Abstract
Bacteroides fragilis, an important component of the human gastrointestinal microbiota, can cause lethal extra-intestinal infection upon escape from the gastrointestinal tract. We demonstrated transfer and recombination of large chromosomal segments from B. fragilis HMW615, a multidrug resistant clinical isolate, to B. fragilis 638R. In one example, the transfer of a segment of ~435 Kb/356 genes replaced ~413 Kb/326 genes of the B. fragilis 638R chromosome. In addition to transfer of antibiotic resistance genes, these transfers (1) replaced complete divergent polysaccharide biosynthesis loci; (2) replaced DNA inversion-controlled intergenic shufflons (that control expression of genes encoding starch utilization system outer membrane proteins) with more complex, divergent shufflons; and (3) introduced additional intergenic shufflons encoding divergent Type 1 restriction/modification systems. Conjugative transposon-like genes within a transferred segment and within a putative integrative conjugative element (ICE5) ~45 kb downstream from the transferred segment both encode proteins that may be involved in the observed transfer. These data indicate that chromosomal transfer is a driver of antigenic diversity and nutrient adaptation in Bacteroides that (1) contributes to the dissemination of the extensive B. fragilis pan-genome, (2) allows rapid adaptation to a changing environment and (3) can confer pathogenic characteristics to host symbionts.
Keywords: Bacteroides fragilis, horizontal transmission, restriction modification system, polysaccharide capsules
Abbreviations
BF, Bacteroides fragilis; CTn, Conjugative Transposon; HGT, Horizontal Gene Transfer; ICE, Integrative Conjugative Element; MGE, Mobile Genetic Element; ori, origin of replication; oriT, origin of transfer; PS, Polysaccharide; R-M, Restriction-Modification.
Data Summary
All supporting data, code and protocols have been provided within the article or through supplementary data files.
Impact Statement
Bacteroides fragilis has a formidable pan-genome which facilitates adaptation to distinct ecological niches, interactions with the host immune system within the GI tract and also enables the transition of the bacterium from symbiont to pathogen. The novel horizontal gene transfer (HGT) event reported here resulted in the insertion of the largest segment of Bacteroides chromosomal DNA ever reported and effected changes in the polysaccharide loci profile, control of nutrient binding systems and phage defense systems. The scale and mechanism of this transfer and potential phenotypic impact has not been demonstrated before and represents a mechanism for dissemination of the Bacteroides pan-genome as well as rapid adaptation to a changing host environment.
Transfer of these chromosomal segments (1) replaced either one (in the case of tetQ2435 and tetQ2356) or two (in the case of tetQ2482) entire micro-capsule operon(s) in B. fragilis 638R with homologous but divergent micro-capsule operons from B. fragilis HMW615, (2) replaced a shufflon that controls nutrient binding and outer membrane protein expression with a divergent and more complex shufflon and (3) introduced a second divergent R-M system gene shufflon, thus contributing to the remarkable redundancy that is characteristic of the B. fragilis genome.
Introduction
Bacteroides fragilis, a Gram-negative anaerobic rod, is a commensal in the human gut [1] that can cause severe infections when it escapes its niche [2]. One of the remarkable traits of B. fragilis is its genetic plasticity, due in part to frequent genetic rearrangements; these include inversions, duplications and horizontal gene transfer (HGT) mediated by a variety of mobile and mobilizable elements [3]. This genetic plasticity facilitates adaptation to distinct ecological niches [3–5] and enhances the spread of antibiotic resistance determinants [6, 7]. Another notable trait of B. fragilis is the extensive redundancy within its genome, including the wide variety of carbohydrate utilization systems that provide factors important both for colonization and energy metabolism [8, 9].
HGT is regarded as a major mechanism of prokaryotic evolution [10–15]; the scale of its importance has been more clearly delineated in recent years. HGT can occur by transformation (acquisition of DNA from the local environment), by phage-mediated transduction and by conjugation via cell-to-cell contact [16–19]. After transfer, the chromosomal segments can integrate into the recipient chromosome through either site-specific or homologous recombination [20]. The nomenclature of these horizontally transferred elements is currently in flux. The most inclusive term, Genomic Islands (GIs), describe HGT-acquired gene clusters that are often associated with adaptations to the niche of the organism and that contribute to evolutionary change [21]. GIs include the group described as integrative and conjugative elements (ICEs) which in turn includes the group of elements described as conjugative transposons (CTns) [17, 22].
In Bacteroides species, integrative and conjugative elements (ICE), including conjugative transposons (CTns), are considered important mediators of horizontal gene transfer (HGT) [23]. CTnDOT, for example, is responsible for transfer of tetracycline resistance. We recently reported a novel Bacteroides CTn, CTnHyb (isolated from B. fragilis HMW615) [24]. CTnHyb is a mosaic of mobile elements from Gram-positive bacteria and transferred tetracycline, kanamycin and metronidazole resistance genes as well as efflux pump genes that may also be associated with antibiotic resistance in B. fragilis [24, 25]. Remarkably, the extensive horizontal gene transfer among Bacteroides and apparent incorporation of genes from other genera occurs despite the diverse DNA restriction and modification systems in Bacteroides genomes. The presence of genes encoding homologues of the anti-restriction protein, ArdA, has been noted within some CTns. These proteins mimic B-form DNA and block binding of Type 1 R-M systems [26] and might help to explain why these elements can be transferred successfully between divergent strains.
The plasticity and ability of B. fragilis to adapt to various niches is due, in part, to modulation of surface antigenicity through variable production of multiple different surface proteins and capsular polysaccharides. Ten or more polysaccharide biosynthesis-associated loci may be present within an individual genome and at least eight of these are associated with production of a microcapsule; the genetic loci encoding these genes can be turned on or off by inversion of promoter sequences [27]. In addition, microcapsule expression is controlled by multiple transcriptional anti-terminators [28]; regulation of these multiple microcapsules is important for colonization of Bacteroides in the host [29, 30]. B. fragilis also exhibits an unprecedented between-strain antigenic diversity of polysaccharide expression as a result of extensive diversity of PS biosynthesis loci among strains; 28 different PS biosynthesis associated loci were identified in just three isolates [28]. This suggests that B. fragilis has a pan-genome with an extensive pool of polysaccharide biosynthesis-associated genes.
The mode of generating polysaccharide diversity between and amongst strains has not been clarified. CTns have been reported to be associated with PS biosynthesis locus duplication within a B. vulgatus genome [31] and CTn and bacteriophage insertion have been observed to disrupt PS synthesis loci [28, 31]. However, the precise mechanism(s) generating the HGT underpinning the observed diversity of PS synthesis loci has, to date, been open to speculation. We show here that large scale chromosomal transfer, likely as a result of conjugation and integration via homologous recombination, mediated replacement of a complete PS biosynthesis region with a divergent PS biosynthesis region, replaced an intergenic shufflon which controls variable transcription of genes encoding outer membrane proteins, and introduced a second shufflon encoding a variable Type 1 restriction and modification (R-M) systems. This transfer helps to explain the remarkable redundancy present in the B. fragilis genome and demonstrates a mechanism for transmission of the pan-genome.
Methods
Bacterial strains and culture conditions
B. fragilis 638R [32] is a plasmid-free, rifampicin resistant B. fragilis that is frequently used for molecular manipulations; rifampicin is used to select against the donor strain in mating experiments. B. fragilis HMW615 is a multidrug resistant clinical isolate from an appendiceal abscess [33]. B. fragilis type strain NCTC 9343 (ATCC 25285) was used as the control for the MIC determinations. All strains of B. fragilis were grown anaerobically (5 % carbon dioxide, 5 % hydrogen, 90 % nitrogen) overnight in supplemented Brain Heart Infusion (BHIS) broth or Agar (Anaerobe systems, CA) at 37 °C. Antibiotics (including rifampicin 10 µg ml−1 or/and tetracycline 1 µg ml−1 [Sigma-Aldrich]) were prepared as directed and used as needed. Prepared media was purchased from Anaerobe Systems.
Mating of B. fragilis 638R and B. fragilis HMW615
B. fragilis 638R and B. fragilis HMW615 were grown anaerobically overnight at 37 °C in BHIS and the mating conducted by described methods [24]. Only one colony from each mating plate was chosen for further analysis to minimize detection of sibling colonies.
To test the effects of tetracycline on transfer rates, B. fragilis HMW615 was either grown overnight in the presence of tetracycline (1 µg ml−1) or grown without antibiotic and then treated with tetracycline (1 µg ml−1) for 1 h at 37 °C. Treated cells were then washed twice with equal volumes of BHIS and the regular mating procedure with B. fragilis 638R was followed. In an alternative method to test effects of tetracycline on mating frequency, B. fragilis 638R and B. fragilis HMW615 were grown separately overnight and 200 µl of each were mixed and plated on BHIS containing 0.001 µg ml−1 of tetracycline, and incubated anaerobically overnight at 37 °C before mating [24].
Molecular techniques
Genomic DNA for PCR analyses was prepared using the DNeasy Blood and Tissue kit (Qiagen). One Taq Polymerase master mix (NEB) was used for the PCR procedures. Amplicons were sequenced using Sanger sequencing (Laragen, Culver City, CA). The primers used are listed in Table S1 (available in the online Supplementary Material).
Genome sequencing of recombinant strains and comparative genome analysis
Genomic DNA was prepared using the Qiagen DNeasy Blood and Tissue Kit and submitted for Mi-Seq analysis (Laragen). The data were analyzed using the DNASTAR Lasergene Genomics Suite (DNASTAR). FastQ files were assembled with the SeqMan NGen module using B. fragilis 638R as the template sequence (638R_NC016776.1; GI : 375356399) and the default parameters. Comparisons of the aligned files were visualized using the SeqMan Pro module and areas of conflict are shown. Detection of prophage sequences within the genome was done with the Phaster tool [34, 35].
DNA alignment and gene comparison
B. fragilis HMW615 was sequenced as part of the Human Microbiome Project, Bacteroides group Sequencing Project, Broad Institute of Harvard and MIT (http://www.broadinstitute.org/) and was given the designation HMPREF1204. B. fragilis HMW615 genes are referred to by HMPREF1204 locus tags which were assigned by the Broad Institute prior to final genome assembly.
The B. fragilis 638R and B. fragilis HMW615 genome sequences were acquired as GenBank files from the NCBI server [36]. The RAST server was used to identify homologs between genomes [37]. Genome comparisons were done using the Double ACT server (http://www.hpa-bioinfotools.org.uk) and viewed with the Artemis Comparison Tool (ACT) viewer [38]. When needed, nucleotide or amino acid sequences were aligned using ClustalW. Based on our genome analysis, we realigned the B. fragilis HMW615 genome using the MAUVE alignment tool [39] and used the predicted origin of replication site (ori) to generate an updated GenBank file which was then used for our subsequent comparative genome analyses with the Artemis and ACT tools [38]. Circular representation of the genome was generated using DNAPlotter [40].
Results and discussion
Mating of the clinical multidrug resistant isolate B. fragilis HMW615 with B. fragilis 638R resulted in the transfer of DNA segments of varying sizes (125 Kb to 482 Kb) from B. fragilis HMW615 to B. fragilis 638R
Originally, twenty independent matings were carried out as described [24] with subsequent selection for rifampicin and tetracycline resistance; 5–10 colonies were isolated on each plate. To reduce the likelihood of detecting siblings, only one colony was chosen from each plate for further analysis. Several individual isolates were chosen for further analysis. B. fragilis HMW615 encodes two tetQ paralogues [HMPREF1204_02983 (tetQ1) and HMPREF1204_03369 (tetQ2)]. Sequencing of the region surrounding the respective tetQ genes in the individual isolates revealed that separate events had occurred, each resulting in one of the two tetQ paralogues being transferred. tetQ1 was contained within a genomic segment that we recently identified as a novel conjugative transposon, CTnHyb, composed of B. fragilis genes as well as a mosaic of genes from various aerobic genera, including several genes encoding resistance determinants [24]. tetQ2 is in a region of the HMW615 chromosome with extensive homology to B. fragilis 638R. We found that transfer of tetQ1 (i.e., the transfer of CTnHyb) was 10X more frequent than the transfer of tetQ2 described in this report. As previously estimated, the frequency of the transfer was 1×10−9 (number of transconjugants [BF638R-CtnHyb]/numbers of donors [BF HMW615]) for tetQ1. Among the 200 to 500 colonies that were seen in each of the mating reactions, approximately every 10th colony was tetQ2 (the others were tetQ1).
The recombinants identified and analyzed in this report are referred to as BF638R :: tetQ2-125 to 482, to reflect the host, the transferred tetQ2 gene and the size of the chromosomal segment that was transferred and integrated into the host genome (the respective chromosomal segments are referred to as tetQ2-125 to 482). We confirmed that the recombinants bearing tetQ2 were of B. fragilis 638R origin. This confirmation was required because tetracycline-resistant, rifampicin-resistant colonies isolated from the mating mix could have arisen as a result of (1) spontaneous mutation of the rpoC RNA polymerase beta subunit gene (conferring rifampicin resistance) in B. fragilis HMW615 or (2) transfer of the mutated rpoC [28] from B. fragilis 638R to B. fragilis HMW615. PCR analysis and sequencing of the recombinants confirmed the presence of BF638R_2089 and BF638R_4382 – two genes with no homologues in B. fragilis HMW615 – thus confirming their B. fragilis 638R origin. Subsequent complete genomic sequencing of the recombinants further confirmed these results.
The mechanism of transfer of the tetQ2-chromosomal segments is not known
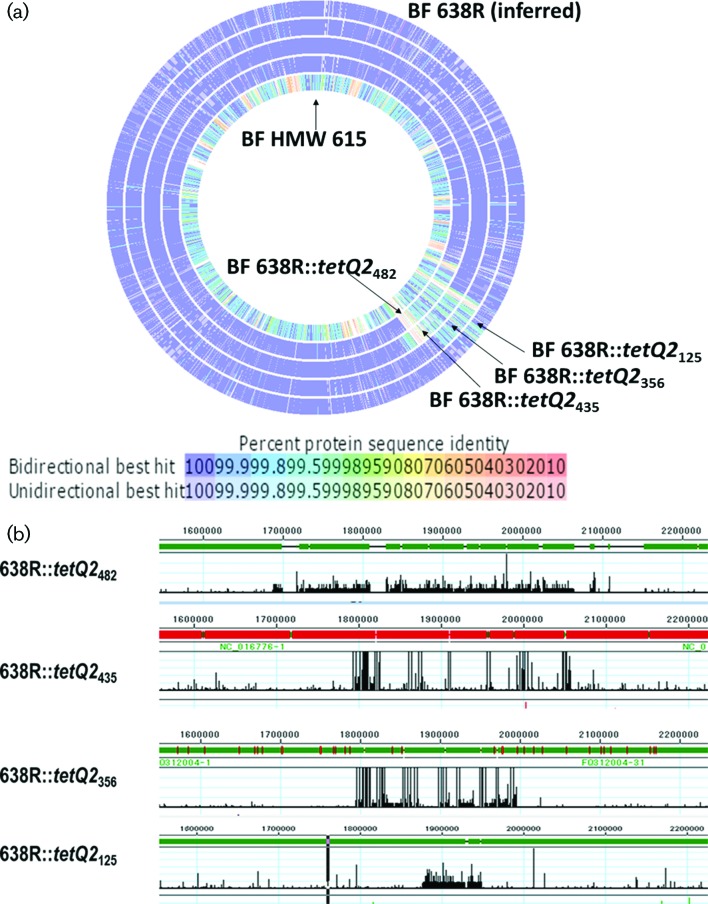
Conjugation is, however, the most likely mechanism of transfer as: (1) there is no evidence that B. fragilis is naturally competent for uptake of DNA (even transformation via electroporation is often inefficient); (2) B. fragilis HMW 615 is plasmid free and thus plasmid-mediated transfer is ruled out; and (3) B. fragilis HMW 615 does not encode a phage genome that could result in transduction. Fig. 1 is a circular representation of the B. fragilis HMW615 chromosome that illustrates the position of the transferred segments within recombinants BF 638R :: tetQ2435 and BF 638R :: tetQ2482. We speculate that co-resident CTns, the CTn341-like region within the transferred segment and/or the CTn86-like ICE5 (49.5 kb downstream of the end of tetQ2435), in B. fragilis HMW 615 provided the functions required for transfer.
Fig. 1.
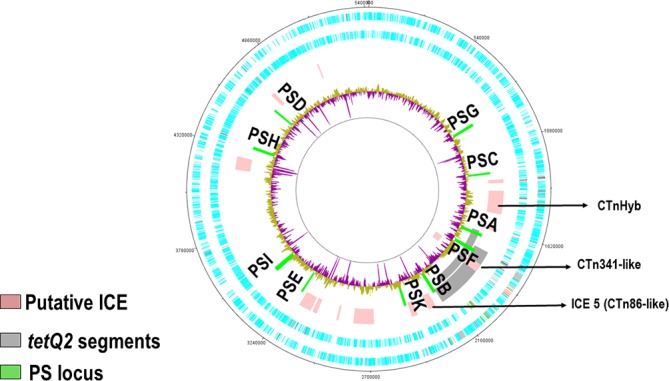
Circular representation of the B. fragilis HMW615 chromosome illustrates the position of tetQ2435, other putative ICEs and polysaccharide (PS) associated loci. From the outside: Circle 1, DNA coordinates; Circle 2 and 3, CDs forward/reverse strands; Circle 4, putative integrative and conjugative elements (ICE) including tetQ2435 (Note: tetQ2435 is marked in this figure as an example of the tetQ2 segment positions); Circle 5, surface polysaccharide synthesis associated loci (PSA-K); Circle 6, graph of %GC content. Above genome average %GC (yellow/green), below average (purple). Note that graph spikes reaching the inner circle relate to %GC sequence assembly gaps.
The transfer of DNA in the tetQ2-chromosomal segments is consistent with an excision-independent mechanism
Since the lengths of DNA transferred were variable, we believe it likely that the transfer of these tetQ2-containing segments did not include excision from the chromosome prior to conjugation. Excision-independent transfers have been described in both Gram-negative and Gram-positive bacteria, with up to 1 Mb of chromosomal DNA being transferred [41, 42]. Transfer of a B. fragilis chromosomal segment mediated by one of the resident elements, such as ICE5, in B. fragilis HMW615 would be consistent with these previous observations. For example, an oriT from the downstream ICE5 could be responsible for cleavage and transfer in an Hfr-like mechanism.
Another CTn-related Hfr-type transfer was described in a Bacteroides strain carrying an excision deficient version of conjugative transposon CTnERL which contained an insertion in the integrase gene [43]. Normal CTnERL excision and integration are independent of homologous recombination. In the case of the integrase deficient CTnERL, however, homologous recombination was needed to ‘rescue’ the segment of CTnERL DNA that had transferred. The authors noted that these were the first studies to show that transfer of a portion of the CTn is possible and that the transferred DNA can be rescued if there is a closely related CTn present in the recipient chromosome. They did not specifically measure the length of DNA transferred, because their studies identified transfer by using a selection marker that was only 20 kb from the oriT region.
Mechanism of integration
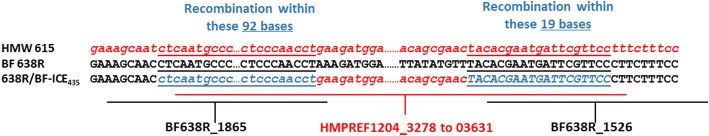
We demonstrated previously that a conjugative transposon (CTnHyb) bearing tetQ1 integrated through site-specific recombination [24]. Analysis of our initial recombinant bearing tetQ2, however, indicated that a large, 435 K bp DNA segment had transferred from B. fragilis HMW615 and replaced a 413 K bp sequence on the B. fragilis 638R chromosome. Subsequently, we isolated three additional recombinants carrying tetQ2. The segments were apparently integrated into the host genome via homologous recombination rather than site specific integration. PCR analysis of the left and right ends of the insertion into B. fragilis 638R :: tetQ2435 delineated the sites of recombination (Fig. 2). On the left end, the recombination occurred between BF638R_1526 and HMPREF1204_03278 and could be localized to a 92 bp region. BF638R_1526 and HMPREF1204_03278 (predicted outer membrane proteins) differ in nine bases, but have only a single amino acid change (V487I change in BF638R_1526). Recombination on the right end occurred between HMPREF1204_03632 and BF638R_1865 (Fig. 2). The 19 bases in which recombination occurred include the start codon of HMPREF1204_03632 (Fig. 2) and the RAST-assigned BF638R_1865 start codon.
Fig. 2.
The recombination of regions of B. fragilis HMW615 and B. fragilis 638R that generated BF638R :: tetQ2435. The italicized (lowercase) sequence is B. fragilis HMW615 and the bold is B. fragilis 638R. Single nucleotide polymorphisms between the B. fragilis 638R and B. fragilis HMW615 genomes allowed localization of the recombination site to small regions, as shown. On the left side, strand exchange within the underlined 92 bp region produces a hybrid gene between BF638R_1526 and HMPREF1204_03278, which has 99 % aa identity to BF638R_1526 and 100 % aa identity to HMPREF1204_03278. On the right side, recombination occurs within the underlined 19 bp region and the hybrid gene created is 100 % homologous to HMPREF1204_03631.
Subsequent whole genome sequence analysis of four recombinants was performed and the edges of the transferred segment and recombination points in B. fragilis 638R were identified. The lengths of the transferred segments ranged from 125 to 482 K bp (Table 1). Fig. 3(a) provides an overall view of the area of insertion into the B. fragilis 638R chromosome, degree of homology between the donor and host genomes and the recombinants resulting from the events described. Fig. 3(b) is a representation of the areas of differing sequence between the recombinants and the B. fragilis 638R genome. The width of the conflict area represents the length of the inserted segment. The genes transferred by the two smaller chromosomal segments, tetQ2125 and tetQ356, were wholly contained within tetQ2435.
Table 1. Characteristics of B. fragilis (BF) 638R recombinants resulting from horizontal transfer of tetQ2-containing chromosomal segments.
| Strain | Size of 638R segment replaced | Size of HMW615 segment transferred | 638R Left end boundary/Locus Tags | 615 Left end boundary/Locus Tags | 615 Right end boundary/Locus Tags | 638R Right end boundary/Locus Tags |
|---|---|---|---|---|---|---|
| BF638R :: tetQ2435 | 412498 | 435461 | BF638R_1526 | HMPREF1204_03278 | HMPREF1204_03631, 03632 | BF638R_1865 |
| BF638R :: tetQ2125 | 70896 | 124549 | BF638R_1604 | HMPREF1204_03357 | HMPREF1204_03467 | BF638R_1665 |
| BF638R :: tetQ2356 | 258992 | 359673 | BF638R_1526 | HMPREF1204_03278 | HMPREF1204_03567 | BF638R_1741 |
| BF638R :: tetQ2482 | 418817 | 482533 | BF638R_1421, 1422, tRNA | HMPREF1204_03179, 03180, tRNA | HMPREF1204_03584 | BF638R_1785 |
Fig. 3.
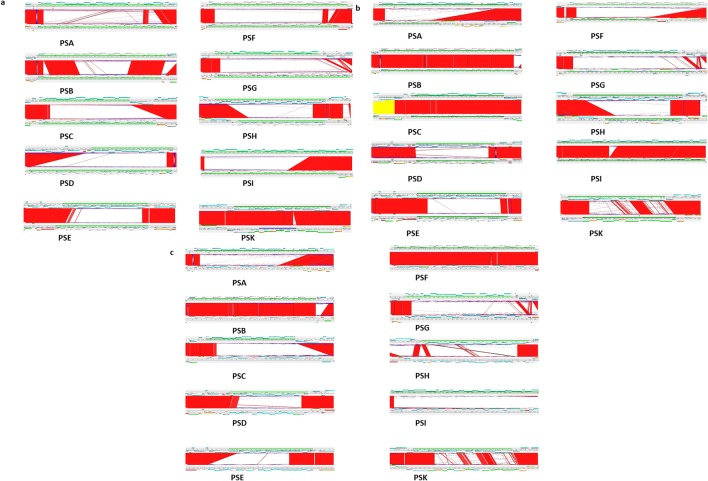
Genomic sequence comparison of B. fragilis 638R, B. fragilis HMW615 and the recombinants. (a) Comparison of the B. fragilis 638R sequence with the four recombinants (B. fragilis 638R :: tetQ2s) and B. fragilis HMW615 using RAST (http://rast.nmpdr.org/). Percent protein sequence identity is indicated by colour of the corresponding gene hit (legend). (b) Comparison after sequencing using MiSeq analysis and assembly with SeqMan Ngen using the B. fragilis 638R sequence as a reference and viewed in SeqMan Pro. The vertical black lines represent conflicts with the reference sequence. The width of the conflict area represents the length of the inserted sequence. As is apparent, there are areas within the insertion where the recombinants are homologous to the B. fragilis 638R sequence.
The reciprocal exchange of loci seen in the recombinant, rather than insertion of the foreign DNA, indicates that recombination most likely occurred through RecA-dependent homologous recombination. In a possible scenario, the early events leading to acquisition of the tetQ2-containing chromosomal segment would have resembled the mechanism of CTn conjugation. In this scenario, B. fragilis HMW615 DNA was nicked and cleaved at the oriT within ICE5 (Fig. 1) and subsequently transferred into B. fragilis 638R. In the recipient, the incoming 5′ single-stranded DNA linked to the relaxosome complex was then replicated by lagging strand synthesis to generate double-stranded DNA. Since the AddAB helicase-nuclease can translocate and degrade DNA up to 45 kb from a double-strand end in vitro [44], it is plausible that exonuclease activity of AddAB could process this DNA segment (prior to RecA-mediated strand invasion), removing evidence of the ICE5 oriT in the recombinant. The 3′ RecA-loaded single strands would then allow duplex invasion in an ‘ends-out’ configuration [45]. This would be similar to the transfer of the pathogenicity locus of C. difficile to non-toxigenic strains. In that study, the pathogenicity locus (PaLoc) could be transferred from the toxin-producing strain to non-toxigenic strains and the recipient strains were positive in assays for cytotoxin B [46]. Because the PaLoc was not contained within a mobile element, the authors concluded that an oriT within the host mediated transfer and the incoming DNA was integrated into the recipient chromosome by homologous recombination. Further, like our variably sized segments, the PaLoc was transferred on variably sized DNA segments (66 034–272 977 bp).
Gene substitutions, additions, deletions or duplications caused by integration of the tetQ2-containing chromosomal segments in B. fragilis 638R
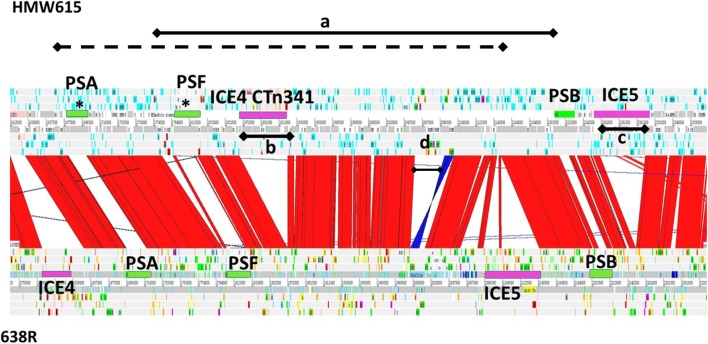
The exact range of genes transferred in the tetQ2-containing chromosomal segments, as well as the genes replaced in B. fragilis 638R, are listed in Table 1. The DNA segments integrated ranged from 124 549 to 482 538 bp and replaced segments of the B. fragilis 638R chromosome ranging from 70 896 bp to 418 817 bp. Fig. 4 is an ACT comparison between B. fragilis HMW615 and B. fragilis 638R of the region encoding polysaccharide microcapsules and highlights several of the important locations in the two largest segments transferred (tetQ2482 and tetQ2435).
Fig. 4.
Artemis Comparison Tool genome comparison illustrating transfer of novel polysaccharide biosynthesis loci. Red colouration between sequences indicates sequence identity, no colour indicates divergent sequence, dark blue indicates inverted sequence. PS biosynthesis associated loci, green; putative integrative and conjugative elements (ICE), light pink: a, BF638R :: tetQ2435 (solid line); BF638R :: tetQ2435 (dashed line) chromosomal segments transferred from HMW615 into 638R; b, CTn341-like element embedded within transferred chromosomal segments; c; CTn86-like element; d, Intergenic shufflon. Transfer of tetQ2482 has introduced two divergent micro-capsule polysaccharide biosynthesis operons, one at the PSA and one at the PSF locus (*).
Tables S2 and S3 compare the gene content of B. fragilis 638R and the B. fragilis 638R :: tetQ2 recombinants using comparative genome analysis by RAST [37]. For example, eighty-four novel genes were introduced from HMW615 to 638R and seventy-nine genes were lost from 638R by the transfer and subsequent recombination of tetQ2435 to B. fragilis 638R.
Integration of the tetQ2s356, 435, and 482 chromosomal segments replaced the B. fragilis 638R PSF micro-capsule locus with a homologous, but not identical, PSF micro-capsule operon
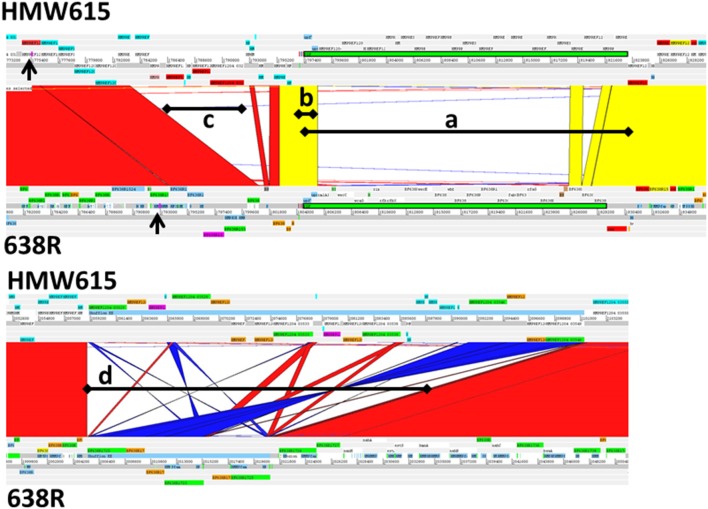
The variably produced and antigenically diverse polysaccharide micro-capsules of B. fragilis are extremely important elements implicated in both normal immune system development and abscess formation. Each B. fragilis strain studied to date contains multiple PS loci controlled by invertible promoters and transcriptional regulators. Comparison of three whole genome sequences revealed an extensive pool of 28 divergent PS biosynthesis loci [47]. Scrutiny of the B. fragilis HMW615 genome has added a further 5 divergent PS loci to the known B. fragilis pan-genome. A comparison of the PSF locus in B. fragilis HMW615 and B. fragilis 638R (Fig. 5, upper panel) shows conservation between the transcriptional regulators but a lack of DNA sequence identity within the polysaccharide synthesis associated genes. PCR analysis with primers specific for B. fragilis 638R PSF and B. fragilis HMW615 PSF and subsequent genomic sequencing confirmed that integration of tetQ2s356, 435 and 482 replaced the complete PSF locus of B. fragilis 638R with the divergent locus present in B. fragilis HMW615 which demonstrates one mechanism for generation of PS diversity; the results of the PCR analysis for B. fragilis 638R :: tetQ2435 are shown as an example in Fig. 6. Remarkably, in the longest transferred segment, tetQ2482, the entire PSA locus is also exchanged, in addition to the PSF locus described earlier. The downstream ICE 5 (Fig. 1) is not seen in any of the recombinants.
Fig. 5.
Genome comparison detail of selected regions within tetQ2435 using the Artemis Comparison Tool. Red/yellow colouration between sequences indicates sequence identity, no colour indicates divergent sequence, dark blue inverted sequence. Left, tetQ2435 recombination region, arrows. Upper panel: (a), Divergent microcapsule PSF biosynthesis associated locus; (b), Conserved PSF regulatory genes and invertible promoter; (c), Type I restriction and modification genes absent in B. fragilis 638R Lower panel: (d), Intergenic shufflon with invertible DNA regions encoding protein pairs of outer membrane associated proteins.
Fig. 6.
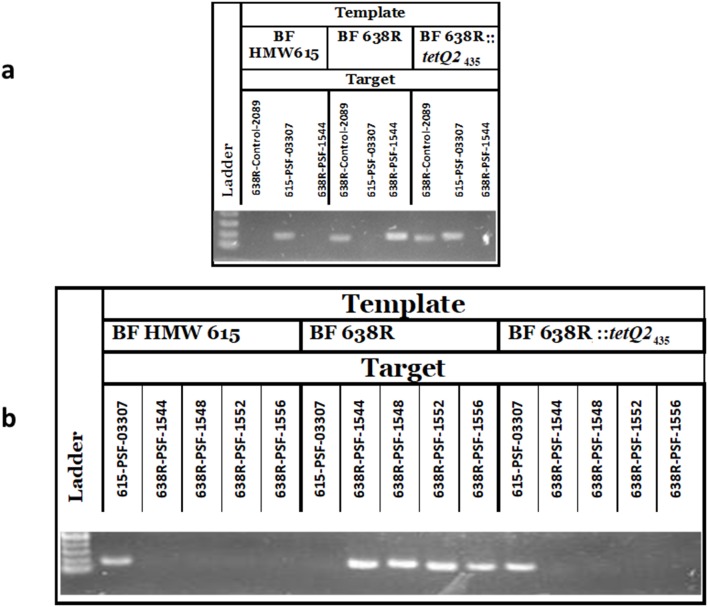
PSF exchange mediated by tetQ2435 and tetQ2482. HMPREF1204_03307, is a gene from the PSF operon of B. fragilis HMW615 (within tetQ2435). BF638R_1544, BF638R_1548, BF638R_1552, and BF638R_1556 are genes within the PSF operon of B. fragilis 638R. BF638R_2089 is a B. fragilis 638R specific gene outside the region replaced by tetQ2435 that is not present on the B. fragilis HMW615 genome. a: Lane 1: MW Standard, Lanes 2–6: Template B. fragilis HMW615. Primers for HMPREF1204_03307, BF638R_1544, BF638R_1548, BF638R_1552, and BF638R_1556, respectively. Lanes 7–11: Template BF 638R. Primers for HMPREF1204_03307, BF638R_1544, BF638R_1548, BF638R_1552, and BF638R_1556, respectively. Lanes 12–16: Template BF638R::tetQ2435. Primers for HMPREF1204_03307, BF638R_1544, BF638R_1548, BF638R_1552, and BF638R_1556, respectively. b: Lane 1: MW Standard. Lanes 2–4: Template B. fragilis HMW615. Primers for BF638R_2089, HMPREF1204_03307, BF638R_1544, respectively. Lanes 5–7: Template B. fragilis 638R. Primers for BF638R_2089, HMPREF1204_03307, BF638R_1544, respectively. Lanes 8–10: BF 638R :: tetQ2435. Primers for BF638R_2089, HMPREF1204_03307, BF638R_1544, respectively.
Strain divergence in polysaccharide synthesis loci was seen in earlier comparisons of B. fragilis 638R, NCTC 9343 (ATCC 25285) and YCH46 [28]. ACT comparison of B. fragilis HMW615 with B. fragilis 638R, B. fragilis ATCC 25285 (NCTC 9343) and B. fragilis YCH46 (Fig. 7) revealed that PSK was similar to the locus in B. fragilis 638R (Fig. 7a) and that only the loci for PSB, C, F and I were similar to loci seen in either B. fragilis ATCC 25285 (NCTC 9343) (Fig. 7b) or YCH46 (Fig. 7c). Thus, B. fragilis HMW615 has five divergent PS biosynthesis loci (PSA, PSD, PSE, PSG, PSH) in addition to the 28 already described for B. fragilis 638R, B. fragilis NCTC9343 (ATCC25285) and B. fragilis YCH46, resulting in 33 divergent biosynthesis loci among the 4 strains. Interestingly, the genes within the PSB locus associated with the formation of a chemically stable carbon-phosphorus bond, aep X,Y and Z [48], are conserved although subsequent genes are divergent. PS locus sequence diversity is reflected by the production of micro-capsules of different antigenicity [28]. The mechanism underpinning this unprecedented level of diversity of PSs among B. fragilis was unknown although the difference in the %GC content from the core genome suggests horizontal gene transfer of these loci (Fig. 1).
Fig. 7.
ACT comparison of PS loci of B. fragilis HMW 615 and B. fragilis 638R (7a), B. fragilis 9343 (7b) and B. fragilis YCH46 (7 c). Note gene conservation at the beginning and end of the PS operon. PSK was similar to the locus in B. fragilis 638R and only the loci for PSB, C, F and I were similar to loci seen in either B. fragilis ATCC 25285 (NCTC 9343) or YCH46.
The replacement of a complete PS biosynthesis locus (or loci) with a complete and divergent PS biosynthesis locus has revealed a mechanism for pan-genome dissemination of PS loci. Interestingly, the PSA of B. fragilis NCTC9343 (ATCC25285), which is linked to the down-regulation of the immune response within the gastro-intestinal tract [49], is not conserved in B. fragilis HMW615 or B. fragilis 638R. Which and how many of the divergent PSs within the B. fragilis pan-genome have similar properties to PSA remains to be determined, but transfer and replacement of PS loci associated with different types of immune system interaction could impact on the dynamics of immune system development. This in turn could alter the balance between immune tolerance and disease.
The transfer of tetQ2435 replaced an intergenic shufflon which controls variable transcription of genes encoding outer membrane proteins
In B. fragilis NCTC 9343, there is a region overlapping the start codon of ragA/susC homologues that is composed of extensive inverted repeats (Region EE, intergenic shufflon) [28]; this entire region is downstream of an invertible promoter. In this system, inversion of large segments of DNA mediated by inverted repeats brings the alternative outer membrane protein genes downstream of this invertible promoter, thus enabling transcription of these genes. These regions encode pairs of genes similar to the ragA/B genes of P. gingivalis [50] and susC/D genes of B. thetaiotaomicron [51]. RagB is a major immunodominant outer membrane protein in P. gingivalis and SusC/D proteins are associated with nutrient binding at the cell surface. Here, tetQ2435 has replaced the equivalent B. fragilis 638R intergenic shufflon with another potentially invertible region containing divergent genes. Comparison of B. fragilis 638R and B. fragilis HMW615 indicates that the tetQ2435 transfer replaced the B. fragilis 638R shufflon containing three potential gene pairs with B. fragilis HMW615 shufflon which contains six genes, five of which are divergent (Fig. 5, lower panel). Thus, transfer of tetQ2435 has not only altered the PS surface antigenicity but also surface protein variability.
The introduction of tetQ2435 introduced a second shufflon encoding a variable Type 1 restriction and modification (R-M) systems
Another highly variable feature of B. fragilis is the diversity of R-M systems, apparent both within a strain as a result of gene shuffling and also between strains. B. fragilis 638R has one R-M shufflon (BF638R 1146–1149). A second shufflon encoding four Type I R-M associated genes that are not present in B. fragilis 638R (HMPREF1204_03284, HMPREF1204_03286, HMPREF1204_03289 and HMPREF1204_03290) are evident at the left end of tetQ2435 (Fig. 5, upper panel, arrow c). HMPREF1204_03284 appears to be an S-subunit, so it could potentially replace HMPREF1204_03286 to generate an alternative DNA-binding specificity. An intervening integrase, HMPREF1204_03285, might be responsible for mediating site-specific recombination to rearrange the genes encoding the DNA-binding S-subunits. This second R-M modification locus was transferred to all but B. fragilis 638R :: tetQ2125 (Table 2).
Table 2. Distinctive characteristics transferred with the B. fragilis (BF) HMW 615 tetQ2 segment to B. fragilis 638R.
| Transferred Element | |||||||
|---|---|---|---|---|---|---|---|
| PSA | PSF | RM-gene shufflon | Shufflon EE | CTn341-like | ICE 5 | ||
| Strain | Size of transfer | ||||||
| BF638R :: tetQ2435 | 435 KB | NO | YES | YES | YES | YES | NO |
| BF638R :: tetQ2125 | 125 KB | NO | NO | NO | YES | YES | NO |
| BF638R :: tetQ2356 | 356 KB | NO | YES | YES | YES | YES | NO |
| BF638R :: tetQ2482 | 482 KB | YES | YES | YES | YES | YES | NO |
The introduction of this segment into the bacterium generated a greater diversity of R-M systems. This new R-M shufflon is immediately adjacent to the PSF operon. We had previously suggested that predation by bacteriophages is an evolutionary driving force for generation of variable polysaccharide and R-M systems in B. fragilis [28]. The close proximity of genes encoding R-M systems and PS biosynthetic enzymes in B. fragilis HMW615 means they will be co-inherited at a high frequency during conjugation. Genetic linkage between these loci suggests that part of the tetQ2 containing segments (except for the smallest tetQ2125) could be classed as a defense island as defined by Makarova et al. [52] and suggests that horizontal transfer in Bacteroides, including the novel HGT mechanism described here, mobilizes genetic features that contribute to enhanced survival in an environment rich in bacteriophages. This transfer and homologous integration may represent a mechanism for dissemination of the multiple polysaccharide loci of B. fragilis as well as other genes important in resistance, virulence, host adaptation and immunogenicity.
The tetQ482 segment is the largest chromosomal segment reported to date that was transferred between Bacteroides strains
The large size of the chromosomal transfer described here invites comparison to pathogenicity islands, which are generally 100 000 to 200 000 bases long and can be transferred due to proximity to a transferable element [20, 53–56]; however, in these events the pathogenicity islands integrate into the host chromosome in a site-specific manner unlike the integration of the chromosomal segments described here. Two examples of excision and mobilization of large genomic islands have been demonstrated in Vibrio spp. and Mesorhizobium loti [57, 58]. In the case of the Vibrio genomic islands, the experiments were conducted using mobile genetic elements (MGIs) and ICEs from Vibrio spp. introduced into various derivatives of E. coli; subsequent manipulations were all done in E. coli and the extent of transfer determined by the inclusion in the transferred segment of a Tn10 element inserted at various distances from the MGI. Subsequent reports of movement of large genomic islands among Vibrio species have been based on bioinformatic analyses of sequenced genomes [59] rather than demonstrations of actual transfer between strains. Additionally, to our knowledge, the Mesorhizobium 502 kb ‘symbiosis island’ is the only described genetic element larger than tetQ2482 in which mating experiments between the same or related species demonstrated mobilization, transfer and integration into the host chromosome [17, 58, 60]. In that case, the ‘symbiosis island’ containing a phage-related integrase at one end could be acquired and integrated into a tRNA gene in a nonsymbiotic Mesorhizobium species and is thus a mechanism by which nonsymbionts could evolve into symbionts [58]. Notably, and in contrast to the HGT event described here, integration was site-specific, required re-circularisation before integration, and the incoming DNA did not replace any of the recipient chromosome.
Proximity of ICEs to the transferred chromosomal segments in B. fragilis HMW 615
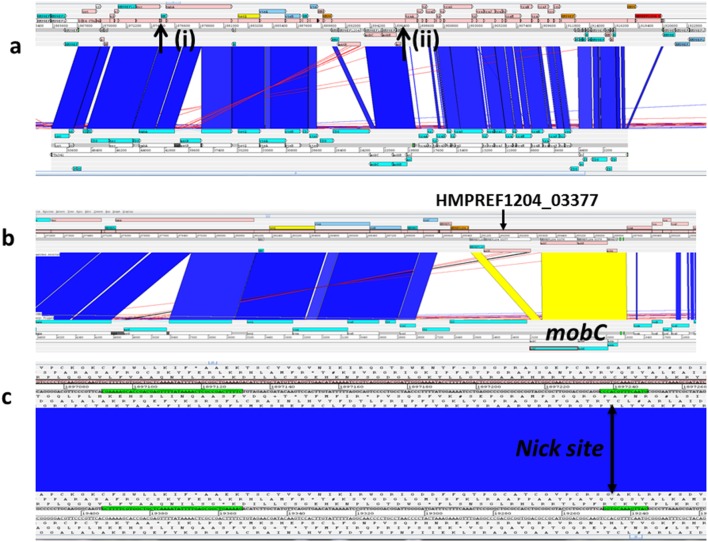
B. fragilis HMW 615 has 13 regions that contain putative ICEs or ICE remnants (Fig. 1, areas indicated by pink bars) including CTnHyb, described earlier [24]. The ICEs relevant to our report include a CTn-like element transferred with tetQ2 and the downstream ICE5, which we suggest is implicated in the transfer described in this report. A 50 000 bp segment (1867893…1920656) of the respective transferred tetQ2-containing segments is nearly identical to two CTns, CTn341 from B. vulgatus (Fig. 8) and CTnYCH46-1 from B. fragilis YCH46 [61, 62]. This CTn-341-like region includes the consensus ends [61, 62], the oriT, the nick site and traA to traQ genes (HMPREF1204_3381 to 3397) that are required to form the core channel apparatus [3, 63]; these genes are not uniformly homologous between the similar CTns. They also encode the gene for tyrosine recombinase, HMPREF1204_3354, with 98 % amino acid identity to the integrase gene in CTn341 and CTnYCH46-1 (known as IntDOT in CTnDOT [64]). An additional tyrosine recombinase gene [HMPREF1204_3285 (xerC) [65]] is present outside the CTn341-like region; this recombinase has only 31 % identity to the other integrases.
Fig. 8.
Genome comparison of the tetQ2435 within B. fragilis HMW615 with B. vulgatus CTn341 using the Artemis Comparison tool. Blue/yellow colouration between sequences indicates sequence identity, no colour indicates divergent sequence. a: Overview of complete CTn341-like region within tetQ2435 in B. fragilis HMW615. Arrow (i) points to the conserved RteR; arrow (ii) points to the conserved oriT site, inverted repeat and CTn341 nick site. b: Mob region. The mobC gene within the CTn341-like ICE in tetQ2435 is truncated by HMPREF1204_3377 (Retron-type RNA-directed DNA polymerase). c: CTn341 nick site. Note sequence conservation between CTn341 and tetQ2435.
Transfer of the B. fragilis HMW615 tetQ2 gene was not induced by tetracycline, in contrast to many CTn-associated-tetQs genes in other strains
In contrast, transfer of two B. fragilis conjugative transposons studied in detail, CTn341 and CTnDot, both encoding tetQ, are induced by tetracycline. In CTnDot, the induction by tetracycline is mediated by the action of RteABC, where RteC activates expression of excision functions, including Xis2c, subsequently leading to activation of genes required for conjugative transfer [66–73]. The tetQ2-chromosomal segments described here do include the rteABC operon (HMPREF1204_03370, 03371, and 03373, respectively) within the CTn341-like region that includes tetQ2. However, the gene encoding the MobC homolog, which is involved in the tetracycline induction of transfer [74], is truncated by HMPREF1204_3377 (Retron-type RNA-directed DNA polymerase) (Fig. 8). Further, CTn86 (similar to the downstream ICE 5), in contrast to most Bacteroides CTns, does not contain the tetracycline gene and excision is not regulated by tetracycline [75]. The estimated frequency of the transfer with tetracycline treatment remained the same as without: 1×10−9 (number of transconjugants [BF638R-CtnHyb]/numbers of donors [BF HMW615]). As in the mating results without tetracycline treatment, every 10th colony was tetQ2 and the others were tetQ1. This strongly suggests that the tetQ2-chromosomal transfer events were not increased in the presence of tetracycline, which would be consistent with involvement of the tetracycline-independent CTn86-like ICE 5 in mediating the transfer.
The HGT event mediated by the transfer of the tetQ2-chromosomal segments strengthens our suggestion that predation by bacteriophages is an evolutionary driving force to generate variability in polysaccharides and R-M systems in B. fragilis
This concept is now strengthened by our data demonstrating the close linkage between the PS, Sus-like outer membrane proteins and R-M systems with the CTn within the larger tetQ2-containing segments; this suggests that CTns actively co-mobilize genetic features that contribute to enhanced survival in an environment rich in bacteriophages and with diverse nutrient availability. Genetic linkage between these loci on these segments indicates that the R-M system genes may contribute to non-adaptive evolution of B. fragilis and hence could be classed as a defense island as defined by Makarova et al. [52]. This would also extend the definition of defense islands to include variable surface structures, such as polysaccharides and outer membrane proteins that may prevent phage adsorption. Thus, our data demonstrate that chromosomal features that contribute to enhanced survival in an environment rich in bacteriophages can be actively mobilized by ICEs on the chromosome, and raises the possibility that ICEs in B. fragilis have evolved to integrate in close proximity to defense islands. This event may represent a mechanism for large scale chromosomal replacement in B. fragilis that could confer significant advantage in adaptation.
Conclusions
The scale of transfer of this novel HGT event has not been demonstrated before in pathogens and provides a remarkable insight into the genomic fluidity of a commensal and opportunistic pathogen. It exemplifies an efficient manner for bacteria to disseminate their pan-genome and provides a novel mechanism for adaptation to rapidly changing environments. Altering the PS surface antigenicity, surface protein variability, and R-M systems of the recipient strain will impact the highly complex and dynamic interactions of B. fragilis and host immune system. The scale of HGT reinforces the paradigm that pathogen characteristics are not restricted to phylogenetic boundaries but includes the genetic source of the immediate environment [14]. This idea supports the current trend toward metagenomic analysis of infectious processes that allows examination of the complex relationships in an entire population (comprising the pathogen and its immediate environment). This may be particularly important in the case of anaerobic infections that are often caused by mixed bacterial populations whose interactions may be an important part of the pathogenic process [76]. Although the selective pressures in the GI tract are the major drivers of the variability of the pan-genome in B. fragilis, this variability is also likely to contribute to the success of B. fragilis as a pathogen. It is also a potentially important mechanism for the spread of antimicrobial resistance within the GI tract microbiota.
Funding information
This work was supported by the Department of Veterans Affairs, Veterans Health Administration, Office of Research and Development, Biomedical Laboratory Research and Development (Grant Number 5IOBX000563 to HMW) and by the National Institute of Allergy and Infectious Diseases at the National Institutes of Health (Grant Number 1R21AI109545 to HMW).
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
There is no human or animal work in this study.
Supplementary Data
References
- 1.Wexler HM. Bacteroides: the good, the bad, and the nitty-gritty. Clin Microbiol Rev. 2007;20:593–621. doi: 10.1128/CMR.00008-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wexler HM. The genus Bacteroides. In: Rosenberg EY, DeLong EF, Thompson F, Lory S, Stackebrandt E, editors. The Prokaryotes: Other Major Lineages of Bacteria and the Archaea. Berlin, Heidelberg: Springer Verlig; 2014. pp. 459–484. et al. (editors) [Google Scholar]
- 3.Nguyen M, Vedantam G. Mobile genetic elements in the genus Bacteroides, and their mechanism(s) of dissemination. Mob Genet Elements. 2011;1:187–196. doi: 10.4161/mge.1.3.18448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salyers AA, Shoemaker NB, Li LY. In the driver's seat: the Bacteroides conjugative transposons and the elements they mobilize. J Bacteriol. 1995;177:5727–5731. doi: 10.1128/jb.177.20.5727-5731.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wozniak RA, Waldor MK. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat Rev Microbiol. 2010;8:552–563. doi: 10.1038/nrmicro2382. [DOI] [PubMed] [Google Scholar]
- 6.Shoemaker NB, Vlamakis H, Hayes K, Salyers AA. Evidence for extensive resistance gene transfer among Bacteroides spp. and among Bacteroides and other genera in the human colon. Appl Environ Microbiol. 2001;67:561–568. doi: 10.1128/AEM.67.2.561-568.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vedantam G, Hecht DW. Antibiotics and anaerobes of gut origin. Curr Opin Microbiol. 2003;6:457–461. doi: 10.1016/j.mib.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 8.Thomas F, Hehemann JH, Rebuffet E, Czjzek M, Michel G. Environmental and gut Bacteroidetes: the food connection. Front Microbiol. 2011;2:93. doi: 10.3389/fmicb.2011.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee SM, Donaldson GP, Mikulski Z, Boyajian S, Ley K, et al. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature. 2013;501:426–429. doi: 10.1038/nature12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aminov RI. Horizontal gene exchange in environmental microbiota. Front Microbiol. 2011;2:158. doi: 10.3389/fmicb.2011.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koonin EV, Wolf YI. Genomics of bacteria and archaea: the emerging dynamic view of the prokaryotic world. Nucleic Acids Res. 2008;36:6688–6719. doi: 10.1093/nar/gkn668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andam CP, Gogarten JP. Biased gene transfer in microbial evolution. Nat Rev Microbiol. 2011;9:543–555. doi: 10.1038/nrmicro2593. [DOI] [PubMed] [Google Scholar]
- 13.Jain R, Rivera MC, Moore JE, Lake JA. Horizontal gene transfer accelerates genome innovation and evolution. Mol Biol Evol. 2003;20:1598–1602. doi: 10.1093/molbev/msg154. [DOI] [PubMed] [Google Scholar]
- 14.Pál C, Papp B, Lercher MJ. Adaptive evolution of bacterial metabolic networks by horizontal gene transfer. Nat Genet. 2005;37:1372–1375. doi: 10.1038/ng1686. [DOI] [PubMed] [Google Scholar]
- 15.Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 16.Dubnau D. DNA uptake in bacteria. Annu Rev Microbiol. 1999;53:217–244. doi: 10.1146/annurev.micro.53.1.217. [DOI] [PubMed] [Google Scholar]
- 17.Burrus V, Pavlovic G, Decaris B, Guédon G. Conjugative transposons: the tip of the iceberg. Mol Microbiol. 2002;46:601–610. doi: 10.1046/j.1365-2958.2002.03191.x. [DOI] [PubMed] [Google Scholar]
- 18.Wilson JW. Genetic exchange in bacteria and the modular structure of mobile DNA elements. In: Nickerson CJ, Schurr MJ, editors. Molecular Paradigms of Infectious Disease (Emerging Infectious Diseases of the 21st Century) Springer; 2006. pp. 34–77. (editors) [Google Scholar]
- 19.Booth SJ, van Tassell RL, Johnson JL, Wilkins TD. Bacteriophages of Bacteroides. Rev Infect Dis. 1979;1:325–336. doi: 10.1093/clinids/1.2.325. [DOI] [PubMed] [Google Scholar]
- 20.Osborn AM, Böltner D. When phage, plasmids, and transposons collide: genomic islands, and conjugative- and mobilizable-transposons as a mosaic continuum. Plasmid. 2002;48:202–212. doi: 10.1016/S0147-619X(02)00117-8. [DOI] [PubMed] [Google Scholar]
- 21.Langille MG, Hsiao WW, Brinkman FS. Detecting genomic islands using bioinformatics approaches. Nat Rev Microbiol. 2010;8:373–382. doi: 10.1038/nrmicro2350. [DOI] [PubMed] [Google Scholar]
- 22.Bi D, Xu Z, Harrison EM, Tai C, Wei Y, et al. ICEberg: a web-based resource for integrative and conjugative elements found in bacteria. Nucleic Acids Res. 2012;40:D621–D626. doi: 10.1093/nar/gkr846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Waters JL, Salyers AA. Regulation of CTnDOT conjugative transfer is a complex and highly coordinated series of events. MBio. 2013;4:e00569-13. doi: 10.1128/mBio.00569-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Husain F, Veeranagouda Y, Boente R, Tang K, Mulato G, et al. The Ellis Island effect: a novel mobile element in a multi-drug resistant Bacteroides fragilis clinical isolate includes a mosaic of resistance genes from gram-positive bacteria. Mob Genet Elements. 2014;4:E29801. doi: 10.4161/mge.29801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Husain F, Veeranagouda Y, Hsi J, Meggersee R, Abratt V, et al. Two multidrug-resistant clinical isolates of Bacteroides fragilis carry a novel metronidazole resistance nim gene (nimJ) Antimicrob Agents Chemother. 2013;57:3767–3774. doi: 10.1128/AAC.00386-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mcmahon SA, Roberts GA, Johnson KA, Cooper LP, Liu H, et al. Extensive DNA mimicry by the ArdA anti-restriction protein and its role in the spread of antibiotic resistance. Nucleic Acids Res. 2009;37:4887–4897. doi: 10.1093/nar/gkp478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cerdeño-Tárraga AM, Patrick S, Crossman LC, Blakely G, Abratt V, et al. Extensive DNA inversions in the B. fragilis genome control variable gene expression. Science. 2005;307:1463–1465. doi: 10.1126/science.1107008. [DOI] [PubMed] [Google Scholar]
- 28.Patrick S, Blakely GW, Houston S, Moore J, Abratt VR, et al. Twenty-eight divergent polysaccharide loci specifying within- and amongst-strain capsule diversity in three strains of Bacteroides fragilis. Microbiology. 2010;156:3255–3269. doi: 10.1099/mic.0.042978-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu CH, Lee SM, Vanlare JM, Kasper DL, Mazmanian SK. Regulation of surface architecture by symbiotic bacteria mediates host colonization. Proc Natl Acad Sci USA. 2008;105:3951–3956. doi: 10.1073/pnas.0709266105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coyne MJ, Chatzidaki-Livanis M, Paoletti LC, Comstock LE. Role of glycan synthesis in colonization of the mammalian gut by the bacterial symbiont Bacteroides fragilis. Proc Natl Acad Sci USA. 2008;105:13099–13104. doi: 10.1073/pnas.0804220105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu J, Mahowald MA, Ley RE, Lozupone CA, Hamady M, et al. Evolution of symbiotic bacteria in the distal human intestine. PLoS Biol. 2007;5:e156. doi: 10.1371/journal.pbio.0050156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Privitera G, Dublanchet A, Sebald M. Transfer of multiple antibiotic resistance between subspecies of Bacteroides fragilis. J Infect Dis. 1979;139:97–101. doi: 10.1093/infdis/139.1.97. [DOI] [PubMed] [Google Scholar]
- 33.Pumbwe L, Chang A, Smith RL, Wexler HM. BmeRABC5 is a multidrug efflux system that can confer metronidazole resistance in Bacteroides fragilis. Microb Drug Resist. 2007;13:96–101. doi: 10.1089/mdr.2007.719. [DOI] [PubMed] [Google Scholar]
- 34.Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. PHAST: a fast phage search tool. Nucleic Acids Res. 2011;39:W347–W352. doi: 10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arndt D, Grant JR, Marcu A, Sajed T, Pon A, et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44:W16–W21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benson DA, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, et al. GenBank. Nucleic Acids Res. 2014;42:D32–D37. doi: 10.1093/nar/gkt1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aziz RK, Bartels D, Best AA, Dejongh M, Disz T, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carver T, Berriman M, Tivey A, Patel C, Böhme U, et al. Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics. 2008;24:2672–2676. doi: 10.1093/bioinformatics/btn529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carver T, Thomson N, Bleasby A, Berriman M, Parkhill J. DNAPlotter: circular and linear interactive genome visualization. Bioinformatics. 2009;25:119–120. doi: 10.1093/bioinformatics/btn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hochhut B, Marrero J, Waldor MK. Mobilization of plasmids and chromosomal DNA mediated by the SXT element, a constin found in Vibrio cholerae O139. J Bacteriol. 2000;182:2043–2047. doi: 10.1128/JB.182.7.2043-2047.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Torres OR, Korman RZ, Zahler SA, Dunny GM. The conjugative transposon Tn925: enhancement of conjugal transfer by tetracycline in Enterococcus faecalis and mobilization of chromosomal genes in Bacillus subtilis and E. faecalis. Mol Gen Genet. 1991;225:395–400. doi: 10.1007/BF00261679. [DOI] [PubMed] [Google Scholar]
- 43.Whittle G, Hamburger N, Shoemaker NB, Salyers AA. A Bacteroides conjugative transposon, CTnERL, can transfer a portion of itself by conjugation without excising from the chromosome. J Bacteriol. 2006;188:1169–1174. doi: 10.1128/JB.188.3.1169-1174.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reuter M, Parry F, Dryden DT, Blakely GW. Single-molecule imaging of Bacteroides fragilis AddAB reveals the highly processive translocation of a single motor helicase. Nucleic Acids Res. 2010;38:3721–3731. doi: 10.1093/nar/gkq100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kogoma T, Cadwell GW, Barnard KG, Asai T. The DNA replication priming protein, PriA, is required for homologous recombination and double-strand break repair. J Bacteriol. 1996;178:1258–1264. doi: 10.1128/jb.178.5.1258-1264.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brouwer MS, Roberts AP, Hussain H, Williams RJ, Allan E, et al. Horizontal gene transfer converts non-toxigenic Clostridium difficile strains into toxin producers. Nat Commun. 2013;4:1–6. doi: 10.1038/ncomms3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patrick S. Bacteroides. In: Tang Y-W, Sussman M, Liu D, Poxton I, editors. Molecular Medical Microbiology. London: Academic Press; 2015. (editors) [Google Scholar]
- 48.Zhang G, Dai J, Lu Z, Dunaway-Mariano D. The phosphonopyruvate decarboxylase from Bacteroides fragilis. J Biol Chem. 2003;278:41302–41308. doi: 10.1074/jbc.M305976200. [DOI] [PubMed] [Google Scholar]
- 49.Rescigno M. Mucosal immunology and bacterial handling in the intestine. Best Pract Res Clin Gastroenterol. 2013;27:17–24. doi: 10.1016/j.bpg.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 50.Hanley SA, Aduse-Opoku J, Curtis MA. A 55-kilodalton immunodominant antigen of Porphyromonas gingivalis W50 has arisen via horizontal gene transfer. Infect Immun. 1999;67:1157–1171. doi: 10.1128/iai.67.3.1157-1171.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cho KH, Salyers AA. Biochemical analysis of interactions between outer membrane proteins that contribute to starch utilization by Bacteroides thetaiotaomicron. J Bacteriol. 2001;183:7224–7230. doi: 10.1128/JB.183.24.7224-7230.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Makarova KS, Wolf YI, Snir S, Koonin EV. Defense islands in bacterial and archaeal genomes and prediction of novel defense systems. J Bacteriol. 2011;193:6039–6056. doi: 10.1128/JB.05535-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paauw A, Leverstein-van Hall MA, Verhoef J, Fluit AC. Evolution in quantum leaps: multiple combinatorial transfers of HPI and other genetic modules in Enterobacteriaceae. PLoS One. 2010;5:e8662. doi: 10.1371/journal.pone.0008662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dobrindt U, Hochhut B, Hentschel U, Hacker J. Genomic islands in pathogenic and environmental microorganisms. Nat Rev Microbiol. 2004;2:414–424. doi: 10.1038/nrmicro884. [DOI] [PubMed] [Google Scholar]
- 55.Schmidt H, Hensel M. Pathogenicity islands in bacterial pathogenesis. Clin Microbiol Rev. 2004;17:14–56. doi: 10.1128/CMR.17.1.14-56.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Groisman EA, Ochman H. Pathogenicity islands: bacterial evolution in quantum leaps. Cell. 1996;87:791–794. doi: 10.1016/S0092-8674(00)81985-6. [DOI] [PubMed] [Google Scholar]
- 57.Daccord A, Ceccarelli D, Burrus V. Integrating conjugative elements of the SXT/R391 family trigger the excision and drive the mobilization of a new class of Vibrio genomic islands. Mol Microbiol. 2010;78:576–588. doi: 10.1111/j.1365-2958.2010.07364.x. [DOI] [PubMed] [Google Scholar]
- 58.Sullivan JT, Ronson CW. Evolution of rhizobia by acquisition of a 500-kb symbiosis island that integrates into a phe-tRNA gene. Proc Natl Acad Sci USA. 1998;95:5145–5149. doi: 10.1073/pnas.95.9.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Spagnoletti M, Ceccarelli D, Rieux A, Fondi M, Taviani E, et al. Acquisition and evolution of SXT-R391 integrative conjugative elements in the seventh-pandemic Vibrio cholerae lineage. MBio. 2014;5:e01356-14. doi: 10.1128/mBio.01356-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sullivan JT, Patrick HN, Lowther WL, Scott DB, Ronson CW. Nodulating strains of Rhizobium loti arise through chromosomal symbiotic gene transfer in the environment. Proc Natl Acad Sci USA. 1995;92:8985–8989. doi: 10.1073/pnas.92.19.8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bacic M, Parker AC, Stagg J, Whitley HP, Wells WG, et al. Genetic and structural analysis of the Bacteroides conjugative transposon CTn341. J Bacteriol. 2005;187:2858–2869. doi: 10.1128/JB.187.8.2858-2869.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kuwahara T, Yamashita A, Hirakawa H, Nakayama H, Toh H, et al. Genomic analysis of Bacteroides fragilis reveals extensive DNA inversions regulating cell surface adaptation. Proc Natl Acad Sci USA. 2004;101:14919–14924. doi: 10.1073/pnas.0404172101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lanka E, Wilkins BM. DNA processing reactions in bacterial conjugation. Annu Rev Biochem. 1995;64:141–169. doi: 10.1146/annurev.bi.64.070195.001041. [DOI] [PubMed] [Google Scholar]
- 64.Malanowska K, Salyers AA, Gardner JF. Characterization of a conjugative transposon integrase, IntDOT. Mol Microbiol. 2006;60:1228–1240. doi: 10.1111/j.1365-2958.2006.05164.x. [DOI] [PubMed] [Google Scholar]
- 65.Blakely G, May G, Mcculloch R, Arciszewska LK, Burke M, et al. Two related recombinases are required for site-specific recombination at dif and cer in E. coli K12. Cell. 1993;75:351–361. doi: 10.1016/0092-8674(93)80076-Q. [DOI] [PubMed] [Google Scholar]
- 66.Whittle G, Shoemaker NB, Salyers AA. Characterization of genes involved in modulation of conjugal transfer of the bacteroides conjugative transposon CTnDOT. J Bacteriol. 2002;184:3839–3847. doi: 10.1128/JB.184.14.3839-3847.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stevens AM, Shoemaker NB, Li LY, Salyers AA. Tetracycline regulation of genes on Bacteroides conjugative transposons. J Bacteriol. 1993;175:6134–6141. doi: 10.1128/jb.175.19.6134-6141.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bonheyo G, Graham D, Shoemaker NB, Salyers AA. Transfer region of a Bacteroides conjugative transposon, CTnDOT. Plasmid. 2001;45:41–51. doi: 10.1006/plas.2000.1495. [DOI] [PubMed] [Google Scholar]
- 69.Whittle G, Hund BD, Shoemaker NB, Salyers AA. Characterization of the 13-kilobase ermF region of the Bacteroides conjugative transposon CTnDOT. Appl Environ Microbiol. 2001;67:3488–3495. doi: 10.1128/AEM.67.8.3488-3495.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cheng Q, Sutanto Y, Shoemaker NB, Gardner JF, Salyers AA. Identification of genes required for excision of CTnDOT, a Bacteroides conjugative transposon. Mol Microbiol. 2001;41:625–632. doi: 10.1046/j.1365-2958.2001.02519.x. [DOI] [PubMed] [Google Scholar]
- 71.Park J, Salyers AA. Characterization of the Bacteroides CTnDOT regulatory protein RteC. J Bacteriol. 2011;193:91–97. doi: 10.1128/JB.01015-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Keeton CM, Hopp CM, Yoneji S, Gardner JF. Interactions of the excision proteins of CTnDOT in the attR intasome. Plasmid. 2013;70:190–200. doi: 10.1016/j.plasmid.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Keeton CM, Park J, Wang G-R, Hopp CM, Shoemaker NB, et al. The excision proteins of CTnDOT positively regulate the transfer operon. Plasmid. 2013;69:172–179. doi: 10.1016/j.plasmid.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Peed L, Parker AC, Smith CJ. Genetic and functional analyses of the mob operon on conjugative transposon CTn341 from Bacteroides spp. J Bacteriol. 2010;192:4643–4650. doi: 10.1128/JB.00317-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Buckwold SL, Shoemaker NB, Sears CL, Franco AA. Identification and characterization of conjugative transposons CTn86 and CTn9343 in Bacteroides fragilis strains. Appl Environ Microbiol. 2007;73:53–63. doi: 10.1128/AEM.01669-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wooley JC, Godzik A, Friedberg I. A primer on metagenomics. PLoS Comput Biol. 2010;6:e1000667. doi: 10.1371/journal.pcbi.1000667. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.