primary AML cell samples were treated with 0.33 μM (AML-1) or 1 μM (AML-2) of DAC for 72 h. The methylation status of CpG sites at the region encompassing exon 3 of NR4A3 in human primary AML samples and control samples (n = 2) was analyzed before and after DAC exposure using bisulfite sequencing. The percentages of methylation at each CpG site are indicated by the black coloring on the pie charts. The percentages of methylation of the total CpG sites in the region of PCR products are also indicated with an average. (B) Two human primary AML cell samples were treated with 0.33 μM (AML-1) or 5 μM (AML-2) of DAC for 72 h. mRNA levels of NR4A3 in primary human cells were evaluated by quantitative real-time PCR and normalized to GAPDH expression. Data are shown as the mean ± S.D. for triplicate analyses. **, P < 0.01, ***, P < 0.001.
3.5. DNA methylation status at the region encompassing exon 3 of NR4A3 shows a trend for survival association
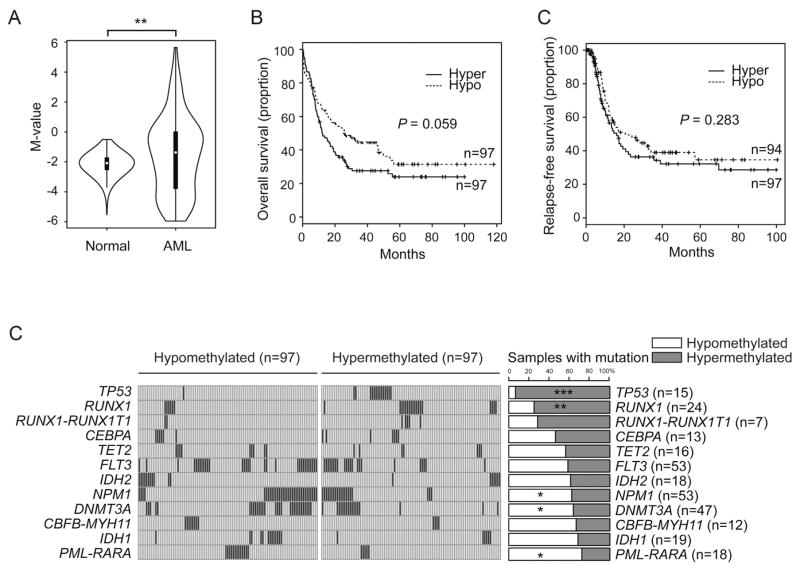
To assess the association between DNA methylation status at the region of exon 3 of NR4A3, and the prognosis and genetic background, we analyzed DNA methylation profile of 194 AML patients from The Cancer Genome Atlas (TCGA) [23]. In line with the findings in the current study, the region encompassing exon 3 of NR4A3 in AML patients was significantly hypermethylated compared with normal controls (Fig. 5A). Although the methylation status was not associated with relapse-free survival, higher methylation at the region encompassing exon 3 of NR4A3 was associated with a trend for poor overall survival (Fig. 5B). Furthermore, increased methylation at the region was significantly associated with mutations in TP53 and RUNX1, and inversely correlated with mutations in NPM1 and DNMT3A, and PML-RARA rearrangements (Fig. 5C).
Fig. 5.
DNA methylation level at exon 3 of NR4A3 influences the prognosis of AML patients, and is associated with genetic abnormalities.
(A) Violin plots showing the M-value distribution of DNA methylation in exon 3 of NR4A3 of 656 healthy controls (GSE40279, GSE35069) [24,25] and of 194 AML patients from The Cancer Genome Atlas (TCGA) database [23]. (B) Overall survival curve (left panel) and relapse-free survival curve (right panel) of TCGA samples classified by median DNA methylation level at exon 3 of NR4A3. (C) Enrichment of specific mutations according to DNA methylation level at exon 3 of NR4A3. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
4. Discussion
AML is a heterogeneous disease with a multitude of chromosomal abnormalities and gene mutations [23,29]. This heterogeneity is a formidable obstacle in the development of therapies applicable for a wide range of AML patients. From this standpoint, transcriptional reactivation of the tumor suppressor NR4A3 is advantageous because its expression has been reported to be commonly silenced in the blasts of patients with AML, irrespective of karyotype [6]. However, the underlying mechanism of the reduced expression of NR4A3 in AML is poorly understood. To our knowledge, this is the first study demonstrating the relationship between NR4A3 silencing and DNA methylation in human AML cells.
In this study, we first confirmed the tumor suppressive function of NR4A3 in human AML cells. To overcome the limitations of standard plasmid transfection approaches for NR4A3, Boudreaux et al. established a tumor suppressive function of NR4A3 in human AML cell lines using in vitro transcribed RNA electroporation system [26]. However, gene expression by electroporation is transient and therefore not suitable for long-term expression in human AML cells. In addition, there are concerns regarding the possibility that electroporation damages the cells and consequently affects the gene expression profile of these cells. In this study, we used a retroviral gene transfer approach for stable and robust expression of NR4A3, and unequivocally demonstrated a tumor suppressive function of NR4A3 in human AML cells.
DNA methylation analysis by bisulfite sequencing in the current study identified hypermethylated CpG sites at an intragenic region that included exon 3 of NR4A3, although not at its promoter region, in both human AML cell lines and primary human AML samples. Notably, the exposure of DAC restored the expression of NR4A3 and was accompanied by DNA demethylation in the region encompassing exon 3. These findings suggest a role of intragenic DNA hypermethylation in the transcriptional repression of NR4A3 in AML. The role of DNA methylation in cancer, particularly in the gene body, is complex. Recently, Yang et al. reported that DAC-induced demethylation of the gene bodies in tumor cells led to the downregulation of a cohort of genes, many of which are involved in metabolic processes regulated by MYC [14]. They also identified a group of genes showing a negative correlation between gene-body methylation and gene expression [14]. These findings suggest that gene-body methylation plays an important role not only in the upregulation of genes related to oncogenesis but also in the transcriptional repression of tumor suppressor genes.
The ChIP-seq data from the ENCODE database showed H3K4me1 and H2A.Z enrichment peaks at the region including exon 3 of NR4A3 in HMECs, human monocytes, and human B cells but not in K562 cells (Fig. S1). These data indicate that histone modifications in the region are replaced by DNA methylation in AML cells. H2A.Z is enriched at active promoter and enhancer regions and is mutually exclusive with DNA methylation [30]. In addition, H2A.Z mediates nucleosome depletion and contributes to an open and accessible chromatin structure in embryonic stem cells, while DNA methylation often occurs at nucleosome-occupied regions [30–36]. Thus, these effects on nucleosome structure by different epigenetic modifications may account for the different expression levels of NR4A3 between normal hematopoietic cells and AML cells. Alternatively, given that H3K4me1 marks the enhancer region, this area may function as an enhancer [28,37]. It is also possible that exon 3 functions as an alternative promoter [15,28]. These aspects should be investigated in future studies of AML.
Notably, NR4A3 is also silenced and functions as a tumor suppressor gene in aggressive B-cell lymphomas. Intriguingly, Deutsch et al. performed DNA methylation analyses of the coding sequences and the regulatory region of NR4A3 in lymphoma cells from patients and did not find any significant alterations [38]. These results may suggest that different mechanisms operate to reduce the expression of NR4A3 in AML and B-cell lymphoma even though NR4A3 functions as a tumor suppressor gene in both diseases.
Despite our findings in the current study, gene-body methylation may not be the only mechanism responsible for silencing of NR4A3 in AML. For instance, NR4A3 has been reported to be a direct, positively regulated transcriptional target of RUNX1 in hematopoietic progenitor cells [39]. Entinostat, a histone deacetylase inhibitor, has also been reported to restore the expression of NR4A3 in AML [40]. Several upregulated microRNAs which target NR4A3 are also reported in primary myelofibrosis [41]. These reports imply that a single therapy with a DNA-demethylating agent is insufficient to restore NR4A3 expression completely in AML cells. In addition, the DNA methylation status at exon 3 of NR4A3 in AML patients is not limited to a single genetic event (Fig. 5C), suggesting that the mechanism of NR4A3 transcriptional repression is likely complex.
In summary, this is the first report showing an inverse relationship between the expression of NR4A3 and intragenic DNA methylation. Our findings suggest the role of intragenic DNA hyper-methylation as a mechanism underlying the reduced expression of NR4A3 in AML. However, the regulatory mechanisms of NR4A3 expression are assumed to be complex and variable depending on the type of AML. As the reactivation of NR4A3 in AML cells is a promising therapeutic strategy, further study is required to unveil the mechanisms that silence NR4A3 in AML.
Supplementary Material
Acknowledgments
This work was supported in part by a Grant-in-aid for Scientific Research (#26870103) and a grant from the Japan Leukemia Research Fund.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.leukres.2016.09.018.
Footnotes
Potential conflict of interest
The authors have no competing financial interests to declare.
Authors contributions
R.S. performed the experiments, analyzed results, created the figures, and wrote the manuscript; T.M. conceived the project, performed the experiments, analyzed the results, secured funding, and wrote the manuscript; A.K. assisted with the experiments; K.C. analyzed TCGA data; M.T. analyzed results and provided critical suggestions to the project; S.K. assisted with the experiments, including gene transduction to leukemia cells; H.N. and I.Y. assisted with the experiments; E.T., C.K., Y.N., S.T., S.S., Y.T., N.M., C.O., E.S. and T.I. collected patient samples; D.S., A.I. and K.Y. provided a critical suggestion to the project; and C.N. directed the project and wrote the manuscript.
References
- 1.Kurakula K, Koenis DS, van Tiel CM, de Vries CJ. NR4A nuclear receptors are orphans but not lonesome. Biochim Biophys Acta. 2014;1843:2543–2555. doi: 10.1016/j.bbamcr.2014.06.010. [DOI] [PubMed] [Google Scholar]
- 2.Safe S, Jin UH, Morpurgo B, Abudayyeh A, Singh M, Tjalkens RB. Nuclear receptor 4A (NR4A) family – orphans no more. J Steroid Biochem Mol Biol. 2015 doi: 10.1016/j.jsbmb.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wenzl K, Troppan K, Neumeister P, Deutsch AJ. The nuclear orphan receptor NR4A1 and NR4A3 as tumor suppressors in hematologic neoplasms. Curr Drug Targets. 2015;16:38–46. doi: 10.2174/1389450115666141120112818. [DOI] [PubMed] [Google Scholar]
- 4.Beard JA, Tenga A, Chen T. The interplay of NR4A receptors and the oncogene-tumor suppressor networks in cancer. Cell Signal. 2015;27:257–266. doi: 10.1016/j.cellsig.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mohan HM, Aherne CM, Rogers AC, Baird AW, Winter DC, Murphy EP. Molecular pathways: the role of NR4A orphan nuclear receptors in cancer. Clin Cancer Res. 2012;18:3223–3228. doi: 10.1158/1078-0432.CCR-11-2953. [DOI] [PubMed] [Google Scholar]
- 6.Mullican SE, Zhang S, Konopleva M, Ruvolo V, Andreeff M, Milbrandt J, et al. Abrogation of nuclear receptors Nr4a3 and Nr4a1 leads to development of acute myeloid leukemia. Nat Med. 2007;13:730–735. doi: 10.1038/nm1579. [DOI] [PubMed] [Google Scholar]
- 7.Milosevic JD, Kralovics R. Genetic, epigenetic alterations of myeloproliferative disorders. Int J Hematol. 2013;97:183–197. doi: 10.1007/s12185-012-1235-2. [DOI] [PubMed] [Google Scholar]
- 8.Yamazaki J, Issa JP. Epigenetic aspects of MDS and its molecular targeted therapy. Int J Hematol. 2013;97:175–182. doi: 10.1007/s12185-012-1197-4. [DOI] [PubMed] [Google Scholar]
- 9.Baylin SB. DNA methylation and gene silencing in cancer. Nat Clin Pract Oncol. 2005;2(Suppl 1):S4–11. doi: 10.1038/ncponc0354. [DOI] [PubMed] [Google Scholar]
- 10.Portela A, Esteller M. Epigenetic modifications and human disease. Nat Biotechnol. 2010;28:1057–1068. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- 11.Jasielec J, Saloura V, Godley LA. The mechanistic role of DNA methylation in myeloid leukemogenesis. Leukemia. 2014;28:1765–1773. doi: 10.1038/leu.2014.163. [DOI] [PubMed] [Google Scholar]
- 12.Schoofs T, Berdel WE, Muller-Tidow C. Origins of aberrant DNA methylation in acute myeloid leukemia. Leukemia. 2014;28:1–14. doi: 10.1038/leu.2013.242. [DOI] [PubMed] [Google Scholar]
- 13.Yang L, Rau R, Goodell MA. DNMT3A in haematological malignancies. Nat Rev Cancer. 2015;15:152–165. doi: 10.1038/nrc3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang X, Han H, De Carvalho DD, Lay FD, Jones PA, Liang G. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell. 2014;26:577–590. doi: 10.1016/j.ccr.2014.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D’Souza C, Fouse SD, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kulis M, Heath S, Bibikova M, Queiros AC, Navarro A, Clot G, et al. Epigenomic analysis detects widespread gene-body DNA hypomethylation in chronic lymphocytic leukemia. Nat Genet. 2012;44:1236–1242. doi: 10.1038/ng.2443. [DOI] [PubMed] [Google Scholar]
- 17.Varley KE, Gertz J, Bowling KM, Parker SL, Reddy TE, Pauli-Behn F, et al. Dynamic DNA methylation across diverse human cell lines and tissues. Genome Res. 2013;23:555–567. doi: 10.1101/gr.147942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aranyi T, Varadi A, Simon I, Tusnady GE. The BiSearch web server. BMC Bioinf. 2006;7:431. doi: 10.1186/1471-2105-7-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumaki Y, Oda M, Okano M. QUMA: quantification tool for methylation analysis. Nucleic Acids Res. 2008;36:W170–5. doi: 10.1093/nar/gkn294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Konuma T, Nakamura S, Miyagi S, Negishi M, Chiba T, Oguro H, et al. Forced expression of the histone demethylase Fbxl10 maintains self-renewing hematopoietic stem cells. Exp Hematol. 2011;39:697–709 (e5). doi: 10.1016/j.exphem.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 21.Iwama A, Oguro H, Negishi M, Kato Y, Morita Y, Tsukui H, et al. Enhanced self-renewal of hematopoietic stem cells mediated by the polycomb gene product Bmi-1. Immunity. 2004;21:843–851. doi: 10.1016/j.immuni.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 22.Ory DS, Neugeboren BA, Mulligan RC. A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc Natl Acad Sci U S A. 1996;93:11400–11406. doi: 10.1073/pnas.93.21.11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.N. Cancer Genome Atlas Research. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49:359–367. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reinius LE, Acevedo N, Joerink M, Pershagen G, Dahlen SE, Greco D, et al. Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS One. 2012;7:e41361. doi: 10.1371/journal.pone.0041361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boudreaux SP, Ramirez-Herrick AM, Duren RP, Conneely OM. Genome-wide profiling reveals transcriptional repression of MYC as a core component of NR4A tumor suppression in acute myeloid leukemia. Oncogenesis. 2012;1:e19. doi: 10.1038/oncsis.2012.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramirez-Herrick AM, Mullican SE, Sheehan AM, Conneely OM. Reduced NR4A gene dosage leads to mixed myelodysplastic/myeloproliferative neoplasms in mice. Blood. 2011;117:2681–2690. doi: 10.1182/blood-2010-02-267906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 29.Naoe T, Kiyoi H. Gene mutations of acute myeloid leukemia in the genome era. Int J Hematol. 2013;97:165–174. doi: 10.1007/s12185-013-1257-4. [DOI] [PubMed] [Google Scholar]
- 30.Zilberman D, Coleman-Derr D, Ballinger T, Henikoff S. Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature. 2008;456:125–129. doi: 10.1038/nature07324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edwards JR, O’Donnell AH, Rollins RA, Peckham HE, Lee C, Milekic MH, et al. Chromatin and sequence features that define the fine and gross structure of genomic methylation patterns. Genome Res. 2010;20:972–980. doi: 10.1101/gr.101535.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiao S, Xie D, Cao X, Yu P, Xing X, Chen CC, et al. Comparative epigenomic annotation of regulatory DNA. Cell. 2012;149:1381–1392. doi: 10.1016/j.cell.2012.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328:916–919. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- 34.Hu G, Cui K, Northrup D, Liu C, Wang C, Tang Q, et al. H2A.Z facilitates access of active and repressive complexes to chromatin in embryonic stem cell self-renewal and differentiation. Cell Stem Cell. 2013;12:180–192. doi: 10.1016/j.stem.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pandey R, Dou Y. H2A.Z sets the stage in ESCs. Cell Stem Cell. 2013;12:143–144. doi: 10.1016/j.stem.2013.01.012. [DOI] [PubMed] [Google Scholar]
- 36.Li Z, Gadue P, Chen K, Jiao Y, Tuteja G, Schug J, et al. Foxa2 and H2A.Z mediate nucleosome depletion during embryonic stem cell differentiation. Cell. 2012;151:1608–1616. doi: 10.1016/j.cell.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stirzaker C, Taberlay PC, Statham AL, Clark SJ. Mining cancer methylomes: prospects and challenges. Trends Genet. 2014;30:75–84. doi: 10.1016/j.tig.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 38.Deutsch AJ, Rinner B, Wenzl K, Pichler M, Troppan K, Steinbauer E, et al. NR4A1-mediated apoptosis suppresses lymphomagenesis and is associated with a favorable cancer-specific survival in patients with aggressive B-cell lymphomas. Blood. 2014;123:2367–2377. doi: 10.1182/blood-2013-08-518878. [DOI] [PubMed] [Google Scholar]
- 39.Bluteau D, Gilles L, Hilpert M, Antony-Debre I, James C, Debili N, et al. Down-regulation of the RUNX1-target gene NR4A3 contributes to hematopoiesis deregulation in familial platelet disorder/acute myelogenous leukemia. Blood. 2011;118:6310–6320. doi: 10.1182/blood-2010-12-325555. [DOI] [PubMed] [Google Scholar]
- 40.Zhou L, Ruvolo VR, McQueen T, Chen W, Samudio IJ, Conneely O, et al. HDAC inhibition by SNDX-275 (Entinostat) restores expression of silenced leukemia-associated transcription factors Nur77 and Nor1 and of key pro-apoptotic proteins in AML. Leukemia. 2013;27:1358–1368. doi: 10.1038/leu.2012.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Norfo R, Zini R, Pennucci V, Bianchi E, Salati S, Guglielmelli P, et al. miRNA-mRNA integrative analysis in primary myelofibrosis CD34+ cells: role of miR-155/JARID2 axis in abnormal megakaryopoiesis. Blood. 2014;124:e21–32. doi: 10.1182/blood-2013-12-544197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.