Abstract
Species of Diaporthe are considered important plant pathogens, saprobes, and endophytes on a wide range of plant hosts. Several species are well-known on citrus, either as agents of pre- or post-harvest infections, such as dieback, melanose and stem-end rot on fruit. In this study we explored the occurrence, diversity and pathogenicity of Diaporthe species associated with Citrus and allied genera in European orchards, nurseries, and gardens. Surveys were carried out during 2015 and 2016 in Greece, Italy, Malta, Portugal, and Spain. A total of 79 Diaporthe strains were isolated from symptomatic twigs, branches and trunks. A multi-locus phylogeny was established based on five genomic loci (ITS, tef1, cal, his3 and tub2), and the morphological characters of the isolates determined. Preliminary pathogenicity tests were performed on lemon, lime, and orange plants with representative isolates. The most commonly isolated species were D. foeniculina and D. baccae, while only four isolates of D. novem were collected. Two new Diaporthe species, described here as D. limonicola and D. melitensis spp. nov. were found associated with a new devastating dieback disease of lemon plants. Furthermore, one cluster of sterile Diaporthe isolates was renamed as D. infertilis. Pathogenicity tests revealed most of the Citrus species as susceptible to D. baccae, D. foeniculina, and D. novem. Moreover, D. limonicola and D. melitensis caused serious cankers affecting all the Citrus species tested. This study is the first report of D. baccae and D. novem on citrus in Europe, and the first detection of a new Diaporthe canker disease of citrus in Europe. However, no isolates of D. citri were found. The study improves our understanding of the species associated with several disease symptoms on citrus plants, and provides useful information for effective disease management.
Keywords: Canker, Citrus, multi-locus sequence typing, pathogenicity
INTRODUCTION
Diaporthe species are present worldwide as plant pathogens and endophytes in healthy leaves, stems, seeds and roots, or as saprobes on decaying tissues of a wide range of hosts (Muralli et al. 2006, Garcia-Reyne et al. 2011, Udayanga et al. 2011). Diaporthe species are well-known as the causal agents of many important plant diseases, including root and fruit rots, dieback, stem cankers, leaf spots, leaf and pod blights, and seed decay (Uecker 1988, Mostert et al. 2001a, b, Van Rensburg et al. 2006, Rehner & Uecker 1994, Santos et al. 2011, Udayanga et al. 2011, Diaz et al. 2017). Species of Diaporthe have also been extensively screened in bioassays for natural products (Isaka et al. 2001, Dai et al. 2005, Kumaran & Hur 2009, Yang et al. 2010), and for the biocontrol of fungal pathogens (Santos et al. 2016).
The generic names Diaporthe and Phomopsis are no longer used to distinguish different morphs of this genus, and recent studies (Rossman et al. 2015) have recommended that Diaporthe be adopted as the correct generic name as it has priority over Phomopsis.
Diaporthe was historically considered monophyletic based on the typical Phomopsis asexual morph and diaporthalean sexual morph (Gomes et al. 2013). However, the paraphyletic nature was recently revealed by Gao et al. (2017), who demonstrated that Ophiodiaporthe (Fu et al. 2013), Pustulomyces (Dai et al. 2014), Phaeocytostroma, and Stenocarpella (Lamprecht et al. 2011), are embedded in Diaporthe s. lat. To address this issue, Senanayake et al. (2017) subsequently named several additional diaporthe-like clades within Diaporthales.
The taxonomy of Diaporthe species has been reviewed in several major studies (Thompson et al. 2011, 2014, Gomes et al. 2013, Udayanga et al. 2014a, b, 2015). Almost 2000 species names are available for both Diaporthe and Phomopsis (Index Fungorum; http://www.indexfungorum.org). The majority of the known species in early literature were described in relation to their host association (Uecker 1988), except for about 150 species that have been described more recently supported by molecular data (Gomes et al. 2013, Lombard et al. 2014, Udayanga et al. 2014a, b, 2015). However, most Diaporthe species can be found on diverse hosts, and can co-occur on the same host or lesion in different life modes (Rehner & Uecker 1994, Mostert et al. 2001a, Guarnaccia et al. 2016). This is demonstrated by D. foeniculina, usually known as an opportunistic pathogen of various herbaceous weeds, ornamentals, and fruit trees including citrus (Santos & Phillips 2009, Udayanga et al. 2014b). However, it has also been isolated from tropical trees as an endophyte, and from herbaceous plants and weeds as a pathogen or saprobe (Udayanga et al. 2014a). As a consequence, identification and description of species based on host association alone is no longer tenable within Diaporthe (Gomes et al. 2013, Udayanga et al. 2014a, b).
Before the molecular era, morphological characters such as immersed ascomata and erumpent pseudostroma with elongated perithecial necks in the sexual morph (Udayanga et al. 2011), and black conidiomata with dimorphic conidia in the asexual morph (Rehner & Uecker 1994), was the basis on which to study the taxonomy of Diaporthe (Van der Aa et al. 1990). Recent studies demonstrated that these characters are not always reliable for species level identification due to their variability under changing environmental conditions (Gomes et al. 2013).
Following the adoption of DNA sequence-based methods, the polyphasic protocols for studying the genus significantly changed the classification and species concepts, resulting in a rapid increase in the description of novelties. Therefore, genealogical concordance methods, based on multi-gene DNA sequence data, provide a much clearer approach to resolving the taxonomy for Diaporthe.
Recent plant pathological studies have shown several Diaporthe species to be particularly important on a wide range of economically significant agricultural crops, such as blueberries, citrus, grapes, oaks, sunflowers, soybeans, tea plants, tropical fruits, vegetables, and various trees (Van Rensburg et al. 2006, Crous et al. 2011a, b, 2016, Thompson et al. 2011, Santos & Phillips 2009, Santos et al. 2011, Grasso et al. 2012, Huang et al. 2013, Lombard et al. 2014, Gao et al. 2015, 2016, Udayanga et al. 2015, Guarnaccia et al. 2016). Furthermore, several Citrus species are colonized and/or affected by different Diaporthe species (Timmer et al. 2000, Huang et al. 2013), which are focussed on here.
BACKGROUND
Citrus represents one of the most important fruit industries worldwide. In the Mediterranean region, Greece, Italy, Portugal, and Spain especially are important producers of citrus fruits, and are the biggest fruit exporter after South Africa (FAO 2016). Therefore, recognizing the pathogens affecting these crops in these countries is imperative.
Diaporthe citri is a well-known pathogen causing melanose and stem-end rot disease of Citrus species in several regions (Timmer 2000, Mondal et al. 2007). Several additional Diaporthe species have been reported associated with Citrus (often as Phomopsis) and have previously been considered as synonyms of D. citri, such as D. citrincola described from the Philippines, P. californica from California, P. caribaea from Cuba, and P. cytosporella from Italy (Rehm 1914, Fawcett 1922). Wehmeyer (1933) also considered D. medusaea, D. californica, P. citri, and P. citrincola as synonyms of Diaporthe citri.
Polyphasic approaches in recent years have revealed many species associated with citrus. Huang et al. (2013) reported D. citri as the predominant species in China and described two new taxa: D. citriasiana and D. citrichinensis. In another study, Huang et al. (2015) identified several Diaporthe species as endophytes of citrus but which had previously been recovered from other hosts, such as D. endophytica, D. eres, D. hongkongensis, D. sojae, and the different taxa clustering in the D. arecae species complex. Moreover, they described D. biconispora, D. biguttulata, D. discoidispora, D. multigutullata, D. ovalispora, D. subclavata, and D. unshiuensis as new species occurring on citrus. Several strains from China, Korea, New Zealand, and the USA have been re-assessed by Udayanga et al. (2014b) within D. citri, which was also epitypified. In the same study, D. cytosporella was recovered from specimens of Citrus limon, C. limonia, and C. sinensis collected respectively in Spain, Italy, and the USA, and D. foeniculina has also been widely associated with citrus.
Diaporthe citri is generally accepted as an important pathogen of citrus, causing stem-end rot and melanose of fruits, young leaf and shoot gummosis, and blight of perennial branches and trunks (Kucharek et al. 1983, Timmer & Kucharek 2001, Mondal et al. 2007, Udayanga et al. 2014b). This species occurs in many citrus growing regions of the world on several Citrus species, including C. limon, C. paradisi, C. reticulata, and C. sinensis (Timmer et al. 2000).
Further infections involving twigs, perennial branches and trunks of citrus are caused by other Diaporthe species, such as cankers developing in woody tissues, often with a gummose exudate, generating serious blight and dieback (Huang et al. 2013, Mahadevakumar et al. 2014). Canker diseases of citrus are also caused by other fungal genera such as Fusarium and Neocosmospora (Sandoval-Denis et al. 2018), and species of Botryosphaeriaceae and Diatrypaceae (Timmer et al. 2000, Polizzi et al. 2009, Mayorquin et al. 2016).
Although the biology and epidemiology of melanose are well studied also with a robust phylogenetic relationship of the causal organisms, genetic variability and population structure (Burnett 1962, Mondal et al. 2004, 2007, Udayanga et al. 2014b), the identification of Diaporthe species associated with citrus cankers and dieback has not been well resolved. Moreover, Gomes et al. (2013) performed a major phylogenetic and morphological study of Diaporthe species and grouped three isolates, one of which was collected from Citrus sinensis in Suriname, under D. citri. However, Udayanga et al. (2014b) re-assessed D. citri based on molecular phylogenetic analysis of conserved ex-type and additional strains collected exclusively from symptomatic citrus tissues in different geographic locations worldwide. Furthermore, according to this latter study, D. citri is unknown in Europe. Because of all these findings, changes in species concepts and poor investigation of Diaporthe on citrus in Europe, new surveys were required to study Diaporthe species diversity related to citrus and their occurrence and association with diseases.
The current study aims to investigate the major citrus production areas in Europe by employing large-scale sampling to isolate Diaporthe strains, and to identify the strains obtained in the light of modern taxonomic concepts via morphological characterization and multi-locus DNA sequence data. In 2015 and 2016, several surveys were conducted in commercial nurseries, citrus orchards, gardens, backyards, and plant collections to determine the occurrence of Diaporthe species associated with Citrus and allied genera (e.g. Microcitrus). In particular the objectives of the present study were to: (1) conduct extensive surveys for sampling symptomatic plant materials; (2) cultivate as many Diaporthe isolates as possible; (3) subject those isolates to DNA sequence analyses combined with morphological characterization; (4) compare the obtained results with the data from other phylogenetic studies on the genus; (5) place three strains previously named as D. citri in the correct taxonomic context based on DNA sequence inference; and (6) evaluate the pathogenicity of the isolated Diaporthe species to citrus plants.
MATERIAL AND METHODS
Sampling and isolation
During 2015 and 2016 many regions of the main citrus-producing area of Europe were surveyed (Guarnaccia et al. 2017a, b). Twig, branch and trunk portions showing cankers and dieback were collected from more than 90 sites in: Andalusia, Valencia, and the Balearic Islands (Spain); Apulia, Calabria, Sicily, and the Aeolian Islands (Italy); Algarve (Portugal); Arta, Crete, Missolonghi, and Nafplio (Greece); and Malta and Gozo (Malta). Investigated species of Citrus and allied genera such as Microcitrus (Rutaceae) included Australasian lime, citrons, kumquat, mandarins, oranges, pumelo, grapefruit, limes, and lemons.
Wood fragments (5 × 5 mm) were cut from the margin between affected and healthy tissues and washed in running tap water. Then, each fragment was surface sterilised by soaking in 70 % ethanol for 5 s, 4 % sodium hypochlorite for 90 s, sterile water for 60 s (Kumaresan & Suryanarayanan 2001) and then dried on sterile filter paper. The fragments were placed on malt extract agar (MEA; Crous et al. 2009) amended with 100 μg / mL penicillin and 100 μg / mL streptomycin (MEA-PS) and incubated at 25 °C until characteristic Diaporthe colonies were observed. In a second procedure, plant material was incubated in moist chambers at room temperature (20 ± 3 °C) for up to 10 d and inspected daily for fungal sporulation. Sporulating conidiomata obtained through both procedures were collected and crushed in a drop of sterile water and then spread over the surface of MEA-PS plates. After 24 h germinating spores were individually transferred onto MEA plates. The isolates used in this study are maintained in the culture collection of the Westerdijk Fungal Biodiversity Institute (CBS), Utrecht, The Netherlands, and in the working collection of Pedro Crous (CPC), housed at the Westerdijk Institute.
DNA extraction, PCR amplification and sequencing
Genomic DNA was extracted using a Wizard® Genomic DNA Purification Kit (Promega, WI) following the manufacturer’s instructions. Partial regions of six loci were amplified. The primers ITS5 and ITS4 (White et al. 1990) were used to amplify the ITS region of the nuclear ribosomal RNA operon, including the 3’ end of the 18S rRNA, the first internal transcribed spacer region, the 5.8S rRNA gene; the second internal transcribed spacer region and the 5’ end of the 28S rRNA gene. The primers EF1-728F and EF1-986R (Carbone & Kohn 1999) were used to amplify part of the translation elongation factor 1-α gene (tef1). Primers CAL-228F and CAL-737R (Carbone & Kohn 1999) or CL1/ CL2A (O’Donnell et al. 2000) were used to amplify part of the calmodulin (cal) gene. The partial histone H3 (his3) region was amplified using CYLH3F and H3-1b primer sets (Glass & Donaldson 1995, Crous et al. 2004a), and the beta-tubulin (tub2) region was amplified using Bt2a and Bt2b primer sets (Glass & Donaldson 1995). The PCR products were sequenced in both directions using the BigDye® Terminator v. 3.1 Cycle Sequencing Kit (Applied Biosystems Life Technologies, Carlsbad, CA), after which amplicons were purified through Sephadex G-50 Fine columns (GE Healthcare, Freiburg) in MultiScreen HV plates (Millipore, Billerica, MA). Purified sequence reactions were analysed on an Applied Biosystems 3730xl DNA Analyzer (Life Technologies, Carlsbad, CA). The DNA sequences generated were analysed and consensus sequences were computed using SeqMan Pro (DNASTAR, Madison, WI).
Phylogenetic analyses
New sequences generated in this study were blasted against the NCBI’s GenBank nucleotide database to determine the closest relatives for a taxonomic framework of the studied isolates. Alignments of different gene regions, including sequences obtained from this study and sequences downloaded from GenBank, were initially performed with the MAFFT v. 7 online server (http://mafft.cbrc.jp/alignment/server/index.html) (Katoh & Standley 2013), and then manually adjusted in MEGA v. 7 (Kumar et al. 2016).
To establish the identity of the isolates at species level, phylogenetic analyses were conducted first individually for each locus (data not shown) and then as combined analyses of five loci. One analysis was performed for all the Diaporthe isolates recovered from samples collected during the surveys conducted for this study. Additional reference sequences were selected based on recent studies of Diaporthe species (Gomes et al. 2013, Huang et al. 2013, Udayanga et al. 2014a, b). Phylogenetic analyses were based on Maximum Parsimony (MP) for all the individual loci and on both MP and Bayesian Inference (BI) for the multi-locus analyses. For BI, the best evolutionary model for each partition was determined using MrModeltest v. 2.3 (Nylander 2004) and incorporated into the analyses. MrBayes v. 3.2.5 (Ronquist et al. 2012) was used to generate phylogenetic trees under optimal criteria per partition. The Markov Chain Monte Carlo (MCMC) analysis used four chains and started from a random tree topology. The heating parameter was set to 0.2 and trees were sampled every 1000 generations. Analyses stopped once the average standard deviation of split frequencies was below 0.01. The MP analyses were done using PAUP (Phylogenetic Analysis Using Parsimony, v. 4.0b10; Swofford 2003). Phylogenetic relationships were estimated by heuristic searches with 100 random addition sequences. Tree bisection-reconnection was used, with the branch swapping option set on “best trees” only with all characters weighted equally and alignment gaps treated as fifth state. Tree length (TL), consistency index (CI), retention index (RI) and rescaled consistence index (RC) were calculated for parsimony and the bootstrap analyses (Hillis & Bull 1993) were based on 1000 replications. Sequences generated in this study are deposited in GenBank (Table 1) and alignments and phylogenetic trees in TreeBASE (www.treebase.org).
Table 1.
Collection details and GenBank accession numbers of isolates included in this study.
| Species | Culture no.1 | Host | Locality | Associated symptoms |
GenBank no.2 |
||||
|---|---|---|---|---|---|---|---|---|---|
| ITS | tub2 | his3 | tef1 | cal | |||||
| D. angelicae | CBS 111592 | Heracleum sphondylium | Austria | - | KC343026 | KC343994 | KC343511 | KC343752 | KC343268 |
| D. arecae | CBS 161.64 | Areca catechu | India | - | KC343032 | KC344000 | KC343516 | KC343758 | KC343274 |
| CBS 535.75 | Citrus sp. | Suriname | - | KC343033 | KC344001 | KC343517 | KC343759 | KC343275 | |
| D. arengae | CBS 114979 | Arenga engleri | Hong Kong | - | KC343034 | KC344002 | KC343518 | KC343760 | KC343276 |
| D. baccae | CBS 136972 | Vaccinium corymbosum | Italy | - | KJ160565 | MF418509 | MF418264 | KJ160597 | - |
| CPC 26170 = CBS 142545 | Citrus sinensis ‘Tarocco Tapi’ | Italy, Catania | Twig dieback | MF418351 | MF418510 | MF418265 | MF418430 | MF418185 | |
| CPC 26465 | Citrus limon | Italy, Catania | Branch canker | MF418352 | MF418511 | MF418266 | MF418431 | MF418186 | |
| CPC 26963 | Citrus paradisi | Italy, Vibo Valentia | Branch canker | MF418353 | MF418512 | MF418267 | MF418432 | MF418187 | |
| CPC 27029 | Citrus sinensis | Italy, Vibo Valentia | Twig dieback | MF418354 | MF418513 | MF418268 | MF418433 | MF418188 | |
| CPC 27075 | Citrus limon | Italy, Vibo Valentia | Twig dieback | MF418355 | MF418514 | MF418269 | MF418434 | MF418189 | |
| CPC 27079 | Citrus limon | Italy, Vibo Valentia | Twig dieback | MF418356 | MF418515 | MF418270 | MF418435 | MF418190 | |
| CPC 27821 | Citrus reticulata ‘Caffin’ | Italy, Cosenza | Trunk canker | MF418357 | MF418516 | MF418271 | MF418436 | MF418191 | |
| CPC 27831 = CBS 142546 | Citrus sinensis | Italy, Catania | Trunk canker | MF418358 | MF418517 | MF418272 | MF418437 | MF418192 | |
| CPC 27834 | Citrus sinensis | Italy, Catania | Trunk canker | MF418359 | MF418518 | MF418273 | MF418438 | MF418193 | |
| CPC 27835 | Citrus sinensis | Italy, Catania | Trunk canker | MF418360 | MF418519 | MF418274 | MF418439 | MF418194 | |
| CPC 27836 | Citrus sinensis | Italy, Catania | Trunk canker | MF418361 | MF418520 | MF418275 | MF418440 | MF418195 | |
| CPC 27837 | Citrus sinensis | Italy, Catania | Trunk canker | MF418362 | MF418521 | MF418276 | MF418441 | MF418196 | |
| CPC 27850 | Citrus sinensis | Italy, Catania | Twig dieback | MF418363 | MF418522 | MF418277 | MF418442 | MF418197 | |
| CPC 27852 | Citrus sinensis | Italy, Catania | Twig dieback | MF418364 | MF418523 | MF418278 | MF418443 | MF418198 | |
| D. biconispora | ICMP20654 | Citrus grandis | China | - | KJ490597 | KJ490418 | KJ490539 | KJ490476 | - |
| D. biguttulata | ICMP20657 | Citrus limon | China | - | KJ490582 | KJ490403 | KJ490524 | KJ490461 | - |
| D. citri | CBS 134237 | Citrus reticulata | China | - | JQ954660 | KC357426 | MF418279 | JQ954676 | KC357465 |
| CBS 134239 | Citrus sinensis | Florida, USA | - | KC357553 | KC357456 | MF418280 | KC357522 | KC357488 | |
| CBS 135422 | Citrus sp. | USA | - | KC843311 | KC843187 | MF418281 | KC843071 | KC843157 | |
| D. citriasiana | CBS 134240 | Citrus unshiu | China | - | JQ954645 | KC357459 | MF418282 | JQ954663 | KC357491 |
| D. citrichinensis | CBS 134242 | Citrus sp. | China | - | JQ954648 | MF418524 | KJ420880 | JQ954666 | KC357494 |
| D. cuppatea | CBS 117499 | Aspalathus linearis | South Africa | - | AY339322 | JX275420 | KC343541 | AY339354 | JX197414 |
| D. cytosporella | CBS 137020 | Citrus limon | Spain | - | KC843307 | KC843221 | MF418283 | KC843116 | KC843141 |
| D. discoidispora | ICMP20662 | Citrus unshiu | China | - | KJ490624 | KJ490445 | KJ490566 | KJ490503 | - |
| D. endophytica | ZJUD73 | Citrus unshiu | China | - | KJ490608 | KJ490429 | KJ490550 | KJ490487 | - |
| D. eres | CBS 439.82 | Cotoneaster sp. | Scotland | - | KC343090 | KC344058 | KC343574 | KC343816 | KC343332 |
| D. foeniculina | CBS 187.27 | Camellia sinensis | Italy | - | KC343107 | KC344075 | KC343591 | KC343833 | KC343349 |
| CBS 111553 | Foeniculum vulgare | Spain | - | KC343101 | KC344069 | KC343585 | KC343827 | KC343343 | |
| CBS 111554 | Foeniculum vulgare | Portugal | - | KC343102 | KC344070 | KC343586 | KC343828 | KC343344 | |
| CBS 123208 | Foeniculum vulgare | Portugal | - | KC343104 | KC344072 | KC343588 | KC343830 | KC343346 | |
| CBS 123209 | Foeniculum vulgare | Portugal | - | KC343105 | KC344073 | KC343589 | KC343831 | KC343347 | |
| CBS 135430 | Citrus limon | USA | - | KC843301 | KC843215 | MF418284 | KC843110 | KC843135 | |
| CPC 26184 | Citrus maxima | Italy, Messina | Branch canker | MF418365 | MF418525 | MF418285 | MF418444 | MF418199 | |
| CPC 26194 | Citrus sinensis ‘Sanguinello’ | Italy, Messina | Branch canker | MF418366 | MF418526 | MF418286 | MF418445 | MF418200 | |
| CPC 26365 | Citrus limon | Italy, Catania | Twig dieback | MF418367 | MF418527 | MF418287 | MF418446 | MF418201 | |
| CPC 26439 | Citrus reticulata | Italy, Catania | Twig dieback | MF418368 | MF418528 | MF418288 | MF418447 | MF418202 | |
| CPC 26441 | Citrus reticulata | Italy, Catania | Twig dieback | MF418369 | MF418529 | MF418289 | MF418448 | MF418203 | |
| CPC 26461 | Citrus reticulata | Italy, Catania | Twig dieback | MF418370 | MF418530 | MF418290 | MF418449 | MF418204 | |
| CPC 26863 | Citrus maxima | Greece, Missolonghi | Branch canker | MF418371 | MF418531 | MF418291 | MF418450 | MF418205 | |
| CPC 26873 | Citrus reticulata | Greece, Arta | Twig dieback | MF418372 | MF418532 | MF418292 | MF418451 | MF418206 | |
| CPC 26883 | Citrus maxima | Greece, Missolonghi | Branch canker | MF418373 | MF418533 | MF418293 | MF418452 | MF418207 | |
| CPC 26885 | Citrus bergamia | Greece, Missolonghi | Branch canker | MF418374 | MF418534 | MF418294 | MF418453 | MF418208 | |
| CPC 26913 | Citrus limon | Greece, Missolonghi | Branch canker | MF418375 | MF418535 | MF418295 | MF418454 | MF418209 | |
| CPC 26923 | Citrus maxima | Greece, Missolonghi | Branch canker | MF418376 | MF418536 | MF418296 | MF418455 | MF418210 | |
| CPC 26927 | Citrus maxima | Greece, Missolonghi | Branch canker | MF418377 | MF418537 | MF418297 | MF418456 | MF418211 | |
| CPC 26953 | Citrus bergamia | Greece, Missolonghi | Branch canker | MF418378 | MF418538 | MF418298 | MF418457 | MF418212 | |
| CPC 26967 | Citrus mitis | Italy, Messina | Twig dieback | MF418379 | MF418539 | MF418299 | MF418458 | MF418213 | |
| CPC 26971 | Citrus mitis | Italy, Messina | Twig dieback | MF418380 | MF418540 | MF418300 | MF418459 | MF418214 | |
| CPC 27027 | Citrus limon | Italy, Cosenza | Branch canker | MF418381 | MF418541 | MF418301 | MF418460 | MF418215 | |
| CPC 27033 | Citrus mitis | Italy, Messina | Twig dieback | MF418382 | MF418542 | MF418302 | MF418461 | MF418216 | |
| CPC 27037 | Citrus paradisi | Italy, Vibo Valentia | Branch canker | MF418383 | MF418543 | MF418303 | MF418462 | MF418217 | |
| CPC 27041 | Citrus sinensis | Italy, Cosenza | Branch canker | MF418384 | MF418544 | MF418304 | MF418463 | MF418218 | |
| CPC 27167 | Citrus paradisi | Italy, Vibo Valentia | Branch canker | MF418385 | MF418545 | MF418305 | MF418464 | MF418219 | |
| CPC 27756 | Citrus limon | Italy, Catania | Trunk canker | MF418386 | MF418546 | MF418306 | MF418465 | MF418220 | |
| CPC 27832 | Citrus sinensis | Italy, Catania | Trunk canker | MF418387 | MF418547 | MF418307 | MF418466 | MF418221 | |
| CPC 27833 | Citrus sinensis | Italy, Catania | Trunk canker | MF418388 | MF418548 | MF418308 | MF418467 | MF418222 | |
| CPC 27859 | Citrus paradisi | Malta, Gozo | Trunk canker | MF418389 | MF418549 | MF418309 | MF418468 | MF418223 | |
| CPC 27877 | Citrus limon | Malta, Gozo | Trunk canker | MF418390 | MF418550 | MF418310 | MF418469 | MF418224 | |
| CPC 27895 | Citrus japonica | Malta, Gozo | Twig dieback | MF418391 | MF418551 | MF418311 | MF418470 | MF418225 | |
| CPC 27896 | Citrus japonica | Malta, Gozo | Twig dieback | MF418392 | MF418552 | MF418312 | MF418471 | MF418226 | |
| CPC 27897 | Citrus japonica | Malta, Gozo | Twig dieback | MF418393 | MF418553 | MF418313 | MF418472 | MF418227 | |
| CPC 27898 | Citrus japonica | Malta, Gozo | Twig dieback | MF418394 | MF418554 | MF418314 | MF418473 | MF418228 | |
| CPC 27901 | Citrus limon | Malta, Gozo | Branch canker | MF418395 | MF418555 | MF418315 | MF418474 | MF418229 | |
| CPC 27903 | Citrus limon | Malta, Gozo | Branch canker | MF418396 | MF418556 | MF418316 | MF418475 | MF418230 | |
| CPC 27945 | Citrus paradisi | Portugal, Faro | Branch canker | MF418397 | MF418557 | MF418317 | MF418476 | MF418231 | |
| CPC 27947 | Citrus sinensis | Portugal, Faro | Branch canker | MF418398 | MF418558 | MF418318 | MF418477 | MF418232 | |
| CPC 27949 | Citrus sinensis | Portugal, Faro | Branch canker | MF418399 | MF418559 | MF418319 | MF418478 | MF418233 | |
| CPC 27950 | Citrus sinensis | Portugal, Faro | Twig dieback | MF418400 | MF418560 | MF418320 | MF418479 | MF418234 | |
| CPC 27959 | Citrus sinensis | Portugal, Faro | Twig dieback | MF418401 | MF418561 | MF418321 | MF418480 | MF418235 | |
| CPC 28033 = CBS 142547 | Citrus sinensis ‘Valencia’ | Portugal, Mesquita | Twig dieback | MF418402 | MF418562 | MF418322 | MF418481 | MF418236 | |
| CPC 28035 | Citrus paradisi | Portugal, Faro | Twig dieback | MF418403 | MF418563 | MF418323 | MF418482 | MF418237 | |
| CPC 28039 | Citrus limon | Portugal, Monchique | Twig dieback | MF418404 | MF418564 | MF418324 | MF418483 | MF418238 | |
| CPC 28041 | Citrus limon | Portugal, Monchique | Twig dieback | MF418405 | MF418565 | MF418325 | MF418484 | MF418239 | |
| CPC 28043 | Citrus limon | Portugal, Monchique | Twig dieback | MF418406 | MF418566 | MF418326 | MF418485 | MF418240 | |
| CPC 28045 | Citrus limon | Portugal, Monchique | Twig dieback | MF418407 | MF418567 | MF418327 | MF418486 | MF418241 | |
| CPC 28047 | Citrus limon | Portugal, Monchique | Twig dieback | MF418408 | MF418568 | MF418328 | MF418487 | MF418242 | |
| CPC 28071 | Citrus limon | Spain, Algemesi | Twig dieback | MF418409 | MF418569 | MF418329 | MF418488 | MF418243 | |
| CPC 28072 | Citrus limon | Spain, Algemesi | Twig dieback | MF418410 | MF418570 | MF418330 | MF418489 | MF418244 | |
| CPC 28073 | Citrus reticulata | Spain, Algemesi | Twig dieback | MF418411 | MF418571 | MF418331 | MF418490 | MF418245 | |
| CPC 28074 | Citrus reticulata | Spain, Algemesi | Twig dieback | MF418412 | MF418572 | MF418332 | MF418491 | MF418246 | |
| CPC 28077 | Citrus limon | Spain, Algemesi | Twig dieback | MF418413 | MF418573 | MF418333 | MF418492 | MF418247 | |
| CPC 28079 | Citrus reticulata | Spain, Algemesi | Twig dieback | MF418414 | MF418574 | MF418334 | MF418493 | MF418248 | |
| CPC 28081 = CBS 142548 | Citrus reticulata | Spain, Algemesi | Twig dieback | MF418415 | MF418575 | MF418335 | MF418494 | MF418249 | |
| CPC 28163 | Microcitrus australasica | Italy, Catania | Twig dieback | MF418416 | MF418576 | MF418336 | MF418495 | MF418250 | |
| CPC 31135 | Citrus limon | Malta, Gozo | Branch canker | MF418417 | MF418577 | MF418337 | MF418496 | MF418251 | |
| CPC 31159 | Citrus sinensis | Malta, Zurrieq | Branch canker | MF418418 | MF418578 | MF418338 | MF418497 | MF418252 | |
| D. helianthi | CBS 344.94 | Helianthus annuus | - | - | KC343114 | KC344082 | KC343598 | KC343840 | KC343356 |
| CBS 592.81 | Helianthus annuus | Serbia | - | KC343115 | KC344083 | KC343599 | KC343841 | JX197454 | |
| D. hongkongensis | CBS 115448 | Dichroa febrifuga | China | - | KC343119 | KC344087 | KC343603 | KC343845 | KC343361 |
| D. inconspicua | CBS 133813 | Maytenus ilicifolia | Brazil | - | KC343123 | KC344091 | KC343607 | KC343849 | KC343365 |
| D. infertilis | CBS 199.39 | Unknown | Italy | - | KC343051 | KC344019 | KC343535 | KC343777 | KC343293 |
| CBS 230.52 | Citrus sinensis | Suriname | - | KC343052 | KC344020 | KC343536 | KC343778 | KC343294 | |
| CPC 20322 | Glycine max | Brazil | - | KC343053 | KC344021 | KC343537 | KC343779 | KC343295 | |
| D. limonicola | CPC 27869 | Citrus limon | Malta, Gozo | Trunk canker | MF418419 | MF418579 | MF418339 | MF418498 | MF418253 |
| CPC 27871 | Citrus limon | Malta, Gozo | Trunk canker | MF418420 | MF418580 | MF418340 | MF418499 | MF418254 | |
| CPC 27879 | Citrus limon | Malta, Gozo | Branch canker | MF418421 | MF418581 | MF418341 | MF418500 | MF418255 | |
| CPC 28200 = CBS 142549 | Citrus limon | Malta, Gozo | Branch canker | MF418422 | MF418582 | MF418342 | MF418501 | MF418256 | |
| CPC 31137 = CBS 142550 | Citrus limon | Malta, Zurrieq | Branch canker | MF418423 | MF418583 | MF418343 | MF418502 | MF418257 | |
| D. melitensis | CPC 27873 = CBS 142551 | Citrus limon | Malta, Gozo | Branch canker | MF418424 | MF418584 | MF418344 | MF418503 | MF418258 |
| CPC 27875 = CBS 142552 | Citrus limon | Malta, Gozo | Branch canker | MF418425 | MF418585 | MF418345 | MF418504 | MF418259 | |
| D. multigutullata | ICMP20656 | Citrus grandis | China | - | KJ490633 | KJ490454 | KJ490575 | KJ490512 | - |
| D. novem | CBS 127270 | Glycine max | Croatia | - | KC343156 | KC344124 | KC343640 | KC343882 | KC343398 |
| CBS 127271 | Glycine max | Croatia | - | KC343157 | KC344125 | KC343641 | KC343883 | KC343399 | |
| CPC 26188 = CBS 142553 | Citrus japonica | Italy, Messina | Twig dieback | MF418426 | MF418586 | MF418346 | MF418505 | MF418260 | |
| CPC 28165 = CBS 142554 | Citrus aurantiifolia | Italy, Catania | Twig dieback | MF418427 | MF418587 | MF418347 | MF418506 | MF418261 | |
| CPC 28167 | Citrus aurantiifolia | Italy, Catania | Twig dieback | MF418428 | MF418588 | MF418348 | MF418507 | MF418262 | |
| CPC 28169 | Citrus aurantiifolia | Italy, Catania | Twig dieback | MF418429 | MF418589 | MF418349 | MF418508 | MF418263 | |
| D. ovalispora | ICMP20659 | Citrus limon | China | - | KJ490628 | KJ490449 | KJ490570 | KJ490507 | - |
| D. pseudomangiferae | CBS 101339 | Mangifera indica | Dominican Republic | - | KC343181 | KC344149 | KC343665 | KC343907 | KC343423 |
| D. pseudophoenicicola | CBS 462.69 | Phoenix dactylifera | Spain | - | KC343184 | KC344152 | KC343668 | KC343910 | KC343426 |
| D. rudis | CBS 113201 | Vitis vinifera | Portugal | - | KC343234 | KC344202 | KC343718 | KC343960 | KC343476 |
| D. saccarata | CBS 116311 | Protea repens | South Africa | - | KC343190 | KC344158 | KC343674 | KC343916 | KC343432 |
| D. sojae | FAU 635 | Glycine max | USA | - | KJ590719 | KJ610875 | KJ659208 | KJ590762 | - |
| D. sojae | ZJUD68 | Citrus unshiu | China | - | KJ490603 | KJ490424 | KJ490545 | KJ490482 | - |
| D. sterilis | CBS 136969 | Vaccinium corymbosum | Italy | - | KJ160579 | KJ160528 | MF418350 | KJ160611 | KJ160548 |
| D. subclavata | ICMP20663 | Citrus unshiu | China | - | KJ490630 | KJ490451 | KJ490572 | KJ490509 | - |
| D. unshiuensis | CGMCC3.17569 | Citrus unshiu | China | - | KJ490587 | KJ490408 | KJ490529 | KJ490466 | - |
| Diaporthella corylina | CBS 121124 | Corylus sp. | China | - | KC343004 | KC343972 | KC343488 | KC343730 | KC343246 |
1 CPC: Culture collection of P.W. Crous, housed at Westerdijk Fungal Biodiversity Institute; CBS: Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands; CGMCC: China, General Microbiological Culture Collection, Beijing, China; FAU: Isolates in culture collection of Systematic Mycology and Microbiology Laboratory, USDA-ARS, Beltsville, MD, USA; ICMP: International Collection of Microorganisms from Plants, Landcare Research, Auckland, New Zealand; ZJUD, Diaporthe strains in Zhejiang University, China. Ex-type and ex-epitype cultures are indicated in bold.
2 ITS: internal transcribed spacers 1 and 2 together with 5.8S nrDNA; tub2: partial beta-tubulin gene; his3: histone3; tef1: partial translation elongation factor 1-α gene; cal: partial calmodulin gene. Sequences generated in this study indicated in italics.
Morphological analyses
Agar plugs (6 mm diam) were taken from the edge of actively growing cultures on MEA and transferred onto the centre of 9 cm diam Petri dishes containing 2 % tap water agar supplemented with sterile pine needles (PNA; Smith et al. 1996), potato dextrose agar (PDA), oatmeal agar (OA) and MEA (Crous et al. 2009), and incubated at 20–21 °C under a 12 h near-ultraviolet light/12 h dark cycle to induce sporulation as described in recent studies (Gomes et al. 2013, Lombard et al. 2014). Colony characters and pigment production on MEA, OA and PDA were noted after 10 d. Colony colours were rated according to Rayner (1970). Cultures were examined periodically for the development of ascomata and conidiomata. Colony diameters were measured after 7 and 10 d. The morphological characteristics were examined by mounting fungal structures in clear lactic acid and 30 measurements at ×1000 magnification were determined for each isolate using a Zeiss Axioscope 2 microscope with interference contrast (DIC) optics. Descriptions, nomenclature and illustrations of taxonomic novelties are deposited in MycoBank (www.MycoBank.org; Crous et al. 2004b).
Pathogenicity
Pathogenicity tests with five Diaporthe species isolated from the European citrus samples were performed to satisfy Koch’s postulates.
Two isolates of each of the five species (D. baccae: CPC 26170, CPC 27831; D. foeniculina: CPC 28033, CPC 28081; D. limonicola: CPC 28200, CPC 31137; D. melitensis: CPC 27873, CPC 27875; and D. novem: CPC 26188, CPC 28165), were inoculated onto potted 2-yr-old healthy plants of lemon (Citrus limon), lime (C. aurantiifolia), mandarin (C. reticulata), and two clones (‘New Hall’ and ‘Tarocco Meli’) of sweet orange (C. sinensis). Three plants per replicate for each isolate were inoculated, each having five wounds on twigs made using a sterile blade. Mycelial plugs (6 mm diam), taken from the margin of actively growing colonies on MEA, were placed on the wound sites on each plant. An equivalent number of plants and inoculation sites were inoculated with sterile MEA plugs and served as controls. The inoculation sites were covered with Parafilm® (American National Can, Chicago, IL). The inoculated plants were incubated with a 16 h photoperiod in a growth chamber at 100 % relative humidity and 25 ± 1 °C. After 2 mo external symptoms were assessed. Twigs were cut and the bark peeled off to check for any internal discolouration.
Small sections (0.5 cm) of symptomatic tissue from the edge of twig lesions were placed on MEA to re-isolate the fungal species, and were identified based on tef1 and tub2 sequencing to fulfil Koch’s postulates.
RESULTS
Isolates
Several shoot blight and canker infections on woody tissue were frequently observed on multiple Citrus species in all countries investigated. Some orchards presented blight of vigorously growing branches and cankers involving both scion branches and rootstock trunks, resulting in a general dieback and tree death (Fig. 1A). Affected trunks and branches appeared cracked, darkly discoloured and/or slightly sunken. Abundant gummosis was frequently associated with the affected tissues (Fig. 1B–D). Twigs showed wilting, typical dieback and wither-tip, and occasionally gummosis (Fig. 1E–F). Under the bark, cankers were reddish brown and variable in shape. Pycnidial formation on dead twig tissue was observed (Fig. 1G). A total of 79 monosporic isolates resembling those of the genus Diaporthe were collected. The Diaporthe isolates were recovered from 10 species of Citrus at 31 sites in different locations of Greece, Italy, Malta, Spain, and Portugal. Among them, 27 isolates were obtained from branch infections, 13 were associated with trunk cankers, and 39 from twig dieback (Table 1).
Fig. 1.

Symptoms on citrus tissues with associated Diaporthe species. A. Commercial lemon orchard infected by D. limonicola and D. melitensis (Malta). B–C. Trunk canker with gummosis of Citrus limon and C. sinensis plants (Malta). D. Branch canker of C. sinensis (Portugal). E–F. Twigs dieback of lemon (Italy). G. Orange twigs wither-tip with Diaporthe pycnidial formation (Italy).
Phylogenetic analyses
Six alignments were analysed representing single gene analyses of ITS, tub2, his3, tef1, cal and a combined alignment of the five genes. The alignments produced topologically similar trees. The combined species phylogeny of the Diaporthe isolates consisted of 123 sequences, including the outgroup sequences of Diaporthella corylina (culture CBS 121124). A total of 3026 characters (ITS: 1–582, tef1: 589–1052, tub2: 1059–1 862, cal: 1869–2484, his3: 2 491–3026) were included in the phylogenetic analysis, 1355 characters were parsimony-informative, 468 were variable and parsimony-uninformative, and 1161 were constant. A maximum of 1000 equally most parsimonious trees were saved (Tree length = 5528, CI = 0.584, RI = 0.868 and RC = 0.507). Bootstrap support values from the parsimony analysis are plotted on the Bayesian phylogenies in Fig. 2. For the Bayesian analyses, MrModeltest suggested that all partitions should be analysed with dirichlet state frequency distributions. The following models were recommended by MrModeltest and used: GTR+I+G for ITS, tef1 and cal, HKY+G for tub2 and GTR+G for his3. In the Bayesian analysis, the ITS partition had 188 unique site patterns, the tef1 partition had 357 unique site patterns, the tub2 partition had 510 unique site patterns, the cal partition had 364 unique site patterns, the his3 partition had 239 unique site patterns and the analysis ran for 1 880 000 generations, resulting in 3762 trees of which 2822 trees were used to calculate the posterior probabilities.
Fig. 2.
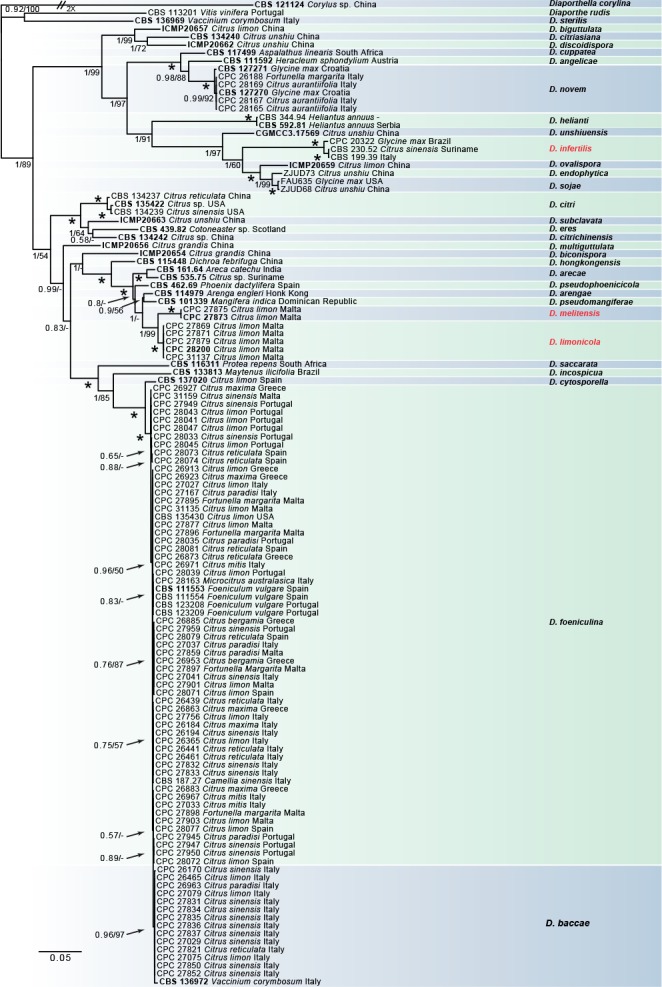
Consensus phylogram of 3 762 trees resulting from a Bayesian analysis of the combined ITS, tub2, his3, tef1 and cal sequence. Bootstrap support values and Bayesian posterior probability values are indicated at the nodes. The asterisk symbol (*) represents full support (1/100). Substrate and country of origin are listed next to the strain numbers. The newly recognized species are in red. The tree was rooted to Diaporthella corylina (CBS 121124).
In the combined analysis, 54 Citrus isolates clustered with five reference strains and the ex-type of D. foeniculina, whilst 14 isolates clustered with the ex-type of D. baccae. Four isolates clustered with the ex-type strain of D. novem. Moreover, five isolates identified as D. limonicola and a further two as D. melitensis, formed two highly supported subclades (1.00/100) embedded in the D. arecae species complex.
The individual alignments and trees of the five single loci used in the analyses, were also compared with respect to their performance in species recognition. D. novem was differentiated by each gene used. Moreover, tef1 and tub2 separated both D. limonicola and D. melitensis from the other species belonging to the D. arecae species complex.
TAXONOMY
Morphological observations, supported by phylogenetic inference, were used to identify three known species (D. baccae, D. foeniculina, and D. novem), and to recognize three new species described here (Table 2). One species (represented by three isolates) was sterile in culture, and is therefore characterized by DNA sequence data (Gomes et al. 2013).
Table 2.
Diaporthe species associated with citrus and their morphological characteristics.
| Species | Conidiomata (μm) | Conidiophores (μm) | Alpha conidia (μm) | Beta conidia (μm) | References |
|---|---|---|---|---|---|
| D. arecae | up to 400 | 15–40 × 1.5–3 | 6–10 × 2–3 | - | Gomes et al. (2013) |
| D. baccae | up to 650 | 20–57 × 2–3 | 7–9 × 2–3 | 20–24 × 1–2 | Lombard et al. (2014) |
| D. biconispora | 145–185 | 12–35.5 × 1.6–2.6 | 6–10.5 × 2–3.5 | - | Huang et al. (2015) |
| D. biguttulata | up to 300 | 5.8–16.9 × 1.3–2.3 | 5.7–7.8 × 2.5–2.9 | 23.7–31.6 × 0.9–1.6 | Huang et al. (2015) |
| D. citri | 200–250 | 10–15 × 1–2 | 7.6–10.2 × 3–4.2 | - | Udayanga et al. (2014b) |
| D. citriasiana | up to 627 | 3.5–10.5 × 1–2 | 10.5–15 × 4–6.5 | 24–42 × 1–2 | Huang et al. (2013) |
| D. citrichinensis | up to 435 | 9–19.5 × 1.5–3 | 5.5–9 × 1.5–2.5 | 27.5–40 × 1–1.5 | Huang et al. (2013) |
| D. cytosporella | 150–200 | 7–18 × 1–2 | 8–9 × 2.6–3.2 | - | Udayanga et al. (2014b) |
| D. discoidispora | 200–118 | 8.9–23.4 × 1.3–2.7 | 5.6–8 × 2.1–3.2 | 21.2–38.7 × 0.9–1.6 | Huang et al. (2015) |
| D. endophytica (sterile) | - | - | - | - | Gomes et al. (2013) |
| D. eres | 200–250 | 10–15 × 2–3 | 6.5–8.5 × 3–4 | 22–28 × 1–1.5 | Udayanga et al. (2014a) |
| D. foeniculina | 400–700 | 9–15(–18) × 1–2 | 8.5–9 × 2.3–2.5 | 22–28 × 1.4–1.6 | Udayanga et al. (2014b) |
| D. hongkongensis | up to 200 | 5–12 × 2–4 | 6–7 × 2.5 | 18–22 × 1.5–2 | Gomes et al. (2013) |
| D. infertilis (sterile) | - | - | - | - | This study |
| D. limonicola | up to 670 | 5–20 × 1.5–4 | 5.5–8.5 × 1.5–2.5 | 15–26.5 × 1–2 | This study |
| D. melitensis | up to 650 | 5–15 × 1.5–5.5 | 4.5–7 × 1.5–3 | - | This study |
| D. multigutullata | up to 358 | 9.8–14.8 × 1.3–3.6 | 8–12.6 × 4.2–6 | - | Huang et al. (2015) |
| D. novem | up to 580 | 5.3–10.4 × 1.9–3.2 | 6.3–8.9 × 1.9–2.5 | 26.4–37.7 × 1–1.3 | Santos et al. (2011) |
| D. ovalispora | up to 242 | 9.5–21.6 × 1.6–3.6 | 6.1–7.9 × 2.7–3.8 | - | Huang et al. (2015) |
| D. sojae | 200–250 | 12–16 × 2–4 | 5.3–7.3 × 2–3 | - | Udayanga et al. (2015) |
| D. subclavata | - | 14.2–27.3 × 1.6–2.6 | 5.5–7.2 × 2.2–2.9 | - | Huang et al. (2015) |
| D. unshiuensis | up to 152 | 14.3–24.2 × 1.4–2.6 | 5.2–7.5 × 2–3.9 | - | Huang et al. (2015) |
Diaporthe infertilis Guarnaccia & Crous, sp. nov.
MycoBank MB821727
(Fig. 3)
Fig. 3.
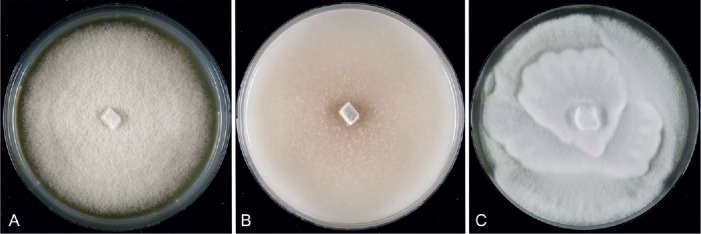
Diaporthe infertilis (CBS 230.52). A–C. Colonies after 7 d at 21 °C on MEA, OA and PDA, respectively.
Etymology: Named after its sterile growth in culture.
Diagnosis: Diaporthe infertilis differs from its closest phylogenetic neighbour, D. ovalispora, in 26 unique fixed alleles in ITS locus, 68 in tef1, 30 in tub2 and 48 in his3 based on the alignments deposited in TreeBASE.
Type: Suriname: Paramaribo, from decaying fruit of Citrus sinensis, Apr. 1932, N.J. van Suchtelen (CBS H-23179 – holotype; CBS 230.52 – culture ex-type).
Description: Culture characteristics: Colony on MEA covering the entire plate after 10 d, pale luteous with abundant white compact aerial mycelium in fluctuating rings. On OA and PDA at first white, becoming cream to yellowish, flat, with dense and felted mycelium, reverse pale brown with brownish dots with age. Cultures sterile.
Notes: Three isolates clustered in a clade distinct from species of Diaporthe known from DNA sequence data. One strain (CPC 20322) was differentiated from the other two (CBS 199.39, CBS 230.52) by unique fixed alleles in four loci based on alignments of the separate loci deposited in TreeBASE: tef1 positions 115 (C), 261 (indel), 314 (G), 395 (C); tub2 positions 123 (C), 631 (G); cal positions 132 (T), 207 (A), 210 (T), 256 (T), 259 (T), 262 (A), 364 (G), 366 (A), 438 (G), 439 (G), 448 (C); his3 positions 201 (A), 438 (A), 448 (T), 450 (A). Gomes et al. (2013) tentatively referred to this clade as D. citri. However, after a molecular re-assessment of many Diaporthe species, D. citri is restricted to a different clade of citrus isolates (Udayanga et al. 2014b). We therefore describe D. infertilis as a new species for this clade.
Additional material examined: Brazil: from seeds of Glycine max, A. Almeida (culture LGMF946 = CPC 20322). – Italy: from unknown host, G. Goidanich (CBS 199.39).
Diaporthe limonicola Guarnaccia & Crous, sp. nov.
MycoBank MB821731
(Fig. 4)
Fig. 4.
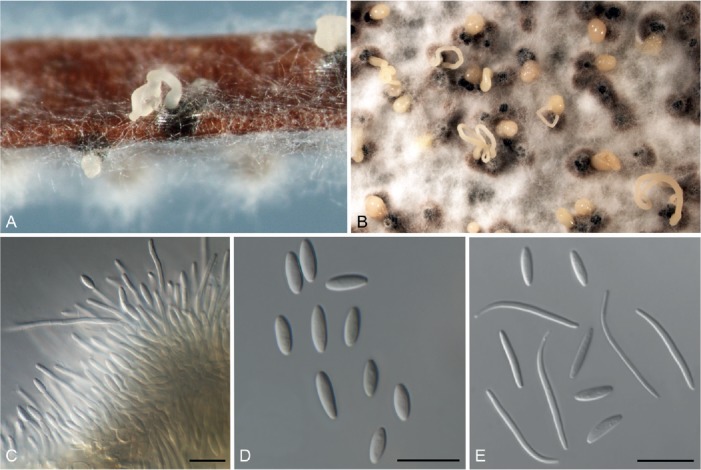
Diaporthe limonicola (CBS 142549). A. Conidiomata sporulating on PNA. B. Conidiomata sporulating on OA. C. Conidiogenous cells. D. Alpha conidia. E. Alpha, beta and gamma conidia. Bars = 10 μm.
Etymology: In reference to the occurrence on Citrus limon.
Diagnosis: Diaporthe limonicola can be distinguished from the closely related D. pseudomangiferae based on tef1, tub2, his3 and cal loci (96 % in tef1, 96 % in tub2, 97 % in his3, and 96 % in cal). Diaporthe limonicola differs from D. pseudomangiferae in the shorter alpha conidia and in producing beta and gamma conidia.
Type: Malta: Gozo, from branch canker of Citrus limon, 11 Jul. 2016, V. Guarnaccia (CBS H-23126 – holotype; CBS 142549 = CPC 28200 – culture ex-type).
Description: Conidiomata pycnidial in culture on PNA, PDA, OA and MEA, solitary or aggregated, deeply embedded in PDA, erumpent, dark brown to black, 250–670 μm diam, whitish translucent to cream conidial drops exuded from the ostioles. Conidiophores hyaline, smooth, 1-septate, densely aggregated, cylindrical, straight, 5–20 × 1.5–4 μm. Conidiogenous cells phialidic, hyaline, terminal, cylindrical, 5–12 × 1–2 μm, tapered towards the apex. Paraphyses intermingled among conidiophores, hyaline, smooth, 1–3-septate, to 90 μm long, apex 1–2 μm diam. Alpha conidia unicellular, aseptate, fusiform, hyaline, mono- to biguttulate and acute at both ends, 5.5–8.5 × 1.5–2.5 μm, mean ± SD = 6.8 ± 0.6 × 2.1 ± 0.3 μm, L/B ratio = 2.8. Beta conidia hyaline, aseptate, eguttulate, filiform, curved, tapering towards both ends, 15–26.5 × 1–2 μm, mean ± SD = 22.7 ± 2.6 × 1.4 ± 0.3 μm, L/B ratio = 16.2. Gamma conidia hyaline, multiguttulate, fusiform to subcylindrical with an acute or rounded apex, 9–15.5 × 1–2 μm, mean ± SD = 10.7 ± 1.6 ×1.4 ± 0.2 μm, L/B ratio 7.6.
Culture characteristics: Colonies covering the medium within 1 wk at 21 °C, surface mycelium flattened, dense and felt-like. Colony on MEA and OA at first white, becoming cream to yellowish, flat, with dense and felted mycelium, reverse pale brown with brownish dots with age, with visible solitary or aggregated conidiomata at maturity. On PDA cream to smoke-grey, reverse pale brown.
Notes: Diaporthe limonicola was isolated from Citrus limon trunk cankers in two different islands of the Malta archipelago, where all the plants were affected. Five strains representing D. limonicola cluster in a well-supported clade, and appear most closely related to D. pseudomangiferae and D. arengae. Diaporthe limonicola can be distinguished based on tef1, tub2, his3 and cal loci from D. pseudomangiferae (96 % in tef1, 96 % in tub2, 97 % in his3, and 96 % in cal), and from D. arengae (97 % in tef1, 98 % in tub2, 98 % in his3, and 96 % in cal). This species is phylogenetically close to but clearly differentiated from D. melitensis (described below) by 22 unique fixed alleles in ITS locus, 2 in tef1 and 47 in tub2.
Morphologically, D. limonicola differs from D. pseudomangiferae in the shorter alpha conidia (5.5–8.5 vs. 7–9 μm) (Gomes et al. 2013) and the production of beta and gamma conidia, which are not known in D. pseudomangiferae (Gomes et al. 2013).
Additional material examined: Malta: Zurrieq, from branch canker of Citrus limon, 11 Jul. 2016, V. Guarnaccia (culture CBS 142550 = CPC 31137).
Diaporthe melitensis Guarnaccia & Crous, sp. nov.
MycoBank MB821732
(Fig. 5)
Fig. 5.
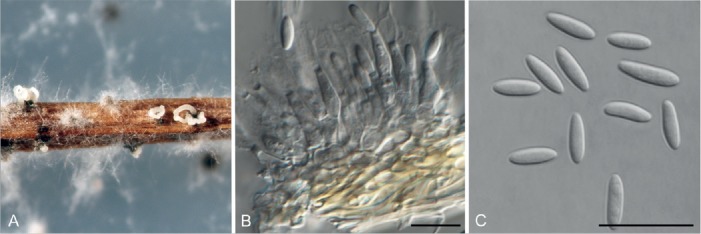
Diaporthe melitensis (CBS 142551). A. Conidiomata sporulating on PNA. B. Conidiogenous cells. C. Alpha conidia. Bars = 10 μm.
Etymology: Named after the country where it was collected, Malta (ancient Latin name, Melita).
Diagnosis: Diaporthe melitensis can be distinguished from the closely related D. pseudomangiferae by the ITS, tef1, tub2, his3 and cal loci (98 % in ITS, 96 % in tef1, 97 % in tub2, 97 % in his3, and 96 % in cal). Diaporthe melitensis also differs from D. pseudomangiferae in the shorter alpha conidia.
Type: Malta: Gozo, from branch canker of Citrus limon, 22 Sep. 2015, V. Guarnaccia (CBS H-23127 – holotype; CBS 142551 = CPC 27873 – culture ex-type).
Description: Conidiomata pycnidial in culture on PNA, PDA, OA and MEA, solitary or aggregated, deeply embedded in the PDA, erumpent, dark brown to black, 250–650 μm diam, whitish translucent to yellowish conidial drops exuded from the ostioles. Conidiophores hyaline, smooth, 1-septate, densely aggregated, cylindrical, straight, 5–15 × 1.5–5.5 μm. Conidiogenous cells phialidic, hyaline, terminal, cylindrical, 6–12 × 1–3 μm, tapered towards the apex. Paraphyses not observed. Alpha conidia unicellular, aseptate, fusiform, hyaline, 1–4-guttulate with acute ends, 4.5–7 × 1.5–3 μm, mean ± SD = 5.9 ± 0.6 × 2.2 ± 0.4 μm, L/B ratio = 2.7. Beta conidia and Gamma conidia not observed.
Culture characteristics: Colonies covering the dish within 1 wk at 21 °C, surface mycelium flattened, dense and felt-like. Colony on MEA and OA at first white, becoming yellowish, flat, with dense and felted mycelium, reverse pale sepia with brownish dots with age, with visible solitary or aggregated conidiomata at maturity. On PDA cream to smoke-grey, reverse pale brown.
Notes: Diaporthe melitensis was isolated from trunk samples of Citrus limon showing serious cankers in Gozo (Malta). The two strains representing D. melitensis cluster in a well-supported clade, and appear closely related to D. pseudomangiferae and D. arengae. This species is phylogenetically closely related to, but clearly differentiated from, D. limonicola (described above) by 22 different unique fixed alleles in ITS, tef1 and tub2 loci (22, 2, and 47 respectively) based on the alignments deposited in TreeBASE.
Morphologically D. melitensis differs from D. pseudomangiferae in the shorter alpha conidia (4.5–7 vs. 7–9 μm) (Gomes et al. 2013).
Additional material examined: Malta: Gozo, from branch canker of Citrus limon, 22 Sep. 2015, V. Guarnaccia (culture CBS 142552 = CPC 27875).
PATHOGENICITY
After 30 d all the isolates of the inoculated species induced lesions on most of the Citrus species tested. The inoculated twigs developed cankers similar to those detected in the field, and the fungi were successfully re-isolated, fulfilling Koch’s postulates (Fig. 6). Cankers and internal discolouration were observed in correspondence to inoculation points. On the contrary, no symptoms were observed on the control plants. Clear differences in aggressiveness among the isolates and susceptibility of the Citrus species were observed: D. limonicola and D. melitensis caused the most serious symptoms with no difference among the hosts. Diaporthe foeniculina was weakly aggressive to each Citrus species. Similarly, D. novem was weakly aggressive on all the hosts except the orange clones, whilst D. baccae caused disease symptoms only on mandarin.
Fig. 6.

Pathogenicity test of selected Diaporthe isolates on citrus plants after 30 d. A. Shoot blight of lime plants inoculated with D. novem (CPC 26188). B–C. Cankers with gummosis of lemon plants caused by D. limonicola and D. melitensis (CPC 28200, CPC 27873). D–E. Internal discoloration of mandarin twigs inoculated respectively with D. melitensis and D. baccae (CPC 27873, CPC 26170). F. Internal lesion of orange branch caused by D. foeniculina (CPC 28081).
DISCUSSION
After a major screening of fungal diseases of citrus in Europe (Guarnaccia et al. 2017a, b, Sandoval-Denis et al. 2018), molecular phylogenetic and morphological analyses were used to evaluate the diversity of Diaporthe species in the Mediterranean basin, focusing on symptomatic plants.
Several Diaporthe species are well established in Europe (Thomidis & Michailides 2009, Santos et al. 2011, Lombard et al. 2014, Guarnaccia et al. 2016). Diaporthe species are also frequently associated with citrus diseases worldwide (Timmer et al. 2000, Huang et al. 2013), such as melanose and stem-end rot. Since the late 18th century these diseases have affected different citrus organs and also cause a sort of wood gummosis (Fawcett 1936, Timmer et al. 2000, Mondal et al. 2007). Diaporthe citri is considered a key pathogen of Citrus species and has been confirmed from Brazil, China, Korea, and New Zealand, and is also reported as widely spread throughout Asia, Australasia, and South America (Timmer et al. 2000, Mondal et al. 2007, Udayanga et al. 2014b). However, D. citri has never been reported from Europe, whilst D. cytosporella and D. foeniculina have been recently isolated from citrus in Spain (Udayanga et al. 2014b).
DNA sequence data are essential in resolving taxonomic questions, redefining species boundaries, and the accurate naming of species required for effective communication about plant pathogens. Thus, during the past decade, a polyphasic approach was used in several Diaporthe studies, revealing new species involved with citrus diseases and as endophytes and plant pathogens (Huang et al. 2013, 2015). Santos et al. (2017a) showed that species separation is better when five loci (ITS, tef1, tub2, his3, and cal) are simultaneously used to build the phylogeny of Diaporthe isolates.
Citrus crops are already compromised by a range of fungal pathogens other than Diaporthe (Vicent et al. 2007, Aiello et al. 2015, Guarnaccia et al. 2017a, Sandoval-Denis et al. 2018). Considering that no surveys for citrus diseases caused by Diaporthe had been performed in Europe, a large-scale investigation of Diaporthe species associated with citrus infections in Europe was needed. This study provides the first molecular characterization of Diaporthe diversity related to citrus production in Europe, combined with morphological characterisation.
Several citrus orchards, plant nurseries, private gardens and collections in five Mediterranean European countries were investigated. We further investigated different host plants in Citrus-allied genera such as Microcitrus, which is also economically important for fruit production.
Canker symptoms were frequently observed on several Citrus species in all countries investigated. Twigs showed wilting, dieback, wither-tip, and gummosis. Some orchards presented branch blight and trunk cankers associated with abundant gummosis. The most critical situation seen was in different lemon orchards in Malta, where the infections led to tree death. Melanose and stem-end rot were never observed.
We collected 79 Diaporthe strains. Phylogenetic analyses based on single and the combined five loci (ITS, tef1, tub2, his3, and cal), as well as morphological characters, revealed five Diaporthe species associated with infections on several Citrus species in Europe. We included in the analysis the closest taxa to the five Diaporthe species recovered in this study, based on BLAST searches of NCBI’s GenBank nucleotide database. The final phylogenetic tree distinguished two newly described species (D. limonicola and D. melitensis) and three known species (D. baccae, D. foeniculina, and D. novem). Moreover, a known clade represented by three strains (CBS 199.39, CBS 230.52, CPC 20322), previously named D. citri, appeared in our final tree. However, this clade also required a separate name as D. citri s. str. is restricted to the pathogen causing melanose and stem-end rot of citrus fruit (Udayanga et al. 2014b). Thus, in this study we have described these three isolates as D. infertilis. Based on sampling in this study, D. citri appears to be absent in Europe as previously reported by Udayanga et al. (2014b).
Huang et al. (2015) obtained two separate groups of citrus isolates within the D. arecae complex, which were either not well supported or non-monophyletic based on a four-locus phylogenetic analysis. However, our analysis based on five loci, combined with morphological observations, clearly separated both D. limonicola and D. melitensis from D. pseudomangiferae and D. areangae, the most closely related species, and from other species in the D. arecae complex such as D. podocarpi-macrophylli and D. xishuangbanica (Gao et al. 2017). Morphologically, D. limonicola and D. melitensis differ from D. pseudomangiferae in the shorter alpha conidia. Moreover, D. limonicola is the only taxon among these species to produces beta and gamma conidia.
Diaporthe foeniculina was the predominant species found in all the Mediterranean countries sampled, but its pathogenicity on Citrus was unknown (Udayanga et al. 2014b). Recently, Lombard et al. (2014) described D. baccae as a new species associated with Vaccinium corymbosum cankers in Italy. Similarly, we found this species associated with twig, branch and trunk cankers of citrus in Italy. Diaporthe novem was isolated for the first time from infected citrus plants in our study, where it was found associated with twig dieback of C. japonica (kumquat) and C. aurantiifolia (lime) in Italy. Moreover, the newly described species were isolated from devastated lemon plants in several orchards on Malta: D. limonicola was recovered from symptomatic trunks and branches, whilst D. melitensis was isolated only from branches. They were isolated separately and from the same affected sample. Colonization of the same host plant by diverse Diaporthe species appears to be frequent as previously reported (Crous & Groenewald 2005, Van Niekerk et al. 2005, Thompson et al. 2011).
Our results reveal a large diversity of Diaporthe species spanning several clades and species complexes, associated with citrus wood cankers in European countries. These include D. baccae, D. infertilis, D. novem, and the two newly described species. In total, 22 Diaporthe species are now confirmed as associated with citrus.
Pathogenicity of the species isolated from citrus samples collected in Europe was tested on healthy plants of lemon, lime, mandarin, and two clones of Citrus sinensis (‘New Hall’ and ‘Tarocco Meli’). All of the Diaporthe species tested caused lesions to develop on twigs. Recently, D. foeniculina (syn. D. neotheicola) has been reported as causing disease in many other hosts: shoot blight of persimmon in Australia (Golzar et al. 2012), kiwi-fruit disease in Greece (Thomidis et al. 2013), and avocado branch cankers (Guarnaccia et al. 2016). This species evidently has the ability to infect a wide range of fruits and plant hosts as an opportunistic pathogen. Diaporthe foeniculina (as “D. foeniculacea” in Gomes et al. 2013) proves to be a pathogen with a broad host range amongst temperate woody plants and fruit trees. In our study, D. foeniculina was isolated from symptomatic plants of eight Citrus species (C. bergamia, C. japonica, C. limon, C. maxima, C. mitis, C. paradisi, C. reticulata, and C. sinensis) and also Microcitrus australasica. In the pathogenicity tests, it was weakly aggressive, but produced lesions on each species tested.
These results demonstrate a cross-infection potential of multiple Diaporthe species on different Citrus species, as previously reported (Lombard et al. 2014, Guarnaccia et al. 2016). Diaporthe limonicola and D. melitensis caused prominent symptoms in all the citrus species inoculated, and because they were isolated from plants with severe disease symptoms, these species can be considered as potentially major new pathogens of Citrus limon. Diaporthe baccae caused symptoms only on mandarin, while D. novem infected lime, lemon, and mandarin plants. Both of these species seemed to be weakly aggressive, with different host susceptibility and known distribution. These fungi merit adding to the list of fungal taxa causing citrus cankers worldwide (Adesemoye et al. 2014, Mayorquin et al. 2016, Sandoval-Denis et al. 2018).
This study provides the first overview of Diaporthe diversity associated with cankers of citrus plants in Europe, and includes information on their pathogenicity. Two of the new species described were established as causal agents of a devastating disease of lemon plants, inducing branch and trunk cankers that lead to plant death. The present study also appears to represent the first reports of D. baccae and D. novem associated with citrus disease in Europe. Despite the worldwide distribution and economical importance of citrus, knowledge of the fungal species associated with Citrus species is still incomplete. Further studies are required in order to fully elucidate the host range, specificity, and global distribution of Diaporthe species, as well as other fungi causing cankers of citrus plants.
Acknowledgments
We are grateful to Arien Van Iperen and Mieke Starink-Willemse for technical assistance, and would like to thank Giancarlo Polizzi (Di3A, University of Catania) for sharing his experience in citrus diseases, and for his kind contribution in performing pathogenicity tests.
REFERENCES
- Adesemoye AO, Mayorquin JS, Wang DH, Twizeyimana S, Lynch C, et al. (2014) Identification of species of Botryosphaeriaceae causing bot gummosis in citrus in California. Plant Disease 98: 55–61. [DOI] [PubMed] [Google Scholar]
- Aiello D, Carrieri R, Guarnaccia V, Vitale A, Lahoz E, et al. (2015) Characterization and pathogenicity of Colletotrichum gloeosporioides and C. karstii causing preharvest disease on Citrus sinensis in Italy. Journal of Phytopathology 163: 168–177. [Google Scholar]
- Burnett HC. (1962) Melanose: Diaporthe citri (Fawc) Wolf. [Plant Pathology Circular no. 2.] Gainesville: Florida Department of Agriculture, Division of Plant Industry. [Google Scholar]
- Carbone I, Kohn LM. (1999) A method for designing primer sets for the speciation studies in filamentous ascomycetes. Mycologia 91: 553–556. [Google Scholar]
- Crous PW, Gams W, Stalpers JA, Robert V, Stegehuis G. (2004b) MycoBank: an online initiative to launch mycology into the 21st century. Studies in Mycology 50: 19–22. [Google Scholar]
- Crous PW, Groenewald JZ. (2005) Hosts, species and genotypes: opinions versus data. Australasian Plant Pathology 34: 463–470. [Google Scholar]
- Crous PW, Groenewald JZ, Risede JM, et al. (2004a) Calonectria species and their Cylindrocladium anamorphs: species with sphaeropedunculate vesicles. Studies in Mycology 50: 415–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Groenewald JZ, Shivas RG, Edwards J, Seifert KA, et al. (2011a) Fungal Planet description sheets: 69–91. Persoonia 26: 108–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Summerell BA, Swart L, Denman S, Taylor JE, et al. (2011b) Fungal pathogens of Proteaceae. Persoonia 27: 20–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Verkley GJM, Groenewald JZ, et al. (2009) Fungal Biodiversity. [CBS Laboratory Manual Series no. 1]. Utrecht: CBS-KNAW Fungal Biodiversity Institute. [Google Scholar]
- Crous PW, Wingfield MJ, Richardson DM, Le Roux JJ, Strasberg D, et al. (2016) Fungal planet description sheets: 400–468. Persoonia 36: 316–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J, Krohn K, Floerke U, Gehle D, Aust HJ, et al. (2005) Novel highly substituted biaryl ethers, phomopsines D-G, isolated from endophytic fungus Phomopsis sp. from Adenocarpus foliolosus. European Journal of Organic Chemistry 23: 5100–5105. [Google Scholar]
- Dai DQ, Wijayawardene NN, Bhat DJ, Chukeatirote E, Bahkali AH, et al. (2014) Pustulomyces gen. nov. accommodated in Diaporthaceae, Diaporthales, as revealed by morphology and molecular analyses. Cryptogamie, Mycologie 35: 63–72. [Google Scholar]
- Díaz GA, Latorre BA, Lolas M, Ferrada E, Naranjo P, et al. (2017) Identification and characterization of Diaporthe ambigua, D. australafricana, D. novem, and D. rudis causing a postharvest fruit rot in kiwifruit. Plant Disease 101: 1402–1410. [DOI] [PubMed] [Google Scholar]
- Diogo ELF, Santos JM, Phillips AJL. (2010) Phylogeny, morphology and pathogenicity of Diaporthe and Phomopsis species on almond in Portugal. Fungal Diversity 44: 107–115. [Google Scholar]
- Fawcett HS. (1912) The cause of stem-end rot of Citrus fruits (Phomopsis citri n. sp.). Phytopathology 2: 109–113. [Google Scholar]
- Fawcett HS. (1922) A Phomopsis of citrus in California. Phytopathology 12: 107. [Google Scholar]
- Fawcett HS. (1936) Citrus Diseases and their Control. New York: McGraw Hill. [Google Scholar]
- Food and Agricultural Organization (2016) Citrus fruits fresh and processed: annual statistics. Rome: Food and Agricultural Organization; http://www.fao.org/3/a-i5558e.pdf [Google Scholar]
- Fu CH, Hsieh HM, Chen CY, Chang TT, Huang YM, et al. (2013) Ophiodiaporthe cyatheae gen. et sp. nov., a diaporthalean pathogen causing a devastating wilt disease of Cyathea lepifera in Taiwan. Mycologia 105: 861–872. [DOI] [PubMed] [Google Scholar]
- Gao Y, Su YY, Sun W, Cai L. (2015) Diaporthe species occurring on Lithocarpus glabra in china, with descriptions of five new species. Fungal Biology 119: 295–309. [DOI] [PubMed] [Google Scholar]
- Gao YH, Liu F, Cai L. (2016) Unravelling Diaporthe species associated with Camellia. Systematics and Biodiversity 14: 102–117. [Google Scholar]
- Gao YH, Lui F, Duan W, Crous PW, Cai L. (2017) Diaporthe is paraphyletic. IMA Fungus 8: 153–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Reyne A, López-Medrano F, Morales JM, Esteban CG, Martín I, et al. (2011) Cutaneous infection by Phomopsis longicolla in a renal transplant recipient from Guinea: first report of human infection by this fungus. Transplant Infectious Disease 13: 204–207. [DOI] [PubMed] [Google Scholar]
- Glass NL, Donaldson GC. (1995) Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Applied and Environmental Microbiology 61: 1323–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golzar H, Tan YP, Shivas RG, Wang C. (2012) First report of shoot blight of persimmon caused by Diaporthe neotheicola in Australia. Australasian Plant Disease Notes 7: 115–117. [Google Scholar]
- Gomes R, Glienke C, Videira S, Lombard L, Groenewald J, et al. (2013) Diaporthe: a genus of endophytic, saprobic and plant pathogenic fungi. Persoonia 31: 1–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasso FM, Marini M, Vitale A, Firrao G, Granata G. (2012) Canker and dieback on Platanus × acerifolia caused by Diaporthe scabra. Forest Pathology 42: 510–513. [Google Scholar]
- Guarnaccia V, Groenewald JZ, Polizzi G, Crous PW. (2017a) High species diversity in Colletotrichum associated with citrus diseases in Europe. Persoonia 39: 32–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarnaccia V, Groenewald JZ, Li H, Glienke C, Carstens E, et al. (2017b) First report of Phyllosticta citricarpa and description of two new species, P. paracapitalensis and P. paracitricarpa, from citrus in Europe. Studies in Mycology 87: 161–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarnaccia V, Vitale A, Cirvilleri G, Aiello D, Susca A, et al. (2016) Characterisation and pathogenicity of fungal species associated with branch cankers and stem-end rot of avocado in Italy. European Journal of Plant Pathology 146: 963–976. [Google Scholar]
- Habib W, Gerges E, Antelmi I, Baroudy F, Choueiri E, et al. (2015) Diaporthe foeniculina associated with severe shoot blight of lemon in Lebanon. Phytopathologia Mediterranea 54: 149–150. [Google Scholar]
- Hillis DM, Bull JJ. (1993) An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Systematic Biology 42: 182–192. [Google Scholar]
- Huang F, Hou X, Dewdney MM, Fu YS, Chen GQ, et al. (2013) Diaporthe species occurring on citrus in China. Fungal Diversity 61: 237–250. [Google Scholar]
- Huang F, Udayanga D, Wang X, Hou X, Mei X, et al. (2015) Endophytic Diaporthe associated with Citrus: A phylogenetic reassessment with seven new species from China. Fungal Biology 119: 331–347. [DOI] [PubMed] [Google Scholar]
- Isaka M, Jaturapat A, Rukseree K, Danwisetkanjana K, Tanticharoen M, et al. (2001) Phomoxanthones A and B, novel xanthone dimers from the endophytic fungus Phomopsis species. Journal of Natural Products 64: 1015–1018. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. (2013) MAFFT Multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucharek T, Whiteside J, Brown E. (1983) Melanose and Stem end rot of Citrus. [Plant pathology fact sheet.] Gainesville: Florida Cooperative Extension Service, Institute of Food and Agricultural Sciences, University of Florida. [Google Scholar]
- Kumar S, Stecher G, Tamura K. (2016) MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaran RS, Hur B. (2009) Screening of species of the endophytic fungus Phomopsis for the production of the anticancer drug taxol. Biotechnology and Applied Biochemistry 54: 21–30. [DOI] [PubMed] [Google Scholar]
- Kumaresan V, Suryanarayanan TS. (2001) Occurrence and distribution of endophytic fungi in a mangrove community. Mycological Research 105: 1388–1391. [Google Scholar]
- Lamprecht SC, Crous PW, Groenewald JZ, Tewoldemedhin YT, Marasas WF. (2011) Diaporthaceae associated with root and crown rot of maize. IMA Fungus 2: 13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard L, Van Leeuwen GCM, Guarnaccia V, Polizzi G, Van Rijswick PC, et al. (2014) Diaporthe species associated with Vaccinium, with specific reference to Europe. Phytopathologia Mediterranea 53: 287–299. [Google Scholar]
- Mahadevakumar S, Yadav V, Tejaswini GS, Sandeep SN, Janardhana GR. (2014) First report of Phomopsis citri associated with dieback of Citrus lemon in India. Plant Disease 98: 1281–1281. [DOI] [PubMed] [Google Scholar]
- Mayorquin JS, Wang DH, Twizeyimana M, Eskalen A. (2016) Identification, distribution, and pathogenicity of Diatrypaceae and Botryosphaeriaceae associated with citrus branch canker in the southern California desert. Plant Disease 100: 2402–2413. [DOI] [PubMed] [Google Scholar]
- Mondal SN, Agostini JP, Zhang L, Timmer LW. (2004) Factors affecting pycnidium production of Diaporthe citri on detached citrus twigs. Plant Disease 88: 379–382. [DOI] [PubMed] [Google Scholar]
- Mondal SN, Vicent A, Reis RF, Timmer LW. (2007) Saprophytic colonization of citrus twigs by Diaporthe citri and factors affecting pycnidial production and conidial survival. Plant Disease 91: 387–392. [DOI] [PubMed] [Google Scholar]
- Mostert L, Crous PW, Kang JC, Phillips AJL. (2001a) Species of Phomopsis and a Libertella sp. occurring on grapevines with specific reference to South Africa: morphological, cultural, molecular and pathological characterization. Mycologia 93: 146–167. [Google Scholar]
- Mostert L, Kang JC, Crous PW, Denman S. (2001b) Phomopsis saccharata sp. nov., causing a canker and die-back disease of Protea repens in South Africa. Sydowia 53: 227–235. [Google Scholar]
- Muralli TS, Suryanarayanan TS, Geeta R. (2006) Endophytic Phomopsis species: host range and implications for diversity estimates. Canadian Journal of Microbiology 52: 673–680. [DOI] [PubMed] [Google Scholar]
- Nylander JAA. (2004) MrModeltest v. 2 Programme distributed by the author. Uppsala: Evolutionary Biology Centre, Uppsala University. [Google Scholar]
- O’Donnell K, Nirenberg HI, Aoki T, Cigelnik E. (2000) A multigene phylogeny of the Gibberella fujikuroi species complex: Detection of additional phylogenetically distinct species. Mycoscience 41: 61–78. [Google Scholar]
- Polizzi G, Aiello D, Vitale A, Giuffrida F, Groenewald JZ, et al. (2009) First report of shoot blight, canker, and gummosis caused by Neoscytalidium dimidiatum on citrus in Italy. Plant Disease 93: 1215. [DOI] [PubMed] [Google Scholar]
- Rayner RW. (1970) A Mycological Colour Chart. Kew: Commonwealth Mycological Institute. [Google Scholar]
- Rehm H. (1914) Ascomycetes Philippinenses VI. Leaflets of Philippine Botany 6: 2258–2281. [Google Scholar]
- Rehner SA, Uecker FA. (1994) Nuclear ribosomal internal transcribed spacer phylogeny and host diversity in the coelomycete Phomopsis. Canadian Journal of Botany 72: 1666–1674. [Google Scholar]
- Ronquist F, Teslenko M, Mark P van der, et al. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman AY, Adams GC, Cannon PF, Castlebury LA, Crous PW. et al. (2015) Recommendations of generic names in Diaporthales competing for protection or use. IMA Fungus 6: 145–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval-Denis M, Guarnaccia V, Polizzi G, Crous PW. (2018) Symptomatic citrus trees reveal a new pathogenic lineage in Fusarium and two new Neocosmospora species. Persoonia 40: 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos L, Alves A, Alves R. (2017a) Evaluating multi-locus phylogenies for species boundaries determination in the genus Diaporthe. PeerJ 5: e3120; DOI 10.7717/peerj.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos JM, Phillips AJL, Crous PW, Alves A. (2017b) Diaporthe species on Rosaceae with descriptions of D. pyracanthae sp. nov. and D. malorum sp. nov. Mycosphere 8: 485–511. [Google Scholar]
- Santos JM, Phillips AJL. (2009) Resolving the complex of Diaporthe (Phomopsis) species occurring on Foeniculum vulgare in Portugal. Fungal Diversity 34: 111–125. [Google Scholar]
- Santos JM, Vrandečicì K, Cìosicì J, Duvnjak T, Phillips AJL. (2011) Resolving the Diaporthe species occurring on soybean in Croatia. Persoonia 27: 9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos PJC, Savi DC, Gomes RR, Goulin EH, da Senkiv C, et al. (2016) Diaporthe endophytica and D. terebinthifolii from medicinal plants for biological control of Phyllosticta citricarpa. Microbiological Research 186–187: 153–160. [DOI] [PubMed] [Google Scholar]
- Senanayake IC, Crous PW, Groenewald JZ, Maharachchikumbura SSN, Jeewon R, et al. (2017) Families of Diaporthales based on morphological and phylogenetic evidence. Studies in Mycology: doi: 10.1016/j.simyco.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H, Wingfield MJ, Crous PW, Coutinho TA. (1996) Sphaeropsis sapinea and Botryosphaeria dothidea endophytic in Pinus spp. and Eucalyptus spp. in South Africa. South African Journal of Botany 62: 86–88. [Google Scholar]
- Swofford DL. (2003) PAUP*: phylogenetic analysis using parsimony (*and other methods), v. 4.0b10. Sunderland, MA: Sinauer Associates. [Google Scholar]
- Thomidis T, Exadaktylou E, Chen S. (2013) Diaporthe neotheicola, a new threat for kiwi fruit in Greece. Crop Protection 47: 35–40. [Google Scholar]
- Thomidis T, Michailides TJ. (2009) Studies on Diaporthe eres as a new pathogen of peach trees in Greece. Plant Disease 93: 1293–1297. [DOI] [PubMed] [Google Scholar]
- Timmer LW, Garnsey SM, Graham JH. (2000) Compendium of Citrus Diseases. 2nd edn St Paul, MN: American Phytopathological Society Press. [Google Scholar]
- Timmer LW, Kucharek TA. (2001) Melanose (revised). [Fact Sheet PP-150.] Gainsville: Plant Pathology Department, Florida Cooperative Extension Service, Institute of Food and Agricultural Sciences, University of Florida. [Google Scholar]
- Thompson S, Tan Y, Young A, Neate S, Aitken E, et al. (2011) Stem cankers on sunflower (Helianthus annuus) in Australia reveal a complex of pathogenic Diaporthe (Phomopsis) species. Persoonia 27: 80–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson S, Tan Y, Shivas R, Neate S, Morin L, et al. (2014) Green and brown bridges between weeds and crops reveal novel Diaporthe species in Australia. Persoonia 35: 39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udayanga D, Castlebury LA, Rossman AY, Chukeatirote E, Hyde KD. (2015) The Diaporthe sojae species complex: Phylogenetic reassessment of pathogens associated with soybean, cucurbits and other field crops. Fungal Biology 119: 383–407. [DOI] [PubMed] [Google Scholar]
- Udayanga D, Castlebury LA, Rossman AY, Chukeatirote E, Hyde KD. (2014a) Insights into the genus Diaporthe: phylogenetic species delimitation in the D. eres species complex. Fungal Diversity 67: 203–229. [Google Scholar]
- Udayanga D, Castlebury LA, Rossman AY, Hyde KD. (2014b) Species limits in Diaporthe: molecular re-assessment of D. citri, D. cytosporella, D. foeniculina and D. rudis. Persoonia 32: 83–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udayanga D, Liu X, McKenzie EHC, Chukeatirote E, Bahkali AHA, et al. (2011) The genus Phomopsis: biology, applications, species concepts and names of common phytopathogens. Fungal Diversity 50: 189–225. [Google Scholar]
- Udayanga D, Xingzhong L, Crous PW, McKenzie EHC, Chukeatirote E, et al. (2012) A multi-locus phylogenetic evaluation of Diaporthe (Phomopsis). Fungal Diversity 56: 157–171. [Google Scholar]
- Uecker FA. (1988) A world list of Phomopsis names with notes on nomenclature, morphology and biology. Mycological Memoirs 13: 1–231. [Google Scholar]
- Van der Aa HA, Noordeloos ME, De Gruyter J. (1990) Species concepts in some larger genera of the coelomycetes. Studies in Mycology 32: 3–19. [Google Scholar]
- Van Niekerk JM, Groenewald JZ, Farr DF, Fourie PH, Halleen F, et al. (2005) Reassessment of Phomopsis species on grapevines. Australasian Plant Pathology 34: 27–39. [Google Scholar]
- Van Rensburg JCJ, Lamprecht SC, Groenewald JZ, Castlebury LA, Crous PW. (2006) Characterization of Phomopsis spp. associated with die-back of rooibos (Aspalathus linearis) in South Africa. Studies in Mycology 55: 65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicent A, Armengol J, García-Jiménez J. (2007) Rain fastness and persistence of fungicides for control of Alternaria brown spot of citrus. Plant Disease 91: 393–399. [DOI] [PubMed] [Google Scholar]
- Wehmeyer LE. (1933) The genus Diaporthe Nitschke and its segregates. University of Michigan Studies, Science Series 9: 1–349. [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor JW. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR Protocols: a guide to methods and applications (Innis MA, Gelfand DH, Sninsky JJ, White TJ, eds): 315–322. San Diego: Academic Press. [Google Scholar]
- Yang J, Xu F, Huang C, Li J, She Z, et al. (2010) Metabolites from the mangrove endophytic fungus Phomopsis sp. (#zsu-H76). European Journal of Organic Chemistry 19: 3692–3695. [Google Scholar]


