Abstract.
Currently, histopathological tissue examination by a pathologist represents the gold standard for breast lesion diagnostics. Automated classification of histopathological whole-slide images (WSIs) is challenging owing to the wide range of appearances of benign lesions and the visual similarity of ductal carcinoma in-situ (DCIS) to invasive lesions at the cellular level. Consequently, analysis of tissue at high resolutions with a large contextual area is necessary. We present context-aware stacked convolutional neural networks (CNN) for classification of breast WSIs into normal/benign, DCIS, and invasive ductal carcinoma (IDC). We first train a CNN using high pixel resolution to capture cellular level information. The feature responses generated by this model are then fed as input to a second CNN, stacked on top of the first. Training of this stacked architecture with large input patches enables learning of fine-grained (cellular) details and global tissue structures. Our system is trained and evaluated on a dataset containing 221 WSIs of hematoxylin and eosin stained breast tissue specimens. The system achieves an AUC of 0.962 for the binary classification of nonmalignant and malignant slides and obtains a three-class accuracy of 81.3% for classification of WSIs into normal/benign, DCIS, and IDC, demonstrating its potential for routine diagnostics.
Keywords: deep learning, convolutional neural networks, breast cancer, histopathology, context-aware CNN
1. Introduction
Breast cancer is the most frequently diagnosed cancer among women worldwide. The most frequent subtype of breast cancer, invasive ductal carcinoma (IDC), accounts for more than 80% of all breast carcinomas. IDC is considered to develop through sequential stages of epithelial proliferation starting from normal epithelium to invasive carcinoma via hyperplasia and ductal carcinoma in situ (DCIS).1 DCIS is the preinvasive stage of breast cancer in which the abnormal cells are confined to the lining of breast ducts. Accurate diagnosis of DCIS and IDC and their discrimination from benign diseases of the breast are pivotal to determine the optimal treatment plan. The diagnosis of these conditions largely depends on a careful examination of hematoxylin and eosin (H&E) stained tissue sections under a microscope by a pathologist.
Microscopic examination of tissue sections is, however, tedious, time-consuming, and may suffer from subjectivity. In addition, due to extensive population-based mammographic screening for early detection of cancer, the amount of data to be assessed by pathologists is increasing. Computerized and computer-aided diagnostic systems can alleviate these shortcomings by assisting pathologists in diagnostic decision-making and improving their efficiency. Computational pathology systems can be used to sieve out obviously benign/normal slides and to facilitate diagnosis by pointing pathologists to regions highly suspicious for malignancy in whole-slide images (WSIs) as well as providing objective second opinions.2,3
Numerous efforts have been undertaken to develop systems for automated detection of breast carcinomas in histopathology images.4–13 Most of the existing algorithms for breast cancer detection and classification in histology images involve assessment of the morphology and arrangement of epithelial structures (e.g., nuclei, ducts). Naik et al.4 developed a method for automated detection and segmentation of nuclear and glandular structures for classification of breast cancer histopathology images. A large set of features describing the morphology of the glandular regions and spatial arrangement of nuclei was extracted for training a support vector machine classifier, yielding an overall accuracy of 80% for classifying different breast cancer grades on a very small dataset containing a total of 21 preselected small regions of interest images. Doyle et al.5 further investigated the use of hand-crafted texture features for grading breast cancer histopathology images. Dundar et al.6 and Dong et al.7 developed automated classification systems based on an initial segmentation of nuclei and extraction of features to describe the morphology of nuclei or their spatial arrangement. While all of the previously mentioned algorithms were designed to manually classify selected regions of interest (mostly selected by expert pathologists), we8 proposed an algorithm based on a multiscale analysis of superpixels14 for automatic detection of DCIS that operates at the whole-slide level and distinguishes DCIS from a large set of benign disease conditions. Recently, Balazsi et al.9 proposed a system for detection of regions expressing IDC in WSIs. This system first divides the WSI into a set of homogeneous superpixels, and subsequently, uses a random forest classifier15 to determine if each region indicates cancer.
Recent advances in machine learning, in particular, deep learning,16,17 have afforded state-of-the-art results in several domains, such as speech18 and image recognition.19,20 Deep learning is beginning to meet the grand challenge of artificial intelligence by demonstrating human-level performance on tasks that require intelligence when carried out by humans.21 Obviating the need for domain-specific knowledge to design features, these systems learn hierarchical feature representations directly from data. On the forefront of methodologies for visual recognition tasks are convolutional neural networks (CNN). A CNN is a type of feed-forward neural network defined by a set of convolutional and fully connected layers. The emergence of deep learning, in particular, CNN, has also energized the medical imaging field22 and enabled development of diagnostic tools displaying remarkable accuracy,23–25 to the point of reaching human-level performance. These motivate the use of CNNs for detection and/or classification of breast cancer in breast histopathology images.
Cruz-Roa et al.10 proposed the first system using a CNN to detect regions of IDC in breast WSIs. In contrast to the modern networks that use very deep architectures to improve recognition accuracy, the utilized network was a three-layer CNN. Due to computational constraints, the model was only trained to operate on images downsampled by a factor of 16. In a recent publication,11 the authors obtained comparable performance when training and validating their system on a multicenter cohort. Rezaeilouyeh et al.12 trained a multistream five-layer CNN taking as input a combination of RGB images, and magnitude and phase of shearlet coefficients. In all these works, the models were evaluated at the patch-level. In a recent work,13 we demonstrated the discriminating power of features extracted from tumor-associated stromal regions identified by a CNN for classifying breast WSI biopsies into invasive or benign.
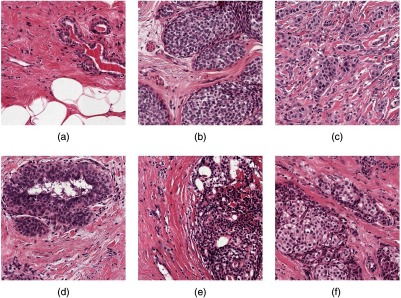
Different from the above-mentioned approaches, the aim of the present study is to develop a system for WSI classification of breast histopathology images into three categories: normal/benign, DCIS, and IDC categories. This problem is particularly difficult because of the wide range of appearances of benign lesions as well as the visual similarity of DCIS lesions to invasive lesions at the cellular level. Figure 1 shows some examples of lesions in our dataset. A system capable of discriminating these three classes, therefore, needs to use high-resolution information for discriminating benign lesions from cancer along with contextual information to discriminate DCIS from IDC. To develop a system that will work in a clinical setting, this study uses WSIs rather than manually extracted regions. Also, the cases in the “nonmalignant” category contained many of the common benign lesions, as they appear in pathology practice.
Fig. 1.
Example of breast tissue structures/lesions. Each image is of size . (a) Normal tissue and benign lesions. Benign breast diseases constitute a heterogeneous group of lesions including developmental abnormalities, inflammatory lesions, epithelial proliferations, and neoplasms. The majority of benign lesions are not associated with an increased risk for subsequent breast cancer. (b) DCIS. In DCIS, the cells lining the ducts inside the breast appear cancerous, but no cancer has spread through the ducts and into the breast tissue. (c) IDC spreads through the wall of the duct into the breast tissue. This invasive carcinoma has the potential to metastasize or spread to other parts of the body through the bloodstream or lymphatic system. The aim of this study is to develop a system for WSI classification of breast histopathology images into three categories: normal/benign, DCIS, and IDC categories.
To this end, we introduce context-aware CNNs for classification of breast histopathology images. First, we use a deep CNN, which uses high-pixel resolution information to classify the tissue into different classes. To incorporate more contexts to the classification framework, we feed a much larger patch to this model at test time. The feature responses generated by this model are then input to a second CNN, stacked on top of the first. This stacked network uses the compact, highly informative representations provided by the first model, which, together with the information from the surrounding context, enables it to learn the global interdependence of various structures in different lesion categories. The performance of our system is evaluated on a large breast histopathology cohort comprising 221 WSIs from 122 patients.
2. Methods
2.1. Overview of the System
The main challenge in the design of our classification framework is that the appearance of many benign diseases of the breast (e.g., usual ductal hyperplasia) mimics that of DCIS, hence requiring accurate texture analysis at the cellular level. Such analysis, however, is not sufficient for discrimination of DCIS from IDC. DCIS and IDC may appear identical on cellular examination but are different in their growth patterns, which can only be captured through the inclusion of larger image patches containing more information about the global tissue architecture. Because of computational constraints, however, it is not feasible to train a deep CNN with large patches at high resolution that contain enough context.
Our method for classification of breast histopathology WSIs overcomes these problems through sequential analysis with a stack of CNNs. The key components of our classification framework, including the CNN used for classification of high-resolution patches, the stacked CNN for producing dense prediction maps, and a WSI labeling module, are detailed in the following sections.
2.2. Deep Convolutional Neural Network for Classification of Small High-Resolution Patches
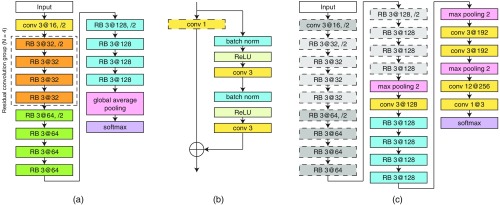
Inspired by the recent successes of deep residual networks26 for image classification, we trained and evaluated the performance of this CNN for classification of small high-resolution patches into normal/benign, DCIS, and IDC. We applied an adaptation of the ResNet architecture called wide ResNet, as proposed by Zagoruyko and Komodakis.27 This architecture has two hyperparameters: and determining the depth and width of the network, respectively. We empirically chose and as a tradeoff between model capacity, training speed, and memory usage. Hereafter, we denote this network as WRN-4-2 (see Fig. 2). This network takes as input patches of size . Zero padding was used before each convolutional layer to keep the spatial dimension of feature maps constant after convolution.
Fig. 2.
Description of the framework for classification of large input patches of breast tissue into benign/normal, DCIS, and IDC. The framework has two main steps. At first, the WRN-4-2 architecture shown in (a) is trained to classify input patches of size . Next, the architecture in (c) is used to classify patches with input size of , which is composed of a CNN stacked on top of the last convolutional layer of the WRN-4-2 architecture. The details of the two architectures are as follows. The WRN-4-2 architecture shown in (a) consists of an initial convolutional layer that is followed by three residual convolution groups (each of size residual blocks), followed by global average pooling and a softmax classifier. Downsampling is performed by the first convolutional layers in each group with a stride of 2 and the first convolutional layer of the entire network. Here, Conv 3 at 32 is a convolutional layer with a kernel size of , and 32 filters. (b) The residual block (RB) used in the WRN-4-2 architecture. Batch normalization and ReLU precede each convolution. indicates an element-wise sum. Note that the convolution layer is only used in the first convolutional layer of each residual convolution group. (c) Architecture of the CAS-CNN with input size of . The weights of the components with dotted outlines are taken from the previously trained WRN-4-2 network and are no longer updated during training.
The goal of this step was to transfer the highly informative feature representations learned by this network produced at its last convolutional layer to a stacked network, which is described next.
2.3. Context-Aware Stacked Convolutional Neural Network
In order to increase the context available for dense prediction, we stack a second CNN on top of the last convolutional layer of the previously trained WRN-4-2 network. The architecture of the stacked network, as shown in Fig. 2, is a hybrid between the wide ResNet architecture and the VGG architecture.28 CAS-CNN is fully convolutional29 and enables fast dense prediction due to reusing of overlapping convolutions during inference. All the parameters of the WRN-4-2 network were fixed during training. Despite being trained with fixed input patches of size , because of being a fully convolutional network, WRN-4-2 can take a larger patch size during training of the stacked network, and consequently, produce feature maps with larger spatial dimensions. Moreover, because of fixing the parameters of WRN-4-2, the intermediate feature maps of this network do not need to be stored during backpropagation of the gradient. This allowed us to train stacked networks with much larger effective patch sizes. Consequently, we trained three networks for classification of breast tissue into normal/benign, DCIS, and IDC with patch sizes of , , and .
To produce the dense prediction map for a given WSI, the stacked network was run over the image using the sliding window approach with a stride of 224. Background regions in the WSI were excluded from the analysis as the scanner automatically skipped scanning of areas with no tissue.
2.4. Whole-Slide Image Labeling
Given a prediction map produced by the stacked network, we extracted a set of features describing global information about the lesions and their architectural distribution for subsequent classification into normal/benign, DCIS, or IDC. To this end, the probability map was transformed into a three label map, by assigning the class with the highest probability for every pixel. The three label map could contain several connected components for different object classes, which were used for extracting features. Next, we describe the set of features extracted for WSI labeling.
2.4.1. Global lesion-based features
Details of the extracted features are presented in Table 1. Global lesion-based features include the total area of pixels classified as benign, DCIS, IDC, or cancerous (DCIS and IDC combined), and their corresponding normalized areas with respect to all nonbackground pixels, along with the fraction of DCIS, and IDC labeled pixels with respect to all cancerous pixels. We additionally computed a convex-hull area feature for IDC detected lesions. IDC lesions usually appear as a large connected mass. As such, we constructed a convex hull of all IDC detected and connected components in the WSI and computed the area ratio between the pixels labeled as IDC and the area of the convex hull. In case multiple tissue sections were present in the WSI, we took the average of these measures over different tissue sections. Note that IDC labeled connected components with an area smaller than were discarded as false positives prior to computation of the convex hull feature. At the end, we computed the mean, median, and standard deviation of the area and eccentricity of DCIS connected components as well as IDC connected components.
Table 1.
Description of the features extracted from the labeled map produced by the CNN for classification of the WSI.
| Feature category | Feature list |
|---|---|
| Global lesion-based features | (1-8) Total area of pixels classified as benign, DCIS, IDC, or cancerous (DCIS and IDC combined) and their corresponding normalized areas with respect to all nonbackground pixels. (9-10) Fraction of DCIS, and IDC labeled pixels with respect to all cancerous pixels. (11) The area ratio between the pixels labeled as IDC and the area of the convex hull including all IDC connected components. (12-17) The mean, median, and standard deviation of the area and eccentricity of DCIS connected components and (18-23) IDC connected components. |
| Architectural features (area-Voronoi diagram) | The eccentricity of the Voronoi region, the area ratio of the Voronoi region and the total tissue area, and the area ratio of the lesion inside the Voronoi region and the Voronoi region itself. (24-41) Per WSI, we computed the mean, median and standard deviation of these Voronoi area metrics for DCIS and IDC lesions, independently. (42-43) The area of the largest Voronoi region for the diagrams built on DCIS and IDC lesions, independently. |
| Architectural features (Delaunay triangulation) | (44-55) The mean, median, and standard deviation of the number of neighbors for each node and the distances of each node with respect to other neighboring nodes, computed independently for DCIS and IDC lesions. (56-57) Highest average node distance for the graphs built on DCIS and IDC lesions. |
2.4.2. Architectural features
These features describe the spatial distribution of DCIS and IDC lesions in the WSI. They were extracted from the area-Voronoi diagram30 and Delaunay triangulation (DT). We built these graphs for DCIS and IDC lesions, independently. The seed points for constructing the graphs were the center of the connected components representing DCIS or IDC lesions.
The set of features computed for each area-Voronoi region includes the eccentricity of the Voronoi region, the area ratio of the Voronoi region and the total tissue area, and the area ratio of the lesion inside the Voronoi region and the Voronoi region itself. As per WSI, we computed the mean, median, and standard deviation of these Voronoi area metrics. Additionally, we added the area of the largest Voronoi region to the feature set.
The features extracted for each of the nodes in the DT include the number of neighbors that are closer than a certain threshold to the node (), and the average distance of these neighbors to the node. We computed the mean, median, and standard deviation of these values as features. Additionally, we added the highest average node distance in the DT to the feature set.
Overall, a total of 57 features were extracted, which were used as input to two random forest classifiers15 with 512 decision trees: one for three class classification of WSIs and the other for binary classification of the WSIs into normal/benign versus cancerous (DCIS and IDC). We tuned the parameters of the classifiers by cross-validation on the combined set of train and validation WSIs.
3. Experiments
3.1. Data
We conducted our study on a large cohort comprising 221 digitized WSIs of H&E stained breast tissue sections from 122 patients, taken from the pathology archive. Ethical approval was waived by the institutional review boards of the Radboud University Medical Center because all images were provided anonymously. All slides were stained in our laboratory and digitized using the 3DHISTECH Pannoramic 250 Flash II digital slide scanner with a objective lens. Each image has square pixels of size in the microscope image plane.
Each slide was reviewed independently by a breast pathologist (P. B.) and assigned a pathological diagnosis. Overall, the dataset contains 100 normal/benign, 69 DCIS, and 55 IDC WSIs. Two human observers (M. B. and M. H.) annotated DCIS and IDC lesions using the automated slide analysis platform.31 All the annotations were verified by the breast pathologist. Note that the slide labels were assigned according to the worst abnormality condition in the WSI. Therefore, a slide with the IDC label may contain both IDC and DCIS lesions.
We split this cohort into three separate sets: one for fitting classification models, one for intermediate validation and model selection, and one set for final evaluation of the system (test set). The training, validation, and test sets had 118 (50 normal/benign, 38 DCIS, and 30 IDC), 39 (19 normal/benign, 11 DCIS, and 9 IDC), and 64 (31 normal/benign, 20 DCIS, and 13 IDC) WSIs, respectively. There was no overlap at the slide- and patient-level between the three sets. The benign/normal category included 15 normal and 85 benign WSIs comprising fibroadenoma (14), ductal hyperplasia (11), adenosis (8), fibrosis (8), fibrocystic disease (8), duct ectasia (7), hamartoma (7), pseudoangiomatous stromal hyperplasia (5), sclerosing lobular hyperplasia (5), and mixed abnormalities (12). The WSIs from these 10 benign categories and the normal class were proportionally distributed in the training, validation, and test sets. Note that the relative occurrence of these lesions in our dataset is comparable to that encountered in routine diagnostics.
3.2. Training Protocols for Convolutional Neural Networks
We preprocessed all the data by scaling the pixel intensities between 0 and 1 for every RGB channel of the image patch and subtracting the mean RGB value that was computed on the training set. The training data were augmented with rotation, flipping, and jittering of the hue and saturation channels in the HSV color model.
Patches were generated on-the-fly to construct mini-batches during training and validation of both WRN-4-2 and CAS-CNN networks, by random selection of samples from points inside the contour of annotations for each class. For each mini-batch, the number of samples per class was determined with uniform probabilities.
Both WRN-4-2 and CAS-CNN were trained using Nesterov accelerated gradient descent. The weights of all trainable layers in the two networks were initialized using He et al.32 initialization. Initial learning rates of 0.05 and 0.005 were used for WRN-4-2 and CAS-CNN, respectively. The learning rates were multiplied by 0.2 after no better validation accuracy was observed for a predefined number of consecutive epochs, which we denote as epoch patience (). The initial value for was set to 8 and increased by 20% (rounded up) after every reduction in learning rate. We used a mini-batch size of 22 for the WRN-4-2 and 18 for the CAS-CNN trained with patches of size and . The network trained on patches had a greater memory footprint and was trained with mini-batches of size 10.
Training of the WRN-4-2 involved one round of hard negative mining. Unlike the annotation of DCIS and IDC regions, the initial manual annotation of normal/benign areas was based on an arbitrary selection of visually interesting areas (e.g., areas that visually resembled cancer). These regions are not necessarily difficult for our network. In addition, some of the more difficult to classify benign regions could be under-represented in our training set. We, therefore, enriched our training dataset by automatically adding all false-positive regions in normal/benign training WSIs resulted by our initially trained WRN-4-2 model.
3.3. Empirical Evaluation and Results
We evaluated the performance of our system for classifying the WSIs into normal/benign, DCIS, and IDC categories using the accuracy measure and Cohen’s kappa coefficient.33 We additionally measured the performance of our system for the binary classification of normal/benign versus cancer (DCIS and IDC combined) WSIs.
As an intermediate evaluation, we began with measuring the performance of the WRN-4-2 for the binary and three-class problems at the patch level (see Table 2). These are only results on the validation set, as this network is not used for producing the dense prediction maps individually. As can be seen, the model performs significantly better for the two class problem with an accuracy of 0.924 compared to the three class accuracy of 0.799 for the three-class problem. This could be explained by the fact that WRN-4-2 only operates on small patches of size and does not have enough contexts for a more accurate discrimination of the three classes.
Table 2.
Patch-level accuracy for different networks on the validation set.
| Classification | Patch size | Architecture | Accuracy |
|---|---|---|---|
| Normal/benign, cancer | WRN-4-2 | 0.9241 | |
| Normal/benign, DCIS, IDC | WRN-4-2 | 0.7995 | |
| Normal/benign, DCIS, IDC | CAS-CNN | 0.8797 | |
| Normal/benign, DCIS, IDC | CAS-CNN | 0.9050 | |
| Normal/benign, DCIS, IDC | CAS-CNN | 0.9135 |
The results for the performance of the CAS-CNN on the validation set for the three-class problem are shown in Table 2. The 3-class accuracy of this network was considerably improved compared to that of the WRN-4-2 at the patch level. We also observe that increasing the training patch-size leads to better performance. Accuracies of 0.872, 0.905, and 0.914 were obtained for the CAS-CNN networks trained on , , and patches, respectively.
Due to heavy computational costs of the network operating on patches, the CAS-CNN network trained on was ultimately selected for producing dense prediction maps. The results of the random forest classifier for WSI classification on the test set of our dataset are presented in Table 3. For the binary classification task, our system achieves an AUC of 0.962 (95% CI, 0.908–0.996). The accuracy and kappa values were 0.891 and 0.781, respectively. The ROC curve of the system for binary classification of WSIs into cancer versus normal/benign is shown in Fig. 3.
Table 3.
Results of WSI label prediction on the test set.
| Labels | Acc | Kappa | AUC |
|---|---|---|---|
| Benign, cancer | 0.891 | 0.781 | 0.962 |
| Benign, DCIS, IDC | 0.813 | 0.700 | — |
Fig. 3.
ROC curve of the proposed system for binary classification of the WSIs in the test set into normal/benign and cancer (DCIS and IDC). The system achieved an AUC of 0.962 (95% CI, 0.908–0.996). The confidence interval for the AUC was obtained using the percentile bootstrap method.34
The system achieves an overall accuracy and kappa value of 0.813 and 0.700 for three-class classification of WSIs. The confusion matrix of the test set predictions is presented in Table 4. Figure 4 presents several examples of correctly and incorrectly classified image patches for different lesion classes. The top-ranked features for both binary and three-class classification tasks, identified based on random forest feature importance analysis, were the total area of IDC regions, the ratio between the total area of IDC regions and the total tissue area in the WSI, the total area of cancerous regions (DCIS and IDC combined), the area ratio between IDC and cancerous regions in the WSI, and the median of the ratios between the area of each IDC connected component and its corresponding Voronoi area.
Table 4.
Confusion matrix of test set predictions.
| Benign | DCIS | IDC | |
|---|---|---|---|
| Benign | 29 | 2 | 0 |
| DCIS | 4 | 12 | 4 |
| IDC | 0 | 2 | 11 |
Fig. 4.
Examples of correctly and incorrectly classified patches for different types of lesions. Each image is of size . (a–c) Correctly classified normal, DCIS, and IDC regions, respectively. (d) A benign lesion (usual ductal hyperplasia) misclassified as DCIS. (e) A DCIS lesion misclassified as normal/benign. (f) IDC misclassified as DCIS.
4. Discussion and Conclusion
In this paper, we presented a context-aware stacked CNN (CAS-CNN) architecture to classify breast WSIs. To the best of our knowledge, this is the first approach investigating the use of deep CNNs for multiclass classification of breast WSIs into normal/benign, DCIS, and IDC categories. CAS-CNN consists of two stages: in the first, we trained a CNN to learn cellular level features from small high-resolution patches and in the second, we stacked a fully convolutional network on top of this to allow for incorporation of global interdependence of structures to facilitate predictions in local regions. Our empirical evaluation demonstrates the efficacy of the proposed approach in incorporating more contexts to afford a high classification performance. CAS-CNN trained on large input patches outperforms the wide ResNet trained with input patches of size by a large margin and consistently yields better results when trained with larger input patches.
Our system achieves an AUC of 0.962 for the binary classification of normal/benign slides from cancerous slides. This is remarkable, given the existence of 10 benign categories in the dataset, demonstrating the potential of our approach for pathology diagnostics. Based on the achieved performance on an independent test set, this system could be used to sieve out of obviously normal/benign slides on our dataset without missing any cancerous slides.
The performance of the system on the three-class classification of WSIs was also very promising. An accuracy of 0.812 and a kappa value of 0.700 were achieved. While discrimination of normal/benign slides from IDC slides was without any misclassification, errors in discriminating between normal/benign slides and DCIS slides, as well as DCIS and IDC slides, were common. We postulate that the reason for these misclassifications is primarily because of the difficulty in discrimination of several benign categories, such as usual ductal hyperplasia from DCIS, which is also a source of subjective interpretation among pathologists. This could, in turn, be alleviated by obtaining more training data for these specific benign classes. The second reason could be the requirement of even larger receptive fields to enable discrimination of DCIS from invasive cancer. As shown in Table 2, the performance of CAS-CNN consistently improved with increasing patch size. However, this came with increased computation time both during training and inference. One major reason for this increase is that larger patch sizes lead to higher computational costs (e.g., larger memory usage). Inference time was increased from to for input patches of size from 4 to 5 h for input of size . In addition, as the patch size increases, more time is required for on-the-fly fetching of multiple patches from the WSI for both training and inference phases. One way to redress the problem could be the inclusion of additional downsampled patches with larger receptive fields as input to a multiscale network35 or using alternative architectures, such as U-net.36,37 The final reason behind these errors lies in the fact that discrimination of certain DCIS patterns from IDC, purely based on H&E staining, can be complex. As such, pathologists may use additional staining, such as myoepithelial markers to differentiate between DCIS and IDC lesions.38
Although the current system learns to exhibit some hue and saturation invariance, specialized stain standardization techniques exist39–41 and have been shown to greatly improve CAD system performance42,43 by reducing the stain variations.44 It is likely that standardizing the WSIs would also improve generalization of the performance of our network.
Although our primary aim was to facilitate pathology diagnostics by discriminating between different breast lesion categories, our system could serve as an important first step for the development of systems that aim at finding prognostic and predictive biomarkers within malignant lesions.45 This will be one of our major directions for future work.
Acknowledgments
The authors wish to acknowledge the financial support by the European Union FP7 funded VPH-PRISM project under grant agreement no. 601040.
Biographies
Babak Ehteshami Bejnordi studied electronics engineering as his undergraduate major at the University of Guilan, Iran. In 2013, he received his MSc in electrical engineering from Chalmers University of Technology, Sweden. In April 2013, he started his PhD at the Diagnostic Image Analysis Group, Radboud University, The Netherlands. His PhD project focuses on the automated detection and characterization of breast cancer in digital pathology images. His research interests are deep learning, machine learning, medical image analysis, and computer vision.
Guido Zuidhof received his BA degree from Radboud University, Nijmegen, The Netherlands, in 2015. He is currently finishing his double master’s degree in artificial intelligence and computing science at this university. His main research interests include machine learning, deep learning, and computer vision.
Maschenka Balkenhol received her MD degree from Maastricht University, The Netherlands, in 2014. In 2015, she started her PhD project at the Department of Pathology, Radboud University Medical Center, Nijmegen, The Netherlands, which aims to construct statistical models for triple negative breast cancer prognosis by using digital image analysis. Since 2016, she also has been a pathology resident in the same department.
Meyke Hermsen received her BSc at the HAN University of applied sciences, with a main focus on cytology, histology, and pathology. After graduating in 2013, she started working as a research technician at the Pathology Department of Radboud University Medical Center in the Computational Pathology and Nephropathology Group. In July 2016, she started her PhD project in the computational pathology group. Her research interests include renal transplantation pathology, transplantation immunology, and image analysis.
Peter Bult has been a breast pathologist at the Radboudumc in Nijmegen, The Netherlands, since 1994. He received his PhD degree in 2009 for his thesis “Prognostic indicators of primary breast cancer in relation to patient’s risk profile.” His research is focused on breast cancer with special attention to the sentinel node procedure, lymph node metastases (isolated tumor cells, micrometastases), hereditary/familial breast cancer and high risk lesions, HER2 testing, and computer-aided detection (CAD) in pathology.
Bram van Ginneken is a professor of functional image analysis at Radboud University Medical Center. He is a cochair of the Diagnostic Image Analysis Group within the Department of Radiology and Nuclear Medicine. He is also a cofounder of Thirona (Nijmegen, The Netherlands). He studied physics at the Eindhoven University of Technology and at Utrecht University. In March 2001, he obtained his PhD at the Image Sciences Institute (ISI) at Utrecht University. He is an associate editor of IEEE Transactions on Medical Imaging and member of the editorial board of Medical Image Analysis.
Nico Karssemeijer is a professor of computer-aided diagnosis at the Radboud University Nijmegen, where he also graduated, and has an MSc degree in physics from Delft University of Technology. He is a member of the editorial boards of Journal of Medical Imaging, Physics in Medicine and Biology, and Medical Image Analysis. In 2012 and 2013, he served as symposium chair of SPIE Medical Imaging. He is also a cofounder of Volpara Solutions (Wellington, New Zealand) and founder and CEO of ScreenPoint Medical (Nijmegen, The Netherlands).
Geert Litjens studied biomedical engineering at Eindhoven University of Technology. Subsequently, he completed his PhD in “Computer-aided detection of prostate cancer in MRI” at the Radboud University Medical Center. He spent 2015 as a postdoctoral researcher at the Tissue Imaging and Analysis Center in Heidelberg. He is currently an assistant professor in computational pathology at the Department of Pathology in the Radboud University Medical Center.
Jeroen van der Laak received his MSc in computer science and PhD in medical science from Radboud University in Nijmegen, The Netherlands. He is currently an associate professor in digital pathology at the Department of Pathology of the Radboud University Medical Center in Nijmegen. His research interest includes deep learning-based analysis of whole-slide images for improvement of routine pathology diagnostics, objective quantification of immunohistochemical markers, and study of imaging biomarkers for prognostics.
Disclosures
The authors have no potential conflicts of interest.
References
- 1.Dupont W. D., et al. , “Breast cancer risk associated with proliferative breast disease and atypical hyperplasia,” Cancer 71, 1258–1265 (1993). [DOI] [PubMed] [Google Scholar]
- 2.Gurcan M. N., et al. , “Histopathological image analysis: a review,” IEEE Rev. Biomed. Eng. 2, 147–171 (2009). 10.1109/RBME.2009.2034865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Veta M., et al. , “Breast cancer histopathology image analysis: a review,” IEEE Trans. Biomed. Eng. 61, 1400–1411 (2014). 10.1109/TBME.2014.2303852 [DOI] [PubMed] [Google Scholar]
- 4.Naik S., et al. , “Automated gland and nuclei segmentation for grading of prostate and breast cancer histopathology,” in 5th IEEE Int. Symp. on Biomedical Imaging: From Nano to Macro (ISBI ’08), pp. 284–287 (2008). 10.1109/ISBI.2008.4540988 [DOI] [Google Scholar]
- 5.Doyle S., et al. , “Automated grading of breast cancer histopathology using spectral clustering with textural and architectural image features,” in 5th IEEE Int. Symp. on Biomedical Imaging: From Nano to Macro (ISBI ’08), pp. 496–499 (2008). 10.1109/ISBI.2008.4541041 [DOI] [Google Scholar]
- 6.Dundar M. M., et al. , “Computerized classification of intraductal breast lesions using histopathological images,” IEEE Trans. Biomed. Eng. 58(7), 1977–1984 (2011). 10.1109/TBME.2011.2110648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dong F., et al. , “Computational pathology to discriminate benign from malignant intraductal proliferations of the breast,” PLoS One 9(12), e114885 (2014). 10.1371/journal.pone.0114885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bejnordi B. E., et al. , “Automated detection of DCIS in whole-slide H&E stained breast histopathology images,” IEEE Trans. Med. Imaging 35, 2141–2150 (2016). 10.1109/TMI.2016.2550620 [DOI] [PubMed] [Google Scholar]
- 9.Balazsi M., et al. , “Invasive ductal breast carcinoma detector that is robust to image magnification in whole digital slides,” J. Med. Imaging 3(2), 027501 (2016). 10.1117/1.JMI.3.2.027501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cruz-Roa A., et al. , “Automatic detection of invasive ductal carcinoma in whole slide images with convolutional neural networks,” Proc. SPIE 9041, 904103 (2014). 10.1117/12.2043872 [DOI] [Google Scholar]
- 11.Cruz-Roa A., et al. , “Accurate and reproducible invasive breast cancer detection in whole-slide images: a deep learning approach for quantifying tumor extent,” Sci. Rep. 7, 46450 (2017). 10.1038/srep46450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rezaeilouyeh H., Mollahosseini A., Mahoor M. H., “Microscopic medical image classification framework via deep learning and shearlet transform,” J. Med. Imaging 3(4), 044501 (2016). 10.1117/1.JMI.3.4.044501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bejnordi B. E., et al. , “Deep learning-based assessment of tumor-associated stroma for diagnosing breast cancer in histopathology images,” in 2017 IEEE 14th Int. Symp. on Biomedical Imaging (ISBI 2017), pp. 929–932 (2017). 10.1109/ISBI.2017.7950668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bejnordi B. E., et al. , “A multi-scale superpixel classification approach to the detection of regions of interest in whole slide histopathology images,” Proc. SPIE 9420, 94200H (2015). 10.1117/12.2081768 [DOI] [Google Scholar]
- 15.Breiman L., “Random forests,” Mach. Learn. 45(1), 5–32 (2001). 10.1023/A:1010933404324 [DOI] [Google Scholar]
- 16.Gu J., et al. , “Recent advances in convolutional neural networks,” Pattern Recognit. (2017) (in press). 10.1016/j.patcog.2017.10.013 [DOI]
- 17.LeCun Y., Bengio Y., Hinton G., “Deep learning,” Nature 521(7553), 436–444 (2015). 10.1038/nature14539 [DOI] [PubMed] [Google Scholar]
- 18.Hinton G., et al. , “Deep neural networks for acoustic modeling in speech recognition: the shared views of four research groups,” IEEE Signal Process. Mag. 29(6), 82–97 (2012). 10.1109/MSP.2012.2205597 [DOI] [Google Scholar]
- 19.Krizhevsky A., Sutskever I., Hinton G., “ImageNet classification with deep convolutional neural networks,” in Advances in Neural Information Processing Systems, pp. 1097–1105 (2012). [Google Scholar]
- 20.Russakovsky O., et al. , “ImageNet large scale visual recognition challenge,” Int. J. Comput. Vision 115(3), 211–252 (2015). 10.1007/s11263-015-0816-y [DOI] [Google Scholar]
- 21.Silver D., et al. , “Mastering the game of go with deep neural networks and tree search,” Nature 529(7587), 484–489 (2016). 10.1038/nature16961 [DOI] [PubMed] [Google Scholar]
- 22.Litjens G., et al. , “A survey on deep learning in medical image analysis,” Med. Image Anal. 42(Suppl. C), 60–88 (2017). 10.1016/j.media.2017.07.005 [DOI] [PubMed] [Google Scholar]
- 23.Gulshan V., et al. , “Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs,” J. Am. Med. Assoc. 316(22), 2402–2410 (2016). 10.1001/jama.2016.17216 [DOI] [PubMed] [Google Scholar]
- 24.Esteva A., et al. , “Dermatologist-level classification of skin cancer with deep neural networks,” Nature 542(7639), 115–118 (2017). 10.1038/nature21056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ehteshami Bejnordi B., van der Laak J., “Camelyon16: grand challenge on cancer metastasis detection in lymph nodes 2016,” https://camelyon16.grand-challenge.org (14 Novermber 2017).
- 26.He K., et al. , “Deep residual learning for image recognition,” in Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition, pp. 770–778 (2016). [Google Scholar]
- 27.Zagoruyko S., Komodakis N., “Wide residual networks,” in Proc. of the British Machine Vision Conf. (BMVC), Wilson R. C., et al., Eds., BMVA Press, p. 87 (2016). 10.5244/C.30.87 [DOI] [Google Scholar]
- 28.Simonyan K., Zisserman A., “Very deep convolutional networks for large-scale image recognition,” CoRR abs/1409.1556 (2014).
- 29.Long J., Shelhamer E., Darrell T., “Fully convolutional networks for semantic segmentation,” in Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition, pp. 3431–3440 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Bejnordi B. E., et al. , “Novel chromatin texture features for the classification of pap smears,” Proc. SPIE 8676, 867608 (2013). 10.1117/12.2007185 [DOI] [Google Scholar]
- 31.Litjens G., “Automated slide analysis platform (ASAP),” https://github.com/GeertLitjens/ASAP (14 November 2017).
- 32.He K., et al. , “Delving deep into rectifiers: surpassing human-level performance on imagenet classification,” in Proc. of the IEEE Int. Conf. on Computer Vision, pp. 1026–1034 (2015). [Google Scholar]
- 33.Cohen J., “A coefficient of agreement for nominal scales,” Educ. Psychol. Meas. 20(1), 37–46 (1960). 10.1177/001316446002000104 [DOI] [Google Scholar]
- 34.Efron B., “Bootstrap methods: another look at the jackknife,” in Breakthroughs in Statistics, pp. 569–593, Springer, New York: (1992). [Google Scholar]
- 35.Ghafoorian M., et al. , “Deep multi-scale location-aware 3D convolutional neural networks for automated detection of lacunes of presumed vascular origin,” Neuroimage: Clin. 14, 391–399 (2017). 10.1016/j.nicl.2017.01.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ronneberger O., Fischer P., Brox T., “U-net: convolutional networks for biomedical image segmentation,” in Int. Conf. on Medical Image Computing and Computer-Assisted Intervention, pp. 234–241, Springer; (2015). [Google Scholar]
- 37.Dalmış M. U., et al. , “Using deep learning to segment breast and fibroglandular tissue in MRI volumes,” Med. Phys. 44(2), 533–546 (2017). 10.1002/mp.12079 [DOI] [PubMed] [Google Scholar]
- 38.Foschini M. P., et al. , “Differential expression of myoepithelial markers in salivary, sweat and mammary glands,” Int. J. Surg. Pathol. 8(1), 29–37 (2000). 10.1177/106689690000800108 [DOI] [PubMed] [Google Scholar]
- 39.Bejnordi B. E., et al. , “Stain specific standardization of whole-slide histopathological images,” IEEE Trans. Med. Imaging 35(2), 404–415 (2016). 10.1109/TMI.2015.2476509 [DOI] [PubMed] [Google Scholar]
- 40.Khan A. M., et al. , “A nonlinear mapping approach to stain normalization in digital histopathology images using image-specific color deconvolution,” IEEE Trans. Biomed. Eng. 61(6), 1729–1738 (2014). 10.1109/TBME.2014.2303294 [DOI] [PubMed] [Google Scholar]
- 41.Macenko M., et al. , “A method for normalizing histology slides for quantitative analysis,” in IEEE Int. Symp. on Biomedical Imaging: From Nano to Macro (ISBI ’09), pp. 1107–1110, IEEE; (2009). 10.1109/ISBI.2009.5193250 [DOI] [Google Scholar]
- 42.Ciompi F., et al. , “The importance of stain normalization in colorectal tissue classification with convolutional networks,” arXiv preprint arXiv:1702.05931 (2017).
- 43.Wang D., et al. , “Deep learning for identifying metastatic breast cancer,” arXiv preprint arXiv:1606.05718 (2016).
- 44.Bejnordi B. E., et al. , “Quantitative analysis of stain variability in histology slides and an algorithm for standardization,” Proc. SPIE 9041, 904108 (2014). 10.1117/12.2043683 [DOI] [Google Scholar]
- 45.Veta M., “Tupac16: tumor proliferation assessment challenge 2016,” http://tupac.tue-image.nl (14 November 2017).