Abstract
Streptococcus (S.) pneumoniae is a major cause of secondary bacterial pneumonia during influenza epidemics. Neuraminidase is a virulence factor of both pneumococci and influenza viruses. Bacterial neuraminidases are structurally related to viral neuraminidases and susceptible to oseltamivir, an inhibitor designed to target viral neuraminidases. This prompted us to evaluate the antipneumococcal potential of two neuraminidase inhibiting natural compounds, the diarylheptanoid katsumadain A and the isoprenylated flavone artocarpin. Chemiluminescence, fluorescence-, and hemagglutination-based enzyme assays were applied to determine the inhibitory efficiency (IC50 value) of the tested compounds towards pneumococcal neuraminidase.
The mechanism of inhibition was studied via enzyme kinetics with recombinant NanA neuraminidase. Unlike oseltamivir, which competes with the natural substrate of neuraminidase, artocarpin exhibits a mixed-type inhibition with a Ki value of 9.70 μM. Remarkably, artocarpin was the only neuraminidase inhibitor for which an inhibitory effect in pneumococcal growth (MIC: 0.99-5.75 μM) and biofilm formation (MBIC: 1.15-2.97 μM) was observable. In addition, we discovered that the bactericidal effect of artocarpin can reduce the viability of pneumococci by a factor of >1000, without obvious harm to lung epithelial cells. This renders artocarpin a promising natural product for further investigations.
Keywords: pneumococci, neuraminidase, NanA, neuraminidase inhibitors, oseltamivir, zanamivir, DANA, katsumadain A, artocarpin, antipneumococcal, biofilm
Introduction
S. pneumoniae is responsible for the majority of pneumonia cases and the death of about 1.2 million young children worldwide each year (18% of all deaths of children under the age of five) (Black et al., 2010; Krzysciak et al., 2013). Spreading of S. pneumoniae in the nasopharynx and surrounding tissues causes the clinical manifestation. The diseases range from mild upper respiratory tract infections, such as acute otitis media, sinusitis, and pneumonia, to severe and potentially life-threatening conditions, such as meningitis and sepsis, by bacterial invasion of the bloodstream (Simell et al., 2012). Additionally, a lethal synergism between pulmonary coinfections with influenza virus and S. pneumoniae has been established, accounting for the excess mortality during influenza epidemics and pandemics whereat pneumococcal NAs were found to support viral release and spread in the lung (Kash et al., 2011; McCullers and Bartmess, 2003).
Pneumococcal NAs (NanA, B and C) belong to a wide range of surface-associated proteins interacting with eukaryotic cells, extracellular matrix proteins, and serum proteins (Lofling et al., 2011). They catalyze the removal of terminal sialic acid residue from various glycoconjugates on cell surface (Taylor, 1996), by which means they reveal receptors for bacterial adhesion (King et al., 2006). and promote upper (Tong et al., 2000) and lower (Orihuela et al., 2004) airway colonization, biofilm formation, and mucosal infection (Brittan et al., 2012; King et al., 2006; Soong et al., 2006). The released sialic acids serve as a carbon source for the bacteria and represent a trigger for biofilm formation (Trappetti et al., 2009). In addition, pneumococcal NAs contribute critically to inflammation and mortality associated with sepsis (Chen et al., 2011).
The essential roles of NAs during coinfection with influenza viruses and in pathogenesis of pneumococcal strains render them an attractive target for therapeutic intervention (Taylor, 1996). Blocking NA activity with small-molecule inhibitors in the intestinal perforation model of sepsis led to a substantial reduction of the inflammatory response and subsequent morbidity (Chen et al., 2011; Paulson and Kawasaki, 2011). Administration of the influenza virus-specific NAI oseltamivir interrupted the lethal synergism between influenza virus and S. pneumoniae and prevented excess mortality from secondary bacterial pneumonia in a mouse model (McCullers and Bartmess, 2003). Currently, there are only two influenza NAIs (zanamivir and oseltamivir) prescribed worldwide for the treatment and control of influenza (Grienke et al., 2012). Their inhibitory potencies are either weak (zanamivir) or medium (oseltamivir) (Gut et al., 2011) to pneumococcal NA.
Recently, we discovered the diarylheptanoid katsumadain A and the isoprenylated flavone artocarpin as novel NAI acting against influenza viruses (Grienke et al., 2010; Kirchmair et al., 2011). In the present study, we evaluated the antipneumococcal potential of both natural product NAI. We analyzed their inhibitory effect on pneumococcal NA and performed enzyme kinetic studies to understand the molecular mechanism of their inhibition of NanA. In addition, we investigated whether these NAI affect the bacterial growth, adsorption, biofilm formation, and viability.
Material and Methods
Compounds
Oseltamivir carboxylate GS4071 (oseltamivir; Roche AG, Basel, Switzerland), zanamivir (GlaxoSmithKline, Brentford, UK), DANA (2,3-dehydro-2-deoxy-N-acetylneuraminic acid), and rifampicin (both purchased from Sigma-Aldrich, Deisenhofen, Germany) were dissolved in water as 10 mM stock solutions. Rifampicin was stored at -20°C. Artocarpin (Quality Phytochemicals LLC, East Brunswick, NJ, USA) (Kirchmair et al., 2011) and katsumadain A, previously isolated from the seeds of Alpinia katsumadai Hayata (Grienke et al., 2010), were dissolved in DMSO as 10 mM stock solutions and stored at 4°C. Their HPLC purity revealed to be >98%.
Bacterial strains, cells, media, and pre-culture conditions
Six S. pneumoniae clinical isolates were collected from patients with different symptoms (Table 1). Two reference strains DSM20566 (serotype 1, ATCC 33400) and DSM14378 (serotype 5, ATCC 6305) were purchased from Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures (Heidelberg, Germany). S. pneumoniae D39 (serotype 2) was kindly provided by ZIK Septomics (Jena, Germany).
Table 1.
The S. pneumoniae strains studied with both genetic and phenotypic identification of NanA activity.
| Strain | Source | nanA | nanB | nanC | NA activitya |
|---|---|---|---|---|---|
| DSM20566 | reference strain | x | x | - | xb |
| Recombinant | NanA of DSM20566 | x | - | - | xb |
| DSM14378 | reference strain | x | x | - | x |
| D39 | reference strain | x | x | - | xb |
| CF6937 | cystic fibrosis | x | - | x | x |
| CF8919 | cystic fibrosis | x | x | - | x |
| BC7326 | blood culture, sepsis | x | x | x | x |
| BC57 | blood culture, sepsis | x | x | - | x |
| PN8828 | pneumonia | x | x | - | x |
| CJ9400 | conjunctivitis | x | x | - | xb |
NA activity was proved in the FL- and CL-based hemagglutination assays.
HA assay was additionally performed to demonstrate the NA activity.
All nine strains used in this study were grown on Columbia blood agar plates supplemented with 5% sheep blood (Becton Dickinson GmbH, Heidelberg, Germany) at 37°C in an atmosphere enriched with 5% CO2 overnight. During pre-cultivation, bacteria were grown in brain heart infusion broth (BHI) with slightly shaking at 37°C for 4 to 5 hours. This incubation time corresponded with the mid-exponential phase of bacterial growth as evaluated exemplarily with two reference strains (data not shown).
To study growth and biofilm inhibition by the test compounds, samples of precultured pneumococci were diluted in BHI to a McFarland of 0.5 (1.5 × 108 cfu/mL). BHI medium was used for all assays, except for determination of biofilm due to the broad usage of tryptic soy broth (TSB) in pneumococcal biofilm research (Oggioni et al. 2006, Trappetti et al. 2009, Yadav et al. 2012).
Human lung carcinoma cells (A549; Institute of Molecular Virology, University of Münster, Germany) were maintained in Dulbecco’s Modified Eagle Medium (Lonza Group Ltd, Basel, Switzerland) supplemented with 10% fetal calve serum (PAA Laboratories GmbH, Cölbe, Germany). Cells were cultivated at 37°C with 5% CO2.
Genomic DNA isolation, nanA gene amplification, and sequencing
Genomic DNA of nine S. pneumoniae strains was isolated from bacterial cells using the High Pure PCR Template Preparation Kit (Roche Applied Science, Mannheim, Germany) according to the manufacturer’s instructions. The ~ 1.5 kb 16S rRNA gene was amplified and sequenced using conventional primers 27F and 1492R devised by Weisburg et al. (Weisburg et al., 1991) to confirm their identity as S. pneumoniae (data not shown). NA gene fragments were amplified with PCR Taq Core Kit 10 (MP Biomedicals, Eschwege, Germany). For nanA amplification primers were designed based on a pneumococcal nanA gene in GenBank (X72967) (Table S1), and for nanB and nanC we used primers described previously (Burnaugh et al., 2008). Amplification conditions were: 5 min initial denaturation, followed by 38 cycles of 30 s at 96°C, 30 s at 50 - 60°C, and 60 s at 72°C. Products were purified with QIAquick PCR Purification Kit (Qiagen, Hilden, Germany) and sequenced by Eurofins Food GmbH (Ebersberg, Germany). The nanA of DSM20566 was completely sequenced (GenBank accession number KJ850445).
Preparation of precipitated pneumococcal total proteins for assays
Out of 10 mL of liquid pneumococcal culture in BHI medium, 200 μL were taken to extract the total proteins from cells. Upon addition of 1.8 mL absolute ethanol, the sample was kept overnight at -20°C. After centrifugation at speed for 10 min, precipitated total protein was washed with ice-cold 70% ethanol and redissolved either in 50 μL PBS buffer for cell-based assay or in 50 μL buffer (32.5 mM MES, 4 mM calcium chloride, pH 6.5) for enzyme-based assays.
Expression and purification of rNanA of S. pneumoniae DSM20566 from E. coli
In order to obtain the nanA from S. pneumoniae strain DSM20566, primer pair NA_116aaNdeI_fw (5’-TGCACGACATATGGAAAATGTC-3’) and NA_Cter_XhoI_rv (5’-TCAAATCTCGAGAATTCTTCTCT-3’) were applied to amplify this gene. The NdeI/XhoI double-digested PCR product was then ligated to the E. coli expression vector pET-28a. The constructed plasmid encodes an N-terminal 6× His-tagged 886aa protein. A 150 mL E. coli culture of BL21(DE3)/pTNA20566-116aa was used to synthesize the enzyme. Gene expression was induced by IPTG at a final concentration of 0.5 mM after a 4 h initial cultivation at 37°C. The broth was further incubated overnight at 25°C before harvested by centrifugation. Cell pellet was subjected to B-PER Protein Extraction Reagents (Thermoscientific) for lysis and protein release. N-terminal His-tagged NanA was purified via HisPur Cobalt Spin Column (Thermoscientific) and desalted by Pierce Concentrators PES 30K MWCO. Protein sample was stored in a 50% glycerol solution at -20°C.
NA activity and inhibition determination by fluorescence (FL)- and chemiluminescence (CL)-based assays
Pneumococcal NA activity was determined by both FL- and CL-based assays (NA-Fluor™ Influenza Neuraminidase Assay Kit and NA-Star® Influenza Neuraminidase Inhibitor Resistance Detection Kit, respectively, Applied Biosystems) according to the manufacturer´s instructions. Some modifications were introduced into the FL-based assay (Okomo-Adhiambo et al., 2010). Briefly, compounds in a ten-fold (FL assay) or half-log (CL assay) serial dilution in the assay buffer (32.5 mM MES, 4 mM calcium chloride, pH 6.5) were mixed (i) with buffer (self-fluorescence control), (ii) a certain dilution of precipitated total protein or (iii) recombinant NanA and incubated at 37°C for 20 min ((Richter et al., 2015). After the addition of 100 μM of MUNANA (2′-(4-methylumbelliferyl)-α-D-N-acetylneuraminic acid sodium salt hydrate) in the FL assay or 5 μL of 500 × diluted NA-Star® in the CL assay, the testing plates were incubated for another 2 h and 30 min, respectively, at 37°C. The reaction was stopped by the addition of 0.1 M glycine, 25% absolute ethanol (pH 10.7) in FL assay. The relative FL of released 4-methylumbelliferone (4-MU) or CL was measured by microplate reader FLUOstar Omega (BMG Labtech GmbH, Ortenberg, Germany). The percentage of self-fluorescence was calculated as described recently (Richter et al., 2015).
At least three individual experiments were performed for the calculation of the IC50 values with the JASPR curve fitting software (Okomo-Adhiambo et al., 2010).
Enzyme kinetics
NA enzymatic parameters were evaluated using the FL-based assay as described above. At least three different inhibitor concentrations were applied in combination with four MUNANA substrate concentrations (30, 60, 120, and 240 μM) to determine the Km value of recombinant NanA of DSM20566 and the Ki value of artocarpin. After 5 min incubation with the enzyme, the reaction was ended by the addition of the stop solution. Relative fluorescence units (RFU) were measured and converted to 4-MU concentration according to the 4-MU standard curve. Velocity (μM/min) of the reaction was represented as the production of 4-MU per minute. The Enzyme Kinetics Module of SigmaPlot 12.0 (Systat Software, San Jose, CA) was applied to fit the data to the Michaelis-Menten equation using nonlinear regression and determine the enzymatic activity (Vmax) and the inhibition constant (Ki) of NAI.
Analysis of NA activity and its inhibition in lectin-based hemagglutination (HA) assay
The HA assay (Nakano et al., 2006; Pereira, 1983; Richter et al., 2015) was adapted to evaluate the pneumococcal NA activity as well as its inhibition by NAI (Richter et al., 2015).
To evaluate NA activity of pneumococci, 5 μL of a 25% human erythrocyte suspension (in PBS buffer) was subjected to 100 μL of serial diluted (dilution factor 2) precipitated pneumococcal total protein in flat-bottom 96-well polystyrene microplates and incubated at 37°C in 5% CO2 for 4 h. Afterwards, 20 μL of this protein-erythrocytes suspension were added to equal volume of Arachis hypogaea (Sigma-Aldrich GmbH) lectin (50 μg/mL) in V-shaped 96-well polystyrene microplates and incubated at 4°C overnight. Thereafter, the lowest concentration of precipitated pneumococcal total protein causing hemagglutination was visually determined (mean value of three independent assays) and used as readout for NA activity.
To analyze NA inhibition by test compounds, 50 μL of the determined bacterial protein dilution was added to 50 μL of serial compound dilutions (dilution factor 2) or PBS (NA activity control) in a flat-bottom 96-well polystyrene microplate. After incubation at 37°C and 5% CO2 for 20 min, 5 μL of 25% erythrocytes solution was supplemented to the protein-compound suspensions, followed by carefully homogenization and incubation under the same condition for 4 h. Then NA activity was determined as described above via hemagglutination. Negative controls included mere erythrocytes in PBS, a mixture of erythrocytes and bacteria proteins, and a mixture of erythrocytes and lectin. In addition, a cocktail of erythrocytes, lectin, and compounds without bacterial protein (one well per compound concentration) was included to confirm that hemagglutination was not caused by the interaction between compounds and erythrocytes. The compound concentration that prevented hemagglutination was defined as minimal NA inhibitory concentration. Each compound was tested at least three times.
Erythrocyte hemolysis assay
A standard hemolysis assay was performed with human erythrocytes (Yu et al., 2011) with some modifications. Briefly, 10 μL of 25% diluted erythrocytes in PBS were incubated with 200 μL of serial diluted artocarpin (diluted in PBS) for 3 h at 37°C. Final concentrations of artocarpin used were 100 μM, 31.6 μM, 10 μM, 3.2 μM, 1 μM, and 0.3 μM. After that, the mixture was gently shaken and centrifuged at 1,500 × g for 10 minutes. 100 μL of supernatant from each sample was transferred to a flat-bottom 96-well plate. The absorbance value of hemoglobin at 577 nm was measured with the reference wavelength at 655 nm. Erythrocytes diluted in PBS or in aqua bidest were used as negative and positive control, respectively. The percent of hemolysis was calculated as: hemolysis % = [(sample absorbance – negative control) / (positive control – negative control)] × 100%.
Determination of MIC90 and MBC
For broth microdilution assay, pneumococci of McFarland 0.5 (1.5 × 108 cfu/mL) were 50 × diluted in BHI, as well as test compound in a two-fold serial dilution in BHI. 100 μL of diluted substance was mixed with 50 μL bacteria in a 96-well V-shape plate in two repetitions and incubated overnight at 37°C with 5% CO2. Wells without compound were used as positive growth control and wells without bacteria as negative control for medium sterility. The maximal concentrations of compounds in test were 200 μM for oseltamivir and DANA, and 50 μM for artocarpin, katsumadain A, and rifampicin. The growth of bacteria was monitored by measuring optical density at 620 nm (OD620) at 0 h and 18 h. Before calculating the minimal inhibitory concentration (MIC90), the OD620-0 h was subtracted from OD620-18 h. The mean value of positive growth control was set as 100%. MIC90 was defined as the drug concentration that reduced 90% turbidity of the untreated bacterial suspension.
Bactericidal effect of the compounds was investigated using the overnight grown MIC plates. 100 μL of suspension from each well was subcultured on blood agar plates without test agent at 37°C with 5% CO2. The minimal bactericidal concentration (MBC) was defined as the lowest concentration that reduced the viability of the initial bacterial inoculum by ≥99.9%. Antibacterial agents are usually regarded as bactericidal if the MBC is not more than four times the MIC (French, 2006).
Biofilm inhibition assay
Broth was diluted as described for broth microdilution assay in TSB and incubated for 2 h at 37°C with 5% CO2 in 96-well F-bottom plates. Then, the supernatant was replaced by 200 μL of fresh medium without compounds as positive control, or by 150 μL of fresh medium plus 50 μL of two-fold serial dilution of compound (maximal 50 μM; each concentration in duplicate). Six wells without bacteria were used as negative controls. After 24 h incubation, plates were washed with water and stained with crystal violet overnight. After rinsing the plates with water, the crystal violet was eluted with 150 μL of lysis buffer (0.898 g of sodium citrate and 1.25 mL of 1 M HCl in 98.05 mL 47.5% ethanol) (Schmidtke et al., 2001), and the optical density of the elution was measured at 550 nm (OD550). Minimal biofilm inhibitory concentration (MBIC90) was defined as the drug concentration reducing the OD550 of untreated controls by 90%.
Adherence inhibition assay
Bacterial adherence was quantified based on Parker’s method (Hsiao et al., 2009). Pulmonary epithelial cell A549 monolayer were grown for 3 days on 24-well tissue culture plates or 8-well plastic slides (Thermoscientific) until confluence (Singer et al., 2010). Suspensions of the strain DSM20566 and PN8828 grown in BHI broth to mid-exponential phase were added (multiplicity of infection (MOI) of 10) to A549 monolayer for 1.5 h at 37°C and 5% CO2 in the absence (control) or presence of the compounds. After vigorous washing for three times with PBS, the cells in the 24-well plate were lysed with 0.5% saponin (Sigma-Aldrich GmbH) for 5 minutes. The cells on the plastic slide were fixed with methanol for 10 minutes. Aliquots of saponin-detached cells were serially diluted and quantitatively plated on Columbia blood agar plates. The numbers of adherent bacteria on the epithelial cells were determined as colony forming units (CFU) from dilution counts. The mean value of CFU from control wells was set as 100%. Adherence inhibitory concentration (AIC50) was defined as the drug concentration reducing the adherence by 50%. In parallel, compound-treated pneumococci were subcultured on blood agar plates to make sure that the reduced adherence was not due to the bactericidal effect of the compounds. The slide was gram-stained according to a standard protocol in the University Hospital Jena and examined with a Zeiss Axio Imager A1 microscope at 100·magnification.
Determination of cytotoxicity of the compounds
The cytotoxicity of compounds was determined with three-day-old confluent A549 cell monolayers grown in 96-well F-bottom plates as described previously (Schmidtke et al., 2001). Briefly, cells were incubated with serial two-fold dilutions of compound at 37°C in 5% CO2 atmosphere for 72 h (each concentration in duplicate); cells without compound treatment served as basic controls. The Dynex Immuno Assay System (DIAS, Guernsey, Great Britain) was afterwards applied to gently wash, fix and stain the cells with crystal violet dye solution in methanol, formalin and water. Subsequently, the crystal violet was eluted by 100 μL of lysis buffer (see above) for the measurement of OD550. Cell viability was evaluated as the percentage of the mean value of optical density resulting from the six cell controls, which was set 100%. The 50% cytotoxic concentrations (CC50) were calculated from the mean dose–response curves of at least three assays each with two parallels.
Results
All S. pneumoniae strains tested encoded NA genes and exhibited NA activity
As shown in Table 1, we successfully amplified and sequenced nanA fragments from the genomic DNAs of the three pneumococcal reference strains and the six clinical isolates tested in this study. All reference strains and five of the six clinical isolates also encoded nanB (with the exception of strain CF6937). In only two of the studied clinical isolates nanC was detected but in none of the reference strains.
In all tested strains, significant NA activity was detected by the CL, FL and HA assays for ethanol extracts of total bacterial protein (Table 1). In order to confirm these results and prepare for subsequent studies on enzyme inhibition, we exemplarily expressed NanA of strain DSM20566 in E. coli, for which we were able to detect activity in all three assays.
Inhibitory effect of investigational NAI on pneumococcal NA
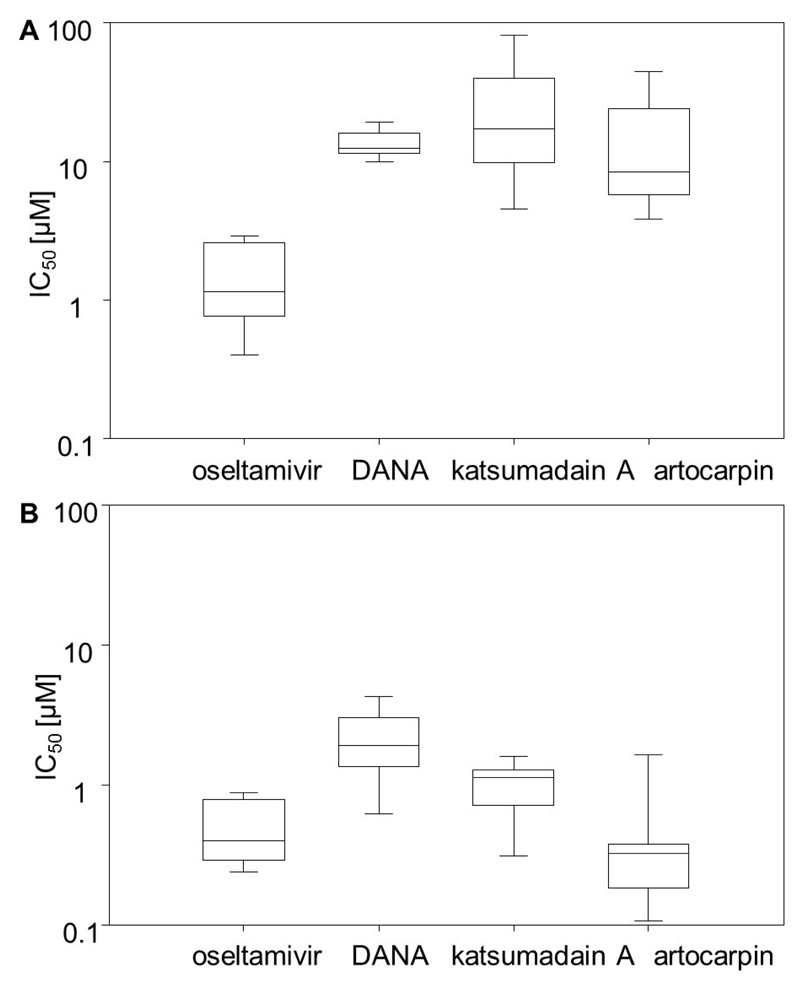
When assessing the inhibition of NA activity of the precipitated total proteins of all strains, we found that oseltamivir and DANA, as well as the two natural compounds artocarpin and katsumadain A, exhibited their inhibitory potential in a dose-dependent manner (Fig. 1A, Table 2). In contrast, zanamivir did not show an effect. The mean IC50 value determined by the FL-based assay for oseltamivir was 1.40 μM, whereas ~ 10-fold higher concentrations of DANA and artocarpin were required to produce a similar effect (Fig. 1A, Table S2). The IC50 values of katsumadain A were in a broad range between 4.50 and 81.03 μM, most probably caused by the strong self-fluorescence determined for this compound (Richter et al., 2015). All other studied compounds did not exhibit self-fluorescence effects (data not shown).
Figure 1.
Box and whisker plots showing distribution of mean 50% inhibitory concentration (IC50) values of NAIs oseltamivir, DANA, katsumadain A, and artocarpin among the nine studied pneumococci strains. After confirming activity of precipitated bacterial proteins, they were used in the (A) FL and (B) CL enzyme inhibitory assay. Mean IC50 values are calculated based on a minimum of three individually performed experiments.
Table 2.
Inhibition of pneumococcal NA activity (HA assay) by total proteins extracted from pneumococcal strains by testing compounds.
| Inhibitory concentration (µM)a | |||
|---|---|---|---|
| compounds | DSM20566 | D39 | CJ9400 |
| oseltamivir | 2.1 ± 1.3 | 10.0 ± 0.0 | 0.3 ± 0.0 |
| zanamivir | >100 | >100 | >100 |
| DANA | 45.3 ± 30.6 | > 100 | 15.4 ± 10.8 |
| katsumadain A | 3.2 ± 0.0 | 1.0 ± 0.0 | 0.7 ± 0.4 |
| artocarpin | 7.7 ± 4.0 | - b | 2.6 ± 1.1 |
Mean inhibitory concentration values were calculated based on three individually performed assays.
The erythrocytes were lysed in the presence of artocarpin at concentrations higher than 10 µM.
The CL-based assay was used to confirm these results. Our results underline susceptibility of NanAs of all nine strains for oseltamivir, DANA, katsumadain A, and artocarpin (Fig. 1B, Table S3), as well as inefficacy of zanamivir (data not shown). Generally, IC50 values determined by CL-based assays are lower than those obtained from FL-based assays. Artocarpin and oseltamivir exhibited mean IC50 values of 0.49 μM and 0.51 μM, respectively. The broad range of inhibitory effects of katsumadain A measured in FL-based assays towards diverse test strains was reduced to 0.31-1.61 μM in the CL-based assays (Fig. 1B).
To validate bacterial enzyme inhibition by the test compounds under more physiological conditions, the cell-based HA assay was performed with three selected pneumococcal strains, DSM20566, D39 as reference strains and one clinical isolate, CJ9400. As shown in Table 2 the total protein extracted from the DSM20566 and CJ9400 strains was susceptible to all interrogated NAIs in this cell-based assay.
With the exception of katsumadain A, the tested NAI showed lower activity against the reference strain D39, or even no activity in the case of DANA and artocarpin. In general, the activities of the cell-based HA assay clearly confirmed the results obtained from the enzyme-based assays (Tables 2 and 3). A 20% hemolysis was detected when 31.60 μM of artocarpin was applied to erythrocytes (Fig. S2).
Table 3.
Mean 90% inhibitory concentration and standard deviation of oseltamivir, DANA, artocarpin, and katsumadain A against three reference strains and six clinical isolates in broth microdilution and biofilm assay as well as bactericidal concentrations.
| 90% minimal inhibitory concentration (µM) a | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| compound | DSM20566 | D39 | DSM14378 | CJ9400 | CF6937 | BC7326 | CF8919 | PN8828 | BC57 |
| rifampicin | 0.041 ± 0.011 | 0.021 ± 0.002 | 0.031 ± 0.023 | 0.032 ± 0.030 | 0.034 ± 0.028 | 0.016 ± 0.005 | 0.009 ± 0.001 | 0.005 ± 0.001 | 0.012 ± 0.002 |
| oseltamivir, zanamivir, DANA, katsumadain A | > 50 | > 50 | > 50 | > 50 | > 50 | > 50 | > 50 | > 50 | > 50 |
| artocarpin | 5.58 ± 0.97 | 1.29 ± 0.31 | 5.75 ± 0.13 | 3.45 ± 2.05 | 3.39 ± 2.10 | 1.12 ± 0.37 | 1.45 ± 0.01 | 0.99 ± 0.48 | 1.45 ± 0.04 |
|
| |||||||||
| 90% minimal biofilm inhibitory concentration (µM) a | |||||||||
| rifampicin | 0.049 ± 0.030 | 0.013 ± 0.002 | 0.014 ± 0.008 | 0.003 ± 0.001 | 0.020 ± 0.002 | 0.016 ± 0.010 | 0.006 ± 0.004 | 0.011 ± 0.007 | 0.012 ± 0.001 |
| oseltamivir, zanamivir, DANA, katsumadain A | > 50 | > 50 | > 50 | > 50 | > 50 | > 50 | > 50 | > 50 | > 50 |
| artocarpin | 2.88 ± 0.07 | 2.22 ± 0.76 | 2.37 ± 0.67 | 2.97 ± 0.13 | 2.73 ± 0.27 | 2.02 ± 0.62 | 1.15 ± 0.34 | 1.40 ± 0.07 | 2.35 ± 0.68 |
|
| |||||||||
| Bactericidal concentration (µM) b | |||||||||
| rifampicin | 1.97 ± 1.23 | 0.22 ± 0.09 | 0.14 ± 0.06 | 0.23 ± 0.16 | 0.05 ± 0.03 | 0.08 ± 0.03 | not studied | not studied | not studied |
| oseltamivir, zanamivir, DANA, katsumadain A | > 50 | > 50 | > 50 | > 50 | > 50 | > 50 | not studied | not studied | not studied |
| artocarpin | 3.13 ± 0.00 | 2.50 ± 0.77 | 4.69 ± 1.98 | 2.74 ± 0.68 | 3.44 ± 1.53 | 3.32 ± 1.94 | not studied | not studied | not studied |
The IC50 values and standard deviations were calculated of at least three independent tests with each concentration tested in duplicate.
The bactericidal concentrations were calculated of at least two independent tests.
NAI susceptibility, kinetic properties, and the mechanism of inhibition of the pneumococcal rNanA
To confirm that the IC50 values determined by using precipitated total pneumococci proteins correspond with inhibition of the ubiquitously expressed pneumococcal NanA, E. coli-derived rNanA of reference strain DSM20566 was tested in the assays described above. As shown in Tables S2 and S3 the susceptibilities of the precipitated total protein and the recombinant NanA towards four test agents were in a comparable range.
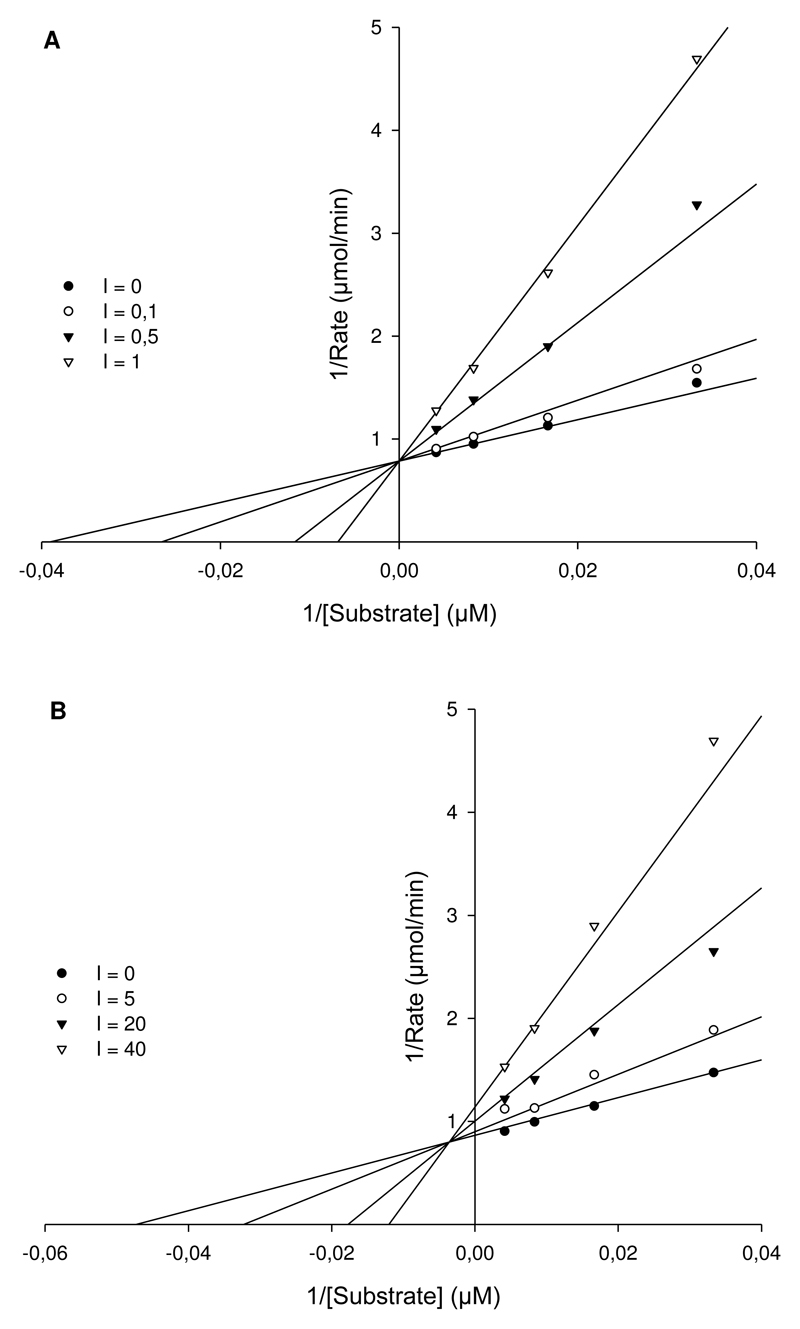
Enzyme kinetics revealed a substrate affinity (Km) of rNanA for 4-MUNANA was 23.60 μM. The inhibition constant of oseltamivir and artocarpin was determined and the mechanism of inhibition modeled. As expected, influenza specific NAI oseltamivir competitively inhibited the activity of rNanA with a Ki value of 0.23 μM (Fig. 2A). According to the SigmaPlot analysis, artocarpin inhibition is of a mixed type with an inhibition constant Ki of 9.70 μM (Fig. 2B). With increasing concentration of artocarpin, the Km of rNanA increases and Vmax drops, indicating that artocarpin binds to the free rNanA and the rNanA-MUNANA complex with different affinity. We were unable to determine the mechanism of inhibition for katsumadain A due to its strong self-fluorescence under the FL-assay conditions (Richter et al., 2015).
Figure 2.
Lineweaver-Burk plot analysis of the inhibition mechanism of NAI. (A) Competitive inhibition of oseltamivir at concentrations of 0.0, 0.1, 0.5 and 1 μM. Regardless of the presence of oseltamivir, Vmax is invariant, however, the increase in concentration of oseltamivir led to the raise of Km. (B) Mixed inhibition of artocarpin at concentrations of 0, 5, 20 and 40 μM. The presence of artocarpin resulted in a decreased Vmax and increased Km. The Ki values of oseltamivir and artocarpin are 0.23±0.04 μM and 9.68±1.49 μM, respectively.
Antipneumococcal effects of artocarpin
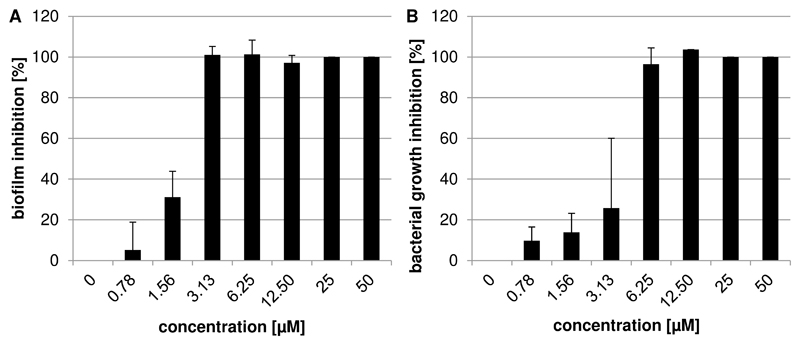
Since NanA was described to be involved in biofilm formation (Brittan et al., 2012; Parker et al., 2009), we investigated the capacity of NAIs to reduce the biofilm production of S. pneumoniae. No significant inhibition on biofilm formation was observed upon addition of oseltamivir, zanamivir, DANA, or katsumadain A up to 50 μM. However, artocarpin exhibited a strong dose-dependent effect, with mean MBIC90 values in the range of 1.15 to 2.97 μM (Table 3). As an example, the dose-response curve of reference strain DSM20566 is reported in Figure 3A.
Figure 3.
Susceptibility of pneumococcal reference strain DSM20566 to artocarpin. (A) The inhibition of biofilm formation, and (B) of bacterial growth by artocarpin is shown. Bars represent the mean and standard deviation of at least three assays each with two parallels per concentration.
In contrast to all other tested NAIs, artocarpin also inhibited planktonic growth of all tested pneumococcal strains (Fig. 3B and Table 3). Its MIC90 values, determined by the broth microdilution assay, ranged from 0.99 to 5.75 μM. The MIC90 values of the antibiotic rifampicin (used as positive control) were in a range from 0.01 to 0.04 μM.
We also detected significant bactericidal activity for artocarpin and the control antibiotic rifampicin. Both compounds reduced the viability of studied pneumococci by a factor of >1000 within 18 h. Neither oseltamivir nor DANA or katsumadain A exhibited inhibitory or bactericidal activity at concentrations up to 50 μM (the maximum tested concentration; Table 3).
Artocarpin prevents pneumococcal adhesion to pulmonary epithelial cells without cytotoxic effects
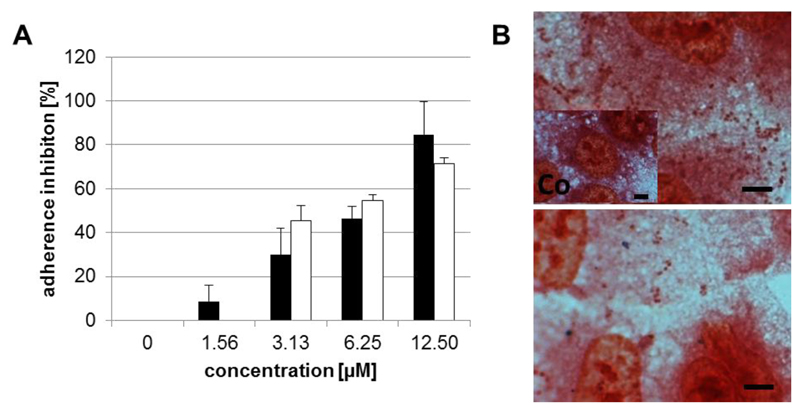
One of the main functions of pneumococcal NA is desialylation of sugar moieties on the cell surface, serving as receptors for bacterial attachment to airway epithelial cells (Brittan et al., 2012). After confirming the inhibitory capacity of viral NAIs for pneumococcal NA, we explored their effect on pneumococcal adherence to pulmonary epithelial A549 cells. This was studied using an adherence assay with DSM20566 and PN8828 (MOI of 10). Artocarpin led to a 50% reduction in adherence levels of the reference strain and clinical isolate to the cell culture at 7.50 and 4.10 μM, respectively (Fig. 4A, Table S4). Similar effects of the reduced bacterial adherence on lung cells with artocarpin were also observed via Gram staining (Fig. 4B). None of the other tested NAIs showed any effect on the adhesion of pneumococci (Table S4).
Figure 4.
Artocarpin inhibits the adherence of pneumococci to A549 cells. (A) Percentage of inhibition of adherence of pneumococcal reference strain DSM20566 (black) and clinical isolate PN8828 (white) by artocarpin in comparison to untreated bacteria. Mean and standard deviation of at least three assays each with two parallels per concentration are shown. (B) Gram staining was used to confirm adhesion of strain DSM20566 in the absence (upper image) and presence (down image) of 12.5 μM artocarpin. The untreated cell control (Co) is shown in the left lower corner of the upper image. Bar represents 5 μm.
To confirm that the adherence inhibitory effect was not a result of the bactericidal potential of the test compounds, DSM20566 was incubated in the absence or presence of artocarpin within 1.5 h and then cultivated on blood agar. No bactericidal effect was observed (results not shown). Furthermore, artocarpin did not affect cell viability.
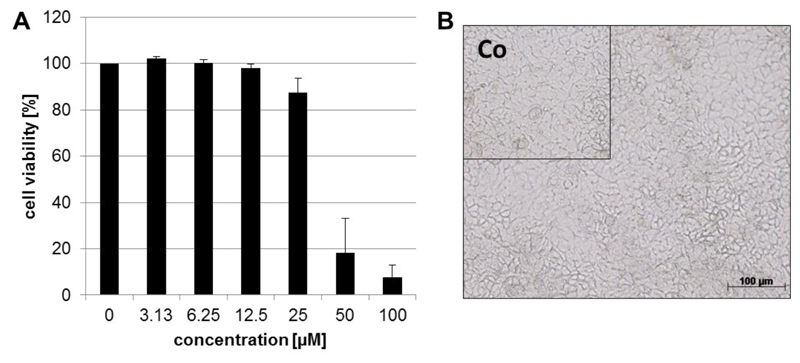
To further analyze the compatibility of artocarpin with A549 cells, we monitored the viability of confluent cell monolayers for 72 h of coincubation with the compound. As reported in Fig. 5A and 5B, artocarpin exhibited a low level of cytotoxicity, with a CC50 of 38.00 μM. At a concentration of 12.50 μM, the effective concentration for adherence inhibition, no cytotoxic effect was observed. This was further confirmed by microscopic analysis (Fig. 5B).
Figure 5.
The influence of artocarpin on the viability of A549 monolayer’s during 72 h of incubation. (A) Mean percentage of cell viablilty (OD of untreated cell control was set 100%) and standard deviation of at least three cytotoxicity assays each with two parallels per concentration are shown.) (B) Photography of phase-contrast microscopy of cells incubated for 72 hours with 12.5 μM artocarpin. The untreated cell control (Co) is shown in the left upper corner. Bar represents 5 μm.
Discussion and Conclusion
Bacterial NAs have been implicated in the pathogenesis of S. pneumoniae and lethal synergism during influenza infections, and were suggested as promising drug targets (Taylor, 1996). Our current work demonstrated katsumadain A and artocarpin as inhibitors of pneumococcal NA. In addition, artocarpin exhibited antipneumococcal activity.
As one of the most conserved gene loci of S. pneumoniae, nanA was detected in all strains described previously (Kelly et al., 1967; Pettigrew et al., 2006) and in all nine strains investigated in the present study. In addition, nanB was detected for eight, and nanC for two strains. In agreement with NA gene detection, pronounced NA activities were observed from the precipitated total protein of all strains and in the E. coli-synthesized recombinant NanA of strain DSM20566. The catalytic activity of all these strains was effectively blocked by oseltamivir and DANA, as well as the influenza A virus NAIs katsumadain A and artocarpin (Grienke et al., 2010; Grienke et al., 2012) at low micromolar concentrations in three different types of NA inhibition assays. However, the inhibitory activity of oseltamivir on bacterial NAs was found to be lower by a factor of 1000 when compared to viral NAs (Ki of 0.32 nM) (Collins et al., 2008). The inhibitory effect of NAI was corroborated using purified recombinant rNanA as target. The IC50 values obtained for the competitive, virus-specific NAI DANA and oseltamivir are in good agreement with published data (Gut et al., 2011).
The enzyme kinetics study with recombinant NanA of strain DSM20566 revealed a competitive mechanism of inhibition for oseltamivir, and a mixed inhibition for artocarpin. Oseltamivir is known to bind the substrate binding site of NanA (Gut et al., 2011), and this is confirmed by the enzyme kinetics measured in this work. The inhibition mechanism of artocarpin as suggested by nonlinear regression (SigmaPlot) is a mixed type, meaning that artocarpin binds, with different affinity, to both free NanA and NanA-sialic acid complexes. This suggests the presence of an alternative binding site for artocarpin, different from the natural substrate recognition site.
Previous reports on the key role of NanA in the bacterial adhesion to host cells, biofilm formation and disease (Nobbs et al., 2009), and the herein reported inhibition of pneumococcal NA by oseltamivir, DANA, katsumadain A and artocarpin prompted us to study their potential to reduce the adhesion of S. pneumoniae to host cells or tissues, and to block the formation of the microbial structured community. Intriguingly, oseltamivir, DANA and katsumadain A did not reduce pneumococcal adherence to A549 cells, and did not inhibit bacterial growth or biofilm formation, despite their significant inhibitory activity on NAs. To our knowledge, this is the first report on antipneumococcal activity of NAIs, although it has previously been demonstrated that N-acetyl neuraminic acid inhibits biofilm formation (Parker et al., 2009). It remains unclear why oseltamivir, DANA, or katsumadain A does not affect pneumococcal adhesion, growth, and/or biofilm formation, despite inhibiting NA activity at low micromolar concentrations. It could be that under physiological conditions compound binding is not strong enough to compete with natural substrates. Another possibility is that blockage of NA activity by binding of the compound to the cleavage site does not result in a measurable inhibitory effect. This would contradict with some previous studies (Brittan et al., 2012; Parker et al., 2009), but be in agreement with the results of King et al. obtained for NA deletion mutants of pneumococci (King et al., 2004; King et al., 2006). King and colleagues did not detect a reproducible difference between the percentage of adherent wildtype and nanA knockout D39 pneumococci, and for the level of persistence of murine colonization.
Artocarpin significantly reduced the adherence to epithelial cells A549 and exhibited antipneumococcal activity on planktonic bacteria as well as biofilm formation at low micromolar concentrations. Unlike the other NAIs tested in this study, but similar to the control antibiotic rifampicin, artocarpin showed bactericidal activity. Thus, in contrast to all other studied NAI, artocarpin exerted antipneumococcal activity. This difference might be based on the mixed binding mode of artocarpin to pneumococcal NA, or activity on a further pneumococcal target(s). There is not enough evidence to conclude whether or not the inhibition of pneumococcal growth, adherence and biofilm formation by artocarpin is to be entirely ascribed to the inactivation of NanA or a result of additional interaction between artocarpin and one or more additional pneumococcal targets.
To the best of our knowledge, this is the first report of a compound with strong inhibitory activity on pneumococcal NAs that in addition is effective in reducing bacterial adherence to pulmonary epithelial cells, biofilm formation, pneumococcal growth, and viability. Strong antipneumococcal and NA inhibitory activity render artocarpin a potentially interesting lead in the search for dual acting antipneumococcal drugs.
Supplementary Material
Acknowledgements
JASPR software was introduced by Larisa V. Gubareva (Centers for Disease Control and Prevention, US) for the determination of IC50 in this study. We are grateful for the critical discussion of docking results with Julian E. Fuchs (University of Innsbruck). We thank NCI/DTP Open Chemical Repository for providing artocarpin (http://dtp.cancer.gov).
Financial disclosure
This work was supported by the European Social Fund (ESF) and the Thuringian Ministry of Economy, Labor and Technology (TMWAT) with the project “Optimization of influenza treatment by keeping into account secondary bacterial infections” (2011FGR0137) and by the Austrian Science Fund (FWF): projects “Targeting Influenza Neuraminidase” (P23051) and “Natural Lead Structures Targeting Influenza” (P24587).
References
- Black RE, Cousens S, Johnson HL, Lawn JE, Rudan I, Bassani DG, Jha P, Campbell H, Walker CF, Cibulskis R, Eisele T, et al. Global, regional, and national causes of child mortality in 2008: a systematic analysis. Lancet. 2010;375:1969–1987. doi: 10.1016/S0140-6736(10)60549-1. [DOI] [PubMed] [Google Scholar]
- Brittan JL, Buckeridge TJ, Finn A, Kadioglu A, Jenkinson HF. Pneumococcal neuraminidase A: an essential upper airway colonization factor for Streptococcus pneumoniae. Molecular Oral Microbiology. 2012;27:270–283. doi: 10.1111/j.2041-1014.2012.00658.x. [DOI] [PubMed] [Google Scholar]
- Burnaugh AM, Frantz LJ, King SJ. Growth of Streptococcus pneumoniae on human glycoconjugates is dependent upon the sequential activity of bacterial exoglycosidases. J Bacteriol. 2008;190:221–230. doi: 10.1128/JB.01251-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GY, Chen X, King S, Cavassani KA, Cheng JS, Zheng XC, Cao HZ, Yu H, Qu JY, Fang DX, Wu W, et al. Amelioration of sepsis by inhibiting sialidase-mediated disruption of the CD24-SiglecG interaction. Nat Biotechnol. 2011;29:428–U234. doi: 10.1038/nbt.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins PJ, Haire LF, Lin YP, Liu JF, Russell RJ, Walker PA, Skehel JJ, Martin SR, Hay AJ, Gamblin SJ. Crystal structures of oseltamivir-resistant influenza virus neuraminidase mutants. Nature. 2008;453:1258–U1261. doi: 10.1038/nature06956. [DOI] [PubMed] [Google Scholar]
- French GL. Bactericidal agents in the treatment of MRSA infections - the potential role of daptomycin. J Antimicrob Chemoth. 2006;58:1107–1117. doi: 10.1093/jac/dkl393. [DOI] [PubMed] [Google Scholar]
- Grienke U, Schmidtke M, Kirchmair J, Pfarr K, Wutzler P, Durrwald R, Wolber G, Liedl KR, Stuppner H, Rollinger JM. Antiviral Potential and Molecular Insight into Neuraminidase Inhibiting Diarylheptanoids from Alpinia katsumadai. J Med Chem. 2010;53:778–786. doi: 10.1021/jm901440f. [DOI] [PubMed] [Google Scholar]
- Grienke U, Schmidtke M, von Grafenstein S, Kirchmair J, Liedl KR, Rollinger JM. Influenza neuraminidase: A druggable target for natural products. Natural Product Reports. 2012;29:11–36. doi: 10.1039/c1np00053e. [DOI] [PubMed] [Google Scholar]
- Gut H, Xu GG, Taylor GL, Walsh MA. Structural Basis for Streptococcus pneumoniae NanA Inhibition by Influenza Antivirals Zanamivir and Oseltamivir Carboxylate. Journal of Molecular Biology. 2011;409:496–503. doi: 10.1016/j.jmb.2011.04.016. [DOI] [PubMed] [Google Scholar]
- Hsiao YS, Parker D, Ratner AJ, Prince A, Tong L. Crystal structures of respiratory pathogen neuraminidases. Biochemical and Biophysical Research Communications. 2009;380:467–471. doi: 10.1016/j.bbrc.2009.01.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kash JC, Walters KA, Davis AS, Sandouk A, Schwartzman LM, Jagger BW, Chertow DS, Li Q, Kuestner RE, Ozinsky A, Taubenberger JK. Lethal synergism of 2009 pandemic H1N1 influenza virus and Streptococcus pneumoniae coinfection is associated with loss of murine lung repair responses. mBio. 2011;2 doi: 10.1128/mBio.00172-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly RT, Farmer S, Greiff D. Neuraminidase Activities of Clinical Isolates of Diplococcus Pneumoniae. J Bacteriol. 1967;94:272. doi: 10.1128/jb.94.1.272-273.1967. &. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King SJ, Hippe KR, Gould JM, Bae D, Peterson S, Cline RT, Fasching C, Janoff EN, Weiser JN. Phase variable desialylation of host proteins that bind to Streptococcus pneumoniae in vivo and protect the airway. Mol Microbiol. 2004;54:159–171. doi: 10.1111/j.1365-2958.2004.04252.x. [DOI] [PubMed] [Google Scholar]
- King SJ, Hippe KR, Weiser JN. Deglycosylation of human glycoconjugates by the sequential activities of exoglycosidases expressed by Streptococcus pneumoniae. Mol Microbiol. 2006;59:961–974. doi: 10.1111/j.1365-2958.2005.04984.x. [DOI] [PubMed] [Google Scholar]
- Kirchmair J, Rollinger JM, Liedl KR, Seidel N, Krumbholz A, Schmidtke M. Novel neuraminidase inhibitors: identification, biological evaluation and investigations of the binding mode. Future Med Chem. 2011;3:437–450. doi: 10.4155/fmc.10.292. [DOI] [PubMed] [Google Scholar]
- Krzysciak W, Pluskwa KK, Jurczak A, Koscielniak D. The pathogenicity of the Streptococcus genus. Eur J Clin Microbiol. 2013;32:1361–1376. doi: 10.1007/s10096-013-1914-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lofling J, Vimberg V, Battig P, Henriques-Normark B. Cellular interactions by LPxTG-anchored pneumococcal adhesins and their streptococcal homologues. Cell Microbiol. 2011;13:186–197. doi: 10.1111/j.1462-5822.2010.01560.x. [DOI] [PubMed] [Google Scholar]
- McCullers JA, Bartmess KC. Role of neuraminidase in lethal synergism between influenza virus and Streptococcus pneumoniae. J Infect Dis. 2003;187:1000–1009. doi: 10.1086/368163. [DOI] [PubMed] [Google Scholar]
- Nakano V, Maria R, Piazza F, Avila-Campos MJ. A rapid assay of the sialidase activity in species of the Bacteroides fragilis group by using peanut lectin hemagglutination. Anaerobe. 2006;12:238–241. doi: 10.1016/j.anaerobe.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Nobbs AH, Lamont RJ, Jenkinson HF. Streptococcus Adherence and Colonization. Microbiol Mol Biol R. 2009;73:407–450. doi: 10.1128/MMBR.00014-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okomo-Adhiambo M, Sleeman K, Ballenger K, Nguyen HT, Mishin VP, Sheu TG, Smagala J, Li Y, Klimov AI, Gubareva LV. Neuraminidase Inhibitor Susceptibility Testing in Human Influenza Viruses: A Laboratory Surveillance Perspective. Viruses-Basel. 2010;2:2269–2289. doi: 10.3390/v2102269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orihuela CJ, Gao GL, Francis KP, Yu J, Tuomanen EI. Tissue-specific contributions of pneumococcal virulence factors to pathogenesis. Journal of Infectious Diseases. 2004;190:1661–1669. doi: 10.1086/424596. [DOI] [PubMed] [Google Scholar]
- Parker D, Soong G, Planet P, Brower J, Ratner AJ, Prince A. The NanA neuraminidase of Streptococcus pneumoniae is involved in biofilm formation. Infect Immun. 2009;77:3722–3730. doi: 10.1128/IAI.00228-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson JC, Kawasaki N. Sialidase inhibitors DAMPen sepsis. Nat Biotechnol. 2011;29:406–407. doi: 10.1038/nbt.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira MEA. A Rapid and Sensitive Assay for Neuraminidase Using Peanut Lectin Hemagglutination - Application to Vibrio-Cholera and Trypanosoma-Cruzi. J Immunol Methods. 1983;63:25–34. doi: 10.1016/0022-1759(83)90206-5. [DOI] [PubMed] [Google Scholar]
- Pettigrew MM, Fennie KP, York MP, Daniels J, Ghaffar F. Variation in the presence of neuraminidase genes among Streptococcus pneumoniae isolates with identical sequence types. Infect Immun. 2006;74:3360–3365. doi: 10.1128/IAI.01442-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter M, Schumann L, Walther E, Hoffmann A, Braun H, Grienke U, Rollinger JM, Von Grafenstein S, Liedl KR, Kirchmair J, Wutzler P, et al. Complementary assays for identifying neuraminidase inhibitors. Future Virology. 2015 doi: 10.2217/FVL.14.97. (in press) [DOI] [Google Scholar]
- Schmidtke M, Schnittler U, Jahn B, Dahse HM, Stelzner A. A rapid assay for evaluation of antiviral activity against coxsackie virus B3, influenza virus A, and herpes simplex virus type 1. J Virol Methods. 2001;95:133–143. doi: 10.1016/s0166-0934(01)00305-6. [DOI] [PubMed] [Google Scholar]
- Simell B, Auranen K, Kayhty H, Goldblatt D, Dagan R, O'Brien KL, Grp PC. The fundamental link between pneumococcal carriage and disease. Expert Rev Vaccines. 2012;11:841–855. doi: 10.1586/erv.12.53. [DOI] [PubMed] [Google Scholar]
- Singer BB, Scheffrahn I, Kammerer R, Suttorp N, Ergun S, Slevogt H. Deregulation of the CEACAM Expression Pattern Causes Undifferentiated Cell Growth in Human Lung Adenocarcinoma Cells. Plos One. 2010;5 doi: 10.1371/journal.pone.0008747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soong G, Muir A, Gomez MI, Waks J, Reddy B, Planet P, Singh PK, Kanetko Y, Wolfgang MC, Hsiao YS, Tong L, et al. Bacterial neuraminidase facilitates mucosal infection by participating in biofilm production. Journal of Clinical Investigation. 2006;116:2297–2305. doi: 10.1172/JCI27920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor G. Sialidases: Structures, biological significance and therapeutic potential. Curr Opin Struc Biol. 1996;6:830–837. doi: 10.1016/s0959-440x(96)80014-5. [DOI] [PubMed] [Google Scholar]
- Tong HH, Blue LE, James MA, DeMaria TF. Evaluation of the virulence of a Streptococcus pneumoniae neuraminidase-deficient mutant in nasopharyngeal colonization and development of otitis media in the chinchilla model. Infect Immun. 2000;68:921–924. doi: 10.1128/iai.68.2.921-924.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trappetti C, Kadioglu A, Carter M, Hayre J, Iannelli F, Pozzi G, Andrew PW, Oggioni MR. Sialic acid: a preventable signal for pneumococcal biofilm formation, colonization, and invasion of the host. J Infect Dis. 2009;199:1497–1505. doi: 10.1086/598483. [DOI] [PubMed] [Google Scholar]
- Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16s Ribosomal DNA Amplification for Phylogenetic Study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T, Malugin A, Ghandehari H. Impact of silica nanoparticle design on cellular toxicity and hemolytic activity. ACS nano. 2011;5:5717–5728. doi: 10.1021/nn2013904. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.