Abstract
Quantitative evaluation of protein expression across multiple cancer-related signaling pathways (e.g. Wnt/β-catenin, TGF-β, receptor tyrosine kinases (RTK), MAP kinases, NF-κB, and apoptosis) in tumor tissues may enable the development of a molecular profile for each individual tumor that can aid in the selection of appropriate targeted cancer therapies. Here, we describe the development of a broadly applicable protocol to develop and implement quantitative mass spectrometry assays using cell line models and frozen tissue specimens from colon cancer patients. Cell lines are used to develop peptide-based assays for protein quantification, which are incorporated into a method based on SDS-PAGE protein fractionation, in-gel digestion, and liquid chromatography-multiple reaction monitoring mass spectrometry (LC-MRM/MS). This analytical platform is then applied to frozen tumor tissues. This protocol can be broadly applied to the study of human disease using multiplexed LC-MRM assays.
Keywords: Liquid chromatography-multiple reaction monitoring mass spectrometry, cancer, signaling, colorectal carcinoma, targeted quantification, protein expression
1. Introduction
Cancer is one of the most prevalent public health concerns in the United States [1]. Novel approaches to further our understanding of basic molecular mechanisms of cancer signaling [2–5], particularly in human tumor tissues, are necessary to improve molecular classification schemes for personalized or precision medicine. With landscapes of tumor types created by genomics [6–9] and discovery proteomics [10] as well as known cancer biology, numerous important biomarkers can be selected to help guide patient treatment. Liquid chromatography-multiple reaction monitoring mass spectrometry (LC-MRM) has a long track record of clinical use and provides a flexible approach to multiplexing numerous protein biomarker assays. This technique is now routinely used for biomarker verification from discovery proteomics datasets [11,12]. Furthermore, the method excels at detection of target proteins in complex matrices [13] as well as high sensitivity and high precision quantification of low abundance proteins that contribute to intricate cancer signaling pathways [14–16]. Finally, this method has the capability to be translated to clinical samples [17–20].
In clinical sample analysis, techniques such as immunohistochemistry [21,22] and more recently matrix assisted laser desorption ionization (MALDI) mass spectrometry [23] or MALDI-MRM imaging [24] have been used for visual elucidation of "molecular images" on the tissue specimens. However, low abundance proteins are not detected with these strategies and require other approaches to quantify their expression in clinical samples. Gel fractionation prior to LC-MRM enables this approach to be compared directly to immunoblotting (Western) [25] and serves as an important pre-fractionation step to enrich the target proteins [26]. A variety of software applications are available to facilitate assay development (MRMer [27], SRM Builder (Thermo), MRMaid [28], Pinpoint (Thermo), and Skyline [29], inter al.) as well as to validate and publish SRM data (MRMAtlas [30], Panorama [31], MRMaid-DB [32], QuAD [33], inter al.), and perform computational processing and statistical validation of the data (e.g. mProphet [34]). These efforts have in turn supported sensitive high-throughput protein detection and quantification in a wide variety of biological samples (e.g. yeast [35–38], worms [39], cell lines [40], plasma [41], and human tissue, inter al.) as well as inter-lab assessment of the precision, portability, and reproducibility of those measurements [42,43].
Experimental Design
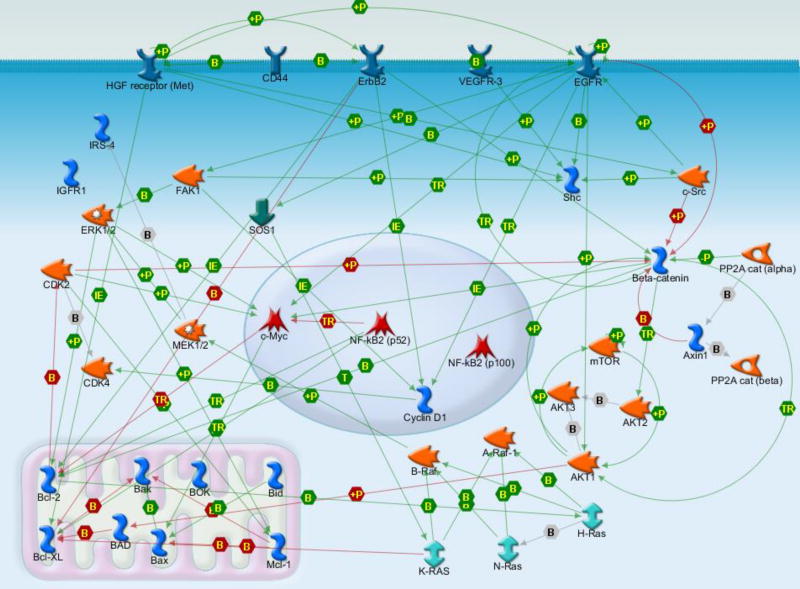
The experimental procedure described in this protocol focuses on selected cancer-related signaling pathways for mechanism elucidation and targeted therapy selection in colon cancer, but could be adapted to investigate other human diseases. The workflow includes: i) hypothesis-driven target selection; ii) SDS-PAGE-LC-MRM assay development with stable isotope-labeled standards; iii) multiplexed LC-MRM assay assembly; iv) frozen tissue evaluation and LC-MRM quantification; and v) data analysis. The rationale for target selection is based on known colon tumor biology and therefore includes low abundance proteins with roles in cancer related signaling pathways (as opposed to targets generated from discovery proteomics). Clinical relevance includes proteins with matched targeted therapeutic agents (e.g. receptor tyrosine kinase or RTK inhibitors). Pathways include Wnt/β-catenin, TGF-β, RTKs, MAP kinases, NF-κB, and apoptosis. The relationships between the proteins in this multiplexed LC-MRM panel have been mapped to illustrate their interactions and begin to generate a system-level view for the LC-MRM results (Figure 1). For assay development, SDS-PAGE protein fractionation is used for enrichment prior to LC-MRM screening. The final platform includes LC-MRM analysis of 5 distinct gel bands and is tested in cell line models prior to analysis of human tumor tissues.
Figure 1. Interaction Map of Selected Targets from Cancer Signaling Pathways.
Selected functional relationships between proteins are displayed using the MapEditor function of GeneGO.
In summary, the goal of this protocol is to enable investigation of cancer-related pathways in human tumors using quantitative mass spectrometry. The potential to achieve this goal is directly linked to recruitment of the scientific team that can contribute the necessary expertise and proper experimental design (see Note 1). Ultimately, we believe that this approach will improve our understanding of disease processes and elucidate mechanisms of response to therapy. The methods described frame the different steps (brief descriptions and estimated timelines are included in Note 2) required to include quantitative proteomics with medical history and histology for comprehensive analysis of tumor tissues.
2. Materials
Prepare all solutions using HPLC grade solvents, ultrapure water and high purity and/or mass spectrometry grade reagents. Follow safety precautions and disposal regulations (see Note 3).
2.1 Cell Culture and Tissue Acquisition
RPMI 1640 medium with L-glutamine and NaHCO3
HyClone Fetal bovine serum abbreviated FBS
Penicillin-streptomycin
0.25% Trypsin-EDTA
Criterion XT 4–12% Bis-Tris precast gel (Bio-Rad)
Gel loading buffer: XT Sample buffer, 4X (Bio-Rad)
Bradford protein assay kit
Coomassie Brilliant Blue G-250 (Bio-Rad)
Control colon adenocarcinoma cell lines: HCT116 and HT29 (American Type Culture Collection, Manassas, VA).
Human colon tumor tissues are obtained from Moffitt’s Total Cancer Care Biorepository with institutional review board approval (Protocol 00001138, University of South Florida).
2.2 Equipment
Source of distilled, deionized (18 MΩ) water
pH Meter (Orion)
Spectrophotometer (UV-Visible, Beckman Coulter)
Microcentrifuge, 14,000 × g.
Sonic dismembrator.
Vacuum concentrator (Speedvac).
Dri-Bath.
Microanalytical balance.
Midi-gel electrophoresis system.
Gel imager.
NanoUPLC system with refrigerated autosampler.
NanoUPLC trap column (PepMap, 100 µm ID × 2 cm, C18, 5 µm particle size, 100 Å pore size).
NanoUPLC separation column (PepMap, 75 µm ID × 25 cm, C18, 2 µm particle size, 100 Å pore size).
Triple quadrupole mass spectrometer with a nanoelectrospray ion source (e.g. TSQ Vantage or Quantiva, Thermo or equivalent).
Electrospray emitters pulled to 10 µm ID tips. For safety precautions, see Note 3.
MALDI MS and MS/MS (e.g. 4700, ABSciex) and semi-preparative HPLC may be required for peptide characterization and purification, unless purchasing the peptides from a vendor that will characterize them prior to delivery.
High-throughput whole slide scanning instrument with whole slide imaging software.
Skyline software or equivalent (https://skyline.gs.washington.edu/labkey/project/home/software/Skyline/begin.view) [29].
GeneGO software (Metacore, Thomson Reuters available at http://portal.genego.com or similar pathway mapping tool).
Matlab R2009b (http://www.mathworks.com).
2.3 Solutions and Buffers
Ammonium bicarbonate: 100 mM solution water. Dissolve 0.79 g ammonium bicarbonate in 80 mL 18 MΩ water, verify pH is 8, and bring volume up to 100 mL total. Dilute as needed with 18 MΩ water to make 50 mM and 30 mM solutions.
Dithiothreitol (DTT): 1.25 M solution in water. Dissolve 192.5 mg DTT in 1 mL water.
Lysis buffer: 8 M urea in 100 mM ammonium bicarbonate buffer, pH 8. Store at room temperature for up to 1 week.
Protein denaturation for SDS-PAGE: Pipette out an aliquot (e.g. 50 µg) of cell lysate or tissue homogenate based on the total protein concentration determined by the Bradford assay. Add appropriate amount of 4X Criterion XT sample buffer, 1 µL of 1.25 M DTT and water to a total volume of 30–45 µL (dependent on the maximum volume per lane). Denature the proteins at 95°C for 10 min and cool on ice prior to loading the gel.
Gel destaining buffer: 10 % v/v methanol, 5 % v/v acetic acid. Add 100 mL methanol and 50 mL glacial acetic acid to distilled, deionized water to make a total amount of 1000 mL.
Gel staining buffer: Dissolve 5 mg Coomassie Brilliant Blue G-250 in 100 mL gel destaining buffer.
Gel slice destaining buffer: 50 mM ammonium bicarbonate, 50 % methanol. Mix 25 mL of 100 mM ammonium bicarbonate and 25 mL of methanol to make 50 mL gel slice destaining buffer.
Disulfide reduction solution (10X stock): 20 mM tris(2-carboxyethyl)phosphine hydrochloride (TCEP) in water. Dissolve 5.7 mg TCEP in 1 mL water. Prepare fresh. Cover with aluminum foil and store at 4°C.
Cysteine alkylation solution (10X stock): 200 mM iodoacetamide (IAA) in water. Dissolve 37 mg IAA in 1 mL water. Prepare fresh. Cover with aluminum foil and store at 4°C.
Trypsin proteolytic digestion solution: 20 ng/µL trypsin. Dissolve 20 µg sequencing grade modified trypsin in 1 mL of aqueous 50 mM acetic acid to prevent autolysis. Aliquot and store at −80 °C until use.
Peptide extraction buffer: 50 % acetonitrile, 0.01 % trifluoroacetic acid (TFA). Add 10 µL TFA and 50 mL acetonitrile to water to make total volume of 100 mL.
LC-MRM solvent A: 2 % acetonitrile, 0.1 % formic acid in HPLC grade or LC-MS grade water.
LC-MRM solvent B: 90 % acetonitrile, 0.1 % formic acid in HPLC grade or LC-MS grade water.
LC loading solvent: 2 % acetonitrile, 0.1% formic acid or 0.04% TFA in HPLC grade or LC-MS grade water.
Autosampler syringe wash solvent: 2 % acetonitrile and 0.1 % formic acid in water. All solvents must be degassed prior to use.
2.4 Instrument Configuration and Data Acquisition Parameters
LC-MRM Instrument Parameters: The UPLC contains a refrigerated autosampler, column compartment with switching valve, two pumps, and degasser/solvent rack. Samples (5 µL) are loaded on the trapping column at 6 µL/min and washed for 5 min using a capillary scale loading pump. Then, the two position six port valve is used to switch the trapping column in-line with the analytical column described above. The LC gradient program is delivered at 300 nL/min by the nanoflow pump according to the program described below (Section 3.2, Step 19).
The UPLC is connected to a triple quadrupole mass spectrometer, equipped with a nanoelectrospray ion source operated in the positive ion mode; specific settings include 2,400 V spray voltage from 10 µm ID spray tips with 250°C transfer tube temperature. The Q1 resolution is set to 0.4 m/z (LC-MRM screening) or 0.7 m/z (for the final scheduled method), and Q3 resolution is set to 0.7 m/z. Fragmentation is obtained with 1.5 mTorr argon. Each transition is monitored for 20 milliseconds.
Software: The instrument vendor data acquisition software (e.g. XCalibur 2.1 and TSQ Vantage) and vendor or academic data analysis software (e.g. Skyline [29]) are required.
3. Methods
3.1 Target Selection and Refinement using Literature Review, Sequence Analysis Tools, and In-House or Publically Available Data
Literature review to determine comprehensive list of proteins associated with pathway or process of interest (e.g. known protein-protein interactions, enzyme-substrate relationships, etc.). See Note 4.
Use visualization software to build the network map of the targets (e.g. Map Editor, GeneGO).
Literature review of relevant biological information about each protein (e.g. highly homologous proteins, isoforms, splice variants, mutations, post-translational modifications, etc.). See Note 4.
Review publically available data about protein separation and available antibody reagents for comparative analysis/verification. Immunoprecipitation and western blot data from literature or vendors are needed to predict the region of protein migration in SDS-PAGE. If needed, adjacent regions are excised and evaluated with LC-MRM.
Obtain the protein sequence from UniProt (http://www.uniprot.org/) and publically available mass spectrometry data from PeptideAtlas (http://www.peptideatlas.org/) [44] or PRIDE/ProteomeXchange (https://www.ebi.ac.uk/pride/archive/) [45,46] inter al. If sufficient MS data are available, steps 6–30 can be omitted.
Use vendor or academic software (e.g. Skyline [29]) to predict tryptic peptides and transitions (precursor peptide and fragment ion pairs) from canonical sequences obtained from UniProt entries. Extensive tutorials are available online (https://skyline.gs.washington.edu/labkey/project/home/software/Skyline/begin.view).
Refine peptide selection to account for isoforms, modifications, and mutations of the proteins, as needed. Select unique peptides by filtering against the relevant background proteome (e.g. most current list of human entries in the UniProt database).
Select doubly charged peptides between 7 to 25 amino acids in length and exclude sequences containing the redox reactive residues, cysteine and methionine, unless there are few other choices. Additional selection criteria may eliminate sequences with adjacent or nearby tryptic cleavages sites or consensus sequences for glycosylation sites. Long peptides may have multiple charge states, which will result in signal splitting and poorer sensitivity; in addition, these peptides have more fragmentation channels and may not have significant amounts of signal in any one transition.
To select transitions for LC-MRM screening, we typically use y ions starting from either y3 or yx > peptide m/z and ending with y(n-1) for initial LC-MRM screens [32,47].
Export the .csv file containing the transition list (precursor m/z, fragment m/z, collision energy, and retention time for scheduling) and import it into the instrument method using the vendor-provided software.
3.2 LC-MRM Assay Development
Select appropriate cell lines with high levels of protein expression for LC-MRM assay development. Acquire cell lines from accredited vendors with specific QA/QC metrics that routinely evaluate each cell line and culture them according to the manufacturer’s instructions. As an example, these colon cancer cell lines are grown to 70 % confluence in RPMI-1640 medium supplemented with 10 % FBS and 1 % penicillin-streptomycin (10,000 U/mL) and incubated in a 5 % CO2 atmosphere at 37 °C.
To provide material for multiple experiments, harvest ten million cells (~1 mg total protein) for each colon cancer cell line from two-dimensional culture after washing with cold phosphate buffered saline (PBS) twice.
Lyse the cells in 8 M urea/100 mM ammonium bicarbonate buffer, pH 8, on ice.
Sonicate twice using 10 second pulses on ice and then centrifuge at 14,000 × g for 10 min at 4 °C.
Pipette the supernatant of the clarified cell lysate into a new microcentrifuge tube and measure the protein concentration by the Bradford assay (or micro-bichinchoninic acid/BCA assay).
Obtain an aliquot of the cell lysate (50 µg) and mix with an appropriate amount of concentrated Criterion XT sample buffer. Boil for 5 min for protein denaturation prior to gel loading.
Choose an appropriate gel to resolve the MW range of interest (see Note 5). In this experiment, separate proteins in 4–12 % Bis-Tris gels (Criterion XT) for ~80 min until dye front reaches bottom of gel at 150 V. The best quality separation is needed to maximize protein enrichment and reduce the potential interference with the peptides selected for the initial LC-MRM screening. For GeLC-MRM analysis of tumor tissues, the gel separation is shortened and 5 regions are excised (see Section 3.3 step 1).
Stain the gel with Coomassie Brilliant Blue G-250 in an aqueous solution of 10 % methanol/5 % acetic acid for 1 h and destain using the aqueous solution of 10 % methanol/5 % acetic acid for at least 3 h.
Excise the gel bands based on the predicted migration of the target proteins, and chop the bands into 1 mm3 pieces.
Add 20 µL of disulfide reduction solution (20 mM TCEP) and 180 µL of 50 mM ammonium bicarbonate to the slices and incubate at 37°C for 15 min, add another aliquot of TCEP and repeat incubation.
Add 20 µL of alkylation solution 200 mM IAA and 180 µL of 50 mM ammonium bicarbonate to the slices and incubate in dark at room temperature for 20 min, add 2nd aliquot of IAA and repeat incubation.
Wash and dehydrate the gel slices with 200 µL 50 mM ammonium bicarbonate/50% methanol.
Add 20 µL of 20 ng/µL trypsin and 200 µL of 30 mM ammonium bicarbonate to each sample and incubate at 37 °C overnight. Check that gel cubes are submerged.
Recover supernatant and extract gel slices twice with 100 µL 50% acetonitrile/0.01%TFA.
Concentrate the digested peptide solutions by vacuum centrifugation.
Resuspend in 30 µL of LC loading solvent for LC-MRM analysis.
Degas solvents A and B for nanoUPLC separation.
Load 5 µL of samples on the trap column at 6 µL/min and wash with loading solvent for 5 min.
- LC gradient program:
Time (min) Duration (min) B (%) 0 0 5 35 35 50 37 2 90 42 5 90 43 1 5 53 10 5 - Triple quadrupole instrument settings:
Nanoelectrospray voltage 2,400 V
Emitter tip 10 µm (ID) 360 µm (OD)
Transfer tube temperature 250 °C
Q1 resolution 0.4 (screening) 0.7 (scheduled)
Q3 resolution 0.7
Collision gas pressure 1.5 mTorr
Scan time/transition 20 ms Evaluate data from LC-MRM screening along with publically available data. As in discovery proteomics, the number of peptides identified in these screens is a direct metric for the confidence that the user should have in the results (see Note 6 for additional details). The highest intensity peptides with multiple strong transitions will produce the most sensitive assays. In addition, the elution position in the LC gradient and observation of potential interferences are also useful in selecting the best peptide candidates for assay development.
Synthesize or purchase at least one stable isotope-labeled standard (SIS) peptide for each target protein based on the signal intensity and interference of the detected peptides. Synthesis of the unlabeled tryptic peptide sequences is useful for assay characterization, but not required (see Note 7).
Estimate the purity and confirm the sequence for each synthetic or SIS peptide with HPLC, MS, and MS/MS. Purify the peptides with a semi-preparative HPLC system, if necessary.
Perform amino acid analysis to quantify concentrations of stocks prepared for the synthetic and SIS peptides.
Perform manual infusion for the synthetic peptides on the triple quadrupole mass spectrometer to determine the charge states observed in MS1, examine the MS/MS fragmentation pattern, choose transitions and optimize collision energy. Select 3 to 6 fragment ions with high signal intensity for each peptide. Compare the selected fragment ions against those observed in the LC-MRM screens described above.
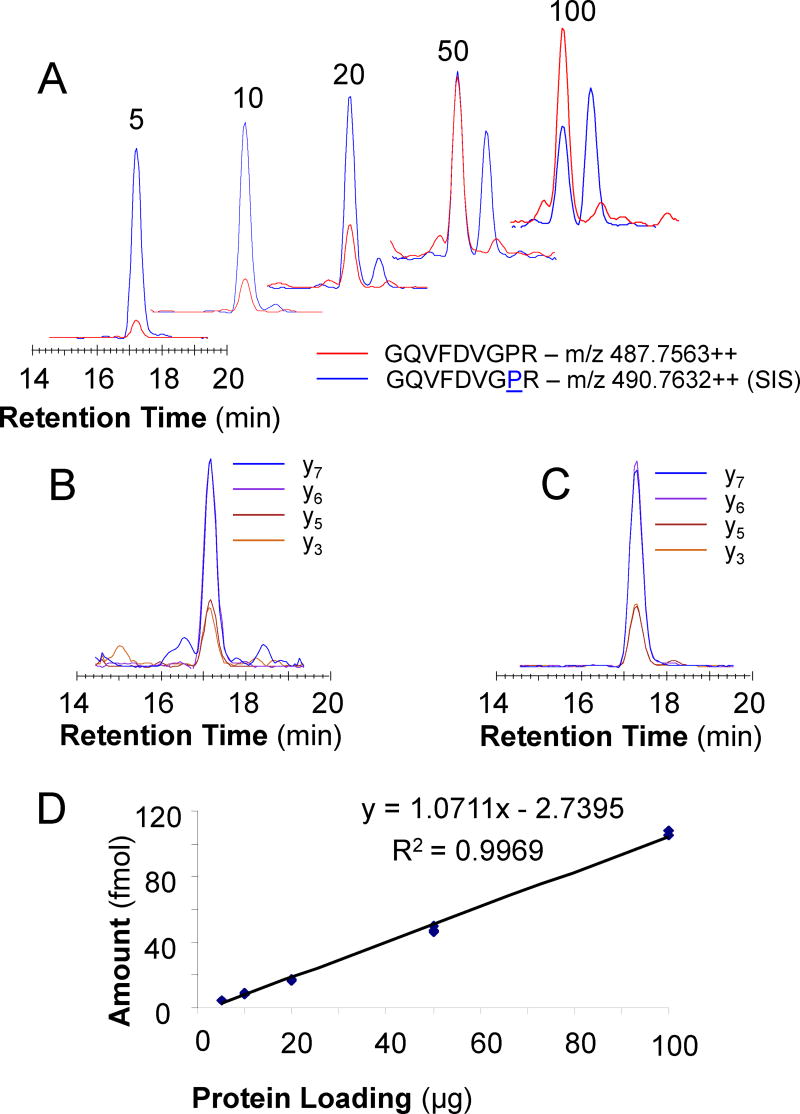
Characterize the peptides. If both the synthetic peptide and the SIS peptide were synthesized, calibration curves in buffer and matrix (preferably not containing the analyte of interest) with blanks and synthetic peptides from 10 amol to 100 fmol can help evaluate the limit of detection (LOD), upper and lower limits of quantification (ULOQ, LLOQ) and to decide on the amount of SIS peptide needed to mix with the biological samples as internal standard for quantification. Therefore, the amounts or concentrations used to make the calibration curve should include amounts expected from the biological samples. For example, ERK2 (MK01_HUMAN) is one of the 32 target proteins in gel region 4. We synthesized the SIS peptide with labeled proline, GQVFDVGPR (P8: 13C515N), for the corresponding proteolytic peptide from the endogenous protein and monitored four transitions, y3, y5, y6, and y7, for both sequences to evaluate the assay performance (Figure 2).
Mix internal standards into higher concentration stock solutions based on the proteins expected to be monitored in each gel fraction. Aliquot based on the expected scale of your batches for the quantitative experiment and store at −80 °C. As an example, if you expect to have 10 clinical samples with 2 controls in each batch, aliquot enough peptide mix for 15 experiments.
Check the stability of the SIS peptides. For example, the peak area of both the total and each transition of the peptides can be measured in 5 freeze-thaw cycle increments to a total of 15 cycles and compared to the fresh aliquot. Long term storage can also be evaluated by comparing results from the standard mix over time against a freshly prepared standard. The Pierce Retention Time Calibration peptide mix is useful for this task. The intensity of the individual peaks should meet specific QC metrics, as should the ratios between the SIS peptide peak intensities. These values will have to be determined for each set of analytes.
Figure 2. MK01 Quantification in HCT116 Cell Lysate Serial Dilutions.
LC-MRM analysis of the GQVFDVGPR peptide (red) and the corresponding SIS peptide (blue) in serial dilutions of HCT116 cell lysate created by loading different amounts of total protein (from 5–100 µg) for SDS-PAGE (A). Relative intensities for each transition (y3, y5-y7) observed for GQVFDVGPR (B) and the SIS peptide (C). Response curve and the equation for quantification of peptide GQVFDVGPR from MK01 (D).
3.3. Assay Assembly and Verification in Cell Lines
The gel separation parameters for the final assay (particularly the number of excised regions or protein fractions) should balance throughput with effective separation. LC-MRM transitions are organized and concatenated into a single method for each SDS-PAGE region. Some proteins are monitored in the adjacent gel sections if they are expected to migrate at the edge of the excised region or have multiple isoforms with different MWs. Rather than excising individual gel bands, the gel run is shortened and 5 regions are excised using the MW markers and overall staining pattern of the cell lysates and tissue homogenates (Figure 3).
In order to sample this number of peptides, the LC-MRM must be scheduled, meaning that peptides are not monitored for the entire experiment only during the expected elution time. If a peptide peak is expected to elute at 30 min, the instrument should be programmed to measure those transitions for a small window around that time. For cell lines, variability is less of an issue, so the time window could be less than 2 min. For complex clinical samples, the time window may need to be as wide as 5 min, depending on the quality of the samples and the reproducibility of the LC. We typically err on the side of expanding the time window, because signals not sampled lead to missingness in the data. In this project, the window is set to 5 min, because that enables sufficient sampling (> 10 points in each peak) even for the largest number of measurements: 1,150 transitions of 236 peptides from 32 target proteins in gel region 3 (MW 70–120, Figure 3).
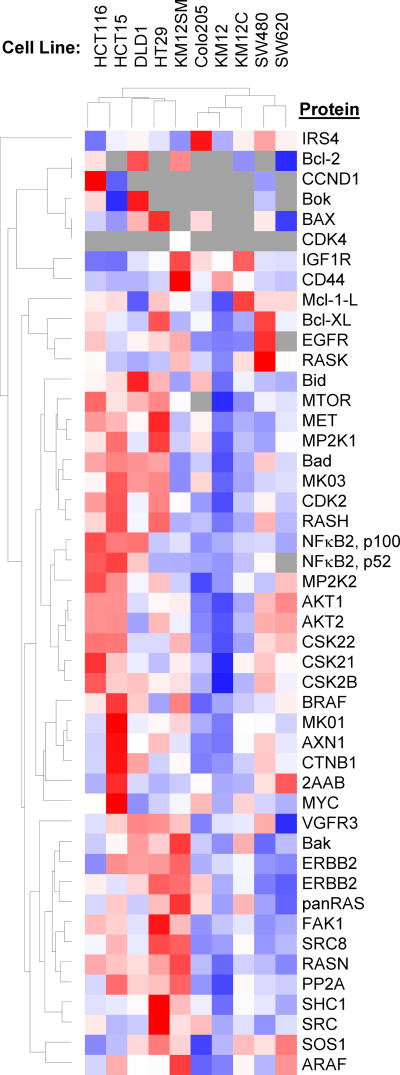
Prepare appropriate concentrations of cell lysate from each selected cell line and load 5, 10, 20, 50 and 100 µg of total protein into different gel lanes for fractionation and excision according to Section 3.3, step 1, following the steps in Section 3.2 to prepare protein fractions for LC-MRM. After concentrating the digested samples, resuspend the dried samples in 30 µL of the LC loading buffer containing the appropriate mix of SIS peptides for each gel fraction. For this project, highly characterized HCT116 and HT29 cell lines were selected for this experiment. Additional cell lines can be selected for larger scale verification of the multiplexed LC-MRM assay performance. We expanded our analysis to include SW620, SW480, HCT15, HCT116, DLD1, HT29, KM12SM, KM12C, Colo205 and KM12 (Figure 4).
Figure 3. Gel Band Excision Pattern using Bio-Rad Pre-stained MW Markers with Corresponding Numbers of Monitored Proteins, Peptides, and Transitions.
Five gel regions are cut with the following MW ranges: 1: > 250 kDa, 2: 120–250 kDa, 3: 70–120 kDa, 4: 30–70 kDa, and 5: < 30 kDa. Guided by the markers and the gel staining, horizontal cuts are made at top of the 250 kDa MW marker, halfway between the 100 and 150 kDa markers, halfway between the 50 and 75 kDa markers, below the 37 kDa marker, and below the dye front, as shown by the red boxes overlaid on the gel image.
Figure 4. Heat Map and Dendrograms for the Comparison of Protein Expression in Colon Cancer Cell Lines.
Lysates from SW620, SW480, HCT15, HCT116, DLD1, HT29, KM12SM, KM12C, Colo205 and KM12 cells were analyzed. Blue indicates lowest expression levels, while red indicates highest protein expression; grey indicates that the peptide was not observed.
3.4 Tumor Tissue Selection, Sectioning, and Evaluation
Obtain the frozen tumor tissues from a tissue bank or via a prospective research protocol. A discussion of sample selection criteria is provided in Note 8. In this particular case for colon cancer research, colon tumor tissues are acquired from Moffitt’s institutional Total Cancer Care Biorepository based on the following criteria: (i) adenocarcinoma of the colon; (ii) stage III or stage IV; (iii) age < 90; (iv) collected at Moffitt; (v) > 90 % tumor; (vi) no prior treatment; (vii) > 10 mg frozen tumor tissue banked; (viii) gene expression profile data complete (n = 372 available); (ix) cellularity > 70 %; (x) tumor/malignancy > 70 %; (xi) processing time/ischemia < 20 min.
Cut 5 serial sections from each frozen tissue block. Full embedding in optimal cutting temperature (OCT) medium is acceptable for these tissue sections, because it is removed prior to processing (see Section 3.5, steps 2–4). Tissue sections 1 and 5 will be prepared at 5 µm thickness and stained with hematoxylin and eosin (H&E) for pathology review to evaluate the consistency of histology though the sectioned depth of the tissue. Tissue sections 2–4 are cut with 25 µm thickness and saved in individual 0.5 mL Eppendorf tubes and stored at −80 °C for GeLC-MRM analysis. Additional sections can be excised based on the total material of the tumor tissue and experimental design for additional confirmatory tests such as immunohistochemistry of specific biomarkers or laser capture microdissection of tumor cells. Remaining material from each tumor could also be used for genomic characterization.
Hematoxylin and Eosin stained (H & E) slides for each tissue specimen must be reviewed by a board certified pathologist (D.C.) of appropriate specialization to confirm the tumor type, grade and other pertinent parameters. Proportions (%) of the following tissue components will be estimated: Malignancy (tumor cells, stroma, necrosis, and pre-neoplastic lesions or adenoma), normal tissue (smooth muscle, colon epithelium), abnormal tissue (acute or chronic inflammation, fibrosis, ulceration, etc.) and benign neoplastic tissue. Evaluate consistency of slides 1 and 5 to make sure that the tissue content and distribution do not change significantly throughout the tissue samples.
Scan the H&E stained tissues with a whole slide scanner (e.g., ScanScope XT, Aperio) for archiving and annotation.
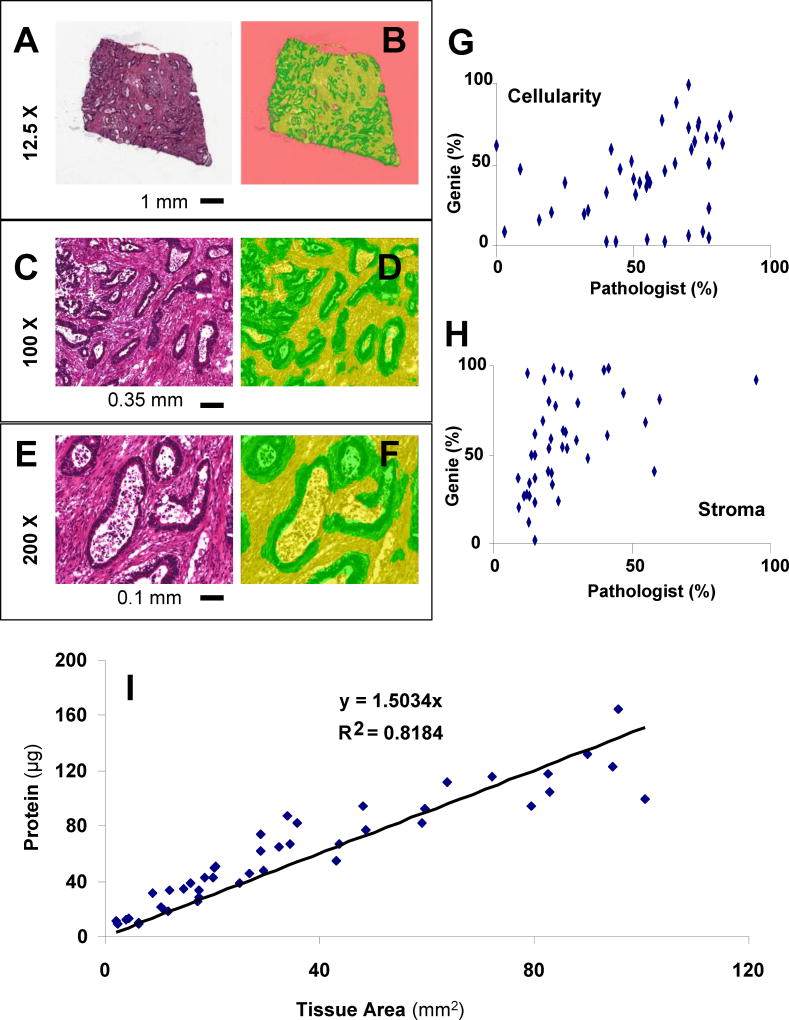
Automated image analysis is optional, but it can be useful to calculate the areas of the tissue components from Section 3.4, Step 3 to supplement the pathologist’s review. In addition, training a pattern recognition algorithm (Genie, Aperio) to differentiate cellular components, stromal components, and blank background enables rapid analysis of tissue content to further select the highest quality specimens. The algorithm has been applied to the entire slide’s digital image to determine the percentage of each tissue type by area (µm2) and to then calculate the amount of protein extracted per unit of tissue area for each specimen (Figure 5).
Figure 5. Example Tissue Image and Evaluation.
An example H&E slide for a colon adenocarcinoma tissue (A) is shown. Automated whole slide tissue evaluation (B) by an in-house algorithm created using Genie (Aperio) indicates areas of cellularity (green), stroma (yellow), and background (pink). Additional views at higher magnification show the relative effectiveness of the assignment (C–F). Correlation plots are shown for comparison of the pathologist’s evaluations of tumor epithelium and stroma and the Genie recognition of cellularity (G) and stromal components (H). Tissue area is predictive of total protein obtained from tumor homogenate in 25 µm sections (I).
3.5 Tumor Tissue Sample Preparation for LC-MRM
In this experiment, individual tumors are assessed, so the clinical samples are randomized. In paired experiment designs (e.g. tumor/normal or pre-/post-treatment samples from the same patient), linked samples should be blocked together and then randomized. Cell lines or tumor tissue pools, when available, (frozen aliquots of the same biological sample) are analyzed with each batch of 10 clinical samples as quality control samples for instrument performance. In general, these experiments should precede clinical sample analysis to enable interruption in the case of errors in sample preparation or poor instrument performance.
Keep the samples on ice to melt optimal cutting temperature (OCT) medium (< 5 minutes). Note that this step should be modified for any measurement other than protein expression, because post-translational modifications may be significantly impacted by this processing.
Centrifuge the samples at 14,000 × g for 10 min.
Discard the OCT supernatant and keep the tissues in original tubes. Additional washes with cold ethanol can further remove OCT without significant loss of protein.
Add 50 µL 8 M urea/100 mM ammonium bicarbonate buffer, pH 8, to each tissue and follow the steps in Section 3.2 to prepare protein fractions for LC-MRM. After concentrating the digested samples, resuspend the dried samples in 30 µL of LC loading buffer with the appropriate SIS peptide mix for each gel fraction.
LC-MRM analysis is performed in triplicate. (See Note 9).
3.6 Quality Control and Data Analysis
Import the raw data to analysis software (e.g. Skyline) and manually evaluate all peak selections.
Compare the retention time and fragmentation pattern (either by transition rank or relative intensity) of proteolytic peptides from the endogenous protein and the corresponding SIS peptides. Retention times should match exactly. Fragment ion ratios should be consistent with specific QC metrics (e.g. 5% variability), but transitions with significantly lower signal intensity may deviate more.
Export the peak area values for individual transitions and the summed totals to a .csv file for analysis.
Plot the peak areas for SIS peptides across the entire sample set. Examine any variations within a batch and between batches. Start by comparing the cell line or pooled standard samples included in each batch and continue to the individual clinical samples. Outliers should be marked and may need to be discarded based on poor instrument performance. If signal for the SIS peptides is less than 50 % of the average in a given sample, it should likely be evaluated thoroughly and potentially removed from the dataset. If trends are observed, investigate possible causes (e.g. loss of instrument sensitivity over long analysis period or selection of samples with poor cellularity concentrated into a batch).
Measure ratios between the peak areas of SIS peptides or rank them by intensity to examine whether any of the SIS peptides is decreasing in ion signal.
Compare the data from cell line QC between batches to examine any changes in the performance of the assays.
Calculate the levels of protein expression using the ratio of proteolytic peptide and SIS peptide ion signals, the amount of the SIS peptide spiked into the sample, the fraction of the sample analyzed by LC-MRM, and the amount of protein loaded into the gel (output value in fmol/µg total protein). Sum the concentrations over multiple gel bands, when the peptide is detected in two GeLC-MRM fractions or treat these two measurements separately, when appropriate. Place values for proteolytic peptide signal, SIS peptide signal, and calculated estimate of protein expression into 3 matrices for statistical analysis.
Using Matlab R2009b or an equivalent software program, examine data sampling distribution using scatter plots and histograms both in original scale of the raw data and after log2 transformation prior to determining whether parametric or nonparametric analyses should be applied.
Batch-to-batch normalization is optional, depending primarily on the data quality. While each SIS peptide is used to calculate the level of protein expression in every individual sample, batch-to-batch variation in the cell line data could be used as another normalizing factor, particularly if SIS peptides are not included or not observed for each analyte. Examine and normalize the clinical samples across multiple batches using internal standards and the peptide concentrations across the same cell line QC samples. These QC cell lines are evaluated in each batch and can be used to make sure the data are comparable between batches.
Lower limits of quantification (LLOQ) determined for each peptide in Section 3.2, step 26 can be used to filter out protein expression levels below the LLOQ, which are detectable but not quantifiable.
Identify outliers from each set of 3 technical replicates for each peptide. A replicate is identified as an outlier using the following criteria: i) the distance from the outlier to the median is over twice the distance of the median from the remaining data point or ii) if the CV is > 20 %. The outliers were removed and treated as missing data.
Flag peptides with reliable detection in <50 % of the (tissue or cell line) samples, as conclusions drawn from these peptides should be interpreted with caution.
Perform quality control for samples (see Note 10). Central tendency, range, variability of all peptide expression levels and also the number of missing peptides are summarized for each sample. Overall expression levels across samples were visualized using boxplots heat maps. As an example, tissue samples with either lower protein expression levels or a high proportion of missing peptides (>30 %) should be flagged for repeat analysis and review of the histology.
Calculate the average of technical triplicates which passed the QC for each biological sample. Log2 average concentration from each biological sample is used in further analyses. Log2 transformed peptide data are often normally distributed (and therefore parametric analyses can be applied). In addition, when comparing the expression levels between two groups, the interpretation is straightforward.
Imputation of missing values is optional; alternatively, analytes with high missingness may need to be discarded or treated as a simple positive/negative readout. For analyses that are not capable of handling missing values, such as cluster analyses, when all three technical replicates of a sample were missing, they are replaced with the overall lowest detected concentration from all samples and peptides. However, other common imputation methods such as K-nearest neighbor method could be performed to impute missing values.
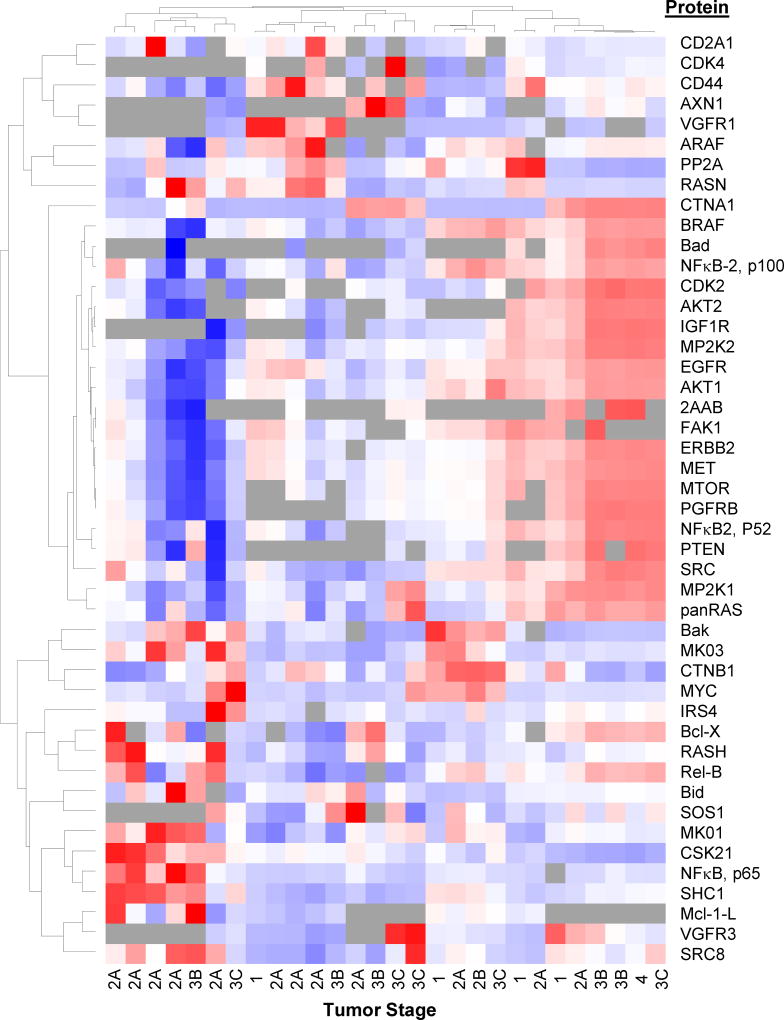
Perform hierarchical clustering analyses of the peptide expression levels and visualize the results in a heat map along with a dendrogram to explore relationships between proteins and tumor samples (Figure 6). This helps to visualize peptides at all levels of expression across tissues/cell lines and examine reproducibility across technical replicates. Cluster 3.0 (http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm) and Java TreeView (http://jtreeview.sourceforge.net/) [48] are available freeware that can be used for this step. TreeView heat maps can be exported as PostScript files (.ps), which can be converted to PDF using GSview (http://pages.cs.wisc.edu/~ghost/gsview/) and then into images with Inkscape (https://inkscape.org/en/).
Perform statistical analyses (e.g., using t-tests or ANOVA) to test specific hypotheses, including, but not limited to mapping protein expression levels against transcript expression levels or known mutational status or relating protein expression levels with patient survival outcomes or responses to therapy. Note 11 includes additional discussion of the expected outcomes.
Figure 6. Heat Map and Dendrograms for Comparison of Protein Expression in Colon Tumor Tissues.
Blue indicates lowest expression levels, while red indicates highest protein expression; grey indicates that the peptide was not observed.
Acknowledgments
This work was supported in part by the following core facilities at Moffitt Cancer Center: Tissue Core, Analytic Microscopy, Proteomics, and Biostatistics; funding provided in part by the Cancer Center Support Grant, P30-CA076292, from the National Cancer Institute. Proteomics instruments are supported by funding from the Moffitt Foundation and shared instrument grants from the Bankhead-Coley Cancer Research program of the Florida Department of Health (06BS-02-9614 and 09BN-14). Project funding was received as subcontract from the Moffitt National Functional Genomics Center funded by the US Army Medical Research and Materiel Command under award DAMD17-02-2-0051. Amino acid analysis was performed by the Protein Chemistry Laboratory at Texas A&M University by Virginia Johnson, MS, and Larry Dangott, PhD.
Footnotes
Assembling the research team with representatives of each type of expertise required to address the research question is critical for success. In this case, the disciplines include colon cancer patient treatment, surgery, and relevant biology (DS), pathology (DC), epidemiology (ES), microscopy (ML), proteomics (YC, EW, and JK), and biostatistics (KF and YAC). Study design is also a critical step. The existing samples and available budget often determine the cohort that will be examined; these practical limitations need to be examined prior to experiment design and evaluated using power calculations as well as knowledge of the patient populations, histological tumor types, and molecular tumor subtypes. Placement of the study in the context of the patients’ medical history information and genomics data is also critical. All previous testing results that are relevant should be accrued at the beginning of the study. Because these tumors are already banked, two to five years of follow-up information could be obtained. This data will enable to combine the results from quantitative proteomics with medical history, histology, and gene expression profiles for comprehensive analysis of tumor tissues with the ultimate goal of establishing a molecular basis for personalized medicine.
Typical times for each step used in our laboratory are: Section 3.1, Selecting target proteins from cancer related signaling pathways and designing the LC-MRM method for peptide screening: iterative and ongoing assessment of the literature is needed, but the initial data can be gathered within a few days. Section 3.2, Selecting appropriate cell line models for assay development and sample preparation: 3 days for sample preparation, including overnight gel destaining and 24 h trypsin digestion. LC-MRM: 2 h per analysis including washes to eliminate carryover. Synthesis and Characterization of Internal Standards: Generally a 2–4 week process, but we have been able to take candidate proteins and have the SIS peptide synthesized and characterized within 1 week. Synthesis requires 1–2 days at a minimum. Further purification and mass analysis can be performed in 1–2 days. Our amino acid analysis has been completed at Texas A&M University (see Acknowledgments), so shipping and turnaround for that data is longer than an in-house service. Calibration curves can be run in 2–3 days on the triple quadrupole mass spectrometer depending on gradient length and number of required data points and technical replicates. Peptide evaluation after freeze-thaw cycles and after long term storage requires additional time. Assay assembly and cell line analysis: With this number of target proteins and excised gel regions, 1 day is needed for gel separation, 2 days for sample preparation, and 2 h of instrument time per LC-MRM analysis (triplicate analysis of 5 gel regions produces 15 LC-MRM datasets for each biological sample). Section 3.3, Sample preparation for multiplexed LC-MRM assay implementation in cell line models: This experiment analyzing 10 cell lines requires 5 days for sample preparation after fresh or frozen lysates are available and then 1.5 days of instrument time per cell line for triplicate LC-MRM analysis of digests of each of the 5 gel regions per cell line (50 samples). Section 3.4, Tumor tissue selection, sectioning and evaluation: Practically, the timeline is dependent on IRB approval, banked sample availability, Tissue Core staff, whole slide scanning, and the pathologist’s schedule. After several discussions, we used ~1 h per sample for pathology review and ~1 h per sample for pattern recognition software evaluation as well as 2 h for data reconciliation and interpretation. Section 3.5, Tumor tissue analysis: For each batch of 10 tumor tissues with 2 cell line controls, 5 days was needed for sample preparation (60 samples total) with 1.5 days of instrument time per tissue/cell line for triplicate LC-MRM analysis. Section 3.6, Quality control and data analysis: For the first project, this process will be iterative and ongoing for weeks to months due to discussion of multiple relevant parameters; timeline is also limited by the other commitments of the biostatisticians. After the research team has accumulated more experience, the time required will significantly decrease.
Safety Concerns: When using toxic, highly flammable or corrosive solvents and reagents, use protective clothing, gloves, safety goggles or eye/face protection as appropriate, and handle the reagent in the fume hood. In addition, special care is needed when handling electrospray emitter tips; safety glasses are recommended. Solvents and waste must be properly disposed or recycled as per relevant local and federal regulations.
For gathering the preliminary information related to proteins, pathways, and biological processes, it is important to understand that our knowledge of biology is continuously growing and evolving. Additional protein interactions or novel post-translational modifications can be discovered that would influence the selection of peptides and the interpretation of the data from these LC-MRM assays. Literature review should be iterative throughout the assay development and implementation process.
The precast gel selection for protein fractionation prior to LC-MRM should be based on the overall list of proteins and expected MW ranges for their separation. This panel included large proteins > 100 kDa (e.g. receptor tyrosine kinases) and small proteins < 25 kDa (e.g. Bcl-2 family regulators of apoptosis), so the gradient gel (4–12 %) was the most appropriate choice.
For evaluation of LC-MRM screening data and other data from publically available sources as part of assay development, single peptide hits have had a poor success rate for generating an assay (~25 % of cases), but the best peptides selected from 5 or more detected sequences almost always (~95 % of cases) did develop into useful assays. Data should also be considered with regard to the number of tryptic peptides that could be detected from a given protein. The apoptosis-regulating proteins in this study would generate only a few potential tryptic peptides amenable to LC-MRM, so the rationale for proceeding with assay development should remain flexible enough to accommodate these issues.
The choice to synthesize the proteolytic peptide from the endogenous protein (without stable isotope-labeling) provides more flexibility and more accurate assessment of the assay performance via calibration curves in buffer and matrix. However, reverse calibration curves (varying the amount of SIS peptide in a sample with a known amount of endogenous protein) can also be used for assay characterization. Note that interference may be more likely in the transitions for the proteolytic peptide from the endogenous protein than in the transitions for the SIS peptide.
Appropriate criteria need to be applied for tissue sample selection. These choices are even more important for evaluation of samples collected across different sites or under different protocols; these projects can include potential confounding variables that are not observed under highly controlled single institution prospective collections under one unified protocol.
Prior to analysis of clinical samples, cleaning, tuning and calibration of the mass spectrometer are recommended. For large scale datasets, the UPLC columns should be replaced, conditioned, and tested prior to sample analysis. Routine injections of the cell lysates samples are ideal for these quality control steps.
| Problem | Solution |
|---|---|
| Pathology report has high stroma/necrosis in tissue sample | Discard the sample as most target proteins are from tumor cellularity. |
| Loss of Signal for SIS peptide | Discard this data or perform relative quantification by normalizing with an adjacent SIS peptide with similar signal intensity |
| Poor data for all proteins in a sample | Check histology of the patient sample and analyze another section |
| Batch-to-batch variability | Normalize data from the tissue samples by using the data from QC cell line samples |
The anticipated results include relative quantification of proteins across cell lines and tumor tissues as well as minimum estimates of protein expression levels in terms of femtomoles per milligram of total protein. The immediate impact of this assay platform and others like it can be in improving our understanding of tumor biology in situ and in ranking the most important proteins to develop as candidate biomarkers for prognosis or prediction of therapeutic response. In addition, these measurements can be included as correlates in clinical trials to examine the mechanisms of drug response and drug resistance. Ultimately, multiplexed LC-MRM has the potential and capability to provide biomarker measurements supporting selection of clinical care regimens.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Taylor IW, Wrana JL. Protein interaction networks in medicine and disease. Proteomics. 2012;12(10):1706–16. doi: 10.1002/pmic.201100594. [DOI] [PubMed] [Google Scholar]
- 3.Koomen JM, Haura EB, Bepler G, Sutphen R, Remily-Wood ER, Benson K, Hussein M, Hazlehurst LA, Yeatman TJ, Hildreth LT, Sellers TA, Jacobsen PB, Fenstermacher DA, Dalton WS. Proteomic contributions to personalized cancer care. Mol Cell Proteomics. 2008;7:1780–94. doi: 10.1074/mcp.R800002-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Myers MV, Manning HC, Coffey RJ, Liebler DC. Protein expression signatures for inhibition of epidermal growth factor receptor-mediated signaling. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.015222. M111.015222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jendrossek V. The intrinsic apoptosis pathways as a target in anticancer therapy. Curr Pharm Biotechnol. 2012;13:1426–38. doi: 10.2174/138920112800784989. [DOI] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–7. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Sousa E, Melo F, Wang X, Jansen M, Fessler E, Trinh A, de Rooij LP, de Jong JH, de Boer OJ, van Leersum R, Bijlsma MF, Rodermond H, van der Heijden M, van Noesel CJ, Tuynman JB, Dekker E, Markowetz F, Medema JP, Vermeulen L. Poor-prognosis colon cancer is defined by a molecularly distinct subtype and develops from serrated precursor lesions. Nat Med. 2013;19:614–8. doi: 10.1038/nm.3174. [DOI] [PubMed] [Google Scholar]
- 8.Sadanandam A1, Lyssiotis CA, Homicsko K, Collisson EA, Gibb WJ, Wullschleger S, Ostos LC, Lannon WA, Grotzinger C, Del Rio M, Lhermitte B, Olshen AB, Wiedenmann B, Cantley LC, Gray JW, Hanahan D. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat Med. 2013;19:619–25. doi: 10.1038/nm.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ren Z, Wang W, Li J. Identifying molecular subtypes in human colon cancer using gene expression and DNA methylation microarray data. Int J Oncol. 2016;48:690–702. doi: 10.3892/ijo.2015.3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang B, Wang J, Wang X, Zhu J, Liu Q, Shi Z, Chambers MC, Zimmerman LJ, Shaddox KF, Kim S, Davies SR, Wang S, Wang P, Kinsinger CR, Rivers RC, Rodriguez H, Townsend RR, Ellis MJ, Carr SA, Tabb DL, Coffey RJ, Slebos RJ, Liebler DC NCI CPTAC. Proteogenomic characterization of human colon and rectal cancer. Nature. 2014;513:382–7. doi: 10.1038/nature13438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turtoi A, Musmeci D, Wang Y, Dumont B, Somja J, Bevilacqua G, De Pauw E, Delvenne P, Castronovo V. Identification of novel accessible proteins bearing diagnostic and therapeutic potential in human pancreatic ductal adenocarcinoma. J Proteome Res. 2011;10:4302–13. doi: 10.1021/pr200527z. [DOI] [PubMed] [Google Scholar]
- 12.Whiteaker JR, Lin C, Kennedy J, Hou L, Trute M, Sokal I, Yan P, Schoenherr RM, Zhao L, Voytovich UJ, Kelly-Spratt KS, Krasnoselsky A, Gafken PR, Hogan JM, Jones LA, Wang P, Amon L, Chodosh LA, Nelson PS, McIntosh MW, Kemp CJ, Paulovich AG. A targeted proteomics-based pipeline for verification of biomarkers in plasma. Nat Biotechnol. 2011;29:625–34. doi: 10.1038/nbt.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Agard NJ, Mahrus S, Trinidad JC, Lynn A, Burlingame AL, Wells JA. Global kinetic analysis of proteolysis via quantitative targeted proteomics. Proc Natl Acad Sci USA. 2012;109:1913–8. doi: 10.1073/pnas.1117158109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi T, Su D, Liu T, Tang K, Camp DG, 2nd, Qian WJ, Smith RD. Advancing the sensitivity of selected reaction monitoring-based targeted quantitative proteomics. Proteomics. 2012;12:1074–92. doi: 10.1002/pmic.201100436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collins BC, Miller CA, Sposny A, Hewitt P, Wells M, Gallagher WM, Pennington SR. Development of a pharmaceutical hepatotoxicity biomarker panel using a discovery to targeted proteomics approach. Mol Cell Proteomics. 2012;11:394–410. doi: 10.1074/mcp.M111.016493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boja ES, Rodriguez H. Mass spectrometry-based targeted quantitative proteomics: Achieving sensitive and reproducible detection of proteins. Proteomics. 2012;12:1093–110. doi: 10.1002/pmic.201100387. [DOI] [PubMed] [Google Scholar]
- 17.Hoofnagle AN, Becker JO, Oda MN, Cavigiolio G, Mayer P, Vaisar T. Multiple-reaction monitoring-mass spectrometric assays can accurately measure the relative protein abundance in complex mixtures. Clin Chem. 2012;58:777–81. doi: 10.1373/clinchem.2011.173856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sprung RW, Martinez MA, Carpenter KL, Ham AJ, Washington MK, Arteaga CL, Sanders ME, Liebler DC. Precision of Multiple Reaction Monitoring Mass Spectrometry Analysis of Formalin-Fixed, Paraffin-Embedded Tissue. J Proteome Res. 2012;11:3498–505. doi: 10.1021/pr300130t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuhn E, Whiteaker JR, Mani DR, Jackson AM, Zhao L, Pope ME, Smith D, Rivera KD, Anderson NL, Skates SJ, Pearson TW, Paulovich AG, Carr SA. Inter-laboratory evaluation of automated, multiplexed peptide immunoaffinity enrichment coupled to multiple reaction monitoring mass spectrometry for quantifying proteins in plasma. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.013854. M111.013854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lam MP, Scruggs SB, Kim TY, Zong C, Lau E, Wang D, Ryan CM, Faull KF, Ping P. An MRM-based workflow for quantifying cardiac mitochondrial protein phosphorylation in murine and human tissue. J Proteomics. 2012;75(15):4602–9. doi: 10.1016/j.jprot.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fonseca-Sánchez MA, Rodríguez Cuevas S, Mendoza-Hernández G, Bautista-Piña V, Arechaga Ocampo E, Hidalgo Miranda A, Quintanar Jurado V, Marchat LA, Alvarez-Sánchez E, Pérez Plasencia C, López-Camarillo C. Breast cancer proteomics reveals a positive correlation between glyoxalase 1 expression and high tumor grade. Int J Oncol. 2012;41:670–80. doi: 10.3892/ijo.2012.1478. [DOI] [PubMed] [Google Scholar]
- 22.Tang A, Li N, Li X, Yang H, Wang W, Zhang L, Li GY, Xiong W, Ma J, Shen S. Dynamic activation of the key pathways: linking colitis to colorectal cancer in a mouse model. Carcinogenesis. 2012;33:1375–83. doi: 10.1093/carcin/bgs183. [DOI] [PubMed] [Google Scholar]
- 23.Casadonte R, Caprioli RM. Proteomic analysis of formalin-fixed paraffin-embedded tissue by MALDI imaging mass spectrometry. Nat Protoc. 2011;6:1695–709. doi: 10.1038/nprot.2011.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clemis EJ, Smith DS, Camenzind AG, Danell RM, Parker CE, Borchers CH. Quantitation of Spatially-Localized Proteins in Tissue Samples using MALDI-MRM Imaging. Anal Chem. 2012;84:3514–22. doi: 10.1021/ac202875d. [DOI] [PubMed] [Google Scholar]
- 25.Tang HY, Beer LA, Barnhart KT, Speicher DW. Rapid verification of candidate serological biomarkers using gel-based, label-free multiple reaction monitoring. J Proteome Res. 2011;10:4005–17. doi: 10.1021/pr2002098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang M, Heo GY, Omarova S, Pikuleva IA, Turko IV. Sample Pre-Fractionation for Mass Spectrometry Quantification of Low-Abundance Membrane Proteins. Anal Chem. 2012;84:5186–91. doi: 10.1021/ac300587v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin DB, Holzman T, May D, Peterson A, Eastham A, Eng J, McIntosh M. MRMer, an interactive open source and cross-platform system for data extraction and visualization of multiple reaction monitoring experiments. Mol Cell Proteomics. 2008;7:2270–8. doi: 10.1074/mcp.M700504-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mead JA, Bianco L, Ottone V, Barton C, Kay RG, Lilley KS, Bond NJ, Bessant C. MRMaid, the web-based tool for designing multiple reaction monitoring (MRM) transitions. Mol Cell Proteomics. 2009;8:696–705. doi: 10.1074/mcp.M800192-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–8. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Picotti P, Lam H, Campbell D, Deutsch EW, Mirzaei H, Ranish J, Domon B, Aebersold RA. database of mass spectrometric assays for the yeast proteome. Nat Methods. 2008;5:913–4. doi: 10.1038/nmeth1108-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharma V, Eckels J, Taylor GK, Shulman NJ, Stergachis AB, Joyner SA, Yan P, Whiteaker JR, Halusa GN, Schilling B, Gibson BW, Colangelo CM, Paulovich AG, Carr SA, Jaffe JD, MacCoss MJ, MacLean B. Panorama: a targeted proteomics knowledge base. J Proteome Res. 2014;13:4205–10. doi: 10.1021/pr5006636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cham JA, Bianco L, Barton C, Bessant C. MRMaid-DB: a repository of published SRM transitions. J Proteome Res. 2010;9:620–5. doi: 10.1021/pr900713u. [DOI] [PubMed] [Google Scholar]
- 33.Remily-Wood ER, Liu RZ, Xiang Y, Chen Y, Thomas CE, Rajyaguru N, Kaufman LM, Ochoa JE, Hazlehurst L, Pinilla-Ibarz J, Lancet J, Zhang G, Haura E, Shibata D, Yeatman T, Smalley KS, Dalton WS, Huang E, Scott E, Bloom GC, Eschrich SA, Koomen JM. A database of reaction monitoring mass spectrometry assays for elucidating therapeutic response in cancer. Proteomics Clin Appl. 2011;5:383–96. doi: 10.1002/prca.201000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reiter L, Rinner O, Picotti P, Hüttenhain R, Beck M, Brusniak MY, Hengartner MO, Aebersold R. mProphet: automated data processing and statistical validation for large-scale SRM experiments. Nat Methods. 2011;8:430–5. doi: 10.1038/nmeth.1584. [DOI] [PubMed] [Google Scholar]
- 35.Picotti P, Bodenmiller B, Mueller LN, Domon B, Aebersold R. Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell. 2009;138:795–806. doi: 10.1016/j.cell.2009.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bluemlein K, Ralser M. Monitoring protein expression in whole-cell extracts by targeted label- and standard-free LC-MS/MS. Nat Protoc. 2011;6:859–69. doi: 10.1038/nprot.2011.333. [DOI] [PubMed] [Google Scholar]
- 37.Picotti P, Rinner O, Stallmach R, Dautel F, Farrah T, Domon B, Wenschuh H, Aebersold R. High-throughput generation of selected reaction-monitoring assays for proteins and proteomes. Nat Methods. 2010;7:43–6. doi: 10.1038/nmeth.1408. [DOI] [PubMed] [Google Scholar]
- 38.Wu R, Haas W, Dephoure N, Huttlin EL, Zhai B, Sowa ME, Gygi SP. A large-scale method to measure absolute protein phosphorylation stoichiometries. Nat Methods. 2011;8:677–83. doi: 10.1038/nmeth.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jovanovic M, Reiter L, Picotti P, Lange V, Bogan E, Hurschler BA, Blenkiron C, Lehrbach NJ, Ding XC, Weiss M, Schrimpf SP, Miska EA, Grosshans H, Aebersold R, Hengartner MO. A quantitative targeted proteomics approach to validate predicted microRNA targets in C. elegans. Nat Methods. 2010;7:837–42. doi: 10.1038/nmeth.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kettenbach AN, Rush J, Gerber SA. Absolute quantification of protein and post-translational modification abundance with stable isotope-labeled synthetic peptides. Nat Protoc. 2011;6:175–86. doi: 10.1038/nprot.2010.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim YJ, Zaidi-Ainouch Z, Gallien S, Domon B. Mass spectrometry-based detection and quantification of plasma glycoproteins using selective reaction monitoring. Nat Protoc. 2012;7:859–71. doi: 10.1038/nprot.2012.023. [DOI] [PubMed] [Google Scholar]
- 42.Addona TA, Abbatiello SE, Schilling B, Skates SJ, Mani DR, Bunk DM, Spiegelman CH, Zimmerman LJ, Ham AJ, Keshishian H, Hall SC, Allen S, Blackman RK, Borchers CH, Buck C, Cardasis HL, Cusack MP, Dodder NG, Gibson BW, Held JM, Hiltke T, Jackson A, Johansen EB, Kinsinger CR, Li J, Mesri M, Neubert TA, Niles RK, Pulsipher TC, Ransohoff D, Rodriguez H, Rudnick PA, Smith D, Tabb DL, Tegeler TJ, Variyath AM, Vega-Montoto LJ, Wahlander A, Waldemarson S, Wang M, Whiteaker JR, Zhao L, Anderson NL, Fisher SJ, Liebler DC, Paulovich AG, Regnier FE, Tempst P, Carr SA. Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nat Biotechnol. 2009;27:633–41. doi: 10.1038/nbt.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prakash A, Rezai T, Krastins B, Sarracino D, Athanas M, Russo P, Ross MM, Zhang H, Tian Y, Kulasingam V, Drabovich AP, Smith C, Batruch I, Liotta L, Petricoin E, Diamandis EP, Chan DW, Lopez MF. Platform for establishing interlaboratory reproducibility of selected reaction monitoring-based mass spectrometry peptide assays. J Proteome Res. 2010;9:6678–88. doi: 10.1021/pr100821m. [DOI] [PubMed] [Google Scholar]
- 44.Desiere F, Deutsch EW, King NL, Nesvizhskii AI, Mallick P, Eng J, Chen S, Eddes J, Loevenich SN, Aebersold R. The PeptideAtlas project. Nucleic Acids Res. 2006;34:D655–8. doi: 10.1093/nar/gkj040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vizcaíno JA, Deutsch EW, Wang R, Csordas A, Reisinger F, Ríos D, Dianes JA, Sun Z, Farrah T, Bandeira N, Binz PA, Xenarios I, Eisenacher M, Mayer G, Gatto L, Campos A, Chalkley RJ, Kraus HJ, Albar JP, Martinez-Bartolomé S, Apweiler R, Omenn GS, Martens L, Jones AR, Hermjakob H. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat Biotechnol. 2014;32:223–6. doi: 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vizcaíno JA, Côté R, Reisinger F, Foster JM, Mueller M, Rameseder J, Hermjakob H, Martens L. A guide to the Proteomics Identifications Database proteomics data repository. Proteomics. 2009;9:4276–83. doi: 10.1002/pmic.200900402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prakash A, Tomazela DM, Frewen B, Maclean B, Merrihew G, Peterman S, Maccoss MJ. Expediting the Development of Targeted SRM Assays: Using Data from Shotgun Proteomics to Automate Method Development. J Proteome Res. 2009;8:2733–2739. doi: 10.1021/pr801028b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20:3246–8. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]