Abstract
Cellular senescence is an antiproliferative response with essential functions in tumor suppression and tissue homeostasis. Here we show that SIX1, a member of the SIX family of homeobox transcriptional factors, is a novel repressor of senescence. Our data show that SIX1 is specifically downregulated in fibroblasts upon oncogenic stress and other pro-senescence stimuli, as well as in senescent skin premalignant lesions. Silencing of SIX1 in human fibroblasts suffices to trigger senescence, which is mediated by p16INK4A and lacks a canonical senescence-associated secretory phenotype. Interestingly, SIX1-associated senescence is further characterized by the expression of a set of development and differentiation-related genes that significantly overlap with genes associated to SIX1 in organogenesis or human tumors, and show coincident regulation in oncogene-induced senescence. Mechanistically, we show that gene regulation by SIX1 during senescence is mediated, at least in part, by cooperation with Polycomb repressive complexes. In summary, our results identify SIX1, a key development regulator altered in human tumors, as a critical repressor of cellular senescence, providing a novel connection between senescence, differentiation, and tumorigenesis.
Keywords: senescence, SIX1, tumor suppression, p16INK4A, differentiation, Polycomb
Introduction
Cellular senescence was initially identified as a consequence of accumulated doublings in cells in culture. However it is now clear that senescence is a wide-ranging antiproliferative response that helps to control cell balance and tissue homeostasis in diverse contexts, including tumor suppression, development, wound healing or fibrosis (reviewed in 1–3). The essential characteristic of senescent cells is cell-cycle arrest. In addition, the senescent phenotype may include specific changes in cell morphology, chromatin and nuclear structure, activation of a DNA-damage response, regulation of autophagy or the activation of a specific secretory phenotype 1, 4. Gene expression during senescence is under strict control, through the coordinated action of transcriptional factors, long non-coding RNAs and miRNAs, as well as gene specific and global chromatin modifications 3, 5, 6. SIX proteins (SIX1 to SIX6 in vertebrates) form a specific class of the superfamily of homeobox transcription factors 7. SIX family factors cooperate with cofactors like EYA or DACH proteins to regulate gene expression, through recognition of specific sequence motifs 8, 9. Work in Drosophila and vertebrates have revealed a critical role for these factors in cell fate specification during organogenesis, often in specific stem/progenitor populations essential for the formation and regeneration of organs and tissues like kidney, skeletal muscle, or inner ear 10–12. In humans, alterations in SIX proteins or cofactors are linked to the BOR syndrome, characterized by renal and otic defects 8, 13. Interestingly, SIX1 is frequently overexpressed in various human tumors, often associated with stem cell phenotypes and increased metastatic potential 7, 14, 15. Here we report a novel function for SIX1 as a repressor of cellular senescence via the control of the cell-cycle inhibitor and senescence regulator p16INK4A, as well as genes related to development and differentiation.
Results
SIX1 is specifically downregulated during senescence
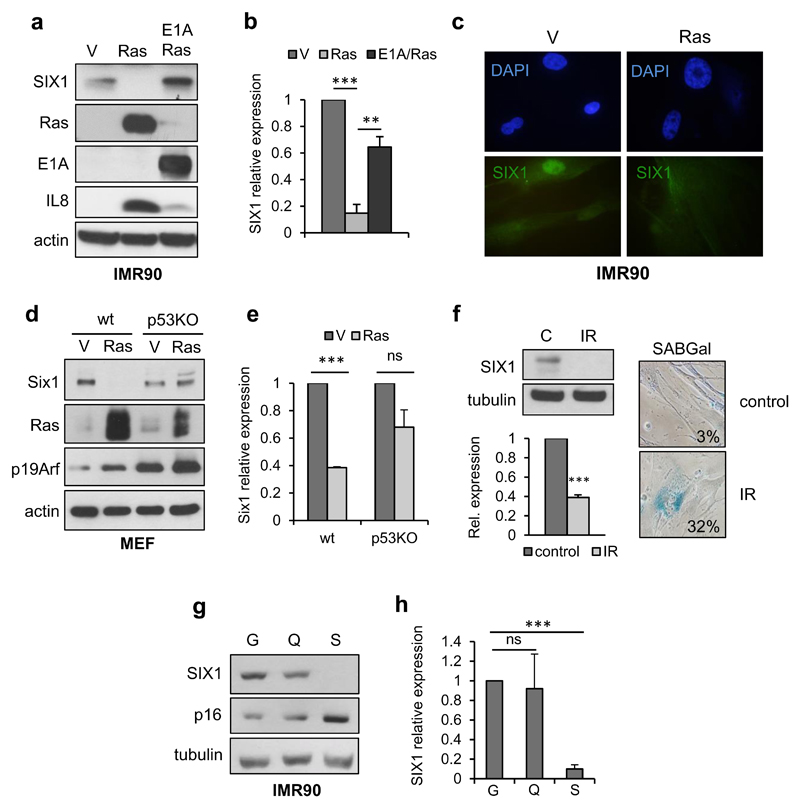
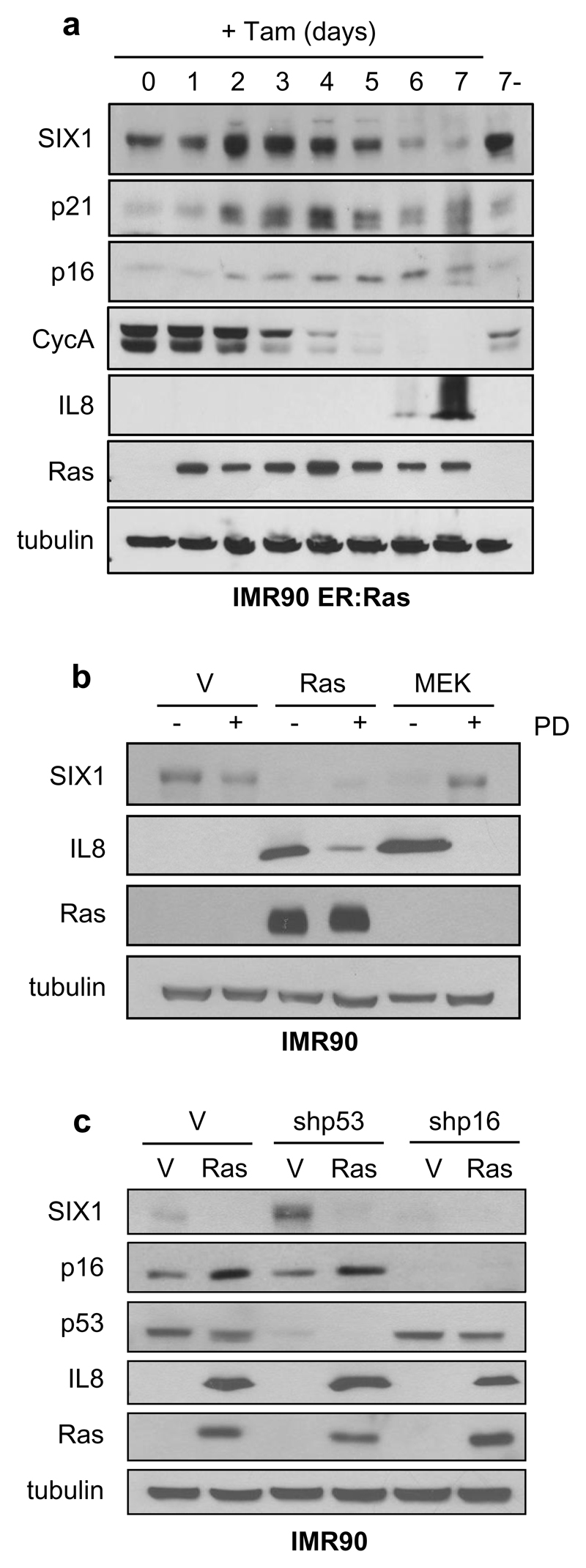
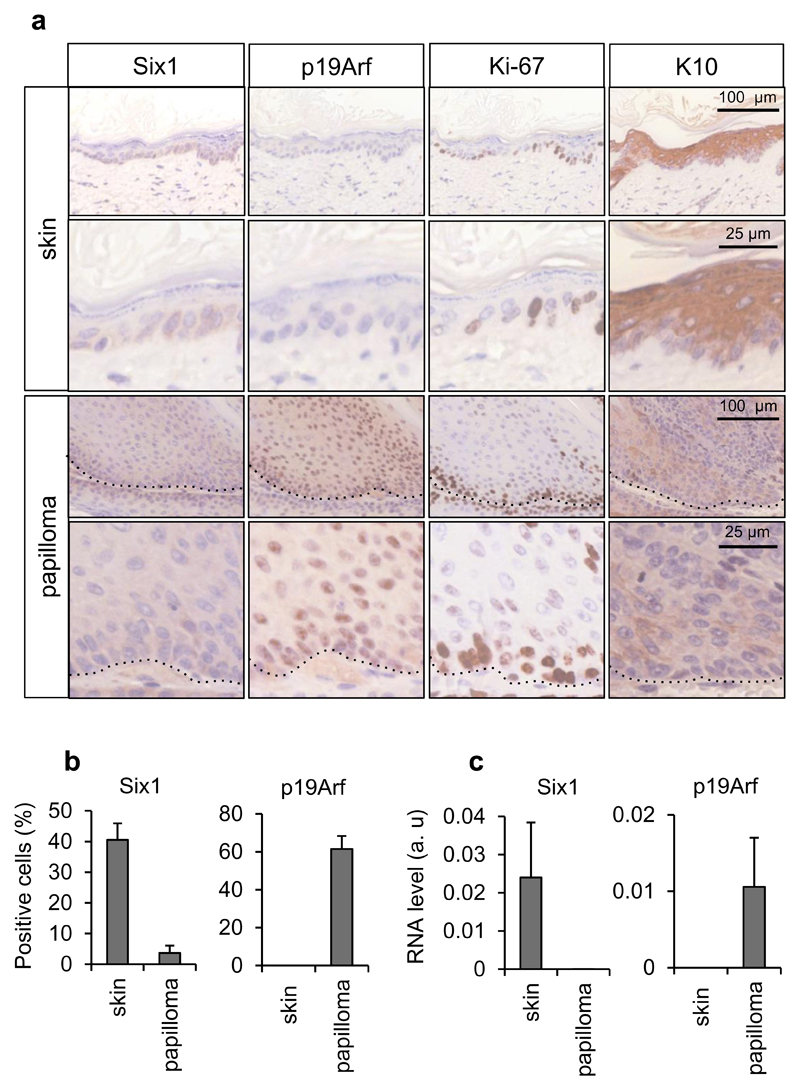
In an expression profiling experiment in IMR90 human primary fibroblasts undergoing oncogene-induced senescence (OIS) 16, we identified the homeoprotein SIX1 as one of the most down-regulated genes in Ras-senescent cells, with its expression being reduced over 20-fold relative to control growing cells. To validate the results from the expression array and test the specificity of SIX1 downregulation, we compared Ras-senescent IMR90 fibroblasts to Ras/E1A fibroblasts where the viral protein E1A prevents senescence by disabling the p53 and Rb pathways 17. We observed that SIX1 transcript and protein were dramatically reduced in Ras-senescent cells, but not in Ras/E1A cells that bypass senescence, (Figure 1a, b, c, Supplementary Figure S1a). SIX1 expression was also investigated in additional senescence cellular models. First, we analyzed Ras-senescent mouse embryo fibroblasts (Figure 1d, e), where inactivation of the Arf/p53 pathway suffices for senescence bypass 18. Consistent with the results in human fibroblasts, Six1 RNA and protein were significantly reduced in wild-type Ras-senescent fibroblasts, but not in p53-null counterparts, which did not enter senescence in response to Ras 17. Similarly, we also observed robust downregulation of SIX1 transcript and protein in IMR90 cells senescent by DNA damage caused by gamma-irradiation, enforced expression of reprogramming factors or accumulation of population doublings (Figure 1f, Supplementary Figure S1b-d). In contrast, SIX1 transcript and protein did not change significantly in quiescent IMR90 fibroblasts (Figure 1g, h), indicating that the downregulation of SIX1 was not merely associated to reduced proliferation, but was instead senescence-specific. Next, to understand the kinetics of SIX1 regulation during senescence, we used a model of inducible OIS, based in the tamoxifen-regulated expression of an ER:RasV12 cassette 19. SIX1 levels were analyzed in these cells over a period of seven days after addition of tamoxifen, in parallel with several well-characterized senescence markers (Figure 2a). In line with the results described above, after an early peak in expression, SIX1 levels were significantly reduced after 5 days of Ras induction. Similar results were obtained in a different fibroblast strain (Supplementary Figure 1e). The decrease in SIX1 occurred with a kinetics similar, although slightly delayed, to the induction of the cell cycle inhibitors and senescence effectors p16INK4A (hereafter p16) and p21CIP1 (p21) and the decrease in the proliferation maker Cyclin A, and it took place before the activation of the Senescence-Associated Secretory Phenotype (SASP), as assessed by IL8 levels 19, 20. We further sought to characterize the signaling pathways responsible for SIX1 regulation during OIS. Ras-induced senescence is mainly relayed by the RAF/MEK promitogenic effector pathway 21. Expression of activated MEK provoked a reduction in SIX1 similar to RasV12, while the addition of the MEK inhibitor PD98059 significantly blunted SIX1 regulation by RasV12 (Figure 2b and QPCR data not shown), suggesting that SIX1 regulation during Ras-induced senescence is mediated mainly by the RAF/MEK pathway 21. Next, we evaluated if SIX1 regulation was dependent on the p53 and p16/Rb tumor suppressive pathways, which play key roles in regulation of senescence in human fibroblasts and also participate directly in transcriptional regulation 5, 22. We observed that the downregulation of SIX1 by Ras was essentially unchanged in cells expressing shRNAs for p53 or p16, indicating that the regulation of SIX1 during OIS occurs independently of the p53 and p16/Rb pathways (Figure 2c and QPCR data not shown), even though some differences in SIX1 basal levels could be observed. It should be noted that independent inactivation of either pathway is not sufficient to blunt induction of Ras-senescence in human fibroblasts 3, 17. IL8 induction was used here as a p53 and p16-independent readout of senescence 23. After characterizing SIX1 regulation in cells in culture, we wished to test if SIX1 underwent similar regulation during senescence in vivo. Cellular senescence is activated in pre-neoplastic lesions where it acts as a critical barrier to prevent tumor progression 22, 24. Thus, we analysed the expression of Six1 in murine skin papillomas induced with a chemical carcinogenesis protocol, a type of benign lesions where senescence has been well characterized 24. Six1-positive cells could be detected by immunohistochemistry in the basal layer of normal epidermis that included Ki-67-positive cells and was negative for the differentiation marker keratin 10. As expected, the senescence marker p19Arf was undetectable in normal skin. In papillomas, some Six1-positive cells could be detected in the basal layer, but they were essentially absent from the keratin 10-positive suprabasal layers that contained senescent cells, as shown by the expression of p19Arf and absence of proliferating Ki-67-positive cells (Figure 3a, 3b). The immunohistochemistry results were confirmed with QPCR data that showed dramatically reduced Six1 expression and increased p19Arf expression in papillomas relative to normal skin (Figure 3c).
Figure 1. SIX1 regulation during senescence.
(a, b) Western Blot analysis of SIX1 and the indicated controls (a) and QPCR analysis of SIX1 transcript (b) in RasV12-senescent and E1A/Ras (senescence escape) IMR90 human fibroblasts. (c) Immunofluorescence of endogenous SIX1 in Ras-senescent and growing (V) IMR90 cells. (d, e) Western Blot analysis of Six1 and the indicated controls (d) and QPCR analysis of Six1 transcript (e) in RasV12-expressing wild-type MEF (senescent), and p53-KO MEF (senescence escape). (f) Western Blot (top left) and QPCR (bottom left) analysis of SIX1 in senescence induced by gamma irradiation in IMR90 fibroblasts. Representative images and percentage of SABGal positive cells (right). (g, h) Western Blot analysis (g) and QPCR analysis of SIX1 (h) in growing (G), quiescent (Q) and Ras-senescent (Ras) IMR90 fibroblasts.
Figure 2. Kinetics and mechanism of SIX1 regulation in oncogene-induced senescence.
(a) Western Blot analysis of SIX1 and the indicated senescence markers in IMR90 cells expressing an ER:RasV12 cassette at the indicated time-points after induction with tamoxifen. 7- indicates 7 days without tamoxifen. (b) Western Blot analysis of the indicated proteins in IMR90 fibroblasts expressing shp53 or shp16 vectors, followed by infection with RasV12 (Ras) or empty vector (V). (c) Western Blot analysis of the indicated proteins in IMR90 fibroblasts expressing RasV12 (Ras), MEKP56 (MEK) or empty vector (V) after incubation with the MEK inhibitor PD98059.
Figure 3. Six1 expression in mouse skin papillomas.
(a, b) Immunohistochemistry analysis of Six1, p19Arf, Ki-67 and Keratin 10 in mouse normal skin and papillomas. (c) QPCR analysis of Six1 and p19Arf in mouse normal skin and papillomas.
SIX1 is a negative regulator of senescence
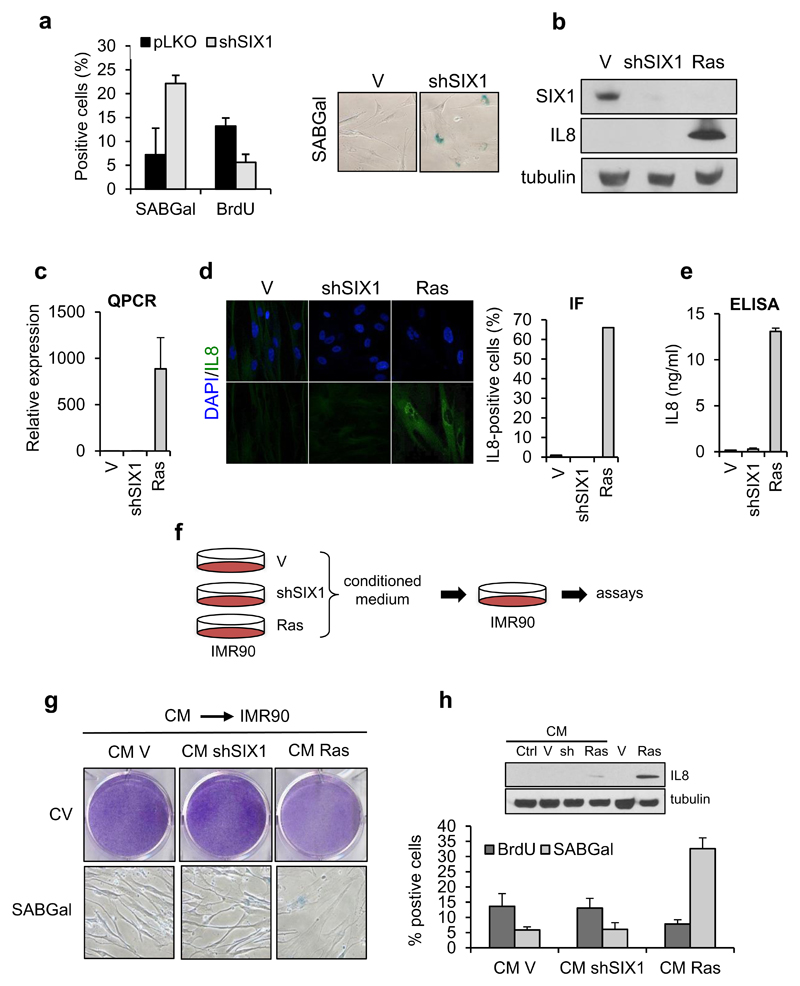
Having characterized the regulation of SIX1 during senescence, we wished to establish the functional relevance of SIX1 downregulation in the implementation of senescence. To this end, we generated IMR90 fibroblasts with stable silencing of SIX1 with shRNA lentiviral vectors, to mimic the changes in SIX1 during senescence. Out of the six members of the human SIX family (ref. 7) only SIX1, SIX4 and SIX5 could be detected in growing IMR90 cells. SIX4 was downregulated during OIS, like SIX1, while SIX5 was unchanged or modestly upregulated (Supplementary Figure S2a). To avoid off-target effects, we used for our experiments an shRNA construct that yielded robust silencing of SIX1 but did not significantly affect other SIX members. Similar results were obtained with additional shSIX1 constructs (Supplementary Figure S2b). Stable silencing of SIX1 in early passage IMR90 cells markedly reduced proliferation, as assessed by BrdU incorporation (Figure 4a, 5e and data not shown). Most importantly, IMR90 shSIX1 cells displayed features of cellular senescence, including senescent morphology with extended vacuolized cytoplasms and prominent nucleoli, and increased number of Senescence-Associated Beta-Galactosidase (SABGal)-positive cells (Figure 4a, 5e; Supplementary Figure S2b and data not shown). Notably, other markers associated with some forms of senescence, such as Senescent-Associated Heterochromatin foci 25, or DNA damage foci 26, were not observed in shSIX1 fibroblasts (Supplementary Figure S3a). Also, during our characterization of the phenotypic effects of SIX1 silencing, we failed to detect any noticeable change in Interleukin 8 (IL8), a key component of the senescent-associated secretory phenotype or SASP 20, in contrast to Ras-induced senescence in the same cells (Figure 4b, c, d, e), suggesting that this part of the senescence phenotype was not activated during shSIX1-senescence. SASP soluble factors have been shown to induce a senescent phenotype in normal recipient fibroblasts 27. To evaluate functionally the effects of SIX1 silencing in this context, we assayed the ability of conditioned medium from shSIX1 senescent IMR90 to trigger senescence in a paracrine fashion (Figure 4f). In agreement with the lack of IL8 induction, the medium from senescent shSIX1 cells failed to induce a senescent phenotype or senescence markers in normal recipient fibroblasts, in sharp contrast to the senescence-promoting effects of conditioned medium from Ras-senescent cells (Figure 4g, h).
Figure 4. Characterization of the SASP in shSIX1-senescent fibroblasts.
(a) Quantification of SABGal and BrdU-positive cells (left) and micrographs showing morphology and SABGal staining (right) in shSIX1-IMR90 cells. (b, c) Western Blot (B) and QPCR analysis (C) of SIX1 and Interleukin 8 (IL8) in IMR90 fibroblasts expressing shSIX1 or RasV12 (Ras). (d) Immunofluorescence of IL8 and quantification of positive cells. (e) ELISA for IL8 with conditioned medium from the indicated cell types. (f) Experimental design of assays with conditioned medium. (g) Crystal violet staining (CV, top) and morphology and SABGal staining (bottom) of parental IMR90 cells with conditioned medium (CM) from the indicated cells. (h) Top: Western blot analysis of IL8 in IMR90 cells exposed to conditioned medium, Ctrl: control non infected cells, sh: shSIX1. Lysates form Ras-senescent and control-infected cells are included as controls. Bottom: Quantification of SABGal and BrdU-positive cells.
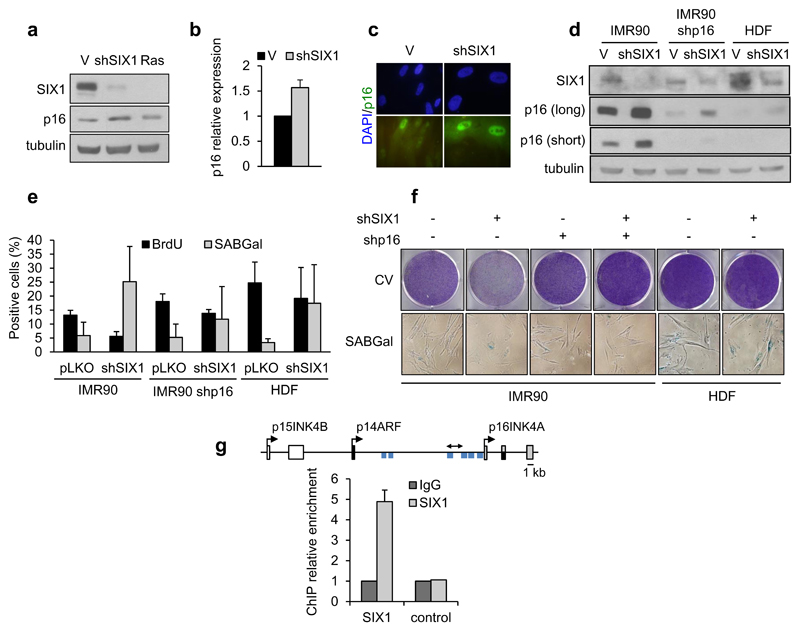
Figure 5. Functional link to p16INK4A.
(a, b) Western Blot analysis (a) of SIX1 and p16 and QPCR analysis (b) of p16 transcript in IMR90 fibroblasts expressing RasV12 (Ras) or shSIX1. (c) Immunofluorescence of p16 in shSIX1-IMR90 cells. (d) Western Blot analysis of SIX1 and p16 after infection with an shSIX1 vector of IMR90 parental fibroblasts, shp16 IMR90 fibroblasts and primary human dermal fibroblasts (HDF). (e) Quantification of SABGal and BrdU-positive cells in the indicated cells. (f) Crystal violet staining (CV, top) and morphology and SABGal staining (bottom). (g) Chromatin immunoprecipitation indicating binding of SIX1 to the INK4A locus in 293T cells with or without ectopic expression of SIX1. Blue bars indicate consensus binding sites for SIX proteins. Double arrow indicates region amplified by PCR.
SIX1 regulates senescence by a p16-dependent mechanism
p16 is a key mediator of the senescent cell cycle arrest, encoded in the INK4A/ARF locus together with ARF 3, 22, 28. In agreement with the inverse correlation between SIX1 and p16 found in ER:Ras induced senescence (Figure 2a), we observed that p16 protein and RNA levels were significantly increased in shSIX1 IMR90 cells, as did the number of p16-positive cells in immunofluorescence assays (Figure 5a, b, c, d). In contrast, p21, also a well-characterized senescence effector, was modestly decreased (Supplementary Figure S3b). To test if the pro-senescence effects of silencing SIX1 could be mediated by p16, we used two strains of human fibroblasts with reduced p16 levels, namely IMR90 fibroblasts with shp16, and primary adult dermal fibroblasts that express low levels of endogenous p16. In both cases, reduced p16 levels resulted in a significant blunting of the pro-senescence effect of shSIX1, as measured by BrdU incorporation, SABGal activity, cell number or morphology (Figure 5d, e, f, and data not shown). Since SIX1 is a transcription factor, we reasoned that these results could reflect direct transcriptional regulation of p16 by SIX1. Chromatin immunoprecipitation (ChIP) revealed specific binding of SIX1 to the human INK4 locus in a region proximal to the p16 transcription start site that contains several consensus binding sites for SIX proteins 29 (Figure 5g), suggesting that the INK4A locus can be a transcriptional target of SIX1. Further supporting the link between p16 and SIX1 in this context, enforced SIX1 expression blunted significantly the induction of p16 by Ras in IMR90 fibroblasts, although it had a limited functional impact, leading to partial reversion of Ras-induced senescent arrest. A similar inverse correlation between SIX1 and p16 was observed in immortalised mouse fibroblasts with ectopic expression of SIX1, supporting the generality of the functional link between both proteins (Supplementary Figure S4a,b, c, d).
SIX1 cooperates with Polycomb to regulate developmental and differentiation genes in senescence
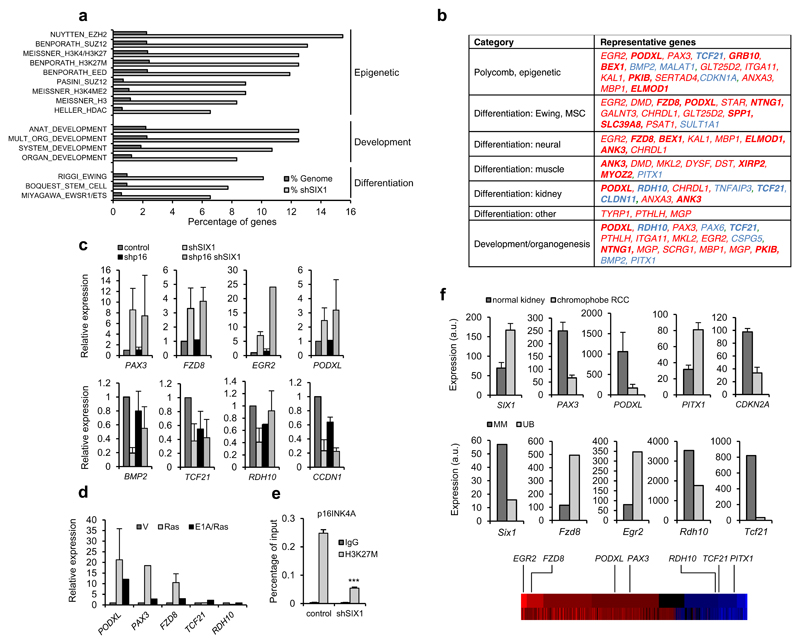
To further characterize the role of SIX1 in senescence, we performed a microarray RNA expression analysis in shSIX1 senescent IMR90 cells. This study identified 435 genes with differential expression in shSIX1 IMR90 cells (at least 2-fold), 300 increased and 135 reduced relative to growing cells. It should be noted that p16 expression could not be analyzed because the probes in the array did not discriminate between the two products of the INK4A/ARF locus. We validated these results for a selection of upregulated and downregulated genes using QPCR (Figure 6c). Interestingly, CCND1, a well-known direct target of SIX1 30 was significantly downregulated in shSIX1 cells, further validating our analysis. Next, we characterized functionally the results, using Gene Set Enrichment Analysis (GSEA). This analysis revealed a significant enrichment of genesets associated to epigenetic regulation, with strong representation of Polycomb repressive complexes (PRC, 9 out of the top 20 genesets, Figure 6a, b, Table S1). PRC complexes (PRC1 and PRC2) are critical epigenetic regulators of pluripotency, differentiation and cancer that lead to transcriptional repression by the establishment and recognition of the histone mark H3K27Me3 (trimethylation of Lys27 of histone H3) 31. Of note, PRC complexes play an essential role in senescence, mainly by derepressing the INK4/ARF locus 5, 28. To test if SIX1 could cooperate with Polycomb complexes in senescence, we analyzed changes in the Polycomb-associated mark H3K27Me3. Chromatin immunoprecipitation showed a clear reduction of H3K27Me3 in the INK4A promoter (approximately 5-fold) in shSIX1-senescent cells (Figure 6e), in the absence of gross changes in H3K27Me3 nuclear level or distribution (Supplementary Figure S3a), similar to other senescence stimuli 6, 32, 33. Similar data was obtained for other SIX1-regulated genes (data not shown), supporting the notion that SIX1 cooperates with Polycomb repressive complexes (PRC) in gene regulation in senescence. Notably, further GSEA analysis of Gene Ontology functional categories in shSIX1 senescence revealed a highly significant enrichment of categories related to development, differentiation and organogenesis. Significant enrichment was also observed for genesets related to mesenchymal stem cells or Ewing sarcoma, which is considered to originate from mesenchymal stem cells and may represent aberrant differentiation events 34 (Figure 6a, b). As mentioned, SIX1 plays a critical role in mammalian organogenesis, and interestingly many of the genes in these categories were specifically associated to organs and tissues where SIX1 is relevant, such as kidney or muscle 12, 35 (Figure 6b). Given the link between SIX1 and p16 described above, we also wished to know if gene regulation by SIX1 during senescence could be mediated by the p16/Rb/E2F pathway. All the genes upregulated with shSIX1 retained similar regulation in the absence of p16. In contrast, the expression of downregulated genes was affected by shp16 to different degrees (Figure 6c). Taken together, the expression profiling data suggests that shSIX1-senescence is accompanied by the activation of a development and differentiation-related gene program that involves Polycomb-mediated gene regulation, and may involve both p16-dependent and independent mechanisms.
Figure 6. Expression signature of shSIX1-senescent fibroblasts.
(a) GSEA analysis of genes differentially expressed in shSIX1-senescent IMR90 fibroblasts. The percentage of genes of each category in the set of shSIX1 genes or total genome is shown. (b) Table showing representative genes of each category. Red, genes upregulated in shSIX1 cells; blue, downregulated; bold, genes with similar regulation in Ras-senescent IMR90 fibroblasts. (c) QPCR analysis of selected targets in IMR90 fibroblasts expressing shSIX1 alone or combined with shp16. Top, upregulated genes; bottom downregulated genes. (d) QPCR analysis of selected targets in Ras-senescent (Ras) or senescence-escape (E1A/Ras) IMR90 fibroblasts. (e) Chromatin immunoprecipitation assay to detect the H3K27M mark in the p16INK4A promoter in shSIX1 IMR90 fibroblasts and controls. (f) Expression of sh-SIX1-senescence genes in human normal kidney and chromophobe renal carcinoma (top) and in metanephric mesenchyme (MM) and ureteric bud (UB) of E11.5 mice (bottom). RNA expression data were retrieved from Oncomine and GUDMAP databases and Gene Expression Omnibus (GSE15641 for kidney tumor and normal tissue, and GSE6290 for mouse developing kidney). For reference, the position of the relevant genes in a heatmap representation of their expression in shSIX1 IMR90 cells is included (red, upregulated; blue, downregulated genes).
To characterize the relevance of SIX1-mediated gene regulation in senescence, we asked whether a similar program was present in other forms of senescence. To this end, we compared the data for shSIX1-senescence with our previous analysis in Ras-senescent cells 16. We found that a significant fraction of differentially expressed genes were similarly regulated in Ras-senescence (Figure 6b), especially within the group of shSIX1 upregulated genes (41 out of 300, 14%). Interestingly, this set of genes showed similar enrichment in Polycomb-related genes, further supporting the notion that SIX1 is relevant for PRC-mediated gene regulation during senescence (Table S1). We further validated the senescence-specific regulation of a selection of genes by QPCR, using Ras/E1A-expressing IMR90 as senescence bypass controls (Figure 6d). SIX1 plays a critical role in organogenesis, and it is also overexpressed in a variety of human tumors 7. To understand the relevance of senescence regulation in SIX1 function in these contexts, we asked if the SIX1- senescence expression module could also be associated to SIX1 in tumors or during organogenesis. Inspection of the Oncomine expression database (https://www.oncomine.org) revealed that SIX1 overexpression was most frequent in kidney tumors, mainly in pediatric Wilms tumors and some types of rare adult renal tumors. We interrogated data sets from kidney tumors with significant SIX1 overexpression, and found significant correlation for some of the most differentially expressed SIX1-senescence genes in Wilms tumors and in adult papillary and chromophobe tumors (Figure 6f, top and Supplementary Figure S5). Similar correlations were found for additional SIX1-overexpressing tumors, including soft tissue sarcomas or breast and lung tumors, although the specific SIX1-senescence genes involved varied depending on the tumor type (Supplementary Figure S5). During kidney development, SIX1 and its relative SIX2 play a critical role in specifying a cell population with stem/progenitor features in the metanephric mesenchyme, whose interaction with the ureteric bud is critical for the correct development of the mature kidney or metanephros 12, 36, 37. We interrogated expression data for these structures of the developing mouse kidney, using the GUDMAP database (http://www.gudmap.org), again finding a good correlation between Six1 expression and a set of the most differentially expressed genes in SIX1-senescence (Figure 6f, bottom). Taken together, these results indicate that similar expression programs are controlled by SIX1 in senescence, cancer and organogenesis, and suggest that regulation of senescence could play an important role in SIX1 function in diverse biological settings.
Discussion
Cellular senescence plays a critical role in a variety of pathological and physiological settings 1, 2. Onset and maintenance of senescence are under strict regulation that includes transcriptional and epigenetic mechanisms 3. In this report, we identify the homeoprotein SIX1 as a novel repressor of cellular senescence, via epigenetic regulation of p16INK4A, as well as the implementation of a development and differentiation-related expression program.
Our data strongly support a model where SIX1 regulates senescence mainly by controlling gene expression, in cooperation with Polycomb repressive complexes. Extensive evidence suggests that PRC complexes play an essential role in the regulation of senescence, by derepressing key senescence effector loci, including the INK4/ARF locus 5, 28, 32. The mechanism responsible for targeting PRC to specific loci is poorly understood, and it could involve sequence-specific transcription factors, as well as non-coding RNAs 38. Our results would indicate that SIX1 is a novel factor involved in targeting PRC complexes to specific loci during senescence. The presence of SIX consensus motifs in gene promoters (data not shown) and the overlap with genes identified in SIX1 ChIP studies in myoblasts 39, suggest that an important fraction of differentially expressed genes are likely direct targets of SIX1, as we have shown for the p16INK4A locus. However, additional indirect mechanisms, including the participation of the p16/Rb/E2F axis, may be involved in gene expression control by SIX1 in senescence. Previously, other homeobox proteins unrelated to the SIX family have been identified in screenings for regulators of senescence and the INK4A locus. However, the in vivo relevance of these factors in senescent fibroblasts is unclear 40, 41. In contrast, our results strongly indicate that SIX1 is a physiologically relevant regulator of p16 during senescence, based on the regulation of endogenous SIX1 during senescence, and the robust functional link between SIX1 and p16. It has been suggested that SIX proteins can activate or repress transcription depending on their interaction with cofactors of the EYA or Groucho and Dachsund families respectively 36. Our current data provides a novel mechanism for SIX1-mediated repression in cooperation with PRC complexes, and it would interesting to determine the possible participation of SIX1 cofactors in this context. In addition to its role in PRC-mediated repression shown here, previous studies have shown PRC-mediated repression of the Six1 locus during differentiation of cardiac progenitors 42, or embryonic stem cells 43. Whether PRC are involved in SIX1 regulation in senescence is currently unclear, but it seems unlikely. First, senescence is generally associated to diminished PRC repressive activity and increased expression of PRC-repressed genes 28, in contrast to SIX1 repression in senescence. Also, clear changes in the H3K27Me3 mark in the SIX1 locus are not apparent in public datasets for Ras-senescent fibroblasts (Kirschner et al, 2015). Further work is needed to define the mechanism responsible for SIX1 regulation during senescence and also to determine potential links between SIX1 and Polycomb in other settings.
A distinct feature of senescence triggered by SIX1 silencing in human fibroblasts was the absence of a canonical SASP. Notably, a similar SASP-independent pro-senescence effect has been reported for human fibroblasts with enforced p16 expression 44–46. These results, together with the link between SIX1 and p16 we have shown, support a model where SIX1 cooperates with p16 in a signaling axis responsible for the senescent cell cycle arrest, while implementation of the SASP occurs through a SIX1-independent pathway that involves additional factors like C/EBP and NF-kappaB 20, 47, 48.
Our results may have important implications in the role of SIX1 in tumorigenesis. SIX1 overexpression has been reported in various human tumors, frequently associated to increased stemness or invasiveness 7, 8. The novel link between SIX1 and senescence that we report here may provide an additional rationale for SIX1 overexpression in tumors. The reduced SIX1 expression in senescent premalignant lesions, together with the inverse-correlated expression of SIX1-senescence genes in tumors with SIX1 overexpression, are consistent with the notion that senescence escape may contribute to the oncogenic function of SIX1 in human cancer. Thus, it can be speculated that, in addition to other pro-tumor effects, SIX1 overexpression may act at early stages of tumorigenesis to blunt the anti-tumor barrier established by senescence, facilitating tumor formation or progression.
Remarkably, SIX1-senescence was characterized by the orchestrated regulation of a large set of genes linked to development, organogenesis and differentiation, specifically associated to organs and tissues where SIX1 is physiologically relevant 12, 35. Interestingly, some of these genes were similarly regulated during oncogene-induced senescence, suggesting the generality of this expression pattern. Of note, SIX1 is highly expressed in a variety of embryo and adult stem/progenitor cells, and its downregulation is often associated to differentiation 36 10, 49–52. Also, in the context of tumors, SIX1 expression is frequently associated to cancer stem cell populations 15, 53, 54. Taken together, our data indicate that regulation of a development and differentiation-associated gene program, with the participation of SIX1, is an integral part of senescence. It is currently unclear how this differentiation/development signature may contribute to the senescent phenotype, but we speculate that senescence may serve to blunt latent plasticity in adult tissues and reinforce differentiation. Such a pro-differentiation effect may have important implications in the role of senescence as an anti-tumor barrier but also in other physiological and pathological settings, and it is in agreement with evidence showing that senescence can oppose pluripotency 55–57. Intriguingly, despite the prevalence of the differentiation signature, some genes linked to embryo or adult stem cells were also increased in senescence, and further studies should clarify how the balance between differentiation and plasticity is integrated during senescence. Interestingly, recent results have shown that cellular senescence plays a physiological role in development 58, 59 and in adult stem cell homeostasis 60. Defective SIX1 function is linked to severe defects in organogenesis and adult tissue regeneration, often linked to altered differentiation 10, 36. It would be interesting to determine if these SIX1-related phenotypes could be related to senescence. In summary, our results identify SIX1 as a novel repressor of senescence, through epigenetic control of p16INK4A-mediated arrest and a developmental and differentiation gene expression program, and as such they highlight the cross-talk between senescence, differentiation and tumorigenesis.
Materials and Methods
Cell Culture
Human primary fibroblasts IMR90 were obtained from ATCC, p53-null Mouse Embryo Fibroblast (MEF) and Human Dermal Fibroblasts (HDF) were obtained from Manuel Serrano, CNIO, Spain. All primary fibroblasts were used at low passage, using DMEM medium (Gibco), with 10% FBS (Gibco), in presence of penicillin and streptomycin. Inducible ER:Ras IMR90 cells were generated using the vector pLNC-ER:RasV12 and treated with 1μM 4-hydroxy-tamoxifen for Ras induction. Quiescent IMR90 were obtained by culturing the cells in serum-free medium for 48 hours. For MEK inhibition, PD98059 was added during retroviral transduction at a final concentration of 20μM.
Retroviral transduction
Retroviral and lentiviral transductions were performed as described 16, 61. shSIX1 vectors were obtained from Sigma (shSIX1a: NM_005982.1-992s1c1, shSIX1b: NM_005982.3-967s21c1, shSIX1d: NM_005982.3-1198s21c1); pRetroSuper human p53, pRetroSuper mouse p53, pRetroSuper human p16 and pLNC-ER:RasV12 were kind gifts of Daniel Peeper and Rene Bernards, NKI, The Netherlands, Manuel Serrano, and Masashi Narita, CRI, UK, respectively. pLPC-SIX1 was cloned during this study using cDNA from IMR90 cells. Additional vectors used were pLPC-RasV12, pLPC-MEKQ56P, pLPC E1A-RasV12 21 and MSCV-OSKM 55. All assays were performed 8 days after infection, unless stated otherwise.
Western blot
Western blot was carried out as described 62. The following primary antibodies were used: panRas (OP-40, Calbiochem), E1A (M-73, Santa Cruz Biotechnology), SIX1 (HPA001893, Sigma), p53 (sc-126, Santa Cruz Biotechnology), p21CIP1 (sc-397, Santa Cruz Biotechnology), p16INK4 (sc-56330, Santa Cruz Biotechnology), IL8 (MAB208, RD Systems), tubulin (T9026, Sigma), Cyclin A (sc-751, Santa Cruz Biotechnology).
Immunofluorescence
Immunofluorescence was performed as described 16. The primary antibodies used were: BrdU (347580, BD Biosciences), tubulin (T9026, Sigma), p16 (sc-56330, Santa Cruz Biotechnology), IL8 (MAB208, RD Systems), SIX1 (HPA001893, Sigma and 10709-1-AP, Proteintech), gamma-H2AX (JBW301 05-636, Upstate) and H3K27Me3 (07-449, Millipore).
Immunohistochemistry
Murine skin papillomas obtained with a DMBA/TPA carcinogenesis protocol and normal skin controls were processed for immunohistochemistry as described 63, using the following antibodies: p19Arf (sc-32748, Santa Cruz), SIX1 (HPA001893, Sigma), Ki-67 (0003110QD, Master Diagnostica) and Keratin 10 (PRB-159P, Covance).
Other cell biology methods
Senescence-Associated Beta-Galactosidase activity and bromodeoxyuridine (BrdU) incorporation were measured as previously described 16; crystal violet staining was performed as described in 61.
Quantitative PCR
Quantitative RT-PCR was performed as described 64, using the following primers: BMP2 (CGGACTGCGGTCTCCTAA, GGAAGCAGCAACGCTAGAAG), CCDN1 (GCCGAGAAGCTGTGCATC, CCACTTGAGCTTGTTCACCA), EGR2 (TTGACCAGATGAACGGAGTG, TGGTTTCTAGGTGCAGAGACG), EYA2 (ACGCTGCTGTGTGGACTCT, AGAGCTGGGACACTCTCAGG), FZD8 (CTCTGCTTCGTGTCCACCTT, GAAGCGCTCCATGTCGAT), IL8 (AGACAGCAGAGCACACAAGC, ATGGTTCCTTCCGGTGGT), p16INK4A (GTGGACCTGGCTGAGGAG, CTTTCAATCGGGGATGTCTG), p19ARF (GGGTTTTCTTGGTGAAGTTCG, TTGCCCATCATCATCACCT), p21CIP1 (GGCACACACACATTAACACACTT, GGTGTGTGAGAGCAATTCTCAG), PAX3 (AGCCGGAGAAAGGACCTC, GAAGGGACGCCAGTAAGATG), PODXL (GCGCTGCTGCTACTGTTGT, CCGTAGTAGTCTGGGTTGCAT), RDH10 (ATCAACACGCAAAGCAACG, TCCTCACCATTCCCAGCTT), human SIX1 (ACCGGAGGCAAAGAGACC, GGAGAGAGTTGGTTCTGCTTGT), mouse SIX1 (ACCGGAGGCAAAGAGACC, GGAGAGAGTTGATTCTGCTTGTT), SIX4 (CCGAGACCCAGTCCAAAAG, CCATATGACTGGAAAGGCTGA), SIX5 (CTGCCAATGTGCACCTCAT, GTTGGCCAGGAGGAAGTTT), TCF21 (CATTCACCCGGTCAACCT, TCAGGTCACTCTCGGGTTTC).
Chromatin immunoprecipitation
Chromatin immunoprecipitation was performed essentially as described 64, using 293T cells transiently transfected with pLPC-SIX1 or IMR90 fibroblasts expressing shSIX1, except that the crosslinking step with DMA was omitted for H3K27Me3 immunoprecipitation. The antibodies used were: anti-SIX1 (HPA001893, Sigma) and anti-H3K27Me3 (07-449, Millipore). The primers used for PCR were CTCAAAGCGGATAATTCAAGAGC, AAGCCTTAAGAACAGTGCCACAC (1050 nt upstream of p16INK4A transcription start site).
Conditioned Medium Assays
IMR90 cells were cultured in 10% serum DMEM medium until they reached 90% confluence. Next, medium was removed, cells were washed 3 times with PBS and serum free DMEM medium was added to cells. Seventy-two hours later the medium was collected, centrifuged and stored at -80ºC. Prior to its use, serum was added at a final concentration of 1%. ELISA for IL8 was performed using the Quantikine ELISA kit from RD Systems.
Microarray gene expression profiling
Microarray experiments were performed essentially as described 16 using Human Whole-Genome array G4112F (Agilent Technologies). Two independent lentiviral infections were performed in early passage IMR-90 fibroblasts with shSIX1 and its empty vector as control. Total RNA was prepared 8 days post-infection, labeled and competitively hybridized to arrays using as a reference a reverse-labeled sample from vector-infected cells. One additional hybridization was performed labeling the RNAs with the reciprocal fluorochromes. Differentially expressed genes in shSIX1 versus vector (two-fold difference in all of the samples and SD <0.5) were identified with GEPAS (Gene Expression Pattern Analysis Suite, http://gepas3.bioinfo.cipf.es). This subset of genes was further analyzed by Gene Set Enrichment Analyses (GSEA) (www.broadinstitute.org/gsea) using collections C2 (Curated Gene Sets, which include expression signatures from published studies) and C5 (Gene Ontology gene sets). The top 20 gene sets with a FDR q-value below 0,05 were chosen.
Statistical Analysis
Statistical significance was calculated using unpaired two-tail Student T test (*** P < 0.001, **P< 0.01, *, P< 0.05). All the graphs show the average and SD from at least two independent experiments.
Supplementary Material
Acknowledgements
We thank Manuel Serrano, Daniel Peeper and René Bernards for cells and reagents, and Masashi Narita for reagents and sharing unpublished data. This work was supported by grants from the Spanish Government (SAF2012-32117) to IP, Instituto de Salud Carlos III (PI13/00132 and RETIC-RD12/0036/0007), and Regional Government of Madrid (S2010/BMD-2303) to G M-B. IA is the recipient of a fellowship from the Spanish Government FPI program.
Footnotes
Conflict of interest:
The authors declare that they have no conflict of interest.
References
- 1.Kuilman T, Michaloglou C, Mooi WJ, Peeper DS. The essence of senescence. Genes & development. 2010;24:2463–2479. doi: 10.1101/gad.1971610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Munoz-Espin D, Serrano M. Cellular senescence: from physiology to pathology. Nature reviews Molecular cell biology. 2014;15:482–496. doi: 10.1038/nrm3823. [DOI] [PubMed] [Google Scholar]
- 3.Salama R, Sadaie M, Hoare M, Narita M. Cellular senescence and its effector programs. Genes & development. 2014;28:99–114. doi: 10.1101/gad.235184.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perez-Mancera PA, Young AR, Narita M. Inside and out: the activities of senescence in cancer. Nature reviews Cancer. 2014;14:547–558. doi: 10.1038/nrc3773. [DOI] [PubMed] [Google Scholar]
- 5.Lanigan F, Geraghty JG, Bracken AP. Transcriptional regulation of cellular senescence. Oncogene. 2011;30:2901–2911. doi: 10.1038/onc.2011.34. [DOI] [PubMed] [Google Scholar]
- 6.Chandra T, Kirschner K, Thuret JY, Pope BD, Ryba T, Newman S, et al. Independence of repressive histone marks and chromatin compaction during senescent heterochromatic layer formation. Molecular cell. 2012;47:203–214. doi: 10.1016/j.molcel.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christensen KL, Patrick AN, McCoy EL, Ford HL. The six family of homeobox genes in development and cancer. Advances in cancer research. 2008;101:93–126. doi: 10.1016/S0065-230X(08)00405-3. [DOI] [PubMed] [Google Scholar]
- 8.Patrick AN, Cabrera JH, Smith AL, Chen XS, Ford HL, Zhao R. Structure-function analyses of the human SIX1-EYA2 complex reveal insights into metastasis and BOR syndrome. Nature structural & molecular biology. 2013;20:447–453. doi: 10.1038/nsmb.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tadjuidje E, Hegde RS. The Eyes Absent proteins in development and disease. Cellular and molecular life sciences : CMLS. 2013;70:1897–1913. doi: 10.1007/s00018-012-1144-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Le Grand F, Grifone R, Mourikis P, Houbron C, Gigaud C, Pujol J, et al. Six1 regulates stem cell repair potential and self-renewal during skeletal muscle regeneration. The Journal of cell biology. 2012;198:815–832. doi: 10.1083/jcb.201201050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xia Y, Nivet E, Sancho-Martinez I, Gallegos T, Suzuki K, Okamura D, et al. Directed differentiation of human pluripotent cells to ureteric bud kidney progenitor-like cells. Nature cell biology. 2013;15:1507–1515. doi: 10.1038/ncb2872. [DOI] [PubMed] [Google Scholar]
- 12.Xu PX. The EYA-SO/SIX complex in development and disease. Pediatric nephrology. 2013;28:843–854. doi: 10.1007/s00467-012-2246-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orten DJ, Fischer SM, Sorensen JL, Radhakrishna U, Cremers CW, Marres HA, et al. Branchio-oto-renal syndrome (BOR): novel mutations in the EYA1 gene, and a review of the mutational genetics of BOR. Human mutation. 2008;29:537–544. doi: 10.1002/humu.20691. [DOI] [PubMed] [Google Scholar]
- 14.Farabaugh SM, Micalizzi DS, Jedlicka P, Zhao R, Ford HL. Eya2 is required to mediate the pro-metastatic functions of Six1 via the induction of TGF-beta signaling, epithelial-mesenchymal transition, and cancer stem cell properties. Oncogene. 2012;31:552–562. doi: 10.1038/onc.2011.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Auvergne RM, Sim FJ, Wang S, Chandler-Militello D, Burch J, Al Fanek Y, et al. Transcriptional differences between normal and glioma-derived glial progenitor cells identify a core set of dysregulated genes. Cell reports. 2013;3:2127–2141. doi: 10.1016/j.celrep.2013.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abad M, Moreno A, Palacios A, Narita M, Blanco F, Moreno-Bueno G, et al. The tumor suppressor ING1 contributes to epigenetic control of cellular senescence. Aging cell. 2011;10:158–171. doi: 10.1111/j.1474-9726.2010.00651.x. [DOI] [PubMed] [Google Scholar]
- 17.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 18.Palmero I, Pantoja C, Serrano M. p19ARF links the tumour suppressor p53 to Ras [letter] Nature. 1998;395:125–126. doi: 10.1038/25870. [DOI] [PubMed] [Google Scholar]
- 19.Young AR, Narita M, Ferreira M, Kirschner K, Sadaie M, Darot JF, et al. Autophagy mediates the mitotic senescence transition. Genes & development. 2009;23:798–803. doi: 10.1101/gad.519709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Acosta JC, O'Loghlen A, Banito A, Guijarro MV, Augert A, Raguz S, et al. Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell. 2008;133:1006–1018. doi: 10.1016/j.cell.2008.03.038. [DOI] [PubMed] [Google Scholar]
- 21.Lin AW, Barradas M, Stone JC, van Aelst L, Serrano M, Lowe SW. Premature senescence involving p53 and p16 is activated in response to constitutive MEK/MAPK mitogenic signaling. Genes & development. 1998;12:3008–3019. doi: 10.1101/gad.12.19.3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collado M, Blasco MA, Serrano M. Cellular senescence in cancer and aging. Cell. 2007;130:223–233. doi: 10.1016/j.cell.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 23.Coppe JP, Patil CK, Rodier F, Sun Y, Munoz DP, Goldstein J, et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS biology. 2008;6:2853–2868. doi: 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Collado M, Gil J, Efeyan A, Guerra C, Schuhmacher AJ, Barradas M, et al. Tumour biology: senescence in premalignant tumours. Nature. 2005;436:642. doi: 10.1038/436642a. [DOI] [PubMed] [Google Scholar]
- 25.Narita M, Nunez S, Heard E, Narita M, Lin AW, Hearn SA, et al. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113:703–716. doi: 10.1016/s0092-8674(03)00401-x. [DOI] [PubMed] [Google Scholar]
- 26.Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–642. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- 27.Acosta JC, Banito A, Wuestefeld T, Georgilis A, Janich P, Morton JP, et al. A complex secretory program orchestrated by the inflammasome controls paracrine senescence. Nature cell biology. 2013;15:978–990. doi: 10.1038/ncb2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gil J, Peters G. Regulation of the INK4b-ARF-INK4a tumour suppressor locus: all for one or one for all. Nature reviews Molecular cell biology. 2006;7:667–677. doi: 10.1038/nrm1987. [DOI] [PubMed] [Google Scholar]
- 29.Kumar JP. The sine oculis homeobox (SIX) family of transcription factors as regulators of development and disease. Cellular and molecular life sciences : CMLS. 2009;66:565–583. doi: 10.1007/s00018-008-8335-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu Y, Davicioni E, Triche TJ, Merlino G. The homeoprotein six1 transcriptionally activates multiple protumorigenic genes but requires ezrin to promote metastasis. Cancer research. 2006;66:1982–1989. doi: 10.1158/0008-5472.CAN-05-2360. [DOI] [PubMed] [Google Scholar]
- 31.Laugesen A, Helin K. Chromatin repressive complexes in stem cells, development, and cancer. Cell Stem Cell. 2014;14:735–751. doi: 10.1016/j.stem.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 32.Barradas M, Anderton E, Acosta JC, Li S, Banito A, Rodriguez-Niedenfuhr M, et al. Histone demethylase JMJD3 contributes to epigenetic control of INK4a/ARF by oncogenic RAS. Genes & development. 2009;23:1177–1182. doi: 10.1101/gad.511109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pemberton H, Anderton E, Patel H, Brookes S, Chandler H, Palermo R, et al. Genome-wide co-localization of Polycomb orthologs and their effects on gene expression in human fibroblasts. Genome biology. 2014;15:R23. doi: 10.1186/gb-2014-15-2-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eid JE, Garcia CB. Reprogramming of mesenchymal stem cells by oncogenes. Seminars in cancer biology. 2014 doi: 10.1016/j.semcancer.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 35.Buckingham M, Rigby PW. Gene regulatory networks and transcriptional mechanisms that control myogenesis. Developmental cell. 2014;28:225–238. doi: 10.1016/j.devcel.2013.12.020. [DOI] [PubMed] [Google Scholar]
- 36.Li X, Oghi KA, Zhang J, Krones A, Bush KT, Glass CK, et al. Eya protein phosphatase activity regulates Six1-Dach-Eya transcriptional effects in mammalian organogenesis. Nature. 2003;426:247–254. doi: 10.1038/nature02083. [DOI] [PubMed] [Google Scholar]
- 37.Laclef C, Souil E, Demignon J, Maire P. Thymus, kidney and craniofacial abnormalities in Six 1 deficient mice. Mechanisms of development. 2003;120:669–679. doi: 10.1016/s0925-4773(03)00065-0. [DOI] [PubMed] [Google Scholar]
- 38.Di Croce L, Helin K. Transcriptional regulation by Polycomb group proteins. Nature structural & molecular biology. 2013;20:1147–1155. doi: 10.1038/nsmb.2669. [DOI] [PubMed] [Google Scholar]
- 39.Liu Y, Chakroun I, Yang D, Horner E, Liang J, Aziz A, et al. Six1 regulates MyoD expression in adult muscle progenitor cells. PloS one. 2013;8:e67762. doi: 10.1371/journal.pone.0067762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Irelan JT, Gutierrez Del Arroyo A, Gutierrez A, Peters G, Quon KC, Miraglia L, et al. A functional screen for regulators of CKDN2A reveals MEOX2 as a transcriptional activator of INK4a. PloS one. 2009;4:e5067. doi: 10.1371/journal.pone.0005067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin N, Popov N, Aguilo F, O'Loghlen A, Raguz S, Snijders AP, et al. Interplay between Homeobox proteins and Polycomb repressive complexes in p16INK(4)a regulation. The EMBO journal. 2013;32:982–995. doi: 10.1038/emboj.2013.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Delgado-Olguin P, Huang Y, Li X, Christodoulou D, Seidman CE, Seidman JG, et al. Epigenetic repression of cardiac progenitor gene expression by Ezh2 is required for postnatal cardiac homeostasis. Nature genetics. 2012;44:343–347. doi: 10.1038/ng.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coppe JP, Rodier F, Patil CK, Freund A, Desprez PY, Campisi J. Tumor suppressor and aging biomarker p16(INK4a) induces cellular senescence without the associated inflammatory secretory phenotype. The Journal of biological chemistry. 2011;286:36396–36403. doi: 10.1074/jbc.M111.257071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Imai Y, Takahashi A, Hanyu A, Hori S, Sato S, Naka K, et al. Crosstalk between the Rb pathway and AKT signaling forms a quiescence-senescence switch. Cell reports. 2014;7:194–207. doi: 10.1016/j.celrep.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 46.Rodier F, Coppe JP, Patil CK, Hoeijmakers WA, Munoz DP, Raza SR, et al. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nature cell biology. 2009;11:973–979. doi: 10.1038/ncb1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuilman T, Michaloglou C, Vredeveld LC, Douma S, van Doorn R, Desmet CJ, et al. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell. 2008;133:1019–1031. doi: 10.1016/j.cell.2008.03.039. [DOI] [PubMed] [Google Scholar]
- 48.Chien Y, Scuoppo C, Wang X, Fang X, Balgley B, Bolden JE, et al. Control of the senescence-associated secretory phenotype by NF-kappaB promotes senescence and enhances chemosensitivity. Genes & development. 2011;25:2125–2136. doi: 10.1101/gad.17276711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu PX, Zheng W, Huang L, Maire P, Laclef C, Silvius D. Six1 is required for the early organogenesis of mammalian kidney. Development. 2003;130:3085–3094. doi: 10.1242/dev.00536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wong EY, Ahmed M, Xu PX. EYA1-SIX1 complex in neurosensory cell fate induction in the mammalian inner ear. Hearing research. 2013;297:13–19. doi: 10.1016/j.heares.2012.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Coletta RD, McCoy EL, Burns V, Kawakami K, McManaman JL, Wysolmerski JJ, et al. Characterization of the Six1 homeobox gene in normal mammary gland morphogenesis. BMC developmental biology. 2010;10:4. doi: 10.1186/1471-213X-10-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McCoy EL, Iwanaga R, Jedlicka P, Abbey NS, Chodosh LA, Heichman KA, et al. Six1 expands the mouse mammary epithelial stem/progenitor cell pool and induces mammary tumors that undergo epithelial-mesenchymal transition. The Journal of clinical investigation. 2009;119:2663–2677. doi: 10.1172/JCI37691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sehic D, Karlsson J, Sandstedt B, Gisselsson D. SIX1 protein expression selectively identifies blastemal elements in Wilms tumor. Pediatric blood & cancer. 2012;59:62–68. doi: 10.1002/pbc.24025. [DOI] [PubMed] [Google Scholar]
- 54.Iwanaga R, Wang CA, Micalizzi DS, Harrell JC, Jedlicka P, Sartorius CA, et al. Expression of Six1 in luminal breast cancers predicts poor prognosis and promotes increases in tumor initiating cells by activation of extracellular signal-regulated kinase and transforming growth factor-beta signaling pathways. Breast cancer research : BCR. 2012;14:R100. doi: 10.1186/bcr3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Banito A, Rashid ST, Acosta JC, Li S, Pereira CF, Geti I, et al. Senescence impairs successful reprogramming to pluripotent stem cells. Genes & development. 23:2134–2139. doi: 10.1101/gad.1811609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li H, Collado M, Villasante A, Strati K, Ortega S, Canamero M, et al. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460:1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W, Walsh RM, et al. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460:1145–1148. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Storer M, Mas A, Robert-Moreno A, Pecoraro M, Ortells MC, Di Giacomo V, et al. Senescence is a developmental mechanism that contributes to embryonic growth and patterning. Cell. 2013;155:1119–1130. doi: 10.1016/j.cell.2013.10.041. [DOI] [PubMed] [Google Scholar]
- 59.Munoz-Espin D, Canamero M, Maraver A, Gomez-Lopez G, Contreras J, Murillo-Cuesta S, et al. Programmed cell senescence during mammalian embryonic development. Cell. 2013;155:1104–1118. doi: 10.1016/j.cell.2013.10.019. [DOI] [PubMed] [Google Scholar]
- 60.Sousa-Victor P, Gutarra S, Garcia-Prat L, Rodriguez-Ubreva J, Ortet L, Ruiz-Bonilla V, et al. Geriatric muscle stem cells switch reversible quiescence into senescence. Nature. 2014;506:316–321. doi: 10.1038/nature13013. [DOI] [PubMed] [Google Scholar]
- 61.Gomez-Cabello D, Adrados I, Gamarra D, Kobayashi H, Takatsu Y, Takatsu K, et al. DGCR8-mediated disruption of miRNA biogenesis induces cellular senescence in primary fibroblasts. Aging cell. 2013;12:923–931. doi: 10.1111/acel.12117. [DOI] [PubMed] [Google Scholar]
- 62.Palmero I, Murga M, Zubiaga A, Serrano M. Activation of ARF by oncogenic stress in mouse fibroblasts is independent of E2F1 and E2F2. Oncogene. 2002;21:2939–2947. doi: 10.1038/sj.onc.1205371. [DOI] [PubMed] [Google Scholar]
- 63.Menendez C, Adrados I, Fernandez-Barral A, Jimenez B, Flores JM, Canamero M, et al. Increased melanoma formation and dissemination in TyrNRas mice deficient in the tumor suppressor Ing1. Pigment Cell Melanoma Res. 2014;27:674–677. doi: 10.1111/pcmr.12241. [DOI] [PubMed] [Google Scholar]
- 64.Gomez-Cabello D, Callejas S, Benguria A, Moreno A, Alonso J, Palmero I. Regulation of the microRNA processor DGCR8 by the tumor suppressor ING1. Cancer research. 2010;70:1866–1874. doi: 10.1158/0008-5472.CAN-09-2088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.