Abstract
Carcinoma of unknown primary (CUP) is a rare and difficult-to-treat malignancy, the management of which might be improved by the identification of actionable driver mutations. We interrogated interrogated 54–70 genes in 442 patients with CUP using targeted clinical-grade, next-generation sequencing (NGS) of circulating tumor DNA (ctDNA). Overall, 80% of patients exhibited ctDNA alterations; 66%, ≥ 1 characterized alteration(s) excluding variants of unknown significance. TP53-associated genes were most commonly altered (37.8%) followed by genes involved in the MAPK pathway (31.2%), PI3K signaling (18.1%) and the cell cycle machinery (10.4%). Distinct genomic profiles were observed in 87.9% of CUP cases with 99.7% exhibiting potentially targetable alterations. An illustrative patient with dynamic changes in ctDNA content during therapy and a responder given a checkpoint inhibitor-based regimen because of a mismatch repair gene anomaly are presented. Our results demonstrate that ctDNA evaluation is feasible in CUP and that most patients harbor a unique somatic profile with pharmacologically actionable alterations, justifying the inclusion of non-invasive liquid biopsies in next-generation clinical trials.
Keywords: Cell-free DNA, liquid biopsy, genomic landscape, next-generation sequencing, carcinoma of unknown primary
INTRODUCTION
Cancer of unknown primary (CUP) is a rare cancer (defined as fewer than 15 cases per 100,000 per year (1)) with an incidence of 7–12 cases per 100,000 per year (2). CUP is defined as metastatic disease with no identifiable primary tumor despite comprehensive evaluation including serum biomarker tests, procedures such as endoscopy and colonoscopy, as well as imaging and histopathological examination with specific immunohistochemistry (2–5). Microarray-based technology to seek the primary site of origin based on gene-expression profile have also been investigated (6). Approximately 60% of CUPs are classified as well/moderately differentiated adenocarcinomas, followed by undifferentiated/poorly differentiated adenocarcinomas (30%), and, less frequently, as squamous cell carcinoma (5%) and neuroendocrine tumor (about 1%) (2,5).
Despite its heterogeneous clinicopathological presentation, the treatment of CUP has primarily been with platinum-based combination chemotherapies. Although such regimens do demonstrate response rates of 20–40%, median survival remains poor at six to eight months (7,8). Novel immunotherapy approaches using pembrolizumab (anti-PD-1 antibody) in patients with rare tumors including CUP are underway (NCT02721732). Targeted therapy regimens have been investigated in the past with the combination of bevacizumab plus erlotinib in non-genomically selected CUP. However, the response rate was 10% and the median survival was 7.4 months (9).
Amongst refractory malignancies, a biomarker-based (personalized) approach matching patients with drugs has shown efficacy (10–14). Further understanding of the underlying genomic alterations among patients with CUP may also prove useful. Previous studies using archival tumor tissues of CUP patients found that TP53 (38–55%), KRAS (18–20%), CDKN2A (19%), MYC (12%), ARID1A (11%) and PIK3CA (9–14%) were frequently altered as assessed by targeted next-generation sequencing (NGS) (15,16). Although genomic sequencing is generally done on archival cancer samples, limitations with the use of tissues include intra-tumor genomic heterogeneity (17) as well as the dynamic mutational processes that can occur along with therapeutic intervention (18).
One approach to overcome these challenges is to investigate circulating tumor DNA (ctDNA). ctDNA is shed into the bloodstream from the cancer cells and can be isolated from a small tube of blood (also known as a “liquid biopsy”) (19). This process was first reported in 1987 (20), and, more recently, the technology has rapidly advanced, and has been applied in the clinic (21–24). Analyzing multiple genes by performing NGS on ctDNA is now feasible (25) (Supplemental Table 1). Here, we provide the first report of a very large series of patients with CUP (N = 442) whose ctDNA derived from blood was interrogated by clinical-grade NGS, and we document illustrative cases of the results of serial ctDNA testing and of matching patients to therapy.
METHODS
Patients
We investigated the genomic alteration status of 442 patients with CUP. Samples were sent to a clinical laboratory improvement amendments (CLIA)-licensed and College of American Pathologist (CAP)-accredited clinical laboratory (Guardant Health, Inc., http://www.guardanthealth.com/) for ctDNA testing using NGS (September 2014 to March 2016). Tumor types were provided by the submitting physicians. The database was de-identified. This study was performed in accordance with UCSD Institutional Review Board guidelines for de-identified databases for any investigational treatments for which patients gave consent. The UCSD IRB follows the Declaration of Helsinki and the Belmont Report guidelines.
Next generation sequencing
ctDNA was extracted from whole blood collected in 10mL Streck tubes, and 5ng–30ng of ctDNA was prepared for sequencing as previously described (25). All ctDNA was sequenced, including the somatic ctDNA and the germline ctDNA that is derived from natural leukocyte lysis. All sequence based mutations were evaluated for allele frequency. Allele frequencies were typically at ~ 100% (homozygous single nucleotide polymorphism), ~50% (heterozygous germline) and <5% (somatic fraction) (Supplemental Figure 1). In addition to the allele frequency, the specific alteration was also evaluated using the Database of Short Genetic Variation (dbSNP) and COSMIC database to differentiate germline from somatic mutation (25). Germline alterations were filtered out and not reported. The fractional concentration or variant allele fraction for a given somatic mutation is calculated as the fraction of ctDNA harboring that mutation in a background of wild-type ctDNA fragments at the same nucleotide position (25). The analytic sensitivity reaches detection of 1–2 single mutant fragments from a 10 ml blood sample (0.1% limit of detection) and analytic specificity is greater than 99.9999% (25). Throughout the timeframe of this study, the ctDNA assay expanded from 54 to 68 to 70 genes (Supplemental Table 1): 13 patients were tested with the 54 gene panel; 207 with the 68 gene panel; and 222 with the 70 gene panel. The assay reports single nucleotide variants in all genes and select copy number amplifications, fusions, and indel events (Supplemental Table 1.) We analyzed only non-synonymous alterations throughout the paper, except in the case report with serial sampling wherein all alterations were followed serially.
Endpoints, statistical methods, and case studies
Demographic information such as age and gender were extracted from the de-identified database. Descriptive statistics were used to summarize the genomic alterations identified in this study. Two case studies (from outside time range of de-identified database) are presented: (i) a patient with five serial ctDNA samples; and (ii) a patient who was successfully matched to an immunotherapy/targeted treatment combination (consent obtained according to the University of California San Diego Internal Review Board guidelines; protocol: I-PREDICT study (NCT02534675)).
RESULTS
Genomic alterations among carcinoma of unknown primary (Table 1, Figures 1,2, Supplemental Table 2 and Supplemental Figure 2)
Table 1.
Clinical characteristics and number of genomic alterations in 442 patients with carcinoma of unknown primary.
| Basic characteristics (N=442) | |
| Age, median (range), year | 65 (20–94) |
| Female, No. | 231 (52.3%) |
| Male, No. | 211 (47.7%) |
| Genomic alterations in 442 cases of carcinoma of unknown primary | |
| Number of alterations* | 1368 |
| Mean number of alterations per patient (range) (includes characterized alterations and VUS) | 3.1 (0–20) |
| Median number of alterations per patient (range) (includes characterized alterations and VUS) | 2 (0–20) |
| Number of characterized alterations | 768 (56.1%) |
| Mean number of characterized alterations per patient (range) | 1.7 (range 0–10) |
| Median number of characterized alterations per patient (range) | 1 (0–10) |
| Substitution, No. (%) | 507 (37.1%) |
| Amplification, No. (%) | 257 (18.8%) |
| Fusion, No. (%) | 3 (0.22%) |
| Indel, No. (%) | 1 (0.07%) |
| Variant of unknown significance, No. (%) | 600 (43.9%) |
| Number of patients with ≥ 1 alteration (%)* | 353 (79.9%) |
| Number of patients with ≥ 1 characterized alteration (%) | 290 (65.6%) |
| Number of patients with ≥ 2 characterized alterations (%) | 194 (43.9%) |
Includes both characterized alterations and variants of unknown significance (VUS).
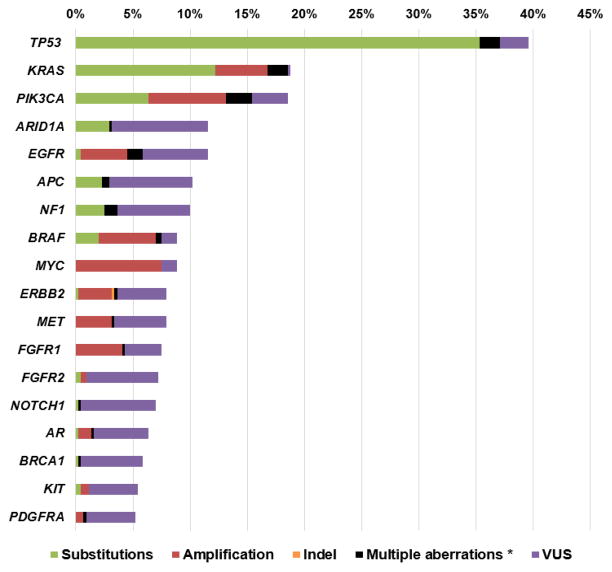
Figure 1. Frequency of genomic alterations among patients with carcinoma of unknown primary (N = 442).
Includes alterations with > 5% frequency. Please see Supplemental Table 1 for complete list of alterations found in this study.
* Multiple alterations indicate that the patient had >1 type of alteration in the same gene (substitutions, amplification, indel, VUS, etc).
Abbreviations: VUS: variants of unknown significance.
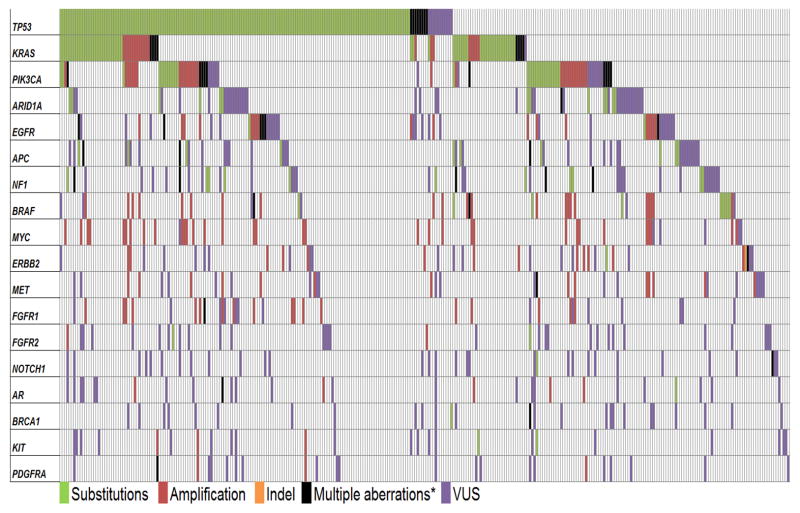
Figure 2. Oncoprint of frequently altered genes in patients with carcinoma of unknown primary.
Of the 442 cases studied, 325 patients had at least one alteration with > 5% frequency, which were included in the figure. Each vertical line represents one patient and the horizontal lines indicate specific genes. Please see Supplemental Figure 2 for the complete oncoprint for 442 cases evaluated in this report.
* Multiple alterations indicate that the patient had >1 type of alteration in the same gene (substitutions, amplification, indel, VUS, etc).
Abbreviations: VUS: variants of unknown significance.
Amongst all patients with CUP (N=442), the median age was 65 years (range, 20–94); 231 cases were women (52.3%) (Table 1). The total number of alterations found amongst 442 patients were 1368 and of these, 768 (56.1%) were characterized alterations including substitutions (N=507 [37.1%]), amplifications (N=257 [18.8%]), fusions (N=3 [0.22%]) and indels (N=1 [0.07%]). Six hundred (43.9%) alterations were variants of unknown significance (VUS). Eighty percent of patients (353/442) had ctDNA alterations detected with 66% (290/442) having at least one characterized alteration and 43.9% (194/442) harboring ≥2 characterized alterations. Amongst all patients (N=442), the median number of alterations per patient was 2 (range, 0–20) and the median number of characterized alterations was 1 (range, 0–10). Amongst patients found to have alterations (N=353), the median number of alterations was 3 (range, 1 to 20) and the median number of characterized alterations was 2 (range, 1–10) (Table 1). Focusing on characterized alterations, the most commonly altered gene was TP53 (37.1% [164/442]) followed by KRAS (18.6% [82/442]), PIK3CA (15.4% [68/442]), BRAF (7.5% [33/442]) and MYC (7.5% [33/442]) (Figures 1, 2, Supplemental and Supplemental Figure 2).
Number of alterations with possible cognate targeted therapies (Tables 1, 2, Supplemental Tables 3 and 4 and Supplemental Figures 3 and 4)
Table 2.
Selected actionable genomic alterations and examples of possible targeted therapies in patients with carcinoma of unknown primary (N = 442).
| Genomic alteration | No. % | Example of possible targeted therapies* |
|---|---|---|
| Tyrosine kinase families (N=79, 17.9%) | ||
| EGFR substitutions/amplification | 26 (5.9%) | Afatinib, cetuximab, erlotinib |
| ERBB2 substitutions/amplification/indel | 16 (3.6%) | Afatinib, trastuzumab, lapatinib |
| FGFR1 amplification | 19 (4.3%) | Lenvatinib |
| FGFR2 substitutions/amplification | 4 (0.9%) | |
| FGFR3 substitutions | 3 (0.7%) | |
| JAK2 substitutions | 1 (0.2%) | Ruxolitinib |
| KIT substitutions/amplification | 5 (1.1%) | Dasatinib, imatinib, sunitinib |
| MET amplification | 15 (3.4%) | Cabozantinib, crizotinib |
| PDGFRA amplification | 4 (0.9%) | Dasatinib, imatinib, sunitinib |
| RET fusion | 3 (0.7%) | Cabozantinib, lenvatinib, vandetanib |
| MAPK signaling (N=138, 31.2%) | ||
| HRAS substitution | 3 (0.7%) | MEK inhibitor (e.g. Trametinib or cobimetinib) |
| KRAS substitution/amplification | 82 (18.6%) | |
| NRAS substitution | 8 (1.8%) | |
| NF1 substitution | 16 (3.6%) | |
| GNAS substitution | 10 (2.3%) | |
| RAF1 substitution/amplification | 8 (1.8%) | |
| MAP2K1 substitution | 2 (0.5%) | |
| BRAF substitution/amplification | 33 (7.5%) | BRAF inhibitor (e.g. Dabrafenib, vemurafenib), MEK inhibitor (e.g. trametinib or cobimetinib) |
| PI3K signaling (N=80, 18.1%) | ||
| PIK3CA substitution/amplification | 68 (15.4%) | mTOR inhibitor (e.g. everolimus, temsirolimus) |
| PTEN substitution | 10 (2.3%) | |
| AKT1 substitution | 2 (0.5%) | |
| STK11 substitution | 4 (0.9%) | |
| TSC1 substitution | 1 (0.2%) | |
| Cell cycle associated genes (N=46, 10.4%) | ||
| CDKN2A substitution | 8 (1.8%) | Cyclin-dependent kinase inhibitor (e.g. Palbociclib) |
| CCND1 substitution/amplification | 3 (0.7%) | |
| CCND2 substitution/amplification | 2 (0.5%) | |
| CDK4 amplification | 5 (1.1%) | |
| CDK6 amplification | 18 (4.1%) | |
| CCNE1 amplification | 16 (3.6%) | Proteasome inhibitor (e.g. Bortezomib) |
| TP53 associated genes (N=167, 37.8%) | ||
| TP53 substitution | 164 (37.1%) | Anti-VEGF (e.g. bevacizumab), WEE1 inhibitor (e.g. AZ1775, NCT01748825) |
| ATM substitution | 4 (0.9%) | PARP inhibitor (e.g. olaparib) |
| Mismatch Repair Gene Alterations (N = 7, 1.6%) | ||
| MLH1 gene** | 7 (1.6%) | Immunotherapy with check point inhibitors (50) |
See Supplemental Table 3 for the rationale for possible targeted therapies.
MLH1 was the only mismatch repair gene tested in the ctDNA assay used.
Of the 768 characterized alterations, 89.6% (688/768) were potentially targetable with FDA-approved agents as off-label use, and an additional 8.5% (65/768) were theoretically targetable with therapies that are currently in clinical trials (Supplemental Tables 3 and 4). Tyrosine kinase families were altered in 17.9% (79/442); mitogen-activated protein kinase (MAPK) signaling pathway was altered in 31.2% (138/442); the phosphoinositide 3 kinase (PI3K) pathway was altered in 18.1% (80/442); cell-cycle associated genes were altered in 10.4% (46/442); and alterations in TP53-associated genes were seen in 37.8% (167/442) of patients with CUP (Table 2). Amongst the 768 characterized alterations, 335 were molecularly distinct alterations (e.g. KRAS and PIK3CA alterations were considered genomically distinct; KRAS G12C and KRAS G12V were considered genomically identical but molecularly distinct). Although infrequent, classically targetable alterations were observed in this CUP cohort, including 9 occurrences of BRAF V600E, 4 occurrences of EGFR L858R, 3 KIF5B-RET fusions, and 1 ERBB2 exon 20 insertion. Altogether, 98.0% (753/768) were theoretically actionable either with agents that are approved by the FDA (albeit off-label) or with agents that are in clinical trials (Supplemental Tables 3 and 4).
Among all 442 patients with CUP, 63.8% (282/442) had theoretically actionable alterations by an FDA-approved agent, and an additional 1.6% (7/442) patients had alterations targetable with investigational agents in clinical trial. The mean number of actionable alterations per patient was 1.7 (range 0 to 10). Altogether, 65.4% (289/442) of patients had theoretically actionable alterations either with FDA-approved or with investigational agents (Supplemental Tables 3 and 4 and Supplemental Figures 3 and 4). The majority of the patients (34.4%) who were deemed to have no targetable alterations had no characterized ctDNA alterations detected.
Distinctness of genomic alterations among 442 patients with CUP (Supplemental Table 4)
Among the 290 patients with at least one characterized genomic alteration, 12.1% [35/290] had identical molecular portfolios (KRAS G12C as a single characterized alteration [ID# 52, 94, 214, 335, 280, 404], KRAS G12V [ID# 54,271, 277]; KRAS G12D [ID# 206, 416]; KRAS G13C, [ID# 81, 225]; TP53 R110P [ID# 30, 248]; TP53 V173L [ID# 318, 386], TP53 R175H [ID# 116,317,418]; TP53 C176Y [ID# 275, 305]; TP53 C275Y [ID# 98,166]; TP53 Y220C [ID# 22,32, 385]; CCNE1 amplification [ID# 395, 127]; GNAS R201H [ID# 46, 105,178]; and IDH1 R132C [ID# 118, 120, 336]) (Supplemental Table 4). However, among the 194 patients who had more than one characterized alteration, no two patients had identical molecular profiles (Supplemental Table 4).
Comparative quantitation of ctDNA in the circulation (Supplemental Figure 5)
The current study calculated the fractional concentration of mutant allele by comparing the concentration of wild-type ctDNA fragments at the same nucleotide position. Among frequently altered genes (Figure 1 and Supplemental Table 1), all characterized mutations had a median ctDNA fraction of less than 5%. There was no clear association between the frequencies of gene alterations and the fraction of ctDNA detected in the blood (Supplemental Figure 5).
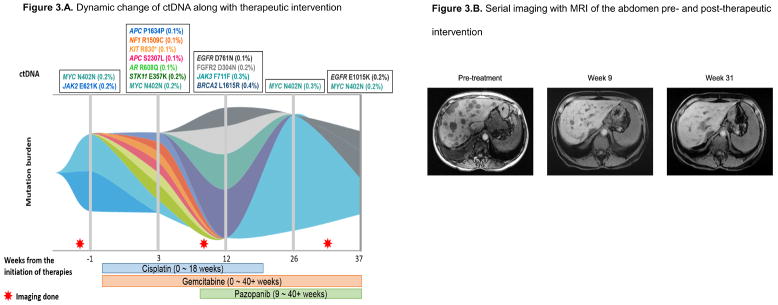
Clinical observation of dynamic change in ctDNA along with therapeutic intervention (Figure 3)
Figure 3. Case report of patient with CUP illustrating dynamic changes in ctDNA that accompanied therapeutic intervention.
Figure 3.A. Dynamic change of ctDNA along with therapeutic intervention.
Figure 3.B. Serial imaging with MRI of the abdomen pre- and post-therapeutic intervention
A 60-year-old woman presented with abdominal bloating. Imaging showed extensive thoracic and retroperitoneal adenopathy and liver metastases. Since the patient had a history of cholangiocarcinoma that was surgically managed eight years prior to the presentation, recurrence of cholangiocarcinoma was suspected. However, biopsy of the liver mass revealed poorly differentiated squamous cell carcinoma that was not consistent with recurrent cholangiocarcinoma. Diagnosis was squamous cell carcinoma of unknown primary. Molecular profiling from archival tissue revealed FGFR2-DDX21 fusion, NF2 E270D, CDKN2A/B loss and PBRM1 Q478* alterations and ctDNA at the time of diagnosis revealed MYC N402N and JAK2 E621K alterations (FGFR2 fusions were not in the panel when the ctDNA analysis was performed) (Figure 3.A.).
The patient was started on chemotherapy with cisplatin and gemcitabine and achieved an excellent response (Figure 3.B.). During the course of therapy, pazopanib was added for the FGFR2-DDX21 fusion, and cisplatin was held after 18 weeks due to toxicity. The patient continued on gemcitabine and pazopanib and has attained on ongoing partial response at 40+ weeks (Figure 3.B.).
During treatment, the patient underwent ctDNA analysis at five different time points, including the one prior to the initiation of therapy (Figure 3.A.). The patient initially had MYC N402N and JAK2 E621K alterations (week -1). After starting cisplatin and gemcitabine, the MYC and JAK2 alterations disappeared, but multiple new alterations started to emerge at week 3, including those in the APC, NF1, KIT, AR and STK11 genes. However, after adding pazopanib, all the previously detected alterations disappeared; even so, new alterations in the EGFR, FGFR2, JAK3 and BRCA2 genes emerged (week 12). Interestingly, after holding the cisplatin, the MYC N402N alteration, which was originally seen in pre-treatment ctDNA and disappeared while on cisplatin, re-appeared. (Figure 3.A.).
Amongst the ctDNA alterations seen in this patient, the following were characterized:
AR R608Q, KIT R830*, EGFR D761N and EGFR E1015K.
The following alterations were VUSs:
JAK2 E621K, STK11 E357K, APC S2307L, NF1 R1509C, BRCA2 L1615R and FGFR2 D304N.
The following alterations were synonymous substitution:
MYC N402N and APC P1634P.
Illustrates mutation % based on maximum ctDNA% detected at each time point. Therefore if two alterations were each detected at 0.2%, the percentage of total ctDNA detected is 0.2%.
To demonstrate the dynamic change seen in ctDNA that accompanies therapeutic intervention, we report a case of 60-year-old woman with squamous CUP who had five ctDNA analyses during the disease course (one ctDNA test before initiation of therapy and four additional analyses during the therapy, Figure 3.A.). The tumor initially demonstrated MYC N402N and JAK2 E621K alterations. Following treatment initiation with cisplatin and gemcitabine, tumor reduction was observed, and the MYC and JAK2 alterations diminished/disappeared from circulation at the second timepoint (Figure 3.B.). However, multiple new alterations emerged including APC, NF1, KIT, AR and STK11 anomalies. Pazopanib was added to the chemotherapy (based on the molecular profiling from archival tissue that revealed FGFR2-DDX21 fusion) and cisplatin was held due to toxicity. Interestingly, after holding cisplatin, the originally observed MYC alteration re-emerged (Figure 3).
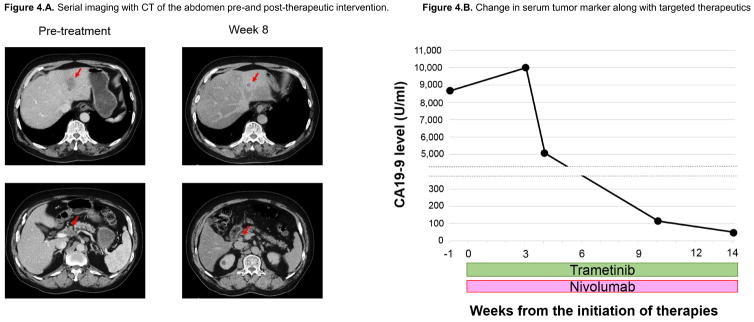
Patient with CUP managed with matched combination of immunotherapy and targeted therapy based on ctDNA (Figure 4)
Figure 4. Case report of patient with CUP harboring MLH1 and KRAS mutations managed with matched targeted therapy approach.
Figure 4.A. Serial imaging with CT of the abdomen pre-and post-therapeutic intervention.
Figure 4.B. Change in serum tumor marker along with targeted therapeutics.
82 year-old-man presented with three months history of worsening right upper quadrant abdominal pain. Imaging showed liver mass along with enlarged abdominal lymph nodes. Tumor marker was positive for CA19-9 (8,667 U/ml, reference range: 30–42 U/ml) but negative for carcinoembryonic antigen (3.9 ng/ml, reference range: <5.5 ng/ml) and alpha-fetoprotein (3 ng/ml, reference range: 0–15 ng/ml). Biopsy of liver mass was consistent with adenocarcinoma. Further analysis with immunohistochemistry were negative for CK7 and CK20, but positive for CK19 and CDX-2. Work up including upper gastrointestinal endoscopy, endoscopic ultrasound of pancreas and colonoscopy were unremarkable. The patient was diagnosed as adenocarcinoma of unknown primary.
Molecular profiling from archival tissue revealed KRAS G12D, MLH1 splice site 1989+1G>T and TP53 R248Q mutations and ctDNA at the time of diagnosis also revealed KRAS G12D, MLH1 R389W, TP53 R248Q as well as CCND2 C46*. Based on the mutation in mismatch repair gene MLH1 and KRAS G12D mutations, patient was initiated on nivolumab (for MLH1) and trametinib (for KRAS).
Four weeks after the initiation of nivolumab and trametinib, patient reported less abdominal pain and stopped his pain medication. Eight weeks after starting treatment, patient achieved partial response (36.4% reduction per RECIST 1.1) by CAT scan imaging (Figure 4.A.). Tumor marker CA19-9 fell markedly (Figure 4.B.). Treatment is ongoing.
An 82 year-old-man with adenocarcinoma of unknown primary with liver and abdominal lymph node metastases harbored KRAS G12D and MLH1 R389W mutations detected by ctDNA (and also by tissue NGS). Matched therapy with a combination of trametinib (a MEK inhibitor targeting downstream of KRAS) and nivolumab (checkpoint inhibitor associated with activity in colorectal cancer with mismatch repair gene anomalies such as MLH1) was initiated (Protocol: I-PREDICT study, NCT02534675). The patient achieved a partial response (36.4% reduction per RECIST 1.1) (Figure 4.A.) at eight weeks, along with rapid decline in the carbohydrate antigen 19-9 (CA-19-9) tumor marker (Figure 4.B). Treatment is ongoing.
DISCUSSION
CUP is a rare malignancy with an incidence of 7–12 cases per 100,000 individuals per year (2). In order to establish the primary site, there is often extensive evaluation, including laboratory, imaging, gene expression profiling, and immunohistochemical testing. Most recently, an algorithm that quantifies the similarity between RNA expression patterns of specimens and tissues on a test panel has shown high reliability in identifying tissue of origin (6). Even so, a single putative primary origin may only be assigned in about 25% of cases of CUP (5). Although empiric platinum-based regimens are generally used in CUP patients with modest response (7,8), overall median survival is poor at six to eight months, and a meta-analysis comparing different chemotherapy regimens demonstrated no statistical difference in overall survival for any treatment group over others (26). Thus, there is an unmet need for novel treatment approaches for patients with CUP.
Accumulating evidence suggests that, amongst patients with refractory cancers, a biomarker-based, personalized approach that matches patients with therapies based on specific genomic or protein markers may be able to improve clinical outcome (10–14). Herein, we report the genomic alterations identified in 442 patients with CUP using targeted NGS that evaluated ctDNA from patient plasma. Overall, 79.9% of CUP patients (353/442) had ctDNA alterations detected and 65.6% (290/442) harbored at least one characterized genomic alteration (Table 1). The most frequent alterations were in the TP53 gene; these alterations were detected in 37.1% of cases, followed by aberrations in KRAS (18.6%), and PIK3CA (15.4%). These results are consistent with previous reports of approximate frequencies of gene alterations in tissue samples from CUP patients (15,16) (Supplemental Figure 6).
There are several advantages to evaluating ctDNA from patient plasma as compared to tissue. First, interrogating ctDNA requires only a small blood sample. ctDNA sampling is therefore safe, easy, and inexpensive when compared to more invasive tissue biopsies. Further, some cancers may involve a difficult-to-biopsy location that limits the availability of tissue samples for molecular profiling (19). Second, since ctDNAs are shed into the bloodstream from multiple sites of metastases, theoretically, ctDNA may attenuate the issues related to assessing tumor heterogeneity (17). This is especially important for patients with CUP, since these individuals often harbor numerous metastases. Third, serial liquid biopsies to identify emergence of resistance mutations that can occur along with therapeutic intervention is feasible (27). Indeed, we have described a patient with CUP who underwent ctDNA analysis at five different time points. This patient demonstrated dynamic changes in ctDNA levels while receiving various therapies (Figure 3). Further understanding of the correlation between genomic alterations and treatment outcomes may help pinpoint response and resistant mechanisms.
Among 290 patients whose tumors carried characterized alterations, most individuals (97.2% [282/290]) had at least one potentially actionable alteration that could be impacted by an FDA-approved agent (albeit off label); an additional seven patients had targetable alterations with agents that are available through clinical trials. Altogether, among patients with characterized genomic alterations, 99.7% (289/290) had anomalies that might be pharmacologically tractable with either FDA-approved or investigational agents (Figures 3, Supplemental Tables 3 and 4 and Supplemental Figure 4).
Interestingly, among these 290 patients, only 12.1% (35/290) had identical molecular portfolios. Among patients harboring more than one genomic alteration, no two patients had an identical molecular portfolio (Supplemental Table 4). Malignant “snowflakes” in advanced cancer are commonly reported in other cancers as well (28,29). Therefore, individualized co-targeting of multiple genomic alterations may be necessary to improve clinical outcome A prospective observation trial with matched targeted therapy based on next-generation sequencing results from archival tissue is ongoing for patients with CUP (NCT02628379).
There were several specific pathway alterations that were potentially attractive for matched targeted therapy (Table 2). TP53-associated genes (TP53 and ATM) were the most commonly altered pathway in patients with CUP (37.8% [167/442]). Although there is no agent that directly targets TP53 alterations, mutations in this tumor suppressor are associated with high VEGF-A levels (30). Consistent with this association, clinical data suggest that patients with TP53 alterations had longer progression-free survival (PFS) (median 11.0 versus 4.0 months [p < 0.0001]) (31) and improved clinical outcome (rate of stable disease [SD] ≧ 6 months/partial response [PR]/complete response [CR]; 31% versus 7%) with anti-VEGF containing regimens (32). However, the same effect was not seen amongst patients with TP53 wild-type malignancies. In addition, another recent study documents that patients with sarcomas who respond to the VEGFR inhibitor pazopanib harbored TP53 mutations (33). TP53 is also potentially targetable with the WEE1 inhibitor currently in clinical development (AZ1775, NCT01748825) (34).
Tyrosine kinase family members were altered in 17.9% (79/442) of CUP cases and, in this pathway, EGFR abnormalities were the most common (5.9% of cases [26/442]) (Table 2). Cancers with EGFR alterations, especially in patients with non-small cell lung cancer, respond well to EGFR inhibitors including afatinib (35) or erlotinib (36). Although previous studies that investigated erlotinib-containing regimens in patients with non-genomically selected CUP only had modest response rates (~10%), further investigations that are focused on CUP with EGFR alterations are required. The ERBB2 gene was altered in 3.6% of our patients (16/442) (Table 2), which is consistent with previous reports (15,16) (Supplemental Figure 6). To our knowledge, there is no report evaluating CUP patients with anti-HER2 therapies. Since ERBB2 alterations are targetable with therapies such as afatinib, lapatinib and trastuzumab (37–39), a clinical trial is warranted.
Alterations in MAPK signaling were also commonly seen (31.2% [138/442]) (Table 2 and Figure 1). KRAS was the most commonly affected gene (18.6% of cases [82/442]) (Table 2 and Supplemental Table 2). Due to limited therapeutic options, RAS-driven cancers are considered to be among the most difficult to treat (40), however, they are potentially targetable with MEK inhibitors (e.g. trametinib and cobimetinib) (41,42). Although less frequent, BRAF V600E mutations were found in 1.6% [7/442] of our CUP patients (Supplemental Table 3). Recent reports suggest that many cancer types, including CUP, with BRAF V600 mutations are targetable with vemurafenib (BRAF inhibitor) (43,44). BRAF alterations are also targetable with trametinib (MEK inhibitor) (42) or with dual suppression by both BRAF and MEK inhibitors (e.g. dabrafenib plus trametinib) (45).
PI3K signaling pathway anomalies involving PIK3CA, PTEN, AKT1, STK11 and TSC1 genes were seen in 18.1% (80/442) of our patients (Table 2). Biomarker analysis of two phase III trials (BOLERO-1 and BOLERO-3) that randomized Her2-positive breast cancer patients to receiving a chemotherapy with and without everolimus revealed that participants with an altered PI3K pathway had a statistically improved PFS with an everolimus-containing regimen (hazard ratio [HR]: 0.67; 95% confidence interval [CI], 0.48–0.93), while there was no benefit from everolimus among patients with wild-type PI3K pathway (HR: 1.19; 95% CI, 0.87–1.62) (46). Janku et al have also reported that, among diverse malignancies, PIK3CA or PTEN alterations were the independent factor predicting response to inhibitors targeting PI3K pathway, including mTor inhibitors, albeit when used in combination therapy rather than as single agents (47).
In the current report, cell cycle-associated genes were altered in 10.4% of patients (46/442) (Table 2). This rate is lower than that in a previous publication which showed 41.6% (112/269) of patients with CUP had alterations in cell-cycle associated genes when archival tumor tissue was interrogated by NGS (48). The difference in frequencies among the studies is possibly due to the different techniques used (e.g. in the previous publication, CDKN2A/B loss was reported in 18% of cases of CUP (48); however, loss of this gene was not assessed in the current study). In terms of therapeutic approach, although there are conflicting data, CDKN2A, CCND1/2 alterations and CDK4/6 amplification may potentially be targetable with a CDK4/6 inhibitor such as palbociclib (49).
Rare alterations may also be important. For instance, MLH1 (mismatch repair gene) abnormalities were found in only 1.6% (7/442) (Supplemental Table 2) of our CUP patients. However, these alterations have been shown to correlate with high tumor mutational burden and response to checkpoint inhibitors in diseases such as colorectal cancer (50). Indeed, our elderly patient with CUP and liver metastases had a rapid response after therapy with a checkpoint inhibitor nivolumab-based regimen (Figure 4).
There are several limitations to the current report. Since the database was de-identified, we were not able to evaluate clinical characteristics such as those related to outcome. Moreover, the cancer diagnosis was reported by the referring physician. Despite these limitations, the current report provides the largest dataset of clinical-grade NGS in ctDNA among patients with CUP.
In conclusion, we have evaluated the genomic landscape of circulating tumor DNA in 442 patients with carcinoma of unknown primary. TP53-associated genes were most commonly altered followed by abnormalities in the MAPK pathway, PI3K signaling and in cell-cycle associated genes. Overall, 80% of CUP patients (353/442) had detectable ctDNA alterations and 66% (290/442) had at least one characterized alteration. Serial ctDNA sampling in a patient revealed significant evolution with therapy (Figure 3). Among the patients in this report, 99.7% (289/290) had an alteration hypothetically targetable with either an FDA-approved or investigational agents. Therefore, patients could potentially be matched to genomically targeted therapy or to immunotherapy or both, as demonstrated by our responding patient who received both a MEK and a checkpoint inhibitor because of the presence of a KRAS and a mismatch repair gene alteration (Figure 4). Previous literature suggests that biomarker-based, matched targeted therapy can improve clinical outcome (10–14). Our current report indicates that the non-invasive liquid biopsy approach merits investigation in next generation clinical trials of cancer of unknown primary.
Supplementary Material
Acknowledgments
Funded in part by the Joan and Irwin Jacobs fund and by National Cancer Institute grant P30 CA016672 (RK)
Footnotes
CONFLICT OF INTEREST
Shumei Kato and Nithya Krishnamurthy do not have conflict of interest.
Razelle Kurzrock receives consultant fees from X-biotech and from Actuate Therapeutics, as well as research funds from Genentech, Pfizer, Sequenom, Guardant, Foundation Medicine and Merck Serono, and has an ownership interest in Novena Inc and CureMatch Inc.
Kimberly C. Banks and Richard B. Lanman are employees of Guardant Health, Inc.
References
- 1.Greenlee RT, Goodman MT, Lynch CF, Platz CE, Havener LA, Howe HL. The occurrence of rare cancers in U.S. adults, 1995–2004. Public Health Rep. 2010;125(1):28–43. doi: 10.1177/003335491012500106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pavlidis N, Pentheroudakis G. Cancer of unknown primary site. Lancet. 2012;379(9824):1428–35. doi: 10.1016/S0140-6736(11)61178-1. [DOI] [PubMed] [Google Scholar]
- 3.Fizazi K, Greco FA, Pavlidis N, Pentheroudakis G, Group EGW. Cancers of unknown primary site: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2011;22(Suppl 6):vi64–8. doi: 10.1093/annonc/mdr389. [DOI] [PubMed] [Google Scholar]
- 4.Massard C, Loriot Y, Fizazi K. Carcinomas of an unknown primary origin--diagnosis and treatment. Nat Rev Clin Oncol. 2011;8(12):701–10. doi: 10.1038/nrclinonc.2011.158. [DOI] [PubMed] [Google Scholar]
- 5.Varadhachary GR, Raber MN. Carcinoma of unknown primary site. N Engl J Med. 2014;371(21):2040. doi: 10.1056/NEJMc1411384. [DOI] [PubMed] [Google Scholar]
- 6.Pillai R, Deeter R, Rigl CT, Nystrom JS, Miller MH, Buturovic L, et al. Validation and reproducibility of a microarray-based gene expression test for tumor identification in formalin-fixed, paraffin-embedded specimens. J Mol Diagn. 2011;13(1):48–56. doi: 10.1016/j.jmoldx.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Briasoulis E, Kalofonos H, Bafaloukos D, Samantas E, Fountzilas G, Xiros N, et al. Carboplatin plus paclitaxel in unknown primary carcinoma: a phase II Hellenic Cooperative Oncology Group Study. J Clin Oncol. 2000;18(17):3101–7. doi: 10.1200/JCO.2000.18.17.3101. [DOI] [PubMed] [Google Scholar]
- 8.Greco FA, Erland JB, Morrissey LH, Burris HA, 3rd, Hermann RC, Steis R, et al. Carcinoma of unknown primary site: phase II trials with docetaxel plus cisplatin or carboplatin. Ann Oncol. 2000;11(2):211–5. doi: 10.1023/a:1008369812295. [DOI] [PubMed] [Google Scholar]
- 9.Hainsworth JD, Spigel DR, Farley C, Thompson DS, Shipley DL, Greco FA, et al. Phase II trial of bevacizumab and erlotinib in carcinomas of unknown primary site: the Minnie Pearl Cancer Research Network. J Clin Oncol. 2007;25(13):1747–52. doi: 10.1200/JCO.2006.09.3047. [DOI] [PubMed] [Google Scholar]
- 10.Jardim DL, Schwaederle M, Wei C, Lee JJ, Hong DS, Eggermont AM, et al. Impact of a Biomarker-Based Strategy on Oncology Drug Development: A Meta-analysis of Clinical Trials Leading to FDA Approval. J Natl Cancer Inst. 2015;107(11) doi: 10.1093/jnci/djv253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwaederle M, Parker BA, Schwab RB, Daniels GA, Piccioni DE, Kesari S, et al. Precision Oncology: The UC San Diego Moores Cancer Center PREDICT Experience. Mol Cancer Ther. 2016;15(4):743–52. doi: 10.1158/1535-7163.MCT-15-0795. [DOI] [PubMed] [Google Scholar]
- 12.Schwaederle M, Zhao M, Lee JJ, Eggermont AM, Schilsky RL, Mendelsohn J, et al. Impact of Precision Medicine in Diverse Cancers: A Meta-Analysis of Phase II Clinical Trials. J Clin Oncol. 2015;33(32):3817–25. doi: 10.1200/JCO.2015.61.5997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsimberidou AM, Iskander NG, Hong DS, Wheler JJ, Falchook GS, Fu S, et al. Personalized medicine in a phase I clinical trials program: the MD Anderson Cancer Center initiative. Clin Cancer Res. 2012;18(22):6373–83. doi: 10.1158/1078-0432.CCR-12-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wheler JJ, Janku F, Naing A, Li Y, Stephen B, Zinner R, et al. Cancer Therapy Directed by Comprehensive Genomic Profiling: A Single Center Study. Cancer Res. 2016;76(13):3690–701. doi: 10.1158/0008-5472.CAN-15-3043. [DOI] [PubMed] [Google Scholar]
- 15.Gatalica Z, Millis SZ, Vranic S, Bender R, Basu GD, Voss A, et al. Comprehensive tumor profiling identifies numerous biomarkers of drug response in cancers of unknown primary site: analysis of 1806 cases. Oncotarget. 2014;5(23):12440–7. doi: 10.18632/oncotarget.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ross JS, Wang K, Gay L, Otto GA, White E, Iwanik K, et al. Comprehensive Genomic Profiling of Carcinoma of Unknown Primary Site: New Routes to Targeted Therapies. JAMA Oncol. 2015;1(1):40–9. doi: 10.1001/jamaoncol.2014.216. [DOI] [PubMed] [Google Scholar]
- 17.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366(10):883–92. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murugaesu N, Wilson GA, Birkbak NJ, Watkins TB, McGranahan N, Kumar S, et al. Tracking the genomic evolution of esophageal adenocarcinoma through neoadjuvant chemotherapy. Cancer Discov. 2015;5(8):821–31. doi: 10.1158/2159-8290.CD-15-0412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kato S, Janku F. Cell-free DNA as a novel marker in cancer therapy. Biomark Med. 2015;9(7):703–12. doi: 10.2217/bmm.15.38. [DOI] [PubMed] [Google Scholar]
- 20.Stroun M, Anker P, Lyautey J, Lederrey C, Maurice PA. Isolation and characterization of DNA from the plasma of cancer patients. Eur J Cancer Clin Oncol. 1987;23(6):707–12. doi: 10.1016/0277-5379(87)90266-5. [DOI] [PubMed] [Google Scholar]
- 21.Janku F, Angenendt P, Tsimberidou AM, Fu S, Naing A, Falchook GS, et al. Actionable mutations in plasma cell-free DNA in patients with advanced cancers referred for experimental targeted therapies. Oncotarget. 2015;6(14):12809–21. doi: 10.18632/oncotarget.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Janku F, Huang HJ, Claes B, Falchook GS, Fu S, Hong D, et al. BRAF Mutation Testing in Cell-Free DNA from the Plasma of Patients with Advanced Cancers Using a Rapid, Automated Molecular Diagnostics System. Mol Cancer Ther. 2016;15(6):1397–404. doi: 10.1158/1535-7163.MCT-15-0712. [DOI] [PubMed] [Google Scholar]
- 23.Schwaederle M, Husain H, Fanta PT, Piccioni DE, Kesari S, Schwab RB, et al. Detection rate of actionable mutations in diverse cancers using a biopsy-free (blood) circulating tumor cell DNA assay. Oncotarget. 2016;7(9):9707–17. doi: 10.18632/oncotarget.7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwaederle M, Husain H, Fanta PT, Piccioni DE, Kesari S, Schwab RB, et al. Use of Liquid Biopsies in Clinical Oncology: Pilot Experience in 168 Patients. Clin Cancer Res. 2016 doi: 10.1158/1078-0432.CCR-16-0318. [DOI] [PubMed] [Google Scholar]
- 25.Lanman RB, Mortimer SA, Zill OA, Sebisanovic D, Lopez R, Blau S, et al. Analytical and Clinical Validation of a Digital Sequencing Panel for Quantitative, Highly Accurate Evaluation of Cell-Free Circulating Tumor DNA. PLoS One. 2015;10(10):e0140712. doi: 10.1371/journal.pone.0140712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Golfinopoulos V, Pentheroudakis G, Salanti G, Nearchou AD, Ioannidis JP, Pavlidis N. Comparative survival with diverse chemotherapy regimens for cancer of unknown primary site: multiple-treatments meta-analysis. Cancer Treat Rev. 2009;35(7):570–3. doi: 10.1016/j.ctrv.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 27.Murtaza M, Dawson SJ, Tsui DW, Gale D, Forshew T, Piskorz AM, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497(7447):108–12. doi: 10.1038/nature12065. [DOI] [PubMed] [Google Scholar]
- 28.Kurzrock R, Giles FJ. Precision oncology for patients with advanced cancer: the challenges of malignant snowflakes. Cell Cycle. 2015;14(14):2219–21. doi: 10.1080/15384101.2015.1041695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wheler J, Lee JJ, Kurzrock R. Unique molecular landscapes in cancer: implications for individualized, curated drug combinations. Cancer Res. 2014;74(24):7181–4. doi: 10.1158/0008-5472.CAN-14-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwaederle M, Lazar V, Validire P, Hansson J, Lacroix L, Soria JC, et al. VEGF-A Expression Correlates with TP53 Mutations in Non-Small Cell Lung Cancer: Implications for Antiangiogenesis Therapy. Cancer Res. 2015;75(7):1187–90. doi: 10.1158/0008-5472.CAN-14-2305. [DOI] [PubMed] [Google Scholar]
- 31.Said R, Hong DS, Warneke CL, Lee JJ, Wheler JJ, Janku F, et al. P53 mutations in advanced cancers: clinical characteristics, outcomes, and correlation between progression-free survival and bevacizumab-containing therapy. Oncotarget. 2013;4(5):705–14. doi: 10.18632/oncotarget.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wheler J, Janku F, Naing A, Li Y, Stephen B, Zinner R, et al. TP53 Alterations Correlate with Response to VEGF/VEGFR Inhibitors: Implications for Targeted Therapeutics. Mol Cancer Ther. 2016 doi: 10.1158/1535-7163.MCT-16-0196. In Press. [DOI] [PubMed] [Google Scholar]
- 33.Koehler K, Liebner D, Chen JL. TP53 mutational status is predictive of pazopanib response in advanced sarcomas. Ann Oncol. 2016;27(3):539–43. doi: 10.1093/annonc/mdv598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mueller S, Haas-Kogan DA. WEE1 Kinase As a Target for Cancer Therapy. J Clin Oncol. 2015;33(30):3485–7. doi: 10.1200/JCO.2015.62.2290. [DOI] [PubMed] [Google Scholar]
- 35.Wu YL, Zhou C, Hu CP, Feng J, Lu S, Huang Y, et al. Afatinib versus cisplatin plus gemcitabine for first-line treatment of Asian patients with advanced non-small-cell lung cancer harbouring EGFR mutations (LUX-Lung 6): an open-label, randomised phase 3 trial. Lancet Oncol. 2014;15(2):213–22. doi: 10.1016/S1470-2045(13)70604-1. [DOI] [PubMed] [Google Scholar]
- 36.Zhou C, Wu YL, Chen G, Feng J, Liu XQ, Wang C, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011;12(8):735–42. doi: 10.1016/S1470-2045(11)70184-X. [DOI] [PubMed] [Google Scholar]
- 37.De Greve J, Teugels E, Geers C, Decoster L, Galdermans D, De Mey J, et al. Clinical activity of afatinib (BIBW 2992) in patients with lung adenocarcinoma with mutations in the kinase domain of HER2/neu. Lung Cancer. 2012;76(1):123–7. doi: 10.1016/j.lungcan.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 38.Geyer CE, Forster J, Lindquist D, Chan S, Romieu CG, Pienkowski T, et al. Lapatinib plus capecitabine for HER2-positive advanced breast cancer. N Engl J Med. 2006;355(26):2733–43. doi: 10.1056/NEJMoa064320. [DOI] [PubMed] [Google Scholar]
- 39.Vogel CL, Cobleigh MA, Tripathy D, Gutheil JC, Harris LN, Fehrenbacher L, et al. Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer. J Clin Oncol. 2002;20(3):719–26. doi: 10.1200/JCO.2002.20.3.719. [DOI] [PubMed] [Google Scholar]
- 40.Stephen AG, Esposito D, Bagni RK, McCormick F. Dragging ras back in the ring. Cancer Cell. 2014;25(3):272–81. doi: 10.1016/j.ccr.2014.02.017. [DOI] [PubMed] [Google Scholar]
- 41.Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. Drugging the undruggable RAS: Mission possible? Nat Rev Drug Discov. 2014;13(11):828–51. doi: 10.1038/nrd4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Falchook GS, Lewis KD, Infante JR, Gordon MS, Vogelzang NJ, DeMarini DJ, et al. Activity of the oral MEK inhibitor trametinib in patients with advanced melanoma: a phase 1 dose-escalation trial. Lancet Oncol. 2012;13(8):782–9. doi: 10.1016/S1470-2045(12)70269-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hyman DM, Puzanov I, Subbiah V, Faris JE, Chau I, Blay JY, et al. Vemurafenib in Multiple Nonmelanoma Cancers with BRAF V600 Mutations. N Engl J Med. 2015;373(8):726–36. doi: 10.1056/NEJMoa1502309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Turski ML, Vidwans SJ, Janku F, Garrido-Laguna I, Munoz J, Schwab R, et al. Genomically Driven Tumors and Actionability across Histologies: BRAF-Mutant Cancers as a Paradigm. Mol Cancer Ther. 2016;15(4):533–47. doi: 10.1158/1535-7163.MCT-15-0643. [DOI] [PubMed] [Google Scholar]
- 45.Robert C, Karaszewska B, Schachter J, Rutkowski P, Mackiewicz A, Stroiakovski D, et al. Improved overall survival in melanoma with combined dabrafenib and trametinib. N Engl J Med. 2015;372(1):30–9. doi: 10.1056/NEJMoa1412690. [DOI] [PubMed] [Google Scholar]
- 46.Andre F, Hurvitz S, Fasolo A, Tseng LM, Jerusalem G, Wilks S, et al. Molecular Alterations and Everolimus Efficacy in Human Epidermal Growth Factor Receptor 2-Overexpressing Metastatic Breast Cancers: Combined Exploratory Biomarker Analysis From BOLERO-1 and BOLERO-3. J Clin Oncol. 2016;34(18):2115–24. doi: 10.1200/JCO.2015.63.9161. [DOI] [PubMed] [Google Scholar]
- 47.Janku F, Hong DS, Fu S, Piha-Paul SA, Naing A, Falchook GS, et al. Assessing PIK3CA and PTEN in early-phase trials with PI3K/AKT/mTOR inhibitors. Cell Rep. 2014;6(2):377–87. doi: 10.1016/j.celrep.2013.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Helsten T, Kato S, Schwaederle M, Tomson BN, Buys TP, Elkin SK, et al. Cell-Cycle Gene Alterations in 4,864 Tumors Analyzed by Next-Generation Sequencing: Implications for Targeted Therapeutics. Mol Cancer Ther. 2016;15(7):1682–90. doi: 10.1158/1535-7163.MCT-16-0071. [DOI] [PubMed] [Google Scholar]
- 49.DeMichele A, Clark AS, Tan KS, Heitjan DF, Gramlich K, Gallagher M, et al. CDK 4/6 inhibitor palbociclib (PD0332991) in Rb+ advanced breast cancer: phase II activity, safety, and predictive biomarker assessment. Clin Cancer Res. 2015;21(5):995–1001. doi: 10.1158/1078-0432.CCR-14-2258. [DOI] [PubMed] [Google Scholar]
- 50.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med. 2015;372(26):2509–20. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.