Abstract
Objective
Smoking patterns and cessation rates vary widely across smokers and can be influenced by variation in rates of nicotine metabolism (i.e. cytochrome P450 2A6, CYP2A6, enzyme activity). There is high heritability of CYP2A6-mediated nicotine metabolism (60–80%) due to known and unidentified genetic variation in the CYP2A6 gene. We aimed to identify and characterize additional genetic variants at the CYP2A6 gene locus.
Methods
A new CYP2A6-specific sequencing method was used to investigate genetic variation in CYP2A6. Novel variants were characterized in a White human liver bank that has been extensively phenotyped for CYP2A6. Linkage and haplotype structure for the novel SNPs were assessed. The association between novel five-SNP diplotypes and nicotine metabolism rate was investigated.
Results
Seven high frequency (MAFs ≥6%) non-coding SNPs were identified as important contributors to CYP2A6 phenotypes in a White human liver bank (rs57837628, rs7260629, rs7259706, rs150298687 [also denoted rs4803381], rs56113850, rs28399453, and rs8192733), accounting for two times more variation in in vitro CYP2A6 activity relative to the four established functional CYP2A6 variants that are frequently tested in Whites (CYP2A6*2, *4, *9, and *12). Two pairs of novel SNPs were in high linkage disequilibrium, allowing us to establish five-SNP diplotypes that were associated with CYP2A6 enzyme activity (rate of nicotine metabolism) in vitro in the liver bank and in vivo among smokers.
Conclusion
The novel five-SNP diplotype may be useful to incorporate into CYP2A6 genotype models for personalized prediction of nicotine metabolism rate, cessation success, and response to pharmacotherapies.
Keywords: CYP2A6, sequencing, nicotine metabolism, genetic variant, SNP, haplotype, diplotype
Introduction
Smoking patterns and rates of smoking cessation vary widely among smokers. Variation in nicotine metabolism rate and clearance is primarily mediated by the hepatic enzyme CYP2A6 (cytochrome P450 2A6)[1,2]. Variation in nicotine metabolism is associated with differences in smoking quantities, dependence, and cessation such that smokers who exhibit a slower rate of nicotine metabolism often smoke less, have lower nicotine dependence scores, and are more likely to quit smoking[3–6]. In addition, nicotine metabolism rate can be used to predict therapeutic response to smoking cessation pharmacotherapies, including behavioral counseling, nicotine replacement therapy, buproprion, and varenicline[4,7,8]. The relationship between the nicotine metabolism rate and smoking quantity stems from smokers’ ability to titrate their nicotine intake from cigarettes in order to maintain consistent nicotine blood levels throughout the day and avoid withdrawal[9]. Nicotine dependence and cessation may be modulated by nicotine metabolism rate such that normal, compared to slow, nicotine metabolizers have greater fluctuation of blood nicotine concentrations, which may result in enhanced reward, altered strength of functional conductivity between rewarding brain regions, and increased conditioned responses to smoking cues[6,10,11].
Heritability estimates suggest that 60–80% of the variation in nicotine metabolism is attributable to genetic influences, with known CYP2A6 genetic variants accounting for approximately 20–30% of the variation[12,13]. Only a small contribution arises from environmental or non-genetic factors; dietary or pharmacological agents, gender and estrogen consumption, age, body mass index, alcohol consumption, and smoking account for approximately 8% of the variation[14–18]. CYP2A6 gene locus polymorphisms are associated with differences in nicotine metabolism in vitro and in vivo[17,19,20], and correspondingly with smoking behaviors[3,21]. In a recent GWAS of CYP2A6 activity, all three independent genome-wide significant signals were within or near the CYP2A6 locus[12]. The CYP2A6 gene is highly polymorphic, with >40 genetic variants characterized to date (http://www.cypalleles.ki.se/cyp2a6.htm). Thus it is likely that genetic variation in CYP2A6 remains unaccounted for, contributing to the high heritability of the nicotine metabolism phenotype.
Part of the difficulty in fully characterizing CYP2A6 genetic variation is high sequence homology of CYP2A6, CYP2A7 and CYP2A13[22]. Consequently, CYP2A6 genotyping typically requires a two-step approach that first ensures specificity to CYP2A6 followed by characterization of the specific variant of interest[23]. However, in order to perform large-scale evaluations of the CYP2A6 locus for identification of novel genetic variation, there is a need for sequencing approaches that both account for high homology with other genes and are high-throughput. In this study we describe a new method for sequencing this gene, examine novel (i.e. uncharacterized) and existing CYP2A6 genetic variation, and characterize seven novel high frequency single nucleotide polymorphisms (SNPs) with respect to CYP2A6 enzyme activity in a human liver bank in vitro assessment, and using an in vivo phenotype measure.
Methods
Overview
Using CYP2A6-specific next-generation sequencing, we identified and characterized novel CYP2A6 genetic variants. Sequencing was performed at the CYP2A6 locus and surrounding region (hg38 40843538–40852095) for White (N=42) and African American (N=170) subjects. The sequencing sample set consisted of DNA from subjects from a human liver bank (N=43)[17], a pharmacokinetic study (N=57)[24], and two smoking cessation clinical trials (KIS2 N=26, and KIS3 N=86)[25, 26]. We prioritized potentially functional SNPs and characterized the association of each SNP with CYP2A6 phenotypes in a human liver bank previously characterized for CYP2A6 (CYP2A6 sequenced, mRNA, protein, and enzyme activity quantified). Finally, we assessed the linkage and haplotype structure of the SNPs, and the impact of resulting diplotypes on in vitro and in vivo CYP2A6 activity, in the liver bank and in a population of treatment-seeking smokers from a smoking cessation clinical trial (Pharmacogenetics of Nicotine and Addiction Treatment clinical trial (PNAT), NCT01314001)[7].
CYP2A6-Specific Sequencing
DNA from N=212 White and African American subjects was sequenced at the CYP2A6 locus, using CYP2A6-specific sequencing, in order to thoroughly interrogate SNPs at this locus for further testing. Library preparation, sequence-capture, sequencing on Illumina MiSeq, and sequence analysis were conducted as outlined in Supplementary Method 1.
Novel CYP2A6 SNPs identified in the region (hg38) 40843538–40852095 (3’-UTR to 2kb 5’ of CYP2A6) were analyzed for potential impact on CYP2A6 using the following bioinformatics tools: including ensembl genome browser[27], GTEx Portal[28], RegulomeDB[29], RegRNA 2.0[30], HaploReg v4.1[31], RESCUE-ESE[32], and FAS-ESS[33]. A final subset of novel SNPs predicted to impact CYP2A6 were further investigated in a human liver bank (described below).
Liver Bank CYP2A6 PGRN-Sequencing and Genotyping
The CYP2A6 locus was previously sequenced in the full human liver bank using PGRN-Seq[34], an earlier sequencing platform prior to further optimization for CYP2A6 (Supplementary Method 2). These CYP2A6 sequence data were one mechanism used to genotype SNP calls in the liver bank for novel SNPs identified by CYP2A6-specific sequencing (described above), and the other method consisted of traditional genotyping approaches. SNP genotype calls for the N=43 liver bank samples included in the original exploratory CYP2A6-specific sequencing were made according to PGRN-Seq or direct genotyping, to remain consistent with genotype calls for the rest of the human liver bank (see Table 1 for summaries of samples used). The majority of SNP genotype calls in the liver bank were readily available from PGRN-seq data; calls were then verified using traditional genotyping approaches (described below).
Table 1.
Summary of the studies that were used for CYP2A6 sequencing and CYP2A6 genotype-phenotype analyses.
| Study Title | Number of subjects analyzed |
Sequencing or Genotyping Approach |
Purpose of sequencing or genotyping |
Reference for original study |
|---|---|---|---|---|
| Liver Bank | 43 | CYP2A6-Specific sequencing | Exploratory sequencing | Tanner, Prasad [17] |
| Pharmacokinetic Study | 57 | CYP2A6-Specific sequencing | Exploratory sequencing | Mwenifumbo, Sellers [24] |
| KIS2 | 26 | CYP2A6-Specific sequencing | Exploratory sequencing | Ho, Mwenifumbo [25] |
| KIS3 | 86 | CYP2A6-Specific sequencing | Exploratory sequencing | Cox, Nollen [26] |
| Liver Bank | 327 | PGRN-Sequencing, and TaqMan SNP genotyping | Identify SNPs for genotype-phenotype analyses | Tanner, Prasad [17] |
| PNAT | 641 | Genotyped via GWAS SNP calls | Identify SNPs for genotype-phenotype analyses | Lerman, Schnoll [7] |
Genotyping for established CYP2A6 genetic variants common among White populations (CYP2A6*2, *4, *9, *12) was performed using two-step allele-specific polymerase chain reaction genotyping or TaqMan SNP genotyping (Applied Biosystems/ThermoFisher Scientific), as reported previously[17, 23]. Concordance between the genotype and PGRN-Seq calls for established SNPs CYP2A6*2 (rs1801272) and *9 (rs28399433) was 99% and 98%, respectively; CYP2A6*4 and *12 are copy number and hybrid variants not identified by this sequencing method. Next, we verified liver bank PGRN-Seq SNP calls with traditional genotyping approaches that consisted of direct genotyping (TaqMan or SYBR Green SNP genotyping assays) for the seven novel SNPs that exhibited significant associations with CYP2A6 phenotypes (Supplementary Table 1). Concordance of genotyping and PGRN-Seq SNP calls was assessed (5/7 SNPs were detected by PGRN-Seq), and was high for all SNPs, ranging from 84–97%. For subsequent genotype-phenotype analyses, calls for the seven novel SNPs were made using PGRN-Seq data (i.e. data from an earlier sequencing platform) unless concordance was <90% (N=3) or was not available (N=2), in which case TaqMan or SYBR Green genotype data were used.
Quantifying In Vitro CYP2A6 Phenotypes
Genotype-phenotype associations for novel SNPs were assessed in White subjects in the full liver bank dataset (N=332). Measurements of CYP2A6 mRNA, protein, and enzyme activity in the liver bank have been described previously[17]. Briefly, CYP2A6 mRNA expression was quantified using RNA sequencing (values expressed as fragments per kilobase per million reads, FPKM). CYP2A6 protein was quantified using a liquid chromatography-tandem mass spectrometry (LC-MS/MS) proteomics method. In vitro CYP2A6 enzyme activity was determined by quantifying the rate of nicotine metabolism to cotinine in liver microsomes.
Clinical Trial CYP2A6 GWAS and Genotyping
A genome-wide association study (GWAS) of nicotine metabolite ratio (NMR) in the PNAT trial was used to ascertain SNP genotype calls for the seven novel SNPs (GWAS methodology described in detail by Chenoweth et al. 2017, manuscript submitted). Briefly, genome-wide SNP genotyping was performed using the Illumina HumanOmniExpressExome-8 v1.2 array (Illumina, San Diego, CA). Genotypes were imputed using IMPUTE2 software, and all variants had a quality score >0.85 and minor allele frequency >1%, and thus were included in analyses. In addition, samples were genotyped for the same established CYP2A6 genetic variants (CYP2A6*2, *4, *9, *12)[23]. GWAS and genotyping SNP calls were 99.8% and 99.2% concordant for *2 and *9, respectively, supporting the concordance of GWAS SNP calls for the novel variants.
Assessing Linkage and Haplotype Structure of Novel SNPs
Using Haploview[35], we estimated the linkage disequilibrium (LD) and haplotype structure of the seven novel SNPs separately in the liver bank and PNAT trial. Analyses were restricted to White subjects not possessing any previously established CYP2A6 genetic variants (i.e. CYP2A6*2, *4, *9, *12; liver bank N=252, PNAT N=536). For SNP pairs in high LD (r2 ≥0.80) in PNAT (the larger study population), one SNP from each pair was selected to include in a simplified haplotype, resulting in a final haplotype comprised of five SNPs. Using the PHASE program[36,37], five-SNP haplotypes were phased (8360bp genomic distance) in order to predict five-SNP diplotypes in both studies.
Clinical Trial CYP2A6 Phenotyping
In vivo CYP2A6 activity was measured in blood at baseline for treatment-seeking smokers in the PNAT trial by quantifying the ratio of nicotine’s metabolites, trans-3’-hydroxycotinine/cotinine, a measure referred to as the NMR, using LC-MS/MS[7], as described previously[38,39]. Limits of quantification for each metabolite were 1ng/ml of blood.
Statistical Analyses
We assessed the association of each of the seven novel SNPs with CYP2A6 mRNA, protein, and enzyme activity in the liver bank using Kruskal Wallis and Dunn’s Multiple Comparisons tests or the Mann-Whitney test. The same tests were used to evaluate the association of the seven- and five-SNP diplotypes with CYP2A6 activity in the liver bank and PNAT trial. We ran nested linear regression models for CYP2A6 activity to calculate the additional proportion of variation that is accounted for by the seven SNPs, or the simplified five-SNP haplotype, relative to what was accounted for by the established CYP2A6 genetic variants alone. Genotypes were the only predictors included in these models. Analyses were performed using GraphPad Prism (v.6.0, LaJolla, CA) and SPSS (v.23; IBM, Armonk, NY), and statistical tests were considered significant for P<0.05.
Results
Seven novel SNPs identified through sequencing
We identified N=229 distinct SNPs total (N=38 5’, N=46 exon, N=133 intron, and N=12 3’-UTR CYP2A6 SNPs) in the region (hg38) 40843538–40852095. Following bioinformatics analyses, N=21 novel SNPs were predicted to impact CYP2A6, including nonsynonymous SNPs, SNPs with a predicted association with CYP2A6 mRNA levels, and SNPs located in an miRNA binding site, an exonic splicing enhancer or silencer site, or a regulatory region (Supplementary Table 1). Of the N=21 novel SNPs, N=13 were sufficiently common to be used in analyses of relationships with in vitro CYP2A6 activity in the liver bank (described below); the remaining N=8 novel SNPs were rare (minor allele frequency, MAF<1% in 1000 Genomes) and we were not powered to investigate these in the liver bank. Of the N=13 common SNPs, N=7 were significantly associated with CYP2A6 activity in the liver bank (described below), and were therefore prioritized for further investigation. Subjects possessing any of the SNPs identified through sequencing (i.e. rare or non-significant SNPs) were included in all of the following analyses.
Of the seven prioritized SNPs, four are located 5’ of CYP2A6 (rs57837628, rs7260629, rs7259706, rs150298687 [also denoted rs4803381]), two are located in introns (rs56113850, rs28399453), and one in the 3’-UTR (rs8192733) (Table 2). The seven SNPs are common in White populations in 1000 Genomes (6−70%), the human liver bank (7−72%), and the PNAT trial (7−69%). The total number of samples from the liver bank and PNAT trial that were included in subsequent analyses is provided in Table 1.
Table 2.
Summary of the seven novel CYP2A6 SNPs of interest
| 1000 Genome | Liver Bank | PNAT Trial | ||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|
||||||
| SNP rs ID | CYP2A6 region |
Hg38 position |
Allele change |
African Allele Freq |
European Allele Freq |
White Allele Freq |
White Allele Freq |
Predicted Functiona |
| rs57837628 | 5’ | 40852005 | A>G | 17% (G) | 54% (G) | 51% (G) | 49% (G) | Increased |
| rs7260629 | 5’ | 40851727 | T>G | 71% (G) | 69% (G) | 72% (G) | 69% (G) | Increased |
| rs7259706 | 5’ | 40851715 | C>T | 73% (T) | 70% (T) | 69% (T) | 69% (T) | Increased |
| rs150298687 (also denoted rs4803381) | 5’ | 40851439 | T>C | 46% (C) | 58% (C) | 63% (C) | 58% (C) | Increased |
| rs56113850 | Intron 4 | 40847202 | T>C | 39% (C) | 59% (C) | 57% (C) | 56% (C) | Increased |
| rs28399453 | Intron 6 | 40845495 | G>A | 0% (A) | 6% (A) | 7% (A) | 7% (A) | Increased |
| rs8192733 | 3’-UTR | 40843645 | G>C | 23% (C) | 48% (C) | 47% (C) | 47% (C) | Increased |
. Function predicted according to bioinformatics tools (e.g. GTEx Portal)
Association of seven novel SNPs with CYP2A6 phenotypes
All seven novel SNPs were significantly associated with higher in vitro CYP2A6 activity, each exhibiting a gene-dose effect (P values provided in Figure 1 and Table 3). Six of the seven SNPs were significantly associated with higher CYP2A6 protein levels (P values 0.49−<0.0001), with the exception of rs28399453 (P=0.11, same direction of effect) (Table 3). None of the seven SNPs significantly altered CYP2A6 mRNA expression (all P>0.11), however all indicated a similar direction of effect. Similar relationships between the novel SNPs and CYP2A6 phenotypes were observed when restricting analyses to CYP2A6*1/*1 genotype liver donors (i.e. excluding donors with established CYP2A6 genetic variants, N=63 White liver donors).
Figure 1.
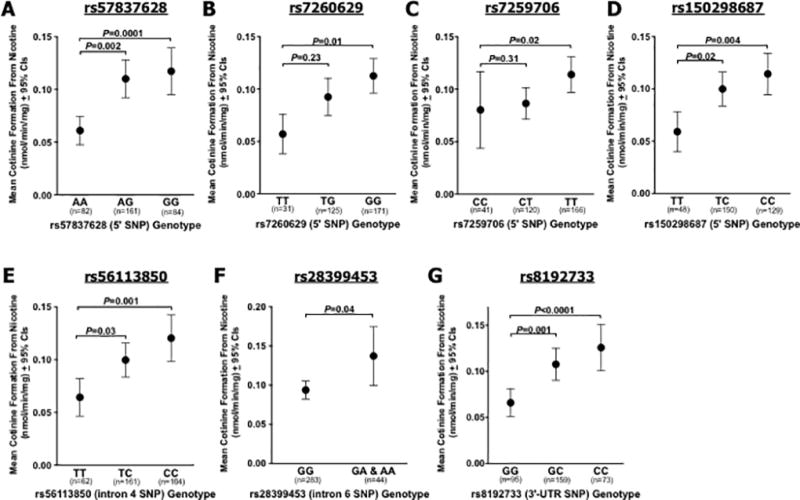
Association of seven novel SNPs with CYP2A6 enzyme activity (rate of cotinine formation from nicotine, nmol/min/mg) among N=327 White liver donors. (A) rs57837628, (B) rs7260629, (C) rs7259706, (D) rs150298687, (E) rs56113850, (F) rs28399453, (G) rs8192733. P values derived from Kruskal Wallis and Dunn’s Multiple Comparisons tests for most SNPs, or Mann-Whitney tests for rs28399453. All White liver bank donors were included in analyses.
Table 3.
Summary of effect sizes (Cohen’s d) and P values for seven novel SNPs with CYP2A6 phenotypes (enzyme activity, protein levels, and mRNA expression) in White liver donors
| SNP rs ID | Genotype groups compared |
CYP2A6 enzyme activity (nmol/min/mg), N=327 |
CYP2A6 protein levels (pmol/mg), N=320 |
CYP2A6 mRNA expression (FPKM values), N=265 |
|||
|---|---|---|---|---|---|---|---|
|
|
|
|
|||||
| Effect size | P value | Effect size | P value | Effect size | P value | ||
| rs57837628 | AA vs. AG | 0.53 | 0.002 | 0.57 | 0.0008 | 0.38 | 0.20 |
| AA vs. GG | 0.67 | 0.0001 | 0.74 | <0.0001 | 0.27 | 0.75 | |
|
| |||||||
| rs7260629 | TT vs. TG | 0.44 | 0.23 | 0.46 | 0.15 | 0.07 | >0.99 |
| TT vs. GG | 0.65 | 0.01 | 0.68 | 0.007 | 0.17 | >0.99 | |
|
| |||||||
| rs7259706 | CC vs. CT | 0.06 | 0.31 | 0.04 | 0.49 | 0.45 | 0.17 |
| CC vs. TT | 0.30 | 0.02 | 0.28 | 0.02 | 0.41 | 0.19 | |
|
| |||||||
| rs150298687 | TT vs. TC | 0.48 | 0.02 | 0.48 | 0.02 | 0.44 | 0.27 |
| TT vs. CC | 0.60 | 0.004 | 0.62 | 0.002 | 0.32 | 0.80 | |
|
| |||||||
| rs56113850 | TT vs. TC | 0.40 | 0.03 | 0.48 | 0.009 | 0.44 | 0.12 |
| TT vs. CC | 0.59 | 0.001 | 0.66 | 0.0003 | 0.32 | 0.56 | |
|
| |||||||
| rs28399453 | GG vs. GA & AA | 0.39 | 0.04 | 0.28 | 0.11 | 0.17 | 0.91 |
|
| |||||||
| rs8192733 | GG vs. GC | 0.44 | 0.001 | 0.49 | 0.002 | 0.25 | >0.99 |
| GG vs. CC | 0.65 | <0.0001 | 0.80 | <0.0001 | 0.21 | >0.99 | |
In order to determine if the novel SNPs collectively accounted for any additional variation in CYP2A6 activity in the liver bank, we performed nested linear regression modeling. The previously established CYP2A6 genetic variants (*2, *4, *9, *12) alone accounted for 2.1% of the variation in CYP2A6 activity (model 1: R2=0.021, P=0.009). When the seven novel SNPs were added to the model, they accounted for an additional 4.5% of CYP2A6 activity variation (model 2: R2=0.066, P=0.005), relative to the established variants (model 1 vs. model 2: R2 change=0.045, P=0.03).
Linkage and haplotype structure for seven novel SNPs
Due to the high frequency of the seven novel SNPs and their similar impact on CYP2A6, we assessed linkage and haplotype structure. Haploview was used to predict r2, D’, and haplotype blocks in liver donors and PNAT treatment-seeking smokers. The seven novel SNPs were not in LD with established CYP2A6 genetic variants (*2, *4, *9, *12) in either the liver bank or PNAT trial (r2 values=0.00−0.07; Supplementary Figure 1). Further analyses were restricted to subjects without established CYP2A6 genetic variants. In the liver bank, of the seven novel SNPs, two SNP pairs were in high LD (r2>0.8, D’>0.9): (1) rs57837628 and rs8192733, and (2) rs56113850 and rs150298687, with the rs7260629 and rs7259706 pair falling just below threshold (r2=0.79) (Figure 2a). In the PNAT trial, two SNP pairs were in high LD: (1) rs57837628 and rs8192733, (2) rs7260629 and rs7259706 (Figure 2b). SNP rs28399453 was not in LD with any of the other six SNPs in either study population (r2 values=0.03−0.07).
Figure 2.
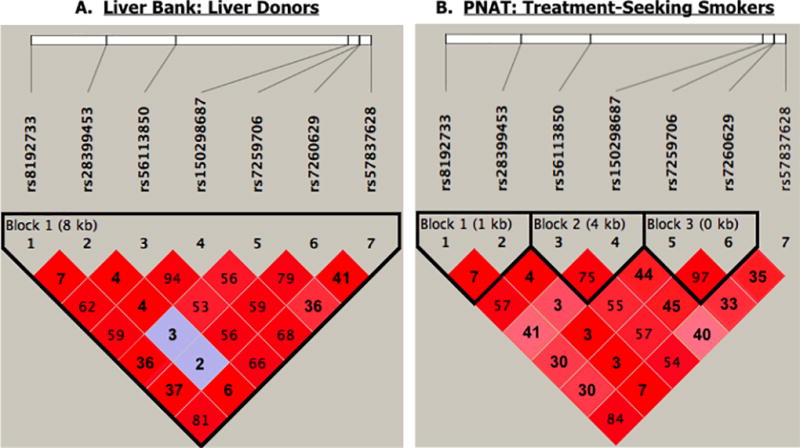
LD plot for seven novel SNPs; r2 values (number) and D’ (shading) were derived using Haploview. Analyses were restricted to White subjects who do not possess any established CYP2A6 genetic variants. (A) Human liver bank. (B) Treatment-seeking smokers from PNAT.
Due to high linkage among the novel SNPs, we chose to exclude one SNP from each of the high LD SNP pairs that were present in PNAT, the larger study population, (i.e. rs57837628 and rs8192733; rs7260629 and rs7259706) and investigate CYP2A6 phenotype associations for a simplified set of five SNPs: rs28399453, rs56113850, rs150298687, rs7260629, and rs57837628. We then quantified the proportion of variation in CYP2A6 activity in the liver bank additionally accounted for by the five SNPs, relative to the established CYP2A6 variants alone, using regression modeling. The five novel SNPs accounted for an additional 3.9% of the variation in CYP2A6 activity (model 2: R2=0.060, P=0.003) compared to the established variants (model 1: R2=0.021, P=0.009; model 1 vs. model 2: R2 change=0.039, P=0.02).
Five-SNP diplotypes in the liver bank and PNAT trial
The PHASE software was used to predict seven- and five-SNP diplotypes. Of note, when phasing the established CYP2A6 genetic variants and the seven novel SNPs, several compound heterozygotes were identified consisting of both established and novel variants; however we were not powered to assess the functional significance of these compound heterozygotes due to low genotype frequencies. As a result, and due to the lack of LD for established CYP2A6 genetic variants with the novel SNPs (r2 values=0.00−0.07; Supplementary Figure 1), subjects possessing established CYP2A6 genetic variants (*2, *4, *9, *12) were therefore excluded from analyses. Summaries of seven-SNP diplotypes in the liver bank and PNAT have been provided in Supplementary Tables 4 and 5, respectively. With respect to the simplified group of five novel SNPs, there were 25 distinct diplotypes total in the liver bank (Table 4) and 40 diplotypes in PNAT (Table 5). The full reference diplotype (GTTTA/GTTTA; reference allele designated according to dbSNP, and corresponds to the CYP2A6*1 allele) was present in 6.6% and 6.4% of samples from the subset of the liver bank and PNAT, respectively. Of note, not all diplotypes observed in the liver bank appeared in PNAT; diplotypes #12, 16–21, and 23–24 (Table 4) were not found among PNAT subjects (Table 5). Furthermore, 24 additional diplotypes were present in PNAT, which were not identified in the liver bank (diplotypes #16–39, Table 5).
Table 4.
Five-SNP phased diplotypes predicted in donors in the liver bank
| Diplotype Group # |
Full Diplotype | N in liver bank | CYP2A6 activity (nicotine metabolism, nmol/min/mg), mean of diplotype group |
|---|---|---|---|
| Reference | G T T T A / G T T T A | 17 | 0.07 |
|
| |||
| 1 | G C C G G / G C C G G | 64 | 0.13 |
| 2 | G T T T A / G C C G G | 60 | 0.09 |
| 3 | G T T G A / G C C G G | 22 | 0.12 |
| 4 | G C C G A / G C C G G | 17 | 0.10 |
| 5 | G T T T A / G T T G A | 15 | 0.08 |
|
| |||
| 6 | G C C G G / A C C G G | 14 | 0.11 |
| 7 | G T T T A / A C C G G | 12 | 0.14 |
| 8 | G T T T A / G C C G A | 8 | 0.08 |
| 9 | G T T G A / G C C G A | 5 | 0.06 |
| 10 | G T T G A / A C C G G | 4 | 0.03 |
| 11 | G C C G A / A C C G G | 3 | 0.31 |
| 12 | G T T G A / G T T G A | 2 | 0.04 |
| 13 | G T T T A / G C C T G | 2 | 0.08 |
| 14 | A C C G G / A C C G G | 1 | 0.03 |
| 15 | G C C G A / G C C G A | 1 | 0.01 |
| 16 | G C T G A / A C C G G | 1 | 0.08 |
| 17 | G C T G A / G C C G G | 1 | 0.01 |
| 18 | G C T T A / G C C G G | 1 | 0.04 |
| 19 | G T C G A / G C C G A | 1 | 0.23 |
| 20 | G T C G A / G C C G G | 1 | 0.03 |
| 21 | G T C G G / A C C G G | 1 | 0.12 |
| 22 | G T C G G / G C C G G | 1 | 0.08 |
| 23 | G T T G A / G T C G G | 1 | 0.19 |
| 24 | G T T T G / G T C G G | 1 | 0.30 |
. Full reference diplotype is bolded
Table 5.
Five-SNP phased diplotypes predicted in treatment-seeking smokers in PNAT trial
| Diplotype Group # |
Full Diplotype | N in PNAT | CYP2A6 activity (nicotine metabolite ratio, NMR), mean of diplotype group |
Corresponding liver bank diplotype group |
|---|---|---|---|---|
| Reference | G T T T A / G T T T A | 41 | 0.3 | Reference Group |
|
| ||||
| 1 | G C C G G / G C C G G | 110 | 0.55 | 1 |
| 2 | G T T T A / G C C G G | 123 | 0.40 | 2 |
| 3 | G T T G A / G C C G G | 43 | 0.46 | 3 |
| 4 | G C C G A / G C C G G | 55 | 0.55 | 4 |
| 5 | G T T T A / G T T G A | 20 | 0.38 | 5 |
|
| ||||
| 6 | G C C G G / A C C G G | 38 | 0.49 | 6 |
| 7 | G T T T A / A C C G G | 29 | 0.42 | 7 |
| 8 | G T T T A / G C C G A | 30 | 0.43 | 8 |
| 9 | G T T G A / G C C G A | 8 | 0.42 | 9 |
| 10 | G T T G A / A C C G G | 8 | 0.54 | 10 |
| 11 | G C C G A / A C C G G | 10 | 0.54 | 11 |
| 12 | G T T T A / G C C T G | 5 | 0.48 | 13 |
| 13 | A C C G G / A C C G G | 3 | 0.45 | 14 |
| 14 | G C C G A / G C C G A | 11 | 0.41 | 15 |
| 15 | G T C G G / G C C G G | 2 | 0.21 | 22 |
| 16 | G C T G G / G C C G G | 23 | 0.54 | N/A |
| 17 | G T T T A / G C T G G | 13 | 0.45 | N/A |
| 18 | G C C G G / A C T G G | 12 | 0.46 | N/A |
| 19 | G T T G G / G C C G G | 11 | 0.46 | N/A |
| 20 | G C C T G / G C C G G | 10 | 0.60 | N/A |
| 21 | G C C G A / A C T G G | 6 | 0.51 | N/A |
| 22 | G C T G G / G C C G A | 5 | 0.54 | N/A |
| 23 | G C T T G / G C C G G | 4 | 0.67 | N/A |
| 24 | G T T T A / G C T G A | 4 | 0.45 | N/A |
| 25 | G T T G A / G T T G G | 2 | 0.35 | N/A |
| 26 | G T T T A / G T T G G | 2 | 0.33 | N/A |
| 27 | G C C T G / A C C G G | 1 | 0.69 | N/A |
| 28 | G T T G G / A C C G G | 1 | 0.35 | N/A |
| 29 | A C T G G / A C T G G | 1 | 0.46 | N/A |
| 30 | G T T T A / A C T G G | 1 | 0.53 | N/A |
| 31 | G C C T G / G C C G A | 1 | 0.67 | N/A |
| 32 | G C C T G / G C C T G | 1 | 0.30 | N/A |
| 33 | G C T T G / G C C T G | 1 | 0.48 | N/A |
| 34 | G C T G A / G C T G A | 1 | 0.61 | N/A |
| 35 | G C T G G / G C T G G | 1 | 0.54 | N/A |
| 36 | G T T G A / G C T G G | 1 | 0.43 | N/A |
| 37 | G T T G G / G C T G G | 1 | 0.33 | N/A |
| 38 | G T T T A / G C T T G | 1 | 0.36 | N/A |
| 39 | G T T T A / G T C G A | 1 | 0.22 | N/A |
. Full reference diplotype is bolded
Association of five-SNP diplotypes and CYP2A6 activity in vitro and in vivo
We compared the rate of in vitro CYP2A6 activity (rate of cotinine formation from nicotine) of the top 5 most frequent diplotypes in the liver bank (diplotypes #1–5, Table 4) to that of the reference diplotype (Table 4), which does not possess the variant form of any of the five SNPs (Figure 3a). Of note, several liver bank donors were missing CYP2A6 activity data while still having genotype/sequence data, and therefore were excluded from phenotype analyses in Figure 3, but were included in the summary of diplotypes in Table 4. Each of the 5 most frequent diplotypes had higher mean CYP2A6 activity compared to the reference diplotype (P=0.23). Additionally, when collapsed together into one group, the 5 most frequent diplotypes collectively had non-significantly higher mean CYP2A6 activity compared to the reference diplotype (P=0.28), which was also observed for the remaining diplotypes combined (diplotypes #6–24, Table 4; P=0.28), and when all variant diplotypes were grouped together (diplotypes #1–24; P=0.26).
Figure 3.
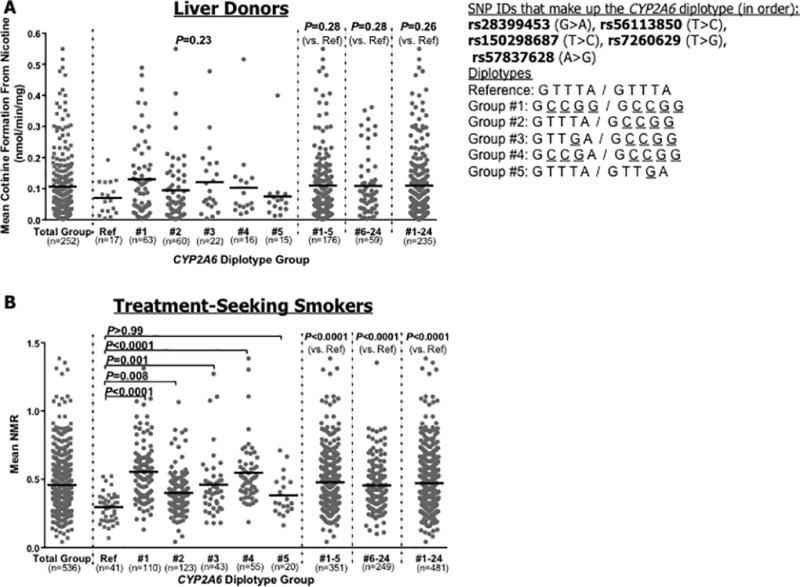
Comparison of CYP2A6 enzyme activity of the full reference diplotype group and top 5 most frequent five-SNP CYP2A6 diplotypes. Data were restricted to White subjects, and to those without established CYP2A6 genetic variants. Enzyme activity for the total group (i.e. before splitting into diplotype groups) has been provided as a reference in each figure (far left). The diplotype group numbers refer to Tables 4 and 5. (A) Data from the human liver bank. In vitro CYP2A6 activity was measured as the rate of cotinine formation (nmol/min/mg) from nicotine [17]. For reference, the mean (±SD) cotinine formation for the pooled established variant group from the liver bank is 0.07 (±0.09), as described previously [17]. (B) Data from the PNAT trial. In vivo CYP2A6 activity was measured using the ratio of nicotine’s metabolites (3HC/COT, nicotine metabolite ratio, NMR) [38, 39]. For reference, the mean (±SD) NMR for the pooled established variant group from PNAT is 0.30 (±0.17). Note: The N=25 diplotypes found in the liver bank (Table 4) were compared to the same diplotypes in PNAT, to maintain consistency of the comparison; it should be noted however that some low frequency diplotypes among the N=25 in the liver bank were not found in PNAT (diplotypes #12, 16–21, and 23–24 from Table 4), and likewise several low frequency diplotypes in PNAT were not found in the liver bank (diplotypes #16–39; Table 5). Therefore, these diplotype groups were excluded from the PNAT phenotype analyses in part B. P values are based on Kruskal Wallis and Dunns Multiple Comparison tests, or Mann-Whitney tests. Among PNAT treatment seeking smokers, the relationship between the reference diplotype and top 5 diplotype groups remained similar and significant when accounting for the non-genetic factors “age” and “gender” (P<0.001), using regression modeling.
To increase our power for assessing associations between diplotypes and CYP2A6 activity (N=252 subjects analyzed in the liver bank), we extended these analyses to the PNAT sample (N=536). In PNAT, we compared the mean NMR between the reference diplotype and the top 5 most frequent diplotypes that were found in the liver bank (diplotypes #1–5, Table 5; Figure 3b). Only diplotypes that were found in the liver bank were included in diplotype-phenotype analyses in PNAT. Compared to the full reference diplotype, diplotypes #1–4 (Table 5) each had significantly higher mean NMR (P values 0.008−<0.0001). Diplotype #5, which only possesses the variant form of one of the five SNPs, exhibited non-significantly higher mean NMR (P>0.99), relative to the reference diplotype. The combined diplotype groups #1–5, #6–24, and #1–24 had significantly higher mean NMR compared to the reference diplotype (all P<0.0001). Analyses of the top five most frequent seven-SNP diplotypes were also conducted in the liver bank and PNAT trial; these illustrated very similar patterns for association of frequent diplotypes with CYP2A6 in vitro and in vivo activity (Supplementary Figure 2) as was observed for the frequent five-SNP diplotypes.
Discussion
We have identified seven novel SNPs, and a resulting simplified five-SNP diplotype, at the CYP2A6 locus, which significantly impact CYP2A6 activity assessed in both a human liver bank in vitro and using the NMR in vivo. All seven novel SNPs characterized here were non-coding variants, present in regulatory and intronic regions of the CYP2A6 gene, with MAFs >1% in European ancestry populations (liver bank: 7−72%, PNAT: 7−69%). Non-coding variants are largely understudied, despite important roles in regulating transcription, chromatin state, splicing, and epigenetic modifications; this work further highlights the utility of full gene or genome sequencing in characterizing complex traits and diseases, such as smoking.
For the novel SNPs characterized here that were in high linkage disequilibrium (LD), allowing for incorporation of only one of the two SNPs into the simplified five-SNP diplotype, LD was similarly moderate-to-high in a GWAS of the NMR in a Finnish population (rs57837628 and rs8192733 r2=0.73, rs7260629 and rs7259706 r2=0.98)[12]. Likewise, associations of CYP2A6 diplotypes with in vitro and in vivo rate of nicotine metabolism were consistent for the seven- and five-SNP diplotypes (see Figure 3 and Supplementary Figure 2), and there was little change in the amount of variation in CYP2A6 enzyme activity accounted for by all seven SNPs compared to only five SNPs (6.6 vs. 6.0%). This supports the use and assessment of the five-SNP diplotype, as opposed to all seven SNPs, in CYP2A6 phenotype association studies and in future genotype-prediction algorithms. As we were able to capture significantly more of the variation in CYP2A6 activity with the five-SNP diplotype (additional 3.9% compared to established variants alone), in the future, as we capture more and more of the heritable variation in CYP2A6, we may be able to use CYP2A6 genotype to tailor smoking cessation. As feasibility of genomics-based treatment optimization increases, implementation of pre-emptive genotyping will require further assessment of both coding- and non-coding genetic variation, and incorporation of genotypes into functionally significant diplotypes, as demonstrated here.
Our in vitro and in vivo functional characterization of novel SNPs expands on previous GWAS’s that have identified significant hits at CYP2A6. Consistent with our prioritization, six of the seven SNPs in our investigation reached genome-wide significance (rs28399453 did not, P=6.42E-06) in the Finnish GWAS of the NMR, with rs56113850 being the top hit (most highly significant, P=5.77E-86)[12]. rs56113850 was also the top hit for association with NMR in a multi-ethnic cohort (MEC) study in which it was globally significantly associated with higher CYP2A6 activity (P=1.19E-50), and it was the second most significant SNP associated with lung cancer risk in the TRICL consortium GWAS (P=5.78E-11)[41]. Similarly, rs56113850 was the top ranked European American SNP in a separate GWAS of NMR (P=3.81E-10)[42]. SNPs rs57837628 and rs8192733 were also globally significant in the MEC (P=6.84E-37 and 2.78E-08, respectively), and were associated with increased lung cancer risk in the TRICL consortium GWAS (P=4.01E-10 and 2.10E-9, respectively)[41]. The remaining SNPs, rs7260629, rs7259706, rs150298687, and rs28399453, were not significant in the MEC or TRICL consortium GWAS’s[41]. Lack of genome-wide significance of rs28399453 in the Finnish GWAS of NMR may result from lower allele frequency (liver bank: 7%, PNAT: 7%, 1000 Genomes: 6% for European ancestry) relative to the other six novel SNPs (liver bank: 47−72%, PNAT: 47−69%, 1000 Genomes: 48−70% for European ancestry), and in the MEC study due to even lower allele frequencies across populations of other ethnic origins (0, 0, and 2% among African, Asian, and Latin American ancestry populations, respectively, from 1000 Genomes).
We also identified novel exon variation in CYP2A6 through sequencing (N=46 exon SNPs); only N=6 missense and N=1 nonsense SNPs were found, all of which were rare (MAF<1% in 1000 genomes). This finding is consistent with the majority of CYP genes where 93% and 83% of coding region SNPs are rare (MAF<1%) or very rare (MAF<0.1%), respectively[43]. Due to the infrequency of nonsynonymous coding region SNPs in the current study, we were not powered to characterize functional impact in the liver bank (Supplementary Table 1). Functional investigation of these SNPs may require the use of an in vitro CYP2A6 cDNA expression system[44], in combination with the BioBin software, which improves power to detect phenotype associations by combining rare variants[45].
In addition to a lack of power for investigating rare coding variation, a study limitation is the assessment of White populations only. Frequency of CYP2A6 genetic variation differs substantially across ethnic groups[46], suggesting that our results may not extend to other ethnic populations. Additionally, due to haplotype and LD structure heterogeneity across different populations[47], the utility of the simplified five-SNP diplotype for predicting CYP2A6 activity and nicotine metabolism should be reassessed independently among other ethnic groups.
Furthermore, our study was limited by the lack of assessment of structural and copy number variants at the CYP2A6 locus. Copy number variants, such as the established CYP2A6*4 deletion allele, while relatively rare, exhibit a significant impact on CYP2A6 activity; PNAT treatment-seeking smokers with the CYP2A6*1/*4 genotype have a 45% lower mean NMR relative to those without the *4 allele (mean NMR: CYP2A6*1/*4, 0.23; CYP2A6*1/*1, 0.42; P<0.0001).
Conclusions
Seven novel high frequency CYP2A6 SNPs were identified as important contributors to CYP2A6 phenotypes, accounting for approximately two times more variation in CYP2A6 activity, compared to established CYP2A6 variants alone. Due to high LD between two pairs of the seven novel SNPs, we established a functional five-SNP haplotype, which, when phased to derive diplotypes, was associated with CYP2A6 enzyme activity in vitro and in vivo in a human liver bank and PNAT clinical trial, respectively. Considering the association of CYP2A6 enzyme activity with smoking behavior and cessation outcomes, it will be important to determine the utility of incorporating the five-SNP diplotype into CYP2A6 genotype models for predicting cessation success and response to pharmacotherapies.
Supplementary Material
Acknowledgments
The authors thank Qian Zhou for conducting the genotyping. We also wish to acknowledge the Director of the BCM-HGSC, Dr. Richard Gibbs, Xiang Qin, Sandra Lee, William Salerno, and the rest of the BCM-HGSC production and informatics staff for sequencing data generation.
sources of funding
Dr. Tyndale has consulted for Apotex. This work was supported by a Canada Research Chair in Pharmacogenomics (R.F.T.), National Institutes of Health (NIH) PGRN grant DA020830 (to R.F.T. and C.L.), U19 GM61388 (all BCM-HGSC personnel), GM092676 (K.E.T.), and P30 ES007033 (K.E.T.), CIHR (grants MOP86471 and TMH-109787 to R.F.T.), the Campbell Family Mental Health Research Institute of CAMH, the CAMH Foundation, the Canada Foundation for Innovation (#20289 and #16014 to R.F.T.) and the Ontario Ministry of Research and Innovation (R.F.T.). Source of liver tissue: Liver Tissue Procurement and Distribution System (National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases Contract HHSN276201200017C) and the Cooperative Human Tissue Network.
Footnotes
Conflicts of interest
The remaining authors declare no conflicts of interest.
Contributor Information
Julie-Anne Tanner, Campbell Family Mental Health Research Institute, Centre for Addiction and Mental Health (CAMH), Toronto, Ontario, M5T1R8, Canada; Department of Pharmacology and Toxicology, University of Toronto, Toronto, Ontario, M5S1A8, Canada.
Andy Z. Zhu, Campbell Family Mental Health Research Institute, Centre for Addiction and Mental Health (CAMH), Toronto, Ontario, M5T1R8, Canada Department of Pharmacology and Toxicology, University of Toronto, Toronto, Ontario, M5S1A8, Canada.
Katrina G. Claw, Department of Pharmaceutics, University of Washington, Seattle, 98195, WA
Bhagwat Prasad, Department of Pharmaceutics, University of Washington, Seattle, 98195, WA.
Viktoriya Korchina, The Baylor College of Medicine Human Genome Sequencing Center, Houston, Texas 77030, USA.
Jianhong Hu, The Baylor College of Medicine Human Genome Sequencing Center, Houston, Texas 77030, USA.
HarshaVardhan Doddapaneni, The Baylor College of Medicine Human Genome Sequencing Center, Houston, Texas 77030, USA.
Donna M. Muzny, The Baylor College of Medicine Human Genome Sequencing Center, Houston, Texas 77030, USA
Erin G. Schuetz, St Jude Children’s Research Hospital, Memphis, TN 38105, USA
Caryn Lerman, Department of Psychiatry, Annenberg School for Communication, and Abramson Cancer Center, University of Pennsylvania, Philadelphia, PA 19104, USA.
Kenneth E. Thummel, Department of Pharmaceutics, University of Washington, Seattle, 98195, WA
Steven E. Scherer, The Baylor College of Medicine Human Genome Sequencing Center, Houston, Texas 77030, USA
Rachel F. Tyndale, Campbell Family Mental Health Research Institute, Centre for Addiction and Mental Health (CAMH), Toronto, Ontario, M5T1R8, Canada Department of Pharmacology and Toxicology, University of Toronto, Toronto, Ontario, M5S1A8, Canada; Department of Psychiatry, University of Toronto, Toronto, Ontario, M5T1R8, Canada.
References
- 1.Messina ES, Tyndale RF, Sellers EM. A major role for CYP2A6 in nicotine C-oxidation by human liver microsomes. The Journal of pharmacology and experimental therapeutics. 1997;282(3):1608–14. [PubMed] [Google Scholar]
- 2.Nakajima M, Yamamoto T, Nunoya K, Yokoi T, Nagashima K, Inoue K, et al. Characterization of CYP2A6 involved in 3'-hydroxylation of cotinine in human liver microsomes. The Journal of pharmacology and experimental therapeutics. 1996;277(2):1010–5. [PubMed] [Google Scholar]
- 3.Wassenaar CA, Dong Q, Wei Q, Amos CI, Spitz MR, Tyndale RF. Relationship between CYP2A6 and CHRNA5-CHRNA3-CHRNB4 variation and smoking behaviors and lung cancer risk. Journal of the National Cancer Institute. 2011;103(17):1342–6. doi: 10.1093/jnci/djr237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patterson F, Schnoll RA, Wileyto EP, Pinto A, Epstein LH, Shields PG, et al. Toward personalized therapy for smoking cessation: a randomized placebo-controlled trial of bupropion. Clinical pharmacology and therapeutics. 2008;84(3):320–5. doi: 10.1038/clpt.2008.57. [DOI] [PubMed] [Google Scholar]
- 5.Chenoweth MJ, O'Loughlin J, Sylvestre MP, Tyndale RF. CYP2A6 slow nicotine metabolism is associated with increased quitting by adolescent smokers. Pharmacogenetics and genomics. 2013;23(4):232–5. doi: 10.1097/FPC.0b013e32835f834d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sofuoglu M, Herman AI, Nadim H, Jatlow P. Rapid nicotine clearance is associated with greater reward and heart rate increases from intravenous nicotine. Neuropsychopharmacology. 2012;37(6):1509–16. doi: 10.1038/npp.2011.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lerman C, Schnoll RA, Hawk LW, Jr, Cinciripini P, George TP, Wileyto EP, et al. Use of the nicotine metabolite ratio as a genetically informed biomarker of response to nicotine patch or varenicline for smoking cessation: a randomised, double-blind placebo-controlled trial. The Lancet Respiratory medicine. 2015;3(2):131–8. doi: 10.1016/S2213-2600(14)70294-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schnoll RA, Patterson F, Wileyto EP, Tyndale RF, Benowitz N, Lerman C. Nicotine metabolic rate predicts successful smoking cessation with transdermal nicotine: a validation study. Pharmacology, biochemistry, and behavior. 2009;92(1):6–11. doi: 10.1016/j.pbb.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McMorrow MJ, Foxx RM. Nicotine's role in smoking: an analysis of nicotine regulation. Psychol Bull. 1983;93(2):302–27. [PubMed] [Google Scholar]
- 10.Tang DW, Hello B, Mroziewicz M, Fellows LK, Tyndale RF, Dagher A. Genetic variation in CYP2A6 predicts neural reactivity to smoking cues as measured using fMRI. Neuroimage. 2012;60(4):2136–43. doi: 10.1016/j.neuroimage.2012.01.119. [DOI] [PubMed] [Google Scholar]
- 11.Li S, Yang Y, Hoffmann E, Tyndale RF, Stein EA. CYP2A6 Genetic Variation Alters Striatal-Cingulate Circuits, Network Hubs, and Executive Processing in Smokers. Biol Psychiatry. 2016 doi: 10.1016/j.biopsych.2016.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loukola A, Buchwald J, Gupta R, Palviainen T, Hallfors J, Tikkanen E, et al. A Genome-Wide Association Study of a Biomarker of Nicotine Metabolism. PLoS Genet. 2015;11(9):e1005498. doi: 10.1371/journal.pgen.1005498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swan GE, Lessov-Schlaggar CN, Bergen AW, He Y, Tyndale RF, Benowitz NL. Genetic and environmental influences on the ratio of 3'hydroxycotinine to cotinine in plasma and urine. Pharmacogenetics and genomics. 2009;19(5):388–98. doi: 10.1097/FPC.0b013e32832a404f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hakooz N, Hamdan I. Effects of dietary broccoli on human in vivo caffeine metabolism: a pilot study on a group of Jordanian volunteers. Curr Drug Metab. 2007;8(1):9–15. doi: 10.2174/138920007779315080. [DOI] [PubMed] [Google Scholar]
- 15.Rae JM, Johnson MD, Lippman ME, Flockhart DA. Rifampin is a selective, pleiotropic inducer of drug metabolism genes in human hepatocytes: studies with cDNA and oligonucleotide expression arrays. The Journal of pharmacology and experimental therapeutics. 2001;299(3):849–57. [PubMed] [Google Scholar]
- 16.Itoh M, Nakajima M, Higashi E, Yoshida R, Nagata K, Yamazoe Y, et al. Induction of human CYP2A6 is mediated by the pregnane X receptor with peroxisome proliferator-activated receptor-gamma coactivator 1alpha. The Journal of pharmacology and experimental therapeutics. 2006;319(2):693–702. doi: 10.1124/jpet.106.107573. [DOI] [PubMed] [Google Scholar]
- 17.Tanner JA, Prasad B, Claw KG, Stapleton P, Chaudhry A, Schuetz EG, et al. Predictors of Variation in CYP2A6 mRNA, Protein, and Enzyme Activity in a Human Liver Bank: Influence of Genetic and Nongenetic Factors. The Journal of pharmacology and experimental therapeutics. 2017;360(1):129–39. doi: 10.1124/jpet.116.237594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chenoweth MJ, Novalen M, Hawk LW, Jr, Schnoll RA, George TP, Cinciripini PM, et al. Known and Novel Sources of Variability in the Nicotine Metabolite Ratio in a Large Sample of Treatment-Seeking Smokers. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2014 doi: 10.1158/1055-9965.EPI-14-0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Al Koudsi N, Hoffmann EB, Assadzadeh A, Tyndale RF. Hepatic CYP2A6 levels and nicotine metabolism: impact of genetic, physiological, environmental, and epigenetic factors. European journal of clinical pharmacology. 2010;66(3):239–51. doi: 10.1007/s00228-009-0762-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Binnington MJ, Zhu AZ, Renner CC, Lanier AP, Hatsukami DK, Benowitz NL, et al. CYP2A6 and CYP2B6 genetic variation and its association with nicotine metabolism in South Western Alaska Native people. Pharmacogenetics and genomics. 2012;22(6):429–40. doi: 10.1097/FPC.0b013e3283527c1c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen LS, Bloom AJ, Baker TB, Smith SS, Piper ME, Martinez M, et al. Pharmacotherapy effects on smoking cessation vary with nicotine metabolism gene (CYP2A6) Addiction. 2014;109(1):128–37. doi: 10.1111/add.12353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernandez-Salguero P, Hoffman SM, Cholerton S, Mohrenweiser H, Raunio H, Rautio A, et al. A genetic polymorphism in coumarin 7-hydroxylation: sequence of the human CYP2A genes and identification of variant CYP2A6 alleles. Am J Hum Genet. 1995;57(3):651–60. [PMC free article] [PubMed] [Google Scholar]
- 23.Wassenaar CA, Zhou Q, Tyndale RF. CYP2A6 genotyping methods and strategies using real-time and end point PCR platforms. Pharmacogenomics. 2016;17(2):147–62. doi: 10.2217/pgs.15.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mwenifumbo JC, Sellers EM, Tyndale RF. Nicotine metabolism and CYP2A6 activity in a population of black African descent: impact of gender and light smoking. Drug Alcohol Depend. 2007;89(1):24–33. doi: 10.1016/j.drugalcdep.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 25.Ho MK, Mwenifumbo JC, Al Koudsi N, Okuyemi KS, Ahluwalia JS, Benowitz NL, et al. Association of nicotine metabolite ratio and CYP2A6 genotype with smoking cessation treatment in African-American light smokers. Clinical pharmacology and therapeutics. 2009;85(6):635–43. doi: 10.1038/clpt.2009.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cox LS, Nollen NL, Mayo MS, Choi WS, Faseru B, Benowitz NL, et al. Bupropion for smoking cessation in African American light smokers: a randomized controlled trial. Journal of the National Cancer Institute. 2012;104(4):290–8. doi: 10.1093/jnci/djr513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yates A, Akanni W, Amode MR, Barrell D, Billis K, Carvalho-Silva D, et al. Ensembl 2016. Nucleic Acids Res. 2016;44(D1):D710–6. doi: 10.1093/nar/gkv1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Consortium GT. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45(6):580–5. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome research. 2012;22(9):1790–7. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang TH, Huang HY, Hsu JB, Weng SL, Horng JT, Huang HD. An enhanced computational platform for investigating the roles of regulatory RNA and for identifying functional RNA motifs. BMC Bioinformatics. 2013;14(Suppl 2):S4. doi: 10.1186/1471-2105-14-S2-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40(Database issue):D930–4. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fairbrother WG, Yeo GW, Yeh R, Goldstein P, Mawson M, Sharp PA, et al. RESCUE-ESE identifies candidate exonic splicing enhancers in vertebrate exons. Nucleic Acids Res. 2004;32(Web Server issue):W187–90. doi: 10.1093/nar/gkh393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Z, Rolish ME, Yeo G, Tung V, Mawson M, Burge CB. Systematic identification and analysis of exonic splicing silencers. Cell. 2004;119(6):831–45. doi: 10.1016/j.cell.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 34.Gordon AS, Fulton RS, Qin X, Mardis ER, Nickerson DA, Scherer S. PGRNseq: a targeted capture sequencing panel for pharmacogenetic research and implementation. Pharmacogenetics and genomics. 2016 doi: 10.1097/FPC.0000000000000202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 36.Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73(5):1162–9. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68(4):978–89. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tanner JA, Novalen M, Jatlow P, Huestis MA, Murphy SE, Kaprio J, et al. Nicotine Metabolite Ratio (3-hydroxycotinine/cotinine) in Plasma and Urine by Different Analytical Methods and Laboratories: Implications for Clinical Implementation. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2015 doi: 10.1158/1055-9965.EPI-14-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dempsey D, Tutka P, Jacob P, 3rd, Allen F, Schoedel K, Tyndale RF, et al. Nicotine metabolite ratio as an index of cytochrome P450 2A6 metabolic activity. Clinical pharmacology and therapeutics. 2004;76(1):64–72. doi: 10.1016/j.clpt.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 40.Levin ED, Conners CK, Silva D, Hinton SC, Meck WH, March J, et al. Transdermal nicotine effects on attention. Psychopharmacology. 1998;140(2):135–41. doi: 10.1007/s002130050750. [DOI] [PubMed] [Google Scholar]
- 41.Patel YM, Park SL, Han Y, Wilkens LR, Bickeboller H, Rosenberger A, et al. Novel Association of Genetic Markers Affecting CYP2A6 Activity and Lung Cancer Risk. Cancer Res. 2016;76(19):5768–76. doi: 10.1158/0008-5472.CAN-16-0446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baurley JW, Edlund CK, Pardamean CI, Conti DV, Krasnow R, Javitz HS, et al. Genome-Wide Association of the Laboratory-Based Nicotine Metabolite Ratio in Three Ancestries. Nicotine & tobacco research : official journal of the Society for Research on Nicotine and Tobacco. 2016;18(9):1837–44. doi: 10.1093/ntr/ntw117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fujikura K, Ingelman-Sundberg M, Lauschke VM. Genetic variation in the human cytochrome P450 supergene family. Pharmacogenetics and genomics. 2015;25(12):584–94. doi: 10.1097/FPC.0000000000000172. [DOI] [PubMed] [Google Scholar]
- 44.Al Koudsi N, Ahluwalia JS, Lin SK, Sellers EM, Tyndale RF. A novel CYP2A6 allele (CYP2A6*35) resulting in an amino-acid substitution (Asn438Tyr) is associated with lower CYP2A6 activity in vivo. Pharmacogenomics J. 2009;9(4):274–82. doi: 10.1038/tpj.2009.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moore CC, Basile AO, Wallace JR, Frase AT, Ritchie MD. A biologically informed method for detecting rare variant associations. BioData Min. 2016;9(1):27. doi: 10.1186/s13040-016-0107-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tanner JA, Chenoweth MJ, Tyndale RF. Pharmacogenetics of nicotine and associated smoking behaviors. Curr Top Behav Neurosci. 2015;23:37–86. doi: 10.1007/978-3-319-13665-3_3. [DOI] [PubMed] [Google Scholar]
- 47.Tishkoff SA, Verrelli BC. Role of evolutionary history on haplotype block structure in the human genome: implications for disease mapping. Curr Opin Genet Dev. 2003;13(6):569–75. doi: 10.1016/j.gde.2003.10.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


