Abstract
Chemical toxicity has a serious impact on public health, and toxicity failures of drug candidates drive up drug development costs. Many in vitro bioassays exist for toxicity screening, and newer versions of these tend to be high throughput or high content assays, some of which rely on electrochemical detection. Toxicity very often results from metabolites of the chemicals we are exposed to, so it is important that assays feature metabolic conversion. Combining bioassays, computational predictions, and accurate chemical pathway elucidation presents our best chance for reliable toxicity prediction. Employing electrochemical and electrochemiluminescent approaches, cell-free microfluidic arrays can measure relative rates of formation of DNA-metabolite adduct formation (a measure of genotoxicity) as well as DNA oxidation levels resulting from enzyme-generated metabolites. Enzymes for several organ types can be studied simultaneously. These arrays can be used to identify the most reactive metabolites, and subsequent mechanistic details can then be investigated with high throughput LC-HPLC using enzyme/DNA-coated magnetic beads.
Keywords: Toxicity, Cytochrome P450s, bioassay, electrochemiluminescence, DNA
Thousands of new chemicals are produced annually, and many find their way into our bodies.1 These may elicit toxicity directly or, most often, via their enzyme-generated metabolites. From a pharmaceutical development standpoint, the cost of developing new drug candidates exceeds US$5 billion; therefore, it is vitally important to predict potential toxicity for compounds designed to be ingested or internalized in some fashion.2 Numerous in vitro high throughput screening methods have been developed to predict toxicity.3–5 New drug candidates are screened using initial bioassay panels followed by animal studies, then human clinical trials. However, 30% of drug candidates fail due to toxicity uncovered only in clinical testing or later.
Animal models are often not very predictive of human responses as toxicity biochemistry is species-dependent and incompletely understood.6 Parent chemicals are converted to metabolites for excretion, but metabolites can be dangerously reactive toward DNA, RNA, and proteins. Metabolic generation of reactive species is called bioactivation, and can result in genotoxic, or covalent adducts, reactive oxidation species (ROS),5,7, or so-called idiosyncratic drug reactions (IDR).8 Major sources of bioactivated metabolites are cytochrome (cyt) P450 enzymes that are present in all human organs and involved in 75% of metabolic reactions of existing drugs.9 Cyt P450s mainly catalyze oxygen transfers while sequential bioconjugation enzymes add hydrophilic moieties, but can also lead to enhanced bioactivation of the parent compound.
Toxicity bioassays are often inaccurate when accounting for reactive metabolites.5,10 Traditional toxicity screens target genotoxicity (related to DNA damage), channel blocking, drug-drug interactions, and metabolite-mediated toxicity.11,12 A common genotoxicity assay is the Ames test,13 which identifies genotoxins based on bacterial growth upon compound exposure.13 High-throughput modifications to this and other established genotoxicity assays have been developed,12,14–18 but sensitivity19,20 and accuracy limitations exist, mainly due to the inability to accurately account for reactive metabolites.12,20,21 New, more predictive toxicity tests are needed as well as elucidation of complex chemical pathways leading to toxicity. Eukaryotic cell assays may provide better accuracy than traditional prokaryotic genotoxicity screens.20 Several genotoxicity assays utilizing electrochemical detection strategies have been implemented and are reviewed elsewhere.22 Additional structure-based toxicity information is desired; however, and if this can be achieved, medicinal chemists may be able to synthetically design toxicity out of specific products while retaining desired therapeutic effects.
There are strong trends toward high content analysis (HCA) or throughput screening (HTS).23,24 Many of these are image-based utilizing light- emitting probes that incorporate into different cellular sites and are multi-color imaged to detect phenotypic changes for monitoring of multiple toxicity endpoints.25 Novel high throughput electrochemical strategies have utilized nanomaterials26 or microdevice fabrication strategies.27 Electrochemical detection can offer rapid, label-free toxicity assessments.27 Carbon fiber microelectrodes implanted in zebrafish embryos have been used to detect reactive oxygen and nitrogen species generation from nanoparticle exposure.28 Microsome metabolism has been monitored with zinc oxide nanowires.26 Electrochemical impedance spectroscopy (EIS) methods have been used extensively to detect toxicity based on altered cellular morphology.29 Novel strategies have utilized EIS to assay pharmaceutical toxicity toward HeLa and fibroblast cells immobilized in a 3D-hydrogel flow array.27
Many HCA assays are limited by two-dimensional cell cultures that may not correlate with in vivo responses.11,30 Recent research has focused on 3-D cell cultures to more accurately model metabolism and toxicity,31–34 but difficulties remain in accurately mimicking human response. Devices that mimic cell environments in one or more organs simultaneously have been described.11,30,35
One key drawback to many established bioassays is that reaction pathways of toxicity are insufficiently addressed. We have attempted to remedy this by first detecting a biological event using electrochemical or electrochemiluminescent arrays and following up using structure-based analysis methods. We developed high throughput sensor arrays designed to first screen for “toxic hits” followed by LC-MS/MS to investigate chemical pathways in more detail.5 These cell-free methods employ high throughput arrays with DNA damage endpoints to reveal the possibility of toxicity-related chemical reactions.3–5 Here, established layer-by-layer (LbL) film fabrication protocols36,37 are utilized to immobilize metabolic enzymes, DNA, and polyions in thin films for screening arrays.5 Similar films are formed on magnetic particles to produce samples for LC-MS/MS.5, The enzymes produce metabolites, and reactive metabolites may damage DNA in the films. DNA and metabolic enzymes have very high relative concentrations in the films, facilitating faster reaction kinetics. Many reactive metabolites form nucleobase adducts that may be stable or may lead to abasic sites.5 DNA strand breaks and oxidation of guanines are also possible.
Electrochemical detection in these arrays is accomplished by oxidation of a catalytic Ru(bipyridyl)-poly(vinylpyridine) (RuPVP) polymer in the films. Reaction with DNA produces enhanced electrochemical and electrochemiluminescent (ECL) signals, both of which have been used for detection.5 The mechanistic details have been covered extensively elsewhere.5,38,39 Damaged DNA disorders the double helix of DNA to provide better access of RuIII sites in the polymer to reactive guanines in DNA, providing larger current signals than for intact DNA.5,40 For LC-MS/MS, similar films without RuPVP are grown on magnetic beads. Metabolic enzyme reactions are performed in 96 well filter plates, and hydrolyzed DNA containing damaged nucleobases is collected by filtration into another plate for LC-MS/MS.5
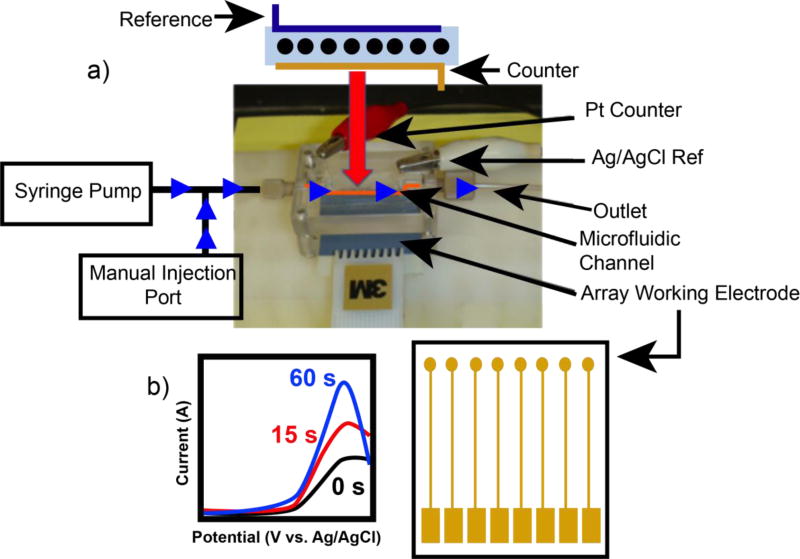
Figure 1 shows a recent electrochemical flow cell array designed for more high-throughput toxicity analysis. If enzyme films contain CPR – e.g. microsomal films – then cyt P450s in the films can be activated via direct CPR reduction by the electrode as opposed to addition of exogenous NADPH. 5,40 This microfluidic array metabolized test molecules in situ to produce reactive metabolites that may damage DNA to produce larger peak currents. These devices have been utilized to screen for pollutant chemicals with known toxicity, to study the interaction of cyt P450s with varied metabolic enzymes,41 and to detect DNA adducts and oxidized DNA in the same array.42 In the latter system, a catalytic osmium metallopolymer (OsPVP) oxidized at lower potentials was utilized, which is selective for 8-oxo-guanine.
Figure 1.
a) Microfluidic electrochemical array system used for detection of reactive metabolites formed by liver enzyme cyt P450s. Flow direction is denotred by blue arrows. b) Simulated SWV data for one electrode in the array showing the increase in oxidative peak current as xenobiotic exposure time increases.
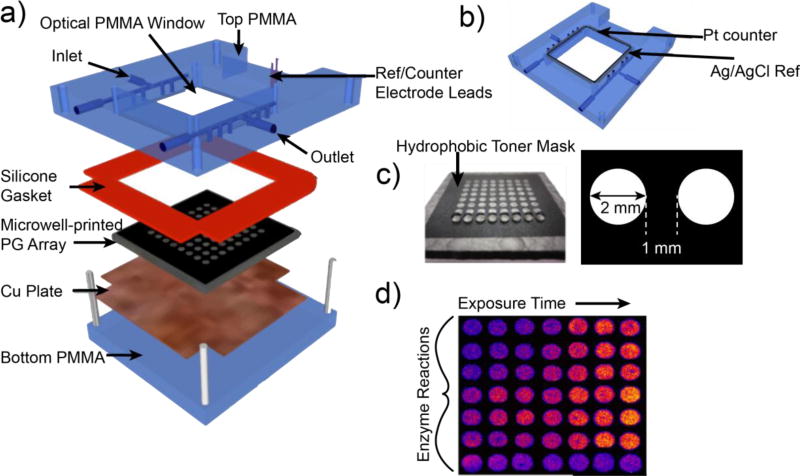
ECL sensor arrays featuring RuPVP have been developed in higher throughput formats.5,37,38 Our most advanced ECL array features a 64-microwell reactor chip and are shown in Figure 2.43 A computer printed pattern of hydrophobic ink is heat transferred to a conductive pyrolytic graphite sheet to form 64 microwells about 10–15 nm deep that are filled with RuPVP/enzyme/DNA films. Reaction media containing the test chemical flows into the reactor where metabolites are generated that can react with DNA. Following a rinsing step, 1.25 V vs. Ag/AgCl is applied for 180 s in a dark box to generate ECL measured with a CCD camera.5, 44 Electrochemical oxidation of RuII in RuPVP initiates a multistep redox pathway involving guanines in DNA as ECL co-reactants to generate electronically excited RuII* sites that decay to emit visible ECL light. Pure human enzymes, enzyme mixtures, and enzyme sources such as microsomes, human liver S9 enzyme fractions (HS9), and supersomes of cyt P450s from various organs can be used in the arrays.44 Array results provide relative rates of DNA damage, as confirmed by LC-MS/MS.5 These arrays have proven effective to establish detailed metabolic pathways utilizing different enzymes, when coupled with LC-MS/MS.5 In this vein, the utility of the ECL sensor/MS approach is not in the analysis and measurement of toxic chemical in samples. Rather, our approach is designed to identify toxic metabolites and pathways involving test compounds potentially leading to toxicity. For instance, we have used ECL arrays and MS to detect reactive metabolites and elucidate how interspecies metabolism differences lead to altered toxicity outcomes from exposure to the cancer-drug tamoxifen.45
Figure 2.
64-microwell ECL chip and the fluidic reaction chamber: a) assembly of the flow cell, b) underside view of reference and counter electrode wires in the top poly(methylmethacrylate) (PMMA) plate, c) pyrolytic graphite (PG) chip with computer-printed microwells. The first row shows 1 µL water droplets on each of the wells. d) Output ECL of the array showing different enzyme reactions and controls. As xenobiotic exposure time increases, ECL becomes more intense.
In summary, the landscape of toxicity screening is rapidly changing and improving in terms of throughput and predictive capabilities. Although not covered here, computational predictions also represent a significant contribution to toxicity prediction. An approach that combines bioassays and computational results with accurate chemical pathway information has an excellent chance to mitigate the harmful consequences of toxicity. We have demonstrated that electrochemical sensor arrays have the potential to rapidly elucidate potential toxicity, particularly metabolite-mediated toxicity. Coupling LC-MS/MS adds structural and pathway information. Overall, while many emerging toxicity bioassays are inherently complex,5 the electrochemical sensor arrays are relatively simple and benefits including ease of setup, speed of analysis, and cost make this an attractive toxicity screening platform for future use in pharmaceutical development.
Highlights.
A discussion of traditional and cutting edge sensor/array chemical toxicity screening methods.
Electrochemical assays provide benefits over established tox-screening methods.
Novel ECL and flow-cell arrays can be used to detect metabolite-mediated toxicity.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1."Tox21". Source: U.S. Environmental Protection Agency. [Last accessed July 21, 2015]. [Google Scholar]
- 2. [last accessed July 21, 2015]; http://www.forbes.com/sites/matthewherper/2013/08/11/how-the-staggering-cost-ofinventing-new-drugs-is-shaping-the-future-of-medicine/ source: Forbes.com.
- 3.Steinberg P, editor. High-Throughput Screening Methods in Toxicity Testing. John Wiley & Sons; Hoboken, NJ: 2013. pp. 433–452. [Google Scholar]
- 4.Lynch AM, Sasaki JC, Elespuru R, Jacobson-Kram D, Thybaud V, et al. New and emerging technologies for genetic toxicity testing. Environ Mol Mutagen. 2011;52:205–223. doi: 10.1002/em.20614. [DOI] [PubMed] [Google Scholar]
- ••5.Hvastkovs EG, Rusling JF. State-of-the-Art Metabolic Toxicity Screening and Pathway Evaluation. Anal. Chem. 2016;88:4584–4599. doi: 10.1021/acs.analchem.5b04772. Recent review of novel advances in toxicity bioassays and chemical pathway elucidation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shanks N, Greek R, Greek J. Are animal models predictive for humans? Philos. Ethics Humanit. Med. 2009;4:2. doi: 10.1186/1747-5341-4-2. (open access) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis CD, Hanumegowda UM. In: Drug Metabolism Handbook. Nasser AF, Hollenberg PF, Scatina J, editors. John Wiley & Sons; Hoboken, NJ: 2009. pp. 561–628. [Google Scholar]
- 8.Claesson A, Spjuth O. On mechanisms of reactive metabolite formation from drugs. Mini Rev. Med.Chem. 2013;13:720–729. doi: 10.2174/1389557511313050009. [DOI] [PubMed] [Google Scholar]
- 9.Liebler DC, Guengerich FP. Elucidating mecahnisms of drug-induced toxicity. Nat. Rev. Drug Discov. 2005;4:410–420. doi: 10.1038/nrd1720. [DOI] [PubMed] [Google Scholar]
- 10.Caldwell GW, Yan Z. Screening for reactive intermediates and toxicity assessment in drug discovery. Curr. Opin. Drug Discov. Dev. 2006;9:47–50. [PubMed] [Google Scholar]
- 11.Roth A, Singer T. The application of 3D cell models to support drug safety assessment: opportunities & challenges. Adv. Drug Deliv. Rev. 2014;69–70:179–189. doi: 10.1016/j.addr.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 12.Kramer JA, Sagartz JE, Morris DL. The application of discovery toxicology and pathology towards the design of safer pharmaceutical lead candidates. Nat. Rev. Drug Discov. 2007;6:636–649. doi: 10.1038/nrd2378. [DOI] [PubMed] [Google Scholar]
- 13.Ames BN, Mcann J, Yamasaki E. Mutat. Res. Methods for detecting carcinogens and mutagens with the Salmonella/mammalian-microsome mutagenicity test. 1975;31:347–364. doi: 10.1016/0165-1161(75)90046-1. [DOI] [PubMed] [Google Scholar]
- 14.Kiskinis E, Suter W, Hartmann A. High throughput Comet assay using 96-well plates. Mutagenesis. 2002;31:37–43. doi: 10.1093/mutage/17.1.37. [DOI] [PubMed] [Google Scholar]
- 15.Flückiger-Isler SM, Baumeister K, Braun V, Gervais N, Hasler-Nguyen R, Reimann J, Van Gompel H-G, Wunderlich G, Engelhardt G. Assessment of the performance of the Ames II assay: a collaborative study with 19 coded compounds. Mutat. Res. 2004;558:181–197. doi: 10.1016/j.mrgentox.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 16.Styles JA, Clark H, Festing MFW, Rew DA. Automation of mouse micronucleus genotoxicity assay by laser scanning cytometry. Cytometry. 2001;44:153–155. doi: 10.1002/1097-0320(20010601)44:2<153::aid-cyto1095>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 17.Shuga J, Zhang J, Samson LD, Lodish HF, Griffith LG. In vitro erythropoiesis from bone marrow-derived progenitors provides a physiological assay for toxic and mutagenic compounds. Proc. Nat. Acad. Sci. 2007;104:8737–8742. doi: 10.1073/pnas.0701829104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flückiger-Isler S, Kamber M. In Genotoxicity and DNA Repair: A Practical Approach. In: Sierra LM, Gaivão I, editors. Genotoxicity and DNA Repair. Springer; New York, NY: 2014. pp. 23–41. [Google Scholar]
- 19.Smith KEC, Heringa MB, Uytewaal M, Mayer P. The dosing determines mutagenicity of hydrophobic compounds in the Ames II assay with metabolic transformation : Passive dosing versus solvent spiking. Mut. Res./Gen. Toxicol. Environ. Mut. 2013;750:12–18. doi: 10.1016/j.mrgentox.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 20.Hastwell PW, Chai L-L, Roberts KJ, Webster TW, Harvey JS, Rees RW, Walmsley RM. High-specificity and high-sensitivity genotoxicity assessment in a human cell line: validation of the GreenScreen HC GADD45a-GFP genotoxicity assay. Mutat. Res. 2006;607:160–175. doi: 10.1016/j.mrgentox.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 21.Oda Y, Nakamura S, Oki I, Kato T, Shinagawa H. Evaluation of the new system (umu-test) for the detection of environmental mutagens and carcinogens. Mutat. Res. 1985;147:212–229. doi: 10.1016/0165-1161(85)90062-7. [DOI] [PubMed] [Google Scholar]
- 22.Fojta M, Danhel A, Havran L, Vyskocil V. Recent progress in electrochemical sensors and assays for DNA damage and repair. Trends Anal. Chem. 2016;79:160–167. [Google Scholar]
- •23.O’Brien PJ. High-content analysis in toxicology: screening substances for human toxicity potential, elucidating subcellular mechanisms and in vivo use as translational safety biomarkers. Basic Clin. Pharmacol. Toxicol. 2014;115:4–17. doi: 10.1111/bcpt.12227. An excellent, throrough review of HCA in toxicity analysis. The author provides an overview of toxicity screening in general and how HCA can be used to improve this landscape. A case study is presented where HCA was used to determine cytotoxicity and mechanistic toxicity details. [DOI] [PubMed] [Google Scholar]
- 24.Judson R, Kavlock R, Martin M, Reif D, Houck K, et al. Perspectives on validation of high-throughput assays supporting 21st century toxicity testing. ALTEX. 2013;30:51–56. doi: 10.14573/altex.2013.1.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garside H, Marcoe KF, Chesnut-Speelman J, Foster AJ, Muthas D, Kenna JG, Warrior U, Bowes J, Baumgartner J. Evaluation of the use of imaging parameters for the detection of compound-induced hepatotoxicity in 384-well cultures of HepG2 cells and cryopreserved primary human hepatocytes, Toxicol. in Vitro. 2014;28:171–181. doi: 10.1016/j.tiv.2013.10.015. [DOI] [PubMed] [Google Scholar]
- •26.Wang N, Gao C, Xue F, Han Y, Li T, Cao X, Zhang X, Zhang Y, Wang ZL. Piezotronic-effect enhanced drug metabolism and sensing on a single ZnO nanowire surface with the presence of human cytochrome P450. ACS Nano. 2015;9:3159–3168. doi: 10.1021/acsnano.5b00142. This paper describes the utilization of ZnO nanowires coated with cyt P450-containing microsomes to sense drug metabolism. The authors utilized cyt P450 2C isoforms and exposed them to different test pharmaceuticals. Upon the introduction of an external stress to the wire, currents are enhanced in the presence of drug due to the piezotronic effect. [DOI] [PubMed] [Google Scholar]
- 27.Tran TB, Cho S, Min J. Hydrogel-based diffusion chip with Electric Cell-substrate Impedance Sensing (ECIS) integration for cell viability assay and drug toxicity screening. Biosens. Bioelectron. 2013;50:453–459. doi: 10.1016/j.bios.2013.07.019. [DOI] [PubMed] [Google Scholar]
- 28.Özel RE, Wallaceb KN, Andreescu S. Alterations of intestinal serotonin following nanoparticle exposure in embryonic zebrafish. Environ. Sci.: Nano. 2014;1:27–36. doi: 10.1039/C3EN00001J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Özel RE, Liu X, Alkasir RSJ, Andreescu S. Electrochemical methods for nanotoxicity assessment. Trends Anal. Chem. 2014;59:112–120. [Google Scholar]
- 30.Astashkina A, Grainger DW. Critical analysis of 3-D organoid in vitro cell culture models for high-throughput drug candidate toxicity assessments. Adv. Drug Deliv. Rev. 2014;69–70:1–18. doi: 10.1016/j.addr.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 31.Hjelm BE, Berta AN, Nickerson CA, Arntzen CJ, Herbst-Kralovetz MM. Development and characterization of a three-dimensional organotypic human vaginal epithelial cell model. Biol. Reprod. 2010;82:617–627. doi: 10.1095/biolreprod.109.080408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Radtke AL, Herbst-Kralovetz MM. Culturing and applications of rotating wall vessel bioreactor derived 3D epithelial cell models. J. Vis. Exp. 2012;62:e3868, 1–10. doi: 10.3791/3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takayama K, Kawabata K, Nagamoto Y, Kishimoto K, Tashiro K, Sakurai F, Tachibana M, Kanda K, Hayakawa T, Furue MK, Mizuguchi H. Biomaterials. 2013;34:1781–1789. doi: 10.1016/j.biomaterials.2012.11.029. [DOI] [PubMed] [Google Scholar]
- 34.Nakamura K, Mizutani R, Sanbe A, Enosawa S, Kasahara M, Nakagawa A, Ejiri Y, Murayama N, Miyamoto Y, Torii T, Kusakawa S, Yamauchi J, Fukuda M, Yamazaki H, Tanoue A. J. Biosci. Bioeng. 2011;111:78–84. doi: 10.1016/j.jbiosc.2010.08.008. [DOI] [PubMed] [Google Scholar]
- ••35.Esch EW, Bahinski A, Huh D. Organs-on-chips at the frontiers of drug discovery. Nat. Rev. Drug Discov. 2015;14:248–260. doi: 10.1038/nrd4539. Recent review focusing on novel organ-on-a-chip applications designed to identify drug targets, study inter-organ interactions, pharmacokinetics, and elucidate possible toxicity. The review discusses the future outlook of these highly sophisticated devices discussing their use in highly specialized fields and areas of medicine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rusling JF, Zhang Z. Designing functional biomolecular films on electrodes. In: Rusling JF, editor. Biomolecular Films: Design, Function and Applications. Marcel Dekker; New York: 2003. pp. 1–64. [Google Scholar]
- 37.Rusling JF, Hvastkovs EG, Hull DO, Schenkman JB. Biochemical applications of ultrathin films of enzymes, polyions and DNA. Chem. Commun. 2008:141–154. doi: 10.1039/b709121b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hvastkovs EG, Schenkman JB, Rusling JF. Annu. Rev. Anal. Chem. 2012;5:79–105. doi: 10.1146/annurev.anchem.111808.073659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Armistead PM, Thorp HH. Modification of Indium Tin Oxide Electrodes with Nucleic Acids: Detection of Attomole Quantities of Immobilized DNA by Electrocatalysis. Anal. Chem. 2000;72:3764–3770. doi: 10.1021/ac000051e. [DOI] [PubMed] [Google Scholar]
- 40.Rusling JF, Wasalathanthri DP, Zhao L, Schenkman JB. Thin multicomponent films for functional enzyme devices and bioreactor particles. Soft Matter. 2014;10:8145–8156. doi: 10.1039/c4sm01679c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wasalathanthri DP, Faria RC, Malla S, Joshi AA, Schenkman JB, Rusling JF. Screening Reactive Metabolites Bioactivated by Multiple Enzyme Pathways Using a Multiplexed Microfluidic System. Analyst. 2013;138:171–178. doi: 10.1039/c2an35993f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •42.Song B, Shen M, Jiang D, Malla S, Mosa IM, Choudhary D, Rusling JF. Microfluidic Array for Simultaneous Detection of DNA Oxidation and DNA-Adduct Damage. Analyst. 2016 doi: 10.1039/c6an01237j. Describes the first electrochemical array to detect oxidized and metabolite-damaged DNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wasalathanthri DP, Malla S, Bist I, Tang CK, Faria RC, Rusling JF. High-Throughput Metabolic Genotoxicity Screening with a Fluidic Microwell Chip and Electrochemiluminescence. Lab on Chip. 2013;13:4554–4562. doi: 10.1039/c3lc50698c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••44.Wasalathanthri DP, Li D, Song D, Zheng Z, Choudhary D, Jansson I, Lu X, Schenkman JB, Rusling JF. Organ-Specific Metabolic Toxicity Chemistry from Electro-Optical Enzyme/DNA Arrays and LC-MS/MS. Chem. Science. 2015;6:2457–2468. doi: 10.1039/c4sc03401e. First report on development of a microfluidic reactor array to explore metabolic genotoxicity reaction chemistry using enzymes from multiple organs simultaneously. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao L, Krishnan S, Zhang Y, Schenkman JB, Rusling JF. Differences in Metabolite-mediated Toxicity of Tamoxifen in Rodent vs. Human Using Electrochemiluminescent Arrays and DNA/microsome Nanoreactors. Chem. Res. Toxicology. 2009;22:341–347. doi: 10.1021/tx8004295. [DOI] [PMC free article] [PubMed] [Google Scholar]