Abstract
The aim of the present study was to measure the level of microRNA (miRNA or miR)-24 in the serum of patients with atherosclerosis and to investigate the effect of miR-24 on the expression of importin-α3 and tumor necrosis factor (TNF)-α, as well as the proliferation and migration of vascular endothelial cells. A total of 30 patients with atherosclerosis admitted to hospital between January and June 2016 were enrolled in the present study; 30 healthy subjects with a similar age range were enrolled as controls. Peripheral blood (10 ml) was collected from all participants. Human umbilical vein endothelial cells (HUVECs) were transfected with miR-24 mimic using Lipofectamine 2000. TargetScan was used to elucidate whether importin-α3 (KPNA4) was a target gene of miR-24. Expression levels of miR-24 and mRNAs were measured using reverse transcription-quantitative polymerase chain reaction, and protein expression was determined using western blotting. Cell Counting Kit 8 assay was used to assess the proliferation of HUVECs, and a Transwell assay was performed to detect the migration of HUVECs. Expression of miR-24 in peripheral blood from patients with atherosclerosis was significantly lower when compared with healthy subjects (P<0.05). Overexpression of miR-24 was demonstrated to significantly inhibit the transcription and translation of the importin-α3 gene (P<0.05) and negatively regulate the expression of endothelial inflammatory factor TNF-α (P<0.05). Furthermore, overexpression of miR-24 significantly inhibited the proliferation and migration of HUVECs (P<0.05), and miR-24 knockdown significantly promoted these processes (P<0.05). The results of the present study suggest that miR-24 exerts its effect in atherosclerosis by blocking the nuclear factor-κB signaling pathway, regulating inflammation in endothelial cells, and inhibiting the proliferation and migration of vascular endothelial cells.
Keywords: atherosclerosis, microRNA, importin-α3
Introduction
Atherosclerosis is the most common lesion that occurs in the cardiovascular system, with a high incidence worldwide (1). The occurrence of atherosclerotic lesions is a chronic and complex process that involves interactions among various factors, such as local hemodynamics (2), arterial wall cells (3), the extracellular matrix (4), the environment (5) and genetics (6). Vascular endothelial injury, phenotypic transformation of smooth muscle cells, lipid absorption by monocytes or macrophages, and inflammatory mediator release are important processes in the development of atherosclerosis (7–9). The nuclear factor κ-light-chain-enhancer of activated B cells (NF-κB) signaling pathway is considered a key signaling pathway that mediates vascular endothelial cell injury and the release of inflammatory mediators, such as vascular cell adhesion molecule-1 (VCAM-1) and intercellular adhesion molecule-1 (ICAM-1) (10). An important step in the development of atherosclerosis is the inflammation of the vascular endothelial cell layer (11). Importin-α3 is a key protein that is associated with the nuclear transfer of NF-κB, which is important in the occurrence of inflammation (12). Importin-α3 was therefore chosen as a target gene in the present study. In addition to being a proinflammatory factor, tumor necrosis factor (TNF)-α also activates NF-κB, which then initiates a cascade reaction of cytokines, further aggravating inflammatory responses and apoptosis in tissues (13).
microRNA (miRNA or miR) are a class of endogenous non-coding RNA that regulate a variety of cellular mechanisms, including inflammation, cytothesis and lipid metabolism, as well as participating in the development of atherosclerosis (14). For example, miR-155 regulates multiple functions of macrophages, including inflammation, lipid absorption and apoptosis (15,16). In addition, low-density lipoprotein and mildly oxidized low-density lipoprotein are able to induce the expression of miR-155 in macrophages (15,16). miR-342-5p promotes the expression of miR-155 via V-protein kinase B murine thymoma viral oncogene homolog 1, and facilitates the expression of inflammatory mediators, such as nitric oxide, TNF-α and interleukin-6 (17). Furthermore, miR-33 (18), miR-122 (19) and miR-27a (20) affect the development of atherosclerosis by participating in lipid metabolism. It has been reported that miR-24 participates in pathophysiological processes, including tumor formation (21) and ischemic reperfusion injury (22). However, the role of miR-24 in atherosclerosis has rarely been reported. Maegdefessel et al (23) reported that miR-24 limits aortic vascular inflammation and murine abdominal aneurysm development. Murata et al (24) demonstrated that miR-24 in plasma may function as a biomarker for rheumatoid arthritis. In the present study, serum expression of miR-24 in elderly patients with atherosclerosis was assessed, and the effect of miR-24 on importin-α3, TNF-α and the proliferation and migration ability of vascular endothelial cells was investigated.
Materials and methods
Patients
A total of 30 patients with atherosclerosis admitted to Cangzhou Central Hospital between January and June 2016 were enrolled in the present study, including 20 males and 10 females (age range, 60–70 years; mean, 64±4.9 years). Inclusion criteria were as follows: Significantly elevated blood lipids and a positive diagnosis of atherosclerotic plaque formation using peripheral vascular ultrasound examinations. A total of 30 healthy subjects were enrolled as controls, including 20 males and 10 females (age range, 60–70 years; mean, 63±5.6 years). Healthy subjects had no history of hypertension, diabetes, atherosclerosis or tumors. Peripheral blood (10 ml) was collected from all patients and healthy subjects, and centrifuged at 1,200 × g and 20–22°C for 8 min to separate serum. All procedures were approved by the Ethics Committee of Cangzhou Central Hospital (Cangzhou, China). Written informed consent was obtained from all participants or their families.
Cells
Human umbilical vein endothelial cells (HUVECs; Cell Bank of Chinese Academy of Sciences, Shanghai, China) were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific, Inc., Waltham, MA, USA) at 37°C in an atmosphere containing 5% CO2. Cells were seeded in culture plates at 3×105 cells/well and cultured for 24 h. HUVECs were randomly divided into the has-miR-24 mimic group (transfected with has-miR-24 mimic; Guangzhou RiboBio Co., Ltd., Guangzhou, China) or negative control group (untransfected). When cells reached 50% confluency they were used for transfection. In the first vial, 7.5 µl small RNA fragments was mixed with 125 µl serum-free DMEM. In the second vial, 7.5 µl liposome (Lipofectamine 2000; Thermo Fisher Scientific, Inc.) was mixed with 125 µl serum-free DMEM. After standing for 5 min, the two vials were combined and left to stand at room temperature for 20 min. The mixtures were subsequently added to cells for incubation at 37°C for 6 h. The medium was subsequently replaced with DMEM containing 10% fetal bovine serum and cultivated at 37°C for 48 h, at which point the cells were collected for further assays.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Serum (1 ml) was mixed with 1 ml TRIzol (Thermo Fisher Scientific, Inc.) for lysis and total RNA was extracted using the phenol chloroform method. The purity of RNA was determined by A260/A280 using ultraviolet spectrophotometry (Nanodrop ND1000; Thermo Fisher Scientific, Inc.). cDNA was obtained by RT using a PrimeScript™ RT reagent kit with gDNA Eraser (Takara Biotechnology Co., Ltd., Dalian, China) from 1 µg RNA and stored at −20°C. The temperature protocol was as follows: 42°C for 2 min, 37°C for 15 min and 85°C for 5 sec.
Expression of miR-24 was determined using a SYBR PrimeScript miRNA RT-PCR Kit (Takara Biotechnology Co., Ltd.), with U6 as an internal reference. The reaction system (25 µl) contained 12.5 µl SYBR Premix Ex Taq, 1 µl of each forward primer (miR-24, 5′-GCAGATGGCTCAGTTCAGCAG-3′; U6, 5′-TGCGGGTGCTCGCTTCGGCAGC-3′), 1 µl Uni-miR qPCR primer (cat no. 638315; Takara Biotechnology Co., Ltd.), 2 µl template and 8.5 µl ddH2O. The reaction protocol was as follows: Initial denaturation at 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min (iQ5; Bio-Rad Laboratories, Inc., Hercules, CA, USA). The 2−ΔΔCq method was used to calculate the relative expression of miR-24 against U6 (25). Each sample was tested in triplicate.
A SYBR-Green RT-qPCR kit (Kapa Biosystems, Inc., Wilmington, MA, USA) was used to detect the mRNA expression of importin-α3 and TNF-α, using GAPDH as an internal reference. The reaction system (20 µl) was composed of 10 µl SYBR Premix Ex Taq, 0.5 µl upstream primer (importin-α3, 5′-CTGTGTACGAGAGCGTGGTT-3′; TNF-α, 5′-GGAGAAGGGTGACCGACTCA-3′; and GAPDH, 5′-AAGGTGAAGGTCGGAGTCA-3′), 0.5 µl downstream primer (importin-α3, 5′-TATCAGCCCCCTGAAGGTGA-3′; TNF-α, 5′-CTGCCCAGACTCGGCAA-3′; and GAPDH, 5′-GGAAGATGGTGATGGGATTT-3′), 1 µl cDNA and 8 µl ddH2O. Thermocycling conditions were as follows: Initial denaturation at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 1 min, annealing at 60°C for 40 sec and elongation at 72°C for 30 sec, and final elongation at 72°C for 1 min. The 2−ΔΔCq method was used to calculate the relative expression of importin-α3 and TNF-α mRNA against GAPDH (25). Each sample was tested in triplicate.
Western blot analysis
HUVECs were seeded into 6-well plates at a density of 1×106 cells/well. At 48 h after transfection, the cells were collected and mixed with 100 µl precooled radioimmunoprecipitation assay lysis buffer (Beyotime Institute of Biotechnology, Haimen, China) containing 1 mM phenylmethylsulfonyl fluoride (Beyotime Institute of Biotechnology) for lysis of 15 min at 4°C. Then, the mixture was centrifuged at 12,000 × g and 4°C for 5 min. The supernatant was used to determine protein concentration using a bicinchoninic acid protein concentration determination kit (RTP7102; Real-Times Biotechnology Co., Ltd., Beijing, China). Protein samples (50 µg) were mixed with 2X SDS loading buffer and denatured in a boiling water bath for 5 min. Following this, the samples (10 µl) were separated by 10% SDS-PAGE at 100 V. Resolved proteins were transferred to polyvinylidene difluoride membranes on ice (300 mA, 1.5 h) and blocked with 5 g/l skimmed milk at room temperature for 1 h. The membranes were subsequently incubated with goat anti-human importin-α3 polyclonal primary antibody (1:1,000; cat no. ab6039), rabbit anti-human TNF-α polyclonal primary antibody (1:1,000; cat no. ab9635) and rabbit anti-human GAPDH primary antibody (1:2,000; cat no. ab9485, all Abcam, Cambridge, UK) at 4°C overnight. Following washing with PBST three times of 15 min, the membranes were incubated with polyclonal goat anti-rabbit horseradish peroxidase-conjugated secondary antibody (1:1,000; cat no. ab205718, Abcam) for 1 h at room temperature. Membranes were washed three times with PBST three times for 15 min and developed using an enhanced chemiluminescence detection kit (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). Image Lab v3.0 software (Bio-Rad Laboratories, Inc.) was used to acquire and analyze imaging signals. Expression of importin-α3 and TNF-α protein was calculated relative to GAPDH.
Cell-Counting Kit 8 (CCK-8) assay
HUVECs were seeded at 5,000 cells/well in 96-well plates for transfection. At 48 h after transfection, HUVECs were subjected to CCK-8 assay for the detection of proliferation. At 24, 48 and 72 h, DMEM (Hyclone; GE Healthcare Life Sciences, Logan, UT, USA) was discarded, the cells were washed with twice with PBS and 10% CCK-8 reaction reagent (Beyotime Institute of Biotechnology) diluted in DMEM medium was added. Following incubation at 37°C for 2 h, the absorbance of each well was measured at 450 nm using 600 nm as a reference for plotting cell proliferation curves. A total of five replicate wells were assayed for each group and the mean values were calculated.
Transwell assay
HUVECs were seeded at 3×105/well into 6-well plates for transfection. At 48 h after transfection, Transwell chambers (8 µm diameter and 24 wells; Corning Inc., Corning, NY, USA) were used to evaluate the migration ability of HUVECs. Transfected cells were collected by trypsin digestion, and resuspended at a density of 1×105 cells/ml using DMEM. The cell suspension (100 µl) was added into the upper chamber. In the lower chamber, 600 µl DMEM medium supplemented with 10% fetal bovine serum was added. Following incubation at 37°C for 24 h, cells in the upper chamber were wiped with a cotton swab. The chamber was subsequently fixed using 95% ethanol for 10 min at room temperature. Following staining with 0.1% crystal violet at 22°C for 30 min, the number of cells in 10 fields were counted under a light microscope (magnification, ×200).
Dual luciferase reporter assay
Bioinformatics prediction is a powerful tool for studying the functions of miRNA. To elucidate whether importin-α3 (KPNA4) was a target gene of miR-24, TargetScan (targetscan.org) was used to predict miRNA molecules that may regulate importin-α3, and miR-24 was identified as a potential regulator of importin-α3. According to the bioinformatics results, wild-type (WT) and mutant seed regions of miR-24 in the 3′-untranslated region (UTR) of importin-α3 gene were chemically synthesized in vitro, added to SpeI and HindIII restriction sites, and cloned into pMIR-REPORT luciferase reporter plasmids (Promega Corporation, Madison, WI, USA). Plasmids (0.5 µg) with WT or mutant 3′-UTR DNA sequences were co-transfected with miR-24 mimic (100 nM; Sangon Biotech Co., Ltd., Shanghai, China) into HEK293T cells (ATCC, Manassas, VA, USA). Following cultivation at 37°C for 24 h, cells were lysed using a dual luciferase reporter assay kit (Promega Corporation) according to the manufacturer's manual, and fluorescence intensity was measured using GloMax 20/20 illuminometer (Promega Corporation). Using Renilla fluorescence activity as an internal reference, the fluorescence values of each group of cells were measured.
Statistical analysis
Statistical analysis was performed using SPSS v16.0 (SPSS, Inc., Chicago, IL, USA). Measurement data were expressed as the mean ± standard deviation. Two groups of data were compared using an independent samples t-test. Single factor analysis of variance was used to compare the means of multiple samples followed by a Tukey's post hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of miR-24 in peripheral blood from patients with atherosclerosis is abnormal
To measure the expression of miR-24 in the peripheral blood, RT-qPCR was performed. The results demonstrated that the level of miR-24 in patients with atherosclerosis was significantly lower than that of the controls (P<0.05; Fig. 1). These findings suggests that miR-24 expression is abnormal in patients with atherosclerosis.
Figure 1.
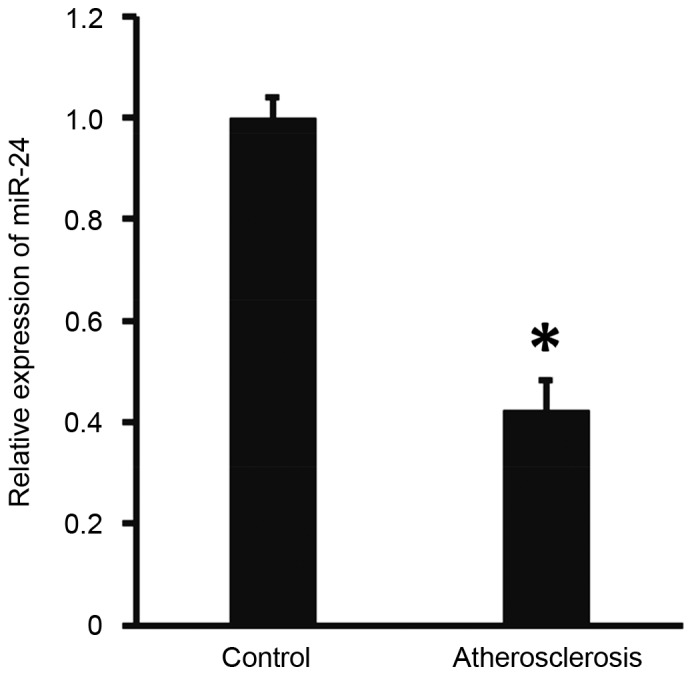
Expression of miR-24 in peripheral blood from healthy control subjects and patients with atherosclerosis. Expression of miR-24 was determined by reverse transcription-quantitative polymerase chain reaction. *P<0.05 vs. control. miR, microRNA.
Overexpression of miR-24 inhibits the transcription and translation of importin-α3 gene
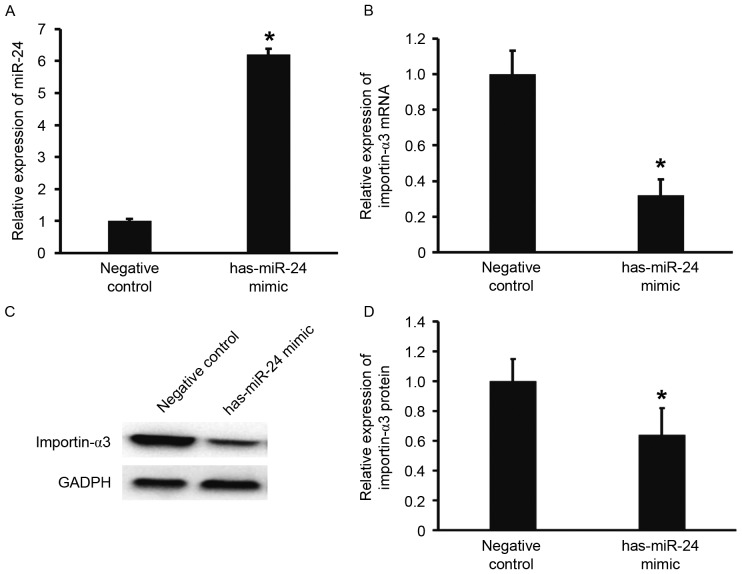
To predict whether miR-24 is able to target importin-α3, TargetScan was used. The data demonstrated that miR-24 is able to bind with the 3′-UTR of miR-24 (Fig. 2). To test whether miR-24 regulates the expression of importin-α3, HUVECs were transfected with miR-24 mimic. The results showed that that HUVECs transfected with miR-24 mimics had significantly increased miR-24 levels when compared with the negative control group (P<0.05; Fig. 3A). In addition, the expression of importin-α3 mRNA and protein in HUVECs transfected with miR-24 mimics was significantly lower than that of the negative control (P<0.05; Fig. 3B-D). These results indicate that overexpression of miR-24 inhibits the transcription and translation of importin-α3 gene.
Figure 2.

Bioinformatics prediction of direct interactions between miR-24 and importin-α3 using TargetScan (targetscan.org). miR, microRNA.
Figure 3.
Effect of miR-24 on the expression of importin-α3 in HUVECs. (A) Relative expression of miR-24, (B) importin-α3 mRNA and (C and D) importin-α3 protein in HUVECs from the negative control and miR-24 mimic groups. Reverse transcription-quantitative polymerase chain reaction was used to determine the expression of miR-24 and importin-α3 mRNA, and western blotting was employed to measure importin-α3 protein expression. *P<0.05 vs. negative control group. miR, microRNA; HUVECs, human umbilical vein epithelial cells.
miR-24 negatively regulates the expression of endothelial inflammatory factor TNF-α
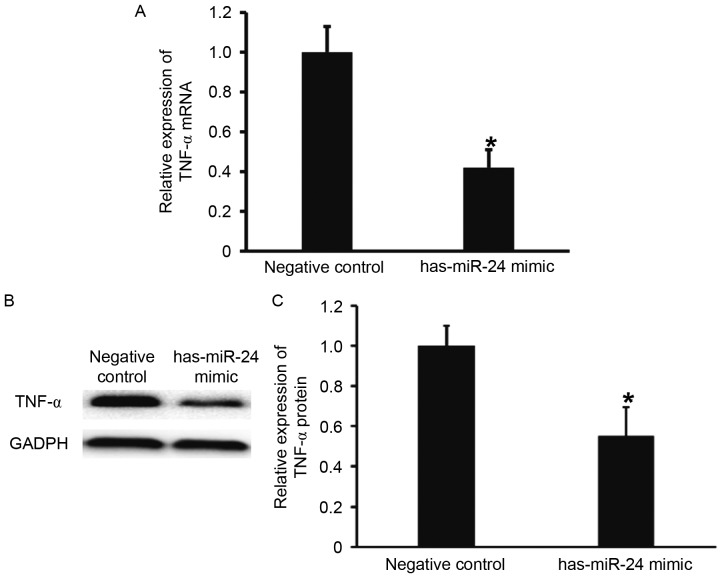
To examine the effect of miR-24 on the expression of TNF-α, RT-qPCR and western blotting were performed. TNF-α mRNA and protein expression levels were significantly lower in HUVECs transfected with miR-24 mimic, when compared with the negative control group (P<0.05; Fig. 4). These results suggest that miR-24 negatively regulates the expression of endothelial inflammatory factor, TNF-α.
Figure 4.
Effect of miR-24 on the expression of TNF-α in HUVECs. (A) Relative expression of TNF-α mRNA in HUVECs of negative control and miR-24 mimic groups. (B and C) Relative expression of TNF-α protein in HUVECs of negative control and miR-24 mimic groups. Reverse transcription-quantitative polymerase chain reaction was used to determine the expression of TNF-α mRNA, and western blotting was employed to measure TNF-α protein expression. *P<0.05 vs. negative control group. miR, microRNA; TNF, tumor necrosis factor; HUVECs, human umbilical vein epithelial cells.
Overexpression of miR-24 inhibits HUVEC proliferation and miR-24 knockdown promotes it
To determine how miR-24 affects the proliferation of HUVECs, a CCK-8 assay was employed. The absorbance of HUVECs transfected with miR-24 mimic was significantly lower than that of the negative control group at 48 and 72 h (P<0.05; Fig. 5). By contrast, the absorbance of HUVECs transfected with miR-24 inhibitor was significantly higher compared with the negative control group at 48 and 72 h (P<0.05; Fig. 5). This suggests that overexpression of miR-24 inhibits the proliferation of endothelial cells, whereas the inhibition of miR-24 expression promotes.
Figure 5.
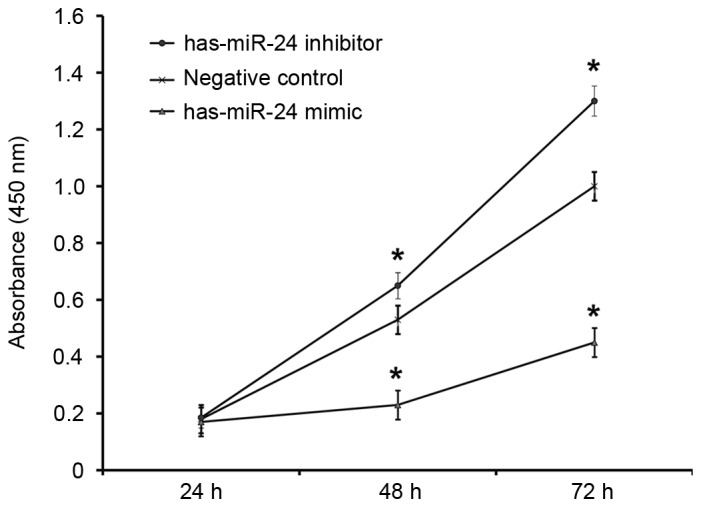
Proliferation of HUVECs at 24, 48, and 72 h after transfection with miR-24 mimics or inhibitors. Cell Counting Kit-8 assay was used to determine the proliferation of the cells. Absorbance of each well was measured at 450 nm with a microplate reader and cell proliferation curves were plotted. *P<0.05 vs. negative control group. miR, microRNA; HUVECs, human umbilical vein epithelial cells.
Overexpression of miR-24 reduces the migration ability of endothelial cells, but inhibition of miR-24 expression promotes it
To examine the effect of miR-24 on the migration ability of HUVECs, a Transwell assay was used. The number of cells that migrated to the lower chamber was significantly lower in the miR-24 mimic group compared with the negative control group (P<0.05; Fig. 6), whereas the number of cells that migrated to the lower chamber in miR-24 inhibitor group was significantly higher (P<0.05; Fig. 6). This suggests that overexpression of miR-24 reduces the migration ability of endothelial cells, whereas miR-24 knockdown promotes it.
Figure 6.
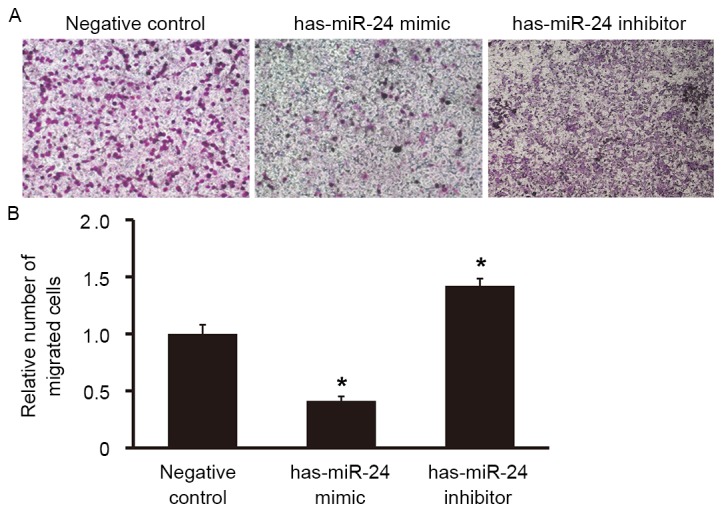
Effect of miR-24 on the migration ability of HUVECs. Transwell assay was used to determine the migration ability of the cells. (A) Representative images the number of migrated cells in the lower chamber (magnification, ×100) and (B) subsequent relative analysis. *P<0.05 vs. negative control group. miR, microRNA; HUVEC, human umbilical vein epithelial cell.
miR-24 downregulates the expression of importin-α3 by binding with the 3′-UTR of importin-α3 gene
To understand whether miR-24 directly targets importin-α3, a dual luciferase reporter assay was performed. Transfection with miR-24 mimics and pMIR-REPORT-wild type importin-α3 led to a significant decrease in fluorescence intensity, as compared with the negative control (P<0.05; Fig. 7). Transfection with miR-24 mimic and pMIR-REPORT-mutant importin-α3 had no significant effect on fluorescence intensity. These results suggests that miR-24 downregulates the expression of importin-α3 by binding with the 3′-UTR of the importin-α3 gene.
Figure 7.
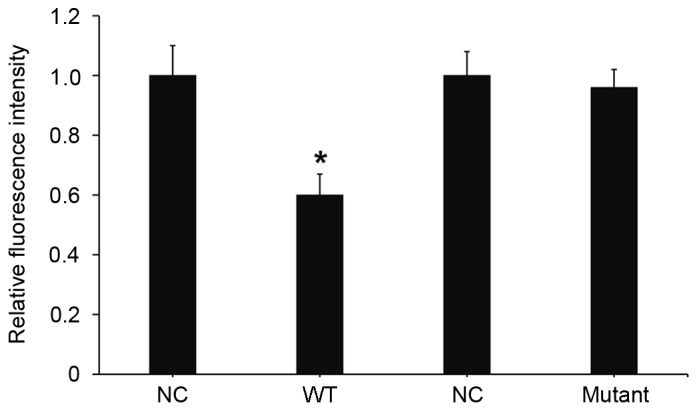
Fluorescence values of HEK293T cells transfected with WT or mutant 3′-untranslated region DNA sequences of importin-α3 and miR-24 mimic. A dual luciferase reporter assay was used to evaluate the interaction between miR-24 and importin-α3. *P<0.05 vs. NC. WT, wild type; miR, microRNA; NC, negative control.
Discussion
Atherosclerosis is the pathological basis of various types of cardiac and cerebral vascular diseases (26). The most important processes in the occurrence and development of atherosclerosis include i) vascular endothelial cell injury and inflammatory activation; ii) the proliferation and migration of endothelial/smooth muscle cells; and iii) the release of inflammatory mediators stimulated by macrophage activation (27–30). It has been reported that miR-24 expression is downregulated in atherosclerotic plaques, and miR-24 participates in the formation of atherosclerosis primarily by targeting and inhibiting matrix metalloproteinase-14 (31). In the present study, it was demonstrated that miR-24 expression is significantly lower in patients with atherosclerosis, as compared with healthy subjects. Bioinformatics revealed that the importin-α3 gene is a potential target gene of miR-24. A previous study reported that the importin protein family (α1, α3, α4, α5, α6 and α7) is an important class of transport proteins associated with the nuclear transfer of NF-κB, which is a key process in the activation of the NF-κB signaling pathway (32). The results of the present study demonstrate that miR-24 inhibits the expression of importin-α3 and NF-κB signaling pathway-mediated inflammatory factor, TNF-α. Naqvi et al (33) demonstrated that TNF-α is also a target gene of miR-24. Combined with the results of the present study, this suggests that miR-24 may regulate inflammatory processes by simultaneously targeting importin-α3 and TNF-α in respect to the mechanism of atherosclerosis. Therefore, miR-24 reduces the activation of vascular endothelial cells and inflammatory responses. Maegdefessel et al (23) reported that miR-24 inhibits the development of aortic vascular inflammation and abdominal aortic aneurysm in mice by targeting chitinase 3-like 1. Xiang et al (34) demonstrated that patients with diabetes have low miR-24 expression, which suggests potential thrombotic events in these patients. The present study indicates that endothelial cell activation and inflammatory responses regulated by miR-24 participate in the development of atherosclerosis.
miRNAs have previously been reported to participate in the regulation of vascular endothelial cell functions, and have important roles in the development of atherosclerosis; for example, enhanced expression of miR-221 and miR-222 in human arterial endothelial cells causes reduced levels and activity of nitric oxide synthase, leading to cell dysfunction (35). In addition, reduced miR-126 and increased miR-125b expression results in increased endothelial inflammatory response protein secretion (36). miR-29 affects the production of collagen and elastin, and has important roles in maintaining arterial structural integrity (37). The results of the present study indicate that overexpression of miR-24 inhibits the proliferation and migration of endothelial cells. Lal et al (38) reported that miR-24 targets E2F2 and inhibits the proliferation of erythroleukemia K562 cells. Similarly, Amelio et al (39) demonstrated that miR-24 regulates the migration of human keratinocytes and mouse epidermal cells, suggesting that miR-24 has important roles in the regulation of proliferation, invasion and migration of cells.
In conclusion, the results of the present study demonstrate that miR-24 inhibits the proliferation and migration of endothelial cells by targeting importin-α3 and regulating inflammatory responses in endothelial cells. Low levels of miR-24 in the peripheral blood are associated with atherosclerosis progression. Furthermore, miR-24 may provide a novel strategy and direction for the clinical treatment and research on atherosclerosis. Further investigation is required to elucidate the underlying mechanism of inflammation in atherosclerosis and to explore the mechanism of abnormal expression of miR-24 in the blood of patients with atherosclerosis.
Acknowledgements
The present study was supported by the Hebei Science and Technology Support Program Key Projects (grant no. 16277707D).
References
- 1.Beckman JA, Creager MA, Libby P. Diabetes and atherosclerosis: Epidemiology, pathophysiology, and management. JAMA. 2002;287:2570–2581. doi: 10.1001/jama.287.19.2570. [DOI] [PubMed] [Google Scholar]
- 2.Malek AM, Alper SL, Izumo S. Hemodynamic shear stress and its role in atherosclerosis. JAMA. 1999;282:2035–2042. doi: 10.1001/jama.282.21.2035. [DOI] [PubMed] [Google Scholar]
- 3.Ross R, Glomset JA. Atherosclerosis and the arterial smooth muscle cell: Proliferation of smooth muscle is a key event in the genesis of the lesions of atherosclerosis. Science. 1973;180:1332–1339. doi: 10.1126/science.180.4093.1332. [DOI] [PubMed] [Google Scholar]
- 4.Katsuda S, Kaji T. Atherosclerosis and extracellular matrix. J Atheroscler Thromb. 2003;10:267–274. doi: 10.5551/jat.10.267. [DOI] [PubMed] [Google Scholar]
- 5.Morland K, Wing S, Roux A Diez. The contextual effect of the local food environment on residents' diets: The atherosclerosis risk in communities study. Am J Public Health. 2002;92:1761–1767. doi: 10.2105/AJPH.92.11.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fox CS, Polak JF, Chazaro I, Cupples A, Wolf PA, D'Agostino RA, O'Donnell CJ. Framingham Heart Study: Genetic and environmental contributions to atherosclerosis phenotypes in men and women: Heritability of carotid intima-media thickness in the Framingham Heart Study. Stroke. 2003;34:397–401. doi: 10.1161/01.STR.0000048214.56981.6F. [DOI] [PubMed] [Google Scholar]
- 7.Fenyo IM, Gafencu AV. The involvement of the monocytes/macrophages in chronic inflammation associated with atherosclerosis. Immunobiology. 2013;218:1376–1384. doi: 10.1016/j.imbio.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 8.Gomez D, Owens GK. Smooth muscle cell phenotypic switching in atherosclerosis. Cardiovasc Res. 2012;95:156–164. doi: 10.1093/cvr/cvs115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Munro JM, Cotran RS. The pathogenesis of atherosclerosis: Atherogenesis and inflammation. Lab Invest. 1988;58:249–261. [PubMed] [Google Scholar]
- 10.de Winther MP, Kanters E, Kraal G, Hofker MH. Nuclear factor kappaB signaling in atherogenesis. Arterioscler Thromb Vasc Biol. 2005;25:904–914. doi: 10.1161/01.ATV.0000160340.72641.87. [DOI] [PubMed] [Google Scholar]
- 11.Rajendran P, Rengarajan T, Thangavel J, Nishigaki Y, Sakthisekaran D, Sethi G, Nishigaki I. The vascular endothelium and human diseases. Int J Biol Sci. 2013;9:1057–1069. doi: 10.7150/ijbs.7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fagerlund R, Kinnunen L, Köhler M, Julkunen I, Melén K. NF-{kappa}B is transported into the nucleus by importin {alpha}3 and importin {alpha}4. J Biol Chem. 2005;280:15942–15951. doi: 10.1074/jbc.M500814200. [DOI] [PubMed] [Google Scholar]
- 13.Bouwmeester T, Bauch A, Ruffner H, Angrand PO, Bergamini G, Croughton K, Cruciat C, Eberhard D, Gagneur J, Ghidelli S, et al. A physical and functional map of the human TNF-alpha/NF-kappaB signal transduction pathway. Nat Cell Biol. 2004;6:97–105. doi: 10.1038/ncb1129. [DOI] [PubMed] [Google Scholar]
- 14.Rayner KJ, Fernandez-Hernando C, Moore KJ. MicroRNAs regulating lipid metabolism in atherogenesis. Thromb Haemost. 2012;107:642–647. doi: 10.1160/TH11-10-0694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koch M, Mollenkopf HJ, Klemm U, Meyer TF. Induction of microRNA-155 is TLR- and type IV secretion system-dependent in macrophages and inhibits DNA-damage induced apoptosis; Proc Natl Acad Sci USA; 2012; pp. E1153–E1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nazari-Jahantigh M, Wei Y, Noels H, Akhtar S, Zhou Z, Koenen RR, Heyll K, Gremse F, Kiessling F, Grommes J, et al. MicroRNA-155 promotes atherosclerosis by repressing Bcl6 in macrophages. J Clin Invest. 2012;122:4190–4202. doi: 10.1172/JCI61716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei Y, Nazari-Jahantigh M, Chan L, Zhu M, Heyll K, Corbalán-Campos J, Hartmann P, Thiemann A, Weber C, Schober A. The microRNA-342-5p fosters inflammatory macrophage activation through an Akt1- and microRNA-155-dependent pathway during atherosclerosis. Circulation. 2013;127:1609–1619. doi: 10.1161/CIRCULATIONAHA.112.000736. [DOI] [PubMed] [Google Scholar]
- 18.Ramírez CM, Goedeke L, Rotllan N, Yoon JH, Cirera-Salinas D, Mattison JA, Suárez Y, de Cabo R, Gorospe M, Fernández-Hernando C. MicroRNA 33 regulates glucose metabolism. Mol Cell Biol. 2013;33:2891–2902. doi: 10.1128/MCB.00016-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu J, Xu Y, Hao J, Wang S, Li C, Meng S. MiR-122 in hepatic function and liver diseases. Protein Cell. 2012;3:364–371. doi: 10.1007/s13238-012-2036-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shirasaki T, Honda M, Shimakami T, Horii R, Yamashita T, Sakai Y, Sakai A, Okada H, Watanabe R, Murakami S, et al. MicroRNA-27a regulates lipid metabolism and inhibits hepatitis C virus replication in human hepatoma cells. J Virol. 2013;87:5270–5286. doi: 10.1128/JVI.03022-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie Y, Tobin LA, Camps J, Wangsa D, Yang J, Rao M, Witasp E, Awad KS, Yoo N, Ried T, Kwong KF. MicroRNA-24 regulates XIAP to reduce the apoptosis threshold in cancer cells. Oncogene. 2013;32:2442–2451. doi: 10.1038/onc.2012.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meloni M, Marchetti M, Garner K, Littlejohns B, Sala-Newby G, Xenophontos N, Floris I, Suleiman MS, Madeddu P, Caporali A, Emanueli C. Local inhibition of microRNA-24 improves reparative angiogenesis and left ventricle remodeling and function in mice with myocardial infarction. Mol Ther. 2013;21:1390–1402. doi: 10.1038/mt.2013.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maegdefessel L, Spin JM, Raaz U, Eken SM, Toh R, Azuma J, Adam M, Nakagami F, Heymann HM, Chernogubova E, et al. miR-24 limits aortic vascular inflammation and murine abdominal aneurysm development. Nat Commun. 2014;5:5214. doi: 10.1038/ncomms6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murata K, Furu M, Yoshitomi H, Ishikawa M, Shibuya H, Hashimoto M, Imura Y, Fujii T, Ito H, Mimori T, Matsuda S. Comprehensive microRNA analysis identifies miR-24 and miR-125a-5p as plasma biomarkers for rheumatoid arthritis. PLoS One. 2013;8:e69118. doi: 10.1371/journal.pone.0069118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Hansson GK. Inflammation, atherosclerosis, and coronary artery disease. N Engl J Med. 2005;352:1685–1695. doi: 10.1056/NEJMra043430. [DOI] [PubMed] [Google Scholar]
- 27.Libby P, Ridker PM, Hansson GK. Leducq Transatlantic Network on Atherothrombosis: Inflammation in atherosclerosis: From pathophysiology to practice. J Am Coll Cardiol. 2009;54:2129–2138. doi: 10.1016/j.jacc.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Packard RR, Libby P. Inflammation in atherosclerosis: From vascular biology to biomarker discovery and risk prediction. Clin Chem. 2008;54:24–38. doi: 10.1373/clinchem.2007.097360. [DOI] [PubMed] [Google Scholar]
- 29.Chachaj A, Drozdz K, Szuba A. Reverse cholesterol transport processes and their role in artherosclerosis regression. Postepy Biochem. 2008;54:301–307. (In Polish) [PubMed] [Google Scholar]
- 30.Hansson GK, Hermansson A. The immune system in atherosclerosis. Nat Immunol. 2011;12:204–212. doi: 10.1038/ni.2001. [DOI] [PubMed] [Google Scholar]
- 31.Di Gregoli K, Jenkins N, Salter R, White S, Newby AC, Johnson JL. MicroRNA-24 regulates macrophage behavior and retards atherosclerosis. Arterioscler Thromb Vasc Biol. 2014;34:1990–2000. doi: 10.1161/ATVBAHA.114.304088. [DOI] [PubMed] [Google Scholar]
- 32.Kelley JB, Talley AM, Spencer A, Gioeli D, Paschal BM. Karyopherin alpha7 (KPNA7), a divergent member of the importin alpha family of nuclear import receptors. BMC Cell Biol. 2010;11:63. doi: 10.1186/1471-2121-11-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naqvi AR, Fordham JB, Ganesh B, Nares S. miR-24, miR-30b and miR-142-3p interfere with antigen processing and presentation by primary macrophages and dendritic cells. Sci Rep. 2016;6:32925. doi: 10.1038/srep32925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiang Y, Cheng J, Wang D, Hu X, Xie Y, Stitham J, Atteya G, Du J, Tang WH, Lee SH, et al. Hyperglycemia repression of miR-24 coordinately upregulates endothelial cell expression and secretion of von Willebrand factor. Blood. 2015;125:3377–3387. doi: 10.1182/blood-2015-01-620278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu XD, Wu X, Yin YL, Liu YQ, Geng MM, Yang HS, Blachier F, Wu GY. Effects of dietary L-arginine or N-carbamylglutamate supplementation during late gestation of sows on the miR-15b/16, miR-221/222, VEGFA and eNOS expression in umbilical vein. Amino Acids. 2012;42:2111–2119. doi: 10.1007/s00726-011-0948-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rippe C, Blimline M, Magerko KA, Lawson BR, LaRocca TJ, Donato AJ, Seals DR. MicroRNA changes in human arterial endothelial cells with senescence: Relation to apoptosis, eNOS and inflammation. Exp Gerontol. 2012;47:45–51. doi: 10.1016/j.exger.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boon RA, Seeger T, Heydt S, Fischer A, Hergenreider E, Horrevoets AJ, Vinciguerra M, Rosenthal N, Sciacca S, Pilato M, et al. MicroRNA-29 in aortic dilation: Implications for aneurysm formation. Circ Res. 2011;109:1115–1119. doi: 10.1161/CIRCRESAHA.111.255737. [DOI] [PubMed] [Google Scholar]
- 38.Lal A, Navarro F, Maher CA, Maliszewski LE, Yan N, O'Day E, Chowdhury D, Dykxhoorn DM, Tsai P, Hofmann O, et al. miR-24 Inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to ‘seedless’ 3′UTR microRNA recognition elements. Mol Cell. 2009;35:610–625. doi: 10.1016/j.molcel.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amelio I, Lena AM, Viticchiè G, Shalom-Feuerstein R, Terrinoni A, Dinsdale D, Russo G, Fortunato C, Bonanno E, Spagnoli LG, et al. miR-24 triggers epidermal differentiation by controlling actin adhesion and cell migration. J Cell Biol. 2012;199:347–363. doi: 10.1083/jcb.201203134. [DOI] [PMC free article] [PubMed] [Google Scholar]