Abstract
MicroRNAs (miRNAs/miRs) are a class of small, non-coding RNA molecules that serve a key function in carcinogenesis and tumor progression. Recent evidence indicates that miRNAs may act as powerful regulators of migration and invasion. The present study aimed to investigate the effect of miR-25 on the invasion and metastasis of KYSE-150 and EC109 esophageal squamous cell carcinoma (ESCC) cells, and predict the mechanism of this effect by bioinformatically analyzing the miR-106b-25 cluster. In order to alter the expression of miR-25 in the two cell lines, a miR-25 inhibitor or mimic were transfected into the cells, which were then studied via Transwell migration and invasion assays. Subsequently, the target genes of the miR-106b-25 cluster were predicted using miRanda, PicTar, TargetScan and miRTarbase, and the functions of the target genes were predicted via Gene Ontology term and Kyoto Encyclopedia of Genes and Genomes pathway enrichment analyses. Then, a protein-protein interaction (PPI) network was produced using the Search Tool for the Retrieval of Interacting Genes. The results revealed that overexpressing miR-25 led to significantly increased cell migration and invasion in KYSE150 and EC109 cells. Suppressing miR-25 resulted in significantly decreased cell migration and invasion in KYSE150 cells, while the result was not significant in EC109 cells. Target genes of the miR-106b-25 cluster were significantly enriched in the biological process regulation of cellular metabolic process and several cancer-associated pathways, such as those for glioma and melanoma. The PPI network revealed that PTEN, TP53, MDM2, E2F1, PRMT5, MCM2, RB1, CDKN1A, SHAD7 and EZH2 may serve core roles within the network and associate with one another during the pathogenesis of ESCC. These results indicate that a high expression of miR-25 promotes the invasion and metastasis of ESCC cells, while the influence of low expression of miR-25 differs with cells with different degrees of differentiation. Invasion and metastasis are not effected in cells with poor differentiation, while they were decreased in well differentiated cells. Furthermore, PTEN, TP53, MDM2, E2F1, PRMT5, MCM2, RB1, CDKN1A, SHAD7 and EZH2 may be targeted by the miR-106b-25 cluster, and act together to regulate the development of ESCC.
Keywords: esophageal cancer/carcinoma, miR-106b-25 cluster, miR-25, miR-106b, miR-93, invasion, metastasis, bioinformatic analysis
Introduction
Esophageal cancer is an important public health problem worldwide, as the eighth most common cancer and sixth most common cause of cancer-associated mortality (1). Esophageal squamous cell carcinoma (ESCC) accounts for 90% of all histological types of esophageal carcinoma in the highest risk area, North-central China, which ranks first in ESCC morbidity and mortality rates (2). Genetic heterogeneity and complex regulatory networks make it difficult to treat ESCC (3). Thus, identifying the regulatory pathways for the stratification of ESCC is of great clinical significance.
MicroRNAs (miRNAs/miRs) are single-stranded RNAs of 18–24 nucleotides in length that post-transcriptionally regulate gene expression by directly binding to the 3′-untranslated region (UTR) of target mRNAs (4). Emerging evidence has revealed that numerous miRs are involved in tumorigenesis and/or tumor progression (5). The involvement of miR regulatory mechanisms in cancer progression may be essential to understand the complete molecular landscape of carcinoma progression. Although the first miRNA, lin-4, was discovered in 1,993 (6) and the second, let-7, in 2,000 (7) in Caenorhabditis elegans, only recently has the study of this class of small regulatory RNAs in humans become more widespread. Experimental evidence that has accumulated has led researchers to notice that unknown molecular factors, particularly non-coding RNAs, serve important roles in carcinogenesis (8). Approximately 70% of the genome has been identified to be transcribed in mammals (9). Over the past decade, miRNAs have emerged as important players in RNA interference-mediated post-transcriptional gene regulation.
miRNAs can be transcribed from individual genes or as clusters (10), with ~30% of miRNAs transcribed as polycistronic clusters (11). miRNA clusters are comprised of several miRNA genes, which are located adjacent to one another on the chromosome and are typically highly related structural genes with similar functions (12). The miRNA genes in these clusters are transcribed as one long pri-miRNA transcript, which is subsequently processed into the individual miRNAs (12). The role of miRNA clusters in tumorigenesis has been extensively studied; Liu et al (13) identified that the cluster of miR-143 and miR-145 served a role in the modulation of ESCC metastases through targeting oncogenic fascin actin-bundling protein 1, thus affecting the risk of ESCC progression. The miR-106b-25 cluster is located in the thirteenth intron of the DNA replication gene minichromosome maintenance complex component 7 (MCM7), which resides on chromosome 7 in humans and encodes three miRNAs: miR-106b, miR-93 and miR-25. A previous study revealed that members of the miR-106b-25 cluster are overexpressed in several types of cancer (14).
miR-25, a member of the miR-106b-25 cluster, has been identified to be overexpressed in a variety of types of cancer and serve functional roles in numerous malignancy-associated processes, including tumorigenesis, and cancer cell proliferation, migration and metastasis (15). Furthermore, diverse molecular signaling pathways appear to be the targets of miR-25 in various types of cancer. For example, in breast cancer miR-25 activates the transforming growth factor (TGF)-β signaling pathway to induce tumorigenesis (15). However, the molecular mechanisms underlying ESCC remain unclear; thus, further studies are required to ascertain a more thorough understanding of the molecular basis of ESCC.
The present study aimed to investigate the effects of miR-25 on the metastasis and invasion of KYSE-150 and EC109 ESCC cells. Furthermore, members of the miR-106b-25 cluster were bioinformatically analyzed using a variety of methods, including Gene Ontology (GO) term and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses. Downstream proteins were analyzed and potential target genes of miR-106b, miR-93 and miR-25 were identified, laying a foundation for further research.
Materials and methods
Cell culture
The human ESCC cell lines KYSE150 and EC109 were purchased from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). KYSE150 is highly differentiated, while EC109 is less differentiated. All cells were cultured in RPMI-1640 (Gibco) supplemented with 10% fetal bovine serum (FBS; Invitrogen) (both Thermo Fisher Scientific, Inc., Waltham, MA, USA) at 37°C with 5% CO2.
Transfection
KYSE150 and EC109 were seeded into 6-well plates (2×105 cells per well) and cultured for 5–10 min prior to transfection. HiPerFect Transfection Reagent (Qiagen GmbH, Hilden, Germany) was used for miRNA and siRNA transfection according to the manufacturer's protocol. miScript miR-25 mimic (MSY0000081), miScript miR-25 inhibitor (MIN0000081) and Allstars negative control siRNA (cat.no. 1027280; all Qiagen GmbH) were transfected into the cells. Transfected cells were then subjected to reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis, and Transwell invasion and migration assays.
RT-qPCR analysis
Total RNA from KYSE150 and EC109 cells was isolated 48 h following transfection using the miRNeasy Mini kit, cDNA was reverse transcribed using the miScript II Reverse Transcription kit and qPCR was performed using the miScript SYBR Green PCR kit (all Qiagen GmbH), according to the manufacturer's protocol. The thermocycling conditions were presented in Table I. The threshold cycle (Cq) was defined as the fractional cycle number at which the fluorescence passed the fixed threshold. The PCR primers were as follows: miR-25, forward, 5′-CGAGCTCTTTTAAGACAGG-3′ and reverse, 5′-GCCGGCTGCATGCCTGTGG-3′; and U6, forward, 5′-GTGTGCTACGGAGTTCAGAGGTT-3′ and reverse, 5′-TGGGGTTATACATTGTGAGAGGA-3′.miRNA quantitation was performed using a TaqMan miRNA assay (Applied Biosystems; Thermo Fisher Scientific, Inc.) in a CFX96 real-time thermal cycler (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The amount of target gene expression was normalized using U6 as the reference gene and the amount of target gene in the control cells was set at 1.0. Following the initial activaton step for 15 min at 95°C, the reaction system entered into the 3 step cycling which included the following steps for 40 cycles: Denaturation for 15 sec at 94°C; annealing for 30 sec at 55°C and extension for 30 sec at 70°C. The relative expression of miR-25 was then calculated using the 2−ΔΔCq method (16). All samples were processed in triplicate.
Table I.
Predicted target Genes of the miR-106b-25 cluster.
| miRNA | Target genes |
|---|---|
| miR-106b | KAT2B, DNAJB9, E2F1, AGFG2, APC, SMAD5, S1PR1, PLEKHM1, C5orf41, ITCH, APP, CDKN1A, RB1, TCEAL1 |
| miR-93 | KAT2B, ENPP5, TP53, EPHA4, ZNF512B, TRIM3, FGD5, OSR1, RAB5B, GIGYF1, PGBD5, NRP2, NKX3-2, FGD1, MAP3K3, PAFAH1B1, NHLH1, SKI, TNRC6A, INP1, CDKN1A, E2F1, ITGB8, TUSC2, PTEN, LATS2 |
| miR-25 | PTEN, ZNF512B, ERC2, KAT2B, MIER3, DMXL1, TP53, EXOC5, PRKCE, PCDH11Y, PDS5B, E2F3, LHFPL2, COL1A2, ARHGEF17, FOXG1, MAP2K4, TMEM87A, PRDM13, ZNF287, TRAK2, ATRX, |
| DNAJB12, KCNK10, C17orf39, GDF11, XYLT2, LATS2, DSCAML1, TOB1, EGR2, ADM, IQGAP2, ADCY3, FNBP4,NAA15, SOBP, RNF44, NLK, BAZ2A, BCL2L11, CDKN1C, WDR4, CDH1, CCL26, MDM2, EZH2, SMAD7, PRMT5, KLF4, FBXW7, NOX4, WWP2, TMEM188 |
miR, microRNA.
Transwell invasion and migration assays
A total of 48 h after transfection the KYSE-150 and EC109 cells were used for Transwell invasion and migration assays. For the invasion assay, the upper chamber of a 24-well Transwell permeable support with 8 um pores (cat. no. 5422) coated with Matrigel (1 mg/ml; Matrigel Basement Membrane Matrix) (both Corning Incorporated, Corning, NY, USA) was loaded with 200 µl of cell suspension (1–4×105 cells/well) and the lower chamber was loaded with 600 µl of RPMI-1,640 containing 10% FBS. The Transwell was then incubated at 37°C with 5% CO2 for 48 h. Subsequently, the cells remaining on the filter surface were fixed with 4% formaldehyde for 15 min at room temperature, stained with 0.5% crystal violet at room temperature for 30 min and examined under a microscope (objective, ×40; magnification, ×100). Cells in at least six random microscopic fields were counted. A total of five wells were tested for each group and all experiments were performed in triplicate. The migration assay was performed in the same way, with the exclusion of the Matrigel.
miR-106b, miR-2 and miR-93 target gene prediction, and GO term and KEGG pathway enrichment analyses
Three target gene prediction databases were utilized to predict the target genes of the three miRs of miR-106b-25 cluster. These were TargetScan (version 6.1; targetscan.org), PicTar (pictar.mdc-berlin.de) and miRanda (microrna.org). The intersection of the predicted targeted genes was taken and then combined with the validated genes that miRTarBase 6.0 (http://mirtarbase.mbc.nctu.edu.tw/php/search.php) provided in order to obtain the target genes. GO analysis is widely used for the functional annotation of genomic data. To explore the functions of miR-106b, miR-2 and miR-93, GO (geneontology.org) (17) was employed to perform biological process term enrichment analysis. In addition, KEGG pathway enrichment analysis was performed. These enrichment analyses were performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID; version 6.7) (17). KEGG can provide a valid estimate of the target genes' P-value by a Monte Carlo simulation procedure (18). The enrichment analysis results with P<0.05 were identified as significant results.
Protein-protein interaction (PPI) network construction
The identified target genes of the miR-106b-25 cluster were placed into the Search Tool for the Retrieval of Interacting Genes (STRING) database (string-db.org), a meta resource that collects available information on protein-protein associations, scores and weights them, and then augments them with predicted interactions and with the results of automatic text-mining searches in order to construct a PPI network (17).
Statistical analysis
Data are expressed as the mean ± standard error. The differences among groups were determined by one-way analysis of variance, which was followed by a post hoc Dunnett's T3 (3) test when multiple groups were being compared using SPSS 20.0 (IBM Corp., Armonk, NY, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of miR-25 in KYSE-150 and EC109 cells after transfection
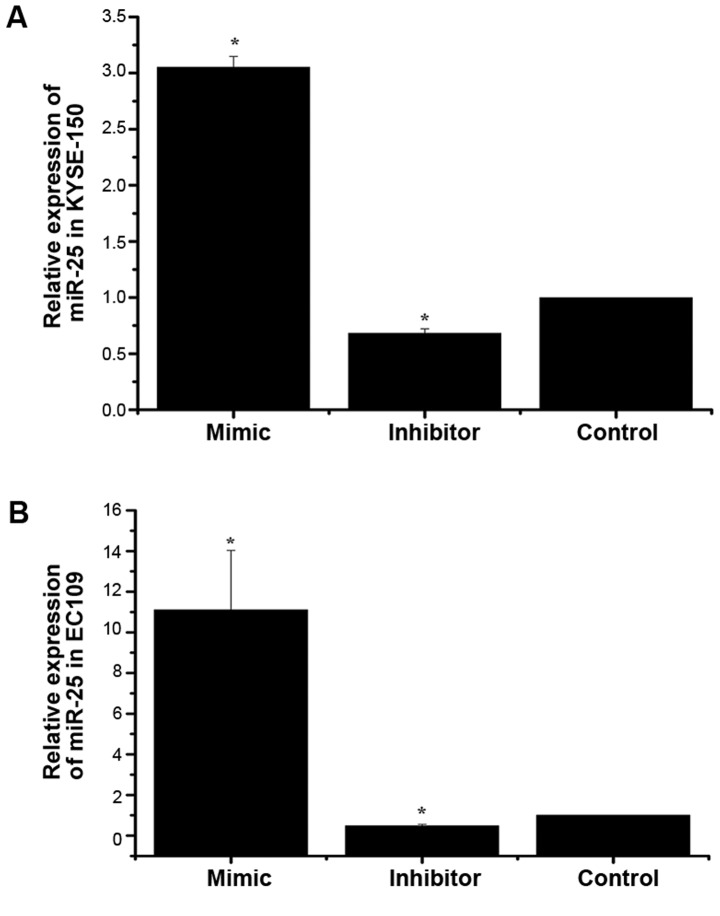
To investigate the impact of miR-25 on the growth of KYSE-150 and EC109 cells, the expression of miR-25 was altered by the transfection of a mimic or inhibitor. A negative control was also transfected into a third group of cells. The expression of miR-25 in the three groups of KYSE-150 and EC109 cells was then detected by RT-qPCR analysis. U6 was used as an internal control. As demonstrated in Fig. 1, the expression of miR-25 was significantly increased in the mimic group compared with the control group, and significantly decreased in the inhibitor group. This indicated that cell models with differential expression of miR-25 were successfully constructed.
Figure 1.
Expression of miR-25 in KYSE-150 and EC109 cells after transfection. The relative expression of miR-25 in (A) KYSE-150 and (B) EC109 cells after transfection of the mimic or inhibitor was measured using reverse transcription-quantitative polymerase chain reaction analysis. *P<0.05 vs. the control group. miR, microRNA.
miR-25 affects the invasion and metastasis of KYSE-150 and EC109 ESSC cells differently
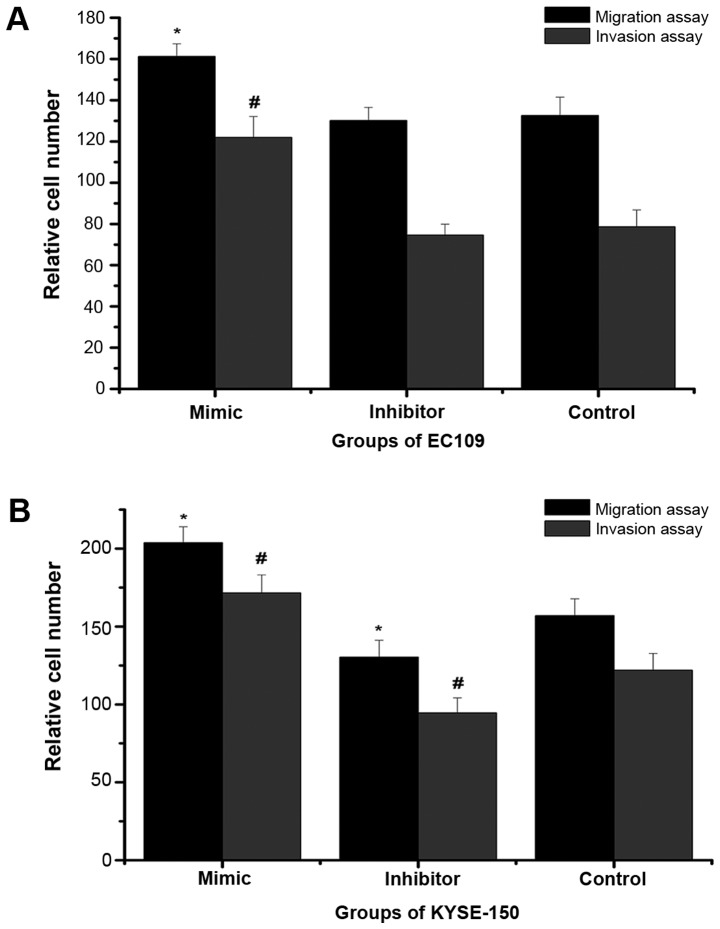
In order to analyze metastasis and invasiveness, which are important features of malignant cells, Transwell migration and invasion assays were performed (Figs. 2 and 3). In the migration and invasion assays, the number of KYSE-150 cells in the mimic group was significantly higher than that in the negative control group, while the number of cells in the inhibitor group was significantly decreased compared with the negative control group. In the EC109 cells, similarly, the number of cells in the mimic group was significantly higher than that in the negative control group, however, there was no significant difference between the numbers of cells in the inhibitor and negative control groups. This indicated that high expression of miR-25 may promote the invasion and metastasis of ESCC cells, while low expression of miR-25 did not necessarily inhibit invasion and metastasis.
Figure 2.
Effect of miR-25 on the migration and invasion of KYSE-150 and EC109 cells. The relative number of cells after Transwell migration and invasion assays were calculated in (A) KYSE-150 and (B) EC109 cells 48 h after transfection of the miR-25 mimic or inhibitor. *P<0.05 vs. the control migration assay group; #P<0.05 vs. the control invasion assay group. miR, microRNA.
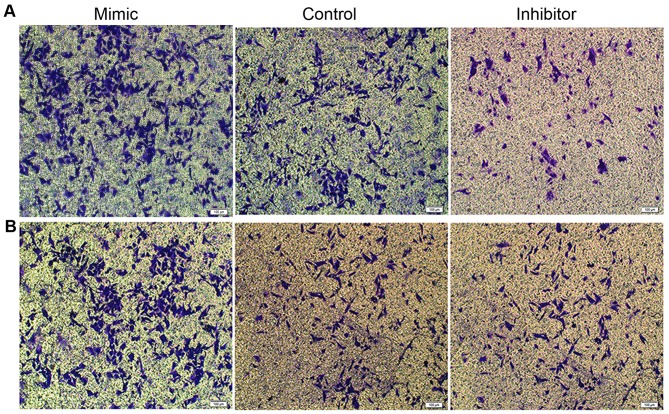
Figure 3.
Microscopy observations of the effect of miR-25 on the migration of KYSE-150 and EC109 cells. Following the Transwell migration assay, the (A) KYSE-150 and (B) EC109 cells transfected with the miR-25 mimic or inhibitor that had migrated were stained with crystal violet and inspected under a microscope. Scale bar, 100 µm. miR, microRNA.
Bioinformatical analysis of the predicted target genes of miR-106b, miR-25 and miR-93
In order to investigate potential regulation mechanisms of the miR-106b-25 cluster in the pathogenesis of ESCC, four online bioinformatics databases (TargetScan, PicTar, miRanda and miRTarbase) were used to identify and validate potential targets of the three members of the cluster. A total of 14 target genes of miR-106b, 25 target genes of miR-93 and 54 target genes of miR-25 were identified (Table I). Next, the biological processes that these 93 targets are involved in were investigated using GO term and KEGG pathway enrichment analyses (Tables II and III, respectively). The first three pathways of the enrichment analysis indicated where the target genes were concentrated and the results demonstrated that they might be important genes in the regulation of macromolecule metabolic processes and cell cycle and may serve a role in Glioma.
Table II.
GO biological process term enrichment analysis of the targets of the miR-106b-25 cluster.
| miRNA | GO no. | Term | P-value |
|---|---|---|---|
| miR-106b | 0010604 | Positive regulation of macromolecule metabolic process | <0.0001 |
| 0051329 | Interphase of mitotic cell cycle | 0.000116 | |
| 0007050 | Cell cycle arrest | 0.000116 | |
| miR-93 | 0030030 | Projection organization | 0,000261 |
| 0032319 | Regulation of Rho GTPase activity | 0.000930 | |
| 0007049 | Cell cycle | 0.001180 | |
| miR-25 | 0008285 | Negative regulation of cell proliferation | <0.0001 |
| 0042127 | Regulation of cell proliferation | <0.0001 | |
| 0030030 | Cell projection organization | <0.0001 |
GO, Gene Ontology; miR, microRNA.
Table III.
Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis of the miR-106b-25 cluster.
| miRNA | Term | P-value |
|---|---|---|
| miR-106b | Bladder cancer | 0.0018 |
| Glioma | 0.0040 | |
| Melanoma | 0.0051 | |
| miR-93 | Glioma | 0.0051 |
| Melanoma | 0.0065 | |
| Prostate cancer | 0.0100 | |
| miR-25 | Melanoma | <0.0001 |
| Glioma | 0.0011 | |
| Prostate cancer | 0.0030 |
miR, microRNA.
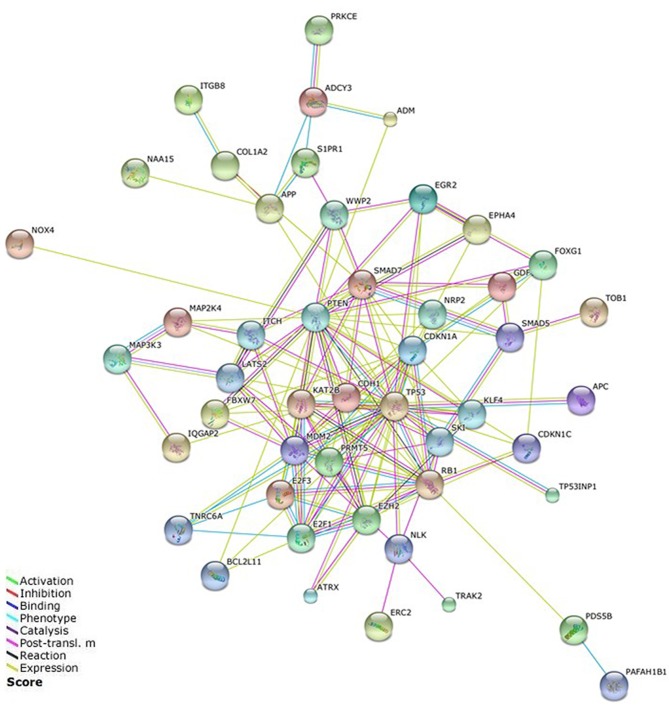
PPI network
A PPI network of the miR-106b-25 cluster target genes was constructed using STRING (Fig. 4). A total of 45 target genes that were identified as being linked to lax isolated nodes and not connected to any other nodes through data analysis were removed. The PPI network created revealed complex interactions, particularly for the target genes PTEN, TP53, MDM2, E2F1, PRMT5, MCM2, RB1, CDKN1A, SMAD7 and EZH2. So these genes were the focus of our study.
Figure 4.
Protein-protein interaction network of the miR-106b-25 cluster. The network was constructed using the Search Tool for the Retrieval of Interacting Genes. Post-transl. m, post-translational modification.
Discussion
Compared with normal cells, cancer cells exhibit reduced cell adhesion, unlimited proliferation and metastasis. Cancer cells may exhibit different degrees of differentiation. Cell differentiation refers to the process undergone by cells of the same origin, which produces cells with different morphological and functional features. Selective expression of the genome in a time- and location-dependent manner causes cell differentiation. In vitro cultured cells grow under specific conditions, and retain the same basic structure and function as cells in vivo. The present study used two ESCC cells, which exhibited different degrees of differentiation. KYSE150 is highly differentiated, while EC109 is less differentiated. The cells were transfected with an miR-25 mimic or inhibitor in order to investigate the role of miR-25 in ESCC. The results indicated that overexpression of miR-25 promoted the invasion and metastasis of ESCC cells regardless of the degree of cell differentiation. However, inhibiting miR-25 had different effects on the invasion and metastasis abilities of ESCC cells of different degrees of differentiation. The invasion and metastasis of cells with a high degree of differentiation was significantly decreased, while the abilities of poorly differentiated cells did not change significantly. Thus, the impact of different degrees of differentiation should be taken into consideration in cancer studies. These results also suggest that clinical treatment should be individualized according to the degree of differentiation; when ESCC is well differentiated, inhibition of miR-25 may be an effective treatment, while if it is not this treatment would be ineffective.
Multiple steps in tumorigenesis, including migration, cell adhesion and immunological escape, are involved in invasion and metastasis, which serve a significant role in the development of ESCC. Complicated processes such as these cannot be regulated by a single molecule and require numerous molecules to interact with one another. Research over the last decades has identified a number of oncogenic and tumor suppressor proteins that are associated with the tumorigenesis of ESCC (13,17,19,20), however, molecular indicators of the origin of cellular deregulation in ESCC have not yet been identified. In order to improve the survival rates of patients with ESCC, it is necessary to understand the molecular etiology of the disease, and identify novel biomarkers for the early detection of pre-malignant lesions and localized carcinoma (17).
miR clusters are closely related structural genes with similar functions. Investigating how miRNA families are expressed in clusters and control cell signaling pathways is likely to increase understanding of cancer progression. Several studies have identified that the polycistronic miR-106b-25 cluster is tumorigenic (21,22). Li et al (21) reported that the miR-106b-25 cluster may have an anti-apoptotic role and could promote cell cycling in vitro. In the present study, bioinformatical analysis of the miR-106b-25 cluster was performed, including target prediction, followed by enrichment analysis these targets and PPI network construction. A total of 93 target genes were predicted for the three members of the cluster, including PTEN, TP53, MDM2, E2F1, PRMT5, MCM2, RB1, CDKN1A, SHAD7 and EZH2. GO term enrichment analysis revealed that the target genes of the cluster were significantly enriched in cell metabolism and the cell cycle. A previous study revealed that inhibiting the expression of the three members of the miR-106b-25 cluster slowed the development of liver cancer, and although the effects of the three members differed from each other, the trend was the same, which suggests that members of this cluster serve a role in tumor development (21). KEGG pathway enrichment analysis in the present study revealed that the gene targets of miR-106b-25 cluster members were concentrated in signaling pathways associated with glioma, melanoma and bladder cancer.
In the present study, a PPI network of the predicted targets of the miR-106b-25 cluster was constructed. PTEN, MDM2 and p53 were located centrally in this network. Research in gastric cancer cells by Petrocca et al (23) demonstrated that the miR-106b-25 cluster was involved in E2F1 post-transcriptional regulation and may serve a key role in the development of TGF-β resistance in gastric cancer. E2F1 is an important transcription factor that serves a role cancer progression (24,25) MDM2, a negative regulator of p53, is an important regulator of the cell cycle and apoptosis. MDM2 serves a pivotal role in p53 stabilization via phosphorylation at serine residues, and overexpression of MDM2 has frequently been observed in solid tumors and is associated with tumor progression (17). TP53 is at the core of the complicated PPI network, since deregulated p53 expression is closely associated with numerous types of cancer (17). p53 acts as a tumor by triggering cell cycle arrest and apoptosis, as well as maintaining genome stability (26). Xu et al (27) revealed that miR-25 promoted cell migration and invasion in ESCC. In prostate cancer and ovarian cancer, miR-93 can bind to and decrease PTEN expression, thereby promoting cancer cell proliferation and invasion (28,29). These results indicate that certain genes in the PPI network serve important roles in tumorigenesis. However, many other genes in the network have not been fully studied yet, including PRMT5, MDM2, RB1, CDKN1A, SMAD7 and EZH2. These genes are located centrally in the PPI network and have numerous links to other genes in the network, so further experiments are required to verify their functions.
The results of the present study and previous research suggest that interactions between the three members of miR-106b-25 cluster regulate multiple downstream target proteins to promote or inhibit carcinogenesis through cell cycle regulation mediated via several tumor-associated signaling pathways. Though several of the target genes of the miR-106-25 cluster have been studied, this research is still limited and knowledge about the interaction between genes is still lacking; thus, further research into the role of the miR-106b-25 cluster is required. This research should include further bioinformatical analysis, which greatly improves the efficiency of miRNA research, as it can provide important information about genes and also provide targets for further experiments.
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2008;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Ning Z, Zhu H, Li F, Liu Q, Liu G, Tan T, Zhang B, Chen S, Li G, Huang D, et al. Tumor suppression by miR-31 in esophageal carcinoma is p21-dependent. Genes Cancer. 2014;5:436–444. doi: 10.18632/genesandcancer.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guohong Z, Min S, Duenmei W, Songnian H, Min L, Jinsong L, Hongbin L, Feng Z, Dongping T, Heling Y, et al. Genetic heterogeneity of oesophageal cancer in high-incidence areas of southern and northern China. PLoS One. 2010;5:e9668. doi: 10.1371/journal.pone.0009668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao Z, Liu J, Wang C, Wang Y, Jiang Y, Guo M. MicroRNA-25 regulates small cell lung cancer cell development and cell cycle through cyclin E2. Int J Clin Exp Pathol. 2014;7:7726–7734. [PMC free article] [PubMed] [Google Scholar]
- 5.Li HL, Xie SP, Yang YL, Cheng YX, Zhang Y, Wang J, Wang Y, Liu DL, Chen ZF, Zhou YN, Wu HY. Clinical significance of upregulation of mir-196a-5p in gastric cancer and enriched kegg pathway analysis of target genes. Asian Pac J Cancer Prev. 2015;16:1781–1787. doi: 10.7314/APJCP.2015.16.5.1781. [DOI] [PubMed] [Google Scholar]
- 6.Lee RC, Feinbaum RL, Ambros V. The C. Elegans Heterochronic Gene lin-4 Encodes Small RNAs with Antisense Complementarity to &II-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 7.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 8.Lee NK, Lee JH, Park CH, Yu D, Lee YC, Cheong JH, Noh SH, Lee SK. Long non-coding HOTAIR promotes carcinogenesis and invasion of gastric adenocarcinoma. Biochem Biophys Res Commun. 2014;451:171–178. doi: 10.1016/j.bbrc.2014.07.067. [DOI] [PubMed] [Google Scholar]
- 9.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 10.Lai EC, Tomancak P, Williams RW, Rubin GM. Computational identification of Drosophila microRNA genes. Genome Biology. 2003;4:R42. doi: 10.1186/gb-2003-4-7-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Axtell MJ, Westholm JO, Lai EC. Vive la différence: Biogenesis and evolution of microRNAs in plants and animals. Genome Biol. 2011;12:221. doi: 10.1186/gb-2011-12-4-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Altuvia Y, Landgraf P, Lithwick G, Elefant N, Pfeffer S, Aravin A, Brownstein MJ, Tuschl T, Margalit H. Clustering and conservation patterns of human microRNAs. Nucleic Acids Res. 2005;33:2697–2706. doi: 10.1093/nar/gki567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu R, Liao J, Yang M, Sheng J, Yang H, Wang Y, Pan E, Guo W, Pu Y, Kim SJ, Yin L. The Cluster of miR-143 and miR-145 affects the risk for esophageal squamous cell carcinoma through co-regulating fascin homolog. PLoS One. 2012;7:e33987. doi: 10.1371/journal.pone.0033987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khuu C, Utheim TP, Sehic A. The Three Paralogous MicroRNA Clusters in Development and Disease, miR-17-92, miR-106a-363, andmiR-106b-25. Scientifica (Cairo) 2016;2016:1379643. doi: 10.1155/2016/1379643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith AL, Iwanaga R, Drasin DJ, Micalizzi DS, Vartuli RL, Tan AC, Ford HL. The miR-106b-25 cluster targets Smad7, activates TGF-β signaling and induces EMT and tumor initiating cell characteristics downstream of Six1 in human breast cancer. Oncogene. 2012;31:5162–5171. doi: 10.1038/onc.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Feber A, Xi L, Luketich JD, Pennathur A, Landreneau RJ, Wu M, Swanson SJ, Godfrey TE, Litle VR. MicroRNA Expression Profiles of Esophageal Cancer. J Thorac Cardiovasc Surg. 2008;135:255–260. doi: 10.1016/j.jtcvs.2007.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Antonov AV, Dietmann S, Mewes HW. KEGG spider: interpretation of genomics data in the context of the global gene metabolic network. Genome Biol. 2008;9:R179. doi: 10.1186/gb-2008-9-12-r179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kimura S, Naganuma S, Susuki D, Hirono Y, Yamaguchi A, Fujieda S, Sano K, Itoh H. Expression of microRNA in squamous cell carcinoma of human head and neck and the esophagus: miR-205 and miR-21 are specific markers for HNSCC and ESCC. Oncol Rep. 2010;23:1625–1633. doi: 10.3892/or_00000804. [DOI] [PubMed] [Google Scholar]
- 20.Matsumura Y, Hiraoka K, Ishikawa K, Shoji Y, Noji T, Hontani K, Itoh T, Nakamura T, Tsuchikawa T, Shichinohe T, Hirano S. CD40 expression in human esophageal squamous cell carcinoma is associated with tumor progression and lymph node metastasis. Anticancer Res. 2016;36:4467–4475. doi: 10.21873/anticanres.10991. [DOI] [PubMed] [Google Scholar]
- 21.Li Y, Tan W, Neo TW, Aung MO, Wasser S, Lim SG, Tan TM. Role of the miR-106b-25 microRNA cluster in hepatocellular carcinoma. Cancer Sci. 2009;100:1234–1242. doi: 10.1111/j.1349-7006.2009.01164.x. [DOI] [PubMed] [Google Scholar]
- 22.Kan T, Sato F, Ito T, Matsumura N, David S, Cheng Y, Agarwal R, Paun BC, Jin Z, Olaru AV, et al. The miR-106b-25 polycistron, activated by genomic amplification, functions as an oncogene by suppressing p21 and Bim. Gastroenterology. 2009;136:1689–1700. doi: 10.1053/j.gastro.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de Martino I, Iliopoulos D, Pilozzi E, Liu CG, Negrini M, et al. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13:272–286. doi: 10.1016/j.ccr.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 24.Spender LC, Inman GJ. TGF-beta induces growth arrest in Burkitt lymphoma cells via transcriptional repression of E2F-1. J Biol Chem. 2009;284:1435–1442. doi: 10.1074/jbc.M808080200. [DOI] [PubMed] [Google Scholar]
- 25.Chen HZ, Tsai SY, Leone G. Emerging roles of E2Fs in cancer: An exit from cell cycle control. Nat Rev Cancer. 2009;9:785–797. doi: 10.1038/nrc2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saha MN, Qiu L, Chang H. Targeting p53 by small molecules in hematological malignancies. J Hematol Oncol. 2013;6:23. doi: 10.1186/1756-8722-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu X, Chen Z, Zhao X, Wang J, Ding D, Wang Z, Tan F, Tan X, Zhou F, Sun J, et al. MicroRNA-25 promotes cell migration and invasion in esophageal squamous cell carcinoma. Biochem Biophys Res Commun. 2012;421:640–645. doi: 10.1016/j.bbrc.2012.03.048. [DOI] [PubMed] [Google Scholar]
- 28.Fu X, Tian J, Zhang L, Chen Y, Hao Q. Involvement of microRNA-93, a new regulator of PTEN/Akt signaling pathway, in regulation of chemotherapeutic drug cisplatin chemosensitivity in ovarian cancer cells. FEBS Lett. 2012;586:1279–1286. doi: 10.1016/j.febslet.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 29.Poliseno L, Salmena L, Riccardi L, Fornari A, Song MS, Hobbs RM, Sportoletti P, Varmeh S, Egia A, Fedele G, et al. Identification of the miR-106b~25 microRNA cluster as a proto-oncogenic PTEN-targeting intron that cooperates with its host gene MCM7 in transformation. Sci Signal. 2010;3:ra29. doi: 10.1126/scisignal.2000594. [DOI] [PMC free article] [PubMed] [Google Scholar]