Abstract
Zika virus (ZIKV) encodes a precursor protein (also called polyprotein) of about 3424 amino acids that is processed by proteases to generate 10 mature proteins and a small peptide. In the present study, we characterized the chemical features, suborganelle distribution and potential function of each protein using Flag-tagged protein expression system. Western blot analysis revealed the molecular weight of the proteins and the polymerization of E, NS1, and NS3 proteins. In addition, we performed multi-labeled fluorescent immunocytochemistry and subcellular fractionation to determine the subcellular localization of these proteins in host cells. We found that 1) the capsid protein colocalizes with 3 different cellular organelles: nucleoli, Golgi apparatus, and lipid droplet; NS2b and NS4a are associated with the Golgi apparatus; 2) the capsid and NS1proteins distribute in both cytoplasm and nucleus, NS5 is a nuclear protein; 3) NS3 protein colocalizes with tubulin and affects Lamin A; 4) Envelope, PrM, and NS2a proteins co-localize with the endoplasmic reticulum; 5) NS1 is associated with autophagosomes and NS4b is related to early endosome; 6) NS5 forms punctate structures in the nucleus that associate with splicing compartments shown by SC35, leading to reduction of SC35 protein level and trafficking of SC35 from the nucleus to the cytoplasm. These data suggest that ZIKV generates 10 functional viral proteins that exhibit distinctive subcellular distribution in host cells.
Keywords: Zika virus (ZIKV), Capsid, Envelope, Nonstructural (NS) proteins, Centrosome, Mitochondria, Endoplasmic reticulum (ER), Golgi apparatus, Endosome, Autophagy
1. Introduction
The recent outbreak of the Zika virus (ZIKV) has attracted attention worldwide and ZIKV infected cases are now spreading from the Americas to many other countries and its infection might be linked to some severe medical sequelae (Mlakar et al., 2016; Petersen et al., 2016; Weaver et al., 2016). The recent reports that Zika virus infection is probably associated with microcephaly of the neonates and Guillain–Barré syndromes (GBS) in adults spurred researchers to seriously reevaluate the medical significance of this agent as a pathogen (Mlakar et al., 2016; Petersen et al., 2016; Costa et al., 2016; Oehler et al., 2013). Since its first isolation from an infected monkey in 1947 in Uganda, only a few studies had been taken before the recent outbreak in Brazil. Detailed information about ZIKV that can be acquired via in depth investigation at the molecular, epidemical, and clinical levels will be critical for the elucidation of its role as a pathogen of serious diseases in humans.
The ZIKV, together with the West Nile virus, Yellow fever virus, Japanese encephalitis virus, Dengue fever virus, and many other viruses, forms the genus Flavivirus that belongs to family Flaviviridae (Wikan and Smith, 2016; Plourde and Bloch, 2016). The family Flaviviridae consists of many other viruses that have been summarized in a 2010 review (Bollati et al., 2010). A growing number of strains of ZIKV have been isolated from > 60 countries (Ramos da Silva and Gao, 2016; Dick et al., 1952). In earlier studies, it was determined that it causes only a mild arthropod-borne disease in humans, known as Zika fever, and so it had been rarely taken into clinical consideration seriously until recent epidemic outbreaks. The first ZIKV was isolated from a monkey and it is known that ZIKV can be transmitted to humans via mosquito bite or occasionally by sexual contact (Ramos da Silva and Gao, 2016; Boorman and Porterfield, 1956; Haddow et al., 1964; Marchette et al., 1969; Musso et al., 2015). It was then isolated from humans in Nigeria many years later (Moore et al., 1975). Since 2007, ZIKV-caused epidemic outbreaks with different scales have occurred in Micronesia, French Polynesia, Cook Island, and Easter Island, ZIKV has become an emerging arbovirus (Musso et al., 2014). More recently, a pandemic of ZIKV infection occurred in South America. ZIKV infection has been related to the increasing number of cases of microcephaly and GBS in the areas of epidemics (Ramos da Silva and Gao, 2016; Stratton, 2016). Recent studies using mouse models demonstrated that ZIKV infection directly inhibited neuron stem cell proliferation, which supports the hypothesis that ZIKV is causatively related to microcephaly (Aliota et al., 2016; Hickman and Pierson, 2016; Lazear et al., 2016; Rossi et al., 2016; Werner et al., 2016). Phylogenetic studies have led to the classification of the ZIKVs into Asian and African lineages (Faye et al., 2014; Lanciotti et al., 2016). The recent cases of microcephaly and GBS linked to ZIKV are mostly, if not all, caused by Asian strains (Weaver et al., 2016). During the evolution of ZIKV, the virus developed new molecular relationships with factors of host cells (Faye et al., 2014; Qin, 2016; Singh et al., 2016; Shen et al., 2016; Wang et al., 2016). It is likely that interactions of viral proteins and viral genomic RNA with host factors determine the fate and/or efficiency of infection, pathogenicity, transmission, and epidemic potential.
This family of viruses has an enveloped, icosahedral capsid that contains a single stranded RNA genome (about 11,000 nucleotides) with positive sense (Faye et al., 2014). Therefore, the infected viral RNA can be directly translated to a large polyprotein precursor, which is co- and post-translationally processed by viral and cellular proteases into structural and nonstructural proteins. The three structural proteins are critical for the formation of envelope and capsid, and the seven nonstructural (NS) proteins play important roles in virus replication. The three structural proteins are envelope, E; membrane precursor, PrM; and capsid C. The seven nonstructural (NS) proteins include NS1, NS2a, NS2b, NS3, NS4a, NS4b, and NS5. The molecular features, subcellular trafficking with host cells and biological functions of each viral protein of ZIKV remain largely unknown. These information are important to understand ZIKV infection/replication and virus-host interactions. Due to the lack of highly specific antibody against each viral protein of ZIKV, the present study aims to clone and express 10 ZIKV viral genes using mammalian Flag-tagged expression vector. Upon overexpression of each FLAG-tagged ZIKV protein, we characterized their chemical features and subcellular localization. We identified some novel functions of the ZIKV proteins.
2. Results
2.1. Molecular cloning and expression of the viral proteins
We selected ZIKV MR766 strain as a representative study because it has been widely used in recent studies. Kuno and Chang performed a comparative analysis of the genomic sequences of MR766 strain together with other flaviviruses and indicated that MR766 genome encodes a protein precursor with a length of 3424 aa that is cleaved to generate 10 functional proteins as shown in Fig. 1A (Kuno and Chang, 2007; Cai et al., 2016; Armstrong et al., 2017). We cloned them into the pcDNA3 vector for their production in mammalian cells. Every ZIKV protein is tagged with FLAG for two reasons: (a) short tag minimizes the risk of affecting the function of the protein, (b) good anti-Flag antibody is available for western blot, immunostaining and immunoprecipitation studies. All the cloned plasmids were confirmed by Sanger sequencing. To determine whether the proteins can be expressed, we transfected the plasmids into HEK293T cell for 24 h, whole cell lysate samples were applied to western blot assay using anti-FLAG antibody. As shown in Fig. 1B, all 10 proteins were detected by anti-FLAG antibody and the molecular weight of each viral protein was mostly close to the predicted size according to the number of the translated amino acids. When we exposed the same membrane of the western blot for a longer time, some of the ZIKV proteins (E, NS1, NS3, and NS5) show bigger or smaller bands that may be caused by post-translational modification and/or potential degradation/cleavage (data not shown).
Fig. 1.
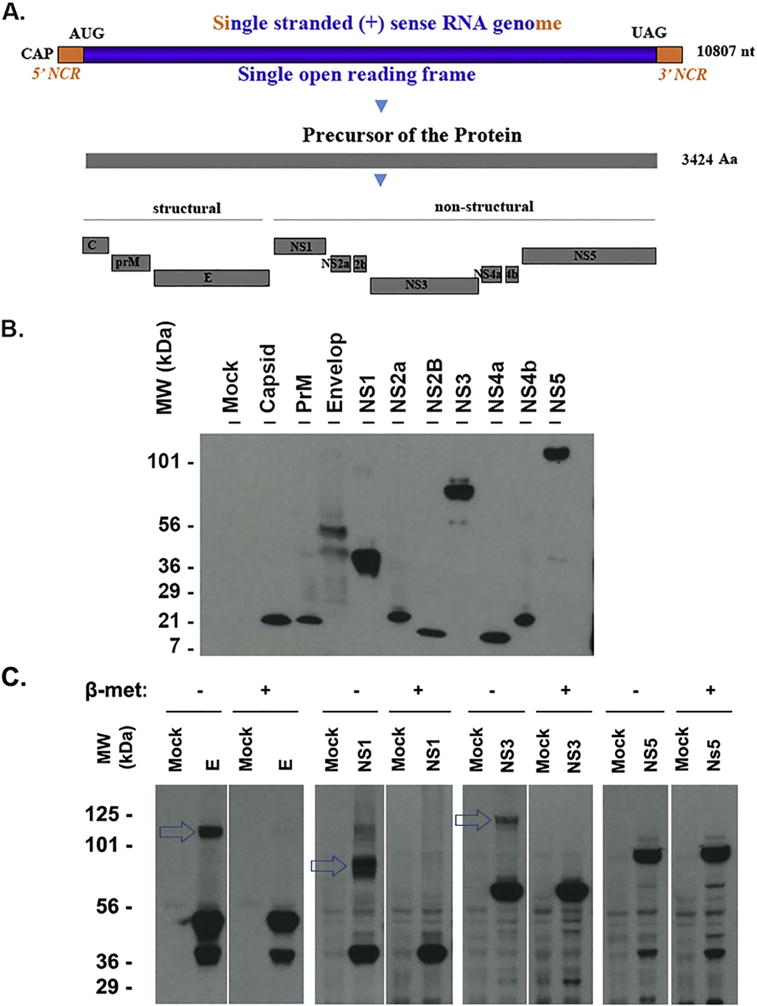
Expression of the ZIKV proteins that are detected by western blot. A. Genomic structure and gene production of ZIKV (MR766 strain, GenBank Sequence Accession: LC002520). AUG: translation start codon; UAG: translation stop codon; NCR: noncoding RNA sequence; nt: nucleotide; Aa: amino acid; B. Western blot assay to examine ZIKV proteins. The ZIKV protein-expressing plasmids were transfected into HEK 293T cells for 24 h. The whole cell lysate samples were applied to run a PAGE and the transferred membrane was blotted with anti-FLAG antibody. The names of the protein were shown on the top and the size marker was shown on the left. C. western blot assay to examine the protein polymerization. HEK293T cells were transfected with the plasmid-expressing ZIKV protein E, NS1, NS3, or NS5 for 24 h. The whole cell lysate samples were collected in a SDS-lysis buffer with or without reducer (beta-mercaptoethanol) and examined for ZIKV protein using anti-FLAG antibody. The experiments have been performed for more than 3 times, one representative is shown.
To determine whether dimerization or polymerization occurred to ZIKV viral proteins, we performed the western blot assay for the samples in SDS-lysis buffer with or without reducer (beta-mercaptoethanol). As shown in Fig. 1C, the upper bands (shown by arrows) of E, NS1, and NS3 can only be seen in the blots of samples without beta-mercaptoethanol. When the reducer (beta-mercaptoethanol) was added in the samples, the upper band disappeared. No such a difference was seen for NS5 (last panel of Fig. 1C). Therefore, E, NS1, NS3, but not NS5 can be polymerized.
2.2. Subcellular localization of ZIKV proteins in Vero cells after transfection
It is important to know the protein’s localization in cells because proteins exert their functions largely in a specific subcellular location. First, we wondered whether the proteins could have any nuclear localization signals (NLS). To address this, we analyzed the aa sequences of ZIKV proteins using a Mapper cNLS software (Kosugi et al., 2009). This bioinformatics analysis resulted in the recognition of NLS in 4 of the ZIKV proteins: capsid, NS1, NS3 and NS5 as listed in Table 1. Next, we transfected the plasmids into Vero cells for 24 h and the cells were fixed and permeabilized for immunocytochemistry (ICC). As shown in Fig. 2, NS5, presented with punctate structures - is only found in the nucleus, which is consistent with a recent report (Grant et al., 2016). Capsid and NS1 are found in both cytoplasm and nucleus. Capsid protein forms domain-like patterns in the nucleus. PrM, envelope, NS2a, NS2b, NS3, NS4a and NS4b distribute only in cytoplasm. NS3 does not go into nucleus despite the presence of putative NLS sequence.
Table 1.
Nuclear localization signals of ZIKV proteins.
| Capsid | RKERKRRGADT |
| NS1 | RLKRAHLIEM |
| NS3 | RRVLPEIVREAIKKRLRTV |
| KYGEKRVLKPRWMDARVCSDHAALKSFKE | |
| NS5 | ELGKRKRPRVCTKEEFINKVRSN |
The Aa sequences of the ZIKV proteins were input into cNLS Mapper software, the NLS sequences with high score > 5 were recorded and listed in the table.
Fig. 2.
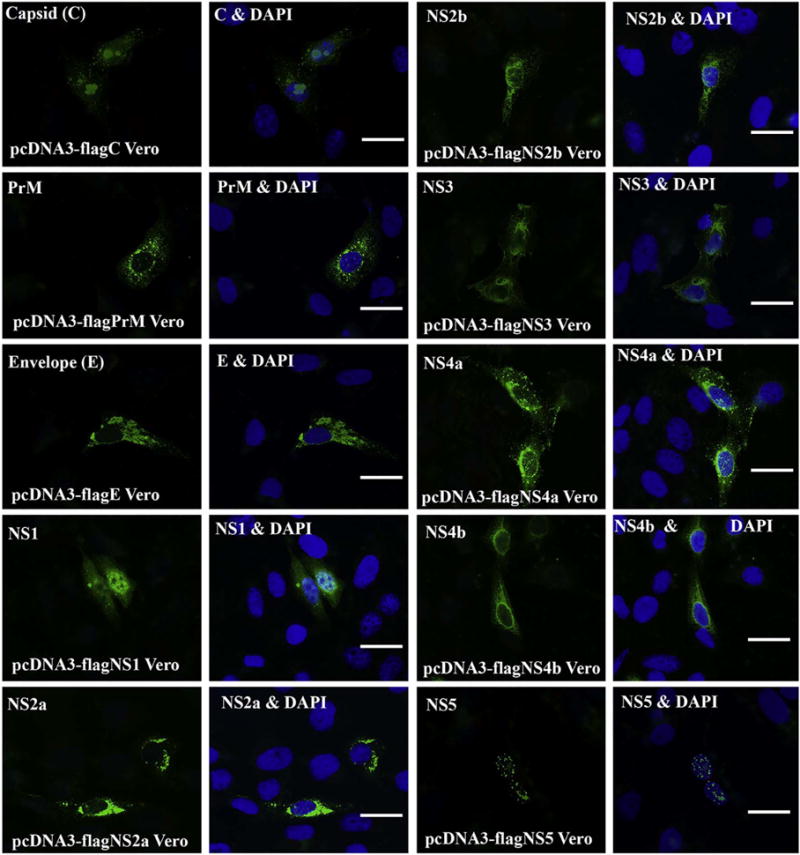
ICC for Visualizing the localization of ZIKV proteins in Vero cells. Vero cells were transfected with the ZIKV protein-expressing plasmid for 24 h. Then, the cells were fixed and permeabilized for ICC. The expressed ZIKV protein was stained with anti-FLAG antibody in green. For each protein, there are two panels, left panel shows the ZIKV protein only in green, and the right panel shows both the nucleus (in blue by DAPI) and the protein. PrM: precursor membrane; C: capsid; E: envelop; NS: nonstructural. Scale bar: 15 μm. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
To further confirm the function of the putative NLS within 4 viral proteins as described above, we performed site-mutagenesis assay and generated pcDNA3-flagCdNLS, pcDNA3-flagNS1dNLS, pcDNA3-flagNS3dNLS1, pcDNA3-flagNS3dNLS2, or pcDNA3-flagNS5dNLS followed by immunocytochemical analysis in Vero cells. As shown in Fig. 3, C protein formed domain-like structure in the cytoplasm, but no longer localized in the nucleus (Fig. 3A); NS5 formed dots only in the cytoplasm (Fig. 3C) and NS1 diffused only in the cytoplasm (Fig. 3B). Again, both wt and mutated NS3 distributed only in the cytoplasm (data not shown). Therefore, the predicted NLSs for C, NS1, and NS5 proteins are correct but the NLS within NS3 does not affect its cytoplasmic distribution.
Fig. 3.
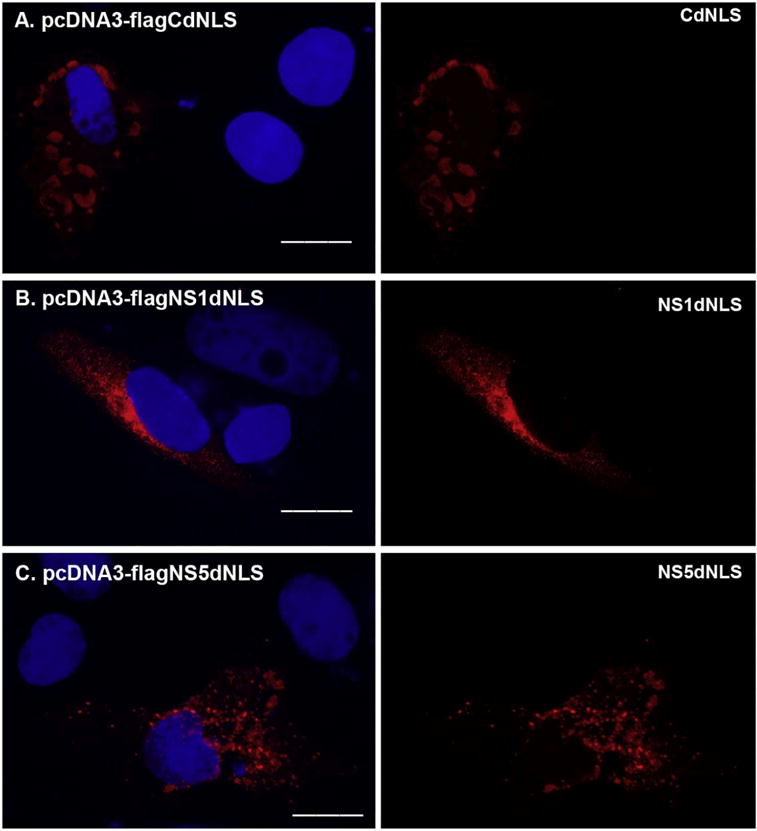
The localization of NLSs mutated ZIKV-encoded proteins. The overlapping PCR method was employed to mutate the putative NLS from protein C, NS1, NS3, or NS5. The primers used for the overlapping PCR were shown in Table 3. The fragments generated by overlapping PCR contain the desired mutation, were gel-purified and cloned into pcDNA3 resulting in pcDNA3-flagCdNLS, pcDNA3-flagNS1dNLS, or pcDNA3-flagNS5dNLS. The plasmid pcDNA3-flagCdNLS (A), pcDNA3-flagNS1dNLS (B), or pcDNA3-flagNS5dNLS (C) was transfected into Vero cells for 24 h. The merged color of the protein (red) and DAPI (blue) was shown in the left panel, and the protein alone (red) was shown in the right panel. ICC was performed to stain the transfected proteins using anti-FLAG antibody in red. DAPI staining was to show the nucleus. Scale bar: 15 μm. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.3. ZIKV C protein associates with Golgi apparatus in the cytoplasm and co-localizes with nucleoli in the nucleus
Recent structural studies on ZIKV reached an agreement that ZIKV virion structure is like that of DENV (Kostyuchenko et al., 2016; Sirohi et al., 2016). According to the structure of the viral particle of DENV (Rey et al., 1995), each viral particle contains three E protein monomers in the asymmetric unit of the virus, and both membrane (M) and E proteins are associated with host-derived lipid bilayer. Multiple copies of C protein form the nucleocapsid core that is located inside the lipid bilayer (Jones et al., 2003). Therefore, C protein may be important for assembly of ZIKV. To characterize the function of the ZIKV C protein, we transfected pcDNA3-flagC into Vero cells. Then, multi-labeled ICC was performed to show the relationship between C protein and host subcellular markers including Calregulin (ER), Giantin (Golgi), SC35 (splicing compartment), and Tubulin (spindle and microtubule). Interestingly, in many cells, the capsid domains co-localize with nucleoli in the nucleus and with Golgi apparatus in the cytoplasm as shown in Fig. 4A–F. To better visualize the relationship of the three components: nucleoli, viral capsid protein, and Golgi apparatus that was stained by Giantin, a matrix protein of Golgi apparatus (Linstedt and Hauri, 1993), we separated and merged different colors. Clearly, the capsid protein colocalizes with the nucleoli (Fig. 4C, F). We can also see that the capsid protein in the cytoplasm partially overlaps with Golgi apparatus. The observation of localization of the C protein with the nucleoli in the nucleus and with Golgi apparatus in the cytoplasm inspires our future investigation of whether the C protein plays roles in viral protein transportation and ribosome assembly. The third pattern distribution of C protein is the dot-like structure in the cytoplasm, which can be seen in all the transfected cells. To determine whether any cellular organelles colocalize with the C protein dots, we co-stained the cells with anti-Flag for C protein in red and with BODIPY 500/510 for lipid droplet (LD) in green (Fig. 4D–F). As can be seen in the Fig. 4D–F, the C protein dots in the cytoplasm all colocalize with LD. We then counted 500C protein-positive cells to examine the rate of association of C protein with the three cellular organelles. As shown in Fig. 4G, C protein colocalizes with LD in all the 500 cells (100%), associate with Golgi apparatus in 350 cells (70%), and colocalize with nucleoli in 450 cells (90%). Therefore, we revealed that ZIKV C protein has three different association patterns in host cells: nucleoli, Golgi apparatus, and LD.
Fig. 4.
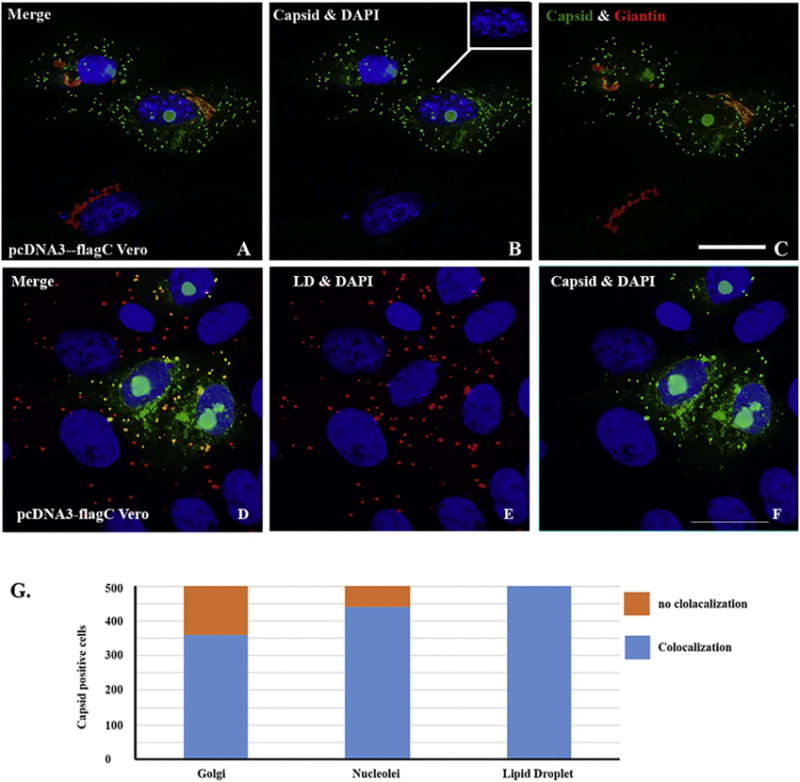
ICC for showing the localization of capsid protein that colocalizes with nucleoli in nucleus and Golgi apparatus or lipid droplet (LD) in cytoplasm. Vero cells were transfected with pcDNA3-flagC. After 24 h, the cells were fixed and ICC was performed to show the relationship between C protein and cellular proteins. A: merged colors of C protein (green), Giantin (Golgi in red) and DAPI (blue); B: merged C (green) and DAPI (blue); C: merged C and (green) and Giantin (red). D: merged C (green), lipid droplet (LD in red) and DAPI (blue); E: merged LD (red) and DAPI (blue); F: merged C protein (green) and DAPI (blue). Scale bar: 10 μm. G. Quantitating the association of C protein with LD, nucleoli, and Golgi apparatus. Five hundred C protein positive cells were counted for the colocalization of C protein with LD, nucleoli, and Golgi apparatus. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.4. ZIKV envelope, PrM, and NS2a proteins co-localize with endoplasmic-reticulum (ER)
ER is the site for synthesis of ZIKV proteins immediately after ZIKV infection. It was also detected by electronic microscopy that ZIKV replicates in enveloped organelles, including mitochondria and ER (Garcez et al., 2016). Before viral replication, several viral proteins must interact with ER. In DENV, NS3, NS4a and NS4b were found to interact with each other in ER (Zou et al., 2015). We wondered which protein of ZIKV locates with ER. To find out, we transfected ZIKV protein-expressing plasmid into Vero cells for 24 h. We simultaneously stained ZIKV protein (in green) and ER protein (Calregulin in red). As shown in Fig. 5, envelope (E) protein, PrM, and NS2a colocalize with ER. However, in another ICC experiment we didn’t observe that NS3, NS4a and NS4b of ZIKV localized with ER (data not shown). Therefore, different flaviviruses have different genomically encoded proteins that associate with the ER.
Fig. 5.
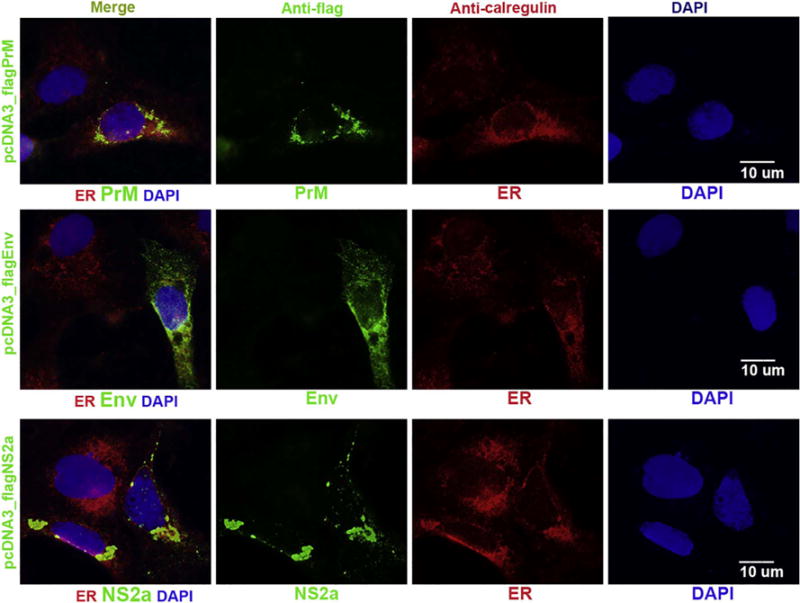
ICC to show that ZIKV PrM, envelope and NS2a proteins in cells after transfection. Vero cells were transfected with pcDNA-flagPrM, pcDNA-flagE, or pcDNA3-flagNS2a for 24 h. The cells were simultaneously stained with anti-FLAG antibody to show ZIKV protein (in green) and ER protein (Calregulin in red). The nuclei were shown by DAPI. Scale bar: 10 μm. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.5. NS1 associates with autophagy, NS2b and NS4a associate with Golgi apparatus, and NS4b is related to early endosome
The same methods were used to determine the subcellular localization of NS1. After having tried many different antibodies for the ICC assays, we observed that NS1 colocalizes with the microtubule associated protein (MAP) light chain 3 (LC3) using anti-LC3beta antibody as shown in Fig. 6A. MAPLC3 is the central protein in the autophagosome biogenesis and an important molecular component of autophagy pathway. Therefore, LC3 is the most widely used maker of autophagosomes (Bernard et al., 2015). Our revelation that NS1 colocalizes with autophagosome implies a biological function of ZIKV NS1 through interaction with autophagy pathway.
Fig. 6.
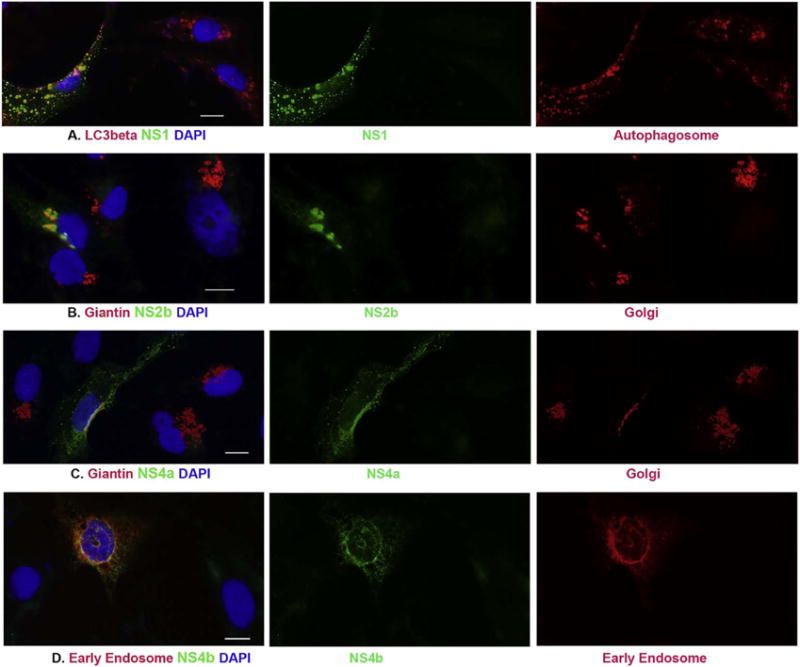
ICC to characterize the localization of NS1, NS2b, NS4a and NS4b localizes in cells after transfection. pcDNA3-flagNS1 (panel A), pcDNA3-flagNS2b (panel B), or pcDNA3-flagNS4a (panel C) was transfected to Vero cells for 24 h. ICC was performed to show the association between ZIKV proteins in green and LC3beta (marker of Autophagosome) or Giantin (marker of Golgi apparatus) in red. D. Vero cells were cotransfected with pcDNA3flagNS4b and pmRFPRab5 for 24 h, anti-FLAG antibody was used show NS4b in green and the early endosome protein Rab5 is in red. The nuclei were shown by DAPI. Scale bar: 10 μm. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
It was previously demonstrated that DENV NS4a protein co-localized with NS4b and NS3 and showed an ER-like staining pattern by ICC (Zou et al., 2015). We performed ICC and demonstrated that there is no association between ZIKV NS4a and ER. Interestingly, we found that NS4a co-localizes with the Golgi apparatus as shown in Fig. 6C. Although the NS4a is a cytoplasmic protein distributing all over the cytoplasm, it is clear that NS4a condensed in the Golgi apparatus and co-localizes with Giantin, a matrix protein of the Golgi apparatus (Linstedt and Hauri, 1993). Our results may suggest that there exist differences between DENV NS4a and ZIKV NS4a. In addition, we also found that NS2b associates with Golgi apparatus (Fig. 6B). Therefore, ZIKV NS2b and NS4a probably play their biological functions through interaction with Golgi components.
We have tried several antibodies to know whether any ZIKV proteins are associated with early endosome, but no conclusion can be drawn due to the antibodies used were not good. Therefore, we used cotransfection of ZIKV protein-expressing plasmid and pmRFP-Rab5 that expresses a protein of early endosome. We only found that the NS4b is associated with Rab5 that is an early endosomal protein as shown in Fig. 6D.
2.6. ZIKV NS3 associates with tubulin and affects Lamin A
Tubulin is one of the important components of microtubule that is a cytoskeleton and essential for cellular mitosis (Weisenberg, 1972). Lamin A is an important component of the nuclear lamina, forming a dense fibrillary network inside the nucleus that not only provides the mechanical support for cells but also regulates important cellular events such as DNA replication and cell division (Gruenbaum et al., 2000). Our ICC experiments for visualizing ZIKV NS3 protein showed that NS3 co-localizes with alpha tubulin in transfected Vero cells, as shown in the left panel of Fig. 7 (A1–A4). The NS3 protein (A1–A3, green) forms a fibrillary structure extended from the centrosome that is shown in red (A1 and A2). As can be seen in the Fig. 7, the centrosome is located near the nucleus (A4) and microtubules (tubulin) extend outward to the cell periphery, suggesting that NS3 affects the distribution of tubulin and centrosome. Another interesting observation is that NS3 affected the nuclear lamina as shown in the right panel of Fig. 7 (B1–B5). In the untransfected Vero cells, the nuclear lamina smoothly surrounds the nuclear membrane. However, in the NS3-expressed cells, the nuclear lamina morphology clearly changed with an extruding site (shown by the arrows in Fig. 7B3) that is close to the centrosome. Our results suggest that NS3 may affect function of centrosome.
Fig. 7.
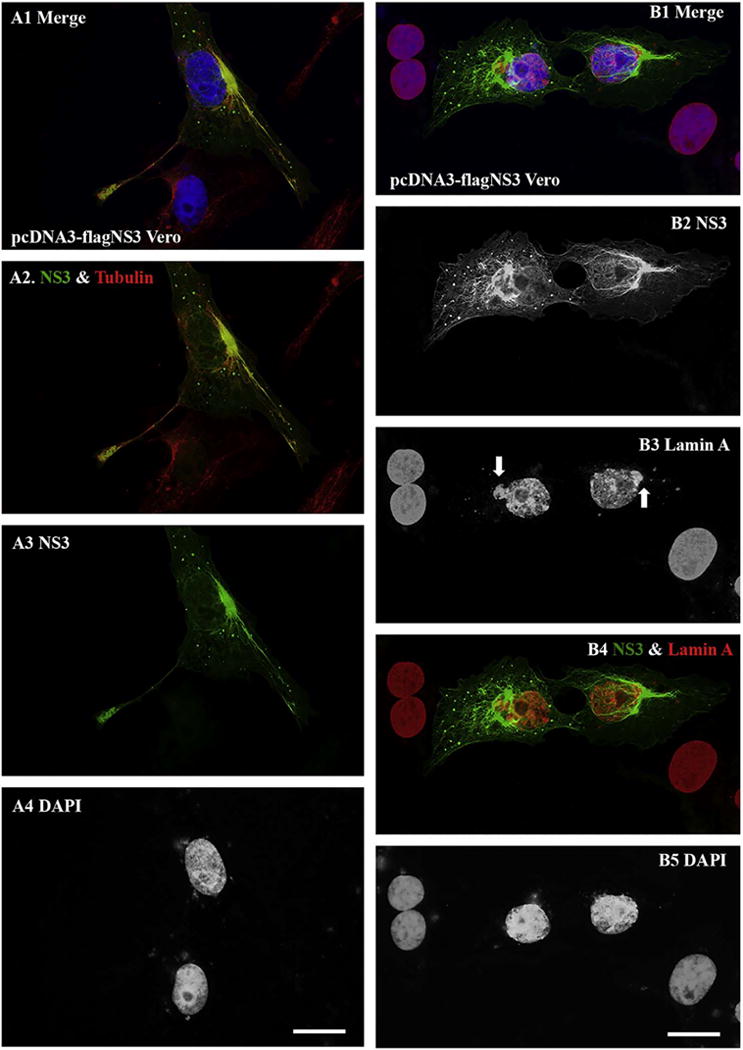
ICC to show the NS3 protein association with tubulin and with Lamin A. A. Vero cells were transfected with pcDNA3-flagNS3 for 24 h. ICC experiments were performed for visualizing ZIKV NS3 protein (in green) and tubulin in red (A1–A4). The nuclei of the cells were shown by staining with DAPI (A1 and A4). B. ICC for showing NS3 in green and the nuclear lamin A in red. The arrows in B3 are to show the sites that were affected by NS3. Scale bar: 10 μm. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.7. NS5 associates with SC35
The nucleus is a cellular organelle that contains many protein-composed dot-like functional domains. Splicing compartments, needed for gene splicing (Fu and Maniatis, 1992; Fu et al., 1992) are one such functional domain. To find out whether any nuclear protein associates with NS5, we transfected pcDNA3-flagNS5 into Vero cells and simultaneously stained NS5 with different subcellular proteins. We found that NS5 closely localizes with SC35 in the nucleus as shown in Fig. 8. We also observed in the NS5-expressing cells that: 1) SC35 density is reduced dramatically compared to the nontransfected cells, and 2) SC35 localized in the cytoplasm as a domain (shown by the arrows in Fig. 8). These interesting observations were further corroborated by nuclear/cytosolic fractionation and western blot analysis. As shown in Fig. 8E, the total SC35 level in NS5-expressed HEK 293 T cells is significantly lower than that in Mock cells. SC35 was completely localized in the nuclear fraction like Lamin A in Mock fractions (Fig. 8F, left) but over 50% SC35 was translocated to cytoplasm in NS5-expresing cells (Fig. 8F, right). Therefore, NS5 not only reduces SC35 level but also translocates SC35 from nucleus to cytoplasm.
Fig. 8.
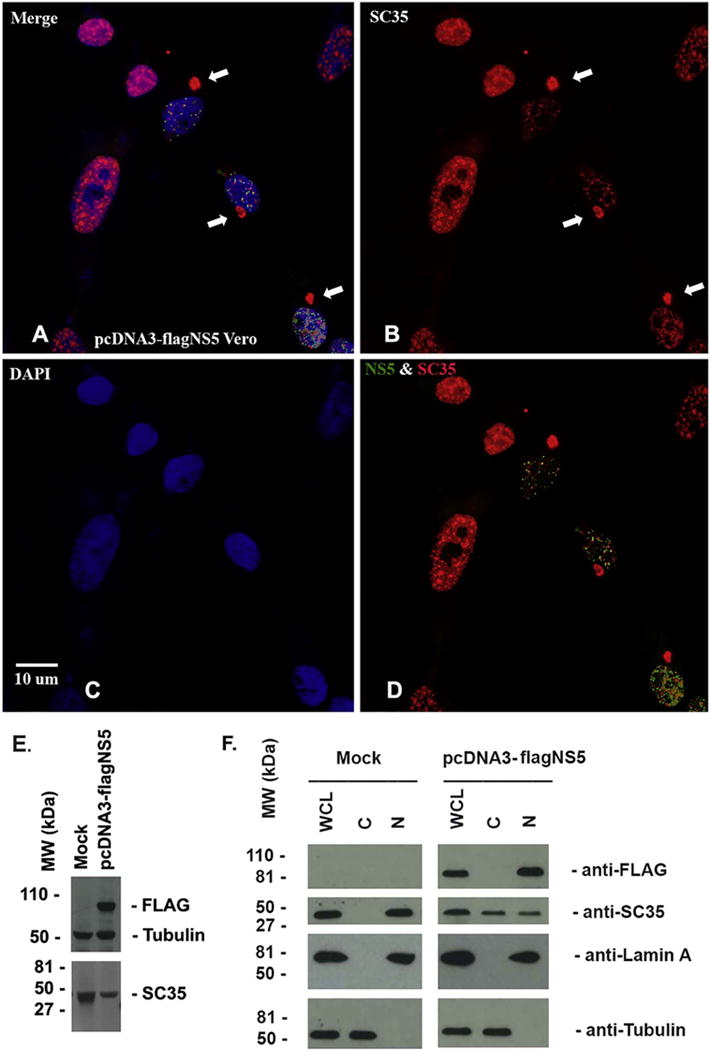
ICC to show that NS5 associates with SC35. pcDNA3-flagNS5 was transfected into Vero cells for 24 h. A–D: ICC was performed to simultaneously stained NS5 in green (A and D) and SC35 in red (A, B, and D). The arrows were to show that SC35 localized to cytoplasm as a domain. The nuclei were shown by DAPI. Scale bar: 10 μm. E. Vero cells were transfected with pcDNA3 (mock) or pcDNA3flagNS5 for 24 h, the whole cell lysates were applied to run western blot assay to determine the level of SC35, NS5, and tubulin. F. Vero cells were transfected with pcDNA3 (mock) or pcDNA3flagNS5 for 24 h, the whole cell lysates and cell fractions (cytoplasm and nuclear plasm) were prepared and were applied to run western blot assay to determine the level of SC35, NS5, Lamin A (nuclear protein) and tubulin (cytoplasmic protein). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3. Discussion
The salient finding of this study is the unique subcellular distribution of the 10 ZIKV proteins (as summarized in Table 2). Using selected markers for subcellular organelles, we found that NS5 is a nuclear protein, that C and NS1 localized in both nucleus and cytoplasm, that E protein, PrM, and NS2a are with ER, NS3 in centrosome, NS4a, NS2b and C protein in Golgi, and that NS1 is associated with autophagosomes while C protein is also in lipid droplet. NS5 promotes the trafficking of the serine/arginine-rich splicing factor SC35 from the nucleus to the cytoplasm and reduces the total level of SC35 protein, suggesting that NS5 may play important role in regulating the gene splicing process in host cells. It has been reported that ZIKV NS5 has proteasome activity, which resulted in degradation of the IFN-regulated transcriptional activator STAT2 from humans (Grant et al., 2016). Further studies are needed to determine whether NS5 also causes degradation of SC35 and to ascertain the consequence of NS5-mediated decrease of SC35 in host cells.
Table 2.
Characterization of ZIKV proteins.
| Name | Size: Aa (kDa) |
Cellular distribution | Subcellular location |
|---|---|---|---|
| C | 122 (14) | Cyto. & Nuc. | Golgi apparatus, LD, and nucleoli |
| PrM | 168 (19) | Cyto. | Endoplasmic reticulum, centrosome |
| sEnv | 504 (54) | Cyto. | Endoplasmic reticulum |
| NS1 | 352 (40) | Cyto. & Nuc. | Autophagosome |
| NS2a | 226 (24) | Cyto. | Endoplasmic reticulum |
| NS2b | 130 (14) | Cyto. | Golgi apparatus |
| NS3 | 617 (69) | Cyto. | Co-localize with Tubulin, centrosome |
| NS4a | 127 (14) | Cyto. | Golgi apparatus |
| NS4b | 251 (27) | Cyto. | Early endosome |
| NS5 | 903 (103) | Nuc. | Co-localize with SC35 |
Env: envelope; C: capsid; LD: lipid droplet; PrM: membrane precursor; NS: nonstructural protein; Cyto: Cytoplasm; Nuc: Nucleus.
Currently, the characterization of ZIKV proteins is usually inferred from other flaviviruses, especially DENV. In the present study, we performed the first cloning of all the ZIKV proteins for their expression in mammalian cells. The western blot assays using highly specific Flag antibody demonstrated that the sizes of most ZIKV proteins are similar to those of DENV (Henchal and Putnak, 1990). Using the cNLS Mapper software (Kosugi et al., 2009), we found that C protein, NS1, NS3 and NS5 have the NLS. Our gene mutation and ICC results also confirmed the predictions for C, NS1 and NS5. NS3 is largely a cytoplasmic protein, but its effects on Lamin A, a nuclear protein, suggest NS3 has activities in nucleus. C protein is structural protein and hence important for viral assembly. We found that it co-localizes with nucleoli in the nucleus and co-localizes with Golgi apparatus in the cytoplasm, suggesting that the C protein is not only a structure of the viron, but also play important roles in ribosome assembly for viral gene translation, in transportation of viral proteins and in viral RNA-protein interactions. These biological activities of C proteins are presently under investigation in our laboratory.
Our ICC results showed that NS3 associates with both tubulin and Lamin A, both of which are important for cell division (Dabauvalle et al., 1999; Kim et al., 2008; Kim et al., 2013). Lamin A is a nuclear protein and connected to the nuclear membrane by an isoprenyl group that is associated with membrane through protein-protein interaction (Reese and Maltese, 1991). Lamin A is required for early biological activities of cell mitosis by depolymerization of lamins (Reese and Maltese, 1991). During mitosis, duplicated centrosomes separate and form mitotic spindles with microtubules. The major component of microtubule is tubulin. NS3 associates with both tubulin and Lamin A, which suggests an important function of NS3 in cell mitosis: inhibitory or enhancive. Most interestingly, NS3 overexpression impaired the structure of centrosome. NS3 may contribute to the centrosome damage induced by ZIKV as reported recently (Gabriel et al., 2017; Wolf et al., 2017).
Cellular organelles such as ER and Golgi apparatus, exhibit different and important functions for viral replication. Viruses need to usurp the cellular machineries for viral biogenesis, which may relate to viral permissiveness (Gabriel et al., 2017; Hou et al., 2017). These procedures should be completed by viral proteins. Interestingly, we found that capsid and NS4a proteins are associated with the Golgi apparatus, while the NS2a and the envelop proteins overlap with ER. The subcellular location of the NS4b has not been characterized yet using our currently used approach. Different location of ZIKV proteins in host cells suggests a dynamic incorporation of each viral protein during ZIKV assemble, trafficking, maturation and production. With the proteins cloned into expressing vectors, we are able to investigate the functions for all the ZIKV proteins, especially their activities in causing defects of stem cell proliferation/differentiation.
Future work related to this study is to demonstrate the protein distribution in the context of ZIKV infection. We are now in the stage of producing specific monoclonal antibodies against each ZIKV protein.
4. Materials and methods
4.1. Cell lines, tissue culture and viruses
Vero cells (ATCC® CCL-81™) and HEK 293T (ATCC® CRL-1573™) were purchased from ATCC. The cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum (FCS) and penicillin (100 IU/ml)-streptomycin (100 μg/ml) and amphotericin B (2.5 μg/ml) (de Bruyn and Knipe, 1988). ZIKV strains MR766 (Kuno and Chang, 2007) was obtained from ATCC.
4.2. Molecular cloning
The complete RNA genome and full cDNA sequences encoding the ZIKV proteins can be seen in the website (GenBank Sequence Accession: LC002520): http://www.viprbrc.org/brc/viprStrainDetails.spg?strainName=MR766-NIID&decorator=flavi. We designed DNA sequences for each ZIKV protein based on the RNA sequence of ZIKV MR766 strain. The FLAG tag (gac tac aaa gac gat gac gac aag) was in-frame fused to the cDNA to form an insert fragment that is flanked by a restriction enzyme site: 1) for PrM, NS1, NS2a, NS2b, NS4a, and NS4b proteins, we added a BamHI site at the 5′ end and an EcoRI site at 3′ end; 2) for C and NS5 proteins, we added an EcoRI site at the 5′ end and an XhoI site at the 3′ end; and 3) for E and NS3 proteins, we added a BamHI site at the 5′ end and an XhoI site at the 3′ end. The DNA fragments were synthesized by Genescript (Piscataway, NJ). After purification by gel extraction, the DNA fragments were cloned into pcDNA3 vector that was cut with BamHI and EcoRI, EcoRI and XhoI, or BamHI and XhoI. The clones were confirmed by DNA sequencing and designated as pcDNA3-flagC (capsid), –flagPrM (precursor), –flagE (envelope), –flagNS1 (nonstructural protein 1), –flagNS2a, –flagNS2b, –flagNS3, –flagNS4a, –flagNS4b, and –flagNS5.
To mutate the putative NLS from protein C, NS1, NS3, or NS5, we employed an overlapping PCR method. Briefly, we designed the primers (Table 3) to amplify two fragments with desired mutation of each protein, the amplified fragments have overlapping at the mutation site. Then the two fragments were gel-purified and used for overlapping PCR to produce the fragment with the desired mutation. The fragment was cloned into pcDNA3 resulting in pcDNA3-flagCdNLS, pcDNA3-flagNS1dNLS, pcDNA3-flagNS3dNLS1, pcDNA3-flagNS3dNLS2, or pcDNA3-flagNS5dNLS.
Table 3.
Primers for the Mutation of Nuclear Localization Signal (NLS) of ZIKV Protein.
| For C protein: |
| ZIKV C EcoRI_Fw: CCG GAATTC ATG GAC TAC AAA GAC GAT GAC |
| ZIKV C XhoI_Rv: CCG CTC GAG TTA TGC CAT GGC TGT |
| ZIKV C NLS del101-104 DN: 5′ AATGCTAGGAAAGAGGGCGCAGACACCAGC 3′ |
| ZIKV C NLS del101-104 UP: 5′ GCTGGTGTCTGCGCCCTCTTTCCTAGCATT 3′ |
| For NS1 protein: |
| ZIKV NS1 BamHI_Fw: CGC GGATCC ATG GAC TAC AAA GAC GAT GAC |
| ZIKV NS1 EcoRI_Rv: CCG GAA TTC TTA CGC TGT CAC CAT TGA |
| ZIKV NS1 NLS del211-214 DN: 5′ GTGAAAAGAATGACACATGGGCCCACCTGATTGAGATG 3′ |
| ZIKV NS1 NLS del211-214 UP: 5′ CATCTCAATCAGGTGGGCCCATGTGTCATTCTTTTCAC 3′ |
| For NS3 protein: |
| ZIKV NS3 BamHI_Fw: CGC GGATCC ATG GAC TAC AAA GAC GAT GAC |
| ZIKV NS3 XhoI_Rv: CCG CTC GAG TTA TCT TTT TCC AGC GGC GAA |
| ZIKV NS3 NLS1 del214-216 DN: 5′ ATAGTCCGTGAAGCCATACTCCGGACAGTGATCTTG 3′ |
| ZIKV NS3 NLS1 del214-216 UP: 5′ CAAGATCACTGTCCGGAGTATGGCTTCACGGACTAT 3′ |
| ZIKV NS3 NLS2 del587-591 DN: 5′ TGGACAAAGTATGGAGAGCCGAGATGGATGGATGCT 3′ |
| ZIKV NS3 NLS2 del578-591 UP: 5′ AGCATCCATCCATCTCGGCTCTCCATACTTTGTCCA 3′ |
| For NS5 protein: |
| ZIKV NS5 EcoRI_Fw: CCG GAATTC ATG GAC TAC AAA GAC GAT GAC |
| ZIKV NS5 XhoI_Rv: CGG CTC GAG TTA CAA CAC TCC GGG TGT GGA |
| ZIKV NS5 NLS del388-391 DN: 5′ CTGTGGAAGGAGCTGGGGCCACGCGTCTGCACCAAA 3′ |
| ZIKV NS5 NLS del388-391 UP: 5′ TTTGGTGCAGACGCGTGGCCCCAGCTCCTTCCACAG 3′ |
C: capsid; NS: nonstructural; DN: down; UP: up.
4.3. Antibodies
Anti-Giantin (ab80864) for visualizing Golgi body, and anti-CoxIV (ab16056) for showing mitochondria were purchased from Abcam (Cambridge, MA). Anti-Calregulin (H-107, sc-11398) for visualizing ER, anti-EEA1 (sc-365652) for showing endosome, anti-MAP LC3beta (sc-271625) to visualizing Autophgosomes, anti-Lamin A (h-102, sc-20680), and anti-Tubulin (4G1, sc-58666) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). The anti-ZIKV antibody for immunocytochemistry (ICC) assay was generated from the hybridoma cell line, D1-4G2-4-15 (ATCC® HB-112™) in our laboratory. The anti-ZIKV antibody for western blot is the immune serum produced from mice in our laboratory. Anti-FLAG antibody (monoclonal), M2, and the anti-SC35 antibody (S4045) were purchased from Sigma.
4.4. Immunocytochemistry (ICC) assay
Immunostaining was performed on cells grown on coverslips after fixation with 1% paraformaldehyde (10 min at room temperature) and permeabilization in 0.2% Triton (20 min on ice) by sequential incubation with primary and Texas red (TR)-labeled secondary antibodies (Vector Laboratories, Burlingame, Calif.) for 30 min each (all solutions in PBS).
To visualize lipid droplet (LD), the fixed cells were incubated with BODIPY 500/510 (Life Technology Corp. cat# B3824) at a final concentration of 1 μg/ml for 1 h at 37 °C.
Finally, cells were equilibrated in PBS, stained for DNA with Hoechst 33258 (0.5 μg/ml), and mounted in Fluoromount G (Fisher Scientific, Newark, Del.).
4.5. Subcellular fractionation
Cells from a 10 cm dish were transferred to 500 μl fractionation buffer [250 mM Sucrose, 20 mM HEHES (pH 7.4), 10 mM KCl, 2 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1 Mm DTT, and PI cocktail] by scrapping. The cell suspension was passed through a 25 gauge needle 10 times using a 1 ml syringe. After sitting on ice for 20 min, the cell suspension was centrifuged at 720 ×g for 5 min. The pellet contains nuclei and the supernatant contains cytoplasm, membrane and mitochondria: (Mlakar et al., 2016) the pellet was resuspended with 500 μl fractionation buffer, passed through a 25 gauge for 10 times, and centrifuged at 720 ×g for 10 min. The supernatant was discarded and the pellet was resuspended in TBS with 0.05% SDS. This is the nuclear extract fraction; (Petersen et al., 2016) the supernatant was used for the cytoplasmic fraction.
4.6. Immunoblot analysis
Proteins were separated by sodium dodecyl sulfate-7.5% poly-acrylamide gel electrophoresis (Munch et al., 1992) (10 to 20 μg loaded in each lane), transferred to nitrocellulose membranes (Amersham Inc., Piscataway, NJ), and blocked with 5% nonfat milk for 60 min at room temperature. Membranes were incubated overnight at 4 °C with primary antibody followed by incubation with a horseradish peroxidase-coupled secondary antibody (Amersham Inc.) and detection with enhanced chemiluminescence (Pierce, Rockford, Ill.), according to standard methods. Membranes were stripped with stripping buffer (100 mM β-mercaptoethanol, 2% SDS, 62.5 mM Tris-HCl, pH 6.8), washed with PBS-0.1% Tween 20, and used to detect additional proteins.
4.7. Confocal microscopy
Cells were examined with a Leica TCS SPII confocal laser scanning system. Two or three channels were recorded simultaneously and/or sequentially and controlled for possible breakthrough between the fluorescein isothiocyanate and Texas Red signals and between the blue and red channels.
Acknowledgments
We acknowledge the instrument support of the PSM Molecular Biology Core Laboratory. This study was supported by an NIH/NIAID SC1AI112785 (Q.T.) and National Institute on Minority Health and Health Disparities of the National Institutes of Health under Award Number G12MD007597. We thank Tommy Tang for the English editing, Drs. Roane and Thompson for the scientific reviewing.
Abbreviation
- ZIKV
Zika virus
- C
Capsid
- Env
Envelope
- NS proteins
nonstructural
- Ctr
Centrosome
- Mito
Mitochondria
- ER
Endoplasmic reticulum
- Golgi
Golgi apparatus
- ICC
Immunocytochemistry
- WB
western blot
Footnotes
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Author contributions
WH and QT designed the study and wrote the paper. RCC made the cloning of all the ZIKV proteins and performed ICC. NA performed the RTqPCR and critically revised the manuscript. LAO performed the western blot, ICC and revised the manuscript. WH performed the RNA ChIP assay and RNA FISH. All authors analyzed the results and approved the final version of the manuscript.
References
- Aliota MT, Caine EA, Walker EC, Larkin KE, Camacho E, Osorio JE. Characterization of lethal Zika virus infection in AG129 mice. PLoS Negl Trop Dis. 2016;10:e0004682. doi: 10.1371/journal.pntd.0004682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong N, Hou W, Tang Q. Biological and historical overview of Zika virus. World J Virol. 2017;6:1–8. doi: 10.5501/wjv.v6.i1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard A, Jin M, Xu Z, Klionsky DJ. A large-scale analysis of autophagy-related gene expression identifies new regulators of autophagy. Autophagy. 2015;11:2114–2122. doi: 10.1080/15548627.2015.1099796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollati M, Alvarez K, Assenberg R, Baronti C, Canard B, Cook S, Coutard B, Decroly E, de Lamballerie X, Gould EA, Grard G, Grimes JM, Hilgenfeld R, Jansson AM, Malet H, Mancini EJ, Mastrangelo E, Mattevi A, Milani M, Moureau G, Neyts J, Owens RJ, Ren J, Selisko B, Speroni S, Steuber H, Stuart DI, Unge T, Bolognesi M. Structure and functionality in flavivirus NS-proteins: perspectives for drug design. Antivir Res. 2010;87:125–148. doi: 10.1016/j.antiviral.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boorman JP, Porterfield JS. A simple technique for infection of mosquitoes with viruses; transmission of Zika virus. Trans R Soc Trop Med Hyg. 1956;50:238–242. doi: 10.1016/0035-9203(56)90029-3. [DOI] [PubMed] [Google Scholar]
- de Bruyn Kops A, Knipe DM. Formation of DNA replication structures in herpes virus-infected cells requires a viral DNA binding protein. Cell. 1988;55:857–868. doi: 10.1016/0092-8674(88)90141-9. [DOI] [PubMed] [Google Scholar]
- Cai HLM, Wang Y, Tang Q. Diversity of and implications from the viral genomes and viral proteins of Zika virus. J Virol Antivir Res. 2016;5 [Google Scholar]
- Costa F, Sarno M, Khouri R, de Paulo, Freitas B, Siqueira I, Ribeiro GS, Ribeiro HC, Campos GS, Alcantara LC, Reis MG, Weaver SC, Vasilakis N, Ko AI, Almeida AR. Emergence of congenital Zika Syndrome: viewpoint from the front lines. Ann Intern Med. 2016 doi: 10.7326/M16-0332. http://dx.doi.org/10.7326/M16-0332. [DOI] [PMC free article] [PubMed]
- Dabauvalle MC, Muller E, Ewald A, Kress W, Krohne G, Muller CR. Distribution of emerin during the cell cycle. Eur J Cell Biol. 1999;78:749–756. doi: 10.1016/S0171-9335(99)80043-0. [DOI] [PubMed] [Google Scholar]
- Dick GW, Kitchen SF, Haddow AJ. Zika virus. I. Isolations and serological specificity. Trans R Soc Trop Med Hyg. 1952;46:509–520. doi: 10.1016/0035-9203(52)90042-4. [DOI] [PubMed] [Google Scholar]
- Faye O, Freire CC, Iamarino A, Faye O, de Oliveira JV, Diallo M, Zanotto PM, Sall AA. Molecular evolution of Zika virus during its emergence in the 20(th) century. PLoS Negl Trop Dis. 2014;8:e2636. doi: 10.1371/journal.pntd.0002636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu XD, Maniatis T. The 35-kDa mammalian splicing factor SC35 mediates specific interactions between U1 and U2 small nuclear ribonucleoprotein particles at the 3′ splice site. Proc Natl Acad Sci U S A. 1992;89:1725–1729. doi: 10.1073/pnas.89.5.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu XD, Mayeda A, Maniatis T, Krainer AR. General splicing factors SF2 and SC35 have equivalent activities in vitro, and both affect alternative 5′ and 3′ splice site selection. Proc Natl Acad Sci U S A. 1992;89:11224–11228. doi: 10.1073/pnas.89.23.11224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel E, Ramani A, Karow U, Gottardo M, Natarajan K, Gooi LM, Goranci-Buzhala G, Krut O, Peters F, Nikolic M, Kuivanen S, Korhonen E, Smura T, Vapalahti O, Papantonis A, Schmidt-Chanasit J, Riparbelli M, Callaini G, Kronke M, Utermohlen O, Gopalakrishnan J. Recent Zika virus isolates induce premature differentiation of neural progenitors in human brain organoids. Cell Stem Cell. 2017;20(397–406):e395. doi: 10.1016/j.stem.2016.12.005. [DOI] [PubMed] [Google Scholar]
- Garcez PP, Loiola EC, Madeiro da Costa R, Higa LM, Trindade P, Delvecchio R, Nascimento JM, Brindeiro R, Tanuri A, Rehen SK. Zika virus impairs growth in human neurospheres and brain organoids. Science. 2016 doi: 10.1126/science.aaf6116. http://dx.doi.org/10.1126/science.aaf6116. [DOI] [PubMed]
- Grant A, Ponia SS, Tripathi S, Balasubramaniam V, Miorin L, Sourisseau M, Schwarz MC, Sanchez-Seco MP, Evans MJ, Best SM, Garcia-Sastre A. Zika virus targets human STAT2 to inhibit type I interferon signaling. Cell Host Microbe. 2016;19:882–890. doi: 10.1016/j.chom.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenbaum Y, Wilson KL, Harel A, Goldberg M, Cohen M. Review: nuclear lamins – structural proteins with fundamental functions. J Struct Biol. 2000;129:313–323. doi: 10.1006/jsbi.2000.4216. [DOI] [PubMed] [Google Scholar]
- Haddow AJ, Williams MC, Woodall JP, Simpson DI, Goma LK. Twelve isolations of Zika virus from Aedes (Stegomyia) Africanus (Theobald) taken in and above a Uganda Forest. Bull World Health Organ. 1964;31:57–69. [PMC free article] [PubMed] [Google Scholar]
- Henchal EA, Putnak JR. The dengue viruses. Clin Microbiol Rev. 1990;3:376–396. doi: 10.1128/cmr.3.4.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman HD, Pierson TC. Zika in the brain: new models shed light on viral infection. Trends Mol Med. 2016 doi: 10.1016/j.molmed.2016.06.004. http://dx.doi.org/10.1016/j.molmed.2016.06.004. [DOI] [PMC free article] [PubMed]
- Hou W, Armstrong N, Obwolo LA, Thomas M, Pang X, Jones KS, Tang Q. Determination of the cell permissiveness spectrum, mode of RNA replication, and RNA-protein interaction of Zika virus. BMC Infect Dis. 2017;17:239. doi: 10.1186/s12879-017-2338-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CT, Ma L, Burgner JW, Groesch TD, Post CB, Kuhn RJ. Flavivirus capsid is a dimeric alpha-helical protein. J Virol. 2003;77:7143–7149. doi: 10.1128/JVI.77.12.7143-7149.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MO, Moon DO, Choi YH, Lee JD, Kim ND, Kim GY. Platycodin D induces mitotic arrest in vitro, leading to endoreduplication, inhibition of proliferation and apoptosis in leukemia cells. Int J Cancer. 2008;122:2674–2681. doi: 10.1002/ijc.23442. [DOI] [PubMed] [Google Scholar]
- Kim YM, Gayen S, Kang C, Joy J, Huang Q, Chen AS, Wee JL, Ang MJ, Lim HA, Hung AW, Li R, Noble CG, Lee le T, Yip A, Wang QY, Chia CS, Hill J, Shi PY, Keller TH. NMR analysis of a novel enzymatically active unlinked dengue NS2B–NS3 protease complex. J Biol Chem. 2013;288:12891–12900. doi: 10.1074/jbc.M112.442723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostyuchenko VA, Lim EX, Zhang S, Fibriansah G, Ng TS, Ooi JS, Shi J, Lok SM. Structure of the thermally stable Zika virus. Nature. 2016;533:425–428. doi: 10.1038/nature17994. [DOI] [PubMed] [Google Scholar]
- Kosugi S, Hasebe M, Tomita M, Yanagawa H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc Natl Acad Sci U S A. 2009;106:10171–10176. doi: 10.1073/pnas.0900604106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuno G, Chang GJ. Full-length sequencing and genomic characterization of Bagaza, Kedougou, and Zika viruses. Arch Virol. 2007;152:687–696. doi: 10.1007/s00705-006-0903-z. [DOI] [PubMed] [Google Scholar]
- Lanciotti RS, Lambert AJ, Holodniy M, Saavedra S, Signor, Ldel C. Phylogeny of Zika virus in western hemisphere 2015. Emerg Infect Dis. 2016;22:933–935. doi: 10.3201/eid2205.160065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazear HM, Govero J, Smith AM, Platt DJ, Fernandez E, Miner JJ, Diamond MS. A mouse model of Zika virus pathogenesis. Cell Host Microbe. 2016 doi: 10.1016/j.chom.2016.03.010. http://dx.doi.org/10.1016/j.chom.2016.03.010. [DOI] [PMC free article] [PubMed]
- Linstedt AD, Hauri HP. Giantin, a novel conserved Golgi membrane protein containing a cytoplasmic domain of at least 350 kDa. Mol Biol Cell. 1993;4:679–693. doi: 10.1091/mbc.4.7.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchette NJ, Garcia R, Rudnick A. Isolation of Zika virus from Aedes aegypti mosquitoes in Malaysia. Am J Trop Med Hyg. 1969;18:411–415. doi: 10.4269/ajtmh.1969.18.411. [DOI] [PubMed] [Google Scholar]
- Mlakar J, Korva M, Tul N, Popovic M, Poljsak-Prijatelj M, Mraz J, Kolenc M, Resman Rus K, Vesnaver Vipotnik T, Fabjan Vodusek V, Vizjak A, Pizem J, Petrovec M, Avsic, Zupanc T. Zika virus associated with microcephaly. N Engl J Med. 2016 doi: 10.1056/NEJMoa1600651. http://dx.doi.org/10.1056/NEJMoa1600651. [DOI] [PubMed]
- Moore DL, Causey OR, Carey DE, Reddy S, Cooke AR, Akinkugbe FM, David-West TS, Kemp GE. Arthropod-borne viral infections of man in Nigeria, 1964–1970. Ann Trop Med Parasitol. 1975;69:49–64. doi: 10.1080/00034983.1975.11686983. [DOI] [PubMed] [Google Scholar]
- Munch K, Messerle M, Plachter B, Koszinowski UH. An acidic region of the 89K murine cytomegalovirus immediate early protein interacts with DNA. J Gen Virol. 1992;73(Pt 3):499–506. doi: 10.1099/0022-1317-73-3-499. [DOI] [PubMed] [Google Scholar]
- Musso D, Nilles EJ, Cao-Lormeau VM. Rapid spread of emerging Zika virus in the Pacific area. Clin Microbiol Infect. 2014;20:O595–596. doi: 10.1111/1469-0691.12707. [DOI] [PubMed] [Google Scholar]
- Musso D, Roche C, Robin E, Nhan T, Teissier A, Cao-Lormeau VM. Potential sexual transmission of Zika virus. Emerg Infect Dis. 2015;21:359–361. doi: 10.3201/eid2102.141363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oehler E, Watrin L, Larre P, Leparc-Goffart I, Lastere S, Valour F, Baudouin L, Mallet H, Musso D, Ghawche F. Zika Virus Infection Complicated by Guillain-Barre Syndrome – Case Report, French Polynesia, December 2013. Euro Surveillpp. 2014;19 doi: 10.2807/1560-7917.es2014.19.9.20720. [DOI] [PubMed] [Google Scholar]
- Petersen E, Wilson ME, Touch S, McCloskey B, Mwaba P, Bates M, Dar O, Mattes F, Kidd M, Ippolito G, Azhar EI, Zumla A. Rapid spread of Zika virus in the Americas - implications for public health preparedness for mass gatherings at the 2016 Brazil olympic games. Int J Infect Dis. 2016;44:11–15. doi: 10.1016/j.ijid.2016.02.001. [DOI] [PubMed] [Google Scholar]
- Plourde AR, Bloch EM. A literature review of Zika virus. Emerg Infect Dis. 2016;22:1185–1192. doi: 10.3201/eid2207.151990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin XF. Understanding the pathogenic evolution of Zika virus. Sci China Life Sci. 2016;59:737–739. doi: 10.1007/s11427-016-5069-7. [DOI] [PubMed] [Google Scholar]
- Ramos da Silva S, Gao SJ. Zika virus: an update on epidemiology, pathology, molecular biology and animal model. J Med Virol. 2016 doi: 10.1002/jmv.24563. http://dx.doi.org/10.1002/jmv.24563. [DOI] [PMC free article] [PubMed]
- Reese JH, Maltese WA. Post-translational modification of proteins by 15-carbon and 20-carbon isoprenoids in three mammalian cell lines. Mol Cell Biochem. 1991;104:109–116. doi: 10.1007/BF00229810. [DOI] [PubMed] [Google Scholar]
- Rey FA, Heinz FX, Mandl C, Kunz C, Harrison SC. The envelope glycoprotein from tick-borne encephalitis virus at 2 a resolution. Nature. 1995;375:291–298. doi: 10.1038/375291a0. [DOI] [PubMed] [Google Scholar]
- Rossi SL, Tesh RB, Azar SR, Muruato AE, Hanley KA, Auguste AJ, Langsjoen RM, Paessler S, Vasilakis N, Weaver SC. Characterization of a novel murine model to study Zika virus. Am J Trop Med Hyg. 2016 doi: 10.4269/ajtmh.16-0111. http://dx.doi.org/10.4269/ajtmh.16-0111. [DOI] [PMC free article] [PubMed]
- Shen S, Shi J, Wang J, Tang S, Wang H, Hu Z, Deng F. Phylogenetic analysis revealed the central roles of two African countries in the evolution and worldwide spread of Zika virus. Virol Sin. 2016;31:118–130. doi: 10.1007/s12250-016-3774-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RK, Dhama K, Malik YS, Ramakrishnan MA, Karthik K, Tiwari R, Saurabh S, Sachan S, Joshi SK. Zika virus - emergence, evolution, pathology, diagnosis, and control: current global scenario and future perspectives - a comprehensive review. Vet Q. 2016;36:150–175. doi: 10.1080/01652176.2016.1188333. [DOI] [PubMed] [Google Scholar]
- Sirohi D, Chen Z, Sun L, Klose T, Pierson TC, Rossmann MG, Kuhn RJ. The 3.8 A resolution cryo-EM structure of Zika virus. Science. 2016;352:467–470. doi: 10.1126/science.aaf5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratton SJ. Zika virus association with microcephaly: the power for population statistics to identify public health emergencies. Prehosp Disaster Med. 2016 doi: 10.1017/S1049023X16000170. http://dx.doi.org/10.1017/S1049023X16000170:1-2. [DOI] [PubMed]
- Wang L, Valderramos SG, Wu A, Ouyang S, Li C, Brasil P, Bonaldo M, Coates T, Nielsen-Saines K, Jiang T, Aliyari R, Cheng G. From mosquitos to humans: genetic evolution of Zika virus. Cell Host Microbe. 2016;19:561–565. doi: 10.1016/j.chom.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver SC, Costa F, Garcia-Blanco MA, Ko AI, Ribeiro GS, Saade G, Shi PY, Vasilakis N. Zika virus: history, emergence, biology, and prospects for control. Antivir Res. 2016;130:69–80. doi: 10.1016/j.antiviral.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisenberg RC. Microtubule formation in vitro in solutions containing low calcium concentrations. Science. 1972;177:1104–1105. doi: 10.1126/science.177.4054.1104. [DOI] [PubMed] [Google Scholar]
- Werner H, Fazecas T, Guedes B, Dos Santos JL, Daltro P, Tonni G, Campbell S, Araujo, Junior E. Intrauterine Zika virus infection and microcephaly: perinatal imaging correlations with 3D virtual physical models. Ultrasound Obstet Gynecol. 2016 doi: 10.1002/uog.15901. http://dx.doi.org/10.1002/uog.15901. [DOI] [PubMed]
- Wikan N, Smith DR. Zika virus: history of a newly emerging arbovirus. Lancet Infect Dis. 2016;16:e119–126. doi: 10.1016/S1473-3099(16)30010-X. [DOI] [PubMed] [Google Scholar]
- Wolf B, Diop F, Ferraris P, Wichit S, Busso C, Misse D, Gonczy P. Zika virus causes supernumerary foci with centriolar proteins and impaired spindle positioning. Open Biol. 2017;7 doi: 10.1098/rsob.160231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Xie X, Wang QY, Dong H, Lee MY, Kang C, Yuan Z, Shi PY. Characterization of dengue virus NS4A and NS4B protein interaction. J Virol. 2015;89:3455–3470. doi: 10.1128/JVI.03453-14. [DOI] [PMC free article] [PubMed] [Google Scholar]


