Abstract
The ability to engineer custom cell-contact-sensing output devices into human non-immune cells would be useful for extending the applicability of cell-based cancer therapies and avoiding risks associated with engineered immune cells. Here, we have developed a new class of synthetic T-cell receptor-like signal-transduction device that functions efficiently in human non-immune cells and triggers release of output molecules specifically upon sensing contact with a target cell. This device employs an interleukin signaling cascade, whose OFF/ON switching is controlled by biophysical segregation of a transmembrane signal-inhibitory protein from the sensor cell/target cell interface. We further showed that designer non-immune cells equipped with this device driving expression of a membrane-penetrator/prodrug-activating enzyme construct could specifically kill target cells in the presence of the prodrug, indicating its potential usefulness for target-cell-specific, cell-based enzyme-prodrug cancer therapy. Our study also contributes to advancement of synthetic biology by extending available design principles to transmit extracellular information to cells.
Devices that can endow mammalian cells with specific-cell-contact-sensing ability are useful to extend the applicability of cell-based cancer therapy. Tumor-specific T cell receptors (TCRs) and chimeric antigen receptors (CAR), which provide T cells with directivity towards target cells1–9, are examples of such devices. Especially, T cells engineered with CAR are the most promising cell-based therapy to date, and several kinds of CAR-T cells are currently under clinical study9. However, there are considerable risks involved in using engineered immune cells, which may cause cytokine release syndrome, macrophage activating syndrome, and neurotoxicity1, 2, 4, 8, 9. Also, current T-cell therapy usually relies on chance encounters between T cells and cancer cells, which is a limiting factor for therapeutic efficacy6, 7, 10, and CAR-T-based therapies have so far been successful only for limited range of cancers.
One approach to overcome these problems would be to engineer non-immune cells that are inherently tumor-tropic for cancer cell ablation. For example, some types of stem cells, including mesenchymal stem cells (MSCs) and neural stem cells (NSCs), are known to be tumor-tropic. Leveraging this characteristic, several researchers have reported applications of these stem cells to cancer therapy by constitutively expressing output molecules that can kill cancer cells, including TNF-related apoptosis-inducing ligand (TRAIL)11, 12, and enzymes that convert anti-cancer prodrugs to active form13–17. In order to maximize therapeutic efficacy while avoiding toxicity derived from constitutive expression of these output molecules, it would be useful to endow such tumor-tropic non-immune cells with a custom cell-contact-sensing ability. However, the CAR-dependent cell-contact-driven gene expression system is not directly portable to non-immune cells, since T-cell signaling is highly specialized to specific cell types, and non-immune cells generally do not express the necessary signaling components for CAR to be functional (including cluster of differentiation 45 (CD45), lymphocyte-specific protein tyrosine kinase (Lck), zeta-chain-associated protein kinase 70 (ZAP70), linker for activation of T cells (LAT), SH2 domain-containing leukocyte protein of 76 kDa (SLP76), and phospholipase Cγ1 (PLCγ1)). So far, few methodologies are available to make non-immune cells responsive to specific cell contact18–20, so there is a need to develop a new class of signaling device for this purpose.
In this study, we show that a new class of T-cell-receptor-like signal transduction device for sensing specific cell contact can be engineered into non-immune cells, including HEK-293T cells and human MSCs (hMSCs). This device employs Janus kinase–signal transducer and activator of transcription (JAK-STAT) signaling mediated by interleukin 4/13 (IL4/13) receptor, with STAT6 as a signaling scaffold, and uses biophysical segregation of a CD45-mimetic molecule upon specific cell contact as an OFF/ON switching mechanism. Further, we show that designer non-immune cells engineered with this cell-contact-sensing device are potentially applicable to target-cell-specific enzyme-prodrug cancer therapy. This was achieved by using a cell-penetrating enzyme that converts 5-fluorocytosine (5-FC, prodrug) into toxic 5-fluorouridine monophosphate (5-FUMP) as an output. The designer cells equipped with this cell-contact-sensing device are expected to be useful for cell-based cancer therapy while avoiding the risks associated with engineered immune cells. Moreover, our work expands the synthetic biology toolbox by demonstrating for the first time that synthetically programmed dynamic movement of a transmembrane protein can be used to transmit extracellular information to cells.
Results
System Design
Reports on the biophysical mechanism of T-cell receptor (TCR) triggering21, 22 indicate that the first step of native TCR signal triggering is cell-contact-induced segregation from the cell interface of transmembrane phosphatase CD45, which negatively regulates signal-initiating kinase Lck. Release of Lck from its suppressor CD45 initiates downstream signaling. On the other hand, CD45 also acts as a phosphatase for JAKs, and negatively regulates cytokine receptor signaling23. Since the JAK-STAT pathway has been functionally rewired to regulate transgene expression in non-immune cells24, we hypothesized that activation of this pathway initiated by cytokine receptors might be controllable by making use of CD45 segregation in response to specific cell contact; i.e. we hypothesized that if we co-express CD45 with interleukin receptors that bear extracellular antigen-recognition moieties, corresponding JAK-STAT signaling mediated by the interleukin receptors would be suppressed by CD45, but the signaling suppression would be released by segregation of CD45 from the cell-cell interface upon recognition of the antigen expressed on a target cell, thereby inducing transgene expression.
CD43ex-45int suppresses JAK-STAT signaling pathways
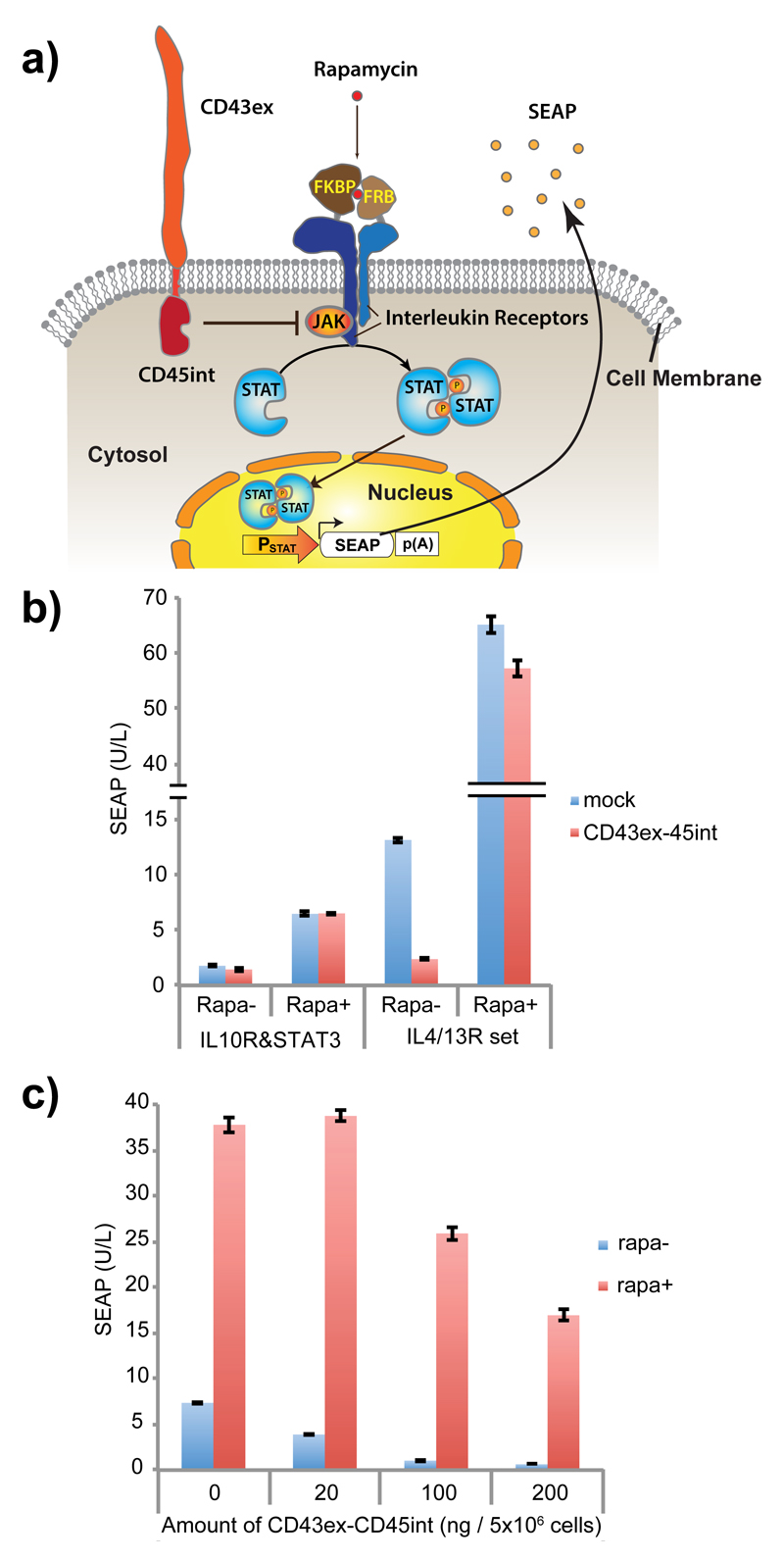
Because reporter constructs of STAT3 signaling and STAT6 are available24, we first set out to screen whether CD43ex-45int could suppress these JAK-STAT pathways. We used CD43ex-45int instead of native CD45, because CD43ex-45int is better expressed than native CD45 in a widely used non-immune cell line, HEK-293T22. As receptor components, we chose IL10 receptor (IL10R, combination of α and β subunits), which triggers STAT3 signaling, and IL4/13 receptors (IL4/13R, hetero dimer of α chain of IL4R (IL4Rα) and α1 chain of IL13R (IL13Rα1), which trigger STAT6 signaling. We expressed CD43ex-45int together with IL10 receptors (IL10Rα and IL10β) or IL4/13 receptors (IL4Rα and IL13Rα1) bearing FKBP (FK506-binding protein) and FRB (FKBP-rapamycin-binding protein) in their extracellular domain, as well as the corresponding STAT (STAT3 for IL10R set, STAT6 for IL4/13R set), so that signaling could be synthetically triggered by addition of rapamycin and transduced through STAT (usually these interleukin receptors trigger downstream signaling by dimerization). Activation of the signaling was monitored with the corresponding STAT reporters expressing secreted alkaline phosphatase (SEAP) (Fig. 1a). As a result, we found that basal (without rapamycin) gene expression of IL4/13 receptor signaling mediated by STAT6 is dose-dependently suppressed by co-expression of CD43ex-45int. (We observed less efficient signal downregulation in the case of artificial signal induction with rapamycin (Fig. 1b,c), suggesting that CD43ex-45int is less able to counteract signaling mediated by strongly bound receptor dimer.) Therefore, we adopted STAT6 signaling mediated by IL4/13 receptor as a scaffold to build a synthetic specific-cell-contact-sensing device.
Figure 1. Evaluation of CD43ex-45int for suppressing cytokine receptor-mediated signaling pathways.
(a) Schematic illustration of the design. In the presence of rapamycin (rapa), Janus kinase (JAK) is activated by receptor dimerization, leading to phosphorylation and dimerization of signal transducer and activator of transcription (STAT). Dimerized STAT translocates to the nucleus, and promotes transgene expression via a STAT-responsive minimal promoter. The effect of co-expression of CD43ex-45int on signaling was determined by quantifying induced expression of a reporter protein, secreted alkaline phosphatase (SEAP). (b) SEAP expression from HEK-293T cells co-transfected with pRK96 (PhCMV-CD43ex-45int-pA) (pA: poly adenylation signal) (or pcDNA3.1(+) for mock) and interleukin receptors together with corresponding STAT and its reporter (IL10R & STAT3 set: pLeo56 (PhCMV-FKBP-IL10Rα-pA), pLeo57 (PhCMV-FRB-IL10Rβ-pA), pLS15 (PhCMV-STAT3-pA) and pLS13 (PSTAT3-SEAP-pA). IL4/13R & STAT6 set: pLeo53 (PhCMV-FKBP-IL4Rα-pA) and pLeo52 (PhCMV-FRB-IL13Rα1-pA), pLS16 (PhCMV-STAT6-pA), and pLS12 (PSTAT6-SEAP-pA)) (± rapa as inducer). (c) Dose-dependency of the inhibitory activity of CD43ex-45int on IL4/13R signaling. SEAP expression with different amounts of pRK96 is shown. All the data are mean ± SEM of three independent experiments measured in triplicate (n=3).
CD43ex-CD45int segregation trigger to sense cell contact
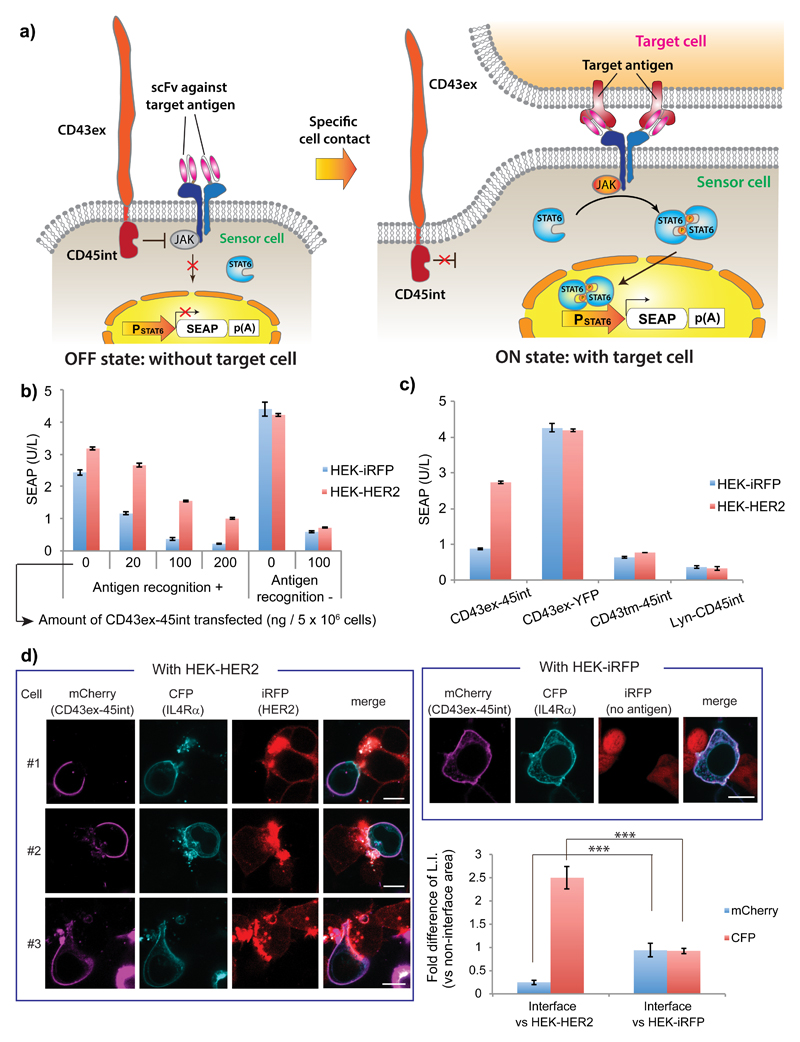
Next, we assessed whether segregation of CD43ex-45int from the cell-cell interface at specific cell-contact locations can be used as a signal trigger. We fused IL4 and IL13 receptor extracellular domain to the single-chain variable fragment (scFv) ML3925, which is directed against a well-known breast cancer marker HER2 (human epidermal growth factor receptor 2), by replacement of FRB and FKBP. Then, we mixed engineered HEK-293T cells co-expressing CD43ex-45int, STAT6, and the STAT6 reporter with model target cells (HEK-293-HER2-iRFP (infrared fluorescent protein), hereinafter referred to as HEK-HER2) or non-target cells (HEK-293-iRFP, hereinafter referred to as HEK-iRFP), and measured expression of SEAP at 24 hours after cell mixing (Fig. 2a). We observed increased transgene expression only when engineered cells expressing the extracellular scFv and an appropriate amount of signal repressor CD43ex-45int were mixed with the target cells (Fig. 2b). This shows that upregulation of reporter gene expression is mediated by antigen recognition and subsequent release of signal suppression by CD43ex-45int. Also, comparison with hetero-type receptors (combinations of ML39-IL4Rα & FRB-IL13Rα1, FKBP-IL4Rα & ML39-IL13Rα1; i.e. only one receptor component bears an antigen recognition moiety) indicates that antigen recognition by IL4Rα is essential, and that antigen recognition by IL13Rα1 improves the performance of the signaling device (Supplementary Results, Supplementary Fig. 1). These findings together with the results in Figure 2b (left column, 0 ng CD43ex-45int) indicate that dimerization of IL4Rα and IL13Rα1 is not the main driving force of the OFF/ON switching of the signaling upon specific cell contact.
Figure 2. Development of the specific-cell-contact-sensing device.
(a) Schematic illustration. Without target cells, JAK is inhibited by CD43ex-CD45int and downstream signaling is shut off. When the sensor cell binds to a target cell via scFv-antigen interaction, CD43ex-45int is segregated from the cell-cell interface, which turns on downstream signaling (translocation of phosphorylated STAT6 dimer to the nucleus, and STAT-6-responsive transgene expression). (b) Evaluation of effect of antigen recognition. Sensor HEK-293T cells were transfected with IL4Rα and IL13Rα1 bearing, or not bearing an antigen recognition moiety, together with various amounts of pRK96 (see Table 1). After mixing them with HEK-HER2 or HEK-iRFP cells, SEAP secreted from the sensor cells was assayed. (c) Assays to verify CD43ex-45int function. Sensor HEK-293T cells were transfected with pRK96, pRK14 (PhCMV-CD43ex-YFP-pA), pRK290 (PhCMV-CD43tm-45int-pA), or pRK291 (PhCMV-Lyn-CD45int-pA) as well as the other signaling devices (see Table 1). Sensor cells were mixed with HEK-HER2 or HEK-iRFP cells, and SEAP secreted from the sensor cells was assayed. Data in Fig. 2b,c are mean ± SEM of three independent experiments measured in triplicate (n=3). (d) Imaging analysis of CD43ex-45int segregation (scale bar: 10 µm). Sensor HEK-293T cells were transfected with pRK293 (PhCMV-ML39-IL10RαΔint-CFP-pA) and pHCM-CD43ex-CD45int-mCherry (LTR-PhCMV-CD43ex-45int-mCherry-LTR), and mixed with HEK-HER2 or HEK-iRFP cells. Statistical analysis of localization of CD43ex-45int-mCherry and ML39-IL10RαΔint-CFP was also conducted. Data represent average fluorescence intensity of each fluorophore at the cell-cell interface (normalized to the intensity at regions of the plasma membrane without cell contacts) ± SEM of 9 different cells. ***P<0.001 (n=9), two-tailed Student’s t-test.
To obtain further evidence that the OFF/ON switching is indeed mediated by segregation of CD43ex-45int and release of JAK-STAT signaling, we prepared and tested the following defective signal repressors: CD43ex-YFP, a conjugate of the transmembrane domain of CD43 and CD45int (CD43tm-CD45int), and Lyn-CD45int (Lyn N-terminal sequence GCIKSKGKDSA has the ability to target proteins to the plasma membrane), Among them, CD43ex-YFP lacks intracellular phosphatase activity, whereas CD43tm-CD45int and Lyn-CD45int lack the extracellular domain while retaining phosphatase activity beneath the plasma membrane. With CD43ex-YFP, overall transgene expression increased, but the whole device lost the ability to sense target cells. With CD43tm-CD45int and Lyn-CD45int, signal suppression was still observed, but again the whole device lost the ability to sense target cells (Fig. 2c). These results indicate that CD43ex-CD45int is indeed segregated in response to close proximity of plasma membranes, via interaction between the scFv on the IL4/13 receptor and target antigen, due to the large extracellular domain of CD43ex-45int, and that this segregation is the main driving force of the OFF/ON switching of the signaling upon specific cell contact.
Moreover, we conducted detailed analysis of the segregation of CD43ex-45int by fluorescence imaging (Fig. 2d). We transfected HEK-293T cells with CD43ex-45int bearing mCherry at its C-terminus (CD43ex-45int-mCherry) as well as ML39-IL4Rα bearing CFP at its C-terminus (replacing intracellular domain of IL4Rα) (ML39-IL4RαΔint-CFP), and conducted quantitative analysis of the fluorescence intensity of mCherry and CFP at the interface and non-interface regions with target (HEK-HER2) or non-target (HEK-iRFP) cells. CD43ex-CD45int-mCherry was segregated from the interface, whereas ML39-IL4RαΔint-CFP accumulated there, only when the cells were mixed with HEK-HER2 cells. This result verifies that CD43ex-45int can be segregated by interaction of the chimeric interleukin receptor and the target antigen, and further supports the conclusion that the cell-contact-sensing device worked as designed.
Specific-cell-contact sensing with truncated IL4Rα
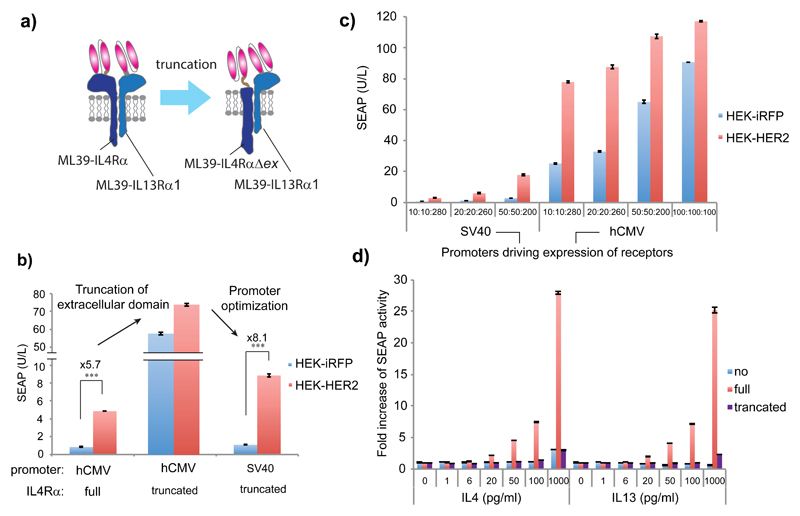
Full output of IL4/13R signaling is usually observed with the native ligands (IL4 and IL13), but we used ligand-free basal signaling for transgene expression. (We further suppressed this basal signaling by ectopic expression of CD43ex-45int, and the signaling was released from the suppression by segregation of CD43ex-45int upon specific cell contact; thus, output gene expression was still at the “basal” gene expression level without ligands.) Therefore, cell-contact-triggered output transgene expression levels were rather low (Fig. 2b,c). To increase cell-contact-triggered transgene expression, we truncated the extracellular domain of IL4Rα (IL4RαΔex to render the receptor hyperactive (Fig. 3a). The hyperactive IL4RαΔex substantially increased cell-contact-triggered transgene expression. Expressing the receptors with a weaker promoter (see Supplementary Fig. 2 for comparison of the promoter strength of human cytomegalovirus (hCMV) immediate early promoter (PhCMV) vs. simian vacuolating virus 40 (SV40) promoter (PSV40). PSV40 is weaker than PhCMV.) further improved the fold induction of transgene expression of engineered cells when mixed with the target cells (Fig. 3b). Using a weaker promoter might be beneficial for balancing the expression levels of the receptors and CD43ex-45int, as signaling mediated by the chimeric interleukin receptors expressed with hCMV promoter could be too strong to be suppressed by CD43ex-45int. This hypothesis is supported by the fact that the output gene expression level could be tuned by changing the dose of each component (Fig. 3c). We examined the effect of modulating sensor cell/target cell ratio with this system, and found that the system showed increased transgene expression when mixed with a larger proportion of target cells (Supplementary Fig. 3). In addition, it should be noted that the use of truncated receptors made the device non-responsive to native ligands IL4 and IL13 (physiological concentration range: <100 pg/mL26) (Fig. 3d), without loss of the ability to sense specific cell contact.
Figure 3. The optimized specific-cell-contact-sensing system.
(a) Schematic illustration of receptor truncation. The receptor set bearing truncated IL4R (IL4RαΔex) does not need ligands for activation. Therefore, output gene expression is increased, and the system is non-responsive to native ligands. (b) Evaluation of the effect of receptor truncation and promoter optimization. The sensor HEK-293T cells were transfected as shown in Table 1. After mixing them with HEK-HER2 or HEK-iRFP cells, SEAP secreted from the sensor cells was assayed. (c) Effect of dose dependency of each component on the system performance. Different amounts of plasmids encoding receptors (driven by either PSV40 or PhCMV) and CD43ex-45int were transfected in sensor HEK-293T cells (See Table 1). Numbers under each column (10:10:280 etc) indicate the amounts (ng/5x105 cells) of transfected plasmids encoding IL4RαΔex (pRK122 or pRK119), IL13Rα1 (pRK123 or pRK114), and CD43ex-45int (pRK96), respectively from left to right. After mixing the sensor cells with HEK-HER2 or HEK-iRFP cells, SEAP secreted from the sensor cells was assayed. (d) System responsiveness to native ligands (IL4, IL13) with different receptor sets. The sensor HEK-293T cells were transfected as shown in Table 1b. SEAP secreted from the sensor cells in the presence of various concentrations of IL4 and IL13 was assayed. All the data are mean ± SEM of three independent experiments measured in triplicate (n=3). Two-tailed Student’s t-tests were conducted for Fig.3b. ***P<0.001.
Generalizability of the system
We confirmed that the device was also functional with DARPin (designed ankyrin repeat proteins; a genetically engineered antibody-mimetic derived from ankyrin proteins)27 as an alternative to the antigen recognition moiety (Supplementary Fig. 4). Further, the device also worked for targeting a different antigen, EpCAM (epithelial cell adhesion molecule), which is a diagnostic marker for various cancers28 (Supplementary Fig. 4). These results support the generalizability of the design principle we employed. This device also enabled hMSCs to respond to coculture with a typical HER2-expressing breast cancer cell line, SKBR3 (Supplementary Fig. 4).
Application for activatable enzyme-prodrug therapy
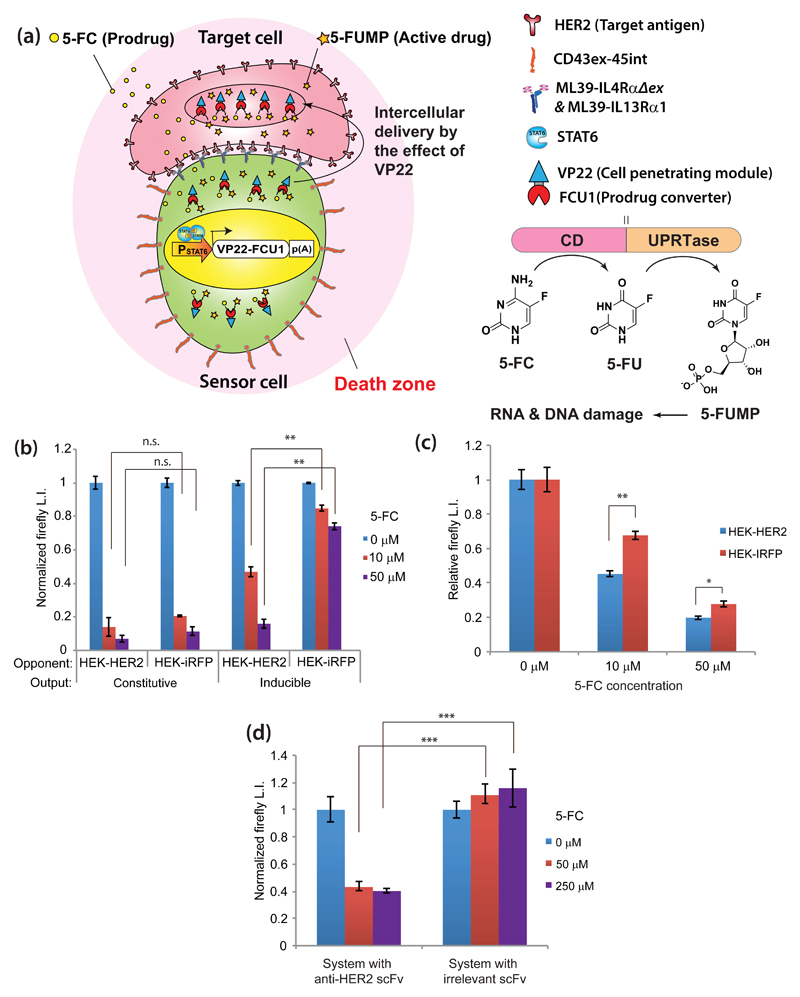
Finally, we examined the potential application of the specific-cell-contact-sensing device for producing an enzyme that selectively catalyzes the formation of an anti-cancer compound from a prodrug when the sensor cells encounter HER2-expressing cells (Fig. 4a). In the context of targeting tumors instead of single cells, it would be beneficial to maximize the bystander effect, i.e., the cell-killing effect of gene-modified cells on surrounding cells due to transfer of toxic metabolites. To maximize this effect, we conjugated VP2229 (major component of the herpes simplex virus type 1 segment that is secreted from cells expressing it through a Golgi-independent non-classical mechanism, and transferred into recipient cells by a unique trafficking pathway involving actin cytoskeleton30–32; protein conjugated to VP22 usually exists mainly in the cytosol of the transfected cells and in the nucleus of adjacent cells) to FCU133,34 (conjugate of cytosine deaminase and uracil phosphoribosyltransferase) and linked it to our cell-contact-sensing device so that VP22-FCU1 would be produced in response to cell contact with HER2-expressing cells. Once expressed, VP22-FCU1 is intercellularly delivered to adjacent cells due to the action of VP22, and promotes conversion of 5-fluorocytosine (5-FC, prodrug) to toxic 5-fluorouridine monophosphate (5-FUMP) (see Supplementary Fig. 5 for the effect of VP22 conjugation). To confirm functionality of the system, we transfected HEK-293T cells with VP22-FCU1 under either a constitutive or a STAT6-responsive minimal promoter, as well as with gene expression components targeting HER2, and mixed the resulting cells with target (HEK-HER2) or non-target (HER2-iRFP) cells expressing firefly luciferase in 3D microtissues (3D cell culture was used to enhance cell-cell contact). Various concentrations of 5-FC were added and the viability of target/non-target cells was assayed. The engineered cells constitutively expressing VP22-FCU1 killed both non-target and target cells, while the engineered cells equipped with the specific cell contact-driven gene expression device showed significant target specificity (Fig. 4b). Further, we tested whether the designer cells could specifically kill target cells in mixed cultures of target and non-target cells. As a result, target selectivity was observed even in the mixed culture (Fig. 4c). Moreover, hMSCs expressing the inducible VP22-FCU1 triggered by the cell-contact-sensing device could kill HER2-positive SKBR3 breast cancer cells in an antigen-recognition-dependent manner (Fig. 4d, irrelevant scFv: SP635(scFv against synthetic hapten 2,4,6-trinitrophenyl)), further supporting the potential value of designer cells equipped with this device for therapeutic applications.
Figure 4. Application to target-cell-specific activatable enzyme-prodrug cancer therapy.
(a) Schematic illustration. Sensor cells express effector protein VP22-FCU1, which is delivered to adjacent cells by VP22 and converts prodrug 5-FC into cytotoxic 5-FUMP via its cytosine deaminase (CD) and uracil phosphoribosyltransferase (UPRTase) activities. This occurs only when sensor cells contact target cells expressing specific antigen. (b) Demonstration of target-specific cell killing. Sensor HEK-293T cells were transfected with pRK96, pRK122 (PSV40-ML39-IL4RαΔex-pA), pRK123 (PSV40-ML39-IL13Rα1-pA), and pLS16, plus either pRK131 (PhCMV-VP22-FCU1-pA) (“constitutive”) or pRK223 (PSTAT6-VP22-FCU1-pA) (“inducible”). Transfected sensor cells were mixed with HEK-HER2-Luc or HEK-iRFP-Luc cells in 3D culture. 5-FC (0-50 µM) was added. After 4 days, viability was measured as luminescence intensity (normalized to no 5-FC condition). (c) Target-specific cell killing in mixed culture. Sensor HEK-293T cells were transfected as in Fig. 4b (inducible), then mixed with 1:1 HEK-HER2-Luc and HEK-iRFP (for measuring HEK-HER2-Luc viability), or 1:1 HEK-HER2 and HEK-iRFP-Luc (for measuring HEK-iRFP-Luc viability) in 3D culture. 5-FC (0-50 µM) was added, and viability was measured as above. (d) SKBR3 cell killing by hMSC-TERT cells equipped with specific-cell-contact-sensing system. hMSC-TERT cells were transfected with the same components as in Fig. 4b (inducible) (system with anti-HER2), or components having irrelevant scFv (pRK145 (PSV40-SP6-IL4RαΔex-pA) and pRK144 (PSV40-SP6-IL13Rα1-pA)), then mixed with SKBR3 cells stably expressing firefly luciferase. After addition of 5-FC (0-250 µM), the same assay as in (b) was conducted. Data are mean ± SEM of three independent experiments measured in triplicate (n=3). Two-tailed Student’s t-test: *P<0.05, **P<0.01, ***P<0.001. n.s: P>0.05.
Discussion
In this study, we have successfully developed a new class of transgene expression systems triggered by specific cell contact, which works in non-immune cells. We also demonstrated a potential application of the system to target-cell-specific activatable enzyme prodrug cancer therapy. A key advantage of the use of smart cell-based therapy with designer non-immune cells is that it would eliminate the risks associated with engineered immune cells.
First, we discovered that ectopically expressed CD43ex-45int could efficiently suppress STAT6-mediated signaling initiated by IL4/13R, but not STAT3-mediated signaling initiated by IL10R. Although native CD45 binds to and dephosphorylates all JAKs (JAK1, JAK2, JAK3, tyrosine kinase 2 (TYK2)) and has negative effects on both STAT3- and STAT6-mediated signaling23, 36, it is possible that CD43ex-45int could not dephosphorylate JAK1 or TYK2, the main upstream component of IL10R-STAT3 signaling37, because of some structural issue in this synthetic setting. Nevertheless, it might be possible to develop effective repressors against other signaling pathways as well (including, but not limited to other JAKs) by further engineering of the signal repressor, which might allow for generalization of the signaling scaffold to transduce information, as well as fine-tuning of the system responsiveness.
Then, we showed that the segregation of CD43ex-CD45int can be used as an OFF/ON switching mechanism for synthetic cytokine signaling mediated by IL4/13 receptor and STAT6 in response to specific cell contact. Notably, the optimized system using a truncated version of IL4Rα (IL4RαΔex) was not responsive to native ligands such as IL4 and IL13. This synthetically created input selectivity (i.e., responsive only to specific contact) would be favorable for future biomedical applications. In addition, tunability of the cell-contact-sensing system by controlling the dose of each component might allow for optimization of output transgene expression level as required for particular applications.
It is noteworthy for possible clinical application that this system is functional in hMSCs. As discussed in the introduction, certain types of stem cells constitutively expressing an enzyme that activates a prodrug have emerged as potential vehicles for cancer therapy13–17. However, the active (= toxic) drug can also be released in non-tumor tissue with this system, which may cause side effects, since stem cells express the enzyme constitutively. Cell-based enzyme-prodrug therapy that is active specifically in tumor tissue should be superior in this respect. We have successfully shown that the specific-cell-contact-sensing device developed in this study could be suitable for this purpose.
From the viewpoint of potential clinical application, it is important that specific cell killing by designer cells equipped with the membrane-penetrator/prodrug-activating enzyme construct also worked in mixed cultures of target- and non-target cells. Although we observed a non-negligible off-target effect in this mixed-culture experiment, this might be partly due to the static in vitro experimental setting, in which the toxic metabolite generated in sensor cells and target cells could remain for a long time and diffuse into non-target cells. It should be noted that the sensor cells themselves were killed by administration of the prodrug in this setting (suicide-type behavior, which is always the case for normal cell-based enzyme prodrug therapy). It might be possible to avoid such sensor cell death and achieve a further increase in therapeutic efficacy by incorporating drug resistance specifically into sensor cells; this could be achieved by expressing multidrug resistance protein (MRP), which is capable of pumping out a wide variety of xenobiotic organic anionic compounds38. It will also be interesting to investigate other choices for output molecules to kill cancer cells, such as FAS ligand and TRAIL, which stimulate apoptosis in adjacent cells in a contact-dependent manner39.
In synthetic biology, mammalian cells have been engineered to respond to changes in the extracellular environment mainly by exploiting natural receptors of soluble molecules5, 6, 40, 41. However, to our knowledge, biological sensors capable of customized modulation of cell-contact-sensing ability have so far been limited to CAR and the recently reported synthetic notch receptor (SynNotch),20 and there is a need for other design principles to transmit cell-contact information. The specific cell-contact-sensing device reported in this study is distinct from the previous devices in that it incorporates a native cytokine signaling pathway controlled by the movement of transmembrane protein as a negative regulator for cell-contact-dependent OFF/ON switching. This design extends the synthetic biology toolbox for the design of artificial cellular functions based on specific cell contact by providing a new class of cell-contact-sensing device that utilizes synthetically programmed dynamic movement of a transmembrane protein to transmit extracellular information to cells. Potential applications include new cell-based cancer therapies.
Online Methods
DNA constructs
Detailed methods of construction of the plasmids are described in Supplementary Table 1.
Cell culture and transfection
HEK-293T cells (DSMZ: ACC-635), HEK-293 cells (DSMZ: ACC-305) and their stable transfected cell lines (HEK-293-HER2-iRFP (designated as HEK-HER2 cells in the main text), HEK-293-iRFP (designated as HEK-iRFP cells in the main text), HEK-293-Epcam-ZsGreen (designated as HEK-Epcam cells), HEK-293-HER2-iRFP-luc-ZsGreen (designated as HEK-HER2-Luc cells in the main text), HEK-293-iRFP-luc-ZsGreen (designated as HEK-iRFP-Luc cells in the main text), and human mesenchymal stem cells transgenic for the catalytic subunit of human telomerase (hMSC-TERT)42 were cultivated in DMEM (Invitrogen) supplemented with 10% (v/v) fetal bovine serum (FBS, Sigma-Aldrich) and 1% (v/v) penicillin/streptomycin solution (Sigma-Aldrich) at 37 °C in a humidified atmosphere containing 5% CO2. For SKBR3 cells (gift from Professor Nancy Hynes, FMI, Basel, Switzerland, HER2 expression was confirmed in Supplementary Fig. 4c) and their transfectant (SKBR3 luc-ZsGreen), DMEM/F12 plus Glutamax (Gibco) was used instead of normal DMEM. 400 µg/ml of G418 (Sigma) was also added to culture HEK-293-HER2-iRFP, HEK-293-iRFP, HEK-293-HER2-iRFP-luc-ZsGreen, and HEK-293-iRFP-luc-ZsGreen cells. For serial passage of these cells, 0.05% Trypsin-EDTA (Gibco) was used. For transfection, 2.5x105 cells/ml of cells (counted with a Casy® TTC Cell Counter) were seeded on a 24-well plate (Thermo Fischer Scientific) (500 µL/well) 24 hours before transfection. DNA-polyethyleneimine (PEI) mixture (50 µL) was produced by incubating 2.5 µL PEI (PEI, 20000 MW, Polysciences; stock solution 1 mg/ml in dH2O) with 500 ng of total DNA (see “Detailed procotol for each figure” section for details), vortexing for 1 s and incubating at r.t. for 15 min. (When necessary, transfection mix and cells used for transfection were scaled up to 12-well plates, 6-well plates, or 10 cm culture dishes. The amounts of DNA and reagent were changed accordingly). Before the transfection, cell culture medium was exchanged to fresh medium containing 10% FBS. The cells were incubated with the transfection mixture for 8-16 hours. Subsequently, the medium was exchanged again to fresh, pre-warmed medium for expression of the gene of interest. For establishing HEK-293-HER2-iRFP and HEK-293-iRFP cells, antibiotic selection (400 µg/ml of G418) was used from 2 days after transfection of pRK21 (PhCMV-HER2-iRFP-pA) or pRK22 (PhCMV-iRFP-pA) into HEK-293 cells. After obtaining bulk stable cell lines, single cell clones were obtained by a limiting dilution method. To establish HEK-293-HER2-iRFP-luc-ZsGreen, HEK-293-iRFP-luc-ZsGreen, HEK-293-Epcam-ZsGreen and SKBR3-luc-Zsgreen cells, lentiviral transduction was used. First, HEK-293T cells were transfected with transfer vector (pRK187 (PEF1α-Epcam-IRES-ZsGreen) for establishing HEK-293-Epcam-ZsGreen, pHIV-luc-ZsGreen (addgene #39196) for establishing HEK-293-HER2-iRFP-luc-ZsGreen, HEK-293-iRFP-luc-ZsGreen, and SKBR3-luc-ZsGreen), psPAX2 (for packaging, addgene #12260), and pMD2.G (for expressing VSV-G, addgene #12259). After 16 hours of transfection, the medium was changed to fresh medium. At 30 hours after the medium change, the medium containing lentivirus was filtered, and used to infect cells. After several passages, ZsGreen-positive cells were sorted by FACS (BD FACS Aria III), and used for the following experiments.
SEAP reporter assay
The supernatant obtained from the transfected cells was incubated at 65°C for 30 min to inactivate endogenous alkaline phosphatase, and 80 µL of the heat-inactivated medium was placed in wells of a transparent 96-well plate. Then, 100 µL of 2 × SEAP buffer (20 mM homoarginine, 1 mM MgCl2, 21% (v/v) diethanolamine, pH 9.8) and 20 µL of p-nitrophenyl phosphate (pNPP) solution (120 mM) were added. The time course of absorbance at 405 nm was measured at 37 °C by using the EnVision 2104 Multilabel Reader. Quantification of SEAP production in U/L was calculated from the slope of the time-dependent increase in absorbance (the equation to convert slope of absorbance and SEAP unit was pre-determined by using a standard), where the time-points for the calculation of the slope were exclusively chosen from the linear part of the increase in order to avoid saturation effects (Figs. 1-3).
Assay of inhibitory effect of CD43ex-45int on JAK-STAT signaling
100 ng of each indicated receptor, 100 ng of pLS15 (PhCMV-STAT3-pA) or pLS16 (PhCMV-STAT6-pA), 100 ng of pLS13 (STAT3 reporter) or pLS 12 (STAT6 reporter), and various amount of pRK96 (PhCMV-CD43ex-45int-pA) (0 or 100 ng for Fig. 1b, and 0-200 ng for Fig. 1c (pcDNA3.1(+) was used as filler to adjust the total amount of plasmids) were transfected into HEK-293T cells (per well of a 24-well plate; approx. 2.5x105 cells/well at the time of transfection). At 16 hours after transfection, the medium was replaced with fresh DMEM (hereinafter, always with 10% FBS + P/S unless otherwise specified) with (50 nM) or without (0 nM) rapamycin. SEAP activity was measured at 24 hours after medium replacement.
Cell-contact-triggered gene expression assay
Sensor HEK-293T cells were transfected with the cell-contact-sensing device (output: SEAP) in wells of a 24-well plate. At 14 hours after transfection, the medium was exchanged. At 24 hours after transfection, the cells were detached with Cell Dissociation Buffer or TryLE Express (Thermo Fisher Scientific), spun down, and suspended in 40 µL of fresh DMEM. The cell suspension was divided into two (2x20 µL), and each part was mixed with another cell suspension (20 µL) containing 2.5 x105 cells of HEK-HER2 or HEK-iRFP cells in 1.5 mL tubes. After incubation for 30 min at 37 °C, the cell suspension was diluted appropriately and seeded on 24-well plates. After 24 hours, SEAP activity was measured. Figure specific conditions are as follows (the plasmid amount is per well of 24-well plate): 100 ng of each receptor, 100 ng of pLS16, 100 ng of pLS12, and various amounts of pRK96 (0-200 ng, pcDNA3.1(+) was used as a filler) were transfected (Fig. 2b). 100 ng of each receptor, 100 ng of pLS16, 100 ng of pLS12, and 100 ng of one of following plasmids were transfected; pRK96 (PhCMV-CD43ex-45int-pA), pRK14 (PhCMV-CD43ex-YFP-pA), pRK290 (PhCMV-CD43tm-45int-pA), or pRK291 (PhCMV-Lyn-CD45int-pA) (Fig. 2c). As shown in Table 1, for hCMV-promoter-driven receptor sets (left, middle), 100 ng of each receptor (PhCMV driven), 100 ng of pLS16, 200 ng of pRK96, and 100 ng of pLS12 were transfected, and for SV40-promoter-driven receptor sets (right), 50 ng of each receptor (PSV40 driven), 100 ng of pLS16, 200 ng of pRK96, and 100 ng of pLS12 were transfected (Fig. 3b). Different amounts of plasmids encoding receptors and CD43ex-45int were transfected as shown in Table 1 (Numbers under each column (10:10:280, 20:20:260, etc.) indicate the amounts (ng/5x105 cells in a 24-well plate) of transfected plasmids encoding IL4RαΔex (pRK122 or pRK119), IL13Rα1 (pRK123 or pRK114), and CD43ex-45int (pRK96), respectively, from left to right. For example, 10:10:280 in the SV40 column: 10 ng of pRK122 (PSV40-ML39-IL4RαΔex-pA), 10 ng of pRK123 (PSV40-ML39-IL13Rα1-pA) and 280 ng of pRK96 were transfected) (Fig. 3c).
Table 1.
Chimeric interleukin receptors (IL4Rα and IL13Rα1) and signal repressors used in this study (for Fig. 2 and Fig. 3). Plasmids encoding each component (expression is driven by either PhCMV or PSV40) were transfected following this table. In addition to these plasmid sets, pLS16 (PhCMV-STAT6-pA) and pLS12 (PSTAT6-SEAP-pA) were always transfected.
| Fig. | Column | Receptor | Repressor | Comment | ||
|---|---|---|---|---|---|---|
| promoter | IL4Rα | IL13α1 | ||||
| 2b | Anti. Recog.+ | hCMV | ML39-IL4Rα (pRK115) | ML39-IL13α1 (pRK114) | pRK96 (CD43ex-45int) | HER2 recognition by ML39 |
| Anti. Recog.- | FKBP-IL4Rα (pLeo53) | FRB-IL13α1 (pLeo52) | No antigen recognition | |||
| 2c | CD43ex-45int | ML39-IL4Rα (pRK115) | ML39-IL13α1 (pRK114) | pRK96 (CD43ex-45int) | Correct repressor | |
| CD43ex-YFP | pRK14 (CD43ex-YFP) | No phosphatase activity | ||||
| CD43tm-45int | pRK290 (CD43tm-45int) | No extracellular domain | ||||
| Lyn-CD45int | pRK291 (Lyn-CD45int) | |||||
| 3b | Left | hCMV | ML39-IL4Rα (pRK115) | ML39-IL13α1 (pRK114) | pRK96 (CD43ex-45int) | Full length receptors expressed by PhCMV |
| Middle | hCMV | ML39-IL4RαΔex (pRK119) | ML39-IL13α1 (pRK114) | Extracellular domain of IL4Rα truncated (PhCMV driven) | ||
| Right | SV40 | ML39-IL4RαΔex (pRK122) | ML39-IL13α1 (pRK123) | Truncated receptor expressed by PSV40. | ||
| 3c | SV40 | SV40 | ML39-IL4RαΔex (pRK122) | ML39-IL13α1 (pRK123) | Different ratio of receptors (PSV40 driven) and CD43ex-45int | |
| hCMV | hCMV | ML39-IL4RαΔex (pRK119) | ML39-IL13α1 (pRK114) | Different ratio of receptors (PhCMV driven) and CD43ex-45int | ||
| 3d | non | Mock (pcDNA3.1+) | No receptor | |||
| full | hCMV | ML39-IL4Rα (pRK115) | ML39-IL13α1 (pRK114) | Full length receptor | ||
| truncated | SV40 | ML39-IL4RαΔex (pRK122) | ML39-IL13α1 (pRK123) | Truncated receptor | ||
Fluorescence imaging for assessing CD43ex-45int segregation
250 ng of pRK292 (PhCMV-ML39-IL4RαΔint-CFP-pA) and 250 ng of pHCM-CD43ex-45int-mCherry (LTR-PhCMV-CD43ex-45int-mCherry-LTR) (LTR: long terminal repeat) were transfected into HEK-293T cells. At 14 hours after transfection, medium was changed to fresh DMEM. At 24 hours after transfection, cells were detached in cell dissociation buffer (Thermo Fisher Scientific), spun down, and suspended in 40 µL of DMEM. The cell suspension was divided into two (2x20 µL), and each part was mixed with another cell suspension (20 µL) containing target (HEK-HER2) or non-target (HEK-iRFP) cells in 1.5 mL tubes. Each cell mixture was incubated at 37 °C for 30 min, and seeded on an Ibidi µ-slide 8 well (Ibidi) after appropriate dilution. Fluorescence was observed with Nikon Confocal A1 microscope. For statistical analysis of CD43ex-45int-mCherry segregation, we picked up cells that were clearly in contact through scFv-antigen interaction, based on observation of accumulation of CFP at the cell-cell interface with HEK-HER2. (In the case of contact with HEK-iRFP, CFP accumulation at the cell interface was never observed, so sensor cells that appeared to be in contact with HEK-iRFP were picked up). We quantified the fluorescence intensity of each fluorophore (CFP or mCherry) at the cell-cell interface, and normalized the fluorescence intensity based on the intensity at the parts of the plasma membrane where there was no cell contact (Fig. 2d).
Assay of system responsiveness to native ligands
As shown in Table 1, sensor HEK-293T cells were transfected with the same components as the experiments for the cell-contact-triggered gene expression assays. (For “non” group, pcDNA3.1(+) was used as a mock plasmid. For “full” group, PhCMV-driven full-length receptor sets were used. For “truncated” group, PSV40-driven receptor sets using truncated IL4Rα were used.) At 14 hours after transfection, the medium was changed to fresh DMEM containing 0-1000 pg/ml of human IL4 or IL13 recombinant proteins (PeproTech). SEAP activity was measured 24 hours later (Fig. 3d).
Cell-contact-triggered cell ablation assay
Sensor cells were transfected with pRK122, pRK123, pRK96, pLS16 and pRK223 (PSTAT6-VP22-FCU1-pA) (for constitutive expression, pRK131 (PhCMV-VP22-FCU1-pA) was used instead of pRK223). At 14 hours after transfection, the medium was changed to fresh medium. At 24 hours after transfection, the cells were detached in cell dissociation buffer, spun down, and suspended in 60 µL of fresh medium. Then, 30 µL of the cell suspension was mixed with another cell suspension (50 µL) containing target or non-target cells. After incubation for 30 min at 37 °C, 500 µL of fresh DMEM was added and cells were seeded on 24-well plates. At 24 hours later, the cells were again detached with cell dissociation buffer, spun down, and suspended in 500 µL of fresh DMEM. This cell suspension was diluted 10-16 times, and seeded on a a 96-well U bottom plate (Nunclon Sphera, Thermofisher Scientific) to form 3D microtissue in the presence of various concentrations of 5-FC (0-250 µM, Apollo Scientific). 4 days later, cell viability was assayed in terms of luminescence. For the firefly luminescence assay, 500 µM D-luciferin (final) was added to the wells containing the cells, and luminescence was measured with an EnVision 2104 Multilabel Reader. The ratio of luminescence intensity to that under the control condition was calculated. Figure specific conditions are as follows (plasmid amount is per well of 24-well plate): Sensor HEK-293T cells were transfected with 50 ng of pRK122, 50 ng of pRK123, 290 ng of pRK96, 100 ng of pLS16, and 10 ng of pRK131 (for constitutive) or pRK 223 (for inducible). DMEM was used for the medium. 2.5 x105 cells of HEK-293-HER2-Luc-ZsGreen and HEK-293-iRFP-Luc-ZsGreen were used as target cells and non-target cells, respectively (Fig. 4b). Sensor hMSC-TERT cells were transfected with 130 ng of pRK122, 130 ng of pRK123, 130 ng of pRK96, 60 ng of pLS16, and 100 ng of pRK223 (for the system with irrelevant scFv, pRK145 and pRK144 were used instead of pRK122 and pRK123). DMEM/F12 was used as medium. 1.0x105 cells of SKBR3-Luc-ZsGreen cells were used as target cells. (Fig. 4d)
Cell ablation assay in mixed culture of target and non-target cells
Suspension of sensor cells was prepared by the same method as for the above cell-contact-triggered cell ablation assay. Then, 30 µL of the cell suspension was mixed with another cell suspension (50 µL) containing the following cells. For monitoring firefly luciferase activity in target cells: mixture of 1.25 x 105 cells of HEK-293-HER2-Luc-ZsGreen and 1.25 x 105 cells of HEK-293-iRFP. For monitoring firefly luciferase activity in non-target cells: 1.25 x 105 cells of HEK-293-HER2 and 1.25 x 105 cells of HEK-293-iRFP-Luc-ZsGreen. After incubating the mixed cell suspension in 1.5 mL tubes for 30 min at 37 °C, 500 µL of fresh DMEM was added to each tube. This cell suspension was diluted 16~30 times, and seeded on a 96-well U bottom plate (Multiwell 96U Nunclon Sphera round bottom plate, ThermoFisher Scientific) in the presence of various concentrations of 5-FC (0-250 µM). 4 days later, firefly luciferase activity was assayed (Fig. 4c).
Statistical analysis and reproducibility
Unless indicated otherwise, the error bars in the figures represent standard error of the mean (SEM) of three independent experiments measured in triplicate. (Sample size was sufficient to establish that the values compared were derived from normal distributions). For statistical analyses, two-tailed Student’s t-tests were conducted to evaluate whether a significant difference exists between two groups of samples after confirming that the variances of the two groups were similar. A P value of less than 0.05 was considered as statistically significant.
Supplementary Material
Acknowledgements
We thank P. Saxena for critical comments on the manuscript, R. Vale, J. James, B. Lindner, J. Schaefer, S. Rosenberg, A. Plückthun, L. Schukur, H. Chassin, and addgene construct suppliers (see SI) for providing plasmids, T. Lopes and V. Jaggin for help with FACS analysis, and E. Montani, A. Ponti, and T. Horn for help with microscopy. This work was supported by the European Research Council (ERC) advanced grant (ProNet, no. 321381) and in part by the National Centre of Competence in Research (NCCR) for Molecular Systems Engineering. R. Kojima was supported by a postdoctoral fellowship by the Human Frontier Science Program (HFSP) (LT000094/2014-L).
Footnotes
Author contributions
R.K. and M.F. designed the project, analyzed the results, and wrote the manuscript. R.K. performed experimental work. L.S. contributed to project design and plasmid generation, and edited the manuscript.
Competing financial interests
The authors declare no competing financial interests.
Data availability
The authors declare that all the data supporting the findings of this study are available within the paper and its supplementary information file. Sequence data of key plasmids have been deposited in GenBank: pRK96 (Genbank, MF770565), pRK122 (MF770566), pRK123 (MF770567), pRK223 (MF770568), pLS12 (MF770569), pLS16 (MF770590).
References
- 1.Almasbak H, Aarvak T, Vemuri MC. CAR T Cell Therapy: A game changer in cancer treatment. J Immunol Res. 2016;2016:5474602. doi: 10.1155/2016/5474602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jackson HJ, Rafiq S, Brentjens RJ. Driving CAR T-cells forward. Nat Rev Clin Oncol. 2016;13:370–383. doi: 10.1038/nrclinonc.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.June CH, Blazar BR, Riley JL. Engineering lymphocyte subsets: tools, trials and tribulations. Nat Rev Immunol. 2009;9:704–716. doi: 10.1038/nri2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kalaitsidou M, Kueberuwa G, Schutt A, Gilham DE. CAR T-cell therapy: toxicity and the relevance of preclinical models. Immunotherapy. 2015;7:487–497. doi: 10.2217/imt.14.123. [DOI] [PubMed] [Google Scholar]
- 5.Kojima R, Aubel D, Fussenegger M. Novel theranostic agents for next-generation personalized medicine: small molecules, nanoparticles, and engineered mammalian cells. Curr Opin Chem Bio l. 2015;28:29–38. doi: 10.1016/j.cbpa.2015.05.021. [DOI] [PubMed] [Google Scholar]
- 6.Kojima R, Aubel D, Fussenegger M. Toward a world of theranostic medication: Programming biological sentinel systems for therapeutic intervention. Adv Drug Deliv Rev. 2016;105:66–76. doi: 10.1016/j.addr.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 7.Zhang H, et al. New Strategies for the Treatment of Solid Tumors with CAR-T Cells. Int J Biol Sci. 2016;12:718–729. doi: 10.7150/ijbs.14405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonifant CL, Jackson HJ, Brentjens RJ, Curran KJ. Toxicity and management in CAR T-cell therapy. Mol Ther Oncolytics. 2016;3:16011. doi: 10.1038/mto.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geyer MB, Brentjens RJ. Review: Current clinical applications of chimeric antigen receptor (CAR) modified T cells. Cytotherapy. 2016;18:1393–1409. doi: 10.1016/j.jcyt.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park JS, et al. Synthetic control of mammalian-cell motility by engineering chemotaxis to an orthogonal bioinert chemical signal. Proc Natl Acad Sci U S A. 2014;111:5896–5901. doi: 10.1073/pnas.1402087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marini I, Siegemund M, Hutt M, Kontermann RE, Pfizenmaier K. Antitumor activity of a mesenchymal stem cell line stably secreting a tumor-targeted TNF-related apoptosis-inducing ligand fusion protein. Front Immunol. 2017;8:536. doi: 10.3389/fimmu.2017.00536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Z, et al. Probe-based confocal laser endomicroscopy for imaging TRAIL-expressing mesenchymal stem cells to monitor colon xenograft tumors in vivo. PLoS ONE. 2016;11:e0162700. doi: 10.1371/journal.pone.0162700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matuskova M, et al. Combined enzyme/prodrug treatment by genetically engineered AT-MSC exerts synergy and inhibits growth of MDA-MB-231 induced lung metastases. J Exp Clin Cancer Res. 2015;34:33. doi: 10.1186/s13046-015-0149-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Metz MZ, et al. Neural stem cell-mediated delivery of irinotecan-activating carboxylesterases to glioma: implications for clinical use. Stem Cells Transl Med. 2013;2:983–992. doi: 10.5966/sctm.2012-0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aboody KS, et al. Neural stem cell-mediated enzyme/prodrug therapy for glioma: preclinical studies. Sci Transl Med. 2013;5 doi: 10.1126/scitranslmed.3005365. 184ra159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amara I, Touati W, Beaune P, de Waziers I. Mesenchymal stem cells as cellular vehicles for prodrug gene therapy against tumors. Biochimie. 2014;105:4–11. doi: 10.1016/j.biochi.2014.06.016. [DOI] [PubMed] [Google Scholar]
- 17.Nouri FS, Wang X, Hatefi A. Genetically engineered theranostic mesenchymal stem cells for the evaluation of the anticancer efficacy of enzyme/prodrug systems. J Controlled Release. 2015;200:179–187. doi: 10.1016/j.jconrel.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morsut L, et al. Engineering Customized Cell Sensing and Response Behaviors Using Synthetic Notch Receptors. Cell. 2016;164:780–791. doi: 10.1016/j.cell.2016.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roybal KT, et al. Precision Tumor Recognition by T Cells With Combinatorial Antigen-Sensing Circuits. Cell. 2016;164:770–779. doi: 10.1016/j.cell.2016.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roybal KT, et al. Engineering T Cells with Customized Therapeutic Response Programs Using Synthetic Notch Receptors. Cell. 2016;167:419–432 e416. doi: 10.1016/j.cell.2016.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang VT, et al. Initiation of T cell signaling by CD45 segregation at 'close contacts'. Nature Immunol. 2016;17:574–582. doi: 10.1038/ni.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.James JR, Vale RD. Biophysical mechanism of T-cell receptor triggering in a reconstituted system. Nature. 2012;487:64–69. doi: 10.1038/nature11220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Irie-Sasaki J, et al. CD45 is a JAK phosphatase and negatively regulates cytokine receptor signalling. Nature. 2001;409:349–354. doi: 10.1038/35053086. [DOI] [PubMed] [Google Scholar]
- 24.Schukur L, Geering B, Charpin-El Hamri G, Fussenegger M. Implantable synthetic cytokine converter cells with AND-gate logic treat experimental psoriasis. Sci Transl Med. 2015;7:318ra201. doi: 10.1126/scitranslmed.aac4964. [DOI] [PubMed] [Google Scholar]
- 25.Song E, et al. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nat Biotechnol. 2005;23:709–717. doi: 10.1038/nbt1101. [DOI] [PubMed] [Google Scholar]
- 26.Kleiner G, Marcuzzi A, Zanin V, Monasta L, Zauli G. Cytokine levels in the serum of healthy subjects. Mediators Inflamm. 2013;2013:434010. doi: 10.1155/2013/434010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pluckthun A. Designed ankyrin repeat proteins (DARPins): binding proteins for research, diagnostics, and therapy. Ann Rev Pharmacol Toxicol. 2015;55:489–511. doi: 10.1146/annurev-pharmtox-010611-134654. [DOI] [PubMed] [Google Scholar]
- 28.Schnell U, Cirulli V, Giepmans BN. EpCAM: structure and function in health and disease. Biochim Biophys Acta. 2013;1828:1989–2001. doi: 10.1016/j.bbamem.2013.04.018. [DOI] [PubMed] [Google Scholar]
- 29.Kerkis A, Hayashi MA, Yamane T, Kerkis I. Properties of cell penetrating peptides (CPPs) IUBMB Life. 2006;58:7–13. doi: 10.1080/15216540500494508. [DOI] [PubMed] [Google Scholar]
- 30.Nishi K, Saigo K. Cellular internalization of green fluorescent protein fused with herpes simplex virus protein VP22 via a lipid raft-mediated endocytic pathway independent of caveolae and Rho family GTPases but dependent on dynamin and Arf6. J Biol Chem. 2007;282:27503–27517. doi: 10.1074/jbc.M703810200. [DOI] [PubMed] [Google Scholar]
- 31.Elliott G, O'Hare P. Intercellular trafficking and protein delivery by a herpesvirus structural protein. Cell. 1997;88:223–233. doi: 10.1016/s0092-8674(00)81843-7. [DOI] [PubMed] [Google Scholar]
- 32.Elliott G, O'Hare P. Intercellular trafficking of VP22-GFP fusion proteins. Gene therapy. 1999;6:149–151. doi: 10.1038/sj.gt.3300850. [DOI] [PubMed] [Google Scholar]
- 33.Erbs P, et al. In vivo cancer gene therapy by adenovirus-mediated transfer of a bifunctional yeast cytosine deaminase/uracil phosphoribosyltransferase fusion gene. Cancer Res. 2000;60:3813–3822. [PubMed] [Google Scholar]
- 34.Ferras C, et al. Abrogation of Microsatellite-instable Tumors Using a Highly Selective Suicide Gene/Prodrug Combination. Mol Ther. 2009;17:1373–1380. doi: 10.1038/mt.2009.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chinnasamy D, et al. Gene therapy using genetically modified lymphocytes targeting VEGFR-2 inhibits the growth of vascularized syngenic tumors in mice. J Clin Invest. 2010;120:3953–3968. doi: 10.1172/JCI43490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shuai K, Liu B. Regulation of JAK-STAT signalling in the immune system. Nat Rev Immunol. 2003;3:900–911. doi: 10.1038/nri1226. [DOI] [PubMed] [Google Scholar]
- 37.Finbloom DS, Winestock KD. IL-10 induces the tyrosine phosphorylation of tyk2 and Jak1 and the differential assembly of STAT1 alpha and STAT3 complexes in human T cells and monocytes. J Immunol. 1995;155:1079–1090. [PubMed] [Google Scholar]
- 38.Chen ZS, Tiwari AK. Multidrug resistance proteins (MRPs/ABCCs) in cancer chemotherapy and genetic diseases. FEBS J. 2011;278:3226–3245. doi: 10.1111/j.1742-4658.2011.08235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wajant H, Gerspach J, Pfizenmaier K. Engineering death receptor ligands for cancer therapy. Cancer Lett. 2013;332:163–174. doi: 10.1016/j.canlet.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 40.Auslander S, Auslander D, Fussenegger M. Synthetic Biology-The Synthesis of Biology. Angew Chem Int Ed. 2017;56:6396–6419. doi: 10.1002/anie.201609229. [DOI] [PubMed] [Google Scholar]
- 41.Haellman V, Fussenegger M. Synthetic Biology-Toward Therapeutic Solutions. J Mol Biol. 2016;428:945–962. doi: 10.1016/j.jmb.2015.08.020. [DOI] [PubMed] [Google Scholar]
- 42.Simonsen JL, et al. Telomerase expression extends the proliferative life-span and maintains the osteogenic potential of human bone marrow stromal cells. Nat Biotechnol. 2002;20:592–596. doi: 10.1038/nbt0602-592. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.