Abstract
The formation of condensed, transcriptionally repressed heterochromatin is essential for controlling gene expression throughout development, silencing parasitic DNA elements, and for genome stability and inheritance. Cells employ diverse mechanisms for controlling heterochromatin states through proteins that modify DNA and histones. An emerging theme is that chromatin-associated RNAs play important roles in regulating heterochromatin proteins by controlling their initial recruitment to chromatin, their stable association with chromatin, their spread along chromatin, or their enzymatic activity. Major challenges for the field include not only identifying regulatory RNAs, but understanding the underlying biochemical mechanisms for how RNAs associate with chromatin, the specificity of interactions between heterochromatin proteins and RNA, and how these binding events manifest in cells to orchestrate RNA-mediated regulation of heterochromatin.
Introduction
A single diploid nucleus in a human zygote gives rise to all the diverse cell types in the body. This is achieved though cell type specific gene expression, which must be established during development and maintained in differentiated somatic cells. The nucleus must also constitutively silence diverse DNA elements — such as selfish transposons and non-coding repetitive sequences — that threaten genome integrity. To achieve both facultative and constitutive transcriptional silencing, eukaryotic organisms regulate DNA through the formation of repressive chromatin states, called heterochromatin.
Heterochromatin regulation begins at the level of the nucleosome — the fundamental basic unit of chromatin — which is comprised of DNA wrapped around an octameric histone core. Cells employ covalent modifications on DNA and histone proteins, as well as specialized histone variants, to regulate several aspects of chromatin behavior, such as transcriptional output, regulatory protein accessibility, compaction, and sub-nuclear localization. Although many protein factors involved in establishing and propagating heterochromatin states have been identified, many large questions remain, including: how do heterochromatin proteins find and modify specific genomic loci? How are improper modifications prevented? And how is heterochromatin regulated throughout development, across cell divisions, and over generations?
Chromatin-associated RNAs have emerged as important regulators of the interactions between chromatin and chromatin-modifying proteins. This idea first gained wide recognition with the characterization of Xist, a non-coding RNA that coats and silences an entire chromosome as part of a dosage compensation mechanism in most mammals to equalize X chromosome expression across the sexes. Since this initial discovery, chromatin-associated RNAs have been shown to influence not only developmentally-controlled facultative heterochromatin, but also the constitutively repressed, gene-poor, repetitive DNA of eukaryotic genomes. These RNAs apparently perform diverse functions, including recruiting factors for establishment of heterochromatin, maintaining heterochromatin over many cell divisions, and/or limiting the activity or spread of heterochromatin proteins.
A complete discussion of RNA-mediated regulation of chromatin is outside the scope of this review (for an excellent in-depth review, see [1]); however, we will focus on recent research on chromatin-associated RNAs that control either facultative or constitutive heterochromatin, with an emphasis on the interactions between RNA and chromatin modifying proteins in mammalian cells. We will discuss challenges in the field for understanding how direct interactions mediate RNA control of heterochromatin, possible models for RNA-dependent chromatin control, and key questions going forward.
Non-coding RNAs that control facultative heterochromatin
As mentioned above, the field of chromatin-associated RNA largely arose with the discovery of Xist RNA and its unique role in X chromosome inactivation in eutherian mammals (for current reviews, see references [2,3]). Although a single Xist locus exists on each X chromosome, Xist RNA is transcribed from only one of the two alleles in XX females, and its localization spreads in cis to cover and inactivate the entire chromosome. Analogous RNAs play similar roles in dosage compensation in other systems, either to repress or upregulate gene expression (e.g. roX1 and roX2 RNA in flies [4], Rsx RNA in metatherians [5]).
Xist RNA is proposed to silence gene expression by directly binding to and recruiting chromatin modifying proteins to chromatin. One of these factors is Polycomb Repressive Complex 2 (PRC2), a multi-subunit complex with H3K27 methyltransferase activity and an essential regulator of gene expression during development and cell differentiation. Though previous work proposed that PRC2 directly and specifically recognizes a stem loop structural element within the Repeat A (RepA) portion of Xist RNA [6–8], recent work has shown PRC2 binding to be promiscuous in vitro and in vivo [9,10], and demonstrated that the RepA stem loop is insufficient for PRC2 recruitment [11•]. In addition, although chromatin immunoprecipitation (ChIP-seq) and capture hybridization analysis of RNA targets (CHART-seq) strategies show high correlation between PRC2, H3K27me3, and Xist RNA localization across the inactive X chromosome [12,13], PRC2 and Xist show weak colocalization by superresolution microscopy [14], and purification of Xist RNA from cells using antisense RNA pull down strategies failed to identify PRC2 as an interactor of Xist RNA in cells [15••,16••]. These data and others point to a model in which PRC2, if it does directly interact with Xist RNA, perhaps does so transiently, and that other interactions may be largely responsible for driving heterochromatin formation on the inactive X, even if PRC2 contributes with other redundant mechanisms to its long-term maintenance [17,18].
A likely candidate for an Xist-recruited heterochromatin initiator is SHARP (SMRT/HDAC1 Associated Repressor Protein), a mammalian homolog of the fly protein SPEN (Split Ends), identified in two independent studies as a in vivo binder of Xist RNA [15••,16••]. SHARP contains three RNA recognition motifs (RRMs), binds to histone deacetylases, and appears to be responsible for early repressive marks on the inactive X and for downstream recruitment of PRC2, making it an attractive candidate for a direct molecular link between Xist localization and transcriptional repression [16••]. Future research will likely explore the RNA binding specificity of SHARP in vitro and in vivo, to determine whether specific Xist RNA sequences or structures may specifically recruit SHARP to the inactive X, or if SHARP exhibits broad, promiscuous RNA binding that may non-specifically stabilize its interactions with chromatin.
Since the discovery of Xist, other chromatin-associated RNAs have been discovered that mediate gene silencing at loci distant from their transcription sites. A landmark example of trans regulation is HOTAIR, a non-coding RNA associated with the HOX gene family, whose precise spatiotemporal regulation is necessary for proper pattern formation during development. Initial experiments performed in human cells indicated that although HOTAIR RNA is transcribed from the HOXC cluster of HOX genes, it localizes to the distant HOXD cluster in trans and represses HOXD transcription by recruiting PRC2 [19]; subsequent studies found this mechanism to be conserved in mice, and observed that loss of HOTAIR leads to developmental abnormalities [20]. However, another study challenged the assertion that this model is conserved and important in mouse development, as HOXD derepression was not detected in mice with the entire HOXC locus deleted [21]. A recent follow-up study has suggested that developmental defects in HOTAIR mutant mice are not due to HOXD derepression in trans, but from an unrelated mechanism of HOXC chromatin disruption in cis [22•]. The trans mechanism is likely to still apply in humans, as HOTAIR was found to localize to the HOXD locus in human cells by ChIRP-seq [23]; nevertheless, its relevance to human development is impossible to test directly. In addition, similar to the recent models for Xist described above, it has been recently suggested that contrary to the original models, HOTAIR can lead to transcriptional repression independent of PRC2 recruitment [24], suggesting a downstream, maintenance role for PRC2.
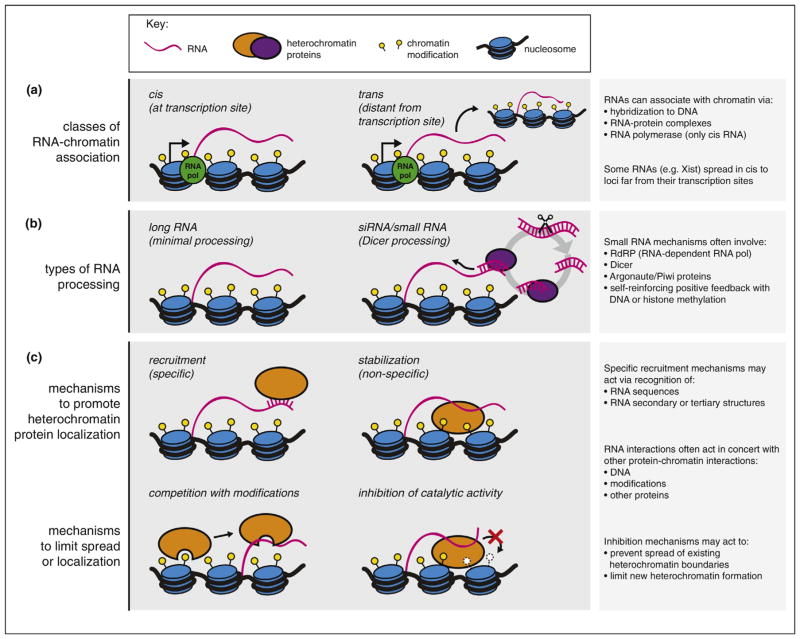
A key objective for those studying HOTAIR or other trans-acting RNAs, in addition to elucidating their biological relevance, is understanding the mechanisms by which RNAs find and associate with their target loci, and how localization specificity is achieved (Figure 1). The development of tools for unbiased discovery and mapping of chromatin-associated RNAs will undoubtedly clarify the prevalence of trans-acting mechanisms, identify classes of RNAs acting by these mechanisms, and begin to dissect their modes of chromatin recognition [25,26].
Figure 1.
General modes of RNA-based heterochromatin regulation. (a) Classes of RNA-chromatin association. Chromatin-associated RNAs may localize mainly at the sites of their transcription (cis), or at sites distant from their transcription sites (trans). (b) Types of RNA processing. Characterized RNA-based mechanisms for heterochromatin regulation fall into two general categories: long, noncoding RNAs or short, siRNA-based mechanisms. (c) Models for how RNA regulates localization of heterochromatin proteins. Chromatin-associated RNAs may act to promote (top) or inhibit (bottom) the localization of heterochromatin proteins. Promotion of protein localization may occur through recruitment through direct recognition of specific RNA sequences and/or structures, or through non-specific interactions that stabilize protein-chromatin associations. Heterochromatin inhibition mechanisms include RNA binding events that compete with other chromatin interactions (such as binding to heterochromatin modifications), or RNA binding that inhibits enzymatic activity.
PRC2: a model chromatin modifying complex regulated by RNA
Because of its established role in facultative heterochromatin — and its proposed association with Xist, HOTAIR, and other RNAs — the PRC2 complex became a model chromatin-modifying complex influenced by RNA binding [27]. PRC2 is a large multi-subunit protein complex with many essential and accessory factors [28]. PRC2 associates with Polycomb Responsive Elements (PREs) in the fly genome, which are necessary for maintenance of silencing over multiple rounds of DNA replication; however, in non-dividing cells, PRC2 localizes independent of these DNA sequences [29,30], and may depend on other interactors such as H3K27 methylation and RNA. Mammalian PRC2 binds RNA through the H3K27 methyltransferase subunit EZH2 and the subunit Eed; but other subunits and modifications can affect RNA binding affinity and specificity [8,31]. Although PRC2 was initially thought to associate with specific RNA sequences through recognition of a stem loop structure [8], recent work has demonstrated that PRC2 is a highly promiscuous RNA binder in cells [9,10] and exhibits largely nonspecific interactions with RNA in vitro [11•]. Several studies have indicated that G-quadraplex RNA structures are preferentially recognized by PRC2; a motif common enough to explain broad binding to many RNAs [32•,33,34].
How RNA binding influences PRC2 localization and silencing at specific loci remains a key question in the field, with several possible models that are not mutually exclusive (Figure 1). One model is that RNA binding is the main determinant of PRC2 recruitment, and this RNA recognition depends on some sequence or structure specificity that may be cryptic [35,36]. Another model is that RNA binding is just one of many biochemical interactions with PRC2 that define its genomic targets — in addition to binding to DNA, H3K27-methylated histones, and other factors [28] — and may either contribute to initial recruitment (increasing on rate) or to post-recruitment stabilization (decreasing off rate). A third model is the RNA association by PRC2 reduces its interaction with non-target loci by clearing PRC2 away from loci that are actively transcribed [9], and/or by inhibiting its methyltransferase activity [34]. Going forward, to clarify the role of each one of these models, it will be essential to systematically combine unbiased in vitro and in vivo RNA binding measurements to understand PRC2 RNA binding specificity in cells, how that relates to its intrinsic RNA binding, what factors influence that specificity, and how PRC2 integrates this information to find target genomic sites.
A role for RNAi in constitutive heterochromatin
In addition to regulating gene expression coupled to organismal development and cell differentiation, non-coding RNAs have also been implicated in the constitutive silencing of gene-poor, repetitive regions of eukaryotic genomes, often concentrated near centromeres and telomeres. Constitutive heterochromatin components include the HP1 (heterochromatin protein 1) and SUV39 (suppressor of variegation 3–9) protein families, which are conserved in a broad range of eukaryotes — including those lacking H3K27-methylated facultative heterochromatin. SUV39 proteins are histone methyltransferases that catalyze the di-methylation and tri-methylation of H3K9; a chromatin mark specifically recognized by both SUV39 and HP1 proteins. HP1 proteins are highly versatile adaptor proteins that can oligomerize and recruit other chromatin factors, resulting in chromatin compaction and transcriptional silencing [37].
Perhaps the most well-studied example of RNA-regulated constitutive heterochromatin is the role for RNAi in pericentric heterochromatin formation and maintenance in fission yeast (Schizosaccharomyces pombe). In this system, RNAs transcribed from pericentric repeat sequences are converted into double-stranded species and processed by Dicer into short siRNAs, which bind to the Argonaute-containing RITS (RNA-induced transcriptional silencing) complex. The RITS complex uses base pairing interactions between the loaded siRNA and nascent transcripts — along with H3K9me2 interactions — to localize to pericentric regions [1]. Localized RITS complexes recruit Clr4/SUV39, leading to H3K9 methylation, Swi6/HP1 recruitment, heterochromatic silencing, and generation of more siRNAs through recruitment of Dicer and an RNA-dependent RNA polymerase (RdRP). In this way, heterochromatin is maintained through a self-reinforcing positive feedback loop; siRNA promotes H3K9 methylation which promotes siRNA production, and both siRNA and H3K9me2 interact with the RITS complex to facilitate its localization to pericentric heterochromatin.
Why would this type of RNA-mediated heterochromatin regulation be beneficial? In contrast to proposed roles of Xist and HOTAIR, fission yeast pericentric RNAs function exclusively at the site of local transcription in cis (Figure 1). This means that RNA transcription is required to effectively silence itself. An attractive advantage of this cis mechanism is that in addition to the positive feedback generation of siRNA, there is negative feedback for RNA transcription, as any defects in transcriptional silencing lead to increased recruitment of heterochromatin factors and reestablishment of repression. In addition, recent studies have shown that a H3K9me2-dependent feedback loop — that is, Clr4/SUV39 recruitment though its own methylation product — on its own is only sufficient for long-term maintenance of heterochromatin in the absence of the endogenous demethylase Epe1 [38••,39••]. Therefore, the siRNA feedback loop contributes an extra layer of regulation that allows for more robust silencing — less vulnerable to an imbalance between demethylation and methylation — than H3K9me2 feedback alone. Finally, these mechanisms are less susceptible to DNA mutations than a mechanism in which heterochromatin factors simply recognize a DNA sequence; which is especially advantageous for regions with high mutation rates, such as centromeres. Consistent with this idea, a heterochromatin region in fission yeast with a lower mutation rate than centromeres — the mating-type locus — relies on specific transcription factor binding DNA sequences for both establishment and maintenance of heterochromatin [40], and only requires RNAi for establishment [41]. In contrast, pericentric heterochromatin requires RNAi for both establishment and maintenance, with no known specific DNA binding factors. This suggests that sequence-specific binding factors may effectively maintain silencing at regions with relatively low mutation rates, but RNAi is a more effective at maintaining quickly evolving regions.
In addition to their role in fission yeast, small RNAs contribute to heterochromatin regulation in other fungi, ciliates, plants, and worms; and like in fission yeast, act through RdRP-dependent, self-reinforcing feedback loops with either histone methylation or DNA methylation systems [1]. How RNAi-based mechanisms contribute to constitutive heterochromatin formation in other eukaryotes such as flies and mammals is less clear, as RdRPs have not been identified in these organisms. RNAi may still play some role, however, as loss of Dicer leads to apparent misregulation of heterochromatin in Drosophila melanogaster cells [42], chicken/human hybrid cells [43], and mouse embryonic stem cells [44] — though small siRNA-sized pericentric RNAs have not been detected in other mammalian cell types [45]. Additionally, injection of pericentric major satellite dsRNAs into mouse embryos was sufficient to rescue phenotypes caused by mutating lysine 27 of histone H3.3 — a critical residue for pericentric heterochromatin establishment during mouse development — suggesting a role for H3.3 in dsRNA generation and RNAi-mediated heterochromatin silencing during this developmental stage [46].
Even though many metazoans lack RdRPs, they employ known germline-specific silencing mechanisms that rely on small RNAs. The largely germline-specific Piwi subfamily of Argonaute proteins and Piwi-interacting RNAs (piRNAs) act both post-transcriptionally and at chromatin to silence transposable elements in the germline and to maintain genome integrity [47]; though it is unclear, particularly in mice, if positive feedback between DNA/histone methylation and piRNA generation occurs.
Direct RNA regulation of constitutive heterochromatin factors HP1 and SUV39
RNA at constitutive heterochromatin can also directly bind HP1 and SUV39 proteins — factors of interest because their high conservation across diverse eukaryotes and their direct role in heterochromatin function. Like PRC2, the RNA binding activity of these proteins may contribute to their recruitment, their stable interaction with chromatin, and/or prevent their mislocalization to ectopic loci.
In addition to recruiting the RITS complex, heterochromatic RNAs in fission yeast have proposed roles in preventing heterochromatin spread. Highly-transcribed clusters of tRNAs genes have long been observed to prevent spread at heterochromatin boundaries; however, a direct role for RNA is unclear. Recently, it has been shown that a non-coding RNA called BORDERLINE, transcribed at a heterochromatin/euchromatin boundary with no tRNA genes, limits heterochromatin spread through a direct interaction with Swi6/HP1 [48]. Interestingly, direct RNA binding by Swi6/HP1 also assists in targeting heterochromatic RNAs for degradation [49]. This involves a mechanism in which RNA and H3K9me2 compete for binding to Swi6/HP1, although RNA and H3K9me2 bind different domains (the hinge and the chromodomain, respectively). Future work is needed to understand Swi6/HP1 RNA binding specificity, how RNA/H3K9me2 competition influences this specificity, and how this impacts heterochromatin regulation by Swi6/HP1.
RNA binding is conserved in at least some mammalian isoforms of HP1. Like Swi6/HP1, mammalian HP1α can directly bind RNA through its hinge domain [50], and in mouse cells, HP1α binds pericentric major satellite RNAs in a manner dependent on HP1α sumoylation [51]. Treatment of mammalian cells with RNase, or inhibition of HP1α sumoylation, reduces HP1α localization [51–53]. These data support a model in which mammalian HP1α, unlike fission yeast Swi6/HP1, relies on direct binding to RNA for its localization to chromatin. Interestingly, the sensitivity of HP1α localization to RNase, even after cells are permeabilized on coverslips, indicates that RNA acts as a structural component necessary for HP1α’s continued stable association with chromatin; as opposed to a dynamic interaction only necessary for recruitment. A recent paper also found that mouse Suv39h1 protein facilitates HP1α sumoylation, and therefore indirectly promotes RNA binding and localization of HP1α [54]. It is unclear whether RNA binding prevents HP1α and heterochromatin spread in mammalian cells, as it does in fission yeast.
Another conserved component of constitutive heterochromatin has been proposed to bind to RNA: the SUV39 methyltransferase responsible for catalyzing H3K9me2/3 formation. In fission yeast, the chromodomain of Clr4/SUV39 was found to bind RNA in vitro only in the presence of H3K9me3 histone tail peptide [55], suggesting a cooperative mechanism of H3K9me3 and RNA binding; however, the biological relevance of this interaction has yet to be explored. Two recent papers have presented evidence that the mammalian homolog SUV39H1 is targeted by RNA-based mechanisms: to telomeres by the non-coding RNA TERRA in human cells [56•], and to the Oct4 locus by an Oct4 pseudogene RNA in mouse cells [57]. It remains unclear how the RNA binding of HP1 and SUV39 isoforms contribute to constitutive heterochromatin in different eukaryotes, including humans (Box 1). Like with PRC2, it will be key to understand RNA binding specificities in vitro and how that relates to interactions and localization in vivo.
Box 1. The complex contributions of α-satellite transcription in human cells.
A unique challenge to studying constitutive heterochromatin and the effects of its transcription in human cells is the repetitive but heterogeneous nature of the α-satellite repeats that underlie both pericentric heterochromatin and core centromere regions of the human genome. Although repeats in mice (minor and major) and fission yeast (imr and otr) are distinctly partitioned to core centromere or pericentric regions, α-satellite RNAs in human cells may come from either region, making it difficult to decouple their contributions to centromere or heterochromatin function. At least some α-satellite sequences are transcribed in human cells, and have been proposed to influence several aspects of centromere biology [59–64]. A study in human cells showed that aberrant overexpression of α-satellite RNA was sufficient to cause genome instability [65], although the mechanisms by which this occurs remain unclear. These data, along with several studies involved targeted repression or expression of centromeric sequences on human artificial chromosome (HACs) [66–68], suggest that some α-satellite transcription may be necessary for proper centromere and/or heterochromatin function, but too much may have damaging effects on the cell. Recent efforts to better classify and map α-satellite repeats of the human genome will greatly improve our understanding of how these sequences contribute to centromere and pericentric heterochromatin function [69].
Concluding remarks
Chromatin-associated RNAs play diverse roles in genome regulation [58], including regulating both facultative and constitutive heterochromatin in a wide variety of eukaryotes. These heterochromatin-controlling RNAs exhibit a range of different behaviors; localizing through either cis or trans mechanisms, using long RNA tethers or short RNA feedback loops, and either inhibiting or promoting heterochromatin function. Key questions going forward in this field include: where do chromatin-associated RNAs interact with chromatin, what heterochromatin proteins do they interact with, and what is the basis of the specificity of these interactions? And how do these biochemical interactions lead to spatially and temporally controlled heterochromatin in cells? The development of new unbiased techniques for assessing RNA binding interactions in vivo and in vitro, in combination with traditional cell-based and biochemical methods, will allow us to further unravel the complex interactions of chromatin-associated RNAs, and fully understand their contributions to the specificity of chromatin-modifying complexes.
Acknowledgments
We thank Bradley French and Shengya Cao for valuable feedback. We apologize to all those whose work was not mentioned here due to space limitations.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Holoch D, Moazed D. RNA-mediated epigenetic regulation of gene expression. Nat Rev Genet. 2015;16:71–84. doi: 10.1038/nrg3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galupa R, Heard E. X-chromosome inactivation: new insights into cis and trans regulation. Curr Opin Genet Dev. 2015;31:57–66. doi: 10.1016/j.gde.2015.04.002. [DOI] [PubMed] [Google Scholar]
- 3.Robert Finestra T, Gribnau J. X chromosome inactivation: silencing, topology and reactivation. Curr Opin Cell Biol. 2017;46:54–61. doi: 10.1016/j.ceb.2017.01.007. [DOI] [PubMed] [Google Scholar]
- 4.Meller VH, Gordadze PR, Park Y, Chu X, Stuckenholz C, Kelley RL, Kuroda MI. Ordered assembly of roX RNAs into MSL complexes on the dosage-compensated X chromosome in Drosophila. Curr Biol. 2000;10:136–143. doi: 10.1016/s0960-9822(00)00311-0. [DOI] [PubMed] [Google Scholar]
- 5.Grant J, Mahadevaiah SK, Khil P, Sangrithi MN, Royo H, Duckworth J, McCarrey JR, VandeBerg JL, Renfree MB, Taylor W, et al. Rsx is a metatherian RNA with Xist-like properties in X-chromosome inactivation. Nature. 2012;487:254–258. doi: 10.1038/nature11171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanhere A, Viiri K, Araújo CC, Rasaiyaah J, Bouwman RD, Whyte WA, Pereira CF, Brookes E, Walker K, Bell GW, et al. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol Cell. 2010;38:675–688. doi: 10.1016/j.molcel.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao J, Sun BK, Erwin JA, Song J-J, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cifuentes-Rojas C, Hernandez AJ, Sarma K, Lee JT. Regulatory interactions between RNA and polycomb repressive complex 2. Mol Cell. 2014;55:171–185. doi: 10.1016/j.molcel.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davidovich C, Zheng L, Goodrich KJ, Cech TR. Promiscuous RNA binding by Polycomb repressive complex 2. Nat Struct Mol Biol. 2013;20:1250–1257. doi: 10.1038/nsmb.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaneko S, Son J, Shen SS, Reinberg D, Bonasio R. PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nat Struct Mol Biol. 2013;20:1258–1264. doi: 10.1038/nsmb.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11•.Davidovich C, Wang X, Cifuentes-Rojas C, Goodrich KJ, Gooding AR, Lee JT, Cech TR. Toward a consensus on the binding specificity and promiscuity of PRC2 for RNA. Mol Cell. 2015;57:552–558. doi: 10.1016/j.molcel.2014.12.017. This study makes significant strides towards understanding the basis of PRC2 RNA binding specificity, and clarifies the role or lack thereof of the RepA two-hairpin motif in PRC2 recruitment. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simon MD, Pinter SF, Fang R, Sarma K, Rutenberg-Schoenberg M, Bowman SK, Kesner BA, Maier VK, Kingston RE, Lee JT. High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature. 2013;504:465–469. doi: 10.1038/nature12719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engreitz JM, Pandya-Jones A, McDonel P, Shishkin A, Sirokman K, Surka C, Kadri S, Xing J, Goren A, Lander ES, et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341:1237973. doi: 10.1126/science.1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cerase A, Smeets D, Tang YA, Gdula M, Kraus F, Spivakov M, Moindrot B, Leleu M, Tattermusch A, Demmerle J, et al. Spatial separation of Xist RNA and polycomb proteins revealed by superresolution microscopy. Proc Natl Acad Sci U S A. 2014;111:2235–2240. doi: 10.1073/pnas.1312951111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15••.Chu C, Zhang QC, da Rocha ST, Flynn RA, Bharadwaj M, Calabrese JM, Magnuson T, Heard E, Chang HY. Systematic discovery of Xist RNA binding proteins. Cell. 2015;161:404–416. doi: 10.1016/j.cell.2015.03.025. This paper, similar to McHugh et al. uses a specific RNA pulldown strategy followed by mass spectrometry to identify in vivo protein interactions of the Xist RNA. SPEN/SHARP is identified as a key Xist interactor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16••.McHugh CA, Chen C-K, Chow A, Surka CF, Tran C, McDonel P, Pandya-Jones A, Blanco M, Burghard C, Moradian A, et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015;521:232–236. doi: 10.1038/nature14443. This paper, similar to Chu et al. uses a specific RNA pulldown strategy followed by mass spectrometry to identify in vivo protein interactions of the Xist RNA. SPEN/SHARP is identified as a key Xist interactor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schoeftner S, Sengupta AK, Kubicek S, Mechtler K, Spahn L, Koseki H, Jenuwein T, Wutz A. Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. EMBO J. 2006;25:3110–3122. doi: 10.1038/sj.emboj.7601187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalantry S, Magnuson T. The polycomb group protein EED is dispensable for the initiation of random X-chromosome inactivation. PLoS Genet. 2006;2:e66–e69. doi: 10.1371/journal.pgen.0020066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li L, Liu B, Wapinski OL, Tsai M-C, Qu K, Zhang J, Carlson JC, Lin M, Fang F, Gupta RA, et al. Targeted disruption of Hotair leads to homeotic transformation and gene derepression. Cell Rep. 2013;5:3–12. doi: 10.1016/j.celrep.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schorderet P, Duboule D. Structural and functional differences in the long non-coding RNA Hotair in mouse and human. PLoS Genet. 2011;7:e1002071–e1002110. doi: 10.1371/journal.pgen.1002071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22•.Amândio AR, Necsulea A, Joye E, Mascrez B, Duboule D. Hotair is dispensible for mouse development. PLoS Genet. 2016;12:e1006232. doi: 10.1371/journal.pgen.1006232. Reanalysis of HOTAIR mutant mice overturns the paradigm that HOTAIR represses the HoxD locus in trans. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Portoso M, Ragazzini R, Brenčič Ž, Moiani A, Michaud A, Vassilev I, Wassef M, Servant N, Sargueil B, Margueron R. PRC2 is dispensable for HOTAIR-mediated transcriptional repression. EMBO J. 2017;36:981–994. doi: 10.15252/embj.201695335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sridhar B, Rivas-Astroza M, Nguyen TC, Chen W, Yan Z, Cao X, Hebert L, Zhong S. Systematic mapping of RNA-chromatin interactions in vivo. Curr Biol. 2017;27:602–609. doi: 10.1016/j.cub.2017.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bell JC, Jukam D, Teran NA, Risca VI, Smith OK, Johnson WL, Skotheim J, Greenleaf WJ, Straight A. Chromatin-associated RNA sequencing (ChAR-seq) maps genome-wide RNA-to-DNA contacts. 2017 doi: 10.7554/eLife.27024. http://dx.doi.org/10.1101/118786. [DOI] [PMC free article] [PubMed]
- 27.Brockdorff N. Noncoding RNA and polycomb recruitment. RNA. 2013;19:429–442. doi: 10.1261/rna.037598.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Margueron R, Reinberg D. The polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laprell F, Finkl K, Müller J. Propagation of polycomb-repressed chromatin requires sequence-specific recruitment to DNA. Science. 2017 doi: 10.1126/science.aai8266. http://dx.doi.org/10.1126/science.aai8266. [DOI] [PubMed]
- 30.Coleman RT, Struhl G. Causal role for inheritance of H3K27me3 in maintaining the OFF state of a Drosophila HOX gene. Science. 2017 doi: 10.1126/science.aai8236. http://dx.doi.org/10.1126/science.aai8236. [DOI] [PMC free article] [PubMed]
- 31.Kaneko S, Li G, Son J, Xu C-F, Margueron R, Neubert TA, Reinberg D. Phosphorylation of the PRC2 component Ezh2 is cell cycle-regulated and up-regulates its binding to ncRNA. Genes Dev. 2010;24:2615–2620. doi: 10.1101/gad.1983810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32•.Wang X, Goodrich KJ, Gooding AR, Naeem H, Archer S, Paucek RD, Youmans DT, Cech TR, Davidovich C. Targeting of polycomb repressive complex 2 to RNA by short repeats of consecutive guanines. Mol Cell. 2017;65:1056–1067.e5. doi: 10.1016/j.molcel.2017.02.003. This analysis of new in vitro and existing in vivo data provides strong evidence that short guanine repeats — which are prone to form G-quadruplex structures — are the basis for PRC2 RNA-binding promiscuity. [DOI] [PubMed] [Google Scholar]
- 33.Wu L, Murat P, Matak-Vinkovic D, Murrell A, Balasubramanian S. Binding interactions between long noncoding RNA HOTAIR and PRC2 proteins. Biochemistry. 2013;52:9519–9527. doi: 10.1021/bi401085h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaneko S, Son J, Bonasio R, Shen SS, Reinberg D. Nascent RNA interaction keeps PRC2 activity poised and in check. Genes Dev. 2014;28:1983–1988. doi: 10.1101/gad.247940.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Betancur G, Tomari YJ. Cryptic RNA-binding by PRC2 components EZH2 and SUZ12. RNA Biol. 2015;12:959–965. doi: 10.1080/15476286.2015.1069463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tu S, Yuan G-C, Shao Z. The PRC2-binding long non-coding RNAs in human and mouse genomes are associated with predictive sequence features. Sci Rep. 2017 doi: 10.1038/srep41669. http://dx.doi.org/10.1038/srep41669. [DOI] [PMC free article] [PubMed]
- 37.Grewal SIS, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 38••.Ragunathan K, Jih G, Moazed D. Epigenetic inheritance uncoupled from sequence-specific recruitment. Science. 2015;348:1258699. doi: 10.1126/science.1258699. Together with Audergon et al. this paper shows that long-term epigenetic maintenance of heterochromatin through Clr4-dependent H3K9 methylation feedback only occurs in the absence of the demethylase Epe1, suggesting H3K9me2-independent mechanisms are also necessary. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39••.Audergon PNCB, Catania S, Kagansky A, Tong P, Shukla M, Pidoux AL, Allshire RC. Epigenetics. Restricted epigenetic inheritance of H3K9 methylation. Science. 2015;348:132–135. doi: 10.1126/science.1260638. Together with Ragunathan et al. this paper shows that long-term epigenetic maintenance of heterochromatin through Clr4-dependent H3K9 methylation feedback only occurs in the absence of the demethylase Epe1, suggesting H3K9me2-independent mechanisms are also necessary. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang X, Moazed D. DNA sequence-dependent epigenetic inheritance of gene silencing and histone H3K9 methylation. Science. 2017;356:88–91. doi: 10.1126/science.aaj2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hall I, Shankaranarayana G, Noma K, Ayoub N, Cohen A, Grewal S. Establishment and maintenance of a heterochromatin domain. Science. 2002;297:2232. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- 42.Pal-Bhadra M, Leibovitch B, Gandhi S, Rao M, Bhadra U, Birchler J, Elgin S. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science. 2004;303:669. doi: 10.1126/science.1092653. [DOI] [PubMed] [Google Scholar]
- 43.Fukagawa T, Nogami M, Yoshikawa M, Ikeno M, Okazaki T, Takami Y, Nakayama T, Oshimura M. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat Cell Biol. 2004;6:784–791. doi: 10.1038/ncb1155. [DOI] [PubMed] [Google Scholar]
- 44.Kanellopoulou C, Muljo S, Kung A, Ganesan S, Drapkin R, Jenuwein T, Livingston D, Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saksouk N, Simboeck E, Déjardin J. Constitutive heterochromatin formation and transcription in mammals. Epigenetics Chromatin. 2015;8:3. doi: 10.1186/1756-8935-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Santenard A, Ziegler-Birling C, Koch M, Tora L, Bannister AJ, Torres-Padilla M-E. Heterochromatin formation in the mouse embryo requires critical residues of the histone variant H3. 3. Nat Cell Biol. 2010;12:853–862. doi: 10.1038/ncb2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656–668. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Keller C, Kulasegaran-Shylini R, Shimada Y, Hotz H-R, Bühler M. Noncoding RNAs prevent spreading of a repressive histone mark. Nat Struct Mol Biol. 2013 doi: 10.1038/nsmb1113-1340. http://dx.doi.org/10.1038/nsmb.2619. [DOI] [PubMed]
- 49.Keller C, Adaixo R, Stunnenberg R, Woolcock KJ, Hiller S, Bühler M. HP1Swi6 mediates the recognition and destruction of heterochromatic RNA transcripts. Mol Cell. 2012 doi: 10.1016/j.molcel.2012.05.009. http://dx.doi.org/10.1016/j.molcel.2012.05.009. [DOI] [PubMed]
- 50.Muchardt C, Guillemé M, Seeler J, Trouche D, Dejean A, Yaniv M. Coordinated methyl and RNA binding is required for heterochromatin localization of mammalian HP1α. EMBO Rep. 2002;3:975–981. doi: 10.1093/embo-reports/kvf194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maison C, Bailly D, Roche D, de Oca RM, Probst AV, Vassias I, Dingli F, Lombard B, Loew D, Quivy J-P, et al. SUMOylation promotes de novo targeting of HP1α to pericentric heterochromatin. Nat Genet. 2011 doi: 10.1038/ng.765. http://dx.doi.org/10.1038/ng.765. [DOI] [PubMed]
- 52.Maison C, Bailly D, Peters A, Quivy J, Roche D, Taddei A, Lachner M, Jenuwein T, Almouzni G. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat Genet. 2002;30:329–334. doi: 10.1038/ng843. [DOI] [PubMed] [Google Scholar]
- 53.Wong L, Brettingham-Moore K, Chan L, Quach J, Anderson M, Northrop E, Hannan R, Saffery R, Shaw M, Williams E. Centromere RNA is a key component for the assembly of nucleoproteins at the nucleolus and centromere. Genome Res. 2007;17:1146. doi: 10.1101/gr.6022807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maison C, Bailly D, Quivy JP, Almouzni G. The methyltransferase Suv39h1 links the SUMO pathway to HP1α marking at pericentric heterochromatin. Nat Commun. 2016;7:12224. doi: 10.1038/ncomms12224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ishida M, Shimojo H, Hayashi A, Kawaguchi R, Ohtani Y, Uegaki K, Nishimura Y, Nakayama J-I. Intrinsic nucleic acid-binding activity of Chp1 chromodomain is required for heterochromatic gene silencing. Mol Cell. 2012:017. doi: 10.1016/j.molcel.2012.05.017. http://dx.doi.org/10.1016/j.molcel.2012.05. [DOI] [PubMed]
- 56•.Porro A, Feuerhahn S, Delafontaine J, Riethman H, Rougemont J, Lingner J. Functional characterization of the TERRA transcriptome at damaged telomeres. Nat Commun. 2014;5:1–13. doi: 10.1038/ncomms6379. This paper first characterizes the binding of mammalian SUV39H1 to RNA, and proposes a role for this interaction in telomere regulation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Scarola M, Comisso E, Pascolo R, Chiaradia R, Marion RM, Schneider C, Blasco MA, Schoeftner S, Benetti R. Epigenetic silencing of Oct4 by a complex containing SUV39H1 and Oct4 pseudogene lncRNA. Nat Commun. 2015;6:7631. doi: 10.1038/ncomms8631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hendrickson D, Kelley DR, Tenen D, Bernstein B, Rinn JL. Widespread RNA binding by chromatin-associated proteins. Genome Biol. 2016 doi: 10.1186/s13059-016-0878-3. http://dx.doi.org/10.1186/s13059-016-0878-3. [DOI] [PMC free article] [PubMed]
- 59.Ferri F, Bouzinba-Segard H, Velasco G, Hube F, Francastel C. Non-coding murine centromeric transcripts associate with and potentiate Aurora B kinase. Nucleic Acids Res. 2009;37:5071–5080. doi: 10.1093/nar/gkp529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ideue T, Cho Y, Nishimura K, Tani T. Involvement of satellite I noncoding RNA in regulation of chromosome segregation. Genes Cells. 2014 doi: 10.1111/gtc.12149. http://dx.doi.org/10.1111/gtc.12149. [DOI] [PubMed]
- 61.Jambhekar A, Emerman AB, Schweidenback CTH, Blower MD. RNA stimulates aurora b kinase activity during mitosis. PLoS ONE. 2014;9:e100748. doi: 10.1371/journal.pone.0100748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mallm J-P, Rippe K. Aurora kinase B regulates telomerase activity via a centromeric RNA in stem cells. Cell Rep. 2015 doi: 10.1016/j.celrep.2015.05.015. http://dx.doi.org/10.1016/j.celrep.2015.05.015. [DOI] [PubMed]
- 63.Quénet D, Dalal Y. A long non-coding RNA is required for targeting centromeric protein A to the human centromere. Elife. 2014;3:e03254. doi: 10.7554/eLife.03254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu H, Qu Q, Warrington R, Rice A, Cheng N, Yu H. Mitotic transcription installs Sgo1 at centromeres to coordinate chromosome segregation. Mol Cell. 2015;59:426–436. doi: 10.1016/j.molcel.2015.06.018. [DOI] [PubMed] [Google Scholar]
- 65.Zhu Q, Pao GM, Huynh AM, Suh H, Tonnu N, Nederlof PM, Gage FH, Verma IM. BRCA1 tumour suppression occurs via heterochromatin-mediated silencing. Nature. 2011;477:179–184. doi: 10.1038/nature10371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cardinale S, Bergmann JH, Kelly D, Nakano M, Valdivia MM, Kimura H, Masumoto H, Larionov V, Earnshaw WC. Hierarchical inactivation of a synthetic human kinetochore by a chromatin modifier. Mol Biol Cell. 2009;20:4194–4204. doi: 10.1091/mbc.E09-06-0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nakano M, Cardinale S, Noskov VN, Gassmann R, Vagnarelli P, Kandels-Lewis S, Larionov V, Earnshaw WC, Masumoto H. Inactivation of a human kinetochore by specific targeting of chromatin modifiers. Dev Cell. 2008;14:507–522. doi: 10.1016/j.devcel.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ohzeki J-I, Bergmann JH, Kouprina N, Noskov VN, Nakano M, Kimura H, Earnshaw WC, Larionov V, Masumoto H. Breaking the HAC barrier: histone H3K9 acetyl/methyl balance regulates CENP-A assembly. EMBO J. 2012 doi: 10.1038/emboj.2012.82. http://dx.doi.org/10.1038/emboj.2012.82. [DOI] [PMC free article] [PubMed]
- 69.Miga KH. Completing the human genome: the progress and challenge of satellite DNA assembly. Chromosome Res. 2015;23:421–426. doi: 10.1007/s10577-015-9488-2. [DOI] [PubMed] [Google Scholar]