Abstract
During surveys of Phytophthora diversity in natural and semi-natural Fagaceae forests in Austria, Italy and Portugal, four new cryptic species were isolated from rhizosphere soil samples. Multigene phylogeny based on nuclear ITS, ß-tubulin and HSP90 and mitochondrial cox1 and NADH1 gene sequences demonstrated that two species, P. tyrrhenica and P. vulcanica spp. nov., belong to phylogenetic Clade 7a, while the other two species, P. castanetorum and P. tubulina spp. nov., clustered together with P. quercina forming a new clade, named here as Clade 12. All four new species are homothallic and have low optimum and maximum temperatures for growth and very slow growth rates at their respective optimum temperature. They differed from each other and from related species by a unique combination of morphological characters, cardinal temperatures, and growth rates. Pathogenicity of all Phytophthora species to the root system of their respective host species was demonstrated in soil infestation trials.
Keywords: Clade 7, cryptic species, evolution, homothallic, phylogeny, Phytophthora quercina, species radiation
INTRODUCTION
The family Fagaceae comprises about 1000 species belonging to eight to ten genera widely distributed across the Northern Hemisphere (Manos & Stanford 2001). In Europe, species from the genera Castanea, Fagus, and Quercus dominate a wide variety of habitats ranging from Mediterranean sclerophyllous evergreen communities to temperate, deciduous lowland and mountainous forests (http://www.euforgen.org). Besides species with wide geographical distributions and ecological amplitudes, such as sweet chestnut (Castanea sativa) and European beech (Fagus sylvatica), some evergreen oaks (e.g. Quercus ilex and Q. suber) are geographically restricted and adapted to particular environmental conditions such as prolonged summer drought and fire (Schirone et al. 2016). Although species in these genera constitute primarily forest resources important for their wide range of uses (biomass, fibre, wood products, cork and food), they are also keystone species in forest ecosystems and considered main drivers of terrestrial biodiversity (Kremer et al. 2012). In some countries, they are major patrimonial and cultural resources (Logan 2005).
Since the early 1990s, numerous surveys in declining and healthy European Fagaceae forests have revealed a wealth of resident Phytophthora species (Jung 2009, Perez-Sierra et al. 2013, Scanu et al. 2013). Several Phytophthora species, including P. cactorum, P. xcambivora (previously known as P. cambivora), P. cinnamomi, P. plurivora, and P. quercina were strongly associated with the decline and dieback of forests, while other species, such as P. cryptogea, P. europaea, P. gallica, P. gonapodyides, P. megasperma, P. pseudosyringae, P. psychrophila, P. syringae, P. uliginosa, P. sp. forestsoil, and P. sp. riversoil, were more cryptic, and their role in forest ecosystems is still not fully understood.
Between 2010 and 2016, surveys of Phytophthora diversity were independently performed in forests in Austria, Italy, and Portugal during which four new cryptic Phytophthora species were recovered from the rhizosphere of Fagaceae species. All four species showed scattered distribution, very slow growth in culture, and were difficult to isolate using traditional soil baiting methods. Preliminary analysis of sequence data from the ITS region of the nrDNA and part of the mitochondrial cox1 gene showed that two taxa were closely related to P. uliginosa from subclade a of phylogenetic Clade 7 (in the following Clade 7a) whereas the other two taxa clustered with P. quercina and the informally designated taxon Phytophthora sp. ohioensis.
In this study, morphological and physiological characteristics are used in combination with DNA sequence data from the ITS, part of the nuclear heat shock protein 90 (HSP90) and β-tubulin (Btub), and the mitochondrial cox1 and NADH1 genes to characterize the four putative new Phytophthora taxa isolated from Fagaceae forests in Austria, Italy, and Portugal, and compare them to their closest relatives. The results of this study are presented and the new taxa described as P. castanetorum, P. tyrrhenica, P. tubulina, and P. vulcanica spp. nov. Pathogenicity of all new Phytophthora species against their respective hosts was also tested in soil infestation trials to confirm Koch’s postulates.
MATERIAL AND METHODS
Phytophthora isolation and culture maintenance
Isolates were obtained from rhizosphere soil of mature trees (Supplementary Table 1) using the sampling and isolation methods described by Jung et al. (1996), Jung (2009), and Scanu et al. (2015). Additional isolates used in the phylogenetic, morphological, and physiological studies and in the pathogenicity tests were sourced from the culture collections of the authors (Supplementary Table 1). In addition, isolates of P. quercina were obtained between 2010 and 2014 from various Quercus species in Portugal and Poland for comparative studies (Supplementary Table 1). For all isolates, single hyphal tip cultures were produced under the stereomicroscope from the margins of fresh cultures on V8-juice agar (V8A; 16 g agar, 3 g CaCO3, 100 mL Campbell’s V8 juice, 900 mL distilled water). Stock cultures were maintained on carrot agar (CA; 16 g agar, 3 g CaCO3, 200 g carrot, 1000 mL distilled water; Scanu et al. 2014a) at 10 °C in the dark. All isolates of the four new species are preserved in the culture collections maintained at Mendel University (BD numbers, referring to the BioDiversA project RESIPATH), the University of Sassari (P and PH numbers, both referring to Phytophthora) and the University of Catania (Roman numbers). Dried culture holotypes were lodged with the CBS Herbarium, and ex-type and isotype cultures were deposited at the Westerdijk Fungal Biodiversity Institute (CBS; Utrecht, The Netherlands) (Supplementary Table 1).
DNA isolation, amplification and sequencing
Extraction of mycelial DNA was performed according to Jung et al. (2017b) using the DNeasy Plant Mini kit (QIAGEN, Hilden) or the E.Z.N.A.® Fungal DNA Mini Kit (OMEGA Biotek, Norcross, GA) following the manufacturer’s instructions and checked for quality and quantity by spectrophotometry. DNA was stored at –20 °C until required. For 29 isolates of the four new Phytophthora species and five isolates of P. quercina, three nuclear and two mitochondrial loci were amplified and sequenced (Supplementary Table 1). The ITS1-5.8S-ITS2 region of the nrDNA was amplified using the primer-pair ITS1/ITS4 (White et al. 1990) and the PCR reaction mixture and cycling conditions described by Nagy et al. (2003) with an annealing temperature of 57 °C for 30 s. Partial heat shock protein 90 (HSP90) gene was amplified with the primer-pair HSP90F1int/HSP90R1 as described previously (Blair et al. 2008). Segments of the β-tubulin (Btub) and the mitochondrial genes cytochrome c oxidase subunit 1 (cox1) and NADH dehydrogenase subunit 1 (NADH1) were amplified using primer-pairs TUBUF2/TUBUR1, COXF4N/COXR4N and NADHF1/NADHR1, respectively, and the PCR reaction mixture and cycling conditions as described by Kroon et al. (2004). Products of Thermo Fisher Scientific (Waltham, MA) and Bio-Rad C1000™ or Applied Biosystems® 2720 Thermal Cyclers were used for the PCR reactions. Amplicons were purified and sequenced in both directions using the primers of the PCR reactions by Macrogen Inc. (Amsterdam) and BMR Genomics DNA sequencing service (www.bmrgenomics.it).
Electrophoregrams were quality checked and forward and reverse reads were compiled using Pregap4 version 1.5 and Gap v.4.10 of the Staden software package (Staden et al. 2000). Heterozygous sites were labelled according to the IUPAC coding system. All sequences derived in this study were deposited in GenBank and accession numbers are given in Supplementary Table 1.
Phylogenetic analysis
DNA sequences generated were combined with sequences of Phytophthora species obtained from GenBank. Sequences of each locus were aligned using the online MAFFT version 7 (http://mafft.cbrc.jp/alignment/server/) (Katoh & Standley 2013) by the E–INS–I strategy (ITS) or the G-INS-i option (all other loci). The phylogenetic analyses of P. tyrrhenica and P. vulcanica from Clade 7a, and of P. castanetorum and P. tubulina from the P. quercina clade were performed separately.
For the phylogenetic analyses of P. tyrrhenica and P. vulcanica all 16 previously known taxa from Clade 7a were included and P. cinnamomi (CBS 144.22) and P. niederhauserii (CBS 124086), both from Clade 7b, were used as outgroups (Abad et al. 2014, Jung et al. 2017b). The datasets of the five loci were first analysed separately and then combined. The 5-loci dataset comprised 4257 characters (ITS=842, Btub=911, HSP90=840, cox1=867, NADH1=797) and included 43 Phytophthora isolates.
To resolve the phylogenetic positions of P. castanetorum and P. tubulina within the P. quercina clade, and of the latter within the genus Phytophthora, 24 isolates from seven putative taxa of the P. quercina clade, including P. castanetorum, P. tubulina, P. quercina, P. versiformis (Paap et al. 2017), and the undescribed taxa P. sp. ohioensis, P. sp. quercina-like (only ITS sequence available), and P. sp. versiformis-like, and representative species from all phylogenetic Phytophthora clades were included in the analyses. The designated ex-type isolate (CBS 142348) of Nothophytophthora amphigynosa, the type species of a new sister genus of Phytophthora (Jung et al. 2017c) was used as outgroup taxon in all analyses. The datasets of the five loci were first analysed separately and then combined. The cox1 sequences of P. sp. ohioensis, P. versiformis and P. sp. versiformis-like from GenBank were generated using primer pair FM77/FM84 (Martin & Tooley 2003) and, hence, had insufficient overlap with those of P. castanetorum, P. quercina and P. tubulina generated in this study with primer pair COXF4N/COXR4N. Therefore, cox1 was excluded from the two multigene analyses of the P. quercina clade. The combined four locus dataset comprised 3539 characters (ITS=992, Btub=915, HSP90=843, NADH1=789). For 11 of the 24 Phytophthora species from other clades no isolates with sequences for all four loci were available at GenBank or at Phytophthora database, hence sequences from two different isolates per species were combined (Supplementary Table 1). Since for P. lilii no NADH1 sequence was available a separate analysis of a combined nuclear ITS-Btub-HSP90 dataset was performed in order to clarify whether P. lilii constitutes a distinct phylogenetic clade as suggested by Rahman et al. (2015) and whether the inclusion of P. lilii has any effect on the grouping of the P. quercina clade. This three locus dataset comprised 2768 characters (ITS=1010, Btub=915, HSP90=843) and included 49 Phytophthora taxa.
For all individual gene and all multigene alignments, both Maximum Lkelihood (ML) phylogenetic and Bayesian Inference (BI) analyses were performed. ML analyses of the datasets were carried out with RAxML (Stamatakis 2014) in the raxmlGUI version 1.5b1 (Silvestro & Michalak 2012) implemented with the GTRGAMMA model. There were 10 runs of the ML and bootstrap (“thorough boostrap”) analyses with 1000 replicates used to test the support of the branches. Phylogenetic BI analyses were performed with MrBayes 3.1.2 (Huelsenbeck & Ronquist 2001, Ronquist & Huelsenbeck 2003) using the GTR+G model. For multigene BI analyses the individual gene alignments were divided into five (Clade 7a) and four or three (P. quercina clade) partitions, respectively. Four Markov chains were run for 10 million generations with three heated chains (temperature = 0.2) and one cold chain. Trees were sampled every 1000 steps after removing the first 6000 generations as burn in. For the multigene BI analysis of Clade 7a 20 million, generations with a burn in of the first 8000 generations were used. Phylogenetic trees were visualized in MEGA6 (Tamura et al. 2013) and edited in figure editor programs. All alignments and trees deriving from BI and ML analyses were deposited in TreeBASE (20982; http://purl.org/phylo/treebase/phylows/study/TB2:S20982).
Morphology of asexual and sexual structures
Morphological features and morphometric data of sporangia, oogonia, oospores, antheridia, chlamydospores, hyphal swellings, and aggregations of the four new species were compared with each other and with those of related species from published studies.
Formation of sporangia was induced by submerging two 12–15 mm square discs cut from the growing edge of a 3-7-d-old V8A colony in a 90 mm diam Petri dish in non-sterile soil extract (50 g of filtered oak forest soil in 1000 mL of distilled water, filtered after 24 h; Jung et al. 1996). The Petri dishes were incubated at 20 °C in natural daylight and the soil extract changed after ca 6 h (Jung et al. 2017b). Shape, type of apex, caducity and special features of sporangia and the formation of hyphal swellings and aggregations were recorded after 24–48 h. For each isolate 50 sporangia were measured at x400 using a compound microscope (Zeiss Imager.Z2), a digital camera (Zeiss Axiocam ICc3) and a biometric software (Zeiss AxioVision).
The formation of gametangia (oogonia and antheridia) and their characteristic features were examined on V8A after 21-30 d growth at 20 °C in the dark. For each isolate each 50 oogonia, oospores and antheridia chosen at random were measured under a compound microscope at x400 as described before. The oospore wall index was calculated according to Dick (1990).
For comparisons, morphometric, morphological and physiological data of P. quercina and Phytophthora species from Clade 7a, respectively, were taken from published descriptions (Jung et al. 1999, 2002, 2017b).
Colony morphology, growth rates and cardinal temperatures
Colony growth patterns of Phytophthora species were described from 15-d-old (Clade 7a species) and 10-d-old (P. castanetorum, P. quercina, and P. tubulina) cultures grown at 20 °C in the dark on CA, V8A, malt-extract agar (MEA; Oxoid, Basingstoke, UK) and potato-dextrose agar (PDA; Oxoid) according to Scanu et al. (2014a), Jung et al. (1999, 2017b), and Erwin & Ribeiro (1996).
For temperature-growth relationships, representative isolates of the four new Phytophthora species and P. quercina (Supplementary Table 1) were subcultured onto 90 mm V8A plates and incubated for 24 h at 20 °C to stimulate onset of growth (Jung et al. 1999). Then, three replicate plates per isolate were transferred to 5, 10, 15, 20, 25, 30, and 35 °C. Radial growth was recorded after 6 d, along two lines intersecting the centre of the inoculum at right angles and the mean growth rates (mm/d) were calculated. Plates showing no growth at 30 or 35 °C were returned to 20 °C to determine isolate viability.
Soil infestation trials
In order to fulfil Koch’s postulates, pathogenicity of all four new Phytophthora species was tested using the soil infestation method described by Jung et al. (1996). Phytophthora tubulina and P. vulcanica were tested in trial 1 with 3-year-old saplings of Fagus sylvatica, while P. castanetorum and P. tyrrhenica were tested in trial 2 with 1-year-old seedlings of Castanea sativa, and Quercus ilex and Q. suber, respectively. Saplings and seedlings were sourced from local nurseries and confirmed to be non-infested by Phytophthora using the soil baiting method described before. Two isolates for each new Phytophthora species were used, and isolates of P. xcambivora and P. cinnamomi (both primary pathogens of Fagaceae) were included as positive controls (Supplementary Table 1). Inocula consisted of 4-wk-old cultures of the respective Phytophthora isolate grown at 20 °C in 500 mL Erlenmeyer flasks on an autoclaved mixture of 250 cm3 of vermiculite and 20 cm3 of millet seeds thoroughly moistened with 175 mL of V8-juice broth (200 mL/L juice, 800 mL/L distilled water amended with 3 g/L CaCO3) (Jung et al. 1996). Before use, the colonized medium was rinsed with distilled water to remove excess nutrients. In trial 1, 12 plants per Phytophthora isolate were planted as pseudo-replicates in 50 x 35 x 27 cm boxes in an autoclaved mixture of peat, vermiculite, and sand (1:1:1 v:v:v). Tubes initially inserted as placeholders in the soil between the individual plants were removed and the holes were filled with the inoculum (ca 40 cm3 of inoculum per plant) (Jung et al. 2017b). Controls received only rinsed non-infested vermiculite/millet seed/V8-juice mixture at the same rate. In trial 2, ten plants of each Phytophthora isolate/ host plant combination were infested in individual pots with the same rate of inoculum as described before. In both trials, plants were incubated for five months in a walk-in growth chamber at 18–20 °C, 65 % relative humidity, a natural daylight regime, and flooded every 3 wk for 72 h. At the end of the trial, roots were thoroughly washed free from soil. Then, specific symptoms such as root and collar rot lesions and chlorosis and wilting of foliage were recorded.
Two different root damage assessments were used. In trial 1 with Fagus sylvatica saplings, the proportion of root damage was assessed visually after spreading the roots uniformly on trays etched with 2 x 2 cm squares according to a scale of five root damage classes: 4 = healthy root system with dense fine root system and well developed tap roots; 3 = < 25 % fine root losses and well developed tap roots; 2 = 26–50 % fine root losses, beginning taproot dieback and small necrotic lesions on woody roots and/or the collar; and 1 = 51–75 % fine root losses, advanced taproot dieback and large necrotic lesions on tap roots and/or the collar; 0 = 76–100 % fine root losses, extensive taproot dieback and girdling necrotic lesions on tap roots and/or the collar. Then roots were dried for 72h at 65 °C and the dry weights of small woody roots (diam 2–10 mm) and fine roots (diam <2 mm) were recorded for each plant. Data were analysed using one-way ANOVA followed by Dunnett’s multiple comparisons test using the programme package Prism 6 (Graphpad, San Diego, CA). In trial 2, Castanea sativa, Quercus ilex, and Q. suber seedlings were removed from the pots and the root system gently washed under tap water (Scanu et al. 2014a). Single roots were then cut off at the collar, scanned and total root length for each plant was calculated using the APS Assess 2.0 software (The American Phytopathological Society, St Paul, MN).
In all cases, re-isolations of Phytophthora from necrotic tissues were made using selective PARPNH agar (Jung 2009). During each flooding cycle, soils were baited using young leaves of Ceratonia siliqua, F. sylvatica and Q. suber as baits in order to test whether the respective Phytophthora species was still active. After each flooding cycle, the water was collected and autoclaved. At the end of the trial boxes were sterilised with bleach and infested substrates and the plants were autoclaved.
RESULTS
Phylogenetic analysis
Sequence datasets of Clade 7a species and species related to Phytophthora quercina were analysed separately. All phylogenetic analyses of individual nuclear (ITS, Btub, and HSP90) and mitochondrial (cox1 and NADH1) DNA sequences resulted in similar overall tree topologies (data not shown), thus indicating that the individual loci could be combined.
To resolve the phylogenetic positions of P. castanetorum and P. tubulina within the P. quercina clade, and of the latter within the genus Phytophthora, a 4-loci dataset (ITS–Btub–HSP90–NADH1) and a nuclear 3-loci dataset (ITS–Btub–HSP90) were analysed separately. Compared to the ML analyses, the BI analyses provided higher support for both terminal clades and the deeper branches. Since the topologies of the BI and ML trees were congruent, the Bayesian trees are presented here (Figs 1–2) with both BI Posterior Probability values and ML bootstrap values shown at the nodes. The resulting phylogenies of the genus Phytophthora were rooted to Nothophytophthora amphigynosa and were in accordance with several published studies (Blair et al. 2008, Martin et al. 2014, Rahman et al. 2015) with the exception that the inclusion of the two new species P. castanetorum and P. tubulina and the three taxa P. sp. ohioensis, P. versiformis, and P. sp. versiformis-like, allowed to resolve the phylogenetic position of the P. quercina clade within Phytophthora. Together with P. quercina, P. sp. ohioensis, P. versiformis, and P. sp. versiformis-like, P. castanetorum and P. tubulina formed a fully supported monophyletic group (BI posterior probability = 1.00, ML bootstrap = 100 %; Fig. 1–2).
Fig. 1.
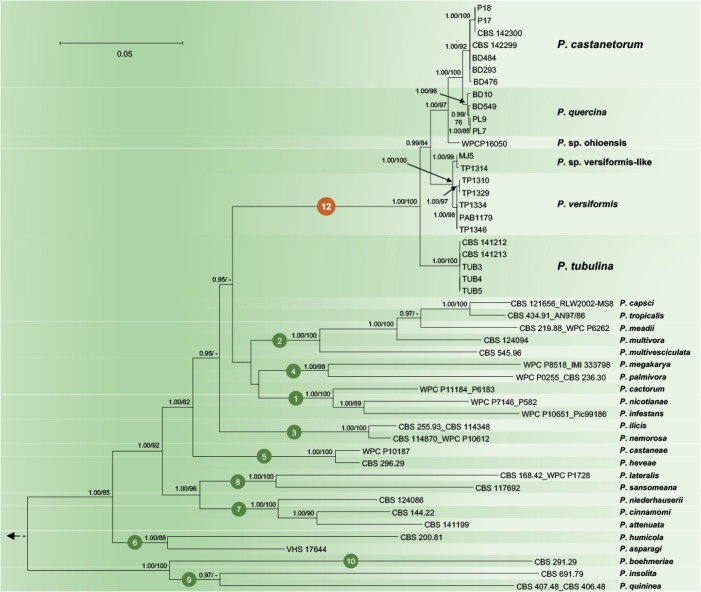
Fifty percent majority rule consensus phylogram derived from Bayesian inference analysis of four–locus (ITS, Btub, HSP90, NADH1) dataset of the Phytophthora Clade 12 and representative species from phylogenetic Clades 1–10. Bayesian posterior probabilities and Maximum Likelihood bootstrap values (in %) are indicated, but not shown below 0.90 and 70 %, respectively. Nothophytophthora amphigynosa was used as outgroup taxon (not shown). Bar = 0.05 expected changes per site per branch.
Fig. 2.
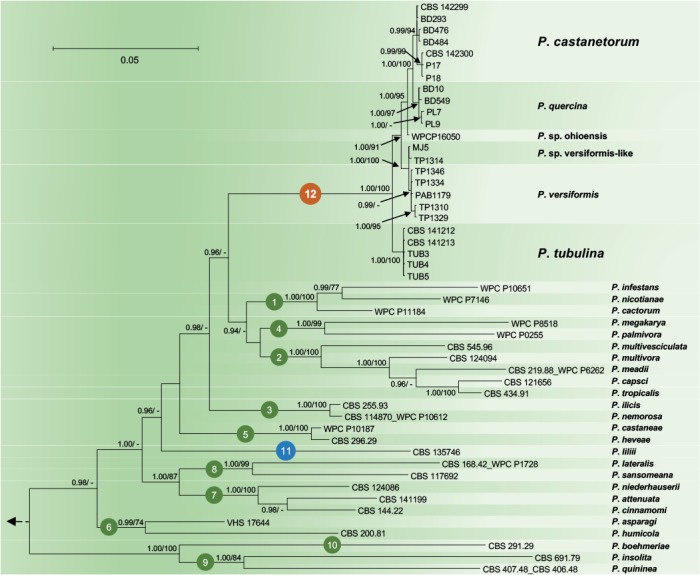
Fifty percent majority rule consensus phylogram derived from Bayesian inference analysis of nuclear three–locus (ITS, Btub, HSP90) dataset of Phytophthora Clade 12 and representative species from phylogenetic Clades 1–11. Bayesian posterior probabilities and Maximum Likelihood bootstrap values (in %) are indicated, but not shown below 0.90 and 70 %, respectively. Nothophytophthora amphigynosa was used as outgroup taxon (not shown). Bar = 0.05 expected changes per site per branch.
Since in the analysis of the nuclear 3-loci dataset P. lilii formed a distinct clade (Fig. 2), confirming Rahman et al. (2015), the names Clade 11 and Clade 12 are proposed here for P. lilii and the new P. quercina clade, respectively. In the analyses of both datasets, Clade 12 formed a fully supported group together with Clades 1, 2 and 4, while Clade 3 was basal to this group (Figs 1–2). Within Clade 12, isolates of P. castanetorum grouped in a well-supported cluster together with P. quercina (posterior probability = 1.00, ML bootstrap = 100 %). Phytophthora sp. ohioensis from North America appeared in a basal position of this cluster, while P. versiformis and P. sp. versiformis-like formed an Australian cluster which grouped in a sister position to the P. quercina–P. castanetorum–P. sp. ohioensis cluster (Figs 1–2). Phytophthora tubulina appeared in a basal position of Clade 12 (Figs 1–2). In the separate BI and ML analyses of an ITS sequence alignment, P. sp. quercina-like, for which only an ITS sequence was available, grouped within Clade 12 (data not shown, but available from TreeBASE: S20982). Phytophthora castanetorum differed from the closest taxon P. quercina and from P. sp. ohioensis in three nuclear (ITS–Btub–HSP90) and the mitochondrial NADH1 gene regions by 9–20 and 23–28 characters, respectively, and from the more distantly related P. versiformis, P. sp. versiformis-like, and P. tubulina by 45–52, 45–51 and 97–102 characters, respectively (Table 1). In addition, in cox1 P. castanetorum showed differences to P. quercina at 5–14 positions (Table 1). Isolates of P. castanetorum showed intraspecific variability, with the Sardinian isolates forming a distinct cluster (Figs 1–2). DNA sequences of Sardinian isolates differed from those of P. castanetorum from Portugal in Btub and NADH1 by 3 bp and 2 bp, respectively (Table 1). In contrast, P. tubulina isolates were identical across all five loci examined. Phytophthora versiformis and P. sp. versiformis-like showed differences to each other at 6–9 positions, and to P. castanetorum and P. quercina at 45–52 and 46–53 positions, respectively (Table 1). The basal species P. tubulina differed from P. castanetorum, P. quercina, P. sp. ohioensis, P. versiformis and P. sp. versiformis-like by 97–102, 92–106, 57, 51–53 and 52–53 characters, respectively (Table 1). In addition, P. tubulina was separated from P. castanetorum and P. quercina in cox1 by 37 and 33–42 characters, respectively (Table 1).
Table 1.
Pairwise numbers of different positions along alignments of ITS (842 bp), Btub (911 bp), HSP90 (840 bp), cox1 (867 bp) and NADH1 (797 bp) among the taxa from Phytophthora Clade 12.
| Phytophthora species | P. castanetorum | P. quercina | P. sp. ohioensis | P. sp. versiformis | P. sp. versiformis-like | P. tubulina | Total no. of unique polymorphisms | ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ITS | Btub | HHSP90 | Cox1 | NADH1 | ITS | Btub | HHSP90 | Cox1 | NADH1 | ITS | Btub | HHSP90 | Cox1 | NADH1 | ITS | Btub | HHSP90 | Cox1 | NADH1 | ITS | Btub | HHSP90 | Cox1 | NADH1 | ITS | Btub | HHSP90 | Cox1 | NADH1 | ITS | Btub | HHSP90 | Cox1 | NADH1 | |
| P. castanetorum | 0 | 0-3 | 0 | 0 | 0-2 | 3 | 0-5 | 2-3 | 5-14 | 4-9 | 1 | 2-5 | 2 | n.a. | 18-20 | 2-4 | 7-10 | 6 | n.a. | 30-32 | 2-3 | 6-9 | 6 | n.a. | 31-33 | 9 | 4-7 | 8 | 37 | 39-41 | 1 | 0-3 | 1 | 5 | 3-5 |
| P. quercina | 3 | 0-5 | 2-3 | 5-14 | 4-9 | 0 | 0-2 | 0-1 | 0-9 | 0-3 | 2 | 2-4 | 2-3 | n.a. | 15-18 | 3-5 | 7-9 | 6-7 | n.a. | 30-32 | 3-4 | 6-8 | 6-7 | n.a. | 32-34 | 10 | 4-6 | 8-9 | 33-42 | 37-39 | 2 | 0-2 | 1-2 | 0-9 | 1-4 |
| P. sp. ohioensis | 1 | 2-5 | 2 | n.a. | 18-20 | 2 | 2-4 | 2-3 | n.a. | 15-18 | 0 | 0 | 0 | n.a. | 0 | 1-3 | 5 | 4 | n.a. | 29 | 1-2 | 4 | 4 | n.a. | 29 | 8 | 3 | 6 | n.a. | 40 | 0 | 0 | 0 | n.a. | 4 |
| P. sp. versiformis | 2-4 | 7-10 | 6 | n.a. | 30-32 | 3-5 | 7-9 | 6-7 | n.a. | 30-32 | 1-3 | 5 | 4 | n.a. | 28 | 0-2 | 0 | 0 | n.a. | 0 | 0-3 | 1 | 0 | n.a. | 5 | 9-11 | 4 | 4 | n.a. | 34 | 0-2 | 1 | 0 | n.a. | 2 |
| P. sp. versiformis-like | 2-3 | 6-9 | 6 | n.a. | 31-33 | 3-4 | 6-8 | 6-7 | n.a. | 32-34 | 1-2 | 4 | 4 | n.a. | 29 | 0-3 | 1 | 0 | n.a. | 5 | 0-1 | 0 | 0 | n.a. | 0 | 9-10 | 5 | 4 | n.a. | 34 | 0-1 | 0 | 0 | n.a. | 3 |
| P. tubulina | 9 | 4-7 | 8 | 37 | 39-41 | 10 | 4-6 | 8-9 | 33-42 | 37-39 | 8 | 3 | 6 | n.a. | 40 | 9-11 | 4 | 4 | n.a. | 34 | 9-10 | 5 | 4 | n.a. | 34 | 0 | 0 | 0 | 0 | 0 | 8 | 1 | 3 | 33 | 23 |
n.a. = not available due to insufficient overlap between cox1 sequences of P. sp. ohioensis, P. versiformis and P. sp. versiformis-like from GenBank and of P. castanetorum, P. quercina and P. tubulina generated in this study.
Nucleotides missing from the terminal part(s) of partial sequences and undetermined bases (N) were not considered as polymorphisms.
Bayesian and ML analyses of the combined five locus dataset for Clade 7a generated trees with essentially the same topology. The Bayesian analysis provided more support for deeper branches, while support for terminal clades and their clustering was equivalent in both analyses. The Bayesian tree is shown in Fig. 3 with both BI posterior probability and ML bootstrap values provided at the nodes (Fig. 3). The resolution was good at terminal clades and allowed for the differentiation of 17 distinct lineages within Clade 7a corresponding to 14 described species, P. attenuata, P. europaea, P. flexuosa, P. formosa, P. intricata, P. fragariae, P. rubi, P. uliginosa, P. uniformis, P. xcambivora, P. xheterohybrida, P. xincrassata, P. xalni / P. xmultiformis, the two informally designated taxa P. sp. xcambivora-like and P. sp. xmultiformis-like, and the two new species P. tyrrhenica and P. vulcanica described in this study (Fig. 3). Isolates of P. tyrrhenica grouped together with P. uliginosa in a well-supported clade (posterior probability = 1.00, ML bootstrap = 100 %), both species being separated by 2, 4 and 1–2 characters in ITS, Btub and HSP90, respectively, and by 22–24 and 8–10 characters in cox1 and NADH1, respectively. The relative phylogenetic position of P. tyrrhenica and P. uliginosa was also well-supported (Fig. 3). The P. tyrrhenica–P. uliginosa clade grouped in a sister position to the remaining in-group taxa representing 15 taxa from Clade 7a (Fig. 3). Small intraspecific variation of up to seven characters across the five gene regions was found amongst P. tyrrhenica isolates, which was related to different oak species and geographic origin. Isolate PH103 (from Quercus suber in Sardinia) represented a distinct basal lineage (Fig. 3). In contrast, P. vulcanica was monomorphic across all five loci examined, suggesting that the tested isolates could belong to the same clone of the species. Across the five DNA regions, P. vulcanica showed differences to its closest relatives P. uliginosa and P. tyrrhenica at 109 and 109–114 positions, respectively, and resided in a well-supported basal position of the whole Clade 7a (Fig. 3).
Fig. 3.
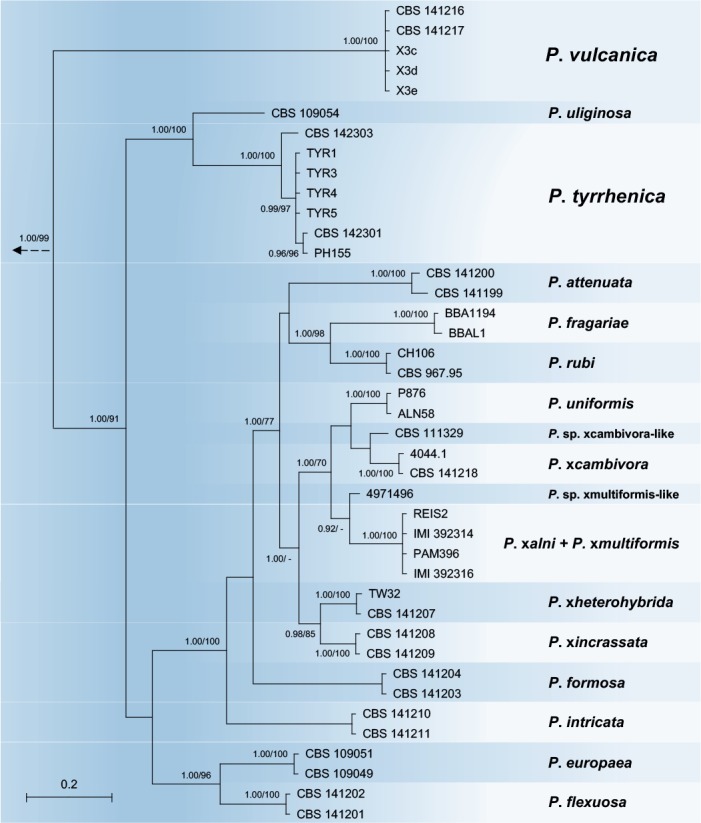
Fifty percent majority rule consensus phylogram derived from Bayesian phylogenetic analysis of five–locus (ITS, Btub, HSP90, cox1, NADH1) dataset of Clade7a. Bayesian posterior probabilities and Maximum Likelihood bootstrap values (in %) are indicated, but not shown below 0.90 and 70 %, respectively. Phytophthora cinnamomi and P. niederhauserii from Clade 7b were used as outgroup taxa (not shown). Bar = 0.05 expected changes per site per branch.
Hosts and geographic distribution
Phytophthora castanetorum was exclusively isolated from Castanea sativa forests in Portugal, at Monchique (N 37o 18.851’, W 8o 32.455’; 496 m above sea level, asl) and in the Parque Natural Serra da Estrela (N 40o 25.489, W 7o 22.523’; 901 m asl), and in Sardinia at Gennargentu montain (N 39o 56.946’, E 9o 11.387’; 858 m asl). It usually co-occurred with P. xcambivora and P. cinnamomi, associated with severe ink disease symptoms. Phytophthora tubulina was isolated alongside P. xcambivora and P. plurivora from mildly declining Fagus sylvatica trees in the Dunkelsteiner Forest in Austria (N 48o 14.561’, E 15o 28.276’; 497 m asl), while P. vulcanica was recovered from relatively healthy F. sylvatica trees at Timpa Rossa on Mount Etna in Sicily (N 37o 48.849’, E 15o 1.407’; 1862 m asl). Phytophthora tyrrhenica was associated with declining Quercus ilex and Q. suber trees in different forest stands on the Italian islands Sardinia (N 40o 52.492’, E 9o 2.720’; 183 m asl) and Sicily (N 37o 54.341’, E 14o 4.490’; 1110 m asl).
Soil infestation trials
Pathogenicity on Fagus sylvatica seedlings (trial 1)
At the end of the trial shoots and root systems of control plants of F. sylvatica were generally healthy and well developed with a fine root/main root weight (frw/mrw) ratio of 2.2 and a root damage class of 3.6 ± 0.67 (Fig. 4). Phytophthora xcambivora was the most aggressive species causing after 5 months 50 % mortality, a frw/mrw ratio of 0.78 ± 0.36 (64.5 % reduction compared to the control), 54.4 % reduction of fine root weight compared to control plants and a root damage class of 1.3 ± 0.14 (i.e. 67.5 % fine root losses and advanced dieback and necrotic lesions of tap roots and root collars) (Fig. 4). Phytophthora tubulina isolates (TUB1= CBS 141212 and TUB2= CBS 141213) caused 8.3 and 16.6 % mortality, a frw/mrw ratio of 1.22 ± 0.49 and 1.39 ± 0.1 (44.5 and 36.8 % reduction compared to the control), and a root damage class of 2.5 ± 1.0 and 2.7 ± 1.0, respectively (Fig. 4). Beech plants in soil infested by isolates X3d and X3e of P. vulcanica had a frw/mrw ratio of 1.55 ± 0.52 and 1.27 ± 0.41 (29.8 and 42.3 % reduction compared to the control), and a root damage class of 2.0 ± 0.95 and 2.4 ± 0.9, respectively (Fig. 4). Both isolates caused 8.3 % mortality and, chlorosis and wilting in 58.3 % (X3d) and 75 % (X3e) of the plants. Differences of root damage class and frw/mrw ratio to the control were statistically significant for P. xcambivora, both isolates of P. vulcanica and isolate TUB1 of P. tubulina (Fig. 4). For P. tubulina isolate TUB 2, the difference to the control was only significant for the frw/mrw ratio but not for the root damage class (Fig. 4).
Fig. 4.
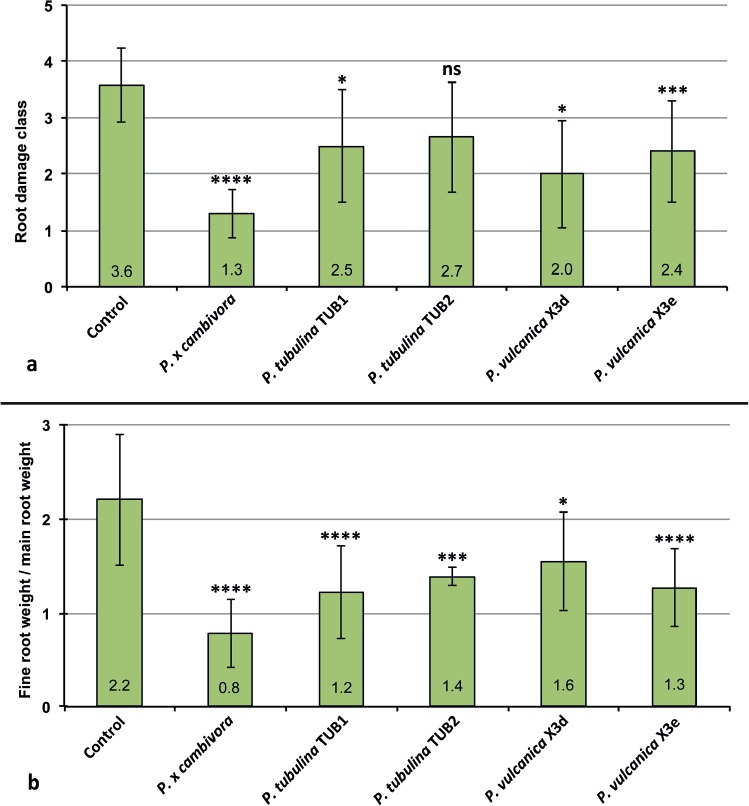
Mean root damage class and mean fine root weight / main root weight ratio of 3–year–old Fagus sylvatica plants after 5 months growth in uninfested soil (control) and in soil infested by Phytophthora xcambivora (CBS 141218), P. tubulina (CBS 141212, CBS 141213) and P. vulcanica (X3d, X3e), respectively. Bars show standard deviations; asterisks represent statistical significances (* = P < 0.05, ** = P < 0.01, *** = P < 0.001, **** = P < 0.0001), ns = not significant.
Pathogenicity on Castanea sativa seedlings (trial 2)
At the end of the experiment seedlings inoculated with P. xcambivora were mostly dead (mortality rate above 80 %) or showed severe wilting associated with collar and root necrosis. In contrast, all seedlings inoculated with P. castanetorum were alive and showed only weak symptoms of wilting and chlorosis of leaves. Control plants did not show any aboveground symptoms and exhibited overall faster growth. Phytophthora xcambivora was extremely aggressive to the root systems causing more than 60 % reduction of total root length (P < 0.0001). In contrast, seedlings inoculated with P. castanetorum showed only mild fine root infections but no symptoms of woody root and collar infections, and total root length did not differ statistically from the control plants (Fig. 5).
Fig. 5.
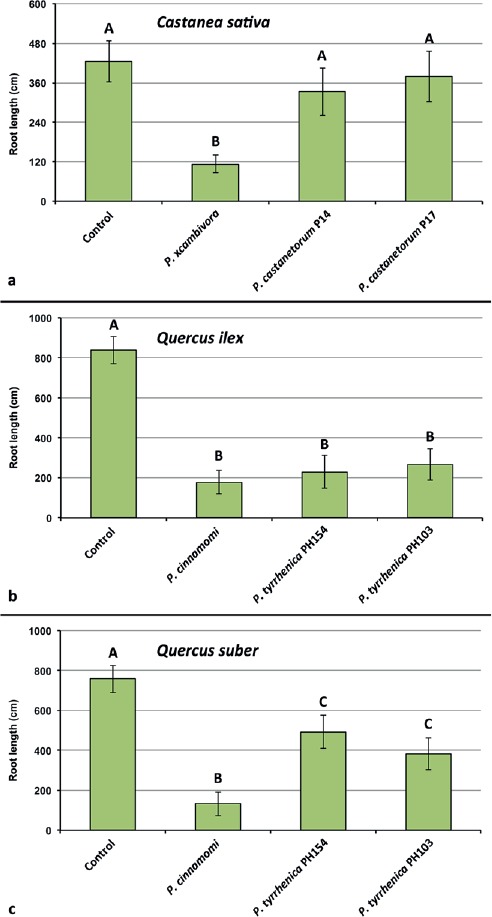
Mean root length of 1–year–old plants of Castanea sativa, Quercus ilex and Q. suber after 5 months growth in uninfested soil (control) and in soil infested by Phytophthora xcambivora (CBS 141218), P. castanetorum (CBS 142300, P17), P. cinnamomi (PH069) and P. tyrrhenica (CBS 142301, CBS 142303), respectively. Bars show standard deviations; asterisks represent statistical significances (* = P < 0.05, ** = P < 0.01, *** = P < 0.001, **** = P < 0.0001), ns = not significant.
Pathogenicity on Quercus ilex and Quercus suber seedlings (trial 2)
Oak seedlings inoculated with P. tyrrhenica started to show symptoms of leaf chlorosis and shoot dieback 45 d post-inoculation. Mortality of seedlings occurred between the third and fourth month after inoculation. Based on mortality rates after five months, P. cinnamomi was more aggressive than P. tyrrhenica on both Q. ilex and Q. suber, killing 80 % and 63.5 % of seedlings, respectively, whereas P. tyrrhenica caused an average mortality on Q. ilex and Q. suber of 48.5 % and 25 %, respectively. On Q. ilex, both Phytophthora species caused fine root losses and necrosis of mother roots, and a significant reduction in root length compared to control seedlings (P < 0.0001) (Fig. 5). Phytophthora cinnamomi caused significantly higher root reduction than P. tyrrhenica for which Dunnett’s test revealed no significant differences in total root length of inoculated seedlings. Also on Q. suber, both P. cinnamomi and P. tyrrhenica caused significant total root length reduction, but the reduction by P. cinnamomi was significantly higher from that caused by both isolates of P. tyrrhenica (P < 0.0001) (Fig. 5).
At the end of all pathogenicity trials, each Phytophthora species could be re-isolated from both infested soils using young leaves of C. siliqua, F. sylvatica and Q. suber as baits and necrotic fine roots or necrotic root lesions by direct plating onto PARPNH agar. No Phytophthora species could be isolated from soil and roots of control plants.
TAXONOMY
Morphological and physiological characters and measurements of the four new Phytophthora taxa and related species are given in the comprehensive Table 2.
Table 2.
Morphological characters and dimensions (μm), cardinal temperatures (°C) and temperature-growth relations (mm/d) on V8A of Phytophthora castanetorum, P. tyrrhenica, P. tubulina, P. vulcanica and their closest relatives in phylogenetic Clades 7a and 12, respectively. Most discriminating characters are highlighted in bold.
| P. castanetorum | P. tubulina | P. quercina | P. tyrrhenica | P. vulcanica | P. uliginosa | P. europaea | P. flexuosa | |
|---|---|---|---|---|---|---|---|---|
| No. of isolates | 9 | 5 | 10 b | 7 | 5 | 3a | 7a | 3a |
| Sporangia | ovoid, subglobose, obpyriform, globose, (ellipsoid, distorted) | ovoid, obpyriform, distorted, (globose, subglobose, ellipsoid, limoniform) | subglobose, ovoid, obpyriform, peanut, ampulliform, distorted | elongated ellipsoid, elongated ovoid, elongated limoniform | ovoid, ellipsoid, (obpyriform, pyriform) | ellipsoid, ovoid, peanut-shaped, (obpyriform, limoniform) | ovoid, obpyriform, ellipsoid, (subglobose), | ovoid, ellipsoid, (obpyriform, limoniform) |
| apex | 77.4 % papillate, 14.9 % semipapillate; 7.7 % nonpapillate, 4.9 % curved | 78 % mono-, bi- or tripapillate; 13.3 % semi-papillate; 8.7 % non-papillate, 4.9 % curved | all papillate, some bi- and tripapillate; often curved | nonpapillate | nonpapillate | nonpapillate | nonpapillate, often pointed | |
| lxb mean | 47.0±5.7 x 36.6±3.7 | 51.6±9.8 x 37.3±7.0 | 42.4±11.5 x 29.3±13.8 | 68.4±1.7 x 28.9±1.9 | 57.3±8.7 x 34.5±4.2 | 67.0±8.5 x 42.4±6.4 | 63.7±16.9 x 44.6±8.3 | 56.1±7.4 x 36.7±5.2 |
| range of isolate means | 44.2–48.1 x 35.4–38.3 | 43.3–57.8 x 31.2–45.0 | 36.0–49.2 x 23.3–33.7 | 66.1–70.0 x 27.5–31.9 | 53.6–61.0 x 31.8–36.4 | 65.7–70.3 x 30.9–44.4 | 50.0–78.9 x 36.7–51.2 | 53.3–58.6 x 33.9–39.4 |
| total range | 27.1–74.5 x 18.1–47.9 | 29.5–98.5 x 19.2–59.2 | 18.8–112.5 x 13.8–47.5 | 45.3–110.2 x 16.2–39.4 | 35.5–80.7 x 22.5–42.9 | 41.1–85.0 x 24.8–56.9 | 23.6–124.3 x 21.9–67.3 | 34.9–74.8 x 22.8–49.7 |
| l/b ratio | 1.29 ± 0.15 | 1.40 ± 0.18 | 1.45 ± 0.37 | 2.40 ± 0.18 | 1.67 ± 0.22 | 1.60 ± 0.19 | 1.42 ± 0.19 | 1.54 ± 0.18 |
| caducity | – | rare | – | – | – | – | – | – |
| lateral insertion | 37.7 % | 22.4 % | frequent | |||||
| exit pores | 7.0 ± 1.1 | 6.4 ± 1.2 | 6.5 ± 1.2 | 12.1 ± 2.4 | 17.0 ± 3.0 | 15.4 ± 3.4 | 19.3 ± 4.7 | 19.7 ± 3.4 |
| zoospore cysts | 9.2 ± 1.0 | 11.7 ± 1.8 | 9.5 (7.1–12.9) | 14.6 ± 2.3 | 10.0 ± 1.2 | 11.9 ± 1.8 | 15.3 ± 2.0 | 13.3 ± 1.3 |
| Breeding system | homothallic | homothallic | homothallic | homothallic | homothallic | homothallic | homothallic | homothallic |
| Oogonia | ||||||||
| mean diam | 32.9 ± 3.0 | 29.2 ± 4.4 | 29.4 ± 4.1 | 37.2 ± 3.3 | 35.8 ± 3.6 | 46.2 ± 7.5 | 37.3 ± 5.4 | 36.8 ± 3.3 |
| range of isolate means | 30.5–34.4 | 25.4-30.1 | 25.8–32.0 | 36.8–40.9 | 33.6–36.5 | 41.6–53.3 | 34.4–39.9 | 36.0-38.3 |
| total range | 24.3–41.7 | 16.5–42.5 | 19.0–45.0 | 25.0–39.3 | 23.0–46.0 | 17.9–65.5 | 19.2–48.4 | 25.7–42.8 |
| tapering base | 61.3 % (40–86 %) | 48.7 % (30–78 %) | n.a. | – | 37.3 % (8–60 %) | – | 60.0 % (52–81 %) | 47.9 % (32–58 %) |
| elongated | 65.4 % (50–82 %) | 59.5 % (36–86 %) | 45 % (10–86 %) | – | 44.8 % (8–80 %) | – | 21.3 % (12–34 %) | 19.7 % (4–36 %) |
| tubular | – | 24.5 % (21–28 %) | – | – | – | – | – | – |
| excentric | 6.7 % (2–20 %) | 5.0 % (2–8 %) | n.a. | – | 20.9 % (9–46 %) | – | 4.0 % (0–8 %) | 5.3 % (2–10 %) |
| smooth-walled | 100 % | 100 % | 100 % | 100 % | 100 % | 100 % | 100 % | 34.4 % (19–48 %) |
| Oospores | ||||||||
| plerotic oospores | 64.3 % (50–83 %) | 44 % (24–76 %) | mostly aplerotic | >99 % (99–100 %) | 27.3 % (10–45 %) | 30.0 % (12-46 %) | 46.3 % (38–53 %) | >99 % (99–100 %) |
| mean diam | 28.7 ± 2.6 | 26.1 ± 3.7 | 24.7 ± 2.6 | 33.7±1.3 | 30.5 ± 3.2 | 41.3±6.1 | 33.2 ± 5.3 | 32.2 ± 2.7 |
| Total range | 21.9–38.8 | 15.7–36.3 | 18.0–30.0 | 24.6–39.9 | 19.7-38.2 | 12.5-55.0 | 16.3–45.1 | 22.7–37.7 |
| wall diam | 3.1 ± 0.49 | 2.0 ± 0.6 | 2.4 ± 0.56 | 2.9 ± 1.5 | 2.4 ± 0.4 | 4.2 ± 0.7 | 2.5 ± 0.7 | 3.1 ± 0.5 |
| oospore wall index | 0.51 ± 0.05 | 0.38 ± 0.08 | 0.44 ± 0.1 | 0.41 ± 0.03 | 0.40 ± 0.06 | 0.49 ± 0.07 | 0.38 ± 0.06 | 0.47 ± 0.06 |
| Abortion rate | 21.5 % (18–29 %) | 72.3 % (40–91 %) | <10 % | 5 % (3–7 %) | 52.8 % (0–80 %) | 6.2 % (3-10 %) | 24.9 % (18–31 %) | 1.7 % (1–2 %) |
| Antheridia | 99 % paragynous | 100 % paragynous | 100 % paragynous | 100 % paragynous | 4.7 % amphigynous | 100 % paragynous | 100 % paragynous | 100 % paragynous |
| size | 14.0±2.3 x 10.2±1.6 | 14.9±3.1 x 10.9±1.7 | 16.6±6.3 x 12.7±2.5 | 16.9±4.6 x 12.8±0.13 | 16.3±3.1 x 12.6±2.3 | 16.9±3.1 x 13.3±2.5 | 13.6±2.6 x 10.8±2.1 | 13.4±2.1 x 10.0±1.7 |
| Chlamydospores | globose | – | rare, in some isolates | – | – | – | – | – |
| mean diam | 29.5 ± 4.9 (17.7–47.0) | 17–35 | ||||||
| Hyphae | Sympodial branching with protruding tip of mother hypha | Sympodial branching with protruding tip of mother hypha | Sympodial branching with protruding tip of mother hypha | Often inflated tubular or club-shabed | Coralloid | |||
| Hyphal aggregations | – | – | – | – | + | (+) | – | – |
| Hyphal swellings | in water; globose, subglobose, limoniform; some catenulate | in water; subglobose, triangular; some catenulate | – | in water; subglobose, irregular, catenulate | in water; subglobose, irregular, catenulate | in water; subglobose, irregular, catenulate | – | – |
| Maximum temperature | 25–<30 | 25–<30 | 27.5 | 25–<30 | 25–<30 | 25 | 30–<35 | 35 |
| Optimum temperature | 20 | 20 | 25 | 25 | 15 | 20 | 25 | 25 |
| Growth rate at optimum | 2.9 ± 0.91 | 1.7 ± 1.0 | 3.7 | 1.1 ± 0.4 | 1.0 ± 0.1 | 1.2 ± 0.2 | 4.9 ± 0.53 | 4.7 ± 0.61 |
| Growth rate at 20°C | 2.7 ± 0.75 | 1.7 ± 1.0 | 3.3 | 1.5 ± 0.3 | 0.9 ± 0.04 | 1.2 ± 0.2 | 4.4 ± 0.59 | 4.4 ± 0.56 |
– = character not observed; + = character observed; n.a. = not available.
a Data from Jung et al. (2017b).
b Morphological and morphometric data from Jung et al. (1999); standard deviations of all morphometric data, oospore wall diameter and oospore wall index calculated from original dataset.
Phytophthora castanetorum T. Jung, M. Horta Jung, Bakonyi & Scanu, sp. nov.
MycoBank MB819699 (Fig. 6)
Fig. 6.
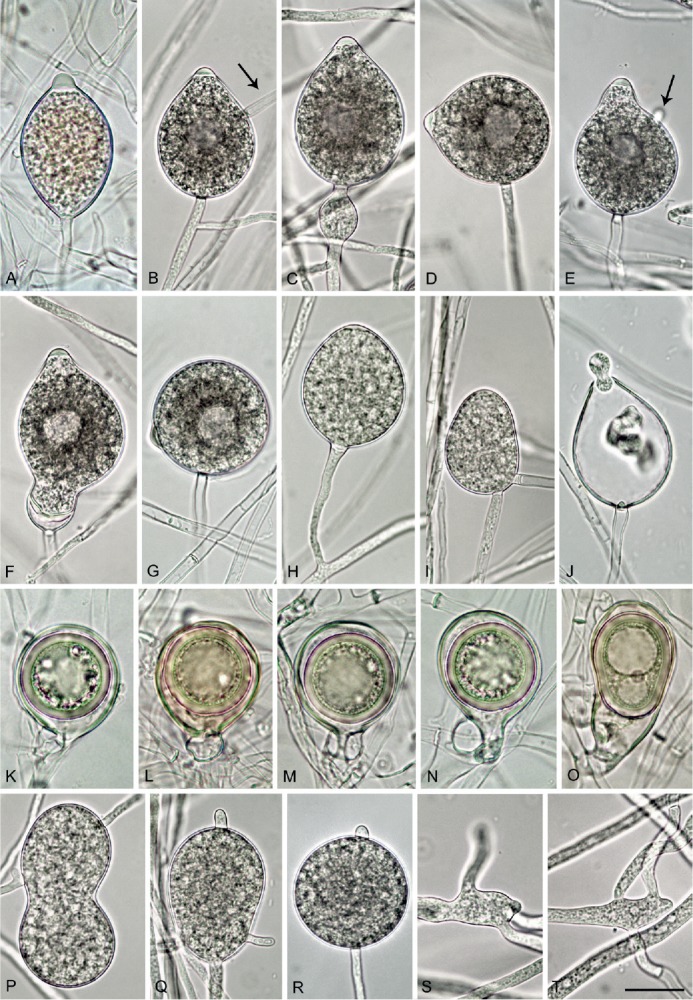
Phytophthora castanetorum. A. papillate limoniform sporangium formed in solid V8 agar (V8A) (CBS 142299 — ex-type). B–J. Sporangia formed on V8A flooded with soil extract. B–G. With vacuole. B. Ovoid, papillate with external proliferation close to sporangial base and hyphal extension (arrow) (BD 484). C. Ovoid, papillate with limoniform swelling before sporangial base (CBS 142299). D. Ovoid, papillate with laterally attached sporangiophore (BD 484). E. Obpyriform, papillate with short hyphal extension (arrow) (CBS 142299). F. Limoniform, papillate with elongated base and conspicuous basal plug (BD 486). G. Globose, papillate with laterally attached sporangiophore (BD 484). H. Ovoid, semipapillate on short lateral hypha (BD 476). I. ovoid, nonpapillate intercalary (BD 336). J. Ovoid sporangium with conspicuous basal plug, releasing zoospores (CBS 142299). K–O. Mature oogonia containing thick-walled oospores with large ooplast, formed in single culture in V8A. K–L. Globose to subglobose with aplerotic oospore and paragynous antheridium (CBS 142299). M. Globose to subglobose with aplerotic oospore and amphigynous antheridium (BD 484). N. Elongated excentric with aplerotic oospore and paragynous antheridium (BD 484). O. Elongated pyriform with paragynous antheridium, curved base and plerotic elongated oospore with two ooplasts (CBS 142299). P–R. Hyphal swellings formed on V8A flooded with soil extract (CBS 142299). P. Peanut-shaped intercalary. Q. Pyriform, terminal with radiating hyphal extensions. R. Globose, terminal with short hyphal extension. S–T. Sympodially branching primary hyphae in solid V8A with the mother hypha ending in a short protruding tip (CBS 142299). Bar A-T = 25 μm.
Etymology: Referring to the association of this species with forests of Castanea sativa (Castanetum is the phytosociological term for a chestnut forest).
Diagnosis: Phytophthora castanetorum differs from all other Phytophthora species from Clade 12 by regularly producing chlamydospores, and from P. quercina also by producing a low proportion of semipapillate and nonpapillate sporangia.
Type: Portugal: Algarve: Monchique, isolated from rhizosphere soil of a mature Castanea sativa tree, March 2015, T. Jung (CBS H-22983–holotype, dried culture on V8A; CBS 142299 = BD 292–ex-type culture). ITS and cox1 sequences GenBank MF036182 and MF036266, respectively.
Description: Sporangia, hyphal swellings and chlamydospores (Fig. 6A–G): Sporangia commonly observed in solid agar of older cultures (Fig. 6A) and produced abundantly in non-sterile soil extract; typically borne terminally on unbranched sporangiophores or in irregular lax or regular dense sympodia, and some formed on short lateral hyphae (Fig. 6H) or intercalary (Fig. 6I); non-caducous, monopapillate (77.4 %; Fig. 6A–G), very rarely bipapillate (over all isolates <1 %), semipapillate (14.9 %; Fig. 6H) or non papillate (7.7 %; Fig. 6I), and often forming a conspicuous basal plug that protrudes into the empty sporangium (Fig. 6J); in solid agar with conspicuously protruding papillae (Fig. 6A); sporangial shapes very variable, ovoid (over all isolates 77.4 %; Fig. 6B–D, I–J) or subglobose to globose (11.7 %; Fig. 6G) to obpyriform (4.6 %; Fig. 6E), ellipsoid (2.0 %), limoniform (1.0 %; Fig. 6A, F), or distorted (3.3 %); unusual features such as lateral attachment of the sporangiophore (over all isolates 37.7 %; Fig. 6D, G), hyphal extensions (9.1 %; Fig. 6B, D), markedly curved apices (4.9 %) or the presence of a vacuole (8.0 %; Fig. 6B–G) common in all isolates. Small subglobose to limoniform hyphal swellings sometimes formed close to the sporangial base (Fig. 6C). Zoospores discharged through an exit pore 4.0–9.5 μm wide (av. 7.0 ± 1.1 μm) (Fig. 6J), limoniform to reniform whilst motile, becoming spherical (av. diam = 9.2 ± 1.0 μm) on encystment, direct germination common in older water cultures (Fig. 6L). Sporangia of seven isolates averaged 47.0 ± 5.7 × 36.6 ± 3.7 μm (overall range 27.0–74.5 × 18.0–48.0 μm) with a range of isolate means of 44.2–48.1 × 35.4–38.3 μm and a length/breadth ratio of 1.29 ± 0.15 (range of isolate means 1.25–1.36). Swellings commonly observed in liquid culture; 27.1 ± 9.9 μm diameter (total range 10.0–47.0 μm), subglobose to globose, liminiform, pyriform or distorted; sometimes catenulate and often with individual or radiating hyphal extensions, (Fig. 6P–Q). Chlamydospores globose, produced on both CA and V8A, 29.5 ± 4.9 μm diam (total range 18.0–47.0 μm) (Fig. 6R).
Oogonia, oospores, antheridia and hyphae (Fig. 6H–O): Gametangia readily produced in single culture by all isolates on V8A within 7-10 d. Oogonia terminal, smooth-walled, and elongated pyriform to ellipsoid (on av. 65.4 %; Fig. 6N–O) or globose to slightly subglobose (34.6 %; Fig. 6K–M); bases often tapering (61.3 %; Fig. 6K, N–O) and sometimes slightly curved (4.9 %; Fig. 6O); mean 32.9 ± 3.0 μm diam (overall range 24.5–41.5 μm and range of isolate means 30.5–34.5 μm) and an average length of 40.5 ± 4.6 μm; almost plerotic (53.1 %) or aplerotic (46.9 %). Oospores usually globose (98.3 %; Fig. 6K–N) but could be slightly elongated in elongated oogonia (1.7 %; Fig. 6O); thick-walled, wall thickness 3.1 ± 0.5 μm (range 1.8–4.5 μm), and oospore wall index 0.51 ± 0.05.; abortion 19-24 % after 4 wk increasing to 58-91 % after 12 months. Antheridia almost exclusively paragynous and club-shaped to subglobose (Fig. 6K–L, N), sometimes with one or more finger-like projections (4.3 %), but a few amphigynous antheridia observed in all isolates (Fig. 6M). Primary hyphae often branched in a mono- or dichasium with the mother hypha ending in a short protruding tip (Fig. 6S–T).
Cultures (Figs 11, 13): Colonies on CA stellate with limited aerial mycelium and on V8A uniform and woolly; on PDA and MEA isolates formed uniform colonies, dense felty on PDA and mostly submerged on MEA. All isolates with very slow growth on PDA and MEA (Fig. 11). Temperature-growth relations are shown in Fig. 13. All six isolates included in the growth test had similar growth rates. The maximum growth temperature was 25–30 °C, with no grow when plates incubated for 5 d at 30 °C were transferred to 20 °C. Average radial growth rate at the optimum temperature of 25 °C 3.7 mm/d.
Fig. 11.
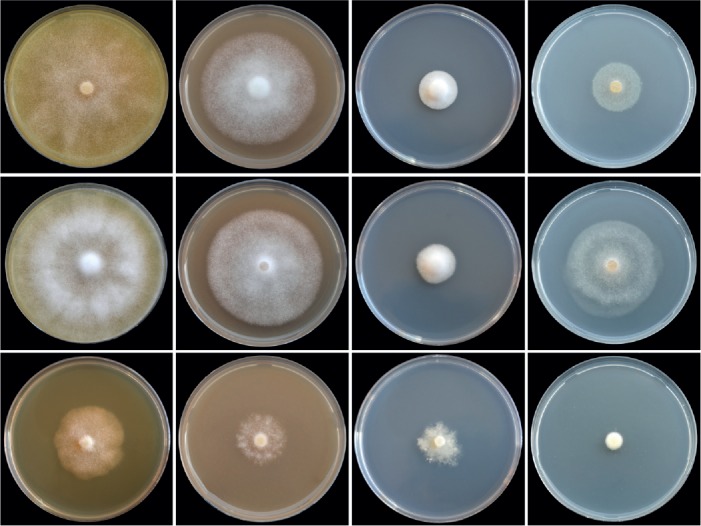
Colony morphology of Phytophthora castanetorum (CBS 142299), P. quercina (Beja 5) and P. tubulina (CBS 141212) (from top to bottom) after 10 d growth at 20 °C on carrot agar, V8 agar, potato-dextrose agar and malt extract agar (from left to right).
Fig. 13.
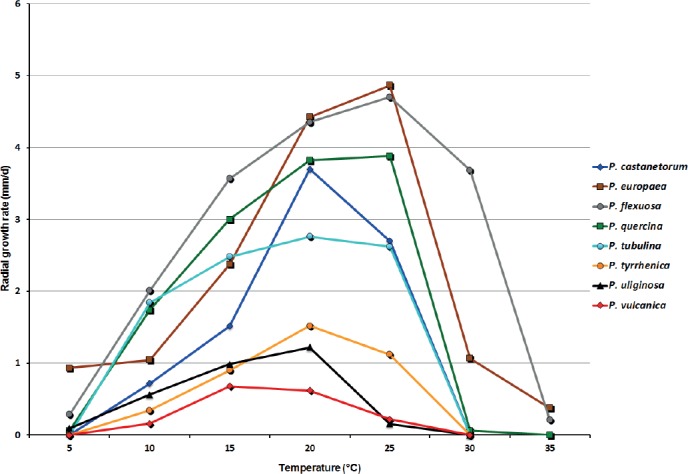
Mean radial growth rates of Phytophthora castanetorum (6 isolates), P. tubulina (5 isolates), P. quercina (5 isolates), P. tyrrhenica (5 isolates), P. vulcanica (5 isolates), P. uliginosa (2 isolates), P. europaea (5 isolates) and P. flexuosa (3 isolates) on V8 agar at different temperatures.
Additional material examined: Portugal: Beira Alta: Parque Natural da Serra da Estrela, isolated from rhizosphere soil of a mature Castanea sativa tree, June 2015, T. Jung (BD 476,484, 485, 486); Algarve: Serra de Monchique, isolated from rhizosphere soil of a mature C. sativa tree, March 2015, T. Jung (BD 293).–Italy: Sardinia; Gennargentu mountain, isolated from rhizosphere soil of a coppiced C. sativa tree, May 2014, B. Scanu & S. Seddaiu (CBS 142300 = P14, P17, P18).
Phytophthora tubulina T. Jung, T. Cech, Scanu, Horta Jung & Bakonyi, sp. nov.
MycoBank MB819701
Fig. 7.
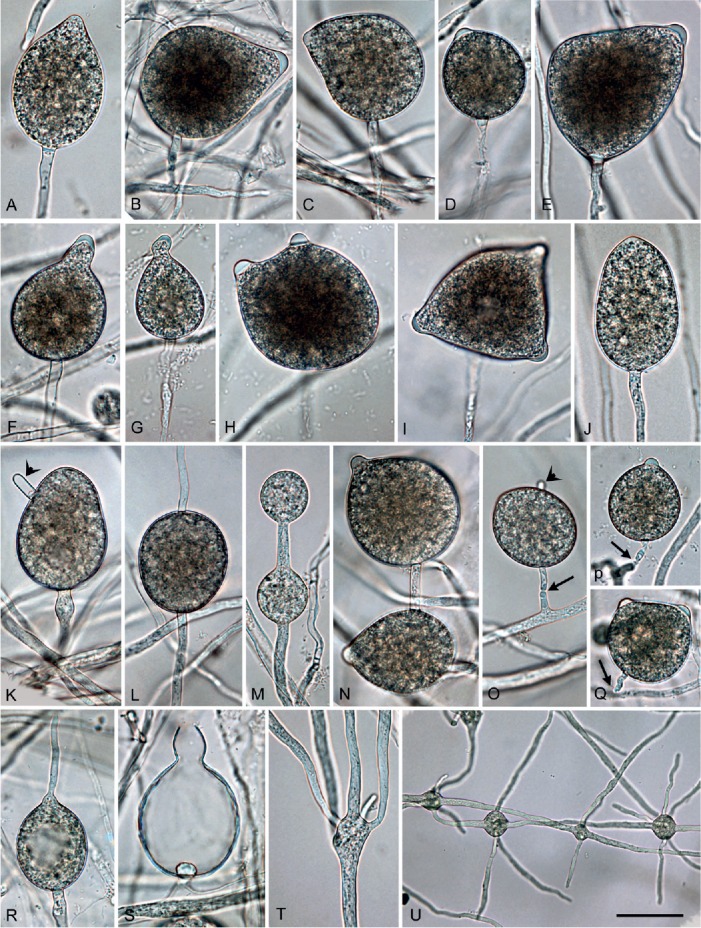
Phytophthora tubulina (CBS 141212 — ex-type), structures formed on V8 agar flooded with soil extract. A–L, N–S. Sporangia. A. Papillate ovoid. B. Papillate obpyriform with laterally attached sporangiophore, on a short lateral hypha. C. Papillate mouse-shaped with curved apex. D. Papillate subglobose. E. Papillate distorted with laterally attached sporangiophore. F–G. Papillate obpyriform with beak-like, curved (G) apex. H. Bipapillate. I. Tripapillate. J. Semipapillate, elongated ovoid. K. Nonpapillate ovoid with hyphal extension. L. Nonpapillate intercalary. M. Subglobose catenulate hyphal swellings. N. Papillate, ovoid intercalary sporangium and papillate subglobose sporangium. O. Nonpapillate ovoid sporangium with short hyphal extension (arrow head), shedding (arrow) from short lateral hypha. P. Subglobose to ovoid, papillate sporangium with constriction of sporangiophore. Q. Bipapillate detached sporangium. R. Direct germination of papillate ovoid sporangium. S. Empty obpyriform sporangium after release of zoospores, with beak-like apex, narrow exitpore and conspicuous basal plug. T. Branching of sporangiophore into multiple sporangiophores. U. Globose to subglobose hyphal swellings with radiating hyphae. Bar A–T = 25 μm, U = 40 μm.
Fig. 8.
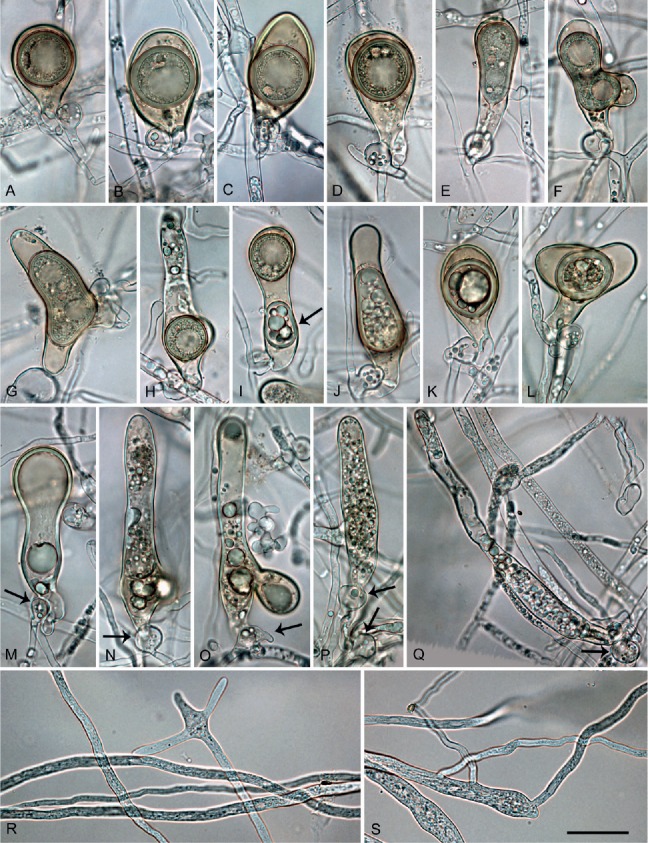
Phytophthora tubulina (CBS 141212 — ex-type), structures formed in solid V8 agar. A–H. Mature oogonia with paragynous antheridia, formed in single culture. A. Subglobose with tapering base and aplerotic, globose thick-walled oospore. B–C. Elongated ellipsoid with tapering bases and aplerotic, globose thick-walled oospores. D. Elongated pyriform with tapering base and aplerotic, globose thick-walled oospore. E. Elongated tubular with tapering base and plerotic, elongated tubular, thick-walled oospore. F–G. Elongated excentric with almost plerotic elongated oospores containing two ooplasts. H. Elongated tubular containing a globose, viable thin-walled oospore. I. Elongated tubular containing a subglobose, viable thin-walled oospore and a second, ellipsoid aborted oospore (arrow). J–L. Oogonia aborted after oospore formation. J. Elongated tubular. K. Elongated pyriform. L. Excentric. M–Q. Extremely elongated, tubular oogonia, aborted before oospore formation, with paragynous antheridia (arrows). R–S. Primary hyphae branching in a dichasium (R) or monochasium (S) with the mother hypha ending in a short protruding tip. Bar A–S = 25 μm.
Etymology: Name refers to the tubular shape of many oogonia (tubulina Lat., tubular).
Diagnosis: Phytophthora tubulina differs from all other known Phytophthora species by having partially extremely elongated, often tubular oogonia with high oospore abortion rates, and from P. quercina by producing a low proportion of semipapillate and nonpapillate sporangia.
Type: Austria: Lower Austria; Dunkelsteiner Forst, isolated from rhizosphere soil of a mature Fagus sylvatica tree, Sept. 2010, T. Jung (CBS H-22557–holotype, dried culture on V8A; CBS 141212 = TUB1–ex-type culture). ITS and cox1 sequences GenBank MF036196 and MF036277, respectively.
Description: Sporangia, hyphal swellings and chlamydospores (Fig. 7A–U): Sporangia infrequently observed in solid agar, abundantly produced in non-sterile soil extract; borne terminally on unbranched sporangiophores, in lax sympodia, on short lateral sporangiophores (Fig. 7B, O) or intercalary (3.4 %; Fig. 7L); sometimes formed on short hyphal appendices growing from mature sporangia (Fig. 7N); usually non-caducous (Fig. 7A–L, N, R–S) but a low proportion (<1 %) of caducous sporangia without preformed pedicels (Fig. 7O-Q) present in most isolates; apices highly variable ranging from monopapillate (56.6 %; Fig. 7A–G, N, P), bi- or tripapillate (21.4 %; Fig. 7H–I, Q), semipapillate (13.3 %; Fig. 7J) to nonpapillate (8.9 %; Fig. 7K–L, O); sometimes curved (4.9 %; Fig. 7C, G); sporangial shapes ranging from ovoid or elongated ovoid (69.4 %; Fig. 7A–B, E), obpyriform (15.3 %; Fig. 7C), limoniform (3.0 %; Fig. 7D), subglobose (3.7 %) or less frequently ellipsoid and globose (2.3 %) to distorted shapes (6.3 %; Fig. 7H–I, Q); hyphal appendices (6.7 %; Fig. 7K, O) and laterally attached sporangiophores (22.4 %; Fig. 7B, E, N–O) commonly observed; dimensions of six isolates of P. tubulina averaging 51.6 ± 9.8 x 37.3 ± 7.0 μm (overall range 29.5–98.5 x 19.0–59.0 μm) with a range of isolate means of 43.5–58.0 x 31.0–45.0 μm; length/breadth ratio 1.4 ± 0.18 with a range of isolate means of 1.27–1.49; sporangial germination directly (Fig. 7R) or more often indirectly with zoospores discharged through an exit pore 4.0–9.0 μm wide (av. 6.4 ± 1.2 μm) (Fig. 7S). Zoospores limoniform to reniform whilst motile, becoming spherical (av. diam = 11.7 ± 1.8 μm) on encystment. Swellings subglobose to liminoform, often catenulate and with radiating hyphae, infrequently produced on sporangiophores (Fig. 7M, T–U). Chlamydospores not observed.
Oogonia, oospores, antheridia and hyphae (Fig. 8A–S): Gametangia readily produced in single culture by all isolates of P. tubulina on V8A within 10 d. Oogonia borne terminally or laterally, with smooth walls and tapering, often long bases (on av. 53.2 %; Fig. 8A–E, G, K); globose to subglobose (35.8 %; Fig. 8A), elongated to tubular (59.2 %; Fig. 8B-K) or excentric (5 %; Fig. 8L); diameters averaging 29.2 ± 4.4 μm (overall range 16.5–42.5 μm and range of isolate means 25.4–30.1 μm). Oogonial length 44.8 ± 11.6 μm; tubular viable oogonia (Fig. 8A–J) up to 109.0 μm long, aborted tubular oogonia (Fig. 8M–Q) reaching lengths of 300.0 μm. Oospores with a mean diameter of 26.1 ± 3.7 μm (total range 16.0–36.5 μm), plerotic or aplerotic, usually globose (89 %) or less frequently elongated (11 %) assuming the shape of the elongated or excentric oogonium (Fig. 8E–H). Some oogonia with two oospores of which one was usually aborted (Fig. 8J). Oospores with medium-thick walls (2.0 ± 0.6 μm) and a mean oospore wall index of 0.38 ± 0.08; containing either one large ooplast (Fig. 8A–D) or several smaller lipid vacuoles (Fig. 8E–H); abortion high (on. av. 72.3 %; 40–91 %) occurring either before (Fig. 8M–Q) or after the formation of the oospore (Fig. 8F, J–L). Antheridia exclusively paragynous (Fig. 8A–Q) averaging 14.9 ± 3.1 x 10.9 ± 1.7 μm, with shapes ranging from subglobose to club-shaped. Primary hyphae often branching in a mono- or dichasium with the mother hypha ending in a short protruding tip (Fig. 8R–S).
Cultures (Figs 11, 13): Colonies of all five P. tubulina isolates similar on the four different types of media (Fig. 11); largely submerged with limited felty aerial mycelium around the inoculum plug on all four agar media, uniform on CA and V8A, and irregular to dendroid on PDA; almost no growth on MEA. Temperature–growth relations on V8A are shown in Fig. 13. All isolates with identical cardinal temperatures and similar growth rates at all temperatures. The maximum growth temperature was between 25 and 30 °C. All isolates with slow growth, unable to grow at 30 °C and not resuming growth when plates incubated for 5 d at 30 °C were transferred to 20 °C. Phytophthora tubulina had a broad growth optimum between 15 and 25 °C with an average radial growth rate of 2.8 ± 1.1 mm/d at 20 °C.
Additional material examined: Austria: Lower Austria: Dunkelsteiner Forest, isolated from rhizosphere soil of a mature Fagus sylvatica tree, Sept. 2010, T. Jung (CBS 141213 = TUB2, TUB3, TUB4, TUB5).
Phytophthora tyrrhenica B. Scanu, S.O. Cacciola, Seddaiu, Bakonyi & T. Jung, sp. nov.
MycoBank MB819700
(Fig. 9)
Fig. 9.
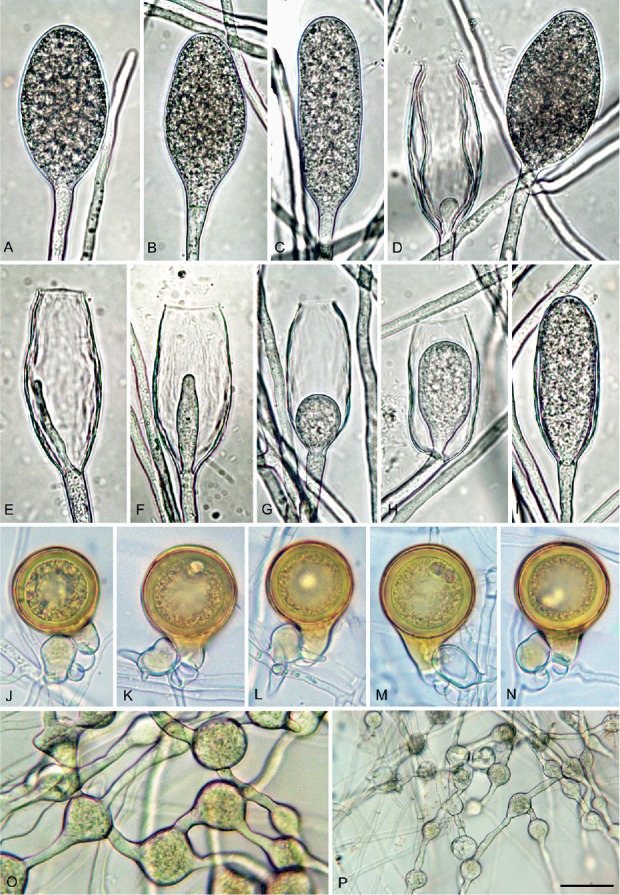
Phytophthora tyrrhenica (CBS 142301 — ex-type). A–I. Sporangia formed on V8 agar (V8A) flooded with soil extract. A–D. Nonpapillate with flat apex. A. Ellipsoid. B. Limoniform with tapering base. C. Elongated ellipsoid. D. Mature ovoid (right) and empty with wide exit pore and internal nested proliferation (left). E–I. Empty with wide exit pores and internal extended (E) or internal nested proliferation (F–I). J–N. Mature, golden-brown, smooth-walled, globose oogonia formed in single culture in V8A, containing thick-walled plerotic oospores with big ooplasts, with paragynous antheridia. K–L. Antheridia with finger-like hyphal extensions. O–P. Globose, limoniform and angular catenulate hyphal swellings formed on V8A flooded with soil extract. Bar A–O = 25 μm, P = 50 μm.
Etymology: Name refers to the origin of all known isolates in the Tyrrhenian islands Sardinia and Sicily (tyrrhenica Lat., Tyrrhenian).
Diagnosis: Phytophthora tyrrhenica is distinguished from its closest relatives P. uliginosa and P. vulcanica by the exclusive production of elongated and mostly ellipsoid sporangia, and from P. vulcanica also by the absence of amphigynous antheridia.
Type: Italy: Sardinia; Tergu, isolated from rhizosphere soil of a mature Quercus ilex tree, May 2012, B. Scanu (CBS H-22984–holotype, dried culture on V8A; CBS 142301 = PH154–ex-type culture). ITS and cox1 sequences GenBank KU899188 and KU899343, respectively.
Description: Sporangia, hyphal swellings and chlamydospores (Fig. 9A–I): Sporangia not observed on solid agar but produced abundantly after 24 h in non-sterile soil extract; borne terminally on unbranched sporangiophores, non-caducous, and non-papillate (Fig. 9A–I); ellipsoid to elongated ellipsoid (55.6 %; Fig. 9A, C, I), limoniform to elongated limoniform (18 %; Fig. 9B), ovoid to elongated ovoid (18.8 %; Fig. 9D) and elongated obpyriform (7.6 %); sporangial proliferation usually internal, mainly in a nested (Fig. 9D, F-J) but also extended way (Fig. 9E); dimensions of five isolates 68.4 ± 1.7 x 28.9 ± 1.9 μm (overall range 45.5–110.0 x 16.0–39.5 μm and range of isolate means 66.1–70.0 x 27.5–31.9), with a length/breadth ratio of 2.4–0.18 and a range of isolate means of 2.1–2.57. Zoospores discharged through an exit pore 9.0–22.0 μm wide (av. 12.1 ± 2.4 μm) (Fig. 9E–H), limoniform to reniform whilst motile, becoming spherical (av. diam = 14.6 ± 2.3 μm) on encystment; cysts usually germinating directly by forming a hypha but diplanetism also observed in all isolates. Catenulate, intercalary, globose to subglobose or irregular hyphal swellings (Fig 9O, P) with an average diameter of 18.6 ± 6.5 μm abundantly produced by most isolates in liquid culture. Chlamydospores not observed.
Oogonia, oospores and antheridia (Fig. 9J–N): Oogonia produced by all five isolates of P. tyrrhenica in single culture on V8A; borne terminally or laterally with globose to subglobose (93.3 %; Fig. 9I–N; P) or slightly excentric (6.7 %; Fig. 9O) shapes, often with a tapering base (52.3 %; Fig. 9I–L, N–P). Oogonial diameters 37.2 ± 3.3 μm (overall range 25.0–39.5 μm and range of isolate means 26.6–44.1 μm). Oospores plerotic, globose and usually containing a large ooplast (Fig. 9J–N); mean diameter 33.7 ± 1.3 μm (total range 24.5–40.0 μm), thick walls (av. 2.9 ± 1.5 μm, total range 1.5–4.5 μm), with a mean oospore wall index of 0.41 ± 0.03. Mean oogonial abortion rate low (5 %). Antheridia formed terminally or laterally (Fig. 9M), exclusively paragynous, averaging 16.9 ± 4.6 x 12.8 ± 0.13 μm, with shapes ranging from clavate, globose to subglobose.
Cultures (Figs 12–13): Colonies uniform with dense-felty to woolly aerial mycelium and regular margin on CA and V8A, and uniform appressed on PDA. All isolates with very limited and irregular growth on MEA (Fig. 12). Temperature-growth relations are shown in Fig. 13. All five isolates included in the growth test with similar growth rates and cardinal temperatures. Maximum growth temperature was 25–30 °C; all isolates resuming growth when plates incubated for 5 d at 30 °C were transferred to 20 °C. Average radial growth rate at the optimum temperature of 20 °C 1.5 ± 0.3 mm/d.
Fig. 12.
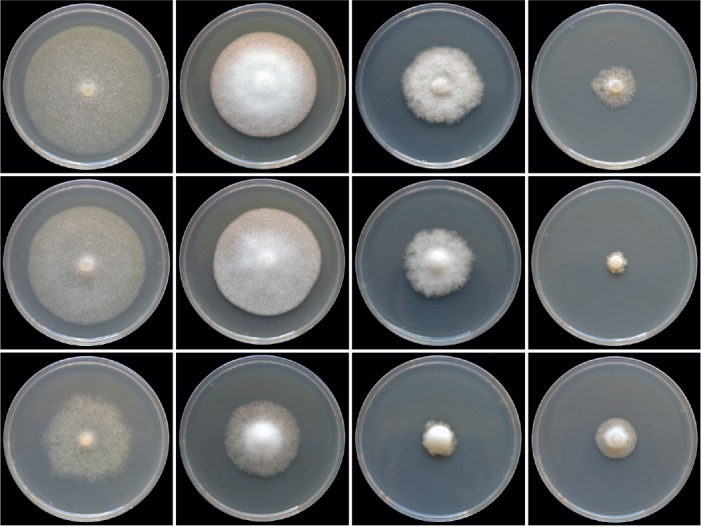
Colony morphology of Phytophthora tyrrhenica from Sardinia (CBS 142301), P. tyrrhenica from Sicily (CBS 142302) and P. vulcanica (CBS 141216) (from top to bottom) after 15 d growth at 20°C on carrot agar, V8 agar, potato-dextrose agar and malt extract agar (from left to right).
Additional material examined: Italy: Sicily; Madonie mountains, isolated from rhizosphere soil of mature Quercus ilex trees, May 2015, T. Jung (TYR1, TYR2, TYR3, TYR4, TYR5, TYR6); Sardinia; Tergu, isolated from rhizosphere soil of a mature Q. ilex tree, May 2012, B. Scanu (PH155); Sardinia; Alà dei Sardi, isolated from rhizosphere soil of a mature Quercus suber tree, June 2012, B. Scanu and S. Seddaiu (CBS 142303 = PH103).
Phytophthora vulcanica T. Jung, M. Horta Jung, Scanu, Bakonyi & Cacciola, sp. nov.
MycoBank MB819702
(Fig. 10)
Fig. 10.
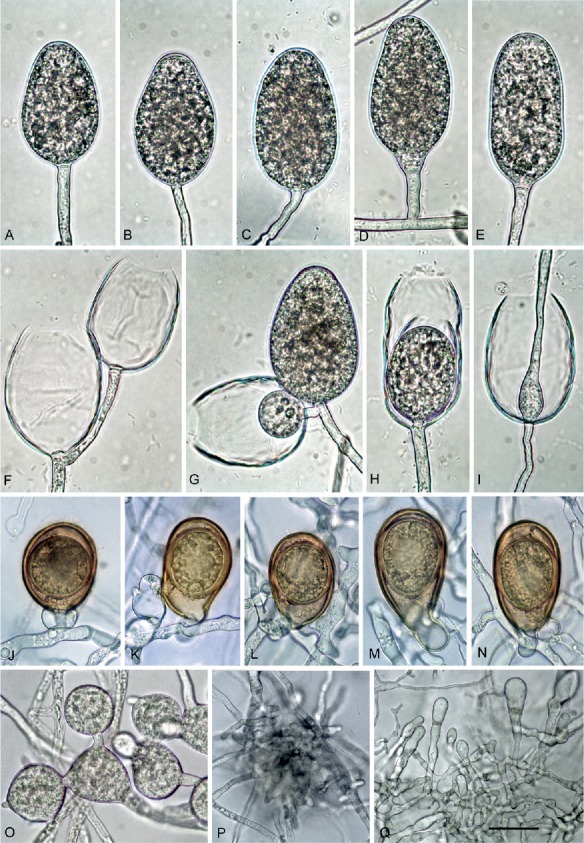
Phytophthora vulcanica (CBS 141216 — ex-type). A–I. Sporangia formed on V8 agar (V8A) flooded with soil extract. A–E. Nonpapillate with flat apex. A–B. Ovoid. C. Elongated ovoid. D. Limoniform, on short lateral hypha. E. Elongated ellipsoid. F. Empty sporangia after release of zoospores through wide exitpores, showing external proliferation. G. Empty sporangium with both internal nested proliferation (arrow) and external proliferation forming a mature ovoid sporangium. H. Empty, elongated ellipsoid sporangium with internal nested proliferation. I. Empty ovoid sporangium with internal extended proliferation. J–N. Mature, golden-brown oogonia formed in single culture in V8A, with medium thick-walled oospores. J. Subglobose with aplerotic globose oospore and amphigynous antheridium. K. Elongated, slightly excentric with aplerotic subglobose oospore and paragynous antheridium. L. Obovoid with aplerotic globose oospore and paragynous antheridium. M. Elongated ellipsoid with aplerotic ellipsoid oospore and paragynous antheridium. N. Elongated pyriform with plerotic ellipsoid oospore. O. Globose and triangular, catenulate hyphal swellings. P. Dense hyphal aggregation. Q. Tubular to elongated club-shaped hyphae. Bar A–O = 25 μm, P–Q = 40 μm.
Etymology: Name refers to the origin of all known isolates from volcanic soil (vulcanica Lat., volcanic).
Diagnosis: Phytophthora vulcanica differs from its closest relatives P. uliginosa and P. tyrrhenica by the production of both paragynous and amphigynous antheridia, by its low optimum temperature of 15 °C and by slower growth rates at 15 and 20 °C.
Type: Italy: Sicily; Mount Etna, isolated from rhizosphere soil of a mature Fagus sylvatica tree, May 2013, T. Jung (CBS H-22556–holotype, dried culture on V8A; CBS 141216 = X3a–ex-type culture). ITS and cox1 sequences GenBank MF036209 and MF036287, respectively.
Description: Sporangia, hyphal swellings and chlamydospores (Fig. 10A–I): Sporangia not formed on solid agar but produced in non-sterile soil extract; non-caducous and non-papillate with a flat apex (Fig. 10A–E, G); borne terminally, on unbranched sporangiophores or in lax sympodia, or less frequently laterally (Fig. 10D), proliferating internally in both a nested (Fig. 10G, H) and extended way (Fig. 10I). Sporangiophores often branching close to the sporangial base forming lax sympodia (Fig. 10F–G). Sporangial shapes ovoid (52.5 %; Fig. 10A–B, F–G, I), elongated ovoid (14.0 %; Fig. 10C), ellipsoid to elongated ellipsoid (32.5 %; Fig. 10E–F, H) or less frequently limoniform (Fig. 10D), pyriform and obpyriform (1.0 %); dimensions of six isolates 57.3 ± 8.7 x 34.5 ± 4.2 μm (overall range 35.5–81.0 x 22.5–43.0 μm) with a range of isolate means of 53.6–61.0 x 31.8–36.4 μm, and a length/breadth ratio of 1.67 ± 0.22 (range of isolate means 1.58–1.72). Zoospores of P. vulcanica discharged through wide exit pores averaging 17.0 ± 3.0 μm (12.0–25.0 μm), limoniform to reniform whilst motile, becoming spherical (av. diam = 10.0 ± 1.2 μm) on encystment; cysts usually germinating directly but diplanetism also observed in all isolates. Swellings formed on sporangiophores by all isolates, subglobose, angular or irregular, often catenulate (Fig. 10O), with an average diameter of 21.1 ± 5.5 μm. Chlamydospores not observed.
Oogonia, oospores, antheridia and hyphae (Fig. 10J–Q): Oogonia produced by all five isolates on V8A in single culture; borne terminally or laterally, with smooth walls and often tapering (37.3 %; Fig. 10L–N) or curved bases (7.2 %; Fig. 10M); elongated, ellipsoid to pyriform or excentric (on av. 65.7 %; Fig. 10K–N) or less frequently globose to subglobose (34.3 %; Fig. 10J), turning golden-brown during ageing (Fig. 10J–N); mean 35.8 ± 3.6 μm diam. with an overall range of 23.0–46.0 μm and isolate means ranging from 33.6–36.5 μm. Oospores with a mean diameter of 30.5 ± 3.2 μm (total range 20.0–38.0 μm), aplerotic (72.7 %) or less frequently plerotic (27.3 %), containing a large ooplast (Fig. 10J–N); medium thick-walled (2.4 ± 0.4 μm), with a mean oospore wall index of 0.40 ± 0.06. High oogonial abortion rate in most isolates (av. 52.8 %; 0–80 %). Antheridia 16.3 ± 3.1 x 12.6 ± 2.3 μm, predominantly paragynous (95.3 %; Fig. 10K–N) but few amphigynous antheridia observed in all isolates (4.7 %; Fig. 10J). Hyphal aggregations regularly formed by all isolates (Fig. 10P). Hyphae often inflated-tubular to club-shaped (Fig. 10Q).
Cultures (Figs 12–13): Colonies of all P. vulcanica isolates uniform felty on CA and V8A , with irregular margins on CA and dome-shaped with regular margins on V8A; very limited and irregular growth on PDA and MEA (Fig. 12). Temperature–growth relations on V8A are shown in Fig. 13. All isolates with very slow growth and similar growth rates at all temperatures; maximum growth temperature between 25 and 30 °C. Isolates with no growth when plates incubated for 5 d at 30 °C were transferred to 20 °C. Average radial growth rate at the optimum temperature of 15 °C 0.7 ± 0.1 mm/d.
Additional material examined: Italy: Sicily; Mount Etna, isolated from rhizosphere soil of mature F. sylvatica trees, May 2013, T. Jung (CBS 141217 = X3b, X3c, X3d, X3e).
DISCUSSION
Four new cryptic Phytophthora species recently isolated from Fagaceae trees in forest stands in Austria, Italy, and Portugal were characterized. Phylogenetic analyses of the DNA sequence data for three nuclear (ITS, Btub and HSP90) and two mitochondrial (cox1 and NADH1) gene regions together with detailed morphological and physiological studies allowed the description of these four taxa as P. castanetorum, P. tubulina, P. tyrrhenica, and P. vulcanica.
Multigene phylogenetic analyses demonstrated that P. castanetorum and P. tubulina are distinct species closely related to P. quercina. In previous studies, the phylogenetic position of P. quercina was ambiguous and, depending on the gene regions analysed, this species loosely clustered with Clade 3 (ITS; Cooke et al. 2000), Clade 4 (seven nuclear loci; Blair et al. 2008), Clade 5 (four mitochondrial loci; Martin et al. 2014), and Clade 1 (seven nuclear loci; Martin et al. 2014). Due to the inclusion of P. castanetorum, P. tubulina and the three taxa P. sp. ohioensis, P. versiformis and P. sp. versiformis-like in the phylogenetic analyses of both a four-locus (ITS, Btub, HSP90, NADH1) and a nuclear three-locus (ITS, Btub, HSP90) dataset in this work the phylogenetic position of the P. quercina clade within Phytophthora could be resolved. Phytophthora quercina, P. castanetorum, P. tubulina, P. sp. ohioensis, P. versiformis and P. sp. versiformis-like, formed a fully supported monophyletic group. Due to the unique phylogenetic position of P. lilii, which was confirmed in both the phylogenetic analysis of the three-locus dataset here, and a new expanded phylogeny of Phytophthora with more than 150 taxa included (Yang et al. 2017), Rahman et al. (2015) proposed that P. lilii constituted an eleventh Phytophthora clade. Consequently, the P. lilii clade and the P. quercina clade are named here as Clades 11 and 12, respectively.
All Clade 12 species in Europe and North America are associated with Fagaceae. Phytophthora quercina is involved in decline syndromes of natural and planted Quercus stands (Q. cerris, Q. coccinea, Q. faginea, Q. ilex, Q. palustris, Q. petraea, Q. pubescens, Q. pyrenaica, Q. robur, Q. rubra, Q. suber, and Q. vulcanica) across Europe causing a progressive destruction of fine root systems, which predisposes affected oaks to climatic extremes and secondary pathogens and pests (Jung et al. 1999, 2000, 2013, 2016, Balci & Halmschlager 2003a, b, Perez-Sierra et al. 2013). Interestingly, P. sp. ohioensis and P. sp. quercina-like were also found associated with oak forests in the USA, the latter taxon with Q. ellipsoides and Q. rubra in Minnesota and Wisconsin, respectively (Balci et al. 2007), and P. sp. ohioensis with declining Q. alba in southern Ohio (Balci et al. 2010). Phytophthora sp. quercina-like was also recovered from oak seedlings in woody ornamental nurseries in Minnesota (Schwingle et al. 2007). Phytophthora castanetorum and P. tubulina were isolated from the other two genera of Fagaceae in Europe, Castanea and Fagus, respectively. Interestingly, in Australia, which lacks any native Fagaceae, P. versiformis and P. sp. versiformis-like were both frequently isolated from the rhizosphere of a Myrtaceae tree (Corymbia calophylla) in remnant bushland, parks and gardens in the southwest of Western Australia (Barber et al. 2013, Paap et al. 2017). In a metagenomic Phytophthora survey of rhizosphere soils, P. versiformis was detected in more than 10 % of the 640 sites sampled across Australia, and due to its consistent association with native vegetation it was considered as a native species (Burgess et al. 2017).
Both P. castanetorum and P. tubulina share basic phenotypic features with their closest relative P. quercina, such as the homothallic breeding system, production of elongated oogonia and persistent papillate sporangia, sympodial hyphal branching with the mother hypha ending with a protruding tip, slow growth in culture and uniform woolly colony growth patterns, but differed in a number of characters (Table 2). Phytophthora castanetorum and P. tubulina produce a low proportion of semipapillate and nonpapillate sporangia, whereas sporangia of P. quercina are exclusively papillate. Phytophthora tubulina also forms markedly longer sporangia than the other two species and partially extremely elongated, often tubular oogonia with high oospore abortion rates, suggesting genetic instability. Phytophthora castanetorum differed from the other two species in the regular production of chlamydospores, and by having on average higher oogonial diameters and a higher percentage of elongated oogonia. Phytophthora quercina grew faster at all temperatures tested and had a higher optimum temperature for growth than P. castanetorum and P. tubulina (25 vs 20 °C).
Concerning P. tyrrhenica and P. vulcanica, multigene phylogenetic analyses placed both species in Clade 7a (Cooke et al. 2000, Blair et al. 2008), with P. tyrrhenica forming a sister clade to P. uliginosa and P. vulcanica residing in a basal position of the subclade. Recently, six new Phytophthora species from Clade 7a occurring in natural ecosystems in Taiwan were described and two other new Clade 7a taxa were informally designated (Jung et al. 2017a, b) expanding the number of extant taxa in Clade 7a to 16. Besides the new Taiwanese species, Clade 7a contains several important plant pathogens like the multivorous P. xcambivora and the host-specific P. fragariae, P. rubi, P. uniformis, P. xalni and P. xmultiformis, but also cryptic species such as P. europaea and P. uliginosa (Jung et al. 2002, 2017b). Their scattered distribution and putative host specialization also suggests a cryptic nature for both P. tyrrhenica and P. vulcanica. Morphologically these two species are similar to their closest relative P. uliginosa (Table 2). The main distinguishing characters are the exclusive production of elongated and mostly ellipsoid sporangia in P. tyrrhenica, the occurrence of both amphigynous and paragynous antheridia in P. vulcanica and the average largest oogonial and oospore diameters in P. uliginosa. All three species show similar slow growth rates, but differ in their optimum temperatures for growth and their growth rates at 25 °C, with P. vulcanica being the most psychrophilic and P. tyrrhenica the most thermophilic species.
The lack of previous records of all four new species from other continents, their apparent absence from European nurseries (Jung et al. 2016) and their exclusive occurrence in natural and semi-natural Fagaceae forest ecosystems in Europe indicate that P. castanetorum, P. tubulina, P. tyrrhenica, and P. vulcanica may be endemic to Europe or were introduced in historic times and have evolved in situ. These hypotheses are also supported by the intraspecific variability in both mitochondrial and nuclear DNA, observed particularly in P. castanetorum and P. tyrrhenica. This is in contrast to genetically uniform populations of recently introduced Phytophthora species which originated from relatively small and genetically impoverished founder populations (Goodwin et al. 1992). One well-known example is P. cinnamomi, whose global invasion was mainly achieved by the clonal spread of two genotypes from the A2 mating type (Oudemans & Coffey 1991, Dobrowolski et al. 2003). The other two new species, P. tubulina and P. vulcanica, were monomorphic across all five loci examined resulting in two clonal population structures. However, this is most likely due to their scattered distribution and the origin of all isolates per species from one site. More isolates from different geographic locations are needed to confirm the endemic origin also for these two species. That the four new species and their closest European relatives P. quercina, P. europaea and P. uliginosa are all exclusively associated with Fagaceae suggests sympatric species radiation driven by the adaptation to different genera of Fagaceae in Europe.
All four new species from Fagaceae forests showed a low maximum temperature for growth and an optimum temperature between 15 °C and 25 °C. This is similar to that of other possibly endemic low-temperature Phytophthora species such as P. ilicis, P. pseudosyringae, P. psychrophila, and P. quercina (Jung et al. 1999, 2002, Pérez-Sierra et al. 2013, Scanu et al. 2014b, Scanu & Webber 2016). All four new species are homothallic, with a high oospore wall index (range 0.4–0.5) enabling them to survive both the long hot and dry summers typical of Mediterranean regions (P. castanetorum, P. tyrrhenica, and P. vulcanica) and cold winters in Austria (P. tubulina), on Mount Etna (P. vulcanica), and in the Gennargentu Mountain and Serra da Estrela (P. castanetorum).
While P. vulcanica was isolated alone and from relatively healthy beech trees, P. castanetorum, P. tyrrhenica, and P. tubulina co-occurred with other Phytophthora species in the rhizosphere of declining trees. Infections by multiple Phytophthora species have been previously reported in Fagaceae (Jung 2009, Jung et al. 2013, Pérez-Sierra et al. 2013, Scanu et al. 2015). Because of their co-occurrence with aggressive pathogens of chestnut, beech, and oaks, such as P. xcambivora, P. cinnamomi, and P. plurivora, it is not possible to establish whether the new species were directly involved in the decline of Fagaceae in Europe. Pathogenicity tests, however, showed that all four new species are able to damage the roots of young seedlings of their respective hosts: beech, chestnut, and oak. In comparison to the aggressiveness of invasive Phytophthora species co-occurring with them, they must nevertheless be considered as relatively weak pathogens. For example, in a soil infestation trial during this study, P. xcambivora caused much higher mortality of chestnut seedlings than P. castanetorum (90 % vs <20 %). In addition, P. tubulina and P. vulcanica killed only a low proportion of beech seedlings in artificially infested soil whereas the mortality rate caused by P. xcambivora exceeded 50 %. The only exception was P. tyrrhenica, which showed considerable aggressiveness to Quercus ilex seedlings, although less virulent than P. cinnamomi. Their relatively low aggressiveness to their native Fagaceae hosts supports the endemism hypothesis.
The continuously increasing diversity of both invasive and potentially endemic Phytophthora species in Mediterranean Fagaceae forests (Scanu et al. 2010, Jung et al. 2013, Peréz-Sierra et al. 2013; this study) necessitates future studies to address the ecology of these cryptic species in Fagaceae ecosystems. Most plausibly, they live as fine root “nibblers” as demonstrated for P. quercina (Jung et al. 1999, 2000, Peréz-Sierra et al. 2013) causing a slow chronic decline which in interaction with climatic extremes and secondary pathogens and pests might cause episodic dieback and mortality (Jung et al. 2000, Jung 2009, Peréz-Sierra et al. 2013). A similar life-style was also suggested for the potentially native P. arenaria and P. constricta in Western Australia (Rea et al. 2011). In addition, the four new species may act as damping-off pathogens affecting seedling recruitment, as reported previously for other Phytophthora species (Cohen & Coffey 1986, Simamora et al. 2015). This is particularly plausible for P. tyrrhenica considering the high susceptibility of germinating Q. ilex acorns to Phytophthora, due to the poor suberisation and lignification of tissues and restricted allocation of defence compounds (Herms & Mattson, 1992, Martín-García et al. 2015, B. Scanu & G. Hardy, unpubl.). However, these hypotheses need to be further investigated, for both the new species described here and other Phytophthora species occurring in Fagaceae forest ecosystems. It would also be important to test whether co-infections of trees by endemic and invasive Phytophthora species are causing synergistic or antagonistic interactions. In a recent study conducted by Corcobado et al. (2017), no synergistic interaction was found when Q. ilex seedlings were grown in soil artificially infested with two different Phytophthora species. On the contrary, mortality of Q. ilex seedlings was delayed if the less virulent and potentially endemic European pathogens P. gonapodyides and P. quercina were inoculated prior to the aggressive invasive P. cinnamomi (Corcobado et al. 2017), most likely due to the priming of induced resistance. The putatively endemic, new species may also have a beneficial effect in maintaining plant diversity in these ecosystems. In the kwongan shrublands in Western Australia, the putatively endemic P. arenaria and P. constricta (Rea et al. 2011) can affect the growth of non-mycorrhizal plants while they do not affect co-occurring ectomycorrhizal (ECM) ones, thus conferring an advantage to ECM species in terms of accessing scarce phosphorus resources (Albornoz et al. 2016). Consequently, the occurrence of P. castanetorum and P. tyrrhenica in old impoverished soils in Portugal and Sardinia that are particularly low in phosphorus might suggest a similar ecological contribution to the maintenance of highly diverse ecosystems by reducing differences in competitiveness between plant species of contrasting nutrient-acquisition strategies.
With the advent of molecular-based techniques, the systematics and evolutionary understanding of the genus Phytophthora has advanced and the detection and description of new Phytophthora species have become a priority. However, more studies on pathogen-host-environment interactions are urgently required to increase our understanding of the ecology of cryptic endemic Phytophthora species in natural forest ecosystems.
Acknowledgments
We are grateful to the Portuguese Science and Technology Foundation (FCT) for co-financing with Portuguese national funds the European BiodivERsA project RESIPATH: Responses of European Forests and Society to Invasive Pathogens (BIODIVERSA/0002/2012), and to the Czech Ministry for Education, Youth and Sports and the European Regional Development Fund for financing the Project Phytophthora Research Centre Reg. No. CZ.02.1.01/0.0/0.0/15_003/0000453. Fieldwork in Portugal had logistic support from the Institute for the Conservation of Nature and Forestry (ICNF). DNA sequencing was partly supported by the Hungarian Scientific Research Fund (OTKA) grant K101914. This work has also received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 635646, POnTE (Pest Organisms Threatening Europe). Part of this study was also financially supported by the Sardinian Regional Government (Accordo di collaborazione tecnico-scientifica tra la Regione autonoma della Sardegna, Assessorato della difesa dell’ambiente, e il Dipartimento di Agraria dell’Università degli studi di Sassari per la realizzazione di attività di ricerca e sviluppo nel campo della pianificazione e programmazione della tutela fitosanitaria delle foreste sarde). We thank Treena Burgess (Murdoch University, Perth) for assistance with Fig. 7 and 8.
Supplementary Material
REFERENCES
- Abad GZ, Ivors KL, Gallup CA, Abad JA, Shew HD. (2011) Morphological and molecular characterization of Phytophthora glovera sp. nov. from tobacco in Brazil. Mycologia 103: 341–350. [DOI] [PubMed] [Google Scholar]
- Abad GZ, Abad JA, Cacciola SO, Pane A, Faedda R, et al. (2014) Phytophthora niederhauserii sp. nov., a polyphagous species associated with ornamentals, fruit trees and native plants in 13 countries. Mycologia 106: 431–447. [DOI] [PubMed] [Google Scholar]
- Albornoz FE, Burgess TI, Lambers H, Etchells H, Laliberté E. (2016) Native soil-borne pathogens equalise differences in competitive ability between plants of contrasting nutrient-acquisition strategies. Journal of Ecology 105: 549–557. [Google Scholar]
- Balci Y, Balci S, Eggers J, MacDonald WL, Juzwik J, et al. (2007) Phytophthora spp. associated with forest soils in Eastern and North-Central U.S. oak ecosystems. Plant Disease 91: 705–710. [DOI] [PubMed] [Google Scholar]
- Balci Y, Halmschlager E. (2003a) Incidence of Phytophthora species in oak forests in Austria and their possible involvement in oak decline. Forest Pathology 33: 157–174. [Google Scholar]
- Balci Y, Halmschlager E. (2003b) Phytophthora species in oak ecosystems in Turkey and their association with declining oak trees. Plant Pathology 52: 694–702. [Google Scholar]
- Balci Y, Long RP, Mansfield M, Balser D, MacDonald WL. (2010) Involvement of Phytophthora species in white oak (Quercus alba) decline in southern Ohio. Forest Pathology 40: 430–442. [Google Scholar]
- Barber P, Paap T, Burgess T, Dunstan W, Hardy GSJ. (2013) A diverse range of Phytophthora species are associated with dying urban trees. Urban Forestry and Urban Greening 12: 569–575. [Google Scholar]
- Bezuidenhout CM, Denman S, Kirk SA, Botha WJ, Mostert L, McLeod A. (2010) Phytophthora taxa associated with cultivated Agathosma, with emphasis on the P. citricola complex and P. capensis sp. nov. Persoonia 25: 32–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair JE, Coffey MD, Park S, Geiser DM, Kang S. (2008) A multilocus phylogeny for Phytophthora utilizing markers derived from complete genome sequences. Fungal Genetics and Biology 45: 266–277. [DOI] [PubMed] [Google Scholar]
- Brazee NJ, Wick RL, Hulvey JP. (2016) Phytophthora species recovered from the Connecticut River Valley in Massachusetts, USA. Mycologia 108: 6–19. [DOI] [PubMed] [Google Scholar]
- Burgess TI, White D, McDougall KM, Garnas J, Dunstan WA, et al. (2017) Distribution and diversity of Phytophthora across Australia. Pacific Conservation Biology 23: 1–13. [Google Scholar]
- Cohen Y, Coffey MD. (1986) Systemic fungicides and the control of Oomycetes. Annual Review of Phytopathology 24: 311–338. [Google Scholar]
- Cooke DEL, Drenth A, Duncan JM, Wagels G, Brasier CM. (2000) A molecular phylogeny of Phytophthora and related oomycetes. Fungal Genetic and Biology 30: 17–32. [DOI] [PubMed] [Google Scholar]
- Corcobado T, Miranda-Torres JJ, Martìn-Garcìa J, Jung T, Solla A. (2017) Early survival of Quercus ilex subspecies from different populations after infections and co-infections by multiple Phytophthora species. Plant Pathology 76: 792–804. [Google Scholar]
- Dick MW. (1990) Keys to Pythium. Reading: University of Reading Press. [Google Scholar]
- Dobrowolski MP, Tommerup IC, Blakeman HD. (2003) Non-mendelian inheritance revealed in a genetic analysis of sexual progeny of Phytophthora cinnamomi with microsatellite markers. Fungal Genetics and Biology 35: 197–212. [DOI] [PubMed] [Google Scholar]
- Erwin DC, Ribeiro OK. (1996) Phytophthora Diseases Worldwide. St Paul, MN: American Phytopathological Society Press. [Google Scholar]
- Gallegly ME, Hong C. (2008) Phytophthora: identifying species by morphology and DNA fingerprints. St Paul, MN: American Phytopathological Society Press. [Google Scholar]
- Goodwin SB, Spielman LJ, Matuszak JM, Bergeron SN, Fry WE. (1992) Clonal diversity and genetic differentiation of Phytophthora infestans populations in northern and central Mexico. Phytopathology 82: 955–61. [Google Scholar]
- Hansen EM, Reeser PW, Davidson JM, Garbelotto M, Ivors K, Douhan L, Rizzo DM. (2003) Phytophthora nemorosa, a new species causing cankers and leaf blight of forest trees in California and Oregon, U.S.A. Mycotaxon 88: 129–138. [Google Scholar]
- Hansen EM, Wilcox WF, Reeser PW, Sutton W. (2009) Phytophthora rosacearum and P. sansomeana, new species segregated from the Phytophthora megasperma ‘‘complex’’. Mycologia 101: 129–135. [DOI] [PubMed] [Google Scholar]
- Henricot B, Pérez-Sierra A, Jung T. (2014) Phytophthora pachypleura sp. nov., a new species causing root rot of Aucuba japonica and other ornamentals in the United Kingdom. Plant Pathology 63: 1095–1109. [Google Scholar]
- Herms DA, Mattson WJ. (1992) The dilemma of plants to grow or defend. Quarterly Review of Biology 67: 283–335. [Google Scholar]
- Huelsenbeck JP, Ronquist F. (2001) MrBayes: Bayesian inference of phylogeny. Bioinformatics 17: 754–755. [DOI] [PubMed] [Google Scholar]
- Husson C, Aguayo J, Revellin C, Frey P, Ioos R, et al. (2015) Evidence for homoploid speciation in Phytophthora alni supports taxonomic reclassification in this species complex. Fungal Genetics and Biology 77: 12–21. [DOI] [PubMed] [Google Scholar]
- Ilieva E, Man In ‘t Veld WA, Veenbaas-Rijks W, Pieters R. (1998) Phytophthora multivesiculata, a new species causing rot in Cymbidium. European Journal of Plant Pathology 104: 677–684. [Google Scholar]
- Jee H-J, Cho W-D, Kim W-G. (1997) Phytophthora diseases of apple in Korea: 2. Occurrence of an unusual fruit rot caused by P. cactorum and P. cambivora. Korean Journal Plant Pathology 13: 145–151. [Google Scholar]
- Józsa A, Bakonyi J, Belbahri L, Nagy ZÁ, Szigethy A, Bohár G, et al. (2010) A new species of Phytophthora reported to cause root and collar rot of common boxwood, Nordmann fir and Port Orford cedar in Hungary. Plant Pathology 59: 1166–1167. [Google Scholar]
- Jung T. (2009) Beech decline in Central Europe driven by the interaction between Phytophthora infections and climatic extremes. Forest Pathology 39: 73–94. [Google Scholar]
- Jung T, Blaschke H, Neumann P. (1996) Isolation, identification and pathogenicity of Phytophthora species from declining oak stands. European Journal of Forest Pathology 26: 253–272. [Google Scholar]
- Jung T, Blaschke H, Oßwald W. (2000) Involvement of soilborne Phytophthora species in Central European oak decline and the effect of site factors on the disease. Plant Pathology 49: 706–718. [Google Scholar]
- Jung T, Chang TT, Bakonyi J, Seress D, Pérez-Sierra A, et al. (2017a) Diversity of Phytophthora species in natural ecosystems of Taiwan and association with disease symptoms. Plant Pathology 66: 194–211. [Google Scholar]
- Jung T, Cooke DEL, Blaschke H, Duncan JM, Oßwald W. (1999) Phytophthora quercina sp. nov., causing root rot of European oaks. Mycological Research 103: 785–798. [Google Scholar]
- Jung T, Hansen EM, Winton L, Oßwald W, Delatour C. (2002) Three new species of Phytophthora from European oak forests. Mycological Research 106: 397–411. [Google Scholar]
- Jung T, Horta Jung M, Scanu B, Seress D, Kovács GM. et al. (2017b) Six new Phytophthora species from ITS Clade 7a including two sexually functional heterothallic hybrid species detected in natural ecosystems in Taiwan. Persoonia 38: 100–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung T, Nechwatal J, Cooke DEL, Hartmann G, Blaschke M, et al. (2003) Phytophthora pseudosyringae sp. nov., a new species causing root and collar rot of deciduous tree species in Europe. Mycological Research 107: 772–789. [DOI] [PubMed] [Google Scholar]
- Jung T, Orlikowski L, Henricot B, Campos P, Aday AG, et al. (2016) Widespread Phytophthora infestations in European nurseries put forest, semi-natural and horticultural ecosystems at high risk of Phytophthora diseases. Forest Pathology 46: 134–163. [Google Scholar]
- Jung T, Scanu B, Bakonyi J, Seress D, Kovács GM, et al. (2017c) Nothophytophthora gen. nov., a new sister genus of Phytophthora from natural and semi–natural ecosystems. Persoonia 39: 143–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung T, Vettraino AM, Cech TL, Vannini A. (2013) The impact of invasive Phytophthora species on European forests. In: Phytophthora: a global perspective (Lamour K, ed.): 146–158. Wallingford: CAB International. [Google Scholar]
- Katoh K, Standley DM. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer A, Abbott AG, Carlson JE, Manos PS, Plomion C, et al. (2012) Genomics of Fagaceae. Tree Genetics and Genomes 8: 583–610. [Google Scholar]
- Kroon LPNM, Bakker FT, van den Bosch GBM, Bonants PJM, Flier WG. (2004) Phylogenetic analysis of Phytophthora species based on mitochondrial and nuclear DNA sequences. Fungal Genetics and Biology 41: 766–782. [DOI] [PubMed] [Google Scholar]
- Logan WB. (2005) Oak: the frame of civilization. New York: W W Norton. [Google Scholar]
- Man in ‘t Veld WA. (2007) Gene flow analysis demonstrates that Phytophthora fragariae var. rubi constitutes a distinct species, Phytophthora rubi comb. nov. Mycologia 99: 222–226 [DOI] [PubMed] [Google Scholar]
- Manos PS, Stanford AM. (2001) The biogeography of Fagaceae: tracking the Tertiary history of temperate and subtropical forests of the Northern Hemisphere. International Journal of Plant Science 162: S77–S93. [Google Scholar]
- Martin FN, Tooley PW. (2003) Phylogenetic relationships among Phytophthora species inferred from sequence analysis of mitochondrially encoded cytochrome oxidase I and II genes. Mycologia 95: 269–284. [PubMed] [Google Scholar]
- Martin FN, Blair JE, Coffey MD. (2014) A combined mitochondrial and nuclear multilocus phylogeny of the genus Phytophthora. Fungal Genetics and Biology 66: 19–32. [DOI] [PubMed] [Google Scholar]
- Martìn-Garcìa J, Solla A, Corcobado T, Siasou E, Woodward S. (2015) Influence of temperature on germination of Quercus ilex in Phytophthora cinnamomi, P. gonapodyides, P. quercina and P. psychrophila infested soils. Forest Pathology 45: 215–223. [Google Scholar]
- Nagy ZÁ, Bakonyi J, Érsek T. (2003) Standard and Swedish variant types of the hybrid alder Phytophthora attacking alder in Hungary. Pest Management Science 59: 484–492. [DOI] [PubMed] [Google Scholar]
- Oudemans P, Coffey MD. (1991) Isozyme comparison within and among worldwide sources of three morphologically distinct species of Phytophthora. Mycological Research 95: 19–30. [Google Scholar]
- Paap T, Croeser L, White D, Aghighi S, Barber P, St. J. Hardy GE, Burgess TI. (2017) Phytophthora versiformis sp. nov., a new species from Australia related to P. quercina. Australasian Plant Pathology: DOI 10.1007/s13313-017-0499-7. [Google Scholar]
- Pánek M, Fér T, Mráček J, Tomšovský M. (2016) Evolutionary relationships within the Phytophthora cactorum species complex in Europe. Fungal Biology 120: 836-851. [DOI] [PubMed] [Google Scholar]
- Pérez-Sierra A, López-García C, León M, García-Jiménez J, Abad-Campos P, et al. (2013) Previously unrecorded low temperature Phytophthora species associated with Quercus decline in a Mediterranean forest in Eastern Spain. Forest Pathology 43: 331–339. [Google Scholar]
- Rahman MZ, Uematsu S, Kimishima E, Kanto T, Kusunoki M, et al. (2015) Two plant pathogenic species of Phytophthora associated with stem blight of Easter lily and crown rot of lettuce in Japan. Mycoscience 56: 419–433. [Google Scholar]
- Rea AJ, Burgess TI, Hardy GEStJ, Stukely MJC, Jung T. (2011) Two novel and potentially endemic species of Phytophthora associated with episodic dieback of kwongan vegetation in the south-west of Western Australia. Plant Pathology 60: 1055–1068. [Google Scholar]
- Robideau GP, de Cock AWAM, Coffey MD, Voglmayr H, Brouwer H, et al. (2011) DNA barcoding of oomycetes with cytochrome c oxidase subunit I (COI) and internal transcribed spacer (ITS). Molecular Ecology Resources 11: 1002–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Heuelsenbeck JP. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Saavedra A, Hansen EM, Goheen DJ. (2007) Phytophthora cambivora in Oregon and its pathogenicity to Chrysolepis chrysophylla. Forest Pathology 37: 409–419. [Google Scholar]
- Scanu B, Linaldeddu BT, Franceschini A. (2010) First report of Phytophthora pseudosyringae associated with ink disease of Castanea sativa in Italy. Plant Disease 94: 1068. [DOI] [PubMed] [Google Scholar]
- Scanu B, Hunter GC, Linaldeddu BT, Franceschini A, Maddau L, et al. (2014a) A taxonomic re-evaluation reveals that Phytophthora cinnamomi and P. cinnamomi var. parvispora are separate species. Forest Pathology 44: 1–20. [Google Scholar]
- Scanu B, Linaldeddu BT, Deidda A, Jung T. (2015) Diversity of Phytophthora species from declining Mediterranean maquis vegetation, including two new species, Phytophthora crassamura and P. ornamentata sp. nov. PLoS ONE 10 (12): e0143234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanu B, Linaldeddu BT, Franceschini A, Anselmi N, Vannini A, et al. (2013) Occurrence of Phytophthora cinnamomi in cork oak forests in Italy. Forest Pathology 43: 340–343. [Google Scholar]
- Scanu B, Linaldeddu BT, Peréz-Sierra A, Deidda A, Franceschini A. (2014b) Phytophthora ilicis as a leaf and stem pathogen of Ilex aquifolium in Mediterranean islands. Phytopathologia Mediterranea 53: 480–490. [Google Scholar]
- Scanu B, Webber JF. (2016) Dieback and mortality of Nothofagus in Britain: ecology, pathogenicity and sporulation potential of the causal agent Phytophthora pseudosyringae. Plant Pathology 65: 26–36. [Google Scholar]
- Schirone B, Radoglou K, Vessella F. (2016) Conservation and restoration strategies to preserve the variability of cork oak Quercus suber–a Mediterranean forest species-under global warming. Climate Research 71 (2): 171–185. [Google Scholar]
- Schwingle BW, Smith JA, Blanchette RA. (2007) Phytophthora species associated with diseased woody ornamentals in Minnesota nurseries. Plant Disease 91: 97–102. [DOI] [PubMed] [Google Scholar]
- Scott PM, Burgess TI, Barber PA, Shearer BL, Stukely MJC, Hardy GEStJ, Jung T. (2009) Phytophthora multivora sp. nov., a new species recovered from declining Eucalyptus, Banksia, Agonis and other plant species in Western Australia. Persoonia 22: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvestro D, Michalak I. (2012) raxmlGUI: a graphical front-end for RAxML. Organisms, Diversity and Evolution 12: 335–337. [Google Scholar]
- Simamora AV, Stukely MJC, Hardy GEStJ, Burgess TI. (2015) Phytophthora boodjera sp. nov., a damping-off pathogen in production nurseries and from urban and natural landscapes, with an update on the status of P. alticola. IMA Fungus 6: 319–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R, Beal KF, Bonfield JK. (2000) The Staden package 1998. Methods in Molecular Biology 132: 115–130. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. (2013) MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Molecular Biology and Evolution 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang QH, Gao F, Li GY, Wang H, Zheng XB, Wang YC. (2010) First report of root rot caused by Phytophthora sansomeana on soybean in China. Plant Disease 94: 378. [DOI] [PubMed] [Google Scholar]
- Weir BS, Paderes EP, Anand N, Uchida JY, Pennycook SR, Bellgard SE, Beever RE. (2015) A taxonomic revision of Phytophthora Clade 5 including two new species, Phytophthora agathidicida and P. cocois. Phytotaxa 205: 21–38. [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR Protocols: a guide to methods and applications (Innis MA, Gelfand DH, Sninsky JJ, White TJ, eds): 315–322. San Diego: Academic Press. [Google Scholar]
- Yang X, Copes WE, Hong C. (2014) Two novel species representing a new clade and cluster of Phytophthora. Fungal Biology 118: 72–82. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


