Abstract
Medication non-adherence is a major concern in the healthcare industry and has led to increases in health risks and medical costs. For many neurological diseases, adherence to medication regimens can be assessed by observing movement patterns. However, physician observations are typically assessed based on visual inspection of movement and are limited to clinical testing procedures. Consequently, medication adherence is difficult to measure when patients are away from the clinical setting. The authors propose a data mining driven methodology that uses low cost, non-wearable multimodal sensors to model and predict patients’ adherence to medication protocols, based on variations in their gait. The authors conduct a study involving Parkinson’s Disease patients that are “on” and “off” their medication in order to determine the statistical validity of the methodology. The data acquired can then be used to quantify patients’ adherence while away from the clinic. Accordingly, this data-driven system may allow for early warnings regarding patient safety. Using whole-body movement data readings from the patients, the authors were able to discriminate between PD patients on and off medication, with accuracies greater than 97% for some patients using an individually customized model and accuracies of 78% for a generalized model containing multiple patient gait data. The proposed methodology and study demonstrate the potential and effectiveness of using low cost, non-wearable hardware and data mining models to monitor medication adherence outside of the traditional healthcare facility. These innovations may allow for cost effective, remote monitoring of treatment of neurological diseases.
Keywords: Parkinson’s disease (PD) diagnosis, smart healthcare system, machine learning, healthcare data, multimodal sensor, healthcare delivery, decision support system, medication adherence
1. Introduction
Medication non-adherence is a widespread problem in the United States, increasing safety risks for patients and placing a significant financial burden on the healthcare industry [1]. Poor adherence to medication often results in substantial worsening of a disease, increased mortality, and unnecessary healthcare costs [2,3]. For the purposes of this paper, we limit the definition of adherence to the medication regimen a physician prescribes, and a patient’s commitment to taking those medications as prescribed. When evaluating patients having neurologically-induced movement disorders, clinicians often rely upon self-reports to determine medication adherence. For example, the patient may be asked whether he or she is taking medication as prescribed or whether he or she is attending physical therapy as planned. In the clinic, adherence can be assessed subjectively by trained healthcare professionals. However, a given patient’s presentation during office visits may not accurately reflect his or her typical daily function. Non-adherence is widespread and can be very difficult to determine during brief clinical visits.
Medication non-adherence is particularly problematic in patients with Parkinson’s disease (PD), a disease that often affects elderly individuals. The cognitive deficits associated with PD may exacerbate this problem. A recent study revealed that 61% of PD patients were non-adherent to their medical prescriptions. Additionally, the average medical cost per non-adherent patient was $15,826, compared with $9,228 for adherent patients [4]. Furthermore, the shortage of movement disorders subspecialists limits the availability of in-office visits for PD patients. Technologies that can help monitor patients’ overall health and motor symptoms may allow for improved quality of care, reduced costs, and more efficient office visits. One existing technology, for example, AutoCITE [5] is used to automate partially constraint-induced movement therapy in stroke victims. However, such approaches are highly customized and are physically invasive because they require patients to use wearable devices. Non-wearable approaches are needed so that physical contact with devices may be reduced or eliminated. Finally, since patients spend the majority of their time away from a healthcare provider’s supervised environment, there is a need for systems that can monitor medication adherence outside the clinic or hospital [6].
In order to overcome these challenges, the authors of this work propose a data mining driven methodology that uses low-cost, non-wearable sensors such as the Microsoft Kinect, to quantitatively evaluate adherence to medication and drug responses among movement disorder patients. The proposed methodology can be adapted easily to a patient’s home setting, allowing for the measurement of adherence in a non-office based setting. The methodology presented in this paper will be solving the following scenario:
Physician-Patient Healthcare Scenario
Parkinson’s disease (PD) patient (i) goes to the hospital to visit his/her physician. Since physicians cannot rely on patients to provide consistent and accurate information pertaining to their medication adherence, the physician prescribes the proposed motion sensor system to the patient with instructions on how to use it. Each sensor system will be customized to a specific patient using their initial setup data as the ground truth. The initial calibration/ground truth data can either be captured at the physician’s office or at home in the patient’s location of choice. Given the baseline/ground truth data, the data mining models will determine anomalous gait patterns, hereby indicating that a patient is non-adherent to his/her medication protocol. The availability of quantifiable evidence of medication adherence through the presence/absence of gait dysfunctions will enable healthcare decision makers to know how adherent their patients are and what intervention strategies enhance/diminish adherence. The physician will also know whether a patient is utilizing the system, based on a time log stored on the system itself, hereby minimizing a patient’s ability to “game the system” by not using it.
The methodology proposed in this work first uses remote sensor hardware to capture patient gait data. Machine learning algorithms are then employed to help healthcare practitioners detect PD patients’ medication status as “taking” (“on”) medication and “not taking” (“off”) medication, based on gait features. It has been reported that visual evidence of gait dysfunction are observed in Parkinson’s disease patients that are “off” medication, compared to when they are “on” medication [7–9]. While these gait dysfunctions are readily observable by a trained medical expert, there exists a knowledge gap in terms of whether these variations in gait (between “on” and “off” medication) can be captured by non-wearable sensors and data mined for patients’ adherence to medication protocols. This works aims to fill this knowledge gap by demonstrating the feasibility of non-wearable data capture and data mining of patient gait during “on” and “off” medication states. The methodology described herein has allowed for high discriminative ability using gait data and, in the future, may be used to predict medication adherence in the home setting.
For the purposes of this paper, we limit the definition of adherence to the medication regimen a physician prescribes and a patient’s commitment to taking those medications as prescribed. This paper advances the field of remote detection of adherence by:
The ability to use low cost, non-wearable data acquisition techniques that concurrently maintain patient privacy by collecting only the three-dimensional (3D) geospatial data pertaining to a patient’s skeletal node location.
The ability to quantify movement abnormalities at a non-office/non-hospital location, allowing for use within the home setting.
The ability to act as both an early warning and active monitoring system that can provide relevant information to patients, clinicians, and healthcare decision makers.
The organization of this paper is as follows. This section provides a brief introduction and motivation pertaining to the problem. Section 2 describes previous work related to the current research. Section 3 outlines our methodology for patient adherence monitoring. A clinical study involving PD patients and controls is presented in Section 4, with the results and discussion. Section 5 concludes the paper and outlines some future work and areas for expansion.
2. Literature Review
2.1 Motor Dysfunction in Neurological Diseases and the Effect of Medication
Certain neurological diseases such as Huntington’s disease and Parkinson’s disease are associated with motor dysfunctions. Movement disorders are often detected and evaluated based upon the typical motor patterns of each disease entity (i.e. hyperkinesia, hypokinesia, rigidity, gait freezing, etc.) [10–12]. Pharmacologic treatment of PD focuses upon restoring dopamine levels (depleted in PD) to the basal ganglia in order to ameliorate PD-related symptoms, such as hypokinesia, rigidity, and impaired postural balance [13]. However, pharmacologic treatments often have complications, such as “wearing off” period at the end of dosing cycles. This reduction in efficacy can cause patients’ motor symptoms to return [10,11,13]. Recent advances in sensor technology can help researchers and clinicians to monitor such drug-related fluctuations in motor function [14]. Moreover, since these fluctuations are related to daily medicine intake [15], they may serve to identify and flag patients that require adjustments to their treatment regimens. Since medication adherence for chronic diseases is a known issue in this population [1], the monitoring system developed by this study may allow for identification of individuals who require help remembering their medications or who might benefit from longer-acting drugs.
2.2 Remote Non-Wearable Disease Monitoring
An important aspect that needs to be addressed is the availability and convenience of healthcare providers. Certain factors, such as distance to adequate facilities, can be hindrances for access. Approaches such as teleconferencing have been explored to overcome these limitations [16]. Encouragingly, stroke victims using AutoCITE that were supervised 25% of the time by a person showed the same progress as patients supervised 100% of the time [1]. These results demonstrate the potential for significantly reducing healthcare provider supervision in some diseases. Additionally, patient evaluations during office visits are limited contextually because they may not represent adequately the patient’s status during everyday tasks [16]. Long-term monitoring approaches have been proposed to overcome the short time duration of healthcare provider visits and the unreliability of existing approaches (i.e. patient diaries and self-reporting) [17]. In addition, phonation-based approaches have been explored for detection of neurological diseases, such as PD, and may be extended to adherence monitoring. However, phonation data is complicated by variations in accents and speech patterns. Furthermore, symptoms have to be very severe for detection [18]. Approaches to monitor movement dysfunctions have been explored by Bonato et al. using accelerometers (ACC) and surface electromyographic sensors (EMG) [14]. Barth et al. used gyroscopes and accelerometers in order to measure the hand motor and gait functions [19]. Finally, ankle mounted sensors have been explored by Moore et al. for long-term monitoring of gait in PD [20]. These techniques require physical contact of sensors with the patient, which can affect movement and reduce adherence. Although these methods provided preliminary concepts, the authors concluded that such methods are still too physically intrusive and inconvenient [19]. Thus, there is still a substantial need for contactless detection and monitoring techniques for movement disorders.
Researchers have begun exploring more contactless detection of neurologically induced movement disorders, in order to mitigate some of the aforementioned limitations of existing non-wearable approaches. For example, Clark et al. investigate the validity of the Microsoft Kinect for assessment of postural control, compared to a benchmark multiple-camera 3D motion analysis system [21]. Their findings concluded that the Kinect system can validly assess kinematic strategies of postural control. Gabel et al. propose a Kinect based approach for measuring stride and arm kinematics in patients and demonstrate the validity of the Kinect system for remote gait capture [22]. Clark et al. propose a Kinect based system for providing lateral trunk lean feedback during gait retraining [23]. The authors conclude that the Kinect based system can be used as a real time biofeedback system, without the need for expensive wearable sensor systems. Specifically relating to neurologically induced movement disorders such as Parkinson’s disease, Summa et al. propose the use of a Kinect based system, in conjunction with virtual reality to implement a technologically assisted version of the Lee Silverman’s Voice Therapy, LSVT®BIG [24]. Takač et al. employ both wearable (smartphone sensors) and non-wearable (Kinect) sensors to detect freezing of gait (FoG), a phenomenon observed in PD patients resulting in a temporary, involuntary inability to initiate or continue movement that lasts for a few seconds or minutes [25].
Literature relating to the capture of patient gait data using non-wearable sensors is promising and demonstrates the feasibility of these techniques. While the literature has investigated the ability of systems such as the Kinect to capture patient gait in a manner comparable to more advanced non-wearable or wearable sensors, there exists a knowledge gap in terms of whether the impact of intervention mechanisms (such as medications) on patients’ gait patterns can be distinguished from patients’ abnormal gait patterns (e.g., in PD patients). This work aims to fill this gap through the use of non-wearable sensors that capture patients’ gait data and machine learning algorithms that discover latent, previously unknown patterns that help distinguish patients “on” and “off” medication states.
2.3 Data Mining for Characterization of Human Movement
Together with the recent availability of low-cost multimodal sensors and the growth of motion capture technologies, data mining has emerged rapidly as a means to characterize human movement in an automated and accurate manner. Data mining techniques can allow for accurate automated classification of human movements and gestures [26–28]. Begg et al., for example, explored the ability for data mining to recognize changes in gait between young and older humans [27]. The data recorded using the PEAK Performance Technology Inc. motion analysis system utilized support vector machine learning to identify 24 gait features and distinguish between age-related gait patterns with 91.7% accuracy. Additionally, using skeletal data captured using the Microsoft Kinect, Celebi et al. were able to use dynamic time warping (a template matching algorithm from speech recognition) to recognize gestures made by humans and classify them according to joint positions [26]. Using the same sensor, Raptis et al. were able to illustrate real time classification of dance gestures using a cascaded correlation based classifier, achieving an average accuracy of 96.9% despite noisy sensor data [28]. Such techniques illustrate the potential of data mining methods for human movement pattern recognition and characterization.
3. Methodology
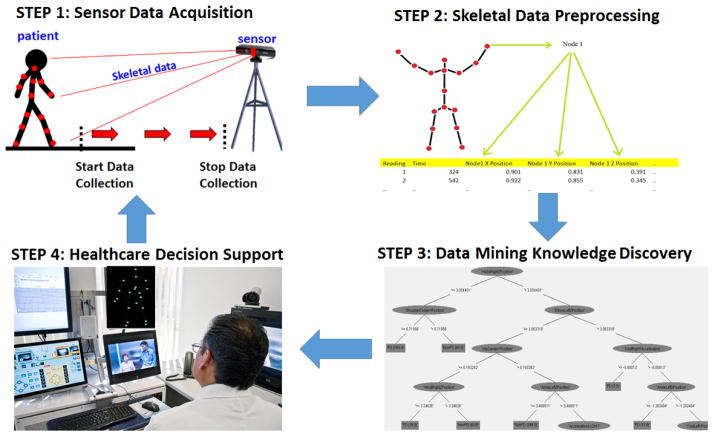
A non-wearable data-mining driven methodology is proposed in this work to differentiate between PD patients who are adherent to their medication and PD patients who are non-adherent. The methodology proposed consists of four steps, as outlined in Figure 1. Step 1 is the usage of non-wearable sensors in a non-hospital environment to gather skeletal joint data from the patient. This allows for patient convenience and remote monitoring of adherence. Step 2 is the processing of the collected skeletal joint data to generate 3D position, velocity, and acceleration values for the skeletal joints of the patient. This data is used to generate a set of identifying features that will be used subsequently to determine whether a patient is adherent to his or her medication. Step 3 employs machine learning algorithms on the generated set of features to classify a patient as adherent to their medication or not. The results are summarized in Step 4 and will serve to inform a physician of a patient’s current PD medication adherence state. This potentially allows the healthcare provider to monitor the patient’s adherence and propose intervention strategies aimed at enhancing patient safety and wellbeing.
Figure 1.
Overview of methodology
3.1 Step 1: Data Acquisition
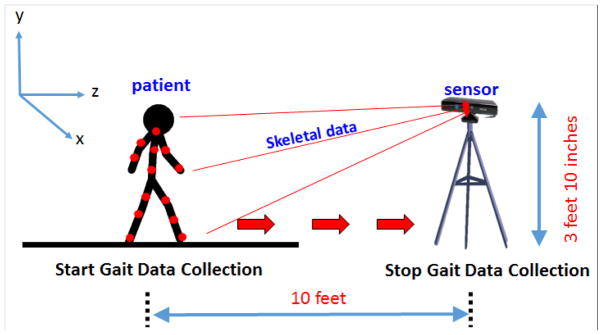
Low cost, off-the-shelf time of flight sensors (e.g., Microsoft Kinect) can be used in a non-clinical environment to capture human gait data. Multimodal time of flight sensors can collect a variety of measurements from video data, similar to regular cameras, with the addition of depth readings of humans and objects. Our methodology relies on collecting 3D coordinate skeletal data of the patient. Time of flight sensors can approximate nodes on a human body without the need of wearable sensors and use these nodes to map the 3D locations of the joints of the skeletal system [29]. For the data collection step, four walking orientations are conducted. I.e., walking towards the sensor (forward), walking away from the sensor (backward), walking to the right of the sensor (RHS) and walking to the left of the sensor (LHS), as seen in Figure 2. With each reading, the sensor collects X, Y, and Z coodinate data for k nodes representing various joints on the human skeleton, as shown in Figure 2. Note that the patient is walking toward the sensor (red arrow). The sensor collects the 3D coordinates of k skeletal joints, represented by the red dots, on the patient in Figure 3. The number of joints tracked, k, varies depending on the sensor used. For the original Kinect sensor employed in this work, the number of joints k tracked is 20, ranging from the head joint (in XYZ space) to the right and left foot joints (in XYZ space), as seen in Figure 3.
Figure 2.
Four different directions of capture
Figure 3.
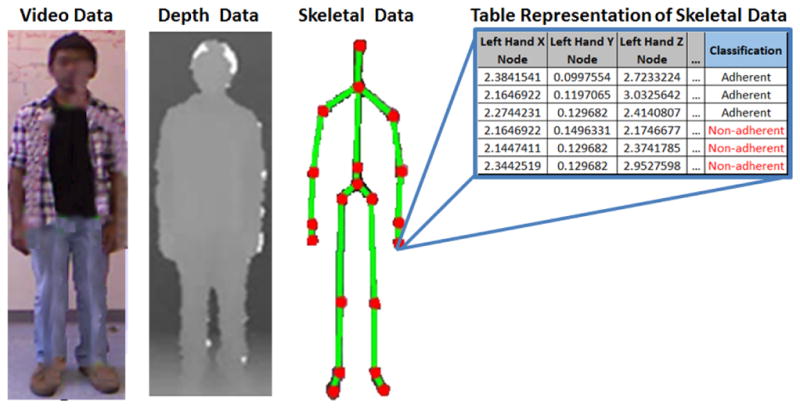
Unprocessed readings from the Kinect Sensor
To take the readings, the sensor should be placed such that the patient is within the range and field of view of the sensor being used. Thereafter, the patient walks in a straight line in one of the following four directions: directly toward the sensor, directly away from the sensor, parallel to the sensor from left to right, and parallel to the sensor from right to left. The sensor captures data as the patient walks in one of the four directions. The amount of skeletal tracking error highly depends on the sensor being employed, the environmental lighting, the sensor’s latency, and a patient’s distance away from the sensor. The Kinect is more accurate when all skeletal joints are being tracked, which is more likely to occur during the front and back orientations. For the left and right walking experiments, there may be instances where certain joints are not observable by the Kinect system due to the orientation of the subject, relative to the patient. Clark et al. performed a study on the accuracy of the Kinect system in providing reliable posture data. Their research found comparable inter-trial reliability (ICC difference = 0.06 ± 0.05; range, 0.00–0.16) and excellent concurrent validity for the majority of measurements (r = 0.96 ± 0.04; range, 0.84–0.99) [21]. In this work, whenever the sensor system is unable to track a joint, this data instance is automatically inferred by the sensor, based on previously known joint data. Given the sampling rate of 30 frames per second, enough data is generated during a 4–6 second walking experiment to offset anomalous joint data. It is important to note that the sensor system tracks skeletal joints, independent of the attire worn by the patient. I.e., a patient could be wearing a business suit or a night gown, as the depth sensor data enables the system to calculate position data (Figure 3). Existing experimental setups found in the literature have proposed collecting data with comparable distance or time constraints [30–33]. However, these data collection techniques typically require extensive human observation or intervention. The proposed methodology employs the use of non-wearable sensors that can capture data at high frequency rates.
3.2 Step 2: Raw Skeletal Data Processing
The raw data aquired by the sensor, is used to generate identifying features that will be subsequently used to distinguish adherent (“on” medication) vs non-adherent (“off medication”) patients. At each time stamp, the sensor captures the 3D coordinate (X, Y, Z) positions for each of the k joints (red circles in Figure 3), hereby generating a 3k dimensional feature space relating to the position data of the k joints. For the Kinect system, the number of joints tracked (k) is 20, although other non wearable sensors may track more or less gait related joints. In addition to joint position data captured by the Kinect system, velocity ( ) and acceleration ( ) are calculated based on the original XYZ position data of the k joints. Given the average frame rate of 30 frames per second, the time between two data points (i.e., ti − ti−1 is approximately 33ms.
In addition to the position, velocity and acceleration features, the ratio between joints are also calculated in order to generate a unit less set of features that accounts for variations in gait across a wide range of individuals and orientations, relative to the sensor system. Therefore by initially tracking k=20 joints, a total of 60 position features (i.e., 20 position features for the X, Y and Z dimensions), 60 velocity features (i.e., 20 velocity features for the X, Y and Z dimensions), 60 acceleration features (i.e., 20 acceleration features pertaining for the X, Y and Z dimensions), in addition to the remaining 1710 ratio features generated using the enumerative combination of each of the XYZ position, velocity and acceleration joints, relative to the other joints. For example, in order to calculate the ratio features for the position dimension in the X direction, each node is compared in a pairwise manner, taking into account symmetry (i.e., the distance from A to B is the same as the distance from B to A and is therefore considered once). Therefore, for the 20 position nodes in the X direction, there are 19+18+17+16.+1 possible ratio features generated, resulting in a total of 190 ratio position features in the X direction. For all ratio positions (i.e., X, Y, and Z), this results in (3*190=570 position ratio features). A similar calculation is performed for the velocity and acceleration features, resulting in a total ratio feature generation (XYZ for position, velocity and acceleration features) of (3*570=1710). The entire feature space containing 1890 features (180 position, velocity and acceleration features +1710 ratio features from the enumerative combinations) serves as the input space, with the class variable being that which we aim to predict (i.e., “adherent” (“on” medication) or “non-adherent” (“off ” medication)).
3.3 Step 3: Data Mining
The objective of the data mining step is to determine the feature combinations that enable the classification of an unseen set of gait readings as “adherent” (“on” medication) or “non-adherent” (“off ” medication), based on the position features captured in Step 1 and the velocity, acceleration and ratio positions generated in Step 2 of the methodology. The elements of this r-tuple contain numeric values for the gait features relating to the binary classifier (i.e., “adherent” or “non-adherent”), thus representing if the reading indicates a person who is adherent to their medication or someone who is nonadherent, for a given time stamp. The binary classifier is trained using a data set of known adherent and non-adherent patients. Each reading is considered an independent sample so as to minimize assumptions about the gait features, their correlations with subsequent samples, and their correlation with the output class variable (label). The resulting data mining model can be retrained and updated if additional gait data is acquired from a patient in order to ensure that evolving gait patterns are captured by the model. Here, the healthcare decision makers can decide the threashold predictive accuracy that a model must fall below in order for the model to be retrained on new patient gait data. If the predictive accuracy of the data mining model falls below this predictive accuracy, then the new patient gait data will be included in the training data, with the model retrained using the same steps originally used during model generation. Manohar and Tucker have demonstrated the feasibility of using non-wearable sensors for modelling human body movement data in order to accurately perform emerging threat detection [34]. Behoora and Tucker have discovered correlations between engineering designers’ body language and their emotional states [35,36]. The authors of this work expand on previous research findings by exploring the ability of the proposed system to accurately predict patients’ “on”/“off” medication states.
In this work, the authors employ the i) C4.5 Decision Tree Classification algorithm, ii) IBK classificaiton algorithm, iii) Naïve Bayes Classification algorithm, iv) Support Vector Machines and v) Random Forest. These five data mining classification algorithms are employed in this work because of their accuracy, complexity, and comprehensibility [37–40,40]. The motivation behind choosing these classifiers is that while quantifying the exact time and space complexity of various algorithms depends on the data set and the implementations details, the above algorithms have been shown to have good real world performance across a wide variety of classification areas and provide a suitable range of classification and learning speeds [41]. For example, C4.5 and Random Forest have good classification and learning speeds. Additionally, the C4.5 can provide insight into the joint positions, velocities, and accelerations that are the most important for classification. SVM’s, conversely, have good classification and computational speed but slow learning speed. Thus, they may be more appropriate in situations when the training data model does not get updated very often. Finally, others such as IBK have slower classification speeds but have been shown to have good performance in a variety of situations [41]. The performance variations of these classifiers across various metrics is summarized in Table 1 below.
Table 1.
Comparison of classifiers used [65], where **** represents the best and * represents the worst performance
| Decision Tree Based Algorithms (C4.5, Random Forest) | SVM | IBk | Naïve Bayes | |
|---|---|---|---|---|
| Model Accuracy | ** | **** | ** | * |
| Training speed | *** | * | **** | **** |
| Classification speed | **** | **** | * | **** |
| Tolerance to noise | ** | ** | * | *** |
3.4 Step 4: Hospital Feedback
Accurate methods for detecting medication non-adherence would provide clinicians with critical information for the management having neurologic movement disorders. Machine learning algorithms may enable individualized monitoring of medication adherence, the lack of which, can exacerbate disease severity. Knowledge of decreasing adherence and worsening disease status would be particularly relevant for the prescribing clinician because these indications may prompt modification of the treatment strategy. For example, cognitive function often declines throughout the progression of PD and is a potential source for reduced medication adherence [42]. Machine learning systems may enable the detection of frequently missed doses, using remotely sensed motion data, and allow for the transmission of important alerts to healthcare providers. Accordingly, providers may utilize this data to evaluate the efficacy of a given treatment plan and adjust medications as necessary. For example, a patient who frequently forgets to take his or her afternoon dose of medication may be placed on a longer acting drug to increase adherence. Finally, the data available to the providers may be customized according to patient consent. Remote visualization of patient-specific gait patterns, for example, would be straightforward to implement and require minimal data transfer using skeletal coordinates and data mining results (Figure 4). The skeletal representation of a patient’s gait provides movement information and is analogous to capturing a video of the patient and uploading it for review by physicians [16]. Interactive video conferences have been shown to have favorable results for remote Parkinson’s disease monitoring. Our approach allows for an analogous review of the data collected by healthcare providers. Simultaneously, our approach has the advantage of only needing to transmit the joint data captured, as opposed to a video thus reducing network bandwidth requirements. Systems such as the Kinect have inbuilt algorithms that separate objects in an environment from actual individuals using computer vision and the depth sensing data streams. For a community deployment (which is planned for future work), an individual would be uniquely identified using facial recognition algorithms and skeletal joint patterns that are unique to each individual such as those proposed by [43–46]. This would help minimize the potential for individuals to “game the system” by having either friends or relatives capture data for them.
Fig 4.
Feedback to healthcare provider
While community based adoption of new systems typically presents new challenges, the “plug and play” nature of the proposed motion sensor system, along with the wide user base of existing users, enhances the ability for researchers to continually make enhancements to the system [47,48]. With the existing Graphical User Interface, a patient would simply go home, plug the Kinect sensor system into their laptop or computer, open the graphical user interface prescribed by their physician, and begin the data capture process, with the system calibrating, based on the height and distance away from the patient. Similar to how a physician instructs a patient to routinely conduct blood pressure or blood sugar tests at home, Step 1 of Figure 1 will be routinely coded by the physician into the instruction manual for the patient. Given that the sensor system collects time stamped data that is connected to the physician’s office, the physician will be flagged if a patient misses a given data collection time period.
4. Parkinson’s Disease Clinical Study
A study involving free walking trials of seven PD patients first off their dopaminergic drugs and thereafter on their prescribed drugs is presented in this section to demonstrate the feasibility of the proposed data mining driven methodology. The details relating to the experimental set up and sensor data acquisition are presented below. For the current study, 7 PD subjects were recruited from a tertiary movement disorders clinic. PD diagnosis was established by a movement disorders specialist (XH) using published criteria [49]. Subjects were confirmed for absence of other major and acute neurological disorders, hypothyroidism, vitamin B12 and folate deficiencies, and kidney and liver disease. All patients were asked to withhold PD medications overnight (> 12 hours) before the off-medication walking trial [50]. Written informed consent was obtained for each subject, in accordance with the Declaration of Helsinki and the research protocol was reviewed and approved by the Penn State Hershey Medical Center Institutional Review Board (IRB protocol number 28989). On-medication trials were recorded after waiting at least one hour from the time of dopaminergic medication administration. The validity of the proposed model will be explored based on the ability of i) the data mining models to provide individually customized monitoring of patient medication adherence and ii) the ability of the data mining models to generalize across PD patients in their “on” and “off” medication states.
4.1 Sensor Data Acquisition
In the experiment, the Microsoft (MS) Kinect multimodal sensor was used to capture 3D skeletal images in a contactless manner. The MS Kinect is an off-the-shelf, low cost sensor that is capable of capturing multimodal motion data and software interface using the Kinect software development kit. The Kinect is capable of tracking up to 20 joints of the human skeleton via its infrared sensor, which captures data at the rate of 30 Hz. The sensor has a resolution of 640 × 480 and the horizontal and vertical fields of view of the sensor are 57 degrees and 43 degrees, respectively. These factors make it suitable for our study [29]. Additionally, while the sensor’s accuracy decreases with distance, the maximum error reaches 4cm. These factors make it suitable for our study [51]. Gait sensor systems such as the Microsoft Kinect are “plug and play”, making them compatible with a wide range of existing laptops and tablets [29,52]. The Kinect sensor has sold over 24 million units as of 2013, in part due to its price and cross platform compatibility (i.e., the ability to use it with the Microsoft gaming platform, along with a wide range of laptops, desktops and tablets).
To record the motion of each subject, the Kinect was configured at an elevation of 3 feet and 10 inches above the floor, as seen in Figure 5. Subsequently, the whole-body representation of the patient was verified and the camera angle calibrated while the patient stood at a distance of 10 feet from the Kinect. Each subject was then instructed to Forward Walk (FW), which took 4–6 seconds per trial, depending on the subject. The subjects repeated the gait data collection by subsequently walking away from the sensor (back) to the left of the sensor (LHS) and to the right of the sensor (RHS) as explained in Figure 2). During each trial, data point sets were recorded every 33 milliseconds, resulting in approximately 100–180 total data point per subject. For example for the forward walking experiment, subjects were asked first to take 2–3 steps backward (4 feet) from the point of camera calibration, while remaining within the distance limit of the device. Subjects were then instructed to walk comfortably towards the Kinect and were not given any specific instructions regarding side of initiation. One trial was recorded during the “off” medication state, mimicking non-adherence. The subsequent trial was recorded one hour after the patient was administered dopaminergic medication (“on” medication, mimicking adherence). During the experiment, only readings where all 20 joints of the subject were located by the Kinect skeletal data tracking were included. For each run, an average of 206 readings were captured per subject. Note that multiple readings were taken per patient and classified to test for accuracy. Each such reading represents an independent sample.
Figure 5.
Example of forward walking experiment
4.2 Data Processing
For the Kinect up to 20 joints can be tracked resulting in k = 20. Once the data collection is complete, the additional features were generated, resulting in a total of 1890 gait related features per patient, per experiment.
Only readings in which the 3D positions of all 20 joints were measured by the sensor were utilized. The remaining readings were discarded. This led to a 60 position features for each complete reading, representing the 3D (X,Y,Z) coordinate data for each joint with the Kinect as the origin and an average of 206 readings per patient.
The velocity and acceleration of each node were computed in X, Y and Z coordinate space using the timestamp generated by the Kinect for each reading. To normalize according to subject-specific body metrics, the ratios between these values were calculated resulting in a total of 1890 features, where 60 features were the 3D position coordinates, 60 were from velocity, 60 from acceleration, and 1710 from the ratios between them.
A data set was generated from the readings acquired by the 7 PD patients while off of their medications. This data set was labeled “non-adherent” and represented non-adherence. Subsequently, each patient was administered his or her prescribed dopaminergic medication. On medication data was collected after one hour from the time of dopaminergic medication administration. These readings were placed into a data set labeled “adherent” and were used to represent medication adherence.
4.3 Application of machine learning algorithms for classification
Given the feature space containing 1890 features, the goal of each data mining algorithm is to first train a model, based on a given training data set. Next, unseen test data is used to quantify the accuracy of the data mining model by classifying unseen instances as either as “adherent” or “non-adherent.” The Waikato Environment for Knowledge Analysis (WEKA) [53] software was used to execute the five data mining algorithms described in the methodology. For the purposes of this study, the default parameters of the WEKA software’s machine learning algorithms were used and summarized in Table 2 below:
Table 2.
Summary of default parameters used in the Data Mining algorithms in WEKA
| Default Weka Parameters | |
|---|---|
| Naïve Bayes | Debug=false, displayModelInOldFormat=False, UseKernelEstimator=False, UseSupervisedDiscretization=False |
| IBK | KNN=1, CrossValidate=False, Debug=False, DistanceWeighting=No Distance Weighting, MeanSquared=False, NearestNeighbourSearchAlgorithm=LinearNNSearch, WindowSize=0 |
| SVM | BuildLogisticModels=False, C=1.0, ChecksTurnedOff=False, Debug=False, Epsilon=1.0E-12, FilterType=Normalize Training Data, Kernel=PolyKernel –C 250007 –E 1.0, NumFolds=−1, RandomSeed=1, ToleranceParameter=0.001 |
| C4.5 (J48) Decision Tree | BinarySplits=False, ConfidenceFactor=0.25, Debug=False, MinNumObj=2, NumFolds=3, ReducedErrorPrunning=False, SaveInstanceData=False, Seed=1, SubtreeRaising=True, Unprunned=False, UseLaplace=False |
| Random Forest | Debug=False, MaxDepth=0, NumFeatures=0, NumTrees=10, Seed=1 |
4.4 Evaluation Metrics for Data Mining Models
In order for the proposed methodology to be viable, the accuracy and robustness of the models needs to be validated. To evaluate the performance of the classifiers, the authors investigated two validation steps: i) Validation of data mining models for providing individually customized monitoring of patient-adherence and ii) Validation of the generalizability of the resulting data mining models to unseen patient data. Each step of the validation is assessed using the statistics presented in Table 3. Table 3 shows how classification results in either true positive (TP), false positive (FP), false negative (FN), and true negative (TN). The four values, when presented together, are termed the “confusion matrix.”
Table 3.
Confusion matrix for adherence
| Actual Adherence Status | Adherence Status Classified as | |
|---|---|---|
| Adherent | Non-adherent | |
| Adherent | True Positive (TP) | False Negative (FN) |
| Non-adherent | False Positive (FP) | True Negative (TN) |
These four outcomes are used to calculate the precision, recall, F-measure, and receiver-operator characteristic (ROC) curve performance metrics of the classifier models [54]. The precision represents the number of patients divided by the total number of elements classified as belonging to the positive class. This measure is indicative of how accurately the methodology predicts an adherent patient between patients “on” and “off” their medication. High precision avoids missed non-adherent patients. Recall represents the number of true positives divided by the total number of elements that actually belong to the positive class. This is indicative of how accurately the methodology correctly identifies a patient on their medication as adherent. High recall avoids false alarms. The F-measure combines precision and recall. The F measure is the harmonic mean of the precision and recall. It represents how well the methodology balances the measures of precision and recall. Accordingly, it represents the degree to which the classifier identifies non-adherent patients while avoiding false alarms. The Receiver Operating Characteristic (ROC) curve measures the performance of a classifier as its discrimination threshold is varied. It is used to characterize the tradeoff between the costs of failing to detect adherence against the costs of raising false alarms. The area under the ROC curve is an indicator of the quality of our classifier.
4.5 Results and Discussion
1) Investigate the ability of the data mining models to provide individually customized monitoring of patient medication adherence
The results presented in Tables 4–10 are quite promising and demonstrate the ability of the proposed methodology to accurately predict the “on” and “off” states of each individual patient. For conciseness, the data mining algorithm that yielded the best performance (compared to the others) are presented for each walking orientation in each of the tables. Out of the 28 experimental walking scenarios spanning the 7 PD patients, it can be seen that the C4.5 (denoted as the J48 algorithm in WEKA) had the most consistent predictive performance, achieving the highest performance for 14 out of the 28 walking experiments (i.e., LHS for patient 113, Back and RHS for patient 118, Back, Front and RHS for patient 162, Back for patient 164, Front, Left and RHS for patient 176, Back and RHS for patient 183, Back and Front for patient 215). The highest predictive accuracy achieved by the C4.5 (J48) model was 100% with patient 162, with the lowest predictive accuracy being 52% with patient 118. The IBK and SVM algorithms performed second and third best across the 28 walking experiments spanning 7 PD patients with 6 and 5 best performances respectively. For patient #164, RHW data was unavailable and denoted as “N/A” in Table 7.
Table 4.
Summary of Data Mining Classification Accuracies for different walking orientations for patient 113
| Patient | Walking Orientation | Algorithm | CCI | ICI | TP Rate | FP Rate | Precision | Recall | F-Measure | ROC Area |
|---|---|---|---|---|---|---|---|---|---|---|
| 113 | Back | SMO | 53.7% | 46.3% | 53.8% | 46.5% | 54.1% | 53.8% | 53.9% | 53.6% |
| Front | IBK | 69.7% | 30.3% | 69.7% | 30.4% | 70.1% | 69.7% | 69.8% | 69.7% | |
| Left (LHS) | J48 | 57.1% | 42.9% | 57.1% | 47.8% | 76.4% | 57.1% | 45.6% | 54.7% | |
| Right (RHS) | IBK | 65.7% | 34.3% | 65.7% | 35.8% | 66.2% | 65.7% | 65.0% | 65.0% |
Table 10.
Summary of Data Mining Classification Accuracies for different walking orientations for patient 215
| Patient | Walking Orientation | Algorithm | CCI | ICI | TP Rate | FP Rate | Precision | Recall | F-Measure | ROC Area |
|---|---|---|---|---|---|---|---|---|---|---|
| 215 | Back | J48 DT | 95.6% | 4.4% | 90.7% | 4.9% | 96.0% | 95.7% | 95.6% | 98.3% |
| Front | J48 DT | 91.7% | 8.3% | 91.8% | 7.5% | 93.0% | 91.8% | 91.7% | 92.1% | |
| Left (LHS) | IBK | 68.3% | 31.70% | 68.4% | 23.1% | 85.1% | 68.4% | 67.7% | 71.1% | |
| Right (RHS) | RF | 66.3% | 3370.0% | 66.3% | 30.1% | 75.3% | 66.3% | 64.0% | 84.6% |
Table 7.
Summary of Data Mining Classification Accuracies for different walking orientations for patient 164
| Patient | Walking Orientation | Algorithm | CCI | ICI | TP Rate | FP Rate | Precision | Recall | F-Measure | ROC Area |
|---|---|---|---|---|---|---|---|---|---|---|
| 164 | Back | J48 DT | 65.3% | 34.7% | 65.3% | 27.5% | 80.5% | 65.3% | 62.3% | 68.9% |
| Front | SVM | 59.5% | 40.5% | 59.5% | 37.9% | 63.1% | 59.5% | 58.1% | 60.8% | |
| Left (LHS) | SVM | 83.8% | 16.20% | 83.8% | 17.1% | 83.8% | 83.8% | 83.8% | 83.4% | |
| Right (RHS) | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
In terms of walking orientation, there does not seem to be a consistent walking direction across patients that represents the best walking experiment to achieve high predictive accuracy. For example, the highest Back walking experiment achieves 100% (patient 118), while the lowest accuracy for a back walking experiment resulted in a 44% accuracy (for patient 183). The data mining models and walking orientations seem to have highest accuracy for patient 162, with three of the orientations achieving an accuracy greater than 99%, with the lowest accuracy for that patient being 85.1%. A potential reason for this could be the severity of this patient’s PD case and the magnitude of the medication effect on the patient’s gait when they are “on” versus “off” medication. The data mining models and walking orientations seem to have the poorest accuracy for patient 176, with three of the four orientations having predictive accuracies in the 60% range and one in the 40% range. This may indicate that the difference between this patient’s gait “on” versus “off” medication, may not be as pronounced as patient 162, hereby making it more difficult for the sensor system and subsequent data mining models to determine differences between the “on” and “off” states.
The next paragraph investigates how the proposed data mining models generalize when multiple patients are used to train the model and a single patient is used to test the model (i.e., leave-one-out-validation). In this work, leave-one-out validation is limited to patients 162 and 176 since they represent the best and worst case predictions in the individual customized model (this section). The generalized model does not quantify the average performance across all patients due to the lack of a superior walking scenario (i.e., front, back, left, right) across all patients, as discussed in this section. Since the focus of this paper is on demonstrating the feasibility of the system to capture, store and mine gait related data for individual patients, future work will explore the generalizability of the proposed methodology across a large sample of patients.
2) Investigate the ability of the data mining models for to generalize across PD patients in their “on” and “off” medication states
The next validation stage explores the generalizability of the proposed methodology in predicting unseen cases of PD patients in their “on” and “off” medication states. In this scenario, the training data consists of a pool of patient gait data (as opposed to individual gait data, as seen in validation stage 1). The test data consists of the gait pattern of an unseen patient in their “on” and “off” medication state. I.e., a patient that was not included in the training data set. To test the generalizable data mining model, the authors select patient 162 (Table 6) to represent the patient with the most distinguishable “on” and “off” medication gait patters, based on the high predictive accuracy of the data mining models in Tables 4–10. The authors select patient 176 (Table 8) to represent the patient with the least distinguishable “on” and “off” medication gait patters, based on the low predictive accuracy of the data mining models in Tables 4–10. Together, patients 162 and 176 represent the best and worst case performance of the non-wearable sensor system and corresponding data mining models presented in validation stage 1. Given that each patient receives his/her own sensor system, the focus on this paper is on individually customized medication adherence models. However, for completeness, the authors include results from the generalizable cases for the patients exhibiting the best and worst predictive performance, as seen in Tables 6 and 8.
Table 6.
Summary of Data Mining Classification Accuracies for different walking orientations for patient 162
| Patient | Walking Orientation | Algorithm | CCI | ICI | TP Rate | FP Rate | Precision | Recall | F-Measure | ROC Area |
|---|---|---|---|---|---|---|---|---|---|---|
| 162 | Back | J48 DT | 100.0% | 0.0% | 100.0% | 0.0% | 100.0% | 100.0% | 100.0% | 100.0% |
| Front | J48 DT | 100.0% | 0.0% | 100.0% | 0.0% | 100.0% | 100.0% | 100.0% | 100.0% | |
| Left (LHS) | IBK | 99.2% | 0.08% | 99.2% | 0.7% | 99.2% | 99.2% | 99.2% | 99.2% | |
| Right (RHS) | J48 DT | 85.1% | 14.9% | 85.1% | 14.7% | 86.9% | 85.1% | 84.9% | 85.2% |
Table 8.
Summary of Data Mining Classification Accuracies for different walking orientations for patient 176
| Patient | Walking Orientation | Algorithm | CCI | ICI | TP Rate | FP Rate | Precision | Recall | F-Measure | ROC Area |
|---|---|---|---|---|---|---|---|---|---|---|
| 176 | Back | Naïve Bayes | 48.7% | 51.3% | 48.7% | 55.2% | 34.3% | 48.7% | 35.5% | 46.7% |
| Front | J48 DT | 65.9% | 34.1% | 65.9% | 35.9% | 75.6% | 65.9% | 61.9% | 64.7% | |
| Left (LHS) | J48 DT | 60.9% | 39.10% | 60.9% | 36.9% | 66.3% | 60.9% | 58.4% | 62.2% | |
| Right (RHS) | J48 DT | 67.7% | 32.3% | 67.7% | 30.4% | 72.1% | 67.7% | 66.7% | 68.7% |
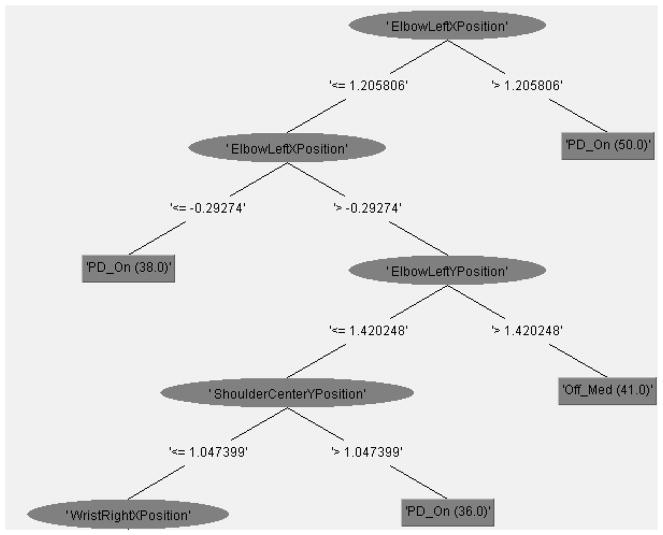
Tables 11 and 12 and Figure 6 present the results of the J48 Data Mining classification model that uses patients 113, 118 164, 176, 183 and 215 as training data to generate the model, with patient 162 as unseen data to test the model. The results in Table 11 reveal a predictive accuracy of 77.9% for the generalized model. It is interesting to note that the model correctly classifies all instances where a patient is “off” their medication (i.e., a precision score of 1), highlighting the system’s ability to accurately detect “off” medication states (Table 11), albeit at times being overly sensitive and in some cases classifying individuals that are “on” medication as being “off” medication. Figure 6 provides us with a detailed understanding of the relevant features of the model that enable the accurate classification of PD “on” and “off” states. Out of the original 1890 features included in the original training data, 139 features are found to be relevant to the Decision Tree model in Figure 6, with the top 10 features (corresponding to their hierarchy in Figure 6) being ElbowLeftXPosition, ElbowLeftYPosition, ShoulderCenterYPosition, WristRightXPosition, KneeLeftXPosition, HeadZPosition, SpineYPosition, KneeLeftXPosition, HipLeftXAcceleration. Positions x2/x16. The importance of the arm features (i.e., elbow joints) is consistent with existing literature that have found that reduced arm swing serves as an identifying feature of PD patients [55]. Beyond arm features, our model discovered relevant features in predicting PD “on” and “off” medication states such as the knee and spine joint locations, hereby expanding on the potential joint locations that physicians can focus on when trying to detect medication adherence in PD patients.
Table 11.
Summary of Data Mining Classification Error Rates for the generalized PD model using patient 162 as the unseen test patient
| Correctly Classified Instances | Incorrectly Classified Instances | Kappa Statistic | Mean Absolute Error | Root Mean Squared Error | Relative Absolute Error | Root Relative Squared Error | Total Number of Instances |
|---|---|---|---|---|---|---|---|
| 127 (77.9%) | 36 (22.1%) | 0.5458 | 0.2236 | 0.4698 | 44.70% | 93.92% | 163 |
Table 12.
Summary of Data Mining Classification Accuracy for the generalized PD model using patient 162 as the unseen test patient
| TP Rate | FP Rate | Precision | Recall | F-Measure | ROC Area | Class | |
|---|---|---|---|---|---|---|---|
| 53.2% | 0.0% | 100.0% | 53.2% | 69.5% | 83.1% | Off_Med | |
| 100.0% | 46.8% | 70.5% | 100.0% | 82.7% | 83.1% | PD_on | |
| Weighted Average | 77.9% | 24.7% | 84.4% | 77.9% | 76.5% | 83.1% |
Figure 6.
Partial Visualization of Weka’s Decision Tree Model [53] for Generalized PD ON-OFF Medication States using patient 162 as the test case
Tables 13 and 14 and Figure 7 present the results of the J48 Data Mining classification model that uses patients 113, 118 162, 164, 183 and 215 as training data to generate the model with patient 176 as unseen data to test the model. The ability of the generalized model to accurately predict patient 176’s “on” and “off” medication state was poor, resulting in only a 50.4% predictive accuracy. Given that patient 176’s “on” and “off” medication states were difficult to predict using both the individually customized approach (validation stage 1) and the generalized approach (validation stage 2) highlights some of the limitations of the current work and outlines potential areas of expansion. Figure 7 presents the J48 Decision Tree classification model with the top 10 attributes being ElbowLeftXPosition, KneeRightYPosition, ElbowRightXPosition, WristRightXPosition, KneeLeftXPosition, HeadZPosition, SpineYPosition, WristLeftXPosition, KneeRightXPosition, AnkleRightXPosition. The size of this tree is 103, slightly smaller than the previous J48 in Figure 6. Similar to the J48 algorithm in Figure 6, the most relevant feature in determining PD “on” and “off” states is the ElbowLeftXPosition feature. The J48 DT model in Figure 7 differs in some of the features such as ankle position, and not containing any velocity or acceleration data in the top 10 attributes.
Table 13.
Summary of Data Mining Classification Error Rates for the generalized PD model using patient 176 as the unseen test patient
| Correctly Classified Instances | Incorrectly Classified Instances | Kappa Statistic | Mean Absolute Error | Root Mean Squared Error | Relative Absolute Error | Root Relative Squared Error | Total Number of Instances |
|---|---|---|---|---|---|---|---|
| 117 (50.4%) | 115 (49.6%) | 0.0152 | 0.4939 | 0.6933 | 98.8% | 138.6% | 232 |
Table 14.
Summary of Data Mining Classification Accuracy for the generalized PD model using patient 176 as the unseen test patient
| TP Rate | FP Rate | Precision | Recall | F-Measure | ROC Area | Class | |
|---|---|---|---|---|---|---|---|
| 43.0% | 41.4% | 53.1% | 43.0% | 47.5% | 52.8% | Off_Med | |
| 58.6% | 57.0% | 48.5% | 58.6% | 53.1% | 52.8% | PD_on | |
| Weighted Average | 50.4% | 48.9% | 50.9% | 50.4% | 50.2% | 52.8% |
Figure 7.
Partial Visualization of Weka’s Decision Tree Model [53] for Generalized PD ON-OFF Medication States using patient 176 as the test case
For completeness, the authors performed an evaluation for each of the PD patients in the study. I.e., for patients 113 (results in Tables 15 and 16), 118 (results in Tables 17 and 18), 164 (results in Tables 19 and 20), 183 (results in Tables 21 and 22) and 215 (results in Tables 23 and 24), a similar experimental setup as described for patients 162 and 176 was performed. The results in Tables 15–24 highlights the challenges of utilizing a generalized PD medication adherence model to predict individual PD gait abnormalities, as each patient may exhibit different gait abnormalities. Furthermore, the effects of PD medication may impact patients differently, hereby making it difficult for gait based anomalies to be the sole differentiating factor for a general population. These results highlight the benefits for individually customized PD medication adherence models, as presented in the first paragraph of section 4.5 (results in Tables 4–10). Given the relatively low cost of the proposed system, each patient could have their own individually customized sensor system and PD medication adherence feedback.
Table 15.
Summary of Data Mining Classification Error Rates for the generalized PD model using patient 113 as the unseen test patient
| Correctly Classified Instances | Incorrectly Classified Instances | Kappa Statistic | Mean Absolute Error | Root Mean Squared Error | Relative Absolute Error | Root Relative Squared Error | Total Number of Instances |
|---|---|---|---|---|---|---|---|
| 86 (46.24%) | 100 (53.76%) | −0.0169 | 0.5375 | 0.7294 | 107.4 % | 145.7 % | 186 |
Table 16.
Summary of Data Mining Classification Accuracy for the generalized PD model using patient 113 as the unseen test patient
| TP Rate | FP Rate | Precision | Recall | F-Measure | ROC Area | Class | |
|---|---|---|---|---|---|---|---|
| 0.824 | 0.842 | 0.452 | 0.824 | 0.583 | 0.496 | Off_Med | |
| 0.158 | 0.176 | 0.516 | 0.158 | 0.242 | 0.496 | PD_On | |
| Weighted Average | 46.2% | 48.0% | 48.7% | 46.2% | 39.8% | 49.6% |
Table 17.
Summary of Data Mining Classification Error Rates for the generalized PD model using patient 118 as the unseen test patient
| Correctly Classified Instances | Incorrectly Classified Instances | Kappa Statistic | Mean Absolute Error | Root Mean Squared Error | Relative Absolute Error | Root Relative Squared Error | Total Number of Instances |
|---|---|---|---|---|---|---|---|
| 141 (42.08%) | 194 (57.91%) | −0.155 | 0.578 | 0.757 | 115.5 % | 151.3 % | 335 |
Table 18.
Summary of Data Mining Classification Accuracy for the generalized PD model using patient 118 as the unseen test patient
| TP Rate | FP Rate | Precision | Recall | F-Measure | ROC Area | Class | |
|---|---|---|---|---|---|---|---|
| 0.357 | 0.512 | 0.421 | 0.357 | 0.386 | 0.429 | Off_Med | |
| 0.488 | 0.643 | 0.421 | 0.488 | 0.452 | 0.429 | PD_On | |
| Weighted Average | 42.1% | 57.6% | 42.1% | 42.1% | 41.8% | 42.9% |
Table 19.
Summary of Data Mining Classification Error Rates for the generalized PD model using patient 164 as the unseen test patient
| Correctly Classified Instances | Incorrectly Classified Instances | Kappa Statistic | Mean Absolute Error | Root Mean Squared Error | Relative Absolute Error | Root Relative Squared Error | Total Number of Instances |
|---|---|---|---|---|---|---|---|
| 102 (52.6%) | 92 (47.4%) | 0.0346 | 0.4567 | 0.6451 | 91.2 % | 128.7 % | 194 |
Table 20.
Summary of Data Mining Classification Accuracy for the generalized PD model using patient 164 as the unseen test patient
| TP Rate | FP Rate | Precision | Recall | F-Measure | ROC Area | Class | |
|---|---|---|---|---|---|---|---|
| 0.442 | 0.407 | 0.463 | 0.442 | 0.452 | 0.564 | Off_Med | |
| 0.593 | 0.558 | 0.571 | 0.593 | 0.582 | 0.564 | PD_On | |
| Weighted Average | 52.6% | 49.1% | 52.4% | 52.6% | 52.4% | 56.4% |
Table 21.
Summary of Data Mining Classification Error Rates for the generalized PD model using patient 183 as the unseen test patient
| Correctly Classified Instances | Incorrectly Classified Instances | Kappa Statistic | Mean Absolute Error | Root Mean Squared Error | Relative Absolute Error | Root Relative Squared Error | Total Number of Instances |
|---|---|---|---|---|---|---|---|
| 43 (42.2%) | 59 (57.8 %) | 0 | 0.5778 | 0.7574 | 115.4 % | 151.2 % | 102 |
Table 22.
Summary of Data Mining Classification Accuracy for the generalized PD model using patient 183 as the unseen test patient
| TP Rate | FP Rate | Precision | Recall | F-Measure | ROC Area | Class | |
|---|---|---|---|---|---|---|---|
| 0 | 0 | 0 | 0 | 0 | 0.492 | Off_Med | |
| 1 | 1 | 0.422 | 1 | 0.593 | 0.492 | PD_On | |
| Weighted Average | 42.2% | 42.2% | 17.8% | 42.2% | 25.0% | 49.2% |
Table 23.
Summary of Data Mining Classification Error Rates for the generalized PD model using patient 215 as the unseen test patient
| Correctly Classified Instances | Incorrectly Classified Instances | Kappa Statistic | Mean Absolute Error | Root Mean Squared Error | Relative Absolute Error | Root Relative Squared Error | Total Number of Instances |
|---|---|---|---|---|---|---|---|
| 83 (36.2%) | 146 (63.8 %) | −0.3031 | 0.6379 | 0.7985 | 127.5 % | 159.6 % | 229 |
Table 24.
Summary of Data Mining Classification Accuracy for the generalized PD model using patient 215 as the unseen test patient
| TP Rate | FP Rate | Precision | Recall | F-Measure | ROC Area | Class | |
|---|---|---|---|---|---|---|---|
| 0.525 | 0.822 | 0.421 | 0.525 | 0.467 | 0.341 | Off_Med | |
| 0.178 | 0.475 | 0.247 | 0.178 | 0.207 | 0.341 | PD_On | |
| Weighted Average | 36.2% | 66.0% | 34.0% | 36.2% | 34.5% | 34.1% |
The proposed evaluation methods would allow healthcare providers to obtain real time and concise feedback regarding patient adherence. Importantly, adherence can be determined in a non-clinical location and transmitted to the healthcare providers remotely. The result of our study is in agreement with the motor function impairment exhibited due to non-adherence and suggests skeletal data mining can be a powerful tool in detecting adherence. Also, although the sensors used have range and accuracy limitations, the study shows using data mining techniques even on such noisy data can provide insight into patient adherence. The methodology proposed herein, accordingly, may provide a feasible and low-cost implementation for remote detection of medication adherence. Additionally, as the sensor used to record data can be turned on, as and when required for remote monitoring and data collection, patient’s privacy concerns are comparable to using video conferencing for medical diagnosis. The sensors proposed for this methodology already have widespread adoption and are present in over 24 million homes [56].
4.6 Improving Medication Adherence in Movement Disorder Patients
Medication adherence among neurologic disease patients is thought to reflect a variety of interconnected factors [57]. Medication non-adherence is common among PD patients, as 51.3% of patients are reported to skip at least one dose per week [58]. These patients may benefit from the proposed methodology as a means to receive self-reminders if non-adherence is detected. Cognitive impairment, a common complication of late-stage PD, may present additional challenges for medication adherence. Indeed, cognitive dysfunction and eventual dementia are known to reduce independence and quality of life substantially among PD patients, often placing heavy burdens on families and caregivers [59,60]. These patients may benefit substantially from remote detection technologies, since repeated missed doses may signal further changes in cognition or independence. Early alerts, received by clinical staff, could prompt reevaluation by the prescribing physician and adjustment of treatment. For example, a patient who repeatedly misses his or her afternoon dose may benefit from longer acting medications or a simpler treatment plan. Notably, substantial efforts have been made to measure accurately medication adherence among PD patients [57]. Our methodology may provide a novel method to monitor drug adherence and has the added ability to prompt accommodations and treatment adjustments for non-adherent patients.
4.7 Limitations of Existing Work
While this work has demonstrated the feasibility of detecting medication adherence in PD patients, several limitations exist that need to be addressed in future work. First, the collection of gait data was conducted in a controlled laboratory setting, in which patient gait dynamics may not represent the entire range of natural walking patterns that might be expected in a free-living environment. Second, this study presented proof of concept for monitoring patient adherence to medication using an off-drug state to represent non-adherence. However, we cannot infer directly that the pattern of gait accompanying medication non-adherence would necessarily be the same as that of an off-drug state. Third, PD patients are known to exhibit increasing gait impairment, including freezing episodes, as the disease progresses [61]. Our study could not capture the wide range of possible gait patterns throughout disease and responsiveness/non-responsiveness of various features to dopaminergic treatment [62]. Nonetheless, our technique was sufficient to detect key differences in the gait of PD subjects between off- and on-drug states. Lastly, our study could not directly address temporal changes of PD-related gait dynamics as a function of fatigue, time of day, or disease subtype [63]. These factors may be important and should be taken into account when designing a broadly generalizable machine learning algorithm to classify PD medication adherence. In order to address these limitations, prolonged clinical studies that utilize a larger sample size and a non-clinical environment are needed in order to assess the propose system’s performance in detecting PD medication adherence across different scenarios.
5. Conclusions and Future Work
In this paper, a remote data mining based methodology is proposed to differentiate between “on” and “off” medication states among PD patients using low-cost, non-wearable sensor hardware. Using whole-body movement data readings from the patients, the authors were able to discriminate between PD patients on and off medication, with accuracies greater than 97% for some patients using an individually customized model and accuracies of 78% for a generalized model containing multiple patient gait data. This shows that remote sensing coupled to data mining approaches may provide feasible techniques for healthcare providers to determine medication adherence among PD subjects from a non-clinical location. Moreover, this system may serve as a platform to provide early warnings if non-adherence is detected. These goals are particularly important among PD patients, since cognitive [64] impairment is common within this population and may increase risk for medication adherence. In addition to their potential role in the classification of medication adherence, machine learning techniques may reveal information regarding highly relevant yet undiscovered motor patterns in neurologic diseases (i.e. specific patterns of joint positions, velocities, and accelerations). Interestingly, the feature selection process yielded a remarkably smaller subset of informative features across all four experiments. This large reduction in dimensionality indicates that relatively few variables are required to discriminate between “off” and “on” medication gait patterns in PD. The relatively low number of variables required for classification is encouraging because it may lead to faster and more accurate learning and classification. Notably, the data collection step for each patient took less than 10 seconds, including calibration of the sensor. The methodology proposed herein, accordingly, may provide a feasible and low-cost implementation for remote detection of medication adherence.
In future work, the proposed methodology can be extended to incorporate measures of movement disorder severity. These data may be of particular use in medication management and adjustment. In addition, the placement of sensors and their directionality in a given room can be optimized to reduce costs and improve detection. Future studies may focus upon the robustness of kinetic measurements across multiple sensors and classification accuracy according to directionality and multi-sensor modalities. Moreover, longitudinal in-home data collection may reveal novel gait features and improve classification accuracy of PD adherence and non-adherence. Additionally, further studies can explore exploiting the temporal nature of the data captured by using techniques such as dynamic time warping to further improve classification. Future work may explore the feasibility of expanding the proposed methodology to include other neurological diseases and movement disorders.
Table 5.
Summary of Data Mining Classification Accuracies for different walking orientations for patient 118
| Patient | Walking Orientation | Algorithm | CCI | ICI | TP Rate | FP Rate | Precision | Recall | F-Measure | ROC Area |
|---|---|---|---|---|---|---|---|---|---|---|
| 118 | Back | J48 DT | 52.1% | 47.9% | 52.1% | 48.9% | 51.8% | 52.1% | 50.8% | 56.5% |
| Front | SVM | 74.3% | 25.7% | 74.3% | 27.1% | 74.2% | 74.3% | 74.2% | 73.6% | |
| Left (LHS) | IBK | 98.1% | 1.9% | 98.1% | 0.0% | 98.1% | 98.1% | 98.1% | 98.1% | |
| Right (RHS) | J48 DT | 61.7% | 38.3% | 61.7% | 37.3% | 78.4% | 61.7% | 55.4% | 62.2% |
Table 9.
Summary of Data Mining Classification Accuracies for different walking orientations for patient 183
| Patient | Walking Orientation | Algorithm | CCI | ICI | TP Rate | FP Rate | Precision | Recall | F-Measure | ROC Area |
|---|---|---|---|---|---|---|---|---|---|---|
| 183 | Back | J48 DT | 44.0% | 56.0% | 44.0% | 41.9% | 56.5% | 44.0% | 32.0% | 51.1% |
| Front | IBK | 63.0% | 37.0% | 63.0% | 39.5% | 62.9% | 63.0% | 62.1% | 61.7% | |
| Left (LHS) | SVM | 56.0% | 64.00% | 56.0% | 42.5% | 87.3% | 56.0% | 55.7% | 56.7% | |
| Right (RHS) | J48 DT | 97.5% | 2.5% | 97.5% | 2.4% | 97.5% | 97.5% | 97.5% | 97.6% |
Acknowledgments
This research is funded through the NSF I/UCRC Center for Healthcare Organization Transformation (CHOT), NSF I/UCRC grant # 1067885.
References
- 1.Cutler DM, Everett W. Thinking outside the pillbox--medication adherence as a priority for health care reform. N Engl J Med. 2010;362:1553–1555. doi: 10.1056/NEJMp1002305. [DOI] [PubMed] [Google Scholar]
- 2.Kripalani S, Yao X, Haynes R. Interventions to enhance medication adherence in chronic medical conditions: A systematic review. Arch Intern Med. 2007;167:540–549. doi: 10.1001/archinte.167.6.540. [DOI] [PubMed] [Google Scholar]
- 3.Osterberg L, Blaschke T. Adherence to Medication. N Engl J Med. 2005;353:487–497. doi: 10.1056/NEJMra050100. [DOI] [PubMed] [Google Scholar]
- 4.Davis KL, Edin HM, Allen JK. Prevalence and cost of medication nonadherence in Parkinson’s disease: Evidence from administrative claims data. Mov Disord. 2010;25:474–480. doi: 10.1002/mds.22999. [DOI] [PubMed] [Google Scholar]
- 5.Taub E, Lum PS, Hardin P, Mark VW, Uswatte G. AutoCITE: automated delivery of CI therapy with reduced effort by therapists. Stroke J Cereb Circ. 2005;36:1301–1304. doi: 10.1161/01.STR.0000166043.27545.e8. [DOI] [PubMed] [Google Scholar]
- 6.Lewis MA, Leake B, Leal-Sotelo M, Clark V. First nursing home admissions: time spent at home and in institutions after discharge. Am J Public Health. 1990;80:22–24. doi: 10.2105/ajph.80.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shults CW, Oakes D, Kieburtz K, Beal MF, Haas R, Plumb S, et al. Effects of coenzyme Q10 in early Parkinson disease: evidence of slowing of the functional decline. Arch Neurol. 2002;59:1541–1550. doi: 10.1001/archneur.59.10.1541. [DOI] [PubMed] [Google Scholar]
- 8.Riley DE, Lang AE. The spectrum of levodopa-related fluctuations in Parkinson’s disease. Neurology. 1993;43:1459–1459. doi: 10.1212/WNL.43.8.1459. [DOI] [PubMed] [Google Scholar]
- 9.Nutt JG, Woodward WR, Hammerstad JP, Carter JH, Anderson JL. The on–off phenomenon in Parkinson’s disease: relation to levodopa absorption and transport. N Engl J Med. 1984;310:483–488. doi: 10.1056/NEJM198402233100802. [DOI] [PubMed] [Google Scholar]
- 10.Lang AE, Lozano AM. Parkinson’s disease. First of two parts. N Engl J Med. 1998;339:1044–1053. doi: 10.1056/NEJM199810083391506. [DOI] [PubMed] [Google Scholar]
- 11.Lang AE, Lozano AM. Parkinson’s disease. Second of two parts. N Engl J Med. 1998;339:1130–1143. doi: 10.1056/NEJM199810153391607. [DOI] [PubMed] [Google Scholar]
- 12.Yamamoto A, Lucas JJ, Hen R. Reversal of Neuropathology and Motor Dysfunction in a Conditional Model of Huntington’s Disease. Cell. 2000;101:57–66. doi: 10.1016/S0092-8674(00)80623-6. [DOI] [PubMed] [Google Scholar]
- 13.Wj W. Motor fluctuations in Parkinson’s disease. Rev Neurol Dis. 2005;3:101–108. [PubMed] [Google Scholar]
- 14.Bonato P, Sherrill DM, Standaert DG, Salles SS, Akay M. Data mining techniques to detect motor fluctuations in Parkinson’s disease. 26th Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2004 IEMBS 04; 2004. pp. 4766–4769. [DOI] [PubMed] [Google Scholar]
- 15.Das S, Amoedo B, De la Torre F, Hodgins J. Detecting Parkinsons’ symptoms in uncontrolled home environments: a multiple instance learning approach. Conf Proc Annu Int Conf IEEE Eng Med Biol Soc IEEE Eng Med Biol Soc Conf. 2012;2012:3688–3691. doi: 10.1109/EMBC.2012.6346767. [DOI] [PubMed] [Google Scholar]
- 16.Hubble JP, Pahwa R, Michalek DK, Thomas C, Koller WC. Interactive video conferencing: a means of providing interim care to Parkinson’s disease patients. Mov Disord Off J Mov Disord Soc. 1993;8:380–382. doi: 10.1002/mds.870080326. [DOI] [PubMed] [Google Scholar]
- 17.Askari S, Zhang M, Won DS. An EMG-based system for continuous monitoring of clinical efficacy of Parkinson’s disease treatments. Conf Proc Annu Int Conf IEEE Eng Med Biol Soc IEEE Eng Med Biol Soc Conf. 2010;2010:98–101. doi: 10.1109/IEMBS.2010.5626133. [DOI] [PubMed] [Google Scholar]
- 18.Athanasios Tsanas MAL. New nonlinear markers and insights into speech signal degradation for effective tracking of Parkinson's disease symptom severity. 2010 [Google Scholar]
- 19.Barth J, Sunkel M, Bergner K, Schickhuber G, Winkler J, Klucken J, et al. Combined analysis of sensor data from hand and gait motor function improves automatic recognition of Parkinson’s disease. 2012 Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. EMBC; 2012. pp. 5122–5125. [DOI] [PubMed] [Google Scholar]
- 20.Moore ST, MacDougall HG, Gracies J-M, Cohen HS, Ondo WG. Long-term monitoring of gait in Parkinson’s disease. Gait Posture. 2007;26:200–207. doi: 10.1016/j.gaitpost.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 21.Clark RA, Pua Y-H, Fortin K, Ritchie C, Webster KE, Denehy L, et al. Validity of the Microsoft Kinect for assessment of postural control. Gait Posture. 2012;36:372–377. doi: 10.1016/j.gaitpost.2012.03.033. [DOI] [PubMed] [Google Scholar]
- 22.Gabel M, Gilad-Bachrach R, Renshaw E, Schuster A. Full body gait analysis with Kinect. Eng. Med. Biol. Soc. EMBC 2012 Annu. Int. Conf. IEEE, IEEE; 2012. pp. 1964–1967. [DOI] [PubMed] [Google Scholar]
- 23.Clark RA, Pua Y-H, Bryant AL, Hunt MA. Validity of the Microsoft Kinect for providing lateral trunk lean feedback during gait retraining. Gait Posture. 2013;38:1064–1066. doi: 10.1016/j.gaitpost.2013.03.029. [DOI] [PubMed] [Google Scholar]
- 24.Summa S, Basteris A, Betti E, Sanguineti V. A feasibility study on using kinect™ for the rehabilitation in persons with Parkinson’s disease. Gait Posture. 2013;37:S15. doi: 10.1016/j.gaitpost.2012.12.040. [DOI] [Google Scholar]
- 25.Takač B, Català A, Martín DR, Van Der Aa N, Chen W, Rauterberg M. Position and orientation tracking in a ubiquitous monitoring system for parkinson disease patients with freezing of gait symptom. JMIR MHealth UHealth. 2013;1 doi: 10.2196/mhealth.2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aydın AS. [accessed March 7, 2014];Gesture Recognition Using Skeleton Data with Weighted Dynamic Time Warping. 2013 http://www.academia.edu/2278042/Gesture_Recognition_Using_Skeleton_Data_with_Weighted_Dynamic_Time_Warping.
- 27.Begg R, Kamruzzaman J. A machine learning approach for automated recognition of movement patterns using basic, kinetic and kinematic gait data. J Biomech. 2005;38:401–408. doi: 10.1016/j.jbiomech.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 28.Raptis M, Kirovski D, Hoppe H. Real-time Classification of Dance Gestures from Skeleton Animation. Proc. 2011 ACM SIGGRAPHEurographics Symp. Comput. Animat; New York, NY, USA: ACM; 2011. pp. 147–156. [DOI] [Google Scholar]
- 29.Zhang Z. Microsoft Kinect Sensor and Its Effect. IEEE Multimed. 2012;19:4–10. doi: 10.1109/MMUL.2012.24. [DOI] [Google Scholar]
- 30.Paul SS, Canning CG, Sherrington C, Lord SR, Close JC, Fung VS. Three simple clinical tests to accurately predict falls in people with Parkinson’s disease. Mov Disord. 2013;28:655–662. doi: 10.1002/mds.25404. [DOI] [PubMed] [Google Scholar]
- 31.Elbers RG, van Wegen EE, Verhoef J, Kwakkel G. Is gait speed a valid measure to predict community ambulation in patients with Parkinson’s disease? J Rehabil Med. 2013;45:370–375. doi: 10.2340/16501977-1123. [DOI] [PubMed] [Google Scholar]
- 32.Tillman A, Muthalib M, Hendy AM, Johnson LG, Rantalainen T, Kidgell DJ, et al. Lower limb progressive resistance training improves leg strength but not gait speed or balance in Parkinson’s disease: a systematic review and meta-analysis. Front Aging Neurosci. 2015;7 doi: 10.3389/fnagi.2015.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Henderson EJ, Lord SR, Close JC, Lawrence AD, Whone A, Ben-Shlomo Y. The ReSPonD trial-rivastigmine to stabilise gait in Parkinson’s disease a phase II, randomised, double blind, placebo controlled trial to evaluate the effect of rivastigmine on gait in patients with Parkinson’s disease who have fallen. BMC Neurol. 2013;13:188. doi: 10.1186/1471-2377-13-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manohar G, Tucker C. A Privacy Preserving Data Mining Methodology for Dynamically Predicting Emerging Human Threats. ASME 2013 Int. Des. Eng. Tech. Conf. Comput. Inf. Eng. Conf; American Society of Mechanical Engineers; 2013. pp. V02AT02A069–V02AT02A069. [Google Scholar]
- 35.Behoora I, Tucker CS. Quantifying Emotional States Based on Body Language Data Using Non Invasive Sensors. ASME 2014 Int. Des. Eng. Tech. Conf. Comput. Inf. Eng. Conf; American Society of Mechanical Engineers; 2014. pp. V01AT02A079–V01AT02A079. [Google Scholar]
- 36.Behoora I, Tucker C. Machine Learning Classification of Design Team Members’ Body Language Patterns For Real Time Emotional State Detection. Des Stud. n.d;39:100–127. doi: 10.1016/j.destud.2015.04.003. [DOI] [Google Scholar]
- 37.D’heygere T, Goethals PLM, De Pauw N. Use of genetic algorithms to select input variables in decision tree models for the prediction of benthic macroinvertebrates. Ecol Model. 2003;160:291–300. doi: 10.1016/S0304-3800(02)00260-0. [DOI] [Google Scholar]
- 38.Ozcift A. SVM feature selection based rotation forest ensemble classifiers to improve computer-aided diagnosis of Parkinson disease. J Med Syst. 2012;36:2141–2147. doi: 10.1007/s10916-011-9678-1. [DOI] [PubMed] [Google Scholar]
- 39.Ramani GS, Geetha R. Parkinson disease classification using data mining algorithms. Int J Comput Appl. 2011;9 [Google Scholar]
- 40.Youn S, McLeod D. A Comparative Study for Email Classification. In: Elleithy K, editor. Adv Innov Syst Comput Sci Softw Eng. Springer; Netherlands: 2007. [accessed March 7, 2014]. pp. 387–391. http://link.springer.com/chapter/10.1007/978-1-4020-6264-3_67. [Google Scholar]
- 41.Kotsiantis SB. Supervised Machine Learning: A Review of Classification Techniques. Proc. 2007 Conf. Emerg. Artif. Intell. Appl. Comput. Eng. Real Word AI Syst. Appl. EHealth HCI Inf. Retr. Pervasive Technol; Amsterdam, The Netherlands, The Netherlands: IOS Press; 2007. [accessed March 7, 2014]. pp. 3–24. http://dl.acm.org/citation.cfm?id=1566770.1566773. [Google Scholar]
- 42.Verbaan D, Marinus J, Visser M, Van Rooden SM, Stiggelbout AM, Middelkoop HAM, et al. Cognitive impairment in Parkinson’s disease. J Neurol Neurosurg Psychiatry. 2007;78:1182–1187. doi: 10.1136/jnnp.2006.112367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Munsell BC, Temlyakov A, Qu C, Wang S. Comput Vision–ECCV 2012 Workshop Demonstr. Springer; 2012. Person identification using full-body motion and anthropometric biometrics from kinect videos; pp. 91–100. [Google Scholar]
- 44.Sinha A, Chakravarty K, Bhowmick B. Person identification using skeleton information from kinect. ACHI 2013 Sixth Int. Conf. Adv. Comput.-Hum. Interact; 2013. pp. 101–108. [Google Scholar]
- 45.Li BY, Mian AS, Liu W, Krishna A. Using kinect for face recognition under varying poses, expressions, illumination and disguise. Appl. Comput. Vis. WACV 2013 IEEE Workshop On; IEEE; 2013. pp. 186–192. [Google Scholar]
- 46.Hossny M, Filippidis D, Abdelrahman W, Zhou H, Fielding M, Mullins J, et al. Low cost multimodal facial recognition via kinect sensors. Proc Land Warf Conf LWC Potent Land Force Jt Marit Strategy Commonw Aust. 2012:77–86. [Google Scholar]
- 47.Webb J, Ashley J. Apress. 2012. Beginning Kinect Programming with the Microsoft Kinect SDK. [Google Scholar]
- 48.Nebeling M, Teunissen E, Husmann M, Norrie MC. XDKinect: development framework for cross-device interaction using kinect. Proc. 2014 ACM SIGCHI Symp. Eng. Interact. Comput. Syst; ACM; 2014. pp. 65–74. [Google Scholar]
- 49.Hughes AJ, Daniel SE, Kilford L, Lees AJ. Accuracy of clinical diagnosis of idiopathic Parkinson’s disease: a clinico-pathological study of 100 cases. J Neurol Neurosurg Psychiatry. 1992;55:181–184. doi: 10.1136/jnnp.55.3.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Langston JW, Widner H, Goetz CG, Brooks D, Fahn S, Freeman T, et al. Core assessment program for intracerebral transplantations (CAPIT) Mov Disord. 1992;7:2–13. doi: 10.1002/mds.870070103. [DOI] [PubMed] [Google Scholar]
- 51.Khoshelham K, Elberink SO. Accuracy and resolution of kinect depth data for indoor mapping applications. Sensors. 2012;12:1437–1454. doi: 10.3390/s120201437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cox D, Wolford J, Jensen C, Beardsley D. An evaluation of game controllers and tablets as controllers for interactive tv applications. Proc. 14th ACM Int. Conf. Multimodal Interact; ACM; 2012. pp. 181–188. [Google Scholar]
- 53.Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten IH. The WEKA Data Mining Software: An Update. SIGKDD Explor Newsl. 2009;11:10–18. doi: 10.1145/1656274.1656278. [DOI] [Google Scholar]
- 54.Fielding AH, Bell JF. A review of methods for the assessment of prediction errors in conservation presence/absence models. Environ Conserv. 1997;24:38–49. [Google Scholar]
- 55.Huang X, Mahoney JM, Lewis MM, Du G, Piazza SJ, Cusumano JP. Both coordination and symmetry of arm swing are reduced in Parkinson’s disease. Gait Posture. 2012;35:373–377. doi: 10.1016/j.gaitpost.2011.10.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chow JC, Lichti DD. Photogrammetric bundle adjustment with self-calibration of the PrimeSense 3D camera technology: Microsoft Kinect. Access IEEE. 2013;1:465–474. [Google Scholar]
- 57.Bainbridge JL, Ruscin JM. Challenges of treatment adherence in older patients with Parkinson’s disease. Drugs Aging. 2009;26:145–155. doi: 10.2165/0002512-200926020-00006. [DOI] [PubMed] [Google Scholar]
- 58.Leopold NA, Polansky M, Hurka MR. Drug adherence in Parkinson’s disease. Mov Disord. 2004;19:513–517. doi: 10.1002/mds.20041. [DOI] [PubMed] [Google Scholar]
- 59.Verbaan D, Marinus J, Visser M, Van Rooden SM, Stiggelbout AM, Van Hilten JJ. Patient-reported autonomic symptoms in Parkinson disease. Neurology. 2007;69:333–341. doi: 10.1212/01.wnl.0000266593.50534.e8. [DOI] [PubMed] [Google Scholar]
- 60.Schrag A. Quality of life and depression in Parkinson’s disease. J Neurol Sci. 2006;248:151–157. doi: 10.1016/j.jns.2006.05.030. [DOI] [PubMed] [Google Scholar]
- 61.Giladi N, Treves TA, Simon ES, Shabtai H, Orlov Y, Kandinov B, et al. Freezing of gait in patients with advanced Parkinson’s disease. J Neural Transm. 2001;108:53–61. doi: 10.1007/s007020170096. [DOI] [PubMed] [Google Scholar]
- 62.Schaafsma JD, Giladi N, Balash Y, Bartels AL, Gurevich T, Hausdorff JM. Gait dynamics in Parkinson’s disease: relationship to Parkinsonian features, falls and response to levodopa. J Neurol Sci. 2003;212:47–53. doi: 10.1016/s0022-510x(03)00104-7. [DOI] [PubMed] [Google Scholar]
- 63.Mitoma H, Yoneyama M, Orimo S. 24-hour recording of parkinsonian gait using a portable gait rhythmogram. Intern Med. 2010;49:2401–2408. doi: 10.2169/internalmedicine.49.3511. [DOI] [PubMed] [Google Scholar]
- 64.Levin BE, Tomer R, Rey GJ. Cognitive impairments in Parkinson’s disease. Neurol Clin. 1992;10:471–485. [PubMed] [Google Scholar]
- 65.Kotsiantis S. [accessed February 15, 2015];Supervised Machine Learning: A Review of Classification Techniques. 2007 http://citeseerx.ist.psu.edu/viewdoc/summary?doi=10.1.1.95.9683.