Abstract
Histone arginine methylation has emerged as an important histone modification involved in gene regulation. Protein arginine methyltransferase (PRMT) 4 and 5 have been shown to play essential roles in early embryonic development and in embryonic stem (ES) cells. Recently, it has been reported that PRMT6-mediated di-methylation of histone H3 at arginine 2 (H3R2me2) can antagonize tri-methylation of histone H3 at lysine 4 (H3K4me3), which marks active genes. However, whether PRMT6 and PRMT6-mediated H3R2me2 play crucial roles in early embryonic development and ES cell identity remain unclear. Here, we have investigated their roles using gain and loss of function studies with mouse ES cells as a model system. We report that Prmt6 and histone H3R2 methylation levels increased when ES cells are induced to differentiate. Consistently, we find that differentiation of ES cells upon upregulation of Prmt6 is associated with decreased expression of pluripotency genes and increased expression of differentiation markers. We also observe that elevation of Prmt6 increases the methylation level of histone H3R2 and decreases H3K4me, Chd1, and Wdr5 levels at the promoter regions of Oct4 and Nanog. Surprisingly, knockdown of Prmt6 also leads to downregulation of pluripotency genes and induction of expression of differentiation markers suggesting that Prmt6 is important for ES cell pluripotency and self-renewal. Our results indicate that a critical level of Prmt6 and histone H3R2me must be maintained in mouse ES cells to sustain their pluripotency.
Introduction
Embryonic stem (ES) cells possess the unique ability to self-renew and give rise to most cell types. A complex transcriptional regulation network consisting of Oct4, Sox2, and Nanog is known to be crucial in maintaining ES cell identity [1]. Further, epigenetic modifications including histone modifications and chromatin remodeling play important roles in regulating ES cell pluripotency [2,3]. Generally, the chromatin of ES cells displays a unique “open” conformation that enables the modifications of histone marks, which in turn modulates gene expression [4]. For example, trimethylation of histone H3 at lysine 4 (H3K4me3) is enriched in the promoters of core transcription factors, Oct4, Sox2, and Nanog, and serves as a gene activation mark by recruiting nucleosome remodeling enzymes [5–7]. By contrast, trimethylation of histone H3 at lysine 27 (H3K27me3) represses the gene expression by promoting a compact chromatin structure [8,9]. Another epigenetic modification that is associated with transcriptionally silenced heterochromatin in ES cells is the methylation of histone H3 at lysine 9 (H3K9me) [10]. It has been reported that the induction of differentiation in ES cells is often accompanied by an increased level of H3K9me [11–13]. It is also accompanied by a decrease in the acetylation levels of histone H3 and H4, modifications that are associated with transcriptionally active euchromatin [12]. Recent studies have revealed that many developmental genes of mouse ES cells contain a “bivalent domain” that harbors both activating H3K4me3 and silencing H3K27me3 marks. These bivalent domains are believed to silence the developmental genes in the pluripotent ES cells but allow them to remain poised for activation upon differentiation [14].
Histone arginine methylation is less well studied, primarily due to the fact that its occurrence is difficult to detect in mammalian histones using standard sequencing [15]. However, arginine methylation has been implicated in a number of basic cellular processes, including RNA processing, transcriptional regulation, signal transduction, and DNA repair [16]. It is noteworthy that histone H3 methylation at arginine 17 and 26 is correlated with cell fate decisions and pluripotency in preimplantation mouse embryos [17] and mouse ES cells [18]. Consequently, the methylation of histone arginine in early embryonic development and ES cells is of great importance, and the mechanisms that determine this methylation require further investigation.
Several studies have demonstrated that dimethylation of histone H3 at arginine 2 (H3R2me2) mediated by protein arginine methyltransferase 6 (PRMT6) antagonizes H3K4 methylation and therefore regulates gene expression [19–21]. Given that H3K4 methylation is the hallmark of gene activation, H3R2 methylation is associated with gene silencing as shown in human HeLa cells [19,21] and in yeast [20]. This raises the question of whether Prmt6 is involved in gene regulation and pluripotency maintenance in ES cells. In this study, we aimed to elucidate the role of Prmt6 in ES cell identity to enable a greater understanding of H3R2 methylation in ES cells.
Materials and Methods
ES cell culture
HM1 mouse ES cells [18] were cultured in GMEM (GIBCO) supplemented with 15% ES-qualified fetal bovine serum (Invitrogen), 1 mM sodium pyruvate (GIBCO), 0.1 mM β-mercaptoethanol, 0.1 mM MEM nonessential amino acids (GIBCO), and 1,000 U/mL of leukemia inhibitory factor (LIF; Millipore) at 37°C and 5% CO2. Alkaline phosphatase staining of ES cells was performed according to the manufacturer's instructions in the Alkaline Phosphatase Detection Kit (Millipore). The ES cells were induced to differentiation using ES culture medium without LIF and 0.1 μm retinoic acid (RA).
Short hairpin RNA-mediated knockdown
siRNA sequences against mouse Prmt6 were designed using Eurofin MWG Operon siRNA design software (5′-gatccctggaaagcatgtagtataattcaagagattatactacatgctttccattttta-3′ and 3′-agcttaaaaatggaaagcatgtagtataatctcttgaattatactacatgctttccagg-5′) and cloned into the pSUPER.puro vector to express short hairpin RNA (shRNA). After transfection with Lipofectamine 2000 (Invitrogen), the cells were selected by puromycin (1 μg/mL) for 3 days before RNA extraction and protein extraction respectively. GFP RNAi served as nontarget RNAi control [22].
Generation of Prmt6-overexpressing ES cells
Mouse Prmt6 was cloned into pCAGIP.puro and transfected into HM1 cells with Lipofectamine 2000 (Invitrogen). After 3 days of selection by puromycin (1 μg/mL), cells were subjected to RNA extraction and protein extraction respectively.
RNA extraction, reverse transcription, and quantitative real-time polymerase chain reaction
Total RNA was isolated using TRIzol Reagent (Invitrogen). Reverse transcription was conducted using SuperScript III Kit (Invitrogen). Quantitative real-time polymerase chain reaction (PCR) analysis was performed on an ABI PRISM 7300 sequence detection system with the use of SYBR Green (Applied Biosystems). Gene expression levels were normalized to beta-actin. The sequences of all real-time PCR primers are available in Supplementary Table S1 (Supplementary Data are available online at www.liebertonline.com/scd).
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed as described previously [22]. HM1 cells and Prmt6-overexpressing cells were cross-linked with 1% formaldehyde for 10 min. Cells were lysed and the chromatin extract was sonicated into the appropriate size (around 500 bp). Immunoprecipitation was carried out with Protein G Sepharose beads (GE Healthcare) coated with 5 μg of antibodies: anti-Prmt6 (Abcam), anti-H3R2me2, anti-H3K4me3 (Abcam), anti-H3K4me2 (Abcam), anti-H3K9me3 (Abcam), anti-Wdr5 (Santa Cruz), and anti-Chd1 (Santa Cruz). ChIP DNA was analyzed by real-time PCR using specific primers. The fold enrichment was calculated by determining the ratios of ChIP-enriched DNA over the input sample and was normalized to the level observed at a control gene region. The sequences of the primers were as follows: Oct4 promoter forward (Chr17: 35,642,963-35,642,989) 5′-GGATTGGGGAGGGAGAGGTGAAACCGT-3′, reverse (Chr17: 35,643,129-35,643,157) 5′-TGGAAGCTTAGCCAGGTTCGAGGATCCAC-3′; Nanog promoter forward (Chr6: 122,657,639-122,657,668) 5′-CTCTTTCTGTGGGAAGGCTGCGGCTCACTT-3′, reverse (Chr6: 122,657,776-122,657,803) 5′-CATGTCAGTGTGATGGCGAGGGAAGGGA-3′.
Western blotting
The primary antibodies (Abcam, unless otherwise indicated) used in this study were the following: anti-Prmt6, anti-H3R2me2 (asymmetric), anti-H3K4me3, anti-H3K4me2, anti-H3K4me, anti-beta actin, anti-histone H3, anti-Carm1, anti-Oct4 (Santa Cruz), anti-Nanog (Santa Cruz), and anti-Prmt5 (Santa Cruz). Appropriate secondary antibodies conjugated with HRP (GE Healthcare) were used. The labeled proteins were visualized using an enhanced chemiluminescence (ECL) detection kit (Amersham Pharmacia Biotech).
Microarray analysis and data selection
Analysis for RNA samples was carried out using Affymetrix Genechip Mouse Gene 1.0ST chips according to the manufacturer's instructions with default settings. For data selection, the probe sets that did not correspond to any known genes were removed from the list. A fold-change of >1.5 for upregulation population and <0.6 for downregulation population were chosen. The total probe sets for analysis were therefore reduced from 35,557 to 2,573 for the data analysis. Hierarchical clustering of these 2,573 probe sets was performed using Cluster version 3.0, applying mean-clustering of genes and average linkage clustering with uncentered correlation. According to the expression profile of the hierarchical cluster, a K-means clustering was performed to define discreet clusters of common gene regulation. The results were visualized using Treeview [23]. All the raw data have been deposited in the MIAME compliant database Gene Expression Omnibus (accession number GSE27685).
Embryoid body formation
Prior to embryoid body (EB) formation, 2–3 confluent 3.5 cm dishes of ES cells (Controls, Prmt6 overexpression, and knockdown respectively) were grown. After 3-day selection by puromycin (1 μg/mL), ES cells were treated with 0.25% trypsin/0.53 mM EDTA. The disassociated cells were plated in Ultra Low Culture Dish (Corning) and cultured with ES medium without LIF. Medium was changed every other day and the EBs were collected at various time points for RNA extraction followed by real-time PCR analysis.
Results
Prmt6 upregulation leads to increased expression of differentiation marker genes
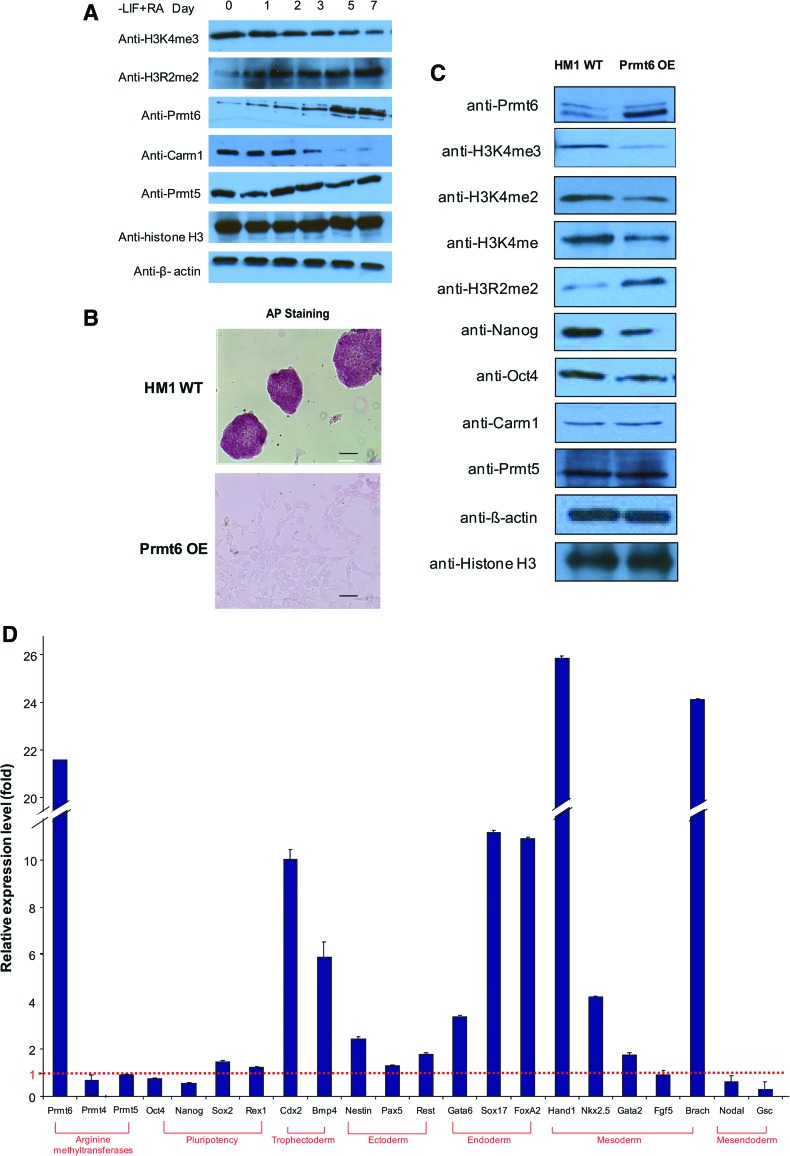
To examine whether Prmt6-mediated histone H3R2me2 also antagonizes H3K4me3 in mouse ES cells and is further associated with ES cell self-renewal and pluripotency, we first investigated the changes of H3R2me2 and H3K4me3 when ES cells were induced to differentiate by LIF removal and RA treatment. We found that Prmt6 and H3R2me2 were increased while H3K4me3 level dropped, while other 2 protein arginine methytransferases Carm1 (Prmt4) and Prmt5 did not show increased level (Fig. 1A). Since this suggested that an elevated level of Prmt6 correlates with ES cell differentiation, we next wished to investigate whether overexpression of Prmt6 would have any effect on maintenance of pluripotency in ES cells. To this end, we transfected the wild-type ES cells with a plasmid leading to Prmt6 overexpression. We also transfected pCAGIP empty vector into wild-type cells. We found that the level of prmt6, Oct4, and Nanog in vector control-transfected cells was almost the same as that in wild-type cells (Supplementary Fig. S1). Interestingly, we observed that Prmt6-overexpressing ES cells lost their ability to form distinct alkaline phosphatase positive colonies (Fig. 1B). Expectedly, Prmt6 overexpression caused upregulation of histone H3 methylation on arginine 2 and led to reduction in histone H3 methylation level on lysine 4 (especially tri-methylation; Fig. 1C). The remaining levels of Prmt4 and Prmt5 suggest that the changes of methylation level on H3R2 and H3K4 is specifically mediated by Prmt6 (Fig. 1C). The protein levels of Oct4 and Nanog were also reduced upon Prmt6 overexpression (Fig. 1C).
FIG. 1.
Prmt6-overexpressing cells undergo differentiation. (A) Immunoblot with cell extracts from HM1 cells treated with leukemia inhibitory factor (LIF) removal and retinoic acid (RA) for different days. Antibodies against Prmt6, histone H3K4me3 and H3R2me2 were used. Beta-actin and histone H3 served as loading controls. (B) Prmt6-overexpressing cells lost the expression marker of undifferentiated embryonic stem (ES) cells, alkaline phosphatase (AP; scale bar=100 μm), compared to the empty vector-transfected cells. Cells were counted using cell counter and result showed that Prmt6-overexpressing cells were 57.3% less than control cells. (C) Immunoblot with chromatin extracts from parental HM1 cells and Prmt6-overexpressing cells using antibodies against Prmt6, histone H3 marks (H3K4 and H3R2) or pluripotency markers (Nanog and Oct4). Beta-actin and histone H3 served as loading controls. (D) Overexpression of Prmt6 induced the high expression levels of certain lineage marker genes, especially endoderm and mesoderm markers. Mean levels expressed relative to the vector control and normalized to beta-actin expression levels. Color images available online at www.liebertonline.com/scd
Quantitative real-time PCR showed that upregulation of Prmt6 in ES cells resulted in a decrease in the expression level of the pluripotency marker Oct4, and a reduction in Nanog level by nearly half. The expression level of other pluripotency markers, Sox2 and Rex1, did not change significantly (1.4 and 1.2-fold respectively). Importantly, overexpression of Prmt6 led to an increased expression of marker genes for specific differentiated lineages: the trophectoderm markers, Cdx2 and BMP4, were increased by 10 and 5.9-fold respectively; the endoderm markers, Gata6, Sox17, and FoxA2, were increased by 3.3-, 11-, and 10-fold respectively; the mesoderm markers, Hand1 and Brachyury, were increased by 25.7- and 24-fold respectively; whereas, Nkx2.5 and Gata2 were increased by 4.2- and 1.7-fold respectively. Fgf5 showed no significant change in its expression level. However, the expression of mesendoderm marker Nodal was decreased by half and the expression of GSC was also significantly diminished, indicating Prmt6 directs cells away from the mesendoderm lineage (Fig. 1D). Taken together, these results indicate that excessive Prmt6 drives the ES cell to differentiate along many pathways.
Global expression of developmental genes upon Prmt6 upregulation
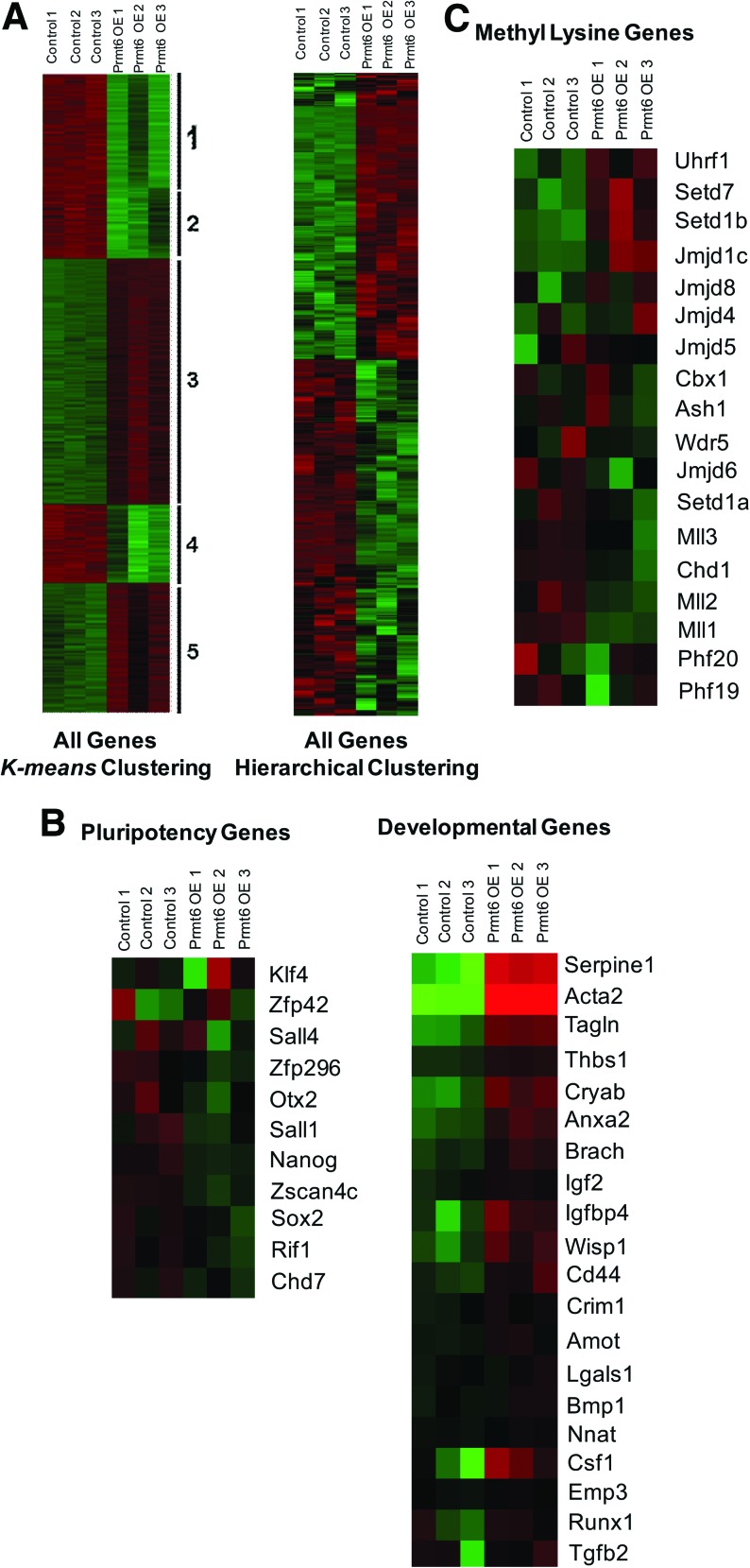
To better understand how Prmt6 overexpression results in ES cell differentiation, we performed whole genome cDNA microarray hybridization to compare the control ES cells and Prmt6-overexpressing cells. We found that 1,706 genes were upregulated and 1,197 genes were downregulated upon Prmt6 overexpression. Microarray data were subjected to K-means clustering to define discreet clusters of common gene regulation (Fig. 2A). This analysis yielded 5 clusters of commonly regulated genes. To examine whether we could define a functional insight between the gene expression differences, genes that are associated with pluripotency and known functions in cell development were selected for hierarchical clustering analysis (Fig. 2B). Overall, most of the known genes that are associated with pluripotency were downregulated upon Prmt6 overexpression. Interestingly, we found that when Prmt6 was overexpressed, a wide range of developmental genes, especially those that are implicated in the cytoskeleton and extracellular matrix formation were upregulated, including Acta2 and Tagln (23- and 9.8-fold increase respectively). This observation highlights that Prmt6 overexpression suppresses some pluripotency genes while activating the developmental genes, triggering the differentiation of ES cells toward the mesoderm lineages (Fig. 2B), which mostly reside in cluster 3 (Fig. 2A). It is noteworthy that most of the genes associated with methyl-lysine, including Ash, Wdr5, and Mll, were downregulated upon overexpression of Prmt6 (Fig. 2C), indicating that genes that are in an activated state by the criterion of these marks may be downregulated.
FIG. 2.
cDNA microarray analysis of gene expression changes upon overexpression of Prmt6. (A) Log2 transformed gene expression of the 3 replicates of both wild-type cells and Prmt6-overexpressing cells were subjected to K-means clustering. Data are shown in thumbnail-dendrogram format, in which downregulated and upregulated genes are shown as green and red tiles respectively. Five clusters of differential gene expression were identified from the 2,573 probe sets, 3 of which represent downregulated genes (cluster 1, 2, and 4) and 2 of which represent upregulated genes (cluster 3 and 5). The gene ontology of these genes is detailed in Supplementary Table S1. (B) Thumbnail-dendrogram of specific gene regulation across the triplicate samples. Genes were selected on the basis of their known roles in either ES cell pluripotency or differentiation to distinct germ layers, and their roles in (C) methylating histone lysine marks. Color images available online at www.liebertonline.com/scd
H3R2 di-methylation levels at the Oct4 and Nanog promoter regions following overexpression of Prmt6
To further dissect the molecular mechanism underlying Prmt6 function in ES cells, we conducted ChIP using anti-Prmt6 antibody in wild-type ES cells and Prmt6-overexpressing cells. Quantitative real-time PCR using specific primers located at the proximal region of the Oct4 and Nanog promoters revealed that in wild-type cells, Prmt6 does not bind to Oct4 promoter (1.3-fold; Fig. 3A). Interestingly, with Prmt6 overexpression, it is enriched significantly by 4.8-fold. Results also showed that Prmt6 binds more to the Nanog promoter (5.9-fold) than to the Oct4 promoter in wild-type cells, but it is not significantly enhanced with Prmt6 overexpression (Fig. 3A). We also performed ChIP experiments using antibodies against other 2 arginine methytransferases: anti-Prmt4 and anti-Prmt5 antibodies. We found that the association of Prmt4 with Oct4 and Nanog promoters was abolished while we did not detect any binding of Prmt5 to the promoters in Prtm6-overexpressing cells and wild-type cells (Supplementary Fig. S2). PRMT6 is known to be responsible for di-methylation of H3R2, which counter-correlates with H3K4 tri-methylation in humans [19,21]. We therefore wished to examine whether Prmt6 overexpression could influence the level of chromatin modifications at its target genes thus regulating their expression. We performed ChIP with anti-H3R2me2 and anti-H3K4me3 antibodies using wild-type ES cells and Prmt6-overexpressing ES cells. Consistent with our Prmt6 ChIP result (Fig. 3A), enrichment of Oct4 promoters was barely detectable (1.5-fold) in control cells in an anti- H3R2me2 ChIP, but their enrichment increased 6.2-fold following Prmt6 overexpression (Fig. 3B). Similarly, we could detect a 3.4-fold enrichment at the Nanog promoter in the anti-H3R2me ChIP in wild-type ES cells, but the enrichment dramatically increased to 11-fold when Prmt6 was overexpressed (Fig. 3B). On the other hand, results revealed significant enrichment of Oct4 and Nanog promoters in the anti-H3K4me3 ChIP from both wild-type and Prmt6-overexpressing cells (Fig. 3C). However, surprisingly no significant difference was found between the 2 cell populations. Thus, these results indicate that upon overexpression of Prmt6, there is an increase in H3R2 di-methylation, but no subsequent decrease in H3K4 tri-methylation.
FIG. 3.
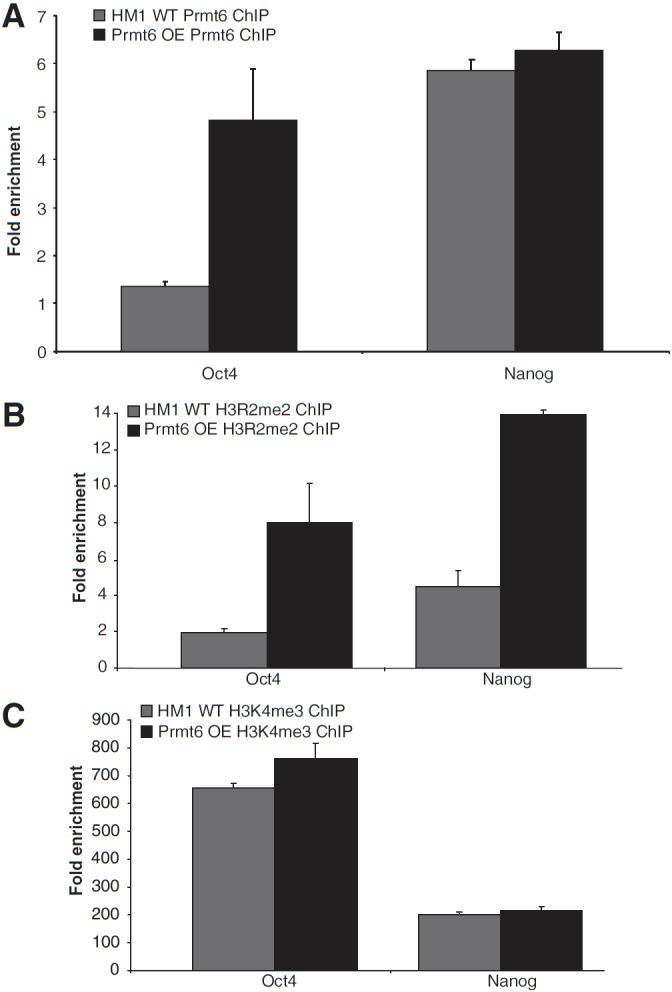
Overexpression of Prmt6 increased H3R2 methylation at promoters of pluripotency genes. Chromatin immunoprecipitation (ChIP) was performed using (A) anti-Prmt6 antibody, (B) anti-H3R2me2 antibody, and (C) anti-H3K4me3 respectively. ChIP DNA was analyzed by quantitative real-time polymerase chain reaction (PCR) with primers located at Oct4 and Nanog promoter regions. Fold enrichments were calculated from the apparent IP efficiency (ratio of ChIP-enriched DNA over input) and normalized to the level at a control region defined as 1.0 for a given extract.
H3K4 mono-methylation levels at the Oct4 and Nanog promoter regions following overexpression of Prmt6
We then sought to understand the mechanism by which Prmt6 regulates mouse ES cell pluripotency. For this, we conducted more ChIPs on histone modification marks in both wild-type and Prmt6-overexpressing cells. Results showed that H3K4me2 is associated with both Oct4 and Nanog promoters (2.9- and 2.5-fold respectively), but no significant difference was found between the wild-type and Prmt6-overexpressing cells (Fig. 4A). On the other hand, H3K4me was enriched in both Oct4 and Nanog promoters in wild-type cells (30.3- and 9.6-fold respectively). When Prmt6 was overexpressed in ES cells, the enrichment of these regions in anti-H3K4me ChIP decreased significantly (Fig. 4B). Interestingly, our results also showed that upon Prmt6 overexpression, the enrichment of Oct4 and Nanog in the anti-H3K9me3 ChIP increased by ∼2-folds (Fig. 4C).
FIG. 4.
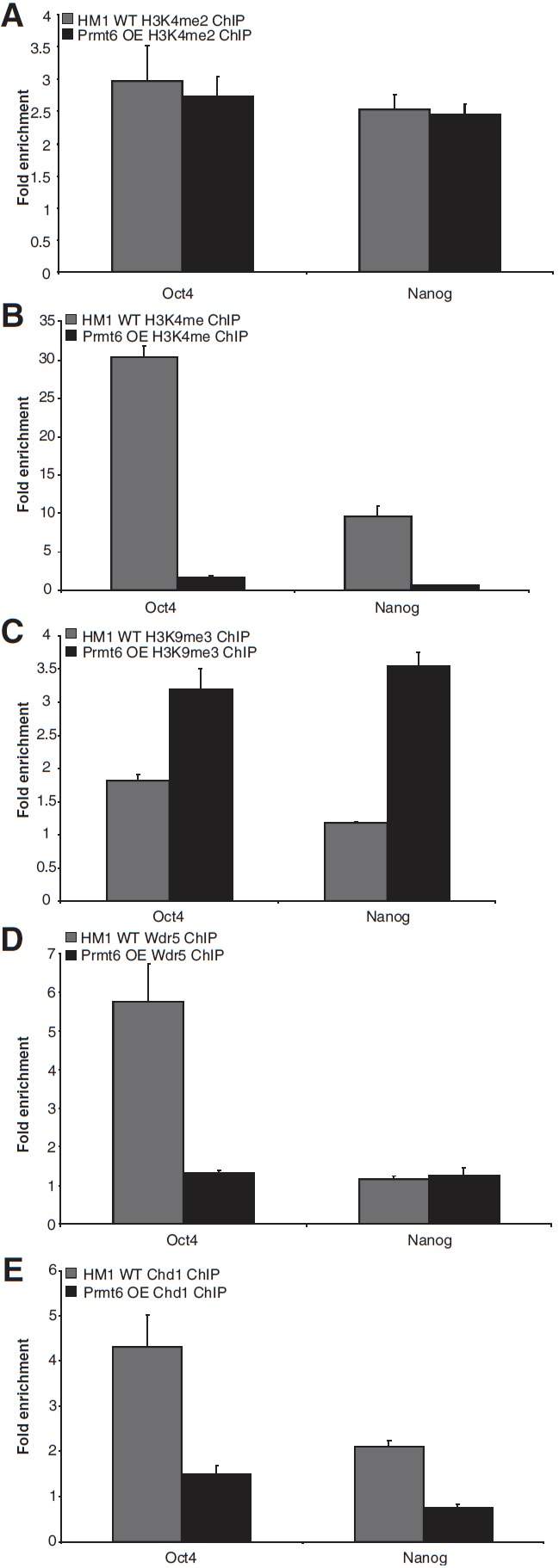
Overexpression of Prmt6 decreased H3K4 mono-methylation, Wdr5, and Chd1 at promoters of pluripotency genes. ChIP was performed using (A) anti-H3K4me2 antibody, (B) anti-H3K4me antibody, (C) anti-H3K9me3 antibody, (D) anti-Wdr5 antibody, and (E) anti-Chd1 antibody respectively. ChIP DNA was analyzed by quantitative real-time PCR with primers located at Oct4 and Nanog promoter regions. Fold enrichments were calculated from the apparent IP efficiency and normalized to the level at a control region defined as 1.0 for a given extract.
We next performed ChIPs using antibodies targeting chromatin-binding proteins, Wdr5, and Chd1 respectively, in wild-type and Prmt6-overexpressing ES cells. Wdr5 was found to be associated with the Oct4 promoter in wild-type ES cells (5.7-fold). Upon Prmt6 overexpression, the enrichment of Oct4 promoter in Wdr5 ChIP decreased significantly (Fig. 4D). However, there was no enrichment of Nanog promoter in Wdr5 ChIP in both wild-type and Prmt6-overexpressing cells (Fig. 4D). We also observed a reduction in Chd1 binding to the promoter regions of Oct4 and Nanog upon Prmt6 overexpression (Fig. 4E). Results thus far have shown that Prmt6 overexpression significantly downregulates H3K4 mono-methylation level at Oct4 and Nanog promoters and also impedes Wdr5 and Chd1 binding at these regions. These suggest that Prmt6 overexpression negatively influences the H3K4 methylation complex at Oct4 and Nanog promoters.
Overexpression of Prmt6 facilitates ES cell differentiation into endoderm lineage
To investigate the role of Prmt6 overexpression in ES cell differentiation, we generated EBs using Prmt6-overexpressing cells and pCAGIP empty vector-transfected cells (control). We then examined the relative expression (compared with control) of various marker genes of 3 germ layers at different stages of EB formation (day 7, 11, and 16; Supplementary Fig. S3). Consistently, the levels of pluripotency markers Oct4, Nanog, and Sox2 were lower in Prmt6-overexpressing EBs than that in control EBs (Supplementary Fig. S3). The levels of endoderm markers (Hnf4a, Gata6, Sox17, FoxA2, Noggin, α-fetoprotein, Vegfr2, and C-kit) were significantly higher in Prmt6 EBs than that in control EBs (from day 7 and throughout day 16 day, as shown in the heat map; Supplementary Fig. S3). Prmt6 expressed lower levels of trophectoderm genes at day 11 and day 16. Interestingly, markers of ectoderm and mesoderm were heterogeneously changed when compared with control EBs (Supplementary Fig. S3). We conclude that elevated level of Prmt6 drives ES cells to differentiate into endoderm lineage and some other specific cell types.
Depletion of Prmt6 induces expression of lineage marker genes
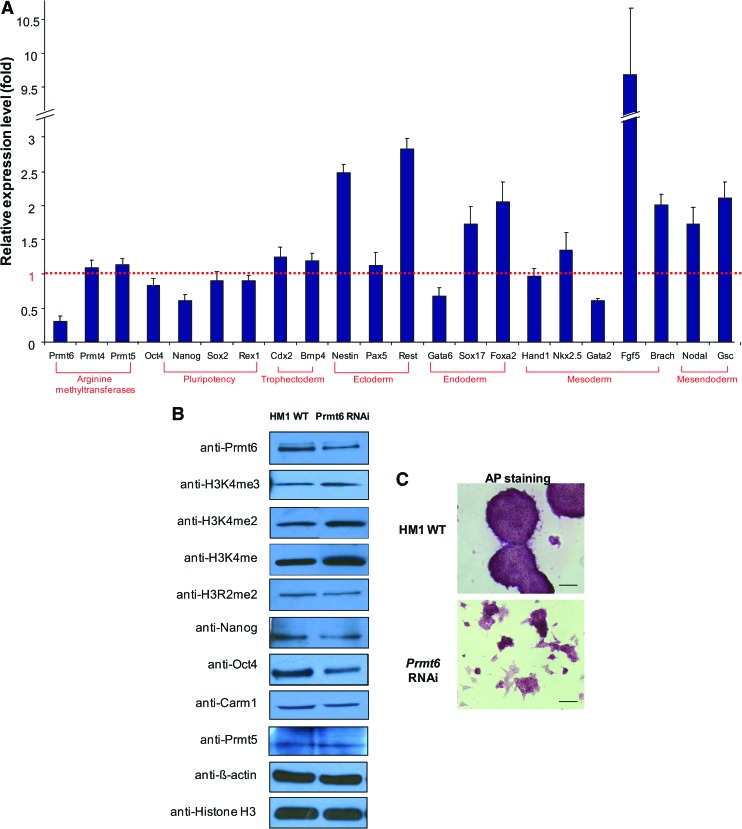
To further investigate the role of Prmt6 in mouse ES cells, we used shRNA-mediated RNAi to knock down Prmt6 in HM1 mouse ES cells. Quantitative real-time PCR using Prmt6 RNAi cDNA revealed that Prmt6 mRNA levels were significantly downregulated to 30% of nontarget RNAi control (GFP RNAi). We found that levels of the pluripotency markers Oct4, Nanog, Sox2, and Rex1 were downregulated (Fig. 5A). The trophectoderm markers Cdx2 and Bmp4, and the ectoderm marker Pax5, endoderm marker Gata6, mesoderm markers Hand1, Nkx2.5, and Gata2 showed no significant change in cDNA levels. On the other hand, the expression of the ectoderm markers Nestin and Rest increased to 2.5 and 2.8-fold respectively. The endoderm markers Sox17 and Foxa2 and mesendoderm markers Nodal and Gsc were elevated to more than 1.5-fold when Prmt6 was depleted. The mesoderm marker Fgf5 was dramatically upregulated by up to 9.7-fold (Fig. 5A). Further, Prmt6 knockdown EBs, compared with mock knockdown EBs, expressed trophectoderm markers (Cdx2 and Eomes), 2 endoderm markers (Vegfr2 and Insulin 1), and several mesoderm markers (Brachyury, Gsc, and Myf5) with higher level (Supplementary Fig. S4), suggesting that depletion of Prmt6 influences directions of ES cell differentiation.
FIG. 5.
Prmt6 RNAi results in ES cell differentiation. (A) Depletion of Prmt6 increases the expression level of certain lineage markers, such as Fgf5. Mean levels are expressed relative to the vector control and normalized to beta-actin expression levels. (B) Immunoblot with chromatin extracts from GFP RNAi cells and Prmt6 RNAi ES cells using antibodies against Prmt6, histone H3 marks (H3K4me3 and H3R2me2), or pluripotency markers (Nanog and Oct4). Beta-actin and histone H3 served as loading controls. (C) Prmt6 RNAi ES cells partially lost the activity of undifferentiated ES cell marker, AP (scale bar=100 μm), compared to the control cells. Color images available online at www.liebertonline.com/scd
In addition, we found that the levels of Prmt6, Oct4, and Nanog protein were consistently lower than the control (Fig. 5B). Further, downregulation of Prmt6 caused an increased level of histone H3K4 methylation. As expected, Prmt6 RNAi resulted in decrease of histone H3R2 methylation (Fig. 5B). The level of Carm1 (Prmt4) and Prmt5 was not affected by Prmt6 RNAi, again suggesting that Carm1 and Prmt5 are not involved in Prtm6 functions. In accord with the increased expression of differentiation markers, we also found that Prmt6-depleted cells largely lost their ability to form typical alkaline phosphatase positive colonies (Fig. 5C). Taken together, our Prmt6 RNAi data suggest that Prmt6 contributes in maintaining the undifferentiated state of ES cells.
Discussion
In recent years histone arginine modifications have emerged as new regulators of pluripotency [24]. CARM1 (PRMT4) has been previously reported to play an important role in maintaining ES cell pluripotency [18]. Loss of CARM1 in ES cells leads to downregulation of Oct4 and Sox2 that ultimately direct ES cells to differentiate into various lineages [18]. A recent Prmt5 knockout study demonstrated that Prmt5 is critical for early inner cell mass formation [25]. These results highlight the importance of PRMT both in the early embryo and in ES cells. PRMT6 was identified during the search for protein arginine methyltransferase (PRMT) family members [26]. PRMT6 was found to have the ability to methylate itself [26]. It is a type I enzyme from the PRMT family, catalyzing asymmetrical arginine di-methylation [16]. PRMT6 is found to be the primary methyltransferase responsible for H3R2 methylation rather than CARM1 [19,27]. PRMT6-mediated H3R2 methylation has a role in antagonizing the methylation of the H3K4 residues [19,20]. Given that H3K4 methylation has long been associated with pluripotency genes, including Nanog and Oct4, in the ES cells, we have analyzed H3R2me2 involvement in ES cell identity maintenance. Using gain and loss of function analysis of Prmt6, we found that Prmt6 depletion affects ES cell identity. Moreover, when Prmt6 is overexpressed in ES cells, H3R2me2 favored enrichment at the Oct4 and Nanog promoters. This in turn might antagonize the deposition of histone H3K4me3 complex at these regions, causing suppression of the pluripotency genes and cell differentiation. It has been shown that H3R2me2 is present throughout the promoter and coding region of inactive genes [20], which supports our finding and explains why ES cells differentiate when Prmt6 is overexpressed.
Our cDNA microarray data show that when Prmt6 is overexpressed, a broad number of genes that are involved in lysine methylation are downregulated. Thus, we propose that like HeLa cells, Prmt6-regulated H3R2 methylation in ES cells inhibits the activity of H3K4 methyltransferase complexes. In fact, our ChIP analysis showed no evidence of reduction in total H3K4me3 at Oct4 and Nanog promoters upon Prmt6 overexpression. This could be because only 3 day selection is not long enough for H3R2me2 to block H3K4me3 sufficiently at Oct4 and Nanog promoters as these genes are robustly expressed in ES cells. Another possible reason is that Prmt6 may function through other substrates. In fact, a recently published article revealed that Prmt6-mediated H2AR29 is important in gene silencing in vivo [28]. However, the decreased association of Wdr5, Chd1, and H3K4me with the Oct4 and Nanog promoters indicates that Prmt6 indeed suppresses lysine methyl transferase activity. It has been demonstrated that H3R2me2 inhibits the activity of the Set1 complex toward H3K4 by regulating the binding of its Spp1 component to the PHD finger [20]. PRMT6-mediated H3R2 di-methylation can also directly inhibit the catalytic activity of H3K4 methyltransferase Set1/mixed lineage leukemia (MLL), and abolish the interaction of WDR5, ASH2, or other components of methyltransferase to the N-terminus of histone H3 [19,29,30], thus affecting their binding to target genes [21]. Interestingly, Hyllus and colleagues reported that pre-existing methylation of H3K4 and H3K9 causes a reduction in PRMT6 activity toward H3 peptides, but the activity is slightly elevated in the presence of H3K27 methylation. This suggests that Prmt6-mediated H3R2me2 might crosstalk with other histone modifications in a delicate interaction to maintain ES cell pluripotency while keeping them poised for differentiation.
Our data indicate that Prmt6 has an impact on a subset of lineage gene markers. cDNA microarray analysis demonstrates that following Prmt6 overexpression, Chd1 is downregulated. In addition, our ChIP data also show that Prmt6 overexpression does indeed inhibit the association of Chd1 with the Oct4 and Nanog promoters. Chd1 is a chromatin remodeling enzyme from a chromodomain (CHD) family that contains an ATPase SNF2-like helicase domain [31]. It is noteworthy that downregulation of Chd1 has been shown to decrease Oct4 expression and also increase the expression of neurogenesis genes [32]. This supports our observation on the expression activation of ectoderm and mesoderm genes upon Prmt6 overexpression, and it indicates a role for Prmt6 in directing ES cells toward specific cell lineages.
The formation of EBs has characteristics similar to embryonic development, making them a valuable tool to study the spontaneous differentiation of ES cells and the interplay of different germ layers during this process [33]. Our data demonstrate that Prmt6 plays a role in cell fate determination by facilitating ES cells to differentiate into endodermal cells when overexpressed or into trophectoderm lineage when depleted. Our EB study further highlights the important role of Prmt6 in ES cell identity and differentiation.
More studies will be needed to examine which subunits of H3K4 methyltransferase complex are being inhibited, and the possible mechanisms that regulate this event. Genome-wide ChIP-seq assay using ChIP-grade anti-Prmt6 and anti-H3R2me2 will allow us to precisely identify the target DNA and determine how histone H3R2me modifications influence the regulation of pluripotency genes. Nonetheless, our results show that Prmt6 plays a role in regulating the undifferentiated/differentiated state when depleted/overexpressed. Our results suggest that Prmt6 plays dual roles in ES cells: suppressing a subset of genes to maintain pluripotency (as indicated by the knockdown study) while driving ES cells toward differentiation when upregulated by some environmental signals (as indicated by the overexpression study).
Supplementary Material
Acknowledgments
We thank Eunice Lin and Soheila Sharghi Namini for critical reading of the article. We thank Ernesto Guccione for valuable discussion and for providing anti-histone H3R2me2 serum. M.Z.G. is supported by the Wellcome Trust. Research in Q. Wu's lab is supported by the National University of Singapore (NUS) and the Singapore Ministry of Education.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Young RA. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Surani MA. Hayashi K. Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128:747–762. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 3.Hemberger M. Dean W. Reik W. Epigenetic dynamics of stem cells and cell lineage commitment: digging Waddington's canal. Nat Rev. 2009;10:526–537. doi: 10.1038/nrm2727. [DOI] [PubMed] [Google Scholar]
- 4.Meshorer E. Yellajoshula D. George E. Scambler PJ. Brown DT. Misteli T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell. 2006;10:105–116. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Santos-Rosa H. Schneider R. Bernstein BE. Karabetsou N. Morillon A. Weise C. Schreiber SL. Mellor J. Kouzarides T. Methylation of histone H3 K4 mediates association of the Isw1p ATPase with chromatin. Mol Cell. 2003;12:1325–1332. doi: 10.1016/s1097-2765(03)00438-6. [DOI] [PubMed] [Google Scholar]
- 6.Pan G. Tian S. Nie J. Yang C. Ruotti V. Wei H. Jonsdottir GA. Stewart R. Thomson JA. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell. 2007;1:299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 7.Zhao XD. Han X. Chew JL. Liu J. Chiu KP. Choo A. Orlov YL. Sung WK. Shahab A, et al. Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell. 2007;1:286–298. doi: 10.1016/j.stem.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 8.Cao R. Wang L. Wang H. Xia L. Erdjument-Bromage H. Tempst P. Jones RS. Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 9.Ringrose L. Ehret H. Paro R. Distinct contributions of histone H3 lysine 9 and 27 methylation to locus-specific stability of polycomb complexes. Mol Cell. 2004;16:641–653. doi: 10.1016/j.molcel.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 10.Mikkelsen TS. Ku M. Jaffe DB. Issac B. Lieberman E. Giannoukos G. Alvarez P. Brockman W. Kim TK, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keohane AM. O'Neill LP. Belyaev ND. Lavender JS. Turner BM. X-Inactivation and histone H4 acetylation in embryonic stem cells. Dev Biol. 1996;180:618–630. doi: 10.1006/dbio.1996.0333. [DOI] [PubMed] [Google Scholar]
- 12.Muller C. Leutz A. Chromatin remodeling in development and differentiation. Cur Opin Genet Dev. 2001;11:167–174. doi: 10.1016/s0959-437x(00)00175-1. [DOI] [PubMed] [Google Scholar]
- 13.Lee JH. Hart SR. Skalnik DG. Histone deacetylase activity is required for embryonic stem cell differentiation. Genesis. 2004;38:32–38. doi: 10.1002/gene.10250. [DOI] [PubMed] [Google Scholar]
- 14.Bernstein BE. Mikkelsen TS. Xie X. Kamal M. Huebert DJ. Cuff J. Fry B. Meissner A. Wernig M, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 15.Gary JD. Clarke S. RNA and protein interactions modulated by protein arginine methylation. Prog Nucleic Acid Res Mol Biol. 1998;61:65–131. doi: 10.1016/s0079-6603(08)60825-9. [DOI] [PubMed] [Google Scholar]
- 16.Bedford MT. Richard S. Arginine methylation an emerging regulator of protein function. Mol Cell. 2005;18:263–272. doi: 10.1016/j.molcel.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 17.Torres-Padilla ME. Parfitt DE. Kouzarides T. Zernicka-Goetz M. Histone arginine methylation regulates pluripotency in the early mouse embryo. Nature. 2007;445:214–218. doi: 10.1038/nature05458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu Q. Bruce AW. Jedrusik A. Ellis PD. Andrews RM. Langford CF. Glover DM. Zernicka-Goetz M. CARM1 is required in ES Cells to maintain pluripotency and resist differentiation. Stem Cells. 2009;27:2637–2645. doi: 10.1002/stem.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guccione E. Bassi C. Casadio F. Martinato F. Cesaroni M. Schuchlautz H. Luscher B. Amati B. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature. 2007;449:933–937. doi: 10.1038/nature06166. [DOI] [PubMed] [Google Scholar]
- 20.Kirmizis A. Santos-Rosa H. Penkett CJ. Singer MA. Vermeulen M. Mann M. Bahler J. Green RD. Kouzarides T. Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature. 2007;449:928–932. doi: 10.1038/nature06160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hyllus D. Stein C. Schnabel K. Schiltz E. Imhof A. Dou Y. Hsieh J. Bauer UM. PRMT6-mediated methylation of R2 in histone H3 antagonizes H3 K4 trimethylation. Genes Dev. 2007;21:3369–3380. doi: 10.1101/gad.447007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loh YH. Wu Q. Chew JL. Vega VB. Zhang W. Chen X. Bourque G. George J. Leong B, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 23.Eisen MB. Spellman PT. Brown PO. Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee YH. Wu Q. Chromatin modification landscape of ES cell identity. Biosci Rep. 2011;31:77–86. doi: 10.1042/BSR20100089. [DOI] [PubMed] [Google Scholar]
- 25.Tee WW. Pardo M. Theunissen TW. Yu L. Choudhary JS. Hajkova P. Surani MA. Prmt5 is essential for early mouse development and acts in the cytoplasm to maintain ES cell pluripotency. Genes Dev. 2010;24:2772–2777. doi: 10.1101/gad.606110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frankel A. Yadav N. Lee J. Branscombe TL. Clarke S. Bedford MT. The novel human protein arginine N-methyltransferase PRMT6 is a nuclear enzyme displaying unique substrate specificity. J Biol Chem. 2002;277:3537–3543. doi: 10.1074/jbc.M108786200. [DOI] [PubMed] [Google Scholar]
- 27.Iberg AN. Espejo A. Cheng D. Kim D. Michaud-Levesque J. Richard S. Bedford MT. Arginine methylation of the histone H3 tail impedes effector binding. J Biol Chem. 2008;283:3006–3010. doi: 10.1074/jbc.C700192200. [DOI] [PubMed] [Google Scholar]
- 28.Waldmann T. Izzo A. Kamieniarz K. Richter F. Vogler C. Sarg B. Lindner H. Young NL. Mittler G. Garcia BA. Schneider R. Methylation of H2AR29 is a novel repressive PRMT6 target. Epigenet Chrom. 2011;4:11. doi: 10.1186/1756-8935-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Couture JF. Collazo E. Trievel RC. Molecular recognition of histone H3 by the WD40 protein WDR5. Nat Struct Mol Biol. 2006;13:698–703. doi: 10.1038/nsmb1116. [DOI] [PubMed] [Google Scholar]
- 30.Han Z. Guo L. Wang H. Shen Y. Deng XW. Chai J. Structural basis for the specific recognition of methylated histone H3 lysine 4 by the WD-40 protein WDR5. Mol Cell. 2006;22:137–144. doi: 10.1016/j.molcel.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 31.Woodage T. Basrai MA. Baxevanis AD. Hieter P. Collins FS. Characterization of the CHD family of proteins. Proc Nat Acad Sci U S A. 1997;94:11472–11477. doi: 10.1073/pnas.94.21.11472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gaspar-Maia A. Alajem A. Polesso F. Sridharan R. Mason MJ. Heidersbach A. Ramalho-Santos J. McManus MT. Plath K. Meshorer E. Ramalho-Santos M. Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature. 2009;460:863–868. doi: 10.1038/nature08212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keller GM. In vitro differentiation of embryonic stem cells. Curr Opin Cell Biol. 1995;7:862–869. doi: 10.1016/0955-0674(95)80071-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.