Abstract
We developed a Python package to help performing drug response experiments in medium- and high-throughput and to evaluate sensitivity metrics from the resulting data. In this protocol, we describe the steps involved in (1) generating files necessary for treating cells with the HP D300 drug dispenser, by pin transfer, or manually; (2) merging the data generated by high-throughput scanners, such as the Perkin Elmer Operetta, with treatment annotations; and (3) analyzing the results to obtain data normalized to untreated controls and sensitivity metrics such as IC50 or newly-developed GR50. These modules are available on GitHub and provide an automated pipeline for the design and analysis of high-throughput drug response experiments, which help to prevent errors that can arise from manually processing large data files.
Keywords: experimental design, drug response, data processing, computational pipeline
Introduction
Biological experiments are increasingly performed in high-throughput and are becoming more reliant on automation. The emphasis in many high-throughput studies is on using the latest reagents and methods (e.g. different types of CRISPR genome editing) and ensuring the availability of state-of-the-art equipment (e.g. robotic plate handlers and screening platforms), while challenges in experimental design and data processing are often under-emphasized. Current approaches rely on manual procedures and extensive human intervention such as manual entry or cut and paste operations in spreadsheets. These operations are not trackable and are susceptible to fragmentation, loss of metadata, and errors arising from forced formatting (which change numerical values and gene names into dates, for example) (Zeeberg et al., 2004; Ziemann et al., 2016). When design and data are stored in disconnected spreadsheets, non-obvious mistakes lead to rapid accumulation of errors (Powell et al., 2008). With increasing attention to data reproducibility and public release, it has become apparent that manual data handling introduces errors at multiple levels and makes suboptimal use of the capabilities of automation.
While standards and tools are increasingly available for the management of data, reproducible and transparent science requires that these data are associated with metadata and linked to a machine-readable experimental design and protocol. Few tools exist for systematic creation, management and logging of experimental designs (Stocker et al., 2009; Wu and Zhou, 2013). The current protocol describes such a tool, with a specific focus on automating the design of drug response experiments performed in multi-well plates. Such assays are sufficiently complex that they warrant dedicated tools and they are paradigmatic of high-throughput experiments in general.
We (Saez-Rodriguez et al., 2008) and others (Stocker et al., 2009) have previously published tools that use graphical user interfaces (UIs) to assist in the layout and analysis of multi-well experiments. Over the years, we have found that UIs are very limiting in this context because they do not easily accommodate different experimental designs or research goals. We have therefore switched from UI-driven software to scripting tools that are run on the command line. This requires a bit more familiarity with programming, but we have found that the closer users are to the actual code, the less likely unintended errors are to occur. Moreover, interactive documents with embedded executable code like Jupyter notebooks facilitate the use of simple executable functions and, once a script is implemented for a particular type of experiment, it can be used repeatedly by individuals with little programming skill.
The tools we described are part of a Python package meant to systematize the design of the experiment and construct digital containers for the resulting metadata and data. Users download the packages and exemplar Jupyter notebooks from our GitHub repository (https://github.com/sorgerlab/datarail) and then modify the scripts as needed to accommodate specific designs, which only requires minimal coding experience. As more example designs become available, it will be increasingly easy to find a notebook that is nearly ready to be used. For example, for a drug dose-response experiment, the user needs to only specify the cell type, drugs to be tested, and drug dose range. Digital designs benefit experimental scientists because they simplify and accelerate plate layouts as well as computational biologists by making data analysis more rapid, transparent and error-free. This protocol is related to the unit to “Measuring and quantifying drug sensitivity in tissue culture” which describes experimental consideration to running drug response experiments. While this protocol can be used by itself, ideally, both protocols should be used in conjunction.
Overview of the Method
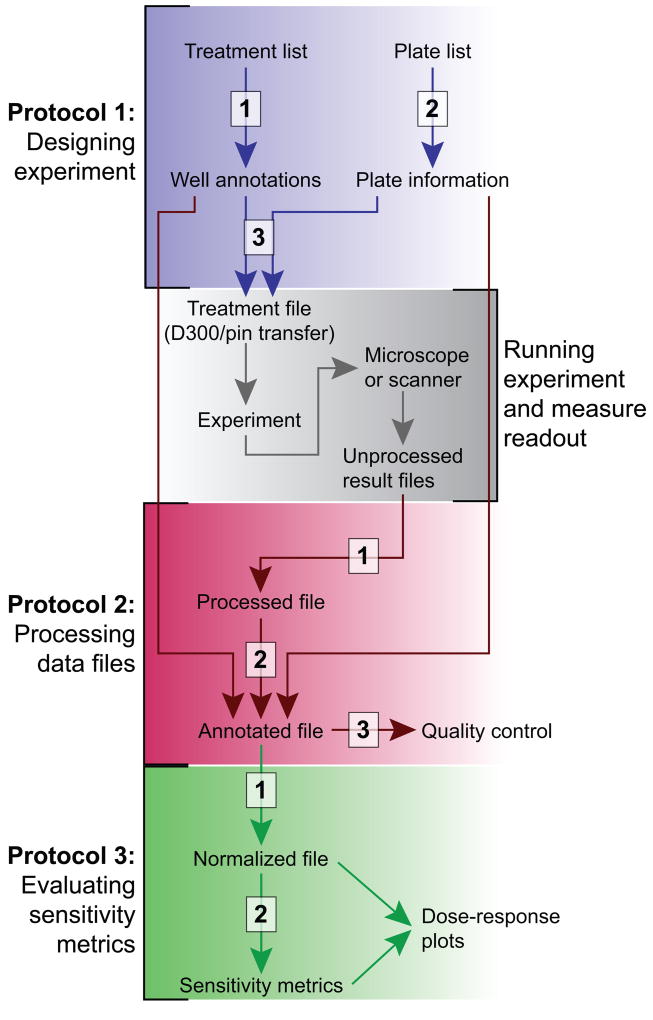
The following protocols are described below: (1) laying out samples and doses across one or more multi-well plates, such that the design document can be used to guide either a human or, preferably, a robot, (2) gathering results from high-throughput instruments and merging them with the underlying metadata in a single data container, and (3) extracting drug response metrics from the experimental results (Figure 1). The response of cells to drugs is processed using the normalized growth rate inhibition (GR) method that corrects for the effects of cell division time on the estimation of drug sensitivity and allows an estimation of time-dependent drug sensitivity (Hafner et al., 2016). In addition to avoiding errors in the design and processing of experiments, our pipeline can fetch references from on-line databases to assist with reagent annotation (including PubChem annotation, internal IDs, and batch numbers) and perform quality control of results. The whole process is written in a documented script that becomes de facto a “literate” (human and machine-readable) archive of the overall experiment, including the design, results, and quality controls.
Figure 1. Pipeline for experimental design and analysis.
The pipeline described in the manuscript comprises three protocols (colored sections), each with a 2–3 steps (white boxes). The datarail package is used in Protocols 1 and 2. Protocol 3 relies on the gr50_tools package.
The pipeline comprises two flexible and modular open-source python packages (https://github.com/sorgerlab/datarail ; https://github.com/sorgerlab/gr50_tools) that can be used for end-to-end processing of drug response experiments or for automating individual steps of such experiments (such as designing the plate layout). The approach assumes a standardized file format and set of key words (such as treatment_duration, cell_count, plate_identifier, well, or concentration) that are used by the system to automate data processing. Data can be stored in long tabular representations as tab separated values (tsv) files or alternatively as more powerful hierarchical data format (HDF5) files. The main application is systematic high-throughput experiments, but the modular implementation makes the pipeline useful for simpler, low-throughput experiments that do not require interfacing with robotic arms or more complex multifactorial experiments for which the low-level functions can be used to generate and store the designs. The modules will continue to evolve as we include new functionality and add contributions from others.
Basic Protocol 1 deals with all steps necessary to set up a multi-well drug response experiment. Basic Protocol 2 describes steps for processing data once an experiment is concluded. Basic Protocol 3 handles the analysis and parameterization of drug response data. We typically use all three protocols in sequence, but individual protocols can be used in combination with personalized scripts. All three protocols can be modified to accommodate other types of studies in which cells are treated with exogenous ligands or small molecules and their state is then measured in plate-based assays. We envision that future iterations of this pipeline will be able to support additional experimental assays and data structures.
STRATEGIC PLANNING
Types of variables
This protocol distinguishes among three classes of variables: model variables, confounder variables, and readout variables.
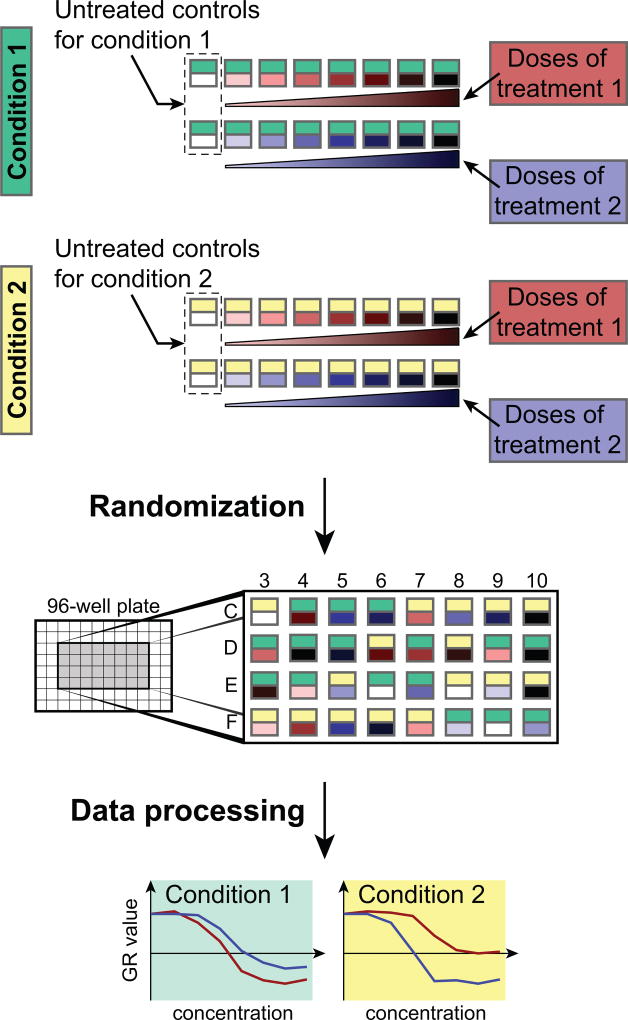
Model variables describe aspects of the experiment that are explicitly changed as part of the experimental design and are expected to affect the values of the readout variables. In the context of drug response assays, we separate model variables into (i) treatment variables that generally refer to agents that perturb cell state, such as drugs or biological ligands and their concentrations and (ii) condition variables that refer to the states of the system at the start of the experiment such as seeding density, serum concentration, or oxygen level, as well as the experiment duration (Figure 2). The distinction between treatment and condition variables is exploited for normalizing readout values. For example, in a multi-factorial design in which the effect of seeding density on drug response is being examined, dose-response curves for different drugs (treatment variable) can be compared at the multiple seeding densities (condition variable).
Figure 2. Illustration of an experimental design including different conditions and treatments.
Serial dilutions are measured for two treatments in two different conditions. Provided scripts help to design the treatments and randomize the conditions for avoiding bias during the experiment. Data processing scripts normalize the different treatments to condition-specific untreated controls and yield the dose-response curves. In the “Randomization” panel, for clarity, only the center wells of a 96-well plate are depicted to illustrate the random distribution of experimental conditions across an assay plate.
Confounder variables are implicit variables that are not intended to affect readouts, but that are recorded to fully document the experiment. These can be media batch number, drug supplier, and assay date. It is frequently observed post facto that assays depend on the value of a particular confounder variable; in this case, forensic analysis might involve a new experiment in which the confounder becomes an explicitly controlled model variable. Among confounder variables, the plate identifier (ID number or barcode) and well number are of particular importance in performing quality control and matching readout and model variables.
Readout variables are values measured while the experiment is underway or when it ends. They constitute the data being collected by the experiment; cell number and fraction of viable or apoptotic cells are typical readout variables for an experiment with anti-cancer drugs. However, readout variables can also be more complex: multi-channel microscope images in the case of high-content experiments, for example. A set of readout variables is typically collected from each well in a multi-well plate. For simplicity, the protocols described below use viable cell number, or a surrogate such as ATP level measured by CellTiter-Glo (Promega), as the readout variable.
Jupyter notebooks and file structure
Proper documentation of the details and rationale behind the design of an experiment is a necessity for reproducibility and transparency. We use Jupyter, which is a web-application that allows snippets of code, explanatory text, and figures to be contained within a single page. We provide templates for commonly used experimental designs to assist in notebook creation. Experimental design notebooks define all model and confounder variables of an experiment such as the layout of drugs, doses, and cell lines on 384-well plates (Figure 3), whereas processing notebooks comprises the commands used to process and analyze the data of an experiment (Figure 4). Recording these commands in a notebook makes it possible to establish the full provenance of each piece of data.
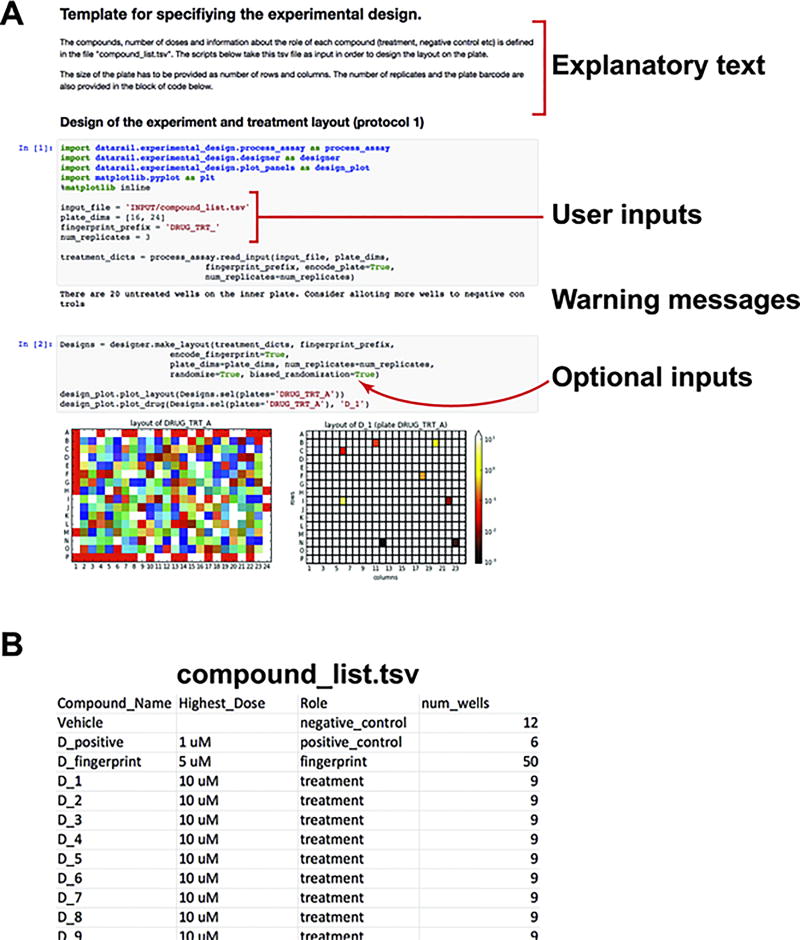
Figure 3. Exemplar Jupyter notebook for the experimental design (protocol 1).
(A) Exemplar experimental design notebook: The Jupyter notebook allows computer code (grey shaded block) and text to be contained in the same document. The user can add or modify the explanatory text portions to describe the rationale of the experiment. In order to generate the experimental design, the user has to only modify parts of the code that specify the input filename, the dimensions and fingerprint of the plate, and the number of replicates in the experiment. The experimental design notebook also provides warning messages if the number of wells for treatments and controls is erroneous or suboptimal. (B) User created tsv file that lists the name and role of compounds to be used in the experiment. The num_wells columns defines the number of doses for compounds that are treatments, or the number of wells reserved for compounds that serve as controls.
Figure 4. Exemplar Jupyter notebook for the processing the data (protocol 2).

(A) Data outputted by Columbus are imported (protocol 2, step 1) in a pandas dataframe. Function input allows selection of the desired readouts and labels them with a standard name (e.g. cell_count). Data are then merged with the design (protocol 2, step 2).
(B) Different quality controls can be performed on the plate (protocol 2, step 3): bias across the plate can be quantified either by distance from the edge, row, or column; difference in the untreated controls can be quantified across plates to test for bias.
For large-scale experiments involving multiple collaborators, it is important to establish a clear file and folder structure for the project so that all investigators can work efficiently together with a minimum of confusion. We suggest creating for each project a set of folders entitled:
SRC (conventional name for folders containing source code) that contains all Juypter notebooks or scripts.
INPUT that stores data and metadata files read by Juypter notebooks or scripts. These files can be created manually by recording measurements or, preferably, generated by assay instruments. In both cases, they should never be modified to ensure data integrity.
OUTPUT that contains all files generated by the Jupyter processing notebook or scripts; these too should not be modified manually.
Experimental design
A major consideration in designing a multi-plate experiment is the number of 96 or 384-well plates that will be used, as this determines the maximum number of samples. The experimental design constitutes the way in which treatments and controls are split among the available number of samples and mapped to each plate. Obtaining reliable results depends on using replicates, including sufficient untreated controls and, in the case of dose-response experiments, assaying an adequate dose-range at an effective step size. The following considerations impact the final design:
For cell lines and experimental setup in which strong edge effects have been observed, we recommend avoiding using the outermost wells. Potential causes and solutions for edge effects have been discussed elsewhere (Tanner, 2001; Lundholt et al., 2003), and are also covered in accompanying protocol. If enough wells are available on each plate to accommodate the experimental design, the scripts we provide automatically avoid using wells at the edge of the plate.
Multiple untreated (or vehicle-treated) controls should be included on each plate. If outermost wells are used, we recommend at least 6 controls on the edges and 6 in the center of the plate to test if edge effects occurred. The scripts we provide will position controls in this manner by default and randomly assign any remaining untreated wells across the plate.
For simple dose-response experiments, we recommend at least three technical replicates. For experiments that aim to identify small differences in GR value (less than 0.2) between treatments or conditions, more technical replicates are generally necessary to properly assess significance. More replicates are also necessary to precisely assess time-dependent GR values, especially for short time intervals or for cell lines growing slowly. We generally recommend 6 replicates if measurements are made four times per division, which corresponds to a population growth of about 20% between measurements.
Technical replicates should spread across multiple plates using different randomization schemes. Default randomization schemes are arranged so that a particular condition (e.g. drug/concentration pair) is not assigned to the same region of the plate across replicates.
For dose-response experiments in which the likely range of cellular sensitivity is unknown, or for drug master plates that will be used to treat multiple cell lines having differing sensitivities, we recommend using a wide range of drug concentrations. In experiments exploring the responses of cancer cells to small molecule drugs we typically use nine doses separated by 3.16-fold and spanning 1 nM to 10 µM (or from 0.1 nM to 1 µM for compounds that are known to be potent).
For evaluating drug response using GR values and metrics, the number of cells present at the time of treatment must be measured. We recommend that an untreated plate, which was prepared in parallel to the plates to be treated, is fixed at the time the other plates are being exposed to drugs.
For evaluating time-dependent GR values and metrics, the number of cells must be measured at multiple time points. This can be done either as a time-course where samples are measured multiple times in a non-destructive manner or with multiple plates fixed at different time points.
Further experimental parameters such as cell plating, recovery time, and control treatments are discussed in an accompanying protocol and should be determined prior to generating treatment files.
The value of automation
Assays in multi-well plates can be performed using a wide variety of instruments including multi-channel pipettes, but even experienced bench scientists are not particularly good at accurately dispensing dilutions across multiple plates. This is particularly true in the case of 384-well plates, because of tight well spacing and small volumes. We therefore strongly advocate the use of automated cell counters, plate fillers, compound dispensers, and plate washers. The Hewlett-Packard D300 Digital Dispenser is particularly valuable for dispensing small volumes of treatment agents and, when combined with the scripts described here, allows samples and controls to be arranged randomly across a plate so that successive doses are not always next to each other. This helps to reduce systematic error arising from edge effects or gradients in cell adherence or growth. All automation instruments that we suggest fit on a benchtop, are simple to operate, and are highly recommended for any laboratory or organization wishing to assay the responses of mammalian cells to perturbation on a regular basis.
OVERVIEW OF THE SOFTWARE
Protocols 1 – 3 describe software routines for specifying the experimental design, processing the readout, and analyzing the results (Figure 1). The two packages used in this protocol are datarail and gr50_tools, which are both open source and can be downloaded directly from our GitHub repositories (github.com/sorgerlab/datarail ; github.com/sorgerlab/gr50_tools). All functions in these packages are written in Python and require the following Python packages: numpy, scipy, sklearn, pandas, matplotlib, and xarray. Submodules in both packages can be used independently of the pipeline and each step can be substituted by a customized function. All inputs and outputs are tsv or HDF5 files and handled internally as pandas tables and xarray datasets. This duality allows more flexibility in interfacing with other scripts or programmatic languages.
As mentioned above, we recommend using Jupyter notebooks (http://jupyter.org/) to describe and generate the experiment design as well as to document and execute the data processing (Figures 3–5). In addition to the blocks of code that execute the commands (grey blocks in Figures 3–5), the experimental design notebook should include the background, rationale, and assumptions of the experiment as explanatory text (white blocks in Figure 3–5). The processing notebook may include an amended version of the protocol to account for any deviation from the intended plan as well as code implementing quality control or data filtering. For data fidelity, it is important to keep the raw data file untouched and filter or correct data only in a programmatic manner and record all operations in the notebook.
Figure 5. Exemplar Jupyter notebook for evaluating sensitivity metrics (protocol 3).
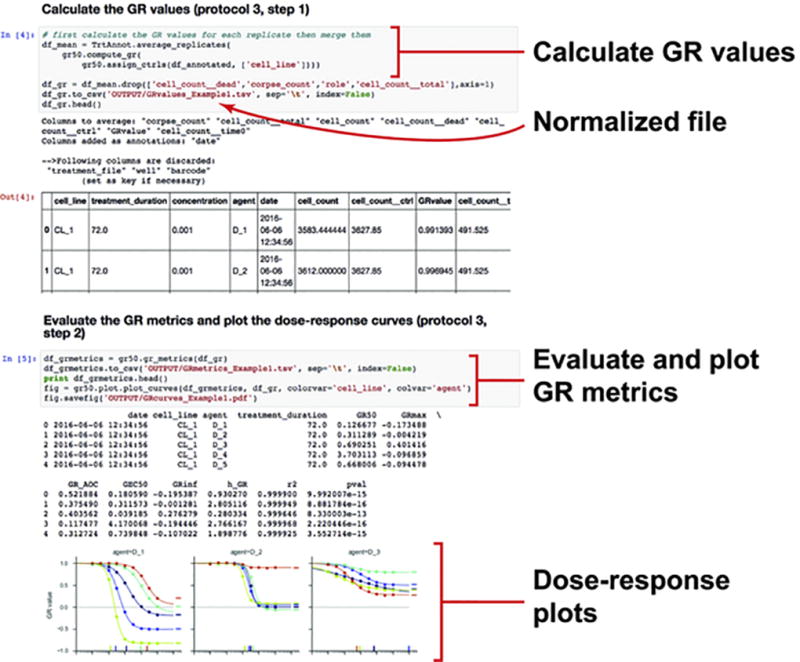
GR values and metrics are calculated using the gr50_tools module (protocol 3, steps 1 and 2). Dose response curves can be plotted and exported as a pdf file.
BASIC PROTOCOL 1: Designing the experiment
Basic Protocol 1 covers experimental design and the generation of files needed to control the robots that dispense treatments. Upon completion of the protocol 1, the user will have:
1 Jupyter notebook that serves as an archive of the design
Several well annotation files describing well-based treatments on a plate basis
1 plate information file listing all plates in the experiments and mapping them to treatments
1 treatment file describing the pin transfer procedure or specifying the actions of the drug dispenser
File types
We split the description of the model variables for a specific experiment into two types of files: well annotation files and plate information files. Well annotation files define the layout of treatments on a plate basis. These files are long tables with one row per treated well and as wide as necessary to describe the treatment with columns such as agent or agent_concentration (see Table 1 for suggested column headers). The second type of file is the plate information file, which associates physical plates based on their plate identifier to a specific well annotation file. The plate information file also contains all the plate-related annotations such a treatment_time, or treatment_duration. Model variables such as cell_line, or seeding_density are recorded either in well annotation files, if they differ from well to well, or in the plate information file if they are constant across a plate. Note that this distinction enhances the clarity of the experimental design, but will not change subsequent analysis. The only hard constraints in the structure of these files are as follows:
A well annotation file needs to have a column whose header is well and that contains the names of each treated well (in conventional alphanumeric plus numerical core - e.g. B04, F12). Well annotation files can be created manually but we strongly recommend scripting this process to avoid errors in parsing names; example scripts are provided in the datarail GitHub repository (github.com/sorgerlab/datarail).
A plate information file needs to have (1) a column plate_identifier containing plate identifiers (barcodes or ID numbers) as they will be reported in the data file (see Protocol 2), (2) a column well_file with the name of a well annotation file (including the appropriate extension such as tsv) or, for untreated plates, an empty field or a dash, and (3), for end-point experiments, a column treatment_duration with the duration of the treatment; a value 0 is used for untreated plates fixed at the time of treatment, or for time-course experiments, a column treatment_time with the date and time of treatment.
A treatment file is generated in the final step of Basic Protocol 1 by merging the two types of files described above into a single document that drives a drug dispenser such as the D300, maps drugs for generating a master plate(s) for pin transfer, or, less desirably, used to guide manual pipetting.
Table 1.
Names and descriptions for typical model variables.
| cell_line | Cell line identity |
| agent | Name of agent used to treat cells; e.g. drug name, ligand name. |
| agent__concentration | Concentration of the agent (µM) |
| agent2 | Name of the second treatment used in a combination |
| agent2__concentration | Concentration of the second agent (µM) |
| seeding_number | Number of cells seeded in a well |
| serum_pct | Percent of serum in the media |
| media | Type of media |
The datarail GitHub repository includes exemplar experimental design notebooks that can be adapted to run specific experiments (Figure 3A). These notebooks are executable and generate the necessary files but also serve as a record of the experimental design. Based on the functions provided in the datarail package, advanced users can write a Python script to generate more complicated designs. Users should explicitly set the seed for the random number generator such that the randomization can be reproduced.
Enforcing standardized names and unique identifiers in design files is necessary for merging datasets collected at different times or by different investigators and prevents mismatches between protocols and actual experiments. We provide a function to check the names of reagents in experimental design notebooks against a reference database and to add unique identifiers (see the function help for more details on the available reference databases).
Materials
- The module experimental_design of the datarail python package (github.com/sorgerlab/datarail). It contains:
-
◦functions to generate various experimental designs such as single drug dose-response or pairwise drug combinations across multiple concentrations
-
◦a function to randomize the positions of the treatments
-
◦functions to plot the experimental design for error checking, trouble shooting, and subsequent data interpretation
-
◦functions to export designs as either tsv files for manual pipetting and pin transfer or as hpdd files (xml format) to drive the D300 drug dispenser
-
◦
- List of treatments including:
-
◦Treatment reagent names and any associated identifiers
-
◦Concentrations for each agent (µM by default)
-
◦Stock concentration for each agent (mM by default)
-
◦Vehicle for dissolution of treatment agent (typically DMSO for small molecules)
-
◦
- Information about plates
-
◦List of plate identifiers (barcodes or ID numbers)
-
◦Any plate-based model variable that should be propagated to the downstream analysis (e.g. cell line, treatment duration)
-
◦Any plate-based confounder variable (e.g. plate model and manufacturer, treatment date)
-
◦
Jupyter notebook (or Python script) called experimental design notebook that will serve as a record of the experimental design. Exemplar notebooks are provided on the datarail GitHub repository (github.com/sorgerlab/datarail).
An external reference database if the user wants to include links to reagent identifiers stored externally (e.g. PubChem https://pubchem.ncbi.nlm.nih.gov or LINCS http://lincs.hms.harvard.edu/db/).
Step 1. Creating well annotation files (step 1)
Step 1A. Select an exemplar experimental design notebook for the desired experiment such as multiple dose-response curves (Figure 3A) or pairwise drug combinations. In the case of a dose-response experiment, create a tsv file with the list of drugs, their highest tested concentration and the number of doses in the dilution series and their role in the experiment (Figure 3B).
Step 1B. Record the treatment conditions and describe the properties of the reagents in the experimental design notebook.
Step 1C. Select the number of randomized replicates. We typically perform three replicates for drug dose-response studies.
Step 1D. Save the treatments as well annotation files (in a tsv file or xarray object).
*Designs can be visualized as images using plotting functions in the module experimental_design.
Alternatively, advanced users can design the treatments using the functions in the module experimental_design. These functions can:
-
◦
Generate single agent dose response
-
◦
Design pairwise combination treatments
-
◦
Specify well-based conditions
-
◦
Generate multiple randomizations of the treatments
-
◦
Stack group of wells with defined pattern to generate full plates
Step 2: Creating plate information files
Step 2A. Create a plate information file (tsv format) either manually or through the experimental design script (recommended). The plate description file should contain a table with the following columns:
-
◦
plate_identifier, matching the plate identifier (barcode or ID number) that will be reported in the scanner output file
-
◦
well_file, that contains the name of a well annotation file used to treat a given plate, or a dash (–) if the plate is not to be exposed to treatments.
-
◦
treatment_duration, duration of the treatment in hours. The untreated plates fixed at the time of treatment should have a value 0.
-
◦Any plate-level model or confounder variables that will be used for downstream analysis such as (see Table 1):
-
▪Identity of the cell line (if not specified in the well annotation file)
-
▪Conditions not specified in the well annotation file (e.g. describing CO2 levels or oxygen tension under which cells were cultured)
-
▪Other metadata and confounder variables such as day and time of plating, plate manufacturer, or batch number
-
▪
Step 3: Creating the treatment files
Step 3A. Create the treatment files for the D300 drug dispenser (hpdd format) using the functions in the module experimental_design to merge the treatment design files and the plate description file.
Step 3B. Create the mapping for master plate dispensing using the functions in the module experimental_design to generate the mapping for transferring drugs from a source plate into master plates. This function takes as inputs the well annotation files and the layout of the treatments to be dispensed. The output will be a two-column table mapping wells from the source plate to the destination plate.
BASIC PROTOCOL 2: Processing data files
Upon completion of protocol 2, a user will have:
1 Jupyter notebook that serves as an record of the data processing procedures
1 result file that comprises model and readout variables for all measurements in the experiment
Quality control reports
Introduction
This unit describes how to process the data file that contains the number of viable cells for each well into a standard table file annotated with the model variables of the experiment. A range of instruments (which we refer to as “scanners”) can be used to collect this data ranging from a plate scanner (for well-average values generated by CellTiter-Glo assays), to high content microscopes. We have written import modules for files that are generated by Columbus Software (Perkin Elmer) and IncuCyte ZOOM in-incubator microscopes (Essen Bioscience), but the output from any image processing software can be converted into a standardized table, called a processed file that can be passed to downstream analysis functions.
The data processing procedure is recorded in a data processing notebook. This notebook can also include any alterations in the original experimental design to account for potential issues that occurred during performance of the experiment, such as failed wells, errors in agent concentrations, etc. For experiments in which data files need to be split or merged, we strongly recommend scripting the procedure in the Jupyter notebook instead of manually handling result files (Figure 4A). We provide exemplar Jupyter notebooks in the GitHub repository (github.com/sorgerlab/datarail).
File types
Protocol 2 converts the output file from the scanner or microscope called unprocessed result file into a standardized file called processed file. The latter is merged with files created in Protocol 1 (well annotation files and plate information file) to generate the annotated file, which contains all model and readout variables from the experiment. The quality control step generates reports in the form of pdf files.
Materials
- The following modules of the datarail python package (github.com/sorgerlab/datarail):
-
◦import_modules: functions to import and convert the outputted data file of a particular scanner into a processed file with standardized format.
-
◦data_processing.drug_response: function to merge the results and the design files into a single annotated result file.
-
◦
Output data from the scanner saved as a tsv file (other formats will require writing new import functions).
The plate description file and well annotation files generated in Protocol 1 (or equivalent files generated manually or through other means).
Jupyter notebook (or Python script) called data processing notebook that will serve as a record of the data processing. Exemplar notebooks are provided on the datarail GitHub repository (github.com/sorgerlab/datarail).
Step 1: Converting the scanner specific data file into a processed file
* This step converts the scanner specific data file into a processed file containing: (1) the column cell_count, which is the readout of interest; (2) the columns plate_identifier (barcodes or ID numbers) and well (e.g. B03, F12), which define uniquely the sample; and (3), for time-course experiments, the column time, which contains the scanning timestamp. Additional readout values such as cell_count__dead or cell_count__dividing can be included based on the readout of the scanner. We have written import modules for files outputted by Columbus Software and IncuCyte microscopes, but the output from any scanner with non-proprietary file format can be converted into a processed file.
Step 1A. Use the functions in the module import_modules to import the file outputted by the scanner. Each scanner-specific function can include processing steps to correct for artefacts. For example, the function to convert the file from Columbus will correct for wells in which fewer fields were analyzed due to scanning errors.
Step 1B. Change the column headers or perform the necessary arithmetic operation to obtain the number of viable cells (or a related measure), labeled as cell_count (see accompanying experimental protocol and exemplar notebooks: github.com/sorgerlab/datarail). This can be done automatically by passing the right arguments to the import function of step 1A.
Step 1C. If necessary, script the operations needed to correct for failure of the experiments in the data processing notebook. For example, this can involve removal of wells in which an experiment failed or correction of barcodes that were misread.
Step 1D. Save the results into a tsv file called processed file.
Step 2: Merging data with treatment design
* This step merges the processed file with the experimental design files generated in Protocol 1. The resulting file, called annotated file, is a simple table in which each row is a measured sample and all model variables describing the experiment (treatment, readout, …) as well as the readout variables are columns. Saved as a tsv file, the annotated file serves as the permanent record of the experimental results.
Step 2A. Use the functions in the module data_processing.drug_response to combine the processed file with the plate description file and the treatment design files generated in Protocol 1 (note that all tables can be kept in memory and passed as arguments instead of as file names). The output is a table that contains the model variables and readout values for each sample. For time-course experiments, it is important to specify the time of treatment in either the plate description file or as input parameter such that the algorithm can properly calculate the treatment time based on the scanner timestamps.
Step 2B. If necessary, script the operations needed to correct for failure of the experiments in the data processing notebook. For example, this can involve changing the name of a reagent that was used by mistake, an incorrect stock concentration for a drug, or the annotation of wells that were not in fact treated.
* For plates in which the outer-most rows and columns are used, bias can be corrected by multiplying the values of wells on the edge by the ratio of the average values for untreated controls on the edge and the average value of the ones in the center of the plate. We suggest correcting a potential bias only if enough controls are positioned on the edge and in the center and that the difference is significant by t-test.
Step 2C. Save the data in a tsv file called annotated file.
Step 3 (optional): Controlling the quality of the data
*These steps use scripts to perform basic quality control analysis on the data (Figure 4B) such as edge bias or other potential artefacts described in the accompanying protocol. If the results of the quality control are not satisfactory, it is up to the user to update step 1C to discard low quality plates or wells. It is important to record all operations in the data processing notebook in particular filtering of low quality results such that the notebook becomes a literate record of how data was processed and thus ensures reproducibility of the analysis.
Step 3A. Test potential bias within each plate by using the functions in data_processing.drug_response. This test is useful for identifying edge effects or issues with cell plating and non-uniform growth across a plate.
Step 3B. Test potential bias across plates based on the untreated controls by using the functions in data_processing.drug_response. This test is useful for identifying differences between replicates and evaluating the variance of the readout.
BASIC PROTOCOL 3: Evaluating sensitivity metrics
Upon completion of protocol 3, a user will have:
1 normalized file which contains the GR values and relative cell count for all treated samples
1 response metric file which contains the drug sensitivity metrics for each dose-response curve
Introduction
This unit provides scripts to normalize drug response values and evaluate sensitivity metrics based on annotated result files. The first step is to normalize the treated samples to the untreated controls. The simplest normalization is to calculate the relative cell count x̄ defined as the cell count value measured in the treated conditions x(c) divided by the cell count value of an untreated (or vehicle-treated) control xctrl: x̄ = x(c)/xctrl. Relative cell count is valid for experiments where the untreated controls do not grow over the course of the experiment, but it is not suitable for comparing cell lines or conditions where the division rate differs (Hafner et al., 2016). An alternative normalization based on growth rate inhibition (GR) corrects such variation:
where x0 is the cell count value measured at the time of treatment. Given the proper controls xctrl and x0 for a set of conditions, both the relative cell count and the GR values are calculated. By default, values for untreated controls are averaged using a 50%-trimmed mean and replicates for treated samples on the same plates are averaged before calculating the normalized values. Values across multiple plates are aggregated to yield the normalized file.
Experiments with multiple time points (either time-course experiments or multiple plates fixed at different time points) can be parameterized using time-dependent GR values defined as:
where x(c,t±Δt) are the measured cell counts after a given treatment at times t+Δt and t−Δt, and where xctrl(t±Δt) are the 50%-trimmed means of the cell count of the negative control wells on the same treated plate at the same times t+Δt and t−Δt. The interval 2 · Δt should be long enough for the negative control to grow of about 20% (a fourth of division time). We found 8 to 12 hours to be a reasonable range for 2·Δt capture adaptive response while keeping experimental noise low.
The second step evaluates sensitivity metrics by fitting a sigmoidal curve to dose-response data:
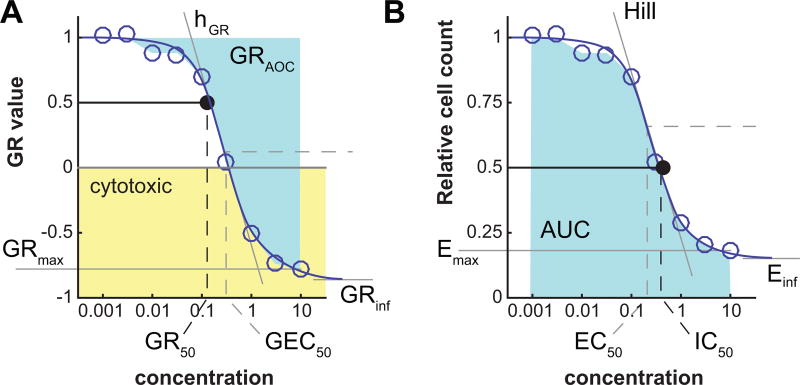
where y corresponds to GR values or the relative cell count across a range of concentrations. By default, fits are performed if there are at least 5 different concentrations for a given treatment and unique set of conditions. The script yields a response metric file which contains, for each dose-response curve, the GR and traditional metrics based on GR values and, respectively, relative cell count values (Figure 6):
GRinf or Einf: the asymptotic value of the sigmoidal fit to the dose-response data (GRinf ≡ GR(c → ∞)), respectively Einf ≡ x̄(c → ∞)). When a drug is potent and tested at a high concentration, GRmax (the highest experimentally tested concentration) and GRinf will be quite similar, but in many cases the convergence to the asymptote occurs only at concentrations well above what is experimentally testable. GRinf lies between –1 and 1; negative values correspond to cytotoxic responses (i.e. cell loss), a value of 0 corresponds to a fully cytostatic response (constant cell number), and a positive value to partial growth inhibition. Einf is bound between 0 and 1; in case of strong toxic response, it can be interesting to study Einf in the log domain to have better resolution of the residual population.
hGR or h: Hill coefficient of the fitted curve. It reflects the steepness of the dose-response curve. By default, the value is constrained between 0.1 and 5.
GEC50 or EC50: drug concentration at half-maximal effect. By default, GEC50 and EC50 values are constrained within two orders of magnitude higher and lower than the tested concentration range.
GR50 or IC50: the concentration of drug at which GR(c = GR50) = 0.5, or respectively x̄(c = IC50) = 0.5. If the GRinf or Einf value is above 0.5, the corresponding GR50 or IC50 value is not defined and therefore set to +∞.
GRmax or Emax: the maximum effect of the drug. For dose-response curves that reach a plateau, the GRmax or Emax value is close to the corresponding GRinf or Einf value. GRmax and Emax can be estimated from the fitted curve or obtained directly from the experimental data; the latter is implemented.
GRAOC or AUC: the area over the GR curve or, respectively, under the relative cell count curve, averaged over the range of concentration values.
Figure 6. Parameters of a dose-response curve.
Parameters of a dose-response curve for GR value (left) or relative cell count (right). Negative GR values correspond to a cytotoxic response (yellow area).
If the fit of the curve fails or is not significantly better than that of a flat curve based on an F-test (see below) with cutoff of p = 0.05, the response is considered flat. In such case, the parameter GEC50 or EC50 is set to 0, the Hill coefficient to 0.01 and the GR50 or IC50 value is not defined. In such case, if GR values are above 0.5, the GR50 or IC50 value is set to +∞ or, if all measured GR values are below 0.5, to –∞.
For experiments with multiple time points, all GR metrics can be evaluated at each time point at which time-dependent GR values were calculated. All considerations discussed above are relevant for time-dependent GR metrics.
File types
From the annotated file generated in Protocol 2, the first step of Protocol 3 generates the normalized file, which contains all result values. Step 2 generates the sensitivity metric file, which contains the parameters of the dose-response fits. Plots of these data can be generated and saved as pdf files.
Materials
The gr50_tools python package (github.com/sorgerlab/gr50_tools).
- Annotated result file, which contains the model and readout variables for each sample. This file can be the output of protocol 2, or a file that has tabular format with the following columns:
-
◦Model variables (descriptions of the perturbation such as drug name and concentration)
-
◦Readout (cell count or a surrogate) in a column labeled cell_count
-
◦
Step 1: Calculating relative cell count and GR values
Use the function add_gr_column.py with the processed data file as input. By default, the match between treated samples and controls is determined based on the model variables: xctrl values have the same model variables as corresponding treated samples, but no drug or reagent (and consequently a value of 0 for concentration); x0 values have same model variables as the corresponding untreated and treated samples but a treatment time of 0 and no drug or reagent (concentration value equals 0). For more complex scheme, the function add_gr_column.py can take as input a table with the controls already assigned to the treated samples in two columns: cell_count__ctrl and cell_count__time0.
For evaluating time-dependent GR values, use the function add_time_gr_column.py with the processed data file as input (Figure 5). By default, the match between treated samples and controls is determined based on the model variables: xctrl values have the same model variables as corresponding treated samples, but no drug or reagent (and consequently a value of 0 for concentration). It is not necessary to have a measurement at time 0 as the time-dependent GR values can be evaluated on any time interval even during treatment.
Technical replicates can be averaged at this stage using the functions in the module data_processing.drug_response. For time-dependent GR values, the function can average technical replicates that were collected at slightly different time points to account for sequential plate scanning.
Step 2: Fitting dose-response curve and evaluating sensitivity metrics
Use the function compute_gr_metrics.py to fit and plot the dose response curves (Figure 5). By default, the fitting function performs a significance test on the fit and replaces non-significant fits by a flat line. More options, such as capping of values or default range for parameters, can be selected by the user.
The significance of the sigmoidal fit is evaluated using an F-test. The sigmoidal fit, which is the unrestricted model, has p2=3 parameters (GRinf, hGR, and GEC50), whereas the flat line, which is the nested model, has p1=1 parameters (GRinf). Given the number of data points n and the residual sums of squares RSS2 and RSS1 of the sigmoidal fit and the flat line respectively, the F statistic is defined as:
The null hypothesis, meaning that the sigmoidal fit is not better than a flat line, is rejected if the value F is greater than a critical value (e.g. 0.95 for a significance of p<0.05) of the cumulative F-distribution with (p2–p1, n–p2) degrees of freedom.
* The interactive website www.grcalculator.org offers the same data analysis and simple visualization widgets and is a good place to explore datasets calculated using the GR method.
* Both the function compute_gr_metrics.py and www.grcalculator.org yield a tabular file with sensitivity metrics and quality of the fit.
COMMENTARY
Importance of processing data using computer scripts
Through this protocol, we emphasize the value of processing data using scripts. Manual steps or cut and paste operations in spreadsheets risk corrupting data. Operations (e.g. transformation, filtering, scaling operations, normalization) that are not scripted are not recorded and therefore not reproducible. While the scripts and functions from the datarail GitHub repository focus on dose-response assays, they are a model for programmatic manipulation of data in other contexts as well. We strongly encourage users to use these functions and Jupyter notebooks for all aspects of data processing.
Critical parameters and limitations in the design and experiment
Four critical parameters must be considered in the design of perturbation experiments:
Control wells: both relative cell count and GR values are sensitive to the value of the negative control (generally untreated or vehicle-treated wells). The experimental design must accommodate sufficient negative controls. We recommend >12 control wells for each 384-well plate (>8 if avoiding the outer-most wells) and >4 for each 96-well plates. For experiments with combinations of conditions and perturbations we recommend >4 control wells per condition independently of plate size.
Number of concentrations for dose-response curves: for proper fitting of a dose response curve, at least 5 different concentrations should be tested, and these must span the high and low end of the phenotypic response. Ideally, the readout at the lowest and highest concentrations should reach plateaus. We suggest using at least 7 doses spanning three orders of magnitude if the response range is known in advance, and at least 9 doses over four orders of magnitude if the response is unknown or if the same range of doses will be used for multiple cell lines across different experiments. We found that a 3.16-fold dilution (i.e. two concentrations per order of magnitude) is a good comprise between range and dose density; however, smaller spacing between doses will allow more precise evaluation of GEC50 or EC50 and GR50 or IC50 values, whereas wider range will improve the precision of the GRinf or Einf values. Note that there is no requirement that the spacing between doses be constant across the entire range; non-uniform spacing is particularly easy to achieve using automated dispenser.
Number of replicates: Readout of cell number or surrogates such as ATP level is relatively noisy when performed in multi-well plates. Replicates are therefore essential for improving the estimation of cell counts and GR values. Repeats are particularly important when assaying small changes such as those resulting from low efficacy drugs or short duration assays. When evaluating time-dependent GR values, it is important that negative controls are sufficiently accurately determined and assay duration long enough such that difference in cell count between time points are larger than the experimental noise.
Edge and plate-based effects: cell viability is frequently altered by differences in temperature and humidity at the edge of the plate. Therefore, we suggest not using the outer-most wells unless preliminary experiments confirm that edge effects are small. Performing multiple technical replicates with different randomization schemes helps to control artefacts arising from non-uniform growth across plates. Our software enables pseudo-randomization to ensure that replicates in different plates are positioned at different distances from the edge to avoid systematic error. If all wells are used in a plate, this also makes it less problematic to disregard some edge wells a posteriori if problems become evident (e.g. low media volume from excess evaporation) since data from an entire condition will not be lost.
Positive controls and fingerprints for master plates
In addition to negative controls (vehicle only wells), we suggest adding positive controls to check that cells are responding to perturbation and that data acquisition and processing steps are performing correctly. We find drugs such as staurosporine or actinomycin D make good positive controls for cytotoxicity. Positive controls can be labeled in the column role with the flag positive_control. The quality control functions in the module data_processing.drug_response will process results for positive controls separately and include them in the quality control report.
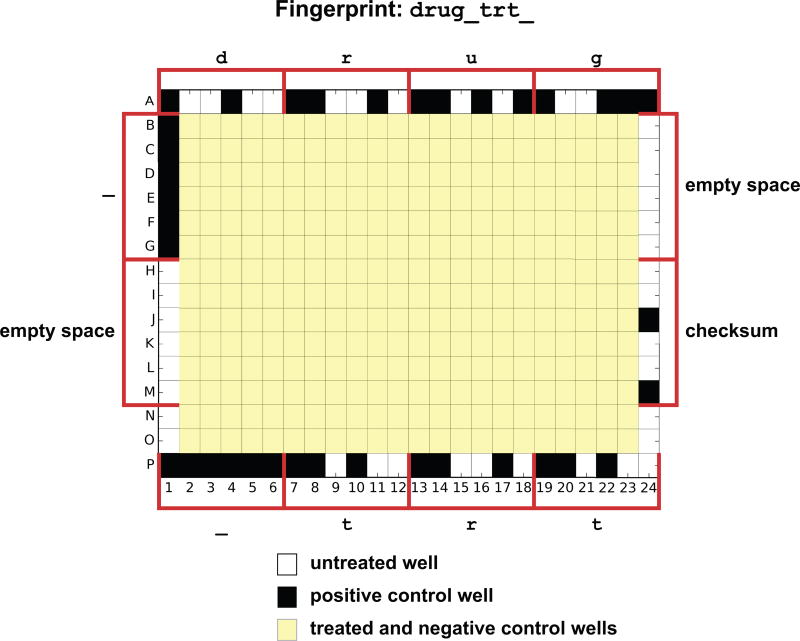
For master drug plates that are meant to be reused over months or years by different experimentalists, we recommend using negative and positive controls to hard-code the identity of the master plate. If the outer-most wells are generally not used for treatments, these wells can include cytotoxic compounds in a unique pattern. Functions in the module experimental_design can encode and decode an 11-character name (5-characters for 96-well plates) in a set of positive and negative controls and position them on the edge on a plate (Figure 7). Treatments used for the fingerprint wells will be labeled with the flag fingerprint as role. Quality control scripts decode the fingerprint on the edge wells and check for consistency with the name of the master plate used for treatment as labeled in the plate information file.
Figure 7. Plate fingerprint.
Positive controls can be used to encode the name of the treatment plate in the edge wells with up to eleven characters for a 384-well plate (5 for a 96-well plate). Each letter in the label ‘ drug_trt_’ is transformed into a list of 6-bit values that defined treatments along row A, row P, column 1 and column 24 (one well per bit, six wells per character). Last value of column 24 is a checksum.
Experimental implementation
By default, only treatment variables are exported in the files used to describe the pin transfer procedure or to control drug dispensers; this is appropriate for experiments in which a single factor is varied from well to well (e.g. drug dose and identity). However, we include a software flag such that condition variables can also be included in the treatment file. This is necessary when two factors are being altered. For example, when exploring the effects of varying the concentration of a soluble ligand on drug response, ligand identity and concentration are a condition variable but flagged as being a reagent for robotic dispensing; drug remains a treatment variable and is thus dispensed by default.
Complex designs
The protocols described above and Jupyter notebooks available on GitHub are designed for common single-agent dose response experiments and pairwise testing of drugs. However, the flexibility of the D300 drug dispenser makes it possible to design much more complex experiments in which wells are exposed to multiple drugs or ligands and multiple cell lines are present per plate. The data structures and packages used within datarail support the design of complex experiments that have two or more treatments per well. The current set of templates does not include complex experiments with multiple conditions or combined treatments. Moreover, our curve fitting algorithm assumes a dilution series across a single variable. In such cases, we leave to the user to write personalized scripts that rely on sub-functions from the package. However, we expect such design and processing scripts to be developed and published on our GitHub repository in the future.
Assignment of control well is based on the model variables
Data processing using the scripts in this protocol relies heavily on keys for identifying the controls necessary to normalize the cell count values. Each treated sample needs to be matched with a negative control xctrl and, in the case of GR calculations, a cell count value at the start of treatment at t=0 (i.e. x0). By default, matching treated samples to control wells is based on the model variables: negative controls have an empty value for reagent and t=0 data have a value of 0 for treatment_duration. Thus, protocol 3 may not be directly usable in complex experiments with multiple reagents and ambiguous normalization. In such cases, users may need to assign control wells to each sample using a custom script.
Troubleshooting and avoiding common problems
We have identified three common pitfalls in this method:
Manual processing steps: Any stage in the generation of the experiment design or in data processing that is not scripted has a high potential to introduce errors that are hard to troubleshoot. In particular, files that are generated by hand are a potential source of mistakes through typos, erroneous formatting, parsing errors or inconsistencies in naming conventions. In addition, we have observed issues with files generated by spreadsheet programs, such as Microsoft Excel, because in certain circumstances, numeric fields are automatically iterated and some names and numbers automatically converted into dates.
Growth of negative controls: Calculation of GR values requires that negative control samples grow over the course of the assay. As the GR acronym indicates, GR values and metrics are meant to study the inhibition of cellular growth. If the study is focused on cell death or growth is minimal during the assay either due to the cell line tested, the incubation conditions, or the length of the assay, relative cell count and metrics such as IC50 and Emax are the correct ones to use since GR values will be undetermined.
Fitting artefacts: The fitting of the dose response curve is subject to several artefacts. First, enough concentrations must be measured for the fit to be reliable. Second, values greater than 1.0 for relative cell count or GR value are not accounted for in the model; this can arise if a drug actually stimulated growth. Third, noisy data may yield a bad fit and the interpolated parameters can have unwanted variability. Fourth, a fair number of drugs are associated with biphasic dose-responses that cannot be properly fitted using a sigmoidal curve. Alternative fitting functions will be implemented in the future to parameterize biphasic dose-responses. In all cases, the goodness of the fit (as defined by the r-squared and the corresponding p-value) reflects how reliable the metrics are. Metrics obtained from low-quality fits should be used carefully.
Companion website
The companion website www.grcalculator.org can be used to replace protocol 3 for simple experiments. It also provides visualization tools and easy comparison between samples and treatments. A manuscript by Clark et al. describes the functionalities of the website and corresponding R package (Clark et al.).
Acknowledgments
This work was funded by NIH P50 GM107618 and NIH LINCS U54-HL127365 grants to PKS and by a Swiss National Science Foundation fellowship P300P3_147876 to MH.
Footnotes
KEY REFERENCES
Paper describing the GR method:
Hafner M, Niepel M, Chung M and Sorger P K, Growth rate inhibition metrics correct for confounders in measuring sensitivity to cancer drugs Nat. Methods (2016) 13 521–7
Companion protocol:
Niepel M, Hafner M, Chung M, and Sorger P K, Measuring cancer drug sensitivity and resistance in cultured cells
- https://github.com/sorgerlab/datarail : GitHub repository with the scripts for the experimental design and data handling
- https://github.com/sorgerlab/gr50_tools : GitHub repository with the scripts for the evaluation of the GR values and metrics.
- http://www.GRbrowser.org : interactive tool to evaluate GR metrics and plot the results.
LITERATURE CITED
- Clark NA, Hafner M, Kouril M, Williams EH, Muhlich JL, Niepel M, Sorger PK, Medvedovic M. GRcalculator: an online tool for calculating and mining drug response data. doi: 10.1186/s12885-017-3689-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Niepel M, Chung M, Sorger PK. Growth rate inhibition metrics correct for confounders in measuring sensitivity to cancer drugs. Nature Methods. 2016;13:521–527. doi: 10.1038/nmeth.3853. Available at: http://dx.doi.org/10.1038/nmeth.3853\ nhttp://www.nature.com/doifinder/10.1038/nmeth.3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundholt BK, Scudder KM, Pagliaro L. A simple technique for reducing edge effect in cell-based assays. Journal of biomolecular screening. 2003;8:566–70. doi: 10.1177/1087057103256465. Available at: http://www.ncbi.nlm.nih.gov/pubmed/14567784. [DOI] [PubMed] [Google Scholar]
- Powell SG, Baker KR, Lawson B. A critical review of the literature on spreadsheet errors. Decision Support Systems. 2008;46:128–138. [Google Scholar]
- Saez-Rodriguez J, Goldsipe A, Muhlich J, Alexopoulos LG, Millard B, Lauffenburger DA, Sorger PK. Flexible informatics for linking experimental data to mathematical models via DataRail. Bioinformatics. 2008;24:840–847. doi: 10.1093/bioinformatics/btn018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker G, Fischer M, Rieder D, Bindea G, Kainz S, Oberstolz M, McNally JG, Trajanoski Z. iLAP: a workflow-driven software for experimental protocol development, data acquisition and analysis. BMC bioinformatics. 2009;10:390–401. doi: 10.1186/1471-2105-10-390. Available at: http://www.ncbi.nlm.nih.gov/pubmed/19941647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner A. Helpful Hints to Manage Edge Effect of Cultured Cells for High Throughput Screening. Corning Cell Culture Application and Technical Notes. 2001:7–8. Available at: http://www.corning.com/Lifesciences/technical_information/techDocs/an_hintsedgeeffecthts.pdf\ nhttp://iccb.med.harvard.edu/screening/helpfulpublications_PDF/an_hintsedgeeffecthts.pdf\ nhttp://www.protocol-online.org/prot/Cell_Biology/Cell_Culture/General_P.
- Wu T, Zhou Y. An Intelligent Automation Platform for Rapid Bioprocess Design. Journal of laboratory automation. 2013;19:381–393. doi: 10.1177/2211068213499756. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4113973&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeberg BR, Riss J, Kane DW, Bussey KJ, Uchio E, Linehan WM, Barrett JC, Weinstein JN. Mistaken identifiers: gene name errors can be introduced inadvertently when using Excel in bioinformatics. BMC bioinformatics. 2004;5:80. doi: 10.1186/1471-2105-5-80. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=459209&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziemann M, Eren Y, El-Osta A, Zeeberg B, Riss J, Kane D, Bussey K, Uchio E, Linehan W, Smedley D, et al. Gene name errors are widespread in the scientific literature. Genome Biology. 2016;17:177. doi: 10.1186/s13059-016-1044-7. Available at: http://genomebiology.biomedcentral.com/articles/10.1186/s13059-016-1044-7. [DOI] [PMC free article] [PubMed] [Google Scholar]