Abstract
Human induced pluripotent stem cells (iPSCs) have been recently found to harbor genomic alterations. However, the integrity of mitochondrial DNA (mtDNA) within reprogrammed cells has yet to be investigated. mtDNA mutations occur at a high rate and are believed to contribute to the pathology of a number of human disorders. Furthermore, lack of mtDNA integrity may alter cellular bioenergetics and limit efficient differentiation. We previously demonstrated that the derivation of iPSCs is associated with mitochondrial remodeling and a metabolic switch towards glycolysis. Here, we aimed to determine the consequences of reprogramming on mtDNA integrity. Massively parallel pyrosequencing of mtDNA revealed that human iPSCs derived from young healthy donors harbored single base mtDNA mutations (substitutions, insertions, and deletions), both homoplasmic (in all mtDNA molecules) and heteroplasmic (in a fraction of mtDNAs). Interestingly, the level of heteroplasmy varied among iPSC lines derived from the same parental fibroblasts. This phenomenon could potentially be exploited for the generation of mtDNA mutation-free iPSCs from patients with mtDNA disorders. By integrating transcriptional, metabolic, and functional bioenergetic data, we unveiled that iPSC lines bearing different mtDNA mutational loads maintained a consistent hESC-like reprogramming of energy metabolism. This included over-expression of glycolytic enzymes, increased amount of G6P, and elevated protein expression of PDK1, which re-routes the bioenergetic flux towards glycolysis. Overall, although the mtDNA mutations within our iPSCs did not affect the reprogramming-associated metabolic modulation, the occurrence of pathogenic mtDNA modifications might be an important aspect to monitor when characterizing iPSC lines.
Keywords: reprogramming, iPS cells, mtDNA, mutations, metabolism, mitochondria, PDK1
Introduction
Defects in mitochondrial DNA (mtDNA) are a frequent cause of genetic disease, as they are estimated to develop in 1/5000 individuals [1]. Unfortunately, due to the paucity of animal and cellular models, the genotype-phenotype correlation of mitochondrial disorders is not yet entirely elucidated [2]. Induced pluripotent stem cells (iPSCs) [3] derived from individuals with mtDNA disorders might then represent an interesting modeling tool, as they would allow to investigate the interplay between mitochondrial and nuclear genomes and the effects of particular mutations on distinct cellular identities. However, before applying iPSC technology in the context of mtDNA diseases, it is essential to determine the effects of the reprogramming process on mitochondrial genome integrity of healthy somatic cells.
A plethora of recent findings revealed the presence of reprogramming-associated genomic alterations [4–8], suggesting that the derivation of iPSCs may carry the risk of inducing mutations. This is particularly relevant for mtDNA, since it is considered to be highly susceptible to mutagenic events due to the lack of protective histones, the poor fidelity of the DNA polymerase gamma, and the proximity to free radical production [9], which has been found increased during the early steps of reprogramming [10].
Mitochondrial genetics is fundamentally different from Mendelian genetics. mtDNA is transmitted along the maternal lineage and its genes are not transcribed individually but rather as polycistronic RNA precursors [11]. Importantly, the mitochondrial genome exhibits a polyploid nature, i.e. it exists in several copies within every single cell. When all copies of mtDNA are identical, the state is defined as homoplasmy, while heteroplasmy describes the situation in which there is a mixture of two mtDNA genotypes. The heteroplasmic level is of critical significance, as the pathogenic phenotypes are believed to occur only when the number of mutated molecules reach a certain threshold [12]. Finally, mtDNA variants can be unequally distributed during proliferative division. This effect, known as replicative segregation, characterizes the mitochondrial genetic bottleneck, a phenomenon in which mothers harboring a mixture of mutated and wild-type mtDNAs transmit varying proportions of mutated and wild-type mtDNA to their offspring [13–14].
The mitochondrial genome encodes thirteen proteins contributing to Complex I, III, IV and V of the electron transport chain (ETC), which generates ATP through oxidative phosphorylation (OXPHOS) [15]. Hence, mtDNA integrity plays a relevant role for cellular bioenergetics. Large-scale deletions are in fact associated with reduced ATP generation [16–17] and mtDNA diseases often affect tissues with high energy requirements, such as the nervous system and skeletal muscle [12].
We previously demonstrated that human somatic mitochondria undergo a complex remodeling within iPSCs, as they adapt their morphology and functionality to the acquired pluripotent state [18]. These mitochondrial changes impacted also the cellular bioenergetic profile, which shifted from OXPHOS to glycolysis upon reprogramming and returned to OXPHOS during subsequent differentiation [18–19]. Other groups confirmed these results and showed that mitochondria within human iPSCs exhibit low oxidative stress [20] and energetic rejuvenation [21]. Moreover, small molecules inducing metabolic switch towards glycolytic metabolism enhanced reprogramming efficiency [22].
In the present work, we sought to investigate mtDNA integrity in healthy human reprogrammed cells. Since the polyploid nature of mitochondrial genome poses a challenge for conventional sequencing, we employed Next Generation Sequencing (NGS) technology, which enables the discovery of heteroplasmy at high resolution [23–25]. We detected several homoplasmic and heteroplasmic mtDNA mutations within distinct iPSC lines compared to their parental fibroblast cells. Importantly, the level of heteroplasmy varied between distinct iPSC lines obtained from the same parental fibroblasts, suggesting that reprogramming might induce replicative segregation of mitochondria in a fashion resembling the mitochondrial genetic bottleneck of embryonic development. This phenomenon could potentially be exploited for the derivation of mutation-free iPSC lines from patients affected by mtDNA disorders. Finally, by combining transcriptional, metabolic, and functional bioenergetic data, we determined that the detected mtDNA variability among our iPSC lines did not affect the reprogramming-associated re-configuration of energy metabolism. iPSC lines harboring different mtDNA mutations exhibited similar hESC-like bioenergetic profiles and, compared to the parental fibroblasts, showed equivalent hESC-like elevation of glucose-6-phosphate (G6P), the largest hub of carbon metabolism [26], and over-expression of PDK1, which inhibits the entry of pyruvate into the mitochondria, thereby re-routing the energetic flux towards glycolysis [27]. Overall, the findings, although conducted on a limited number of human iPSC lines, indicate that cellular reprogramming might be associated with modifications within the mitochondrial genome and thus suggest an additional aspect to monitor during the production of clinical-grade iPSCs.
Materials and Methods
Culture conditions and derivation of iPSCs
hESC lines H1 and H9 (WiCell) and iPSCs were cultured as previously described [18]. Neonatal foreskin fibroblasts BJ and HFF1 were bought from ATCC (#CRL-2522 and # SCRC-1041, respectively). Adult NFH2 fibroblasts were obtained from a healthy 84 year-old woman, after approval of the study protocol by the ethic committee and written informed consent, at the University Hospital of Dessau (Prigione A, Hossini AM et al, unpublished results). All cultures were kept in a humidified atmosphere of 5% CO2 at 37°C under atmospheric oxygen condition.
HFF1-iPSCs (lines iPS2 and iPS4) were previously generated using the Yamanaka retroviral cocktail [18]. BJ-iPSCs (lines iB4 and iB5) were obtained using the same approach. Induced pluripotency in iB4 and iB5 was confirmed by employing embryoid body (EB)-based in vitro differentiation and the in vivo teratoma assay. Teratoma experiments were carried out by EPO-Berlin Gmbh and histological analysis was carried out by a certified pathologist. Fingerprinting analysis was assayed to verify somatic origin of iPSCs using the primer sets D10S1214 and D21S2055 [18]. For detection of possible karyotype abnormalities in iPSCs, chromosomal analysis after GTG-banding was executed at the Human Genetic Center of Berlin. Derived fibroblasts (DF) were obtained from hESCs and iPSCs as previously indicated [18].
Next Generation Sequencing (NGS) of mtDNA
Generation of an amplicon library
Total DNA was isolated with FlexiGene DNA kit (Qiagen) from BJ (passage 4), HFF1 (passage 4), BJ-derived iB4 and iB5 (both passage 18), and HFF1-derived iPS2 and iPS4 (both passage 18). 50 ng of DNA was used as PCR template. PCR reactions contained 1x Phusion HF Buffer, 3% DMSO and 0.6 Units of proof-reading Phusion Hot Start II High Fidelity Polymerase (all from Finnzymes, Espoo, Finland). The reactions were carried out using Dyad Thermal Cycler (BioRad, Hercules, CA, USA) according to the following program: 98°C for 30s, followed by 30 cycles of 98°C for 10s / 58°C for 30s / 72°C for 30s, and a final extension step at 72°C for 5min. Primers sequences are reported in Supp. Table 1.
454 Sequencing and data analysis
PCR fragments were purified, pooled and universal sequencing adaptors ligated. After emulsion and template annealing, beads were incubated with Bst DNA polymerase, apyrase, and single-stranded binding proteins. Pyrosequencing was performed on a 70x75 mm picotiter plate using a gasket with distinct compartments for physical separation of the samples. The picotiter plate was inserted in the flow cell and subjected to pyrosequencing on the Genome Sequencer FLX instrument (454/Roche), as previously described [28]. For each dNTP flow, a single image was captured by a charge-coupled device (CCD) camera on the sequencer. The images were first analyzed to identify DNA bead-containing wells and to compute associated signal intensities and then processed for chemical and optical crosstalk, phase errors, and read quality, before base calling was carried out for each template bead. After default raw data processing, a re-sequencing trimming filter was applied to increase the data output (parameters doValleyFilterTrimBack = false, vfBadFlowThreshold = 6, vfLastFlowToTest = 168, errorQscoreWindowTrim = 0.01).
Mapping 454 reads to reference genome and detection of variants
0.8 million sequences were generated for a total of 662 million bases. The revised Cambridge Reference Sequence (rCRS) (GenBank accession NC_012920) was used as reference for alignment and detection of variants was performed using the GS Reference Mapper Version 2.3 (Roche). Only the HCDiff (high confidence differences) of the GS Mapper software were used as basis for variant detection. HCDiff callings presumed at least three reads with the variant with both forward and reverse reads. Alternatively, the quality scores at the variable positions must be over 20 (or over 30 if a homopolymer of five or more bases was involved). As additional quality criteria, we used only variants with a coverage >10X of high quality reads. With respect to rCRS, a variant was defined heteroplasmic when its frequency was ≥10% and homoplasmic if its frequency was ≥90% [25]. The following databases were utilized to identify mtDNA mutations and mtDNA haplogroups: mitomap (http://www.mitomap.org), mtDB (http://www.genpat.uu.se/mtDB/), and phyotree (http://www.phylotree.org/).
Global gene expression analysis
Total RNA was quality-checked by Nanodrop analysis (Nanodrop Technologies, Wilmington, DE, USA) and a quantity of 500ng was used as input. Illumina BeadStation 500 platform (Illumina, San Diego, CA, United States) was employed for hybridizations, washing, Cy3-streptavidin staining and scanning. cRNAs derived from the following samples were hybridized in duplicate onto Illumina human-8 BeadChips version 3: BJ, HFF1, H1, H9, iB4, iB5, PS2, and iPS4. Data analysis was carried out using the BeadStudio software 3.0. Genes were considered significantly expressed with detection p values ≤ 0.01, differential p values ≤ 0.01 (Illumina custom method), fold change ratio > 1.5. Heatmaps were generated using Microarray Software Suite TM4 (TMEV.bat). The list of genes involved in cellular bioenergetics was derived from SA Biosciences PCR arrays (Human Glucose Metabolism PCR Array, www.sabiosciences.com). Gene expression results have been deposited in the GEO database (record GSE26575). The following link has been created to allow access: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=jvqjzogammmgkdu&acc=GSE26575.
Liquid chromatography tandem mass spectrometry (LC-MS/MS)
Intermediates of glycolysis and the Pentose Phosphate Pathway (PPP) were quantified by LC-MS/MS as described earlier [29]. In brief, metabolites were extracted in Hanks balanced salt solution (HBSS) containing 2% perchloric acid, and spiked with an isotope-labeled internal standard (13C6-glucose-6P). Proteins were precipitated after neutralization with a phosphate buffer. 10 µL from the clear supernatant was injected onto the LC column. Intermediates of the PPP were separated on a C18 column using a linear water/acetonitrile gradient containing octylammonium acetate (500 mg/l, pH 7.5) as ion pairing reagent, with a flow rate of 1 ml/min. Metabolite detection was performed using an API-3000 tandem mass spectrometer (AB/Sciex), equipped with a Turbo Ion electrospray source operating in the negative mode. Data were obtained by operating the tandem mass spectrometer in the Multiple Reaction Monitoring mode employing (Q1/Q3) transitions as previously indicated [29].
Quantitative real-time PCR
Real-Time PCR was performed in 384 or 96 Well Optical Reaction Plates (Applied Biosystems, Foster City, CA, United States) using SYBR®Green PCR Master Mix (Applied Biosystems). Reactions were carried out on the ABI PRISM 7900HT Sequence Detection System (Applied Biosystems). Triplicate amplifications were carried out for each target gene with three wells serving as negative controls. Quantification was carried out using the comparative Ct method (ABI instruction manual), normalized over GAPDH, and presented as a log2 values with respect to the biological controls. Human Embryonic Stem Cells StellARray™ qPCR Array (Lonza, Cologne, Germany) was utilized used to confirm the expression of key pluripotency-associated genes in BJ-iPSC lines. Values of fold change ratio over somatic fibroblasts were obtained by GPR analysis. mtDNA copy number was quantified using ND5 and CF gene targets for mitochondrial and nuclear genome, respectively, as previously described [30]. The list of all primers used for real-time PCR experiments is reported in Supp. Table 2.
Bioenergetic profiling
Live quantification of cellular and mitochondrial bioenergetics was performed using Seahorse XF96 extra-cellular flux analyzer (Seahorse Bioscience, www.seahorsebio.com), as previously described [31]. Preliminary experiments were carried out to prove that hESCs and iPSCs maintained the expression of key pluripotency-associated proteins when grown on matrigel-coated XF96 well plates (data not shown). Assays were initiated by removing the growth medium and replacing it with unbuffered media. The cells were incubated at 37°C for 30 min to allow media temperature and pH to reach equilibrium before starting the baseline measurement. After having obtained the basal oxygen consumption rate (OCR) and extracellular acidification rate (ECAR), the cells were metabolically perturbed by the additions of three different compounds in succession: oligomycin (1µM), FCCP (0.3 µM), and rotenone (1µM) (all from Sigma). At the end of each assay, cells were trypsinized and the number of viable cells was counted.
Cellular ATP content was determined using the ATPLite bioluminescence luciferase-based assay (Perkin Elmer), as previously described [18]. Briefly, 100.000 cells were used as input and luminescence was quantified with a luminometer (Berthold Technologies), following the manufacturer’s manual. Every sample was measured in triplicate and the results are presented as nM of ATP per cell.
Immunofluorescence and alkaline phosphatase staining
Immunocytochemistry and alkaline phosphatase (AP) (Millipore #SCR004) staining were performed as previously indicated [18]. Primary antibodies included SSEA1, SSEA4, TRA-1-60 and TRA-1-81 from the ESC characterization tool (all 1:100, Millipore #SCR004), NANOG (1:100, Abcam #ab62734), Smooth Muscle Actin (SMA) (1:100, Dako Cytomation #M0851), Alpha Feto Protein (AFP) (1:100, Sigma #WH0000174M1), SOX17 (1:50, R&D #AF1924), PAX6 (1:300, Covance #PRB-278P), NESTIN (1:200, Chemicon #MAB5326), TUJ-1 (1:1000, Sigma #T8660). Secondary antibodies were conjugated with either Alexa 488 or Alexa 594 (Invitrogen #A11001, A11055, A21201, A21468, A11005, A21442). Coverslips were mounted using Dako fluorescent mounting medium (Dako #S3023) and visualized using a confocal microscope LSM 510 (Zeiss).
Western blotting analysis
Total cell protein extracts were obtained using a modified RIPA buffer (50 mM Tris pH 7.4, 100 mM NaCl, 10mM EDTA, 1mM PMSF, 1% IGEPAL) supplemented with complete protease inhibitor cocktail (Roche) added just before use. Protein concentration was determined according to the Bradford method. Equal quantities of proteins were separated by electrophoresis in a 10% SDS-polyacrylamide gel (BioRad, Hercules, CA). Primary antibodies anti-PDK1 (1:1000, Assay Design #KAP-PK112) and GAPDH (1:5000, Ambion #4300) were used with the suitable HRP-conjugated secondary antibodies. Bound antibodies on nitrocellulose membrane (GE Healthcare) were detected using the chemiluminescent substrate ECL (GE Healthcare) and membranes were stripped using Restore Western Blot (Thermo Scientific).
Results
Next Generation Sequencing (NGS) of mtDNA in healthy human iPSCs
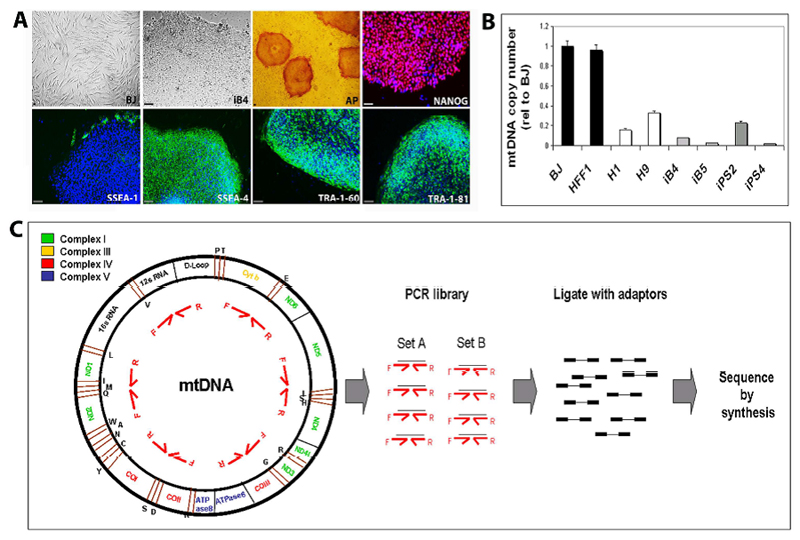
Two sets of healthy human iPSCs, derived from the neonatal foreskin fibroblasts HFF1 and BJ, were employed in this study. HFF1-iPSCs (lines iPS2 and iPS4) were previously generated via retroviral transduction of the Yamanaka cocktail of factors [18]. BJ-iPSCs (lines iB4 and iB5) were obtained in a similar fashion and underwent full characterization. BJ-iPSCs expressed hESC-specific protein markers (Fig. 1A), were karyotypically normal (Supp. Fig.1A), exhibited the same genetic fingerprinting pattern of BJ fibroblasts (Supp. Fig.1B), and efficiently differentiated into the three germ layers both in vitro (Supp. Fig.1C) and in vivo (Supp. Fig. 1D). BJ-iPSCs also acquired a transcriptional signature similar to that of the hESC lines H1 and H9 (Pearson correlation value: 0.8309) and distinct from that of fibroblasts (Pearson correlation value: 0.6660) (Supp. Fig. 2A), and expressed key pluripotency-associated genes (Supp. Fig. 2B and 2C). Finally, before analyzing the mtDNA sequence of all iPSC lines and corresponding fibroblasts, we confirmed that BJ-iPSCs, like HFF1-iPSCs, displayed a decreased mtDNA copy number compared to the parental cells, in agreement with previous findings [18, 20] (Fig. 1B).
Figure 1. Derivation of BJ-iPSCs and mtDNA sequencing.
(A) Two BJ-iPSC lines (iB4 and iB5) were generated from neonatal foreskin BJ fibroblasts by transduction of the OKSM retroviral cocktail. iB4 and iB5 exhibited hESC morphology, expression of alkaline phosphatase (AP), NANOG, SSEA-4, TRA-1-60, TRA-1-81, and lack of expression of SSEA-1. Scale bars represent 10µm (B) Quantification of mtDNA copy number in neonatal human fibroblasts (BJ and HFF1), hESCs (H1 and H9), BJ-iPSCs (iB4 and iB5), and HFF1-iPSCs (iPS2 and iPS4). Error bars indicate standard deviation. (C) Schematic steps of the NGS-based approach employed for mtDNA sequencing of fibroblasts (HFF1 and BJ), HFF1-derived iPSCs (iPS2 and iPS4), and BJ-derived iPSCs (iB4 and iB5).
mtDNA sequencing was carried out employing Roche 454 sequencing, which showed similar reliability coupled with higher sensitivity for the discovery of low level heteroplasmies in comparison to Sanger sequencing [25]. An amplicon library was first created using two sets of 25 primer pairs (Primer Set A and B, Supp. Table 1), each set designed to encompass the entire mitochondrial genome [23]. All primers successfully generated single amplicons of about 650 base pair (Supp. Fig.3). PCR fragments were then purified, pooled, ligated with universal sequencing adaptors, and subjected to massively parallel pyrosequencing (Fig. 1C). The 454 sequencing output had an average read length of 405 bases per read and an average sequencing content of 96.5 megabases per mtDNA genome. More than 99.6 % of all reads mapped to the mitochondrial reference sequence (revised Cambridge Reference Sequence, rCRS) with a total average coverage per base of 5937 X. This led to a single continuous sequence contig of 16560 bases per sample, thus reaching complete covering of the mitochondrial target region (Supp. Table 3). After having determined all mtDNA variants in the individual samples using the rCRS as a reference, we compared each iPSC line to the respective fibroblasts (BJ for iB4 and iB5 and HFF1 for iPS2 and iPS4) in order to identify mtDNA changes occurring upon reprogramming.
Homoplasmic and heteroplasmic mtDNA mutations in human iPSCs
The mitochondrial genome of the four iPSC lines did not exhibit signs of large-scale re-arrangement, such as long insertions or long deletions. However, several point mutations were detected within iPSCs compared to their parental cells (Table 1). Interestingly, the two sets of iPSCs showed a different pattern of mutations and we could not identify any variant consistently present in all reprogrammed cells and not in fibroblasts (Supp. Table 4 and 5). The number of mtDNA mutations also differed between the two sets, being about 20 in BJ-iPSCs and around 80 in HFF1-iPSCs (Table 1). The reason for this variability is not clear and could be due to features of the cell type of origin or to reprogramming-related factors, since the two sets of iPSCs were derived at different times with different retroviral batches. Future studies are warranted to determine whether this mtDNA sequence variability is a common aspect of all iPSCs or it is influenced by differences in the cellular source or in the reprogramming strategy employed.
Table 1. Summary of mtDNA mutations identified within iPSCs.
| iB4 | iB5 | iPS2 | iPS4 | |||
|---|---|---|---|---|---|---|
| Frequency | ||||||
| Homoplasmies | ||||||
| Base substitution | 2 | 1 | 53 | 55 | ||
| Single insertion | 0 | 1 | 3 | 2 | ||
| Single deletion | 0 | 1 | 3 | 3 | ||
| Total | 2 | 3 | 59 | 60 | ||
| Heteroplasmies | ||||||
| Maintained (+/- 5%) | 4 | 5 | 0 | 2 | ||
| Increased | 7 | 6 | 9 | 8 | ||
| Decreased | 6 | 9 | 12 | 12 | ||
| Total | 17 | 20 | 21 | 22 | ||
| TOTAL | 19 | 23 | 80 | 82 | ||
| Nature | ||||||
| Non coding | ||||||
| Control region | 1 | 1 | 20 | 19 | ||
| tRNAs, sRNAs | 9 | 10 | 16 | 17 | ||
| Total | 10 | 11 | 36 | 36 | ||
| Synonymous | ||||||
| Complex I | 2 | 2 | 10 | 11 | ||
| Complex III | 1 | 1 | 3 | 3 | ||
| Complex IV | 0 | 0 | 7 | 7 | ||
| Complex V | 0 | 0 | 2 | 2 | ||
| Total | 3 | 3 | 22 | 23 | ||
| Non-synonymous | ||||||
| Complex I | 2 | 5 | 13 | 14 | ||
| Complex III | 0 | 0 | 2 | 2 | ||
| Complex IV | 2 | 2 | 4 | 4 | ||
| Complex V | 2 | 2 | 3 | 3 | ||
| Total | 6 | 9 | 22 | 23 | ||
| TOTAL | 19 | 23 | 80 | 82 | ||
| Relevance | ||||||
| Reported | ||||||
| Disease associated | 2 | 3 | 0 | 0 | ||
| Cancer associated | 3 | 3 | 4 | 4 | ||
| Common variants | 12 | 13 | 72 | 73 | ||
| Not reported | 7 | 10 | 8 | 9 | ||
| TOTAL | 19 | 23 | 80 | 82 | ||
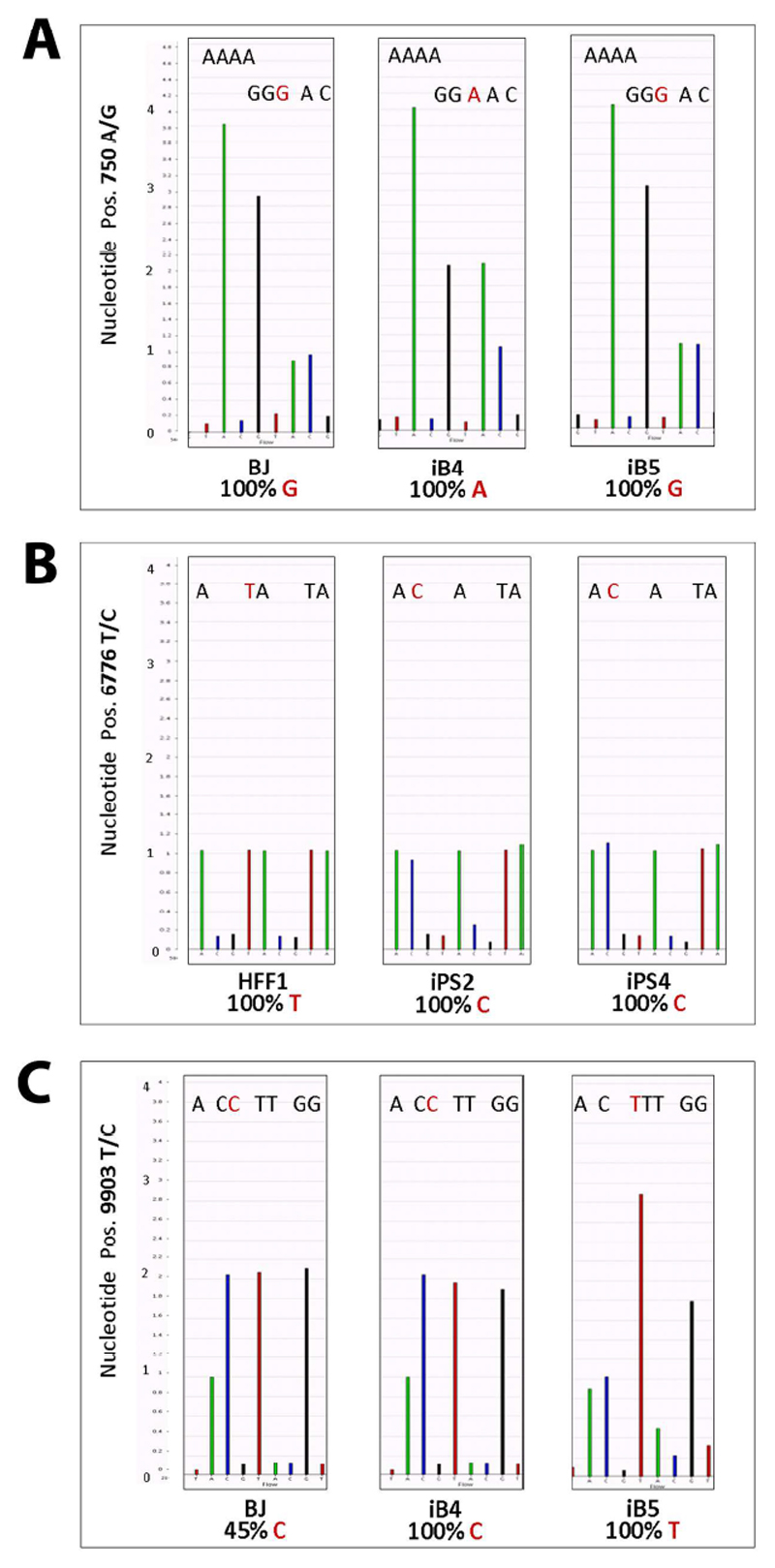
The various mtDNA mutations were classified according to their frequency in homoplasmic and heteroplasmic mutations (Table 1). Homoplasmic variants included single base substitutions, single base insertions, and single base deletions (Supp. Table 4). Base substitutions were highly frequent within HFF1-iPSCs while the other types of homoplasmic or heteroplasmic variants occurred at approximately equal numbers in all iPSC lines (Table 1). In some cases, such as position 750 for the BJ set, fibroblasts contained mtDNA variants with respect to the rCRS and, upon reprogramming, these variants could be either maintained or lost in different iPSC lines (Fig. 2A and Supp. Table 4). In other cases, such as position 6776 for the HFF1 set, iPSCs acquired homoplasmic mtDNA mutations not present in the parental somatic cells (Fig. 2B and Supp. Table 4). Interestingly, base substitutions affected also nucleotides critical for the assignment to specific mitochondrial haplogroups [32]. For example, in BJ fibroblasts, whose haplogroup was calculated to be K1c1, the nucleotide in the haplogroup-defining position 3480 was G, while it became A in iB4 and was deleted in iB5 (Supp. Table 4).
Figure 2. Homoplasmic and heteroplasmic mtDNA mutations within reprogrammed cells.
Exemplary modifications of the mitochondrial genome are reported. (A) The nucleotide at position 750 was homoplasmic for G in BJ fibroblasts. This represented a variant with respect to the rCRS, which exhibits an A. Upon reprogramming, this variant was maintained in the iPSC line iB5. Conversely, a base substitution occurred in iB4, as the reprogrammed cells lost the variant and re-acquired the wild-type nucleotide A. (B) The mtDNA sequence of HFF1 fibroblasts contained a T in the nucleotide position 6776, similarly to the rCRS. However, both HFF1-derived iPSCs displayed a homoplasmic T>C mutation. (C) BJ fibroblasts exhibited a heteroplasmic T>C variant (45% C and 55% T) at the nucleotide position 9903. The level of heteroplasmy varied among the BJ-derived iPSC lines, as it increased in iB4 (100% C and 0% T) and decreased in iB5 (0% C and 100% T). Thus, this latter iPSC line lost the heteroplasmic mtDNA variant originally present in the fibroblasts and acquired a mtDNA sequence profile similar to the rCRS.
The level of heteroplasmy of heteroplasmic variants within fibroblasts remained unaltered (+/-5%) in the reprogrammed cells only in a small percentage of cases, while most frequently it sustained a consistent change (Table 1 and Supp. Table 5). For example, at nucleotide position 9903, BJ fibroblasts showed a heteroplasmic variant whose level of heteroplasmy was 45%. This level increased to 100% in iB4 and decreased to 0% in iB5 (Fig.2C and Supp. Table 5). In addition, iPSCs exhibited heteroplasmic mutations that were not contained in the original fibroblast samples. At the nucleotide at position 16126, for example, HFF1 fibroblasts were homoplasmic for T, while HFF1-iPSCs acquired heteroplasmic T>C mutations (48% C and 50% C in iPS2 and iPS4, respectively) (Supp. Table 5).
mtDNA mutations within iPSCs affected both non-coding genes, including components of the control region (CR) or of the translational machinery, such as tRNA, 12sRNA, and 16sRNA, and genes encoding for proteins contributing to the ETC (Table 1, Supp. Table 4 and 5). In all iPSC lines, a number of coding mutations were synonymous, thus not functionally relevant. The non-synonymous mutations, which gave rise to an amino acid change, affected genes encoding for proteins part of all the four mitochondrial-encoded ETC complexes (Table 1). The non-synonymous/synonymous ratio was 2 for iB4, 3 for iB5, and 1 for iPS2 and iPS4 (Table 1, Supp. Table 4 and 5).
The majority of mtDNA variants within our reprogrammed cells are considered to be ´common variants` in the general populations. Of these, only a very small number appeared to be associated with diseases or cancer, based on literature (Table 1, Supp. Table 4 and 5). On the other hand, some mutations, mostly heteroplasmies, could not be found in any of the mtDNA databases listing all possible mtDNA sequence variants. Future studies will have to address the possible relevance of these unknown mutations in cellular reprogramming.
mtDNA sequence variability does not affect the hESC-like reprogramming of energy metabolism
We previously demonstrated that the process of deriving iPSC induces mitochondria remodeling and metabolic re-configuration from OXPHOS to glycolysis [18]. In view of this, we aimed to determine whether the differences in the mtDNA profile among iPSC lines could affect the reprogramming of energy metabolism.
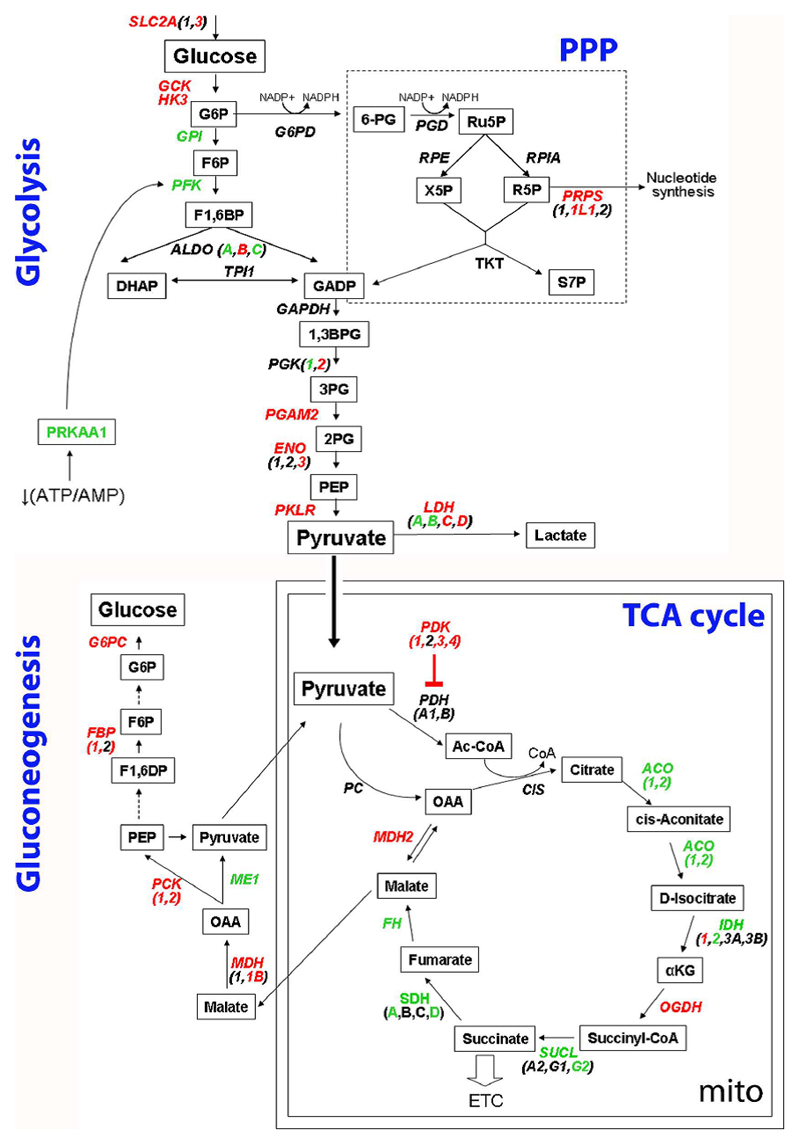
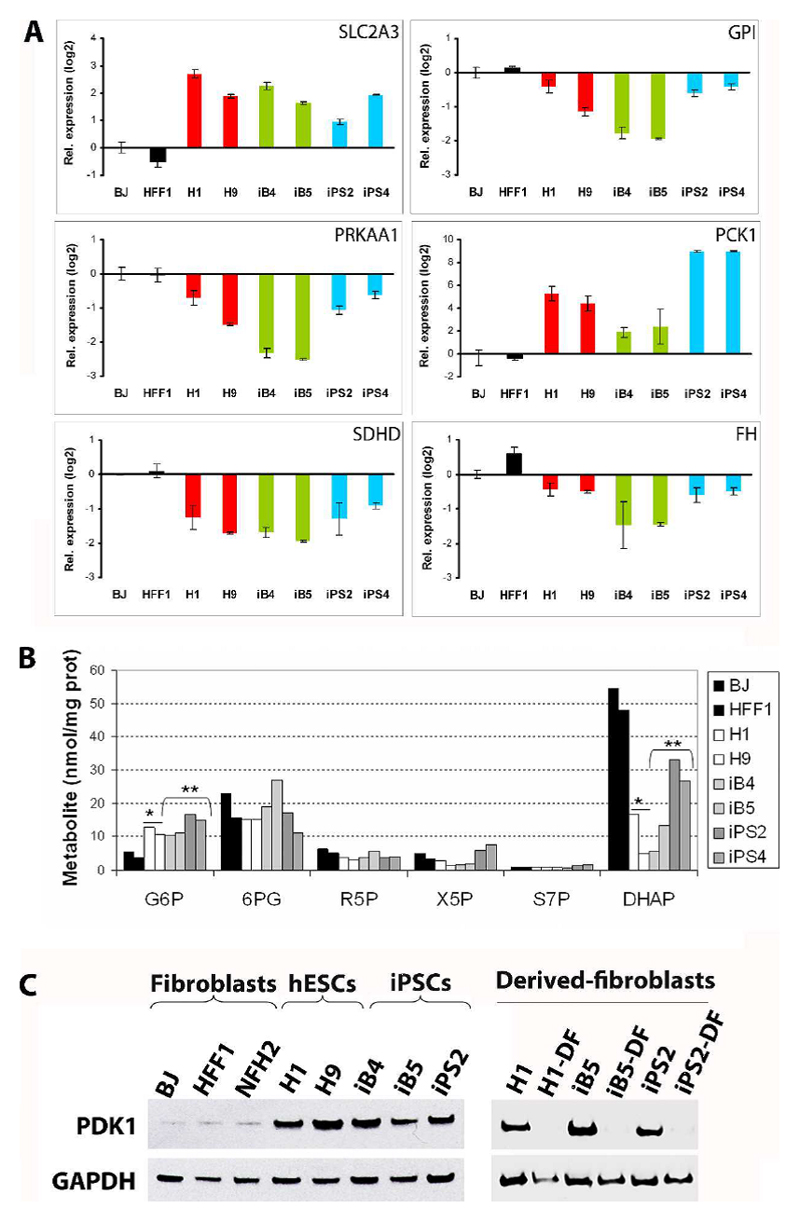
At the transcriptional level, we detected a similar up-regulation of genes regulating the first steps of the glycolytic cascade in all iPSCs and hESCs in comparison to fibroblasts. These included SLC2A3, encoding GLUT3 which is responsible for the up-take of glucose into the cell (Fig. 3, Fig. 4A, Supp. Fig.4), GCK and HK3, whose products are enzymes phosphorylating glucose to glucose-6-phosphate (G6P) (Fig. 3, Supp. Fig.4). Genes of the last steps of glycolysis, such as PGAM2, ENO, PKLR, and LDH, were also mainly up-regulated in both hESCs and all iPSCs (Fig. 3, Supp. Fig.4). On the other hand, the expression of G6P downstream glycolytic enzymes, including GPI (Fig. 3, Fig. 4A, Supp. Fig.4), PFK, and ALDO (Fig. 3, Supp. Fig.4), was reduced in pluripotent stem cells compared to fibroblasts. These results might imply that in hESCs and iPSCs, regardless of the presence of mtDNA mutations, there might be a parallel induction of the first and last steps of glucose metabolism and a possible accumulation of G6P, which may be diverted into the pentose phosphate pathway (PPP) [33].
Figure 3. Similar ESC-like transcriptional reprogramming of energy metabolism in iPSCs regardless of mtDNA variability.
Transcriptional re-configuration of glucose metabolism was determined by interrogating the microarray data for genes differentially regulated in hESCs or in iPSCs compared to fibroblasts (fold change 1.5, detection p value ≤0.01, and differential p value ≤0.01). Genes encoding for enzymes and proteins involved in the metabolic cascade are shown in italics. Upon its uptake into the cell, glucose can follow the glycolytic pathway or can be diverted into the pentose phosphate pathway (PPP). Once pyruvate has been generated, it can either be converted into lactate or enter the mitochondria to take part into the tricarboxylic acid cycle (TCA) cycle and mitochondrial respiration. Finally pyruvate or malate can be transferred outside the mitochondria to generate new glucose. Genes consensually up-regulated in all hESCs and iPSC lines compared to fibroblasts are indicated in red, whereas genes down-regulated in all pluripotent stem cells are presented in green.
Figure 4. Comparable reprogramming of energy metabolism in iPSCs harboring various mtDNA mutations at the transcriptional, metabolic, and protein level.
(A) Quantitative real-time PCR experiments were carried out to detect the expression levels of key genes involved in energy metabolism. The results confirmed the microarray analysis and showed consensual hESC-like regulation in all iPSC lines compared to fibroblasts. Data are presented as log2 relative expression with respect to BJ fibroblasts. Error bars indicate standard deviation. (B) LC-MS/MS- based quantification of sugar phosphate intermediates of the glycolytic cascade and PPP: glucose-6-phosphate/fructose 6-phosphate (G6P), 6-phosphogluconate (6PG), ribose-5-phosphate (R5P), xylulose-5-phosphate/ribulose 5-phosphate (X5P), sedoheptulose-7-phosphate (S7P), and dihydroxyacetone phosphate (DHAP). *two-tailed Unpaired Student’s t test between hESCs and fibroblasts: p=0.029 for G6P and 0.026 for DHAP. **two-tailed Unpaired Student’s t test between iPSCs and fibroblasts: p=0.018 for G6P and 0.030 for DHAP. (C) PDK1 protein levels were monitored by western-blot using GAPDH as loading control protein. PDK1 expression was low in neonatal fibroblasts (BJ and HFF1) and adult fibroblasts (NFH2), but increased in hESCs (H1 and H9), BJ-iPSCs (iB4, iB5), and HFF1-iPSCs (iPS2). Subsequent differentiation into fibroblast-like cells was carried out for H1, iB5 (bearing low mtDNA mutational load), and iPS2 (harboring high levels of mtDNA variants). All derived-fibroblasts (DF) showed reduced PDK1 protein expression compared to cells in the undifferentiated state.
Liquid chromatography-tandem mass spectrometry (LC-MS/MS) was employed to simultaneously analyze the intracellular sugar phosphate intermediates of the glycolytic cascade and PPP. G6P, the largest hub of carbon metabolism, was confirmed to be significantly increased in all hESCs and iPSCs compared to fibroblasts (Fig. 4B). In addition, dihydroxyacetone phosphate (DHAP) was reduced in all pluripotent stem cells, possibly implying an increased flux towards pyruvate generation (Fig. 4B).
Most of the genes encoding for enzymes of the gluconeogenesis pathway were up-regulated in all pluripotent stem cells, including MDH1B, FBP1, G6PC (Fig. 3, Supp. Fig.4) and PCK1 (Fig. 3, Fig. 4A, Supp. Fig.4). Conversely, genes of the mitochondrial tricarboxylic acid cycle (TCA) cycle, such as ACO, IDH2, SUCLG2, SDH (A and D) (Fig. 3, Supp. Fig.4), and FH (Fig. 3, Fig. 4A, Supp. Fig.4) were mainly down-regulated in hESCs and iPSCs in comparison to fibroblasts.
Among the regulators of the bioenergetic metabolism, we observed that all pluripotent stem cells showed down-regulation of PRKAA1 (Fig. 3, Fig. 4A, Supp. Fig.4), which encodes for the catalytic subunit of AMP-activated protein kinase (AMPK) and has been found induced in response to decreased ATP content in mitochondrial-defective cells [16]. It is tempting to speculate that pluripotent stem cells might suppress the activation of AMPK, a metabolic checkpoint switching-off biosynthetic pathways, in order to avoid anabolic inhibition, similarly to cancer cells [26, 34].
A second metabolic regulator, pyruvate dehydrogenase kinase (PDK1, 3, and 4), was up-regulated in hESCs and all iPSCs despite mtDNA sequence variants (Fig. 3, Supp. Fig.4). PDK1 is of particular interest since it inhibits pyruvate dehydrogenase (PDH), thus limiting the pyruvate entry into the TCA cycle [27]. Western blot analysis confirmed increased PDK1 protein expression in hESCs and iPSCs compared to neonatal fibroblasts (HFF1 and BJ) and to adult fibroblasts (NFH2) (Fig. 4C). Moreover, PDK1 protein levels decreased during subsequent differentiation into derived fibroblasts (DF) obtained from hESCs (H1 line), from iPSCs with a low number of mtDNA mutations (iB5 line), and from iPSCs with a high number of mtDNA variants (iPS2 line) (Fig. 4C).
Taken together, regardless of variations within mtDNA sequence, all iPSCs acquired a hESC-like transcriptional and metabolic signature indicative of a metabolic transition from respirative to glycolytic metabolism.
Similar ESC-like bioenergetic profiles in iPSCs harboring different mtDNA mutations
Finally, we asked whether iPSC lines with comparable metabolic reprogramming could be bioenergetically similar despite differences in mtDNA mutational load.
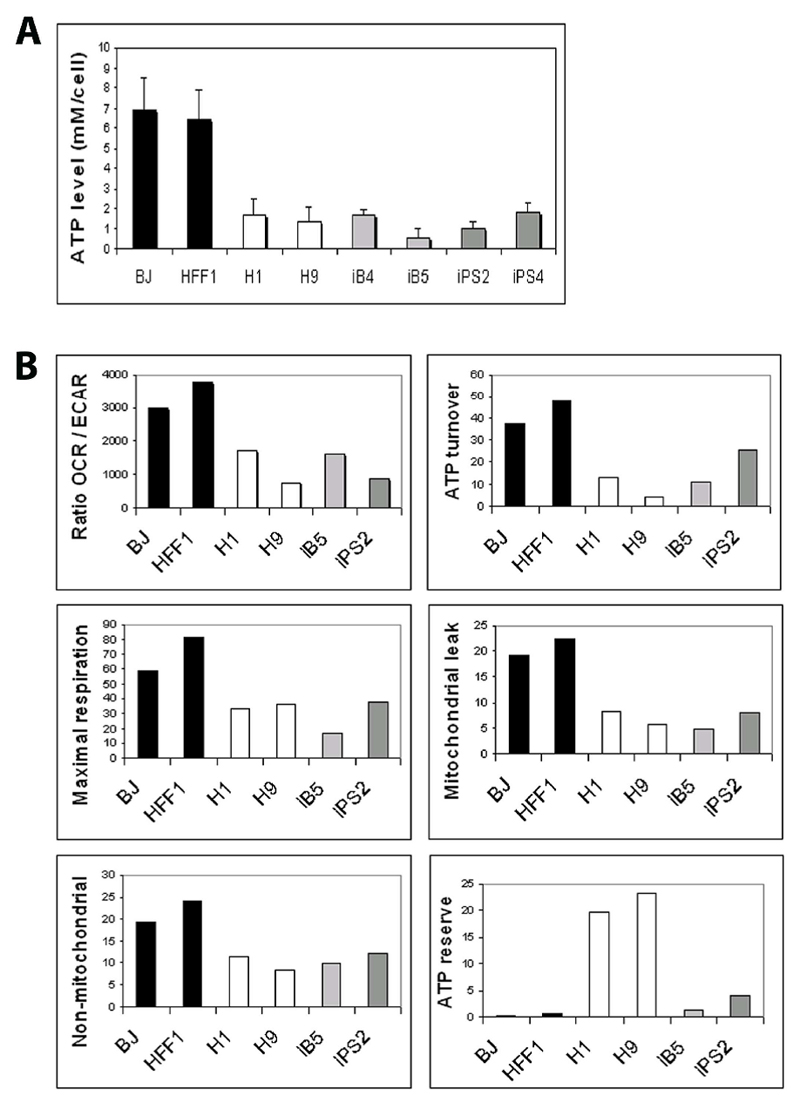
The level of total cellular ATP in all iPSCs was equal to that of hESCs and consensually reduced compared to fibroblasts (Fig. 5A), in agreement with previous findings [18]. Simultaneous quantification of mitochondrial respiration (oxygen consumption rate, OCR) and glycolysis (extracellular acidification rate, ECAR) revealed that hESCs and iPSCs with either low (iB5) or high (iPS2) mitochondrial mutational load (Table 1) exhibited an equivalent glycolytic switch, as indicated by the reduction of the OCR/ECAR ratio in comparison to fibroblast cells (Fig. 5B).
Figure 5. Bioenergetic profile of iPSCs bearing different mtDNA mutational load.
(A) Total cellular ATP in fibroblasts, hESCs, and iPSCs was quantified with a bioluminescence luciferase-based assay. All iPSC lines showed a similar hESC-like reduction of ATP level compared to the parental fibroblasts. Error bars indicate standard deviation (B) Bioenergetic profiling was performed using Seahorse XF96 analyzer. A reduction of the ratio between oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) in hESCs and iPSCs indicated a parallel shift from respirative to fermentative metabolism in all pluripotent stem cells compared to fibroblasts. By monitoring the OCR values upon metabolic perturbations with subsequent introduction of three mitochondrial inhibitors (oligomycin, FCCP, and rotenone), we derived estimates of the following parameters of mitochondrial bioenergetics: ATP turnover, maximal respiration capacity, mitochondrial leak, non-mitochondrial oxygen consumption, and ATP reserve. All parameters, except ATP reserve were similar in all hESCs and iPSCs and distinct from fibroblasts.
Metabolic perturbation with subsequent application of different mitochondrial inhibitors was carried out to shift the energy profile and obtain estimates of different parameters of mitochondrial bioenergetics (Supp. Fig. 5). The ATP turnover, the maximal respiration rate, the magnitude of the proton leak, the amount of non-mitochondrial oxygen consumption, and the level of ATP reserve were all equivalent among iPSC lines bearing different mtDNA mutation levels (Fig. 5B). Interestingly, we observed that the acidification rate increased in fibroblasts but not in hESCs and iPSCs, following the exposure to the respiration-blocker oligomycin. Hence, pluripotent stem cells might fully utilize their glycolytic resources and may not be able to further increase glycolysis upon the blockage of mitochondrial activity (Supp. Fig.3).
Overall, the bioenergetic profile of iPSCs appeared highly similar to the one of hESCs and drastically distinct from their parental somatic cells (Fig. 5B). Nonetheless, the level of ATP reserve in iPSC was comparable to the one of fibroblasts and much lower than the one exhibited by hESCs (Fig. 5B), potentially suggesting that hESCs and iPSCs might exhibit a different ability in responding to increased energy demands.
Discussion
Genetic defects of the human mitochondrial genome form the basis of classical mitochondrial syndromes [12]. In addition, growing evidence demonstrates their involvement in the pathology of several other conditions, including diabetes, aging, and neurodegeneration [2, 35–36]. mtDNA mutations have also been associated with oncogenic transformation [37] and malignant progression [38], although their overall role in cancer remains controversial [39–40]. In this work, we have investigated whether cellular reprogramming might induce mitochondrial mutagenesis in healthy human somatic cells.
We found that distinct iPSC lines harbored various mtDNA point mutations not present in parental fibroblasts. The ratio of non-synonymous versus synonymous mutations was always equal to one or higher, possibly implying positive selection. Most iPSC-specific mutations were listed as ´common variants` in the general population. Cancer-associated mutations showed similar features and have been suggested to represent mild mtDNA alterations helping the cells to adapt to a new environment, as they have been adaptive during population migration [39]. Accordingly, we found that variations in iPSCs could also occurr at haplogroup-defining nucleotide positions.
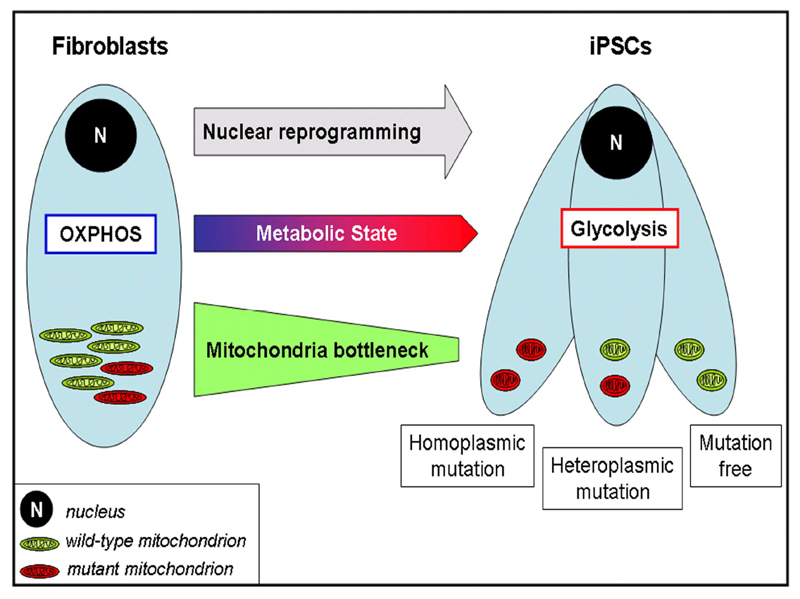
The heteroplasmy level varied among iPSC lines derived from the same parental cells. Hence, during the cellular replication events accompanying reprogramming, a mitochondrial segregation might occur and the derived cells could harbor a reduced number of mtDNA molecules with variable ratio between wild-type and mutant (Fig. 6). This bottleneck effect might potentially allow the derivation of mtDNA mutation-free iPSC lines from individuals affected by mtDNA disorders. However, to achieve this, it might be necessary to screen a vast number of cell lines and confirm that new mutations are not introduced upon long-term culturing.
Figure 6. mtDNA segregation during iPSC derivation.
Generation of iPSCs reprograms somatic nuclei to an embryonic-like epigenetic state. Concurrently, the metabolic state is re-configured and displays a switch from OXPHOS to glycolysis. During this process, mitochondria undergo replicative segregation, as they decrease in number and show random distribution of wild-type and mutant mtDNA molecules within the daughter cells (bottleneck effect). The resulting iPSCs could contain similar levels of mtDNA variants as the parental somatic cells or, alternatively, increased or reduced mtDNA mutational load. Hence, in the case of somatic cells harboring pathogenic mtDNA mutations, the random drift of mitochondria associated with the reprogramming-induced bottleneck might allow the derivation of iPSC lines free of mutations.
Genome-wide sequencing of protein coding regions recently revealed the presence of various point mutations in human iPSCs [7]. Given that mitochondrial genome undergo higher mutation rates compared to chromosomal DNA [41], it is perhaps not surprising that we have detected mtDNA mutations within different human iPSC lines. At the same time, it is necessary to investigate the functional significance of these mutations. Our data, combining transcriptional, metabolic, and functional data, revealed that the mtDNA mutations within our iPSCs did not affect their bioenergetic properties. Nonetheless, given the random drift of mitochondria and the consequent variance in the substitution profile, it is likely that other iPSC lines might harbor different alterations of the mitochondrial genome. This is relevant, since pathogenic mtDNA mutations and lack of mtDNA integrity can affect cellular energy capacities [16–17] and compromise efficient stem cell differentiation [42–43].
By monitoring the energetic consequences of mtDNA sequence variability in iPSCs, we gained new insights into the reprogramming-associated re-configuration of the metabolic state (Fig. 6). First, G6P was increased in hESCs and iPSCs in comparison to fibroblasts, which might suggest that pluripotent stem cells divert their energy flux from the glycolytic path into the pentose phosphate pathway (PPP). This may have significant advantages, since the PPP not only provides nucleotide precursors essential for anabolic growth, but also maintains cellular antioxidant defenses [33, 44]. Furthermore, the protein expression level of PDK1, which promotes glycolytic metabolism by inhibiting the pyruvate entry in the TCA cycle [27], was consensually elevated in undifferentiated hESCs and iPSCs. In accordance, recent data showed that small molecules activating PDK1 significantly enhanced cellular reprogramming [22].
Mitochondria within all pluripotent stem cells possessed high coupling efficiency, implying that, although reduced in number and less employed for cellular energetics, they are still capable of competent energy generation. This is in agreement with recent findings showing that mitochondrial OXPHOS is required for the proliferation of self-renewing hESCs [45–46]. On the other hand, iPSCs showed reduced levels of ATP reserve in comparison to hESCs. Thus, iPSCs might not acquire the same ability of hESCs to respond to increased energy demands, which could occur during differentiation. Indeed, differentiated cells from iPSCs exhibited high variability and premature senescence [47–48]. Our data, demonstrating heterogenic mtDNA rearrangement coupled with a lack of acquisition of high ATP reserve within iPSCs, may help to explain this variability. Perhaps reprogrammed cells might retain not only an epigenetic [49–50] but also a bioenergetic memory of their somatic origin. Accordingly, somatic cell nuclear transfer (SCNT), which does not involve somatic mitochondria and does not require bioenergetic reprogramming, showed low heterogeneity and a more faithful establishment of pluripotency [49, 51].
Overall, our findings demonstrate the presence of various homoplasmic and heteroplasmic mtDNA mutations within human iPSCs and suggest the integrity of the mitochondrial genome as an additional aspect to be assessed before employing iPSC lines for modeling or clinical applications.
Supplementary Material
Acknowledgements
The authors would like to thank all colleagues in the Adjaye lab for fruitful discussion, A. Sabha and C. Vogelgesang at the microarray facility, Dr. A. Wulf-Goldenberg and M. Keil (EPO-Berlin Gmbh) for the teratoma assay, Dr. G. Thiel (Human Genetic Center of Berlin) for the karyotypic analysis, and E. Jansen and Prof. C. Jakobs from VUMC for help with mass spectrometry analysis. We are grateful to Dr. D. Ferrick, Dr. J. Dunn, and H. Hedeby from Seahorse Bioscience for scientific and technical support.The authors declare no competing financial or commercial interests and acknowledge support from the BMBF (01GN0807) and the Max Planck Society.
Footnotes
Disclaimers and acknowledgment: The authors declare no competing financial or commercial interests and acknowledge support from the BMBF (01GN0807) and the Max Planck Society.
References
- 1.Schaefer AM, McFarland R, Blakely EL, et al. Prevalence of mitochondrial DNA disease in adults. Ann Neurol. 2008;63:35–9. doi: 10.1002/ana.21217. [DOI] [PubMed] [Google Scholar]
- 2.Taylor RW, Turnbull DM. Mitochondrial DNA mutations in human disease. Nat Rev Genet. 2005;6:389–402. doi: 10.1038/nrg1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 4.Laurent LC, Ulitsky I, Slavin I, et al. Dynamic changes in the copy number of pluripotency and cell proliferation genes in human ESCs and iPSCs during reprogramming and time in culture. Cell Stem Cell. 2011;8:106–18. doi: 10.1016/j.stem.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mayshar Y, Ben-David U, Lavon N, et al. Identification and classification of chromosomal aberrations in human induced pluripotent stem cells. Cell Stem Cell. 2010;7:521–31. doi: 10.1016/j.stem.2010.07.017. [DOI] [PubMed] [Google Scholar]
- 6.Pasi CE, Dereli-Oz A, Negrini S, et al. Genomic instability in induced stem cells. Cell Death Differ. 2011 doi: 10.1038/cdd.2011.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gore A, Li Z, Fung HL, et al. Somatic coding mutations in human induced pluripotent stem cells. Nature. 2011;471:63–7. doi: 10.1038/nature09805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hussein SM, Batada NN, Vuoristo S, et al. Copy number variation and selection during reprogramming to pluripotency. Nature. 2011;471:58–62. doi: 10.1038/nature09871. [DOI] [PubMed] [Google Scholar]
- 9.Brown WM, George M, Jr, Wilson AC. Rapid evolution of animal mitochondrial DNA. Proc Natl Acad Sci U S A. 1979;76:1967–71. doi: 10.1073/pnas.76.4.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Esteban MA, Wang T, Qin B, et al. Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell Stem Cell. 2010;6:71–9. doi: 10.1016/j.stem.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 11.Clayton DA. Replication and transcription of vertebrate mitochondrial DNA. Annu Rev Cell Biol. 1991;7:453–78. doi: 10.1146/annurev.cb.07.110191.002321. [DOI] [PubMed] [Google Scholar]
- 12.DiMauro S, Schon EA. Mitochondrial respiratory-chain diseases. N Engl J Med. 2003;348:2656–68. doi: 10.1056/NEJMra022567. [DOI] [PubMed] [Google Scholar]
- 13.Cree LM, Samuels DC, de Sousa Lopes SC, et al. A reduction of mitochondrial DNA molecules during embryogenesis explains the rapid segregation of genotypes. Nat Genet. 2008;40:249–54. doi: 10.1038/ng.2007.63. [DOI] [PubMed] [Google Scholar]
- 14.Wai T, Teoli D, Shoubridge EA. The mitochondrial DNA genetic bottleneck results from replication of a subpopulation of genomes. Nat Genet. 2008;40:1484–8. doi: 10.1038/ng.258. [DOI] [PubMed] [Google Scholar]
- 15.Anderson S, Bankier AT, Barrell BG, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–65. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 16.Prigione A, Cortopassi G. Mitochondrial DNA deletions induce the adenosine monophosphate-activated protein kinase energy stress pathway and result in decreased secretion of some proteins. Aging Cell. 2007;6:619–30. doi: 10.1111/j.1474-9726.2007.00323.x. [DOI] [PubMed] [Google Scholar]
- 17.Porteous WK, James AM, Sheard PW, et al. Bioenergetic consequences of accumulating the common 4977-bp mitochondrial DNA deletion. Eur J Biochem. 1998;257:192–201. doi: 10.1046/j.1432-1327.1998.2570192.x. [DOI] [PubMed] [Google Scholar]
- 18.Prigione A, Fauler B, Lurz R, et al. The senescence-related mitochondrial/oxidative stress pathway is repressed in human induced pluripotent stem cells. Stem Cells. 2010;28:721–33. doi: 10.1002/stem.404. [DOI] [PubMed] [Google Scholar]
- 19.Prigione A, Adjaye J. Modulation of mitochondrial biogenesis and bioenergetic metabolism upon in vitro and in vivo differentiation of human ES and iPS cells. Int J Dev Biol. 2010;54:1729–41. doi: 10.1387/ijdb.103198ap. [DOI] [PubMed] [Google Scholar]
- 20.Armstrong L, Tilgner K, Saretzki G, et al. Human induced pluripotent stem cell lines show stress defense mechanisms and mitochondrial regulation similar to those of human embryonic stem cells. Stem Cells. 2010;28:661–73. doi: 10.1002/stem.307. [DOI] [PubMed] [Google Scholar]
- 21.Suhr ST, Chang EA, Tjong J, et al. Mitochondrial rejuvenation after induced pluripotency. PLoS One. 2010;5:e14095. doi: 10.1371/journal.pone.0014095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu S, Li W, Zhou H, et al. Reprogramming of human primary somatic cells by OCT4 and chemical compounds. Cell Stem Cell. 2010;7:651–5. doi: 10.1016/j.stem.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He Y, Wu J, Dressman DC, et al. Heteroplasmic mitochondrial DNA mutations in normal and tumour cells. Nature. 2010;464:610–4. doi: 10.1038/nature08802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li M, Schonberg A, Schaefer M, et al. Detecting heteroplasmy from high-throughput sequencing of complete human mitochondrial DNA genomes. Am J Hum Genet. 2010;87:237–49. doi: 10.1016/j.ajhg.2010.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zaragoza MV, Fass J, Diegoli M, et al. Mitochondrial DNA variant discovery and evaluation in human Cardiomyopathies through next-generation sequencing. PLoS One. 2010;5:e12295. doi: 10.1371/journal.pone.0012295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tennant DA, Duran RV, Gottlieb E. Targeting metabolic transformation for cancer therapy. Nat Rev Cancer. 2010;10:267–77. doi: 10.1038/nrc2817. [DOI] [PubMed] [Google Scholar]
- 27.Kim JW, Tchernyshyov I, Semenza GL, et al. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006;3:177–85. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 28.Querings S, Altmuller J, Ansen S, et al. Benchmarking of mutation diagnostics in clinical lung cancer specimens. PLoS One. 2011;6:e19601. doi: 10.1371/journal.pone.0019601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ralser M, Wamelink MM, Latkolik S, et al. Metabolic reconfiguration precedes transcriptional regulation in the antioxidant response. Nat Biotechnol. 2009;27:604–5. doi: 10.1038/nbt0709-604. [DOI] [PubMed] [Google Scholar]
- 30.Wong A, Cortopassi G. Reproducible quantitative PCR of mitochondrial and nuclear DNA copy number using the LightCycler. Methods Mol Biol. 2002;197:129–37. doi: 10.1385/1-59259-284-8:129. [DOI] [PubMed] [Google Scholar]
- 31.Wu M, Neilson A, Swift AL, et al. Multiparameter metabolic analysis reveals a close link between attenuated mitochondrial bioenergetic function and enhanced glycolysis dependency in human tumor cells. Am J Physiol Cell Physiol. 2007;292:C125–36. doi: 10.1152/ajpcell.00247.2006. [DOI] [PubMed] [Google Scholar]
- 32.Wallace DC. Mitochondrial DNA sequence variation in human evolution and disease. Proc Natl Acad Sci U S A. 1994;91:8739–46. doi: 10.1073/pnas.91.19.8739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ralser M, Wamelink MM, Kowald A, et al. Dynamic rerouting of the carbohydrate flux is key to counteracting oxidative stress. J Biol. 2007;6:10. doi: 10.1186/jbiol61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gruning NM, Lehrach H, Ralser M. Regulatory crosstalk of the metabolic network. Trends Biochem Sci. 2010;35:220–7. doi: 10.1016/j.tibs.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 35.Bender A, Krishnan KJ, Morris CM, et al. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat Genet. 2006;38:515–7. doi: 10.1038/ng1769. [DOI] [PubMed] [Google Scholar]
- 36.Cortopassi GA, Shibata D, Soong NW, et al. A pattern of accumulation of a somatic deletion of mitochondrial DNA in aging human tissues. Proc Natl Acad Sci U S A. 1992;89:7370–4. doi: 10.1073/pnas.89.16.7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petros JA, Baumann AK, Ruiz-Pesini E, et al. mtDNA mutations increase tumorigenicity in prostate cancer. Proc Natl Acad Sci U S A. 2005;102:719–24. doi: 10.1073/pnas.0408894102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ishikawa K, Takenaga K, Akimoto M, et al. ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science. 2008;320:661–4. doi: 10.1126/science.1156906. [DOI] [PubMed] [Google Scholar]
- 39.Brandon M, Baldi P, Wallace DC. Mitochondrial mutations in cancer. Oncogene. 2006;25:4647–62. doi: 10.1038/sj.onc.1209607. [DOI] [PubMed] [Google Scholar]
- 40.Chatterjee A, Mambo E, Sidransky D. Mitochondrial DNA mutations in human cancer. Oncogene. 2006;25:4663–74. doi: 10.1038/sj.onc.1209604. [DOI] [PubMed] [Google Scholar]
- 41.Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120:483–95. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 42.Wang W, Osenbroch P, Skinnes R, et al. Mitochondrial DNA integrity is essential for mitochondrial maturation during differentiation of neural stem cells. Stem Cells. 2010;28:2195–204. doi: 10.1002/stem.542. [DOI] [PubMed] [Google Scholar]
- 43.Kirby DM, Rennie KJ, Smulders-Srinivasan TK, et al. Transmitochondrial embryonic stem cells containing pathogenic mtDNA mutations are compromised in neuronal differentiation. Cell Prolif. 2009;42:413–24. doi: 10.1111/j.1365-2184.2009.00612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Filosa S, Fico A, Paglialunga F, et al. Failure to increase glucose consumption through the pentose-phosphate pathway results in the death of glucose-6-phosphate dehydrogenase gene-deleted mouse embryonic stem cells subjected to oxidative stress. Biochem J. 2003;370:935–43. doi: 10.1042/BJ20021614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mandal S, Lindgren AG, Srivastava AS, et al. Mitochondrial Function Controls Proliferation and Early Differentiation Potential of Embryonic Stem Cells. Stem Cells. 2010 doi: 10.1002/stem.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Birket MJ, Orr AL, Gerencser AA, et al. A reduction in ATP demand and mitochondrial activity with neural differentiation of human embryonic stem cells. J Cell Sci. 2011;124:348–58. doi: 10.1242/jcs.072272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feng Q, Lu SJ, Klimanskaya I, et al. Hemangioblastic derivatives from human induced pluripotent stem cells exhibit limited expansion and early senescence. Stem Cells. 2010;28:704–12. doi: 10.1002/stem.321. [DOI] [PubMed] [Google Scholar]
- 48.Hu BY, Weick JP, Yu J, et al. Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc Natl Acad Sci U S A. 2010;107:4335–40. doi: 10.1073/pnas.0910012107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim K, Doi A, Wen B, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–90. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wolfrum K, Wang Y, Prigione A, et al. The LARGE principle of cellular reprogramming: lost, acquired and retained gene expression in foreskin and amniotic fluid-derived human iPS cells. PLoS One. 2010;5:e13703. doi: 10.1371/journal.pone.0013703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stadtfeld M, Apostolou E, Akutsu H, et al. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010;465:175–81. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.