Abstract
Metabolomics, which is the profiling of metabolites in biofluids, cells and tissues, is routinely applied as a tool for biomarker discovery. Owing to innovative developments in informatics and analytical technologies, and the integration of orthogonal biological approaches, it is now possible to expand metabolomic analyses to understand the systems-level effects of metabolites. Moreover, because of the inherent sensitivity of metabolomics, subtle alterations in biological pathways can be detected to provide insight into the mechanisms that underlie various physiological conditions and aberrant processes, including diseases.
Metabolites are the substrates and products of metabolism that drive essential cellular functions, such as energy production and storage, signal transduction and apoptosis. In addition to being produced directly by the host organism, metabolites can derive from microorganisms, as well as from xenobiotic, dietary and other exogenous sources1.
The biochemical actions of metabolites are far-reaching. To start, metabolites can regulate epigenetic mechanisms and maintain the pluripotency of embryonic stem cells (ES cells)2–6. It has also been well established that metabolites such as ATP, acetyl-CoA, NAD+, and S–adenosyl methionine (SAM) can function as co–substrates, regulating post-translational modifications that affect protein activity7,8. In addition, fatty acids and hormones can interact with plasma proteins to enable their transport in the bloodstream9,10. Furthermore, metabolite–protein interactions can aid in facilitating cellular responses by initiating signalling cascades, thus evidencing the role of metabolites in signal transduction11,12.
Indirectly, metabolites affect the environment in which they are produced. Under normal conditions, homeostatic controls exist to counteract any adverse biological consequences of such effects. For example, acidic metabolites decrease the pH of the microenvironment13,14, and high concentrations of these acidic metabolites are found, for instance, in the colon, owing to bacterial fermentation of dietary carbohydrates that leads to the production of short-chain fatty acids. These are, however, efficiently neutralized by mucosal production of bicarbonate. Notably, such homeostatic controls can be compromised with age and during disease, leading to functional decline and a failure to return to steady state. In addition, the adaptation of aberrant glycolytic cancer cells to the large amounts of lactate and protons that they produce occurs through modification of the activity of transporters, exchangers, pumps and carbonic anhydrases, which all help to maintain the intracellular pH and enable cells to survive the acidic microenvironment15. Thus, as metabolites can have a wide range of functions in the cell and organism, there is growing motivation to better ascertain their specific functions, as well as to understand their physiological roles. This can be done by implementing various metabolomic approaches to identify metabolites and metabolic pathways that are associated with particular phenotypes, and then integrating this knowledge with functional and mechanistic biological studies.
The main methodologies that are used for metabolite recovery and identification are untargeted (global) and targeted mass spectrometry-based metabolomics, which are discussed in more detail in BOX 1. Untargeted metabolomics aims to measure the broadest range of metabolites present in an extracted sample without a priori knowledge of the metabolome. The types of metabolites that are recovered are influenced by the extraction and analytical method of choice, but they result in a complex data set that requires computational tools to identify and correlate metabolites between samples and to examine their interconnectivity in metabolic pathways in relation to the phenotype or aberrant process (see BOX 2 and Supplementary information S1 (box)). By contrast, targeted metabolomics provides higher sensitivity and selectivity than untargeted metabolomics, but metabolites are analysed on the basis of a priori information, whereby methods are developed and optimized for the analysis of specific metabolites and metabolic pathways of interest. Targeted analysis also constitutes an important part of a metabolomics workflow to validate and expand upon results from untargeted analysis16.
Box 1. Mass spectrometry in metabolomics.
Mass spectrometry
Mass spectrometry is an excellent analytical platform for metabolomic analysis, as it provides high sensitivity, reproducibility and versatility. It measures the masses of molecules and their fragments to determine their identity. This information is gained by measuring the mass–to–charge ratio (m/z) of ions that are formed by inducing the loss or gain of a charge from a neutral species. The sample, comprising a complex mixture of metabolites, can be introduced into the mass spectrometer either directly or preceded by a separation approach (using liquid chromatography or gas chromatography). Direct injection has been successfully implemented for high-throughput metabolomics. However, as thousands of ions can be present in metabolomic experiments, chromatographic separation before entering the mass spectrometer minimizes signal suppression and allows for greater sensitivity, and — by providing a retention time identifier — it can further aid metabolite identification. In addition to m/z and retention time information, the identification of an ion is facilitated by fragmentation pattern information that is acquired by tandem mass spectrometry83.
Untargeted metabolomics
Untargeted or global metabolomic analysis allows for an assessment of the metabolites extracted from a sample and can reveal novel and unanticipated perturbations. Untargeted analyses are most effective when implemented in a high-resolution mass spectrometer, to facilitate structural characterization of the metabolites. Its primary advantage is that it offers an unbiased means to examine the relationship between interconnected metabolites from multiple pathways. However, it is not yet possible to obtain all metabolite classes simultaneously, as many factors affect metabolite recovery, depending on the functional group of the metabolite. In addition, there are a large number of unknown metabolites that remain unannotated in metabolite databases35. Thus, depending on the pH, solvent, column chemistry and ionization technique used, untargeted metabolomics can provide a detailed assessment of the metabolites in a sample, revealing a wide range of metabolite classes.
Targeted metabolomics
Targeted metabolomic analyses measure the concentrations of a predefined set of metabolites. A standard curve for a concentration range of the metabolite of interest is prepared, so that accurate quantification can be gained. This type of analysis can be used to obtain exact concentrations of metabolites identified by untargeted metabolomics, providing analytical validation.
Imaging metabolomics
It is also possible to reveal the localization of selected metabolites within a tissue sample using imaging mass spectrometry techniques, such as matrix-assisted laser desorption ionization (MALDI)84, nanostructure-imaging mass spectrometry (NIMS)70,85, desorption electrospray ionization mass spectrometry (DESI)86 and secondary ion mass spectrometry (SIMS)87, among others. NIMS and DESI are especially suited to the analysis of small molecules.
Box 2. Computational tools in metabolomics.
Metabolomic analyses, and untargeted metabolomics in particular, result in the generation of complex data sets; therefore, computational tools are crucial to process and interpret these results. The problems associated with big data processing, statistical analyses, metabolite identification and biological interpretation are not trivial, but there are now some novel tools available that accelerate and automate the computational workflows, providing user-friendly tools for both novice and expert bioinformaticians (for further details, refer to Supplementary information S1 (box)).
Data processing and statistical analysis
After data upload, mass spectral peaks are picked, realigned and annotated. The data is deconvoluted using computational tools to remove instrumental and chemical noise, thus providing only the biologically relevant information.
The types of statistical analyses that can be implemented for metabolomics data are vast, and choosing the correct test can be challenging. Online tools such as XCMS Online42, DeviumWeb MetaboAnalyst43 and many others give researchers the ability to carry out a wealth of tests. Some of the most recent advances are tools that provide false discovery rate measurements to ensure that the data have statistical power. Other concepts that are especially useful for finding biologically relevant metabolites are multi-group and meta-analyses, which can reveal shared metabolic changes across multiple experiments88.
Metabolite identification and databases
Initial putative metabolite identifications can be made on the basis of the accurate mass–to–charge ratio (m/z) of the mass spectral ion. This is aided by the use of comprehensive metabolite databases such as METLIN89, HMDB90, MassBank91 and GMD26,92. Tandem mass spectrometry experiments can then be carried out on the isolated ion, followed by matching with an authentic standard, in order to obtain characteristic fragments and retention time information to distinguish the ion from structural isomers. In silico prediction tools provide further insight into metabolite identification when a particular m/z or tandem mass spectrometry fragmentation pattern does not provide a match24,93. A recent innovation in ion mobility mass spectrometry, the rotationally averaged cross-collisional section (CCS), provides another level of metabolite identification, and databases containing CCS information are currently in the early stages of development94. Despite all of these innovations, some metabolite features cannot be assigned to a molecular structure. It is therefore important that they are published (databases for these have already been set up on METLIN) to aid in their future identification and correlation to phenotypes.
Biological interpretation
Network modelling and pathway-mapping tools can help us to understand the parts that metabolites play in relation to each other and in biological aberrations. Thereafter, metabolites can be placed into context with upstream genes and proteins to lead mechanistic investigations47. As well as the established and comprehensive metabolic network resources Kegg95, Recon1 (REF. 34) and Biocyc96, there are several recently developed programs that use novel methods to find pathway connectivity, as well as aiding in metabolite identification. These include mummichog46 and metabolite set enrichment analysis (MSEA)97. In addition, stable isotope metabolomics56,57 and omics-scale big data integration can reveal interconnectivity between metabolites and their relationships with genes and proteins (see also main text).
The types of samples that can be analysed using metabolomics are wide-ranging and include tissues, cells and biofluids. Tissue analysis, in particular, is perhaps the most powerful approach for studying localized and specific responses to stimuli and pathogenesis, yielding explicit biochemical information about the mechanisms of disease. Traditionally, tissue analysis involves extraction of the complete tissue material into a liquid form, from which the metabolite changes are averaged across the different cell types and regions of the analysed organ. In addition to this total tissue analysis, subregional, cellular and even subcellular metabolite profiles can provide further insight into structure-to–function relationships; this is particularly valuable in the case of heterogeneous tissues such as brain and cancers17. Simultaneous sampling of arterial blood (entering the organ) and venous blood (draining the organ), followed by paired analysis, can also have value in the investigation of tissue metabolic activity16. This paired arteriovenous approach provides information about the metabolite uptake and release patterns across the tissue of interest and therefore gives insight into tissue metabostasis. The power of this paired analysis allows for the measurement of metabolite arteriovenous differences or ratios and offers a compelling compromise with sampling effort, compared to the traditional approach of venous blood analysis.
During the past few years, substantial progress has been made in metabolomic analysis by improving instrument performance, experimental design and sample preparation, ultimately facilitating broader analytical capabilities. Moreover, the surge in new chemoinformatic (computational approaches for handling chemical information) and bioinformatic (computational approaches for handling biological information) tools has provided extensive support for data acquisition, analysis and integration. This has greatly enhanced our ability to identify metabolites in various samples and allowed us to correlate these metabolites with particular phenotypes, thus establishing useful biomarkers that are indicative of particular physiological states or aberrations. The ultimate challenge now is to move beyond simply identifying metabolites and using them as biomarkers, and to start establishing the direct physiological roles of metabolites and their involvement in metabolic networks, as well as determining how changes in their levels are implicated in different phenotypic outcomes. This Innovation article focuses on how this most relevant hurdle for metabolomics can be overcome. We describe how advances in technologies that are used in metabolite identification and analysis, experimental design and pathway mapping are helping us to gain more meaningful data, revealing important nodes for further investigation. We also discuss how this information, when combined with traditional biological methods, can enable us to ascertain molecular mechanisms and begin to infer biological causality.
Current challenges in metabolomics
During the past few years, metabolomics has evolved considerably to overcome challenges that initially confounded analysis18. A major challenge still exists for the identification of metabolites and validation of metabolites in human populations. However, the most important challenge is to develop workflows for assigning biological meaning to metabolites and to move towards finding mechanisms of disease.
Metabolite identification and validation
The initial focus of metabolomics has been on biomarker discovery, with the aim of identifying metabolites that are correlated with various diseases and environmental exposures. This has, for example, led to the identification of plasma trimethylamine N–oxide (TMAO) and urinary taurine as markers of cardiovascular disease (CVD)19 and ionizing radiation exposure20–22, respectively. In order to correlate metabolites with a phenotype, the two biggest hurdles faced are metabolite identification and biomarker validation. In any given untargeted metabolomics experiment, only a subset of all metabolite features present can be positively identified. This has been facilitated by novel in silico tools23–25 (see below, as well as BOX 2 and Supplementary information S1 (box)), the expansion and development of metabolite databases26 (see BOX 2 and Supplementary information S1 (box)) and the synthesis of previously unattainable standard compounds that can confirm the identification of the metabolite (these standards are either novel compounds or were previously not available in an isotope-labelled form)27.
Biomarker validation can be challenging, owing to difficulties in measuring subtle differences in metabolite concentrations between control and aberrant conditions, and because of the lack of follow–up with targeted metabolomic experiments (BOX 1). These follow–up experiments should be carried out in an additional cohort of biological samples for validation of the metabolite changes with the phenotype. Moreover, one of the largest challenges to biomarker validation is overcoming inter-individual metabolite variation, which arises owing to differences in genetic factors and environmental exposures. All of these influences result in significantly different metabolic responses in population studies1, making it extremely difficult to pinpoint metabolites that are correlated with a particular condition and, ultimately, to provide clinical biomarkers. This is the case especially when examining a multifaceted disease such as cancer. There are a number of methods that can be applied before and after analysis to overcome some of the biological variation associated with human studies. Establishing appropriate experimental design and statistical power for the study, and using patient questionnaires with subsequent population stratification, as well as regression modelling, can allow for the extraction of important metabolites28. These types of approaches can remove confounding samples from the analysis and help to streamline the data to identify metabolites that are correlated with the biological stimulus and not another influence. In addition, using appropriate metabolite normalization strategies, such as analysing metabolite ratios or normalizing to creatinine in urine studies, may help. Developing databases to collect data on the normal fluctuations in metabolite concentration ranges that occur in response to factors such as diet29, age, gender, circadian rhythm and exercise, which are frequent causes of sample-to-sample variability, would also be useful. Indeed, some databases that contain information on specific metabolite concentration ranges in human biofluids and in dietary components — the Human Metabolome Database (HMDB)30 and FooDB, respectively — have already been developed.
Functional analysis of metabolites
Perhaps the largest challenge that metabolomic researchers face in any study is relating the identified metabolites to their biological roles, which is a necessary step for moving beyond biomarkers and towards mechanisms. Biomarkers obtained from human population studies can provide a starting point for finding links between diseases and metabolic pathways31, and further mechanistic work can be carried out using in vitro and animal-based studies, as previously shown32. Furthermore, patient-derived primary cell lines and xenografts can provide more reliable models for finding relatable data, as such samples make it possible to control for genetic and environmental influences.
However, to evaluate the biological roles of one or several metabolites (a metabolic signature), one first has to determine their functions in metabolic pathways and their interconnectivity, and, more broadly, determine which metabolic pathways are perturbed by the aberrant condition33. Only such a multi-level analysis can provide a comprehensive understanding of the systemic biological changes that are associated with particular metabolites and potentially direct further mechanistic studies. Determining the interactions of metabolites in metabolic pathways is particularly challenging. Metabolic pathway maps currently include ~2,000 metabolites; however, similar to metabolite databases, they are somewhat incomplete, as some metabolites have not yet been characterized34,35. Novel molecules are regularly being discovered, adding to the pool of known metabolites22,36. Multi-layered approaches that integrate metabolomic and other ‘omics’ data (see below) acquired from the same samples provide an opportunity to investigate the system-wide changes in a disease and to delve further into metabolic pathway interactions and the mechanisms of disease development and progression37,38. In addition, novel experimental approaches, such as stable isotope-assisted analysis (see below), can trace metabolite utilization in pathways in a temporal manner.
Recent technical advancements
Developments in innovative informatics strategies have been a major driver in overcoming some of the challenges presented with metabolomic analysis33. Advances in data processing, statistical analysis and metabolite characterization have enabled the identification of more metabolites that are associated with a particular phenotype than was ever previously achievable. Moving towards mechanistic investigations, novel metabolic pathway analysis tools that assess the interconnectivity of these metabolites can provide important insights, particularly when paired with advanced metabolomic techniques such as stable isotope tracing and integration with other orthogonal data sets, ultimately providing systems-level analyses (FIG. 1).
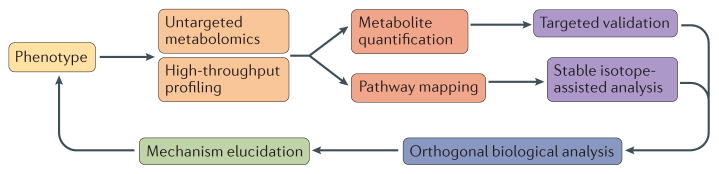
Figure 1. From metabolites to pathways and mechanisms.
The workflow outlines a holistic approach that begins with high-throughput untargeted metabolite profiling. Analysis of biofluids, cells or tissues reveals quantitative metabolite changes (as a result of a stimulus) that can be validated further. Metabolites can be mapped and analysed within metabolic pathways to relate the metabolites to each other, and within interconnected biological pathways, providing potential targets for further mechanistic studies. The combination of metabolomic, orthogonal biological analysis and isotope–assisted deciphering of pathways allows the mechanism of the aberrant phenotype to be ascertained.
Informatics
The development of computational and chemoinformatic tools for metabolomics can effectively support experimental data upload, processing, statistical analyses and metabolite identification, and, when used in conjunction with bioinformatic tools, can place metabolites into biological context (see BOX 2 and Supplementary information S1 (box)). Metabolomic data sets obtained by mass spectrometry (BOX 1) contain information on thousands of ions that are generated in the mass spectrometer from each sample, in which the ions represent the precursor intact metabolite or its fragments, adducts or isotopes. Computational tools are thus essential for reducing the redundancy in these complex data sets and facilitating identification of the most relevant metabolites.
For researchers in the field of metabolomics, computational resources are growing at a rapid rate, and many of these have been discussed in detail elsewhere33,39. However, metabolomic analysis remains a time-consuming process, and metabolite identification is still a limiting factor. Therefore, computational workflows that significantly speed up the process of data upload and data mining, with novel methods for automated or in silico metabolite identification and biological interpretation, are needed. Such automated computational workflows — allowing data streaming from the instrument to the software, automated qualitative and quantitative metabolite characterization, calculation of fold change and statistical significance, and, importantly, metabolite pathway analysis — have recently been developed (for more detail, see Supplementary information S1 (box)).
As metabolomics is highly interdisciplinary, and not all laboratories have personnel that are specialized in all areas of the experimental workflow, it is often the case that some of these computational tools are out of reach for those not specialized in informatic approaches or new to the metabolomics field. Fortunately, this is beginning to change, with several resources provided through the US National Institutes of Health (NIH) Common Fund Metabolomics Program. This programme funds six regional comprehensive metabolomic resource cores, a data repository and a coordination centre, to enable hands–on and online training in a range of areas, including data processing and interpretation. Another initiative, the Coordination of Standards in Metabolomics (COSMOS), is also helping to promote the standardization of metabolomics, by providing both experimental and data sharing, thus aiding new researchers in the field40 (see Supplementary information S1 (box)). There are several tools, including the workflows mentioned, that are user-friendly but have advanced parameters for expert users, thus providing a resource for all levels of expertise41,42. Some of these are available as part of the mass spectrometry vendor software, whereas other tools are provided as open-access software that can be utilized from data upload through to the metabolite pathway analysis42,43. These tools have already been successfully used to correlate single or multiple validated metabolites to a biological aberration. For example, MZmine 2 was used to show the interaction between dietary lipids and gut microbiota for regulating cholesterol metabolism44, and metabolomic analysis using both XCMS Online and MetaboAnalyst revealed metabolic dysregulation in ischaemic retinopathy45.
As discussed above, to move from using metabolites as predictive biomarkers to leading mechanistic investigations, the metabolites need to be put into their biological context by identifying their roles in metabolic pathways, their interconnectivity with other metabolites, and their relationships to upstream genes and proteins. Informatics approaches can greatly facilitate these analyses and can help to reveal broad potential metabolite activity across multiple metabolites and pathways46, and can also provide big data integration across different-omics technologies (see below)47 such as the systems biology approach recently developed on XCMS Online. As an example, a recent study took advantage of various bioinformatics tools to analyse genetic influences on metabolites in human blood. For this, a network of genetic–metabolic interactions was generated, first using Gaussian graphical models to connect biochemically related metabolites and then connecting metabolites with genetic loci from a genome-wide association study38. Novel concepts such as these have maximized the ability to extract important biological information from metabolites.
Stable isotope-assisted metabolomics
One of the most promising ways to ascertain the roles of metabolites in metabolic pathways is to track their utilization with stable isotope tracers. These experiments make use of commercially available metabolites labelled with stable isotopes such as carbon (13C), nitrogen (15N) or deuterium (2H). The design of stable isotope-assisted experiments is based on a priori information for a particular metabolite or metabolic pathway of interest; these studies can thus be led by information obtained from untargeted metabolomic analysis (BOX 1).
The results from targeted and/or untargeted metabolomic analysis do not provide information on intracellular metabolic rates and relative pathway activities, and, for example, increased levels of one metabolite can be caused by increased activity of metabolite-producing enzymes or decreased activity of metabolite-consuming enzymes49. Following up with stable isotope-labelling experiments provides additional information on how a particular compound (nutrient or substrate) is metabolized with respect to a particular phenotype and can help to identify the pathways that contribute the most to substrate utilization. Thus, stable isotope-assisted tracing of a labelled substrate can reveal its metabolic fate.
There are several ways to carry out a stable isotope-assisted experiment. In metabolic steady state experiments, the measured metabolite pools (or levels) are equilibrated, and fluxes (or conversion rates) are roughly constant35. In addition, the labelling enrichment becomes stable over time (from a labelled nutrient into a given metabolite) to reach the isotopic steady state. The interpretation of isotope-enriched data in such conditions can provide information on relative pathway activity, such as the relationship between metabolites, and it also allows quantification of nutrient contributions to the production of different metabolites49. By contrast, in kinetic (or dynamic) flux experiments, the system has yet to reach steady state, and flux refers to the in vivo velocities of the individual metabolic reactions35. Thus, kinetic flux analysis provides dynamic labelling patterns, which allow quantification of metabolite flux when combined with intracellular metabolite concentrations48,49. As a notable example, kinetic flux revealed mechanisms for NADPH metabolism, including the contribution of the 10–formyl-tetrahydrofolate pathway to NADPH production50. Steady state flux analyses have also contributed to revealing important substrate utilization, with a recent clinical example uncovering selective activation of pyruvate carboxylase over glutaminase 1 in early-stage non-small-cell lung cancer51.
Stable isotope-assisted metabolomics can be used to calculate flux within a specific set of related pathways — or, on a larger scale, it can encompass multiple metabolites, labelled precursors and pathways. However, such analyses are computationally highly complex for dynamic experiments, leading to a decrease in accuracy35. In order to overcome this, algorithms have recently been developed that combine both stable isotope analysis and untargeted metabolomics52–55. This technology, called global isotope metabolomics, provides comprehensive differential labelling between two biological conditions, offering further understanding of metabolism at a systems level. Even though untargeted stable isotope metabolomics is a relatively new tool, its value has already been demonstrated in several studies56,57. It also provides yet another example of the power of informatics in metabolomic analyses.
Orthogonal approaches for mechanistic studies
Owing to the fact that transcript and protein levels have only a modest correlation with each other, and that metabolites can be further modified by enzymatic processes and can originate from and be modified by various internal and external stimuli, it is necessary to introduce metabolomic analysis approaches that provide big data integration across different-omics (genomics, epigenomics, proteomics and transcriptomics) in order to comprehensively determine the consequences of all metabolites on biology (FIG. 2). Such integrative approaches can help to determine the relationships between gene and protein expression and metabolite concentrations, and the balance between production and consumption of metabolites58. As an example, by combining metabolomics with metagenomics and metatranscriptomics data, it was possible to elucidate the origins and roles of bacteria-derived metabolites59,60. A recent study also revealed that gut bacteria transplanted from thin or obese people recapitulated the respective phenotypes in gnotobiotic mice, with changes to microbial genes and concomitant downstream metabolites60. In addition, it was possible to demonstrate that individuals from rural African and African American populations that exchanged diets underwent large changes in their metagenome and metabolome, and this altered their cancer risk61.
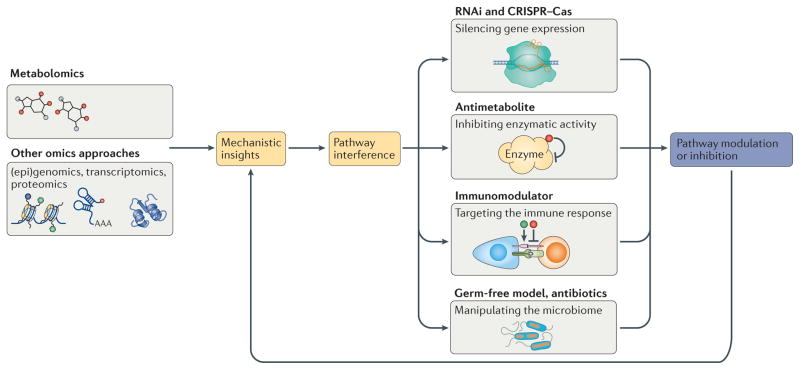
Figure 2. Controlling and influencing metabolism: perspectives from metabolomics.
Using various orthogonal techniques, targets identified with metabolomics can be further verified and investigated in more detail. For instance, other ‘omics’ approaches, including (epi)genomics, transcriptomics and proteomics, can reveal further mechanistic insights into phenotypical changes associated with the metabolite. Various orthogonal techniques also allow targeting of metabolic pathways and can be used to influence metabolite levels and to interfere with metabolic pathways. These approaches can be directed at the gene level and aimed at silencing gene expression, with techniques like CRISPR–Cas-mediated knock outs or RNA interference (RNAi). Alternatively, metabolic pathways can be influenced at at the protein level with the use of antimetabolites. Manipulating sources of exposure to different stimuli can also influence the metabolome, providing further mechanistic insights. For instance, using antibiotics or germ-free models with species-specific inoculation reveals the direct effect of the microbiome on metabolite production. Similarly, immunomodulators can be used to change the efficacy of the host immune system to respond to both the resident microbiota and pathogens, and their metabolic products. This collectively opens up possibilities for better understanding and, eventually, controlling metabolism.
Leading on from multi-layered omics approaches, there are a number of additional orthogonal techniques that can be used to further investigate the biological relationships between metabolites, proteins and genes (FIG. 2). At the gene level, RNA interference (RNAi) or CRISPR–Cas systems can be used to modulate gene expression, and this can help to determine how genes directly affect enzyme activity and metabolite production. Similarly, at the protein level, structural analogues of essential metabolites — so-called antimetabolites — can be used to inhibit a specific metabolic process and attenuate metabolite production or transportation from the cell62, thereby allowing investigation of the function and importance of specific metabolites63. Other approaches that can be used are those that directly change the host metabolome, for instance, through modulating the exposure of the organism to certain stimuli. For example, manipulating the microbiome using germ-free models, antibiotics or immunomodulators (which can change the host response to the resident microbiota) can reveal how bacteria and their metabolites affect the host and their metabolism and can allow us to link these changes to susceptibility to certain diseases60. As an example, it has been shown recently that the microbiome is important for the efficacy of immunotherapeutics used in cancer therapy, and that only in individuals harbouring certain bacterial species can these compounds lead to efficient stimulation of cancer-fighting T cells64,65. Of note, T cells are known to have distinct energy requirements depending on their activation status, with naive T cells utilizing oxidative phosphorylation for ATP generation, and effector (activated) T cells consuming glucose by aerobic glycolysis and glutaminolysis to support cell growth, in a similar manner to cancer cells66,67. Altogether, targeted manipulation of the local cellular environment to affect cellular energy status, in concert with modulation of the microbiome, opens up interesting possibilities to influence the survival of both effector T cells and cancer cells68.
Novel biological insights
Advances in metabolomic analysis have allowed us to gain a novel understanding of metabolism for various states, processes and diseases, and a few of the most recent studies exemplifying the novel biological insights that can be gained with the use of metabolomics are discussed below. These studies collectively show how information at the metabolite level, particularly when combined with other techniques, can lead to successful association of metabolites with phenotypical causality, thus bringing us closer to a mechanistic understanding of metabolism.
Role of bacterial biofilms in cancer
A recent study carried out on a patient population investigated in more detail a previously validated biomarker for colon cancer, N1, N12–diacetylspermine (DAS)69. In this study, a multidisciplinary approach was used that combined four different metabolomic tools with traditional biochemical techniques. First, it revealed that only DAS, and not its precursors, was correlated with biofilm presence as well as with colon cancer, and that DAS is probably a metabolic end-product of polyamine metabolism. The metabolomic approaches used included untargeted analysis (BOX 1) to compare normal tissues to the tumour tissues, both of which were either associated with or devoid of biofilms. This was followed by a targeted validation step (BOX 1) to confirm the fold change in metabolites and expand the analysis to other metabolites in related pathways. Nanostructure-imaging mass spectrometry (NIMS)70 (BOX 1) revealed the in situ localization of DAS in the mucosal layer of the colon where the biofilms resided. Global isotope metabolomics was further used to investigate the metabolic fate of a stable isotope of DAS in colon cancer cell lines, confirming that it is indeed an end-product of metabolism and is not involved in any other metabolic pathways.
In order to determine the source of the metabolite (the patient versus the biofilm), patients were treated with antibiotics to remove the biofilms (this was confirmed by fluorescent in situ hybridization (FISH) analysis), and their samples were analysed for the presence of DAS. In these tissues, DAS concentrations were similar to those previously measured in biofilm-negative patients, showing that the elevated DAS levels seen in biofilm-positive patients originated from the biofilms. In line with this, immunohistochemical analysis of patient samples did not show any change in protein levels of enzymes involved in DAS production. As DAS is a metabolite of polyamine precursors, and polyamines have been associated with various cellular responses including increased cellular proliferation, the propensity of colon cells to overproliferate in the presence of biofilms was investigated and confirmed by immunohistochemistry. In addition, immunofluorescence revealed the presence of pro-inflammatory cytokines in biofilm-covered tissues. This inflammatory state was observed in normal-looking tissues that were associated with biofilms, suggesting that such tissues might be in a pro-carcinogenic state and that biofilm formation indeed promotes colon tumorigenesis71. In sum, this example shows how a combination of several metabolomic approaches with orthogonal biological techniques can be used for the initial metabolite discovery, leading to the elucidation of the potential role of biofilms in colon carcinogenesis (FIG. 3). According to this study, colonic bacteria utilize polyamines to build biofilms (producing DAS), and this biofilm formation induces pro-inflammatory and pro-carcinogenic effects in the host tissues, increasing the risk of tumour formation. Interestingly, some metabolomic studies have associated DAS with other cancers, including cancers of the lung72, breast73, blood74 and bladder75, as well as identifying it as a dietary metabolite76. Thus, further studies assessing the roles of diet and bacteria in cancers are of the utmost importance.
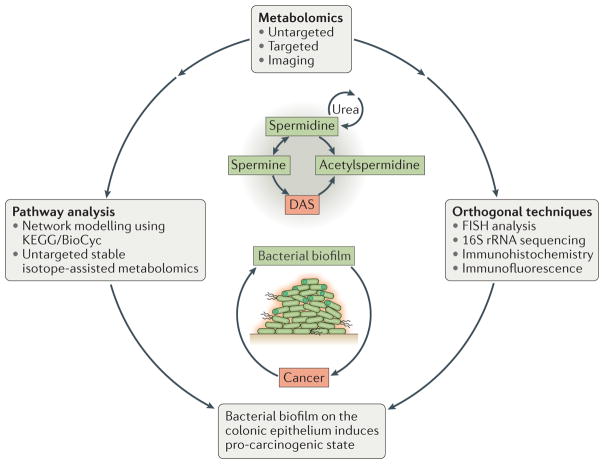
Figure 3. Novel biological insights.
Diacetylspermine (DAS) has a role in biofilm-associated colon cancer. Various metabolomic and orthogonal biological techniques contributed to the association of DAS with bacterial biofilms and their role in the pathology of cancer. Fluorescence in situ hybridization (FISH) analysis and 16S rRNA sequencing identified the presence of bacterial species and biofilms on colon tissues. Untargeted and targeted metabolomics identified and validated the association of polyamine metabolites with colon cancer tissues. Stratification by biofilm status showed that DAS was upregulated primarily in biofilm-associated tissues, which was confirmed by mass spectrometry imaging. Network modelling using the KEGG and BioCyc databases, and pathway analysis using untargeted stable-isotope assisted metabolomics, showed that DAS is an end-product of polyamine metabolism. For further analysis, orthogonal techniques were used. Immunohistochemistry and immunofluorescence revealed increased cellular proliferation and pro-inflammatory cytokines in biofilm-associated tissues. The combination of these techniques led to the conclusion that bacterial biofilms induce a pro-carcinogenic state in the colon epithelium.
Metabolic regulation of cell pluripotency
At the epigenetic level, metabolites have been shown to regulate pluripotency in human ES cells, with a recent study revealing a metabolic switch during the transition between human naive and primed ES cells2. It has been found that this switch is regulated by nicotinamide N–methyltransferase (NNMT), which controls SAM levels that are required for histone methylation. Analysis of oxygen consumption rates revealed that primed human ES cells have a lower mitochondrial respiration capacity than naive human ES cells, and transcriptomic analysis confirmed a downregulation of mitochondrial electron transport chain genes in the primed state. The transition from naive to primed human ES cells also involved reduced WNT signalling and increased hypoxia-inducible factor 1α (HIF1α) stabilization (shown by proteomic analysis). Untargeted and targeted metabolomics based on gas chromatography and liquid chromatography mass spectrometry (GC–MS and LC–MS) (BOX 1) revealed concomitant changes in metabolic pathways, including glycolysis, fatty acid β-oxidation and lipid biosynthesis. Transcriptomic and genomic analyses showed that the genes involved in these pathways were also changed. The use of WNT inhibitors and the generation of HIF1α-knockout cells by CRISPR–Cas gene editing further demonstrated that WNT activity is required for the naive state, and that HIF1α is required for human ES cell transition to the primed state. Furthermore, the loss of NNMT in naive human ES cells was associated with an increase in repressive histone marks (histone 3 Lys27 trimethylation; H3K27me3) in developmental and metabolic genes that regulate the metabolic switch in naive to primed cells. Collectively, this comprehensive analysis showed that both NNMT and the metabolic state regulate ES cell development.
Novel therapy for cardiovascular disease
Another example shows how using metabolomics, together with other techniques, can lead to the establishment of a new therapeutic approach — in this case, for decreasing the risk of CVD. Initially, using untargeted metabolomics and then targeted metabolomics for validation and quantification (BOX 1), an association between an increased risk of CVD and plasma concentrations of choline, betaine and TMAO was established19,77,78. This was further replicated in apolipoprotein E−/− mice, a mouse model that is highly susceptible to the formation of atherosclerotic plaques — the primary cause of CVD — that were fed high-choline and high-TMAO diets, showing a significant correlation between plasma TMAO and the formation of atherosclerotic plaques. Functional experiments revealed that trimethylamine (TMA)-containing nutrients such as choline, phosphatidylcholine and carnitine are dietary precursors for TMAO, and that liver flavin monooxygenases (FMOs; primarily FMO3) are responsible for converting TMA to TMAO. Analysis of antibiotic-treated mice, together with the observation that the risk of CVD was transmissible upon microbial transfer, led to the conclusion that the microbiome generates TMA. As inhibition of FMO3 can produce side-effects and thus does not provide a sustainable therapy, the next step was to search for an inhibitor of microbial TMA production and investigate its potential as a therapeutic for CVD. Using a structural analogue to choline, 3,3–dimethyl-1–butanol (DMB), found in extra-virgin olive oil, it was possible to inhibit microbial TMA lyases, which are responsible for TMA formation. In vivo experiments showed that TMAO levels were indeed reduced in mice fed with high-choline or high-carnitine diets when these mice were simultaneously treated with DMB. Treatment with DMB also prevented atherosclerotic lesion development in apolipoprotein E−/− mice on a choline-enhanced diet79. Altogether, this work led to the proposal of a novel therapy for CVD, which bypasses the issues that arise when using inhibitors targeted to a patient’s own proteins — an approach potentially resulting in various side-effects for the patient. Instead, this study showed that harmful metabolites can be inhibited at their earliest production, by ‘drugging’ the gut microbiome, which in the case of CVD is the source of the metabolite contributing to the disease.
Metabolite-driven regulation of β–cells
An important metabolite, 3–carboxy-4–methyl-5–propyl-2–furanpropanoic acid (CMPF), was recently identified in the plasma of humans with gestational diabetes, as well as in those with impaired glucose tolerance and type 2 diabetes80. CMPF was identified by untargeted and targeted metabolomic analysis (BOX 1), with further validation by enzyme-linked immunosorbent assay (ELISA).
Mice treated with CMPF at doses comparable to levels found in human individuals with diabetes developed glucose intolerance and impaired insulin secretion after an oral glucose-tolerance test. This was monitored using targeted mass spectrometry and ELISA to measure plasma and tissue CMPF concentrations, and also by glucose-stimulated insulin secretion (GSIS) tests. Mechanistically, CMPF was shown to impair mitochondrial function, decrease glucose-induced ATP synthesis and induce oxidative stress, as assessed by measuring mitochondrial membrane potential and with fluorescence- and bioluminescence-based assays, as well as gene expression analysis. Inhibitors of organic anion transporters (OAT), which are responsible for the clearance of CMPF, blocked the transportation of CMPF into β–cells of the pancreas and prevented β–cell dysfunction. In line with this, treatment of pancreatic islets isolated from OAT3-knockout mouse models with CMPF had no effect on insulin content or GSIS. Altogether, the metabolite CMPF, identified by metabolomic analysis, provides a mechanistic link between β–cell dysfunction and diabetes and has been shown to function through impairing mitochondrial function and inhibiting insulin biosynthesis.
Mechanism of ischaemia–reperfusion injury
Steady state flux analysis was recently used to help to identify the mechanisms of ischaemia–reperfusion injury, which is a type of tissue damage resulting from oxidative stress and generation of reactive oxygen species (ROS) following the return of circulation to tissue regions previously deprived of oxygen. It was revealed that succinate, which is a metabolite of the tricarboxylic acid (TCA) cycle, is the driver of ROS generation, which can lead to heart attack and stroke following ischaemia–reperfusion injury81. The authors also used a combination of untargeted and targeted metabolomics (BOX 1) to reveal an elevation of succinate levels across several organs in a mouse model of ischaemia. Mechanistic studies involving in silico modelling, mitochondrial membrane potential measurements, ratiometric assessment and fluorescence assays revealed that in ischaemia, succinate dehydrogenase (SDH) functions in reverse, accumulating succinate from fumarate. Upon reperfusion, succinate is oxidized and drives electrons back through the mitochondrial complex I, thus generating ROS. Together, these findings indicated that SDH could be a target for the prevention of ROS accumulation following reperfusion of ischaemic tissue. Accordingly, antimetabolite inhibitors of SDH prevented succinate accumulation, inhibiting electron flow through complex I and subsequent ROS production, and thereby providing protection from ischaemia–reperfusion injury.
Regulation of cancer cell metabolism
In addition to the previous example, metabolic flux analysis was recently used to investigate the role of mitochondrial enzyme serine hydroxymethyltransferase (SHMT2) in human glioblastoma cells. Specifically, the roles of SHMT2 in central carbon metabolism and in regulating pyruvate kinase M2 (PKM2) activity were investigated and were further linked to glioma cell survival82. In these experiments, SHMT2-knockdown cells were treated with uniformly labelled 13C-glucose and showed increased flux from pyruvate to lactate, citrate and alanine, with a concomitant increase in PKM2 activity and oxygen consumption rate. In addition, overexpression of RNAi-resistant SHMT2 cDNA reverted these effects, confirming that SHMT2 negatively affects PKM2. Thus, the stable isotope analysis showed that SHMT2 expression changes the metabolism of cancer cells and limits carbon flux into the TCA cycle via suppression of PKM2. This has been further shown to improve the survival of cells in ischaemic tumour regions. In addition, the study showed that the survival of cancer cells with high SHMT2 expression can be impaired if glycine decarboxylase is inhibited, as this causes accumulation of glycine, which then contributes to the production of toxic metabolites. Altogether, this series of experiments provided novel insights into cancer cell metabolism and demonstrated how metabolic changes can affect cell properties and responses — in this case, cell survival.
Future perspectives
Metabolomics is an exciting and evolving research area, with numerous success stories demonstrating that its power extends from biomarker discovery to understanding the mechanisms that underlie phenotypes. This step towards mechanistic understanding has been made possible by advances in analytical technologies and informatics, and the combination of these tools has generated novel insights into chemical physiology. It has also been made possible as metabolomics has become more widely used in combination with orthogonal technologies, such as genomics, proteomics, structural biology and imaging, as well as with various techniques that allow us to modify gene expression, enzymatic activity, cell signalling or whole metabolic pathways, including the contribution of the naturally occurring microbiota. Thus, the future prospects of metabolomics lie not only in the unique information it provides, but in its integration into systems biology.
Supplementary Material
Acknowledgments
The authors would like to thank Nadine Levin at UCLA for her comments on the manuscript. Funding for this work was supported by US National Institutes of Health (NIH) grants R01 GM114368 and PO1 A1043376–02S1.
Footnotes
Competing interests statement
The authors declare no competing interests.
FURTHER INFORMATION
Common Fund Metabolomics Program: https://commonfund.nih.gov/metabolomics
Coordination of Standards in Metabolomics (COSMOS): http://www.cosmos-fp7.eu/
DeviumWeb: https://github.com/dgrapov/DeviumWeb
MetaboAnalyst: http://www.metaboanalyst.ca/
MZmine 2: http://mzmine.github.io/
XCMS Online: https://xcmsonline.scripps.edu/
Contributor Information
Caroline H. Johnson, Department of Environmental Health Sciences, Yale School of Public Health, Yale University, 60 College Street, New Haven, Connecticut 06520, USA
Julijana Ivanisevic, Metabolomics Research Platform, Faculty of Biology and Medicine, University of Lausanne, Rue du Bugnon 19, 1005 Lausanne, Switzerland.
Gary Siuzdak, Scripps Center for Metabolomics and Mass Spectrometry, Departments of Chemistry, Molecular and Computational Biology, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, California 92037, USA.
References
- 1.Johnson CH, Patterson AD, Idle JR, Gonzalez FJ. Xenobiotic metabolomics: major impact on the metabolome. Annu Rev Pharmacol Toxicol. 2012;52:37–56. doi: 10.1146/annurev-pharmtox-010611-134748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sperber H, et al. The metabolome regulates the epigenetic landscape during naive–to–primed human embryonic stem cell transition. Nat Cell Biol. 2015;17:1523–1535. doi: 10.1038/ncb3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yanes O, et al. Metabolic oxidation regulates embryonic stem cell differentiation. Nat Chem Biol. 2010;6:411–417. doi: 10.1038/nchembio.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karlic H, et al. Inhibition of the mevalonate pathway affects epigenetic regulation in cancer cells. Cancer Genet. 2015;208:241–252. doi: 10.1016/j.cancergen.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carey BW, Finley LW, Cross JR, Allis CD, Thompson CB. Intracellular α-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature. 2015;518:413–416. doi: 10.1038/nature13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ulanovskaya OA, Zuhl AM, Cravatt BF. NNMT promotes epigenetic remodeling in cancer by creating a metabolic methylation sink. Nat Chem Biol. 2013;9:300–306. doi: 10.1038/nchembio.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wellen KE, et al. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakahata Y, et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gornall AG, editor. Applied Biochemistry of Clinical Disorders. Lippincott Williams & Wilkins; 1986. [Google Scholar]
- 10.Richieri GV, Kleinfeld AM. Unbound free fatty–acid levels in human serum. J Lipid Res. 1995;36:229–240. [PubMed] [Google Scholar]
- 11.Li X, Gianoulis TA, Yip KY, Gerstein M, Snyder M. Extensive in vivo metabolite-protein interactions revealed by large-scale systematic analyses. Cell. 2010;143:639–650. doi: 10.1016/j.cell.2010.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hubbard TD, et al. Adaptation of the human aryl hydrocarbon receptor to sense microbiota-derived indoles. Sci Rep. 2015;5:12689. doi: 10.1038/srep12689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharma M, Astekar M, Soi S, Manjunatha BS, Shetty DC. pH gradient reversal: an emerging hallmark of cancers. Recent Pat Anticancer Drug Discov. 2015;10:244–258. doi: 10.2174/1574892810666150708110608. [DOI] [PubMed] [Google Scholar]
- 14.Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol. 2014;12:661–672. doi: 10.1038/nrmicro3344. [DOI] [PubMed] [Google Scholar]
- 15.Brahimi-Horn MC, Laferriere J, Mazure N, Pouyssegur J. In: Tumor Angiogenesis: Basic Mechanisms and Cancer Therapy. Marme D, Fusenig N, editors. Springer-Verlag; Berlin Heidelberg: 2008. p. 186. [Google Scholar]
- 16.Ivanisevic J, et al. Arteriovenous blood metabolomics: a readout of intra-tissue metabostasis. Sci Rep. 2015;5:12757. doi: 10.1038/srep12757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ivanisevic J, et al. Brain region mapping using global metabolomics. Chem Biol. 2014;21:1575–1584. doi: 10.1016/j.chembiol.2014.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson CH, Gonzalez FJ. Challenges and opportunities of metabolomics. J Cell Physiol. 2012;227:2975–2981. doi: 10.1002/jcp.24002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koeth RA, et al. Intestinal microbiota metabolism of l-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med. 2013;19:576–585. doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pannkuk EL, Laiakis EC, Authier S, Wong K, Fornace AJ., Jr Global metabolomic identification of long-term dose-dependent urinary biomarkers in nonhuman primates exposed to ionizing radiation. Radiat Res. 2015;184:121–133. doi: 10.1667/rr14091.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson CH, et al. Radiation metabolomics. 5 identification of urinary biomarkers of ionizing radiation exposure in nonhuman primates by mass spectrometry-based metabolomics. Radiat Res. 2012;178:328–340. doi: 10.1667/rr2950.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson CH, et al. Radiation metabolomics. 4 UPLC-ESI-QTOFMS-based metabolomics for urinary biomarker discovery in gamma-irradiated rats. Radiat Res. 2011;175:473–484. doi: 10.1667/RR2437.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamdalla MA, Ammar RA, Rajasekaran S. A molecular structure matching approach to efficient identification of endogenous mammalian biochemical structures. BMC Bioinformatics. 2015;16:S11. doi: 10.1186/1471-2105-16-S5-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolf S, Schmidt S, Muller-Hannemann M, Neumann S. In silico fragmentation for computer assisted identification of metabolite mass spectra. BMC Bioinformatics. 2010;11:148. doi: 10.1186/1471-2105-11-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ridder L, et al. Automatic chemical structure annotation of an LC–MSn based metabolic profile from green tea. Anal Chem. 2013;85:6033–6040. doi: 10.1021/ac400861a. [DOI] [PubMed] [Google Scholar]
- 26.Vinaixa M, et al. Mass spectral databases for LC/MS-and GC/MS–based metabolomics: state of the field and future prospects. Trends Analyt Chem. 2015 http://dx.doi.org/10.1016/j.trac.2015.09.005.
- 27.Rocca-Serra P, et al. Data standards can boost metabolomics research, and if there is a will, there is a way. Metabolomics. 2016;12:14. doi: 10.1007/s11306-015-0879-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ellis JK, et al. Metabolic profiling detects early effects of environmental and lifestyle exposure to cadmium in a human population. BMC Med. 2012;10:61. doi: 10.1186/1741-7015-10-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scalbert A, et al. The food metabolome: a window over dietary exposure. Am J Clin Nutr. 2014;99:1286–1308. doi: 10.3945/ajcn.113.076133. [DOI] [PubMed] [Google Scholar]
- 30.Wishart DS, et al. HMDB 3.0 — the human metabolome database in 2013. Nucleic Acids Res. 2013;41:D801–D807. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ji Y, et al. Glycine and a glycine dehydrogenase (GLDC) SNP as citalopram/escitalopram response biomarkers in depression: pharmacometabolomics-informed pharmacogenomics. Clin Pharmacol Ther. 2011;89:97–104. doi: 10.1038/clpt.2010.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sreekumar A, et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457:910–914. doi: 10.1038/nature07762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 33.Johnson CH, Ivanisevic J, Benton HP, Siuzdak G. Bioinformatics: the next frontier of metabolomics. Anal Chem. 2015;87:147–156. doi: 10.1021/ac5040693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thiele I, et al. A community-driven global reconstruction of human metabolism. Nat Biotechnol. 2013;31:419–425. doi: 10.1038/nbt.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zamboni N, Saghatelian A, Patti GJ. Defining the metabolome: size, flux, and regulation. Mol Cell. 2015;58:699–706. doi: 10.1016/j.molcel.2015.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mathe EA, et al. Noninvasive urinary metabolomic profiling identifies diagnostic and prognostic markers in lung cancer. Cancer Res. 2014;74:3259–3270. doi: 10.1158/0008-5472.CAN-14-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu Y, et al. Multilayered genetic and omics dissection of mitochondrial activity in a mouse reference population. Cell. 2014;158:1415–1430. doi: 10.1016/j.cell.2014.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shin SY, et al. An atlas of genetic influences on human blood metabolites. Nat Genet. 2014;46:543–550. doi: 10.1038/ng.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Misra BB, van der Hooft JJ. Updates in metabolomics tools and resources: 2014–2015. Electrophoresis. 2016;37:86–110. doi: 10.1002/elps.201500417. [DOI] [PubMed] [Google Scholar]
- 40.Salek RM, et al. COordination of Standards in MetabOlomicS (COSMOS): facilitating integrated metabolomics data access. Metabolomics. 2015;11:1587–1597. doi: 10.1007/s11306-015-0810-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pluskal T, Castillo S, Villar-Briones A, Oresic M. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics. 2010;11:395. doi: 10.1186/1471-2105-11-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tautenhahn R, Patti GJ, Rinehart D, Siuzdak G. XCMS online: a web-based platform to process untargeted metabolomic data. Anal Chem. 2012;84:5035–5039. doi: 10.1021/ac300698c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xia JG, Psychogios N, Young N, Wishart DS. MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009;37:W652–W660. doi: 10.1093/nar/gkp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Caesar R, Nygren H, Oresic M, Backhed F. Interaction between dietary lipids and gut microbiota regulates hepatic cholesterol metabolism. J Lipid Res. 2016 doi: 10.1194/jlr.M065847. http://dx.doi.org/10.1194/jlr.M065847. [DOI] [PMC free article] [PubMed]
- 45.Paris LP, et al. Global metabolomics reveals metabolic dysregulation in ischemic retinopathy. Metabolomics. 2016;12:15. doi: 10.1007/s11306-015-0877-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li S, et al. Predicting network activity from high throughput metabolomics. PLoS Comput Biol. 2013;9:e1003123. doi: 10.1371/journal.pcbi.1003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cottret L, et al. MetExplore: a web server to link metabolomic experiments and genome-scale metabolic networks. Nucleic Acids Res. 2010;38:W132–W137. doi: 10.1093/nar/gkq312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Metallo CM, Vander Heiden MG. Understanding metabolic regulation and its influence on cell physiology. Mol Cell. 2013;49:388–398. doi: 10.1016/j.molcel.2013.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Buescher JM, et al. A roadmap for interpreting 13C metabolite labeling patterns from cells. Curr Opin Biotechnol. 2015;34:189–201. doi: 10.1016/j.copbio.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fan J, et al. Quantitative flux analysis reveals folate-dependent NADPH production. Nature. 2014;510:298–302. doi: 10.1038/nature13236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sellers K, et al. Pyruvate carboxylase is critical for non-small-cell lung cancer proliferation. J Clin Invest. 2015;125:687–698. doi: 10.1172/JCI72873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang X, et al. X13CMS: global tracking of isotopic labels in untargeted metabolomics. Anal Chem. 2014;86:1632–1639. doi: 10.1021/ac403384n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bueschl C, et al. A novel stable isotope labelling assisted workflow for improved untargeted LC–HRMS based metabolomics research. Metabolomics. 2014;10:754–769. doi: 10.1007/s11306-013-0611-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Creek DJ, et al. Stable isotope-assisted metabolomics for network-wide metabolic pathway elucidation. Anal Chem. 2012;84:8442–8447. doi: 10.1021/ac3018795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Capellades J, et al. geoRge: a computational tool to detect the presence of stable isotope labeling in LC/MS–based untargeted metabolomics. Anal Chem. 2016;88:621–628. doi: 10.1021/acs.analchem.5b03628. [DOI] [PubMed] [Google Scholar]
- 56.Chen YJ, et al. Differential incorporation of glucose into biomass during Warburg metabolism. Biochemistry. 2014;53:4755–4757. doi: 10.1021/bi500763u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Creek DJ, et al. Probing the metabolic network in bloodstream-form Trypanosoma brucei using untargeted metabolomics with stable isotope labelled glucose. PLoS Pathog. 2015;11:e1004689. doi: 10.1371/journal.ppat.1004689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zelezniak A, Sheridan S, Patil KR. Contribution of network connectivity in determining the relationship between gene expression and metabolite concentration changes. PLoS Comput Biol. 2014;10:e1003572. doi: 10.1371/journal.pcbi.1003572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hsiao EY, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155:1451–1463. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ridaura VK, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341:1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.O’Keefe SJ, et al. Fat, fibre and cancer risk in African Americans and rural Africans. Nat Commun. 2015;6:6342. doi: 10.1038/ncomms7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Woolley DW. A Study of Antimetabolites. John Wiley & Sons; 1952. [Google Scholar]
- 63.Johnson CH, et al. Alterations in spinal cord metabolism during treatment of neuropathic pain. J Neuroimmune Pharmacol. 2015;10:396–401. doi: 10.1007/s11481-015-9624-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sivan A, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti–PD–L1 efficacy. Science. 2015;350:1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vetizou M, et al. Anticancer immunotherapy by CTLA–4 blockade relies on the gut microbiota. Science. 2015;350:1079–1084. doi: 10.1126/science.aad1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Frauwirth KA, Thompson CB. Regulation of T lymphocyte metabolism. J Immunol. 2004;172:4661–4665. doi: 10.4049/jimmunol.172.8.4661. [DOI] [PubMed] [Google Scholar]
- 67.van Stipdonk MJB, et al. Dynamic programming of CD8+ T lymphocyte responses. Nat Immunol. 2003;4:361–365. doi: 10.1038/ni912. [DOI] [PubMed] [Google Scholar]
- 68.Mockler MB, Conroy MJ, Lysaght J. Targeting T cell immunometabolism for cancer immunotherapy; understanding the impact of the tumor microenvironment. Front Oncol. 2014;4:107. doi: 10.3389/fonc.2014.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Johnson CH, et al. Metabolism links bacterial biofilms and colon carcinogenesis. Cell Metab. 2015;21:891–897. doi: 10.1016/j.cmet.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Northen TR, et al. Clathrate nanostructures for mass spectrometry. Nature. 2007;449:1033–1036. doi: 10.1038/nature06195. [DOI] [PubMed] [Google Scholar]
- 71.Dejea CM, et al. Microbiota organization is a distinct feature of proximal colorectal cancers. PNAS. 2014;111:18321–18326. doi: 10.1073/pnas.1406199111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wikoff WR, et al. Diacetylspermine is a novel prediagnostic serum biomarker for non-small-cell lung cancer and has additive performance with pro-surfactant protein B. J Clin Oncol. 2015;33:3880–3886. doi: 10.1200/JCO.2015.61.7779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Umemori Y, et al. Evaluating the utility of N1,N12-diacetylspermine and N1,N8-diacetylspermidine in urine as tumor markers for breast and colorectal cancers. Clin Chim Acta. 2010;411:1894–1899. doi: 10.1016/j.cca.2010.07.018. [DOI] [PubMed] [Google Scholar]
- 74.Lee SH, Suh JW, Chung BC, Kim SO. Polyamine profiles in the urine of patients with leukemia. Cancer Lett. 1998;122:1–8. doi: 10.1016/s0304-3835(97)00399-6. [DOI] [PubMed] [Google Scholar]
- 75.Stejskal D, et al. Evaluation of urine N1,N12–diacetylspermine as potential tumor marker for urinary bladder cancer. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2006;150:235–237. doi: 10.5507/bp.2006.033. [DOI] [PubMed] [Google Scholar]
- 76.Vargas AJ, Ashbeck EL, Thomson CA, Gerner EW, Thompson PA. Dietary polyamine intake and polyamines measured in urine. Nutr Cancer. 2014;66:1144–1153. doi: 10.1080/01635581.2014.949801. [DOI] [PubMed] [Google Scholar]
- 77.Wang Z, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tang WH, et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med. 2013;368:1575–1584. doi: 10.1056/NEJMoa1109400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang Z, et al. Non-lethal inhibition of gut microbial trimethylamine production for the treatment of atherosclerosis. Cell. 2015;163:1585–1595. doi: 10.1016/j.cell.2015.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Prentice KJ, et al. The furan fatty acid metabolite CMPF is elevated in diabetes and induces β cell dysfunction. Cell Metab. 2014;19:653–666. doi: 10.1016/j.cmet.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 81.Chouchani ET, et al. Ischaemic accumulation of succinate controls reperfusion injury through mitochondrial ROS. Nature. 2014;515:431–435. doi: 10.1038/nature13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kim D, et al. SHMT2 drives glioma cell survival in ischaemia but imposes a dependence on glycine clearance. Nature. 2015;520:363–367. doi: 10.1038/nature14363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Siuzdak G. The Expanding Role of Mass Spectrometry in Biotechnology. MCC Press; 2006. [Google Scholar]
- 84.Tanaka K, et al. Protein and polymer analyses up to m/z 100 000 by laser ionization time–of–flight mass spectrometry. Rapid Commun Mass Spectrom. 1988;2:151–153. [Google Scholar]
- 85.Siuzdak GE, Buriak J, Wei J. Desorption/ionization of analytes from porous light-absorbing semiconductor. 6288390 B1. US Patent. 2000
- 86.Wiseman JM, Ifa DR, Song Q, Cooks RG. Tissue imaging at atmospheric pressure using desorption electrospray ionization (DESI) mass spectrometry. Angew Chem Int Ed Engl. 2006;45:7188–7192. doi: 10.1002/anie.200602449. [DOI] [PubMed] [Google Scholar]
- 87.Kraft ML, Weber PK, Longo ML, Hutcheon ID, Boxer SG. Phase separation of lipid membranes analyzed with high-resolution secondary ion mass spectrometry. Science. 2006;313:1948–1951. doi: 10.1126/science.1130279. [DOI] [PubMed] [Google Scholar]
- 88.Gowda H, et al. Interactive XCMS Online: simplifying advanced metabolomic data processing and subsequent statistical analyses. Anal Chem. 2014;86:6931–6939. doi: 10.1021/ac500734c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Smith CA, et al. METLIN — a metabolite mass spectral database. Ther Drug Monit. 2005;27:747–751. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- 90.Wishart DS, et al. HMDB: the human metabolome database. Nucleic Acids Res. 2007;35:D521–D526. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Horai H, et al. MassBank: a public repository for sharing mass spectral data for life sciences. J Mass Spectrom. 2010;45:703–714. doi: 10.1002/jms.1777. [DOI] [PubMed] [Google Scholar]
- 92.Kopka J, et al. GMD@CSB.DB: the Golm Metabolome Database. Bioinformatics. 2005;21:1635–1638. doi: 10.1093/bioinformatics/bti236. [DOI] [PubMed] [Google Scholar]
- 93.Gerlich M, Neumann S. MetFusion: integration of compound identification strategies. J Mass Spectrom. 2013;48:291–298. doi: 10.1002/jms.3123. [DOI] [PubMed] [Google Scholar]
- 94.Paglia G, et al. Ion mobility derived collision cross sections to support metabolomics applications. Anal Chem. 2014;86:3985–3993. doi: 10.1021/ac500405x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ogata H, et al. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Karp PD, et al. Expansion of the BioCyc collection of pathway/genome databases to 160 genomes. Nucleic Acids Res. 2005;33:6083–6089. doi: 10.1093/nar/gki892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xia J, Wishart DS. MSEA: a web-based tool to identify biologically meaningful patterns in quantitative metabolomic data. Nucleic Acids Res. 2010;38:W71–W77. doi: 10.1093/nar/gkq329. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.