Summary
Plants fine-tune their sophisticated immunity systems in response to pathogen infections. We previously showed that AtlsiRNA-1, a bacteria-induced plant endogenous small interfering RNA, silences the AtRAP gene, which encodes a putative RNA binding protein.
In this study, we demonstrate that AtRAP functions as a negative regulator in plant immunity by characterizing molecular and biological responses of the knockout mutant and overexpression lines of AtRAP upon bacterial infection.
AtRAP is localized in chloroplasts and physically interacts with Low Sulfur Upregulated 2 (LSU2), which positively regulates plant defense. Our results suggest that AtRAP negatively regulates defense responses by suppressing LSU2 through physical interaction. We also detected downregulation of the transcription factor GOLDEN2-LIKE 1 (GLK1) in atrap-1 using microarray analysis. The glk1 glk2 double mutant showed enhanced resistance to Pseudomonas syringae pv. tomato, which is consistent with a previous study showing enhanced resistance of a glk1 glk2 double mutant to Hyaloperonospora arabidopsidis.
Taken together, our data suggest that silencing of AtRAP by AtlsiRNA-1 upon bacterial infection triggers defense responses through regulation of LSU2 and GLK1.
Keywords: AtRAP, GLK1, glk1 glk2, LSU2, plant defense
Introduction
Plants have evolved complex immune systems to defend against pathogens (Dodds & Rathjen, 2010; Schwessinger & Ronald, 2012; Dangl et al., 2013). In particular, pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) provides the first line of defense to protect the plants from infection of the vast majority of potential pathogens. In addition, plants have evolved a second line of defense to perceive pathogen effector proteins that suppress PTI. This second layer of protection is mediated by specific components, such as resistance (R) proteins that are involved in the recognition of effectors to activate effector-triggered immunity (ETI) (Chisholm et al., 2006; Jones & Dangl, 2006; Dodds & Rathjen, 2010; Schreiber et al., 2011; Dangl et al., 2013). These plant defense responses are orchestrated by a complex transcriptional reprogramming of host cells that regulates activation of various defense-related genes, accumulation of plant hormones and production of reactive oxygen species (ROS) (Grant & Jones, 2009). To benefit plants, the pathways involved in defense responses need to be inactivated under normal growth conditions, but should be activated quickly upon pathogen attack. Thus, activation of the plant defense responses is controlled by complex interconnected signaling networks regulating various plant functions (Feys & Parker, 2000; Katagiri, 2004; Pieterse et al., 2009).
Increasing evidence shows that small RNAs (sRNAs) have pivotal roles in regulating the complex defense signaling network (Ruiz-Ferrer & Voinnet, 2009; Katiyar-Agarwal & Jin, 2010; Seo et al., 2013). sRNAs are short, noncoding RNA molecules that guide transcriptional and post-transcriptional silencing of gene expression (Baulcombe, 2004). Regulatory sRNAs are divided into two classes: microRNAs (miRNAs) and small interfering RNAs (siRNAs). Various miRNAs and siRNAs, as well as some sRNA pathway components are specifically induced by infections of different pathogens, and regulate host PTI and/or ETI pathways, as well as pathogen virulence (Ruiz-Ferrer & Voinnet, 2009; Zhang et al., 2011; Weiberg et al., 2013, 2014; Niu et al., 2016). We previously identified a novel class of long siRNAs (lsiRNAs), which were induced by pathogen infection (Katiyar-Agarwal et al., 2006). One of the identified lsiRNAs, AtlsiRNA-1, targets the Arabidopsis thaliana protein containing a RNA-binding domain abundant in Apicomplexans (AtRAP) gene (Katiyar-Agarwal et al., 2006; Kleinknecht et al., 2014). Because the knockout mutant of AtRAP (atrap-1) showed increased resistance to Pseudomonas syringae pv. tomato (Pst) infection, it was suggested that AtRAP negatively regulates plant defense responses (Katiyar-Agarwal et al., 2007).
In this study, we further investigated the molecular mechanism underlying the function of AtRAP in plant disease resistance. We show that representative defense-related genes and immune responses were activated in the atrap-1 mutant. We demonstrate that AtRAP protein directly interacts with Low Sulfur Upregulated 2 (LSU2) protein, which positively regulates plant defense. Microarray analyses revealed a set of genes that were differentially expressed in the atrap-1 mutant as compared with the wild type, including the transcription factor GOLDEN2-LIKE 1 (GLK1), another negative regulator of plant defense.
Materials and Methods
Plant materials and growth conditions
Arabidopsis thaliana L. Heynh. plants were grown in a controlled growth room at 23°C, with a 12 h : 12 h, light : dark photoperiod. The Arabidopsis knockout mutants atrap-1 (CS_844807), lsu2 (SALK_031648) and glk1 glk2 (Atglk1.1; Atglk2.1) were obtained from the Arabidopsis Biological Resource Center (ABRC). The double mutant atrap lsu2 was generated by crossing the single mutants atrap-1 and lsu2. The double mutant was selected by PCR of the genomic DNA for the homologous T-DNA insertion and confirmed by reverse transcription polymerase chain reaction (RT-PCR) to show no or very low expression of AtRAP and LSU2, respectively, in the double mutant.
Pathogen inoculation
Pathogen inoculations were performed as previously described (Katiyar-Agarwal et al., 2007). In brief, for RNA, protein and hypersensitive response (HR) analysis, plants were infiltrated with Pst DC3000 carrying an empty vector (EV) (pVSP61) or effector gene avrRpt2 at a concentration of 2 × 107 colony-forming units (CFU) ml −1. For bacterial growth assays, plants were infiltrated with Pst (EV) or Pst (avrRpt2) at a concentration of 2 × 105 CFU ml −1. At least six leaf disks were collected at each time point for 0 and 3 d post-inoculation (dpi) by a cock borer. The 0 dpi samples were immediately collected after inoculation. Bacterial titers were measured by grinding, diluting, plating, culturing and counting colonies. Student’s t-test (two samples) and ANOVA test (when comparing more than two samples) were used for significance difference calculation between Col-0 WT and mutant plants, or Col-0 WT and overexpression lines.
RNA extraction, quantitative real-time PCR and Northern blot analysis
Total RNA extraction was performed using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions, and mRNA extraction was carried out using the Oligotex mRNA Mini Kit (Qiagen).
To check the expression levels of PR1 (AT2G14610), PR2 (AT3G57260), RBOHd (AT5G47910), RBOHf (AT1G64060) and PDF1.2 (AT5g44420), cDNA was synthesized from 500 ng of mRNA using Superscript III (Invitrogen). The resulting cDNA was subjected to quantitative real-time PCR using an iCycler iQ5 real-time PCR detection system (Bio-Rad) with specific primers. The sequences of specific primers are given in Supporting Information Table S1.
Northern blot analysis was performed as previously described with minor modifications (Seo et al., 2009). Briefly, 10 µg of total RNA was fractionated by electrophoresis and transferred to positively charged nylon membranes (Amersham). Hybridization was performed using ULTRAhyb-Oligo hybridization buffer (Ambion) and 32P end-labeled DNA oligonucleotide probes specific for each target gene, according to the manufacturer’s protocol. The sequences of probes are given in Table S2.
DAB staining
3,3′-Diaminobenzidine (DAB) staining was performed to detect ROS as described previously (ThordalChristensen et al., 1997). Briefly, leaves were stained with 1 mg ml −1 DAB (Sigma) by vacuum infiltration, and destained with 90% ethanol. The destained samples were mounted in 50% glycerol and observed under a light microscope.
Plasmid DNA constructs and generation of transgenic Arabidopsis plants
The full-length AtRAP coding sequence (CDS) was cloned into a p35SGATFH destination vector with a C-terminal Flag tag. The construct was transformed into atrap-1 by Agrobacterium tumefaciens strain GV3101. The full-length AtRAP CDS was cloned into pEarlyGate101 with a yellow fluorescent protein (YFP) tag. The full-length LSU2 CDS was cloned into pEarlyGate102 or pEarlyGate202, with a cyan fluorescent protein (CFP) or Flag tag, respectively. The vectors were transformed into A. tumefaciens strain GV3101 and used for transient expression of recombinant proteins.
Confocal microscopy
Subcellular localization of AtRAP-YFP and LSU2-CFP was observed by confocal microscopy using a Leica SP2/SP5 laser-scanning confocal microscope (Leica, Wetzlar, Germany), equipped with a specific laser/filter combination to detect CFP (excitation at 458 nm), YFP (excitation at 514 nm) and red fluorescent protein (RFP) (excitation at 594 nm).
Yeast two-hybrid screening
Yeast two-hybrid screening was performed using a cDNA library (generously provided by T. Eulgem) of pooled RNAs from 2-wk-old Arabidopsis Col-0 WT seedlings constructed using the HybriZAP-2.1 library construction system (Stratagene, la Jolla, CA, USA). The Arabidopsis library cDNAs (1.3 × 106) were screened by transformation into the AH109 yeast strain (Clontech, Palo Alto, CA, USA) expressing AtRAP-DNA-binding domain (BD) bait fusion protein generated by the GAL4 system (Stratagene,), as described by the manufacturer. Plasmids were isolated from the positive clones and transformed into Escherichia coli DH5α to amplify for sequencing. The plasmids from the positive clones were retransformed into the yeast transformant expressing AtRAP-BD to verify the interaction. The interactions between SV40 large T antigen (84–708) (pTD1-1) and either murine p53(72–390) (pVA3-1) or human lamin C(66–230) (pLAM5′-1) served as positive and negative controls, respectively.
Co-immunoprecipitation assay
Total protein extracts were prepared from the Nicotiana benthamiana leaves infiltrated with A. tumefaciens containing the AtRAP-YFP and/or LSU2-FLAG constructs. At 3 dpi, infiltrated leaves were homogenized in three volumes of protein extraction buffer (20mM Tris–HCl at pH 7.5, 300mM NaCl, 5 mM MgCl2, 5mM dithiothreitol, 0.5% Triton X-100, proteinase inhibitor cocktail; Sigma). Cell debris was removed by centrifugation at 18 000 g for 20 min at 4°C. The resulting supernatants were incubated with anti-YFP antibody-conjugated agarose beads (Roche) for 8 h at 4°C. The immunocomplexes were then precipitated by centrifugation for 1 min at 8200 g, and washed three times in 1 ml phosphate-buffered saline (PBS: 0.1M NaCl, 90mM sodium phosphate, pH 7.0). The resulting samples were analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), followed by immunoblot analysis using anti-YFP (Roche) and anti-FLAG (Sigma) antibodies.
Microarray analysis
For Affymetrix GeneChip array analysis, we normalized expression profiles with the Robust multiarray analysis (RMA) method (Irizarry et al., 2003). A list of genes with statistically significant changes in expression between the genotypes was generated by the significance analysis of microarrays (SAM) method in which multiple testing was taken into account through q-value (Siggenes, R package v.1.50.0). We used a threshold q-value of 0.05 to select the genes whose expression levels are statistically significantly changed.
Accession number
The microarray data of this paper are deposited at NCBI (GSE98376).
Results
Knocking out AtRAP gene activates defense-related genes and increases ROS accumulation
We previously demonstrated that the AtRAP knockout mutant is more resistant to both virulent Pst carrying an empty vector (EV) and avirulent Pst (avrRpt2) than to the Col-0 wild-type (WT) plants (Katiyar-Agarwal et al., 2007), suggesting that AtRAP has a negative role in defense against bacteria. Infection of Arabidopsis with Pst triggers transcriptional reprogramming of a variety of defense-related genes (Dong et al., 1991; Panstruga et al., 2009). To determine whether expression of the defense-related genes was altered in the atrap mutant, the leaf samples infiltrated with Pst (EV) or Pst (avrRpt2) were collected at different time points, and total RNAs were extracted and subjected to quantitative real-time RT-PCR. We first investigated the induction levels of two representative antimicrobial transcripts: PATHOGENESIS-RELATED PROTEIN 1 (PR1, AT2G14610) and PR2 (AT3G57260). PR1 and PR2 were induced to a higher level in the atrap mutant than in Col-0 WT plants at 10 h post-inoculation (hpi) with both Pst (EV) and Pst (avrRpt2) strains (Fig. 1a,b). PR1 and PR2 transcript levels were also slightly higher in the atrap mutant at 0 hpi than in the Col-0 WT plants (Fig. 1a,b), suggesting that the atrap mutant has an elevated basal defense level. Since PR1 and PR2 are two salicylic acid (SA)-dependent marker genes and the SA pathway is antagonistic to the jasmonic acid (JA) pathway (Kunkel & Brooks, 2002; Anderson et al., 2004), we next examined the expression level of a JA-responsive marker gene: PDF1.2. As shown in Fig. S1(a), the expression of PDF1.2 transcripts was significantly suppressed in the atrap mutant compared with Col-0 WT plants before and after infection.
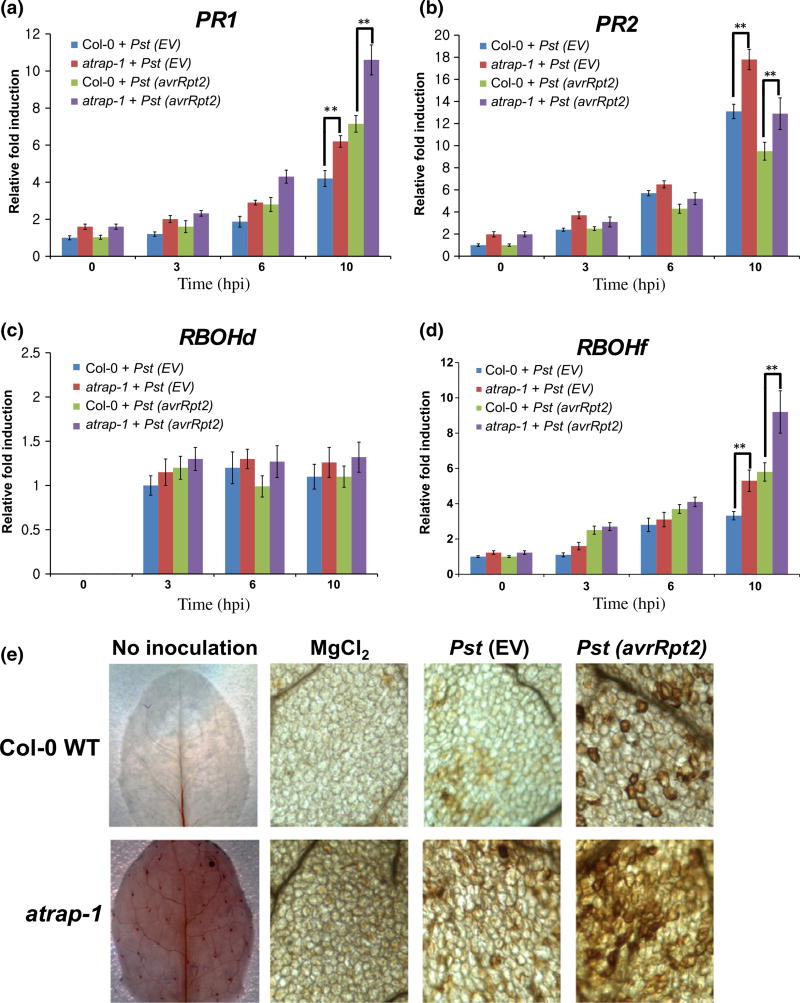
Fig. 1.
Effect of AtRAP knockout mutation on defense responses. Five-week-old Arabidopsis Col-0 wild-type (WT) and the atrap mutant plants were infiltrated with Pseudomonas syringae pv. tomato (Pst) (EV) or Pst (avrRpt2) at a concentration of 2 ×107 colony-forming units (CFU) ml−1. Total RNAs were extracted from the infiltrated leaves at the times indicated on the horizontal axes. Relative accumulation levels of (a) PR1, (b) PR2, (c) RBOHd and (d) RBOHf transcripts were quantified by quantitative real-time RT-PCR. Error bars represent the ±SEM from three independent experiments. **, P < 0.01 (determined by Student’s t-test). (e) The atrap-1 mutant exhibits increased ROS accumulation. Five-week-old Arabidopsis Col-0 WT and the atrap mutant plants were infiltrated with MgCl2, Pst (EV), or Pst (avrRpt2) at a concentration of 2 ×107 CFU ml−1. DAB staining was performed to visualize reactive ROS accumulation at 6 hpi.
Pst (avrRpt2) induces the HR in Col-0 WT plants that carry the cognate resistance gene RPS2 (Kunkel et al., 1993; Mackey et al., 2003). When we examined the HR in the atrap mutant, we observed an earlier and more severe induction of cell death (half leaf inoculation) than in the Col-0 WT plants (Fig. S1b). During the HR progression, the production of ROS is known to trigger programmed cell death and subsequent defense responses (Torres et al., 2006). It has been demonstrated that plant NADPH oxidase genes, including RBOHd (AT5G47910) and RBOHf (AT1G64060), play crucial roles in regulating the generation of ROS in plant defense (Torres et al., 2002; Miller et al., 2009; Vellosillo et al., 2010). We found that RBOHf was induced to a higher level in the atrap mutant than in the Col-0 WT in response to Pst (EV) and Pst (avrRpt2) infection, while no significant alteration of RBOHd mRNA expression was observed in the atrap mutant (Fig. 1c,d). This suggests that RBOHf may play a more important role than RBOHd in regulating ROS production and HR cell death in the atrap mutant upon pathogen attack.
Since we observed increased induction of RBOHf in the atrap mutant upon infection with Pst (avrRpt2) (Fig. 1d), we examined whether there is an increase in ROS accumulation in the atrap mutant. To detect ROS, DAB staining was performed on leaves of Col-0 WT and the atrap mutant plants 6 h post inoculation (hpi) with MgCl2, Pst (EV) or Pst (avrRpt2). DAB staining was visualized as red–brown precipitates. We detected more intense staining in the atrap mutant infiltrated with Pst (avrRpt2) than in the Col-0 WT (Fig. 1e). Our results indicate that atrap more sensitively reacts to Pst to induce ROS production.
Overexpression of AtRAP results in reduced resistance to Pst
The atrap mutant is smaller and displays a virescent phenotype (Katiyar-Agarwal et al., 2007). To determine whether the elevated defense responses observed earlier in the atrap mutant were due to the secondary effect from its developmental defect, or whether AtRAP is directly involved in plant defense responses, we generated overexpression lines of AtRAP in the atrap mutant background. The expression of AtRAP fused with a FLAG tag at the C-terminus was driven by the constitutive 35S promoter. The overexpression lines fully complemented the retarded growth and the virescent phenotype of the atrap mutant, and did not show any distinguishable phenotypes when compared with Col-0 WT plants (Fig. 2a). A transgenic line with a high expression level of AtRAP (#34) and a line with a medium expression level of AtRAP (#21) (Fig. 2b) were chosen and subjected to pathogen growth analysis. These overexpression lines exhibited enhanced disease susceptibility to both virulent Pst (EV) and avirulent Pst (avrRpt2) in proportion to the expression levels of AtRAP-FLAG (Fig. 2c,d). Our results suggest that AtRAP directly and negatively regulates plant defense responses. Taken together with our previous study (Katiyar-Agarwal et al., 2006), the downregulation of AtRAP by AtlsiRNA-1-mediated silencing could be a regulatory mechanism to promptly elevate disease resistance in plants upon bacterial infection.
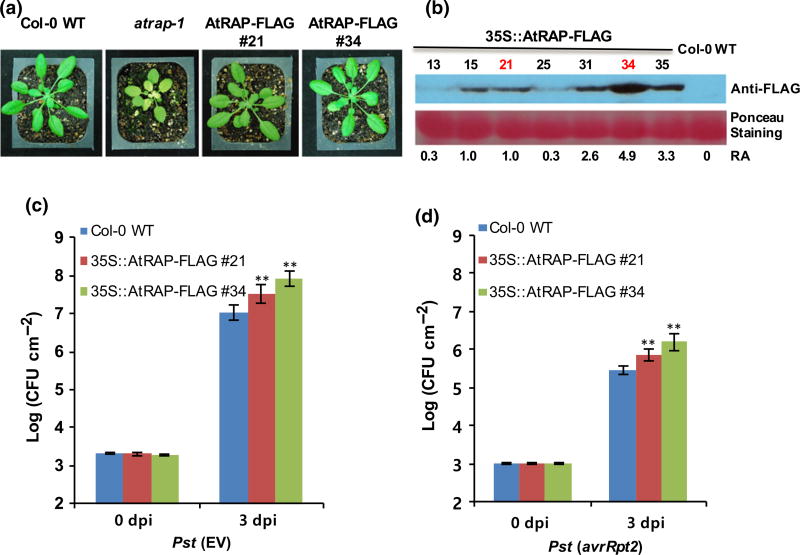
Fig. 2.
Effects of AtRAP overexpression on disease resistance to Pseudomonas syringae pv. tomato (Pst) in Arabidopsis. (a) Phenotypes of the AtRAP knockout mutant and overexpression plants. (b) Immuno-detection of AtRAP-FLAG with anti-FLAG antibody in extracts from seven independent AtRAP overexpression lines. (c, d) Growth of Pst (EV) and Pst (avrRpt2) in two independent AtRAP overexpression lines at 0 and 3 dpi. Arabidopsis plants were infiltrated with Pst (EV) or Pst (avrRpt2) at a concentration of 2 ×105 CFU ml−1. At least six leaf disks were collected at 0 and 3 dpi by a cock borer. The 0 dpi samples were immediately collected after inoculation. Bacterial titers were measured by grinding, plating, culturing and counting colonies. An ANOVA test was used for significance difference calculation between Col-0 wild-type (WT) and overexpression lines. Error bars represent ±SD of more than six replicates. Similar results were obtained from three independent experiments. **, P < 0.01 (determined by Student’s t-test).
AtRAP interacts with LSU2, a positive regulator of plant resistance
To understand how AtRAP regulates plant immunity, we attempted to identify interacting partner proteins of AtRAP using yeast two-hybrid screening. We screened a total of 1.3 × 106 independent yeast transformants using AtRAP as bait and identified several positive candidates. Further analyses revealed that one of the positive candidates encodes LSU2 (AT5G24660) (Fig. 3a), which is known to be highly expressed under sulfur deprivation (13.6-fold upregulation) according to the GENEVESTIGATOR database (Zimmermann et al., 2004).
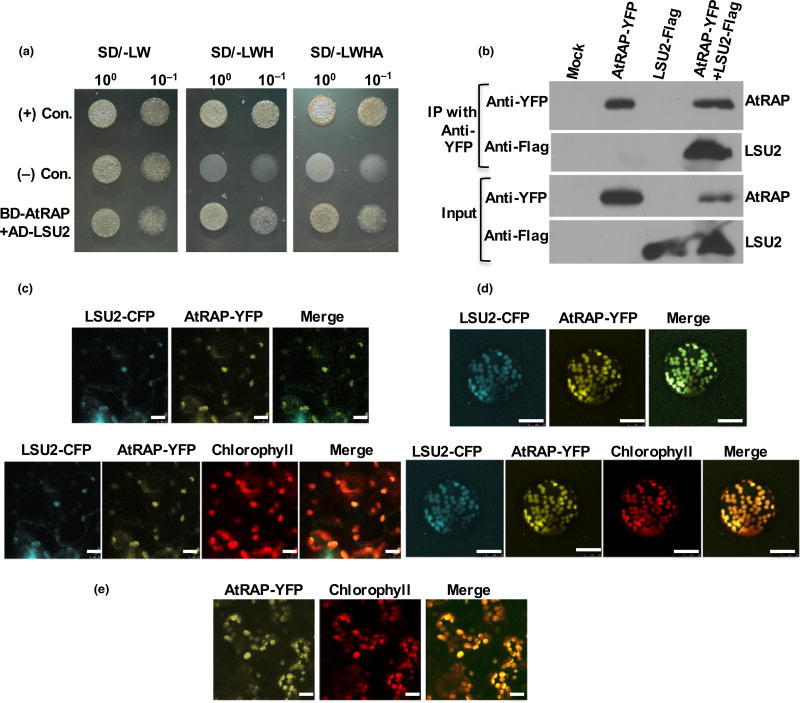
Fig. 3.
AtRAP interacts with LSU2. (a) Interaction between AtRAP and LSU2 in yeast two-hybrid assay. Full-length LSU2 was fused downstream of GAL4-AD in pACT2. Full-length AtRAP was fused downstream of GAL4-BD of pAS2-1. Yeast cells co-transformed with pACT2 and pAS2-1 fusion derivatives were selected on Synthetic Defined (SD) lacking leucine, tryptophan and histidine (SD/-LWH) and SD laking leucine, tryptophan, histidine and adenine (SD/-LWHA) agar media. (b) Co-immunoprecipitation assay in Nicotiana benthamiana leaves. AtRAP and LSU2, tagged with YFP and FLAG, respectively, were coexpressed in N. benthamiana leaves by agro-infiltration and immunoprecipitated with anti-YFP antibody-conjugated agarose beads. Expression of AtRAP-YFP and LSU2-FLAG in total protein extracts was confirmed by Western blotting using anti-YFP and anti-FLAG antibodies, respectively. Total protein extract from leaves infiltrated with the infiltration buffer was used as a negative control (mock). (c, d) AtRAP is co-localized with LSU2 in chloroplasts. AtRAP and LSU2, which were respectively tagged with YFP and CFP, were coexpressed in N. benthamiana leaves by agro-infiltration. The fluorescence signals of YFP and CFP are shown in the yellow and cyan channels, respectively. Both the (c) infiltrated leaves and (d) protoplasts of N. benthamiana were subjected to confocal microscopy. Chloroplasts emit red fluorescence. (e) AtRAP-YFP localizes in chloroplasts from transgenic plants expressing AtRAP-YFP under the 35S promoter. The fluorescence signal of YFP is shown in the yellow channel. Chloroplasts emit red fluorescence. Bars: (c, e) 10 µm; (d) 25 µm.
To confirm the interaction between AtRAP and LSU2, we performed a co-immunoprecipitation assay (Co-IP). AtRAP and LSU2, which were respectively tagged with YFP and FLAG, were coexpressed in N. benthamiana leaves and the results shown in Fig. 3(b) confirmed that LSU2 is an interacting partner of AtRAP.
A recent study demonstrated that AtRAP is localized in chloroplast in N. benthamiana using a transient expression assay (Kleinknecht et al., 2014). Because LSU2 interacts with AtRAP, we hypothesized that LSU2 is at least partially localized in the chloroplasts and interacts with AtRAP. The advanced protein subcellular localization prediction tool WoLF PSORT also predicted LSU2 to be present in the chloroplast/plastid (http://www.genscript.com/wolf-psort.html). To test our hypothesis, AtRAP tagged with YFP (AtRAP-YFP) at its C-terminal end and LSU2 with a C-terminal CFP (LSU2-CFP) were coexpressed in N. benthamiana. As expected, LSU2-CFP co-localized with AtRAP-YFP in the chloroplast (Fig. 3c,d). We then generated transgenic plants using the same construct to express the AtRAP-YFP in the atrap mutant background. These transgenic lines complemented the retarded growth and the virescent phenotype of atrap (Fig. S2), and showed expression of AtRAP-YFP in the chloroplasts (Fig. 3e). This result further confirmed our finding.
Next, to assess the biological function of LSU2, we examined the loss-of-function mutant lsu2 in disease resistance to Pst. The lsu2 mutant was developmentally and phenotypically indistinguishable from Col-0 WT plants. Pathogen growth assays revealed that the lsu2 mutant expresses an enhanced disease susceptibility to both Pst (EV) and Pst (avrRpt2) compared with the Col-0 WT plants (Fig. 4a,b), indicating that LSU2 is a positive regulator of plant defense. Furthermore, the LSU2 transcript level was slightly upregulated upon Pst (avrRpt2) infection in Col-0 WT (Fig. 4c), whereas the atrap mutant exhibited increased expression of LSU2 transcripts, and significant upregulation of LSU2 transcripts upon Pst infection (Fig. 4c). To confirm whether expression levels of AtRAP really affect the level of LSU2 transcripts, we performed Northern blot analysis to examine LSU2 RNA abundance in the AtRAP overexpression line using Col-0 WT and atrap-1 as controls. As expected, the LSU2 transcript level was downregulated in the AtRAP-overexpressed line (Fig. S3), suggesting that AtRAP negatively regulates LSU2 transcript level.
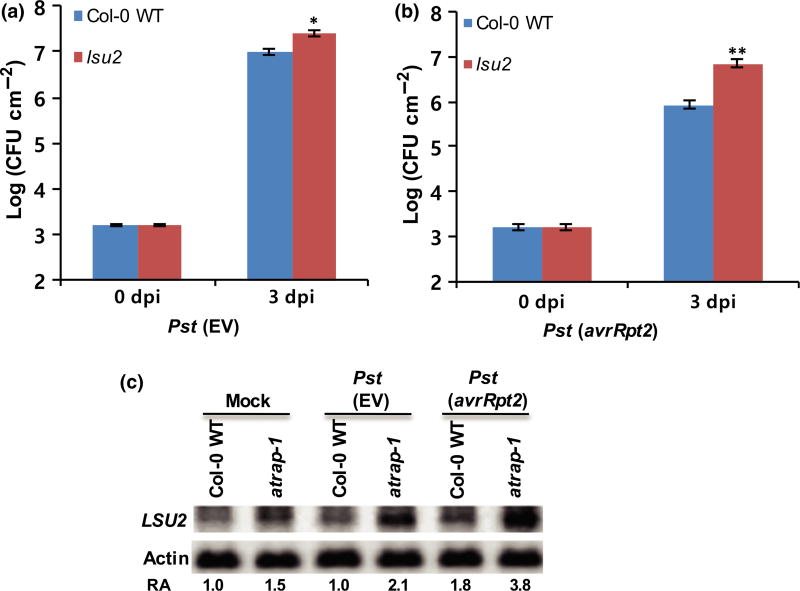
Fig. 4.
LSU2 is a positive regulator of plant defense against Pseudomonas syringae pv. tomato (Pst) in Arabidopsis. (a, b) Growth of Pst (EV) and Pst (avrRpt2) in the lsu2 mutant plants at 0 and 3 dpi. Error bars represent ±SD of more than six replicates. Similar results were obtained from three independent experiments. **, P < 0.01; *, P < 0.05 (determined by Student’s t-test). (c) Detection of LSU2 mRNA in the Col-0 wild-type (WT) and the atrap mutant plants. Five-week-old Arabidopsis Col-0 WT and the atrap mutant plants were infiltrated with Pst (EV) or Pst (avrRpt2) at a concentration of 2 ×107 CFU ml −1. Total RNAs were extracted from the infiltrated leaves at 8 hpi and subjected to Northern blot analysis.
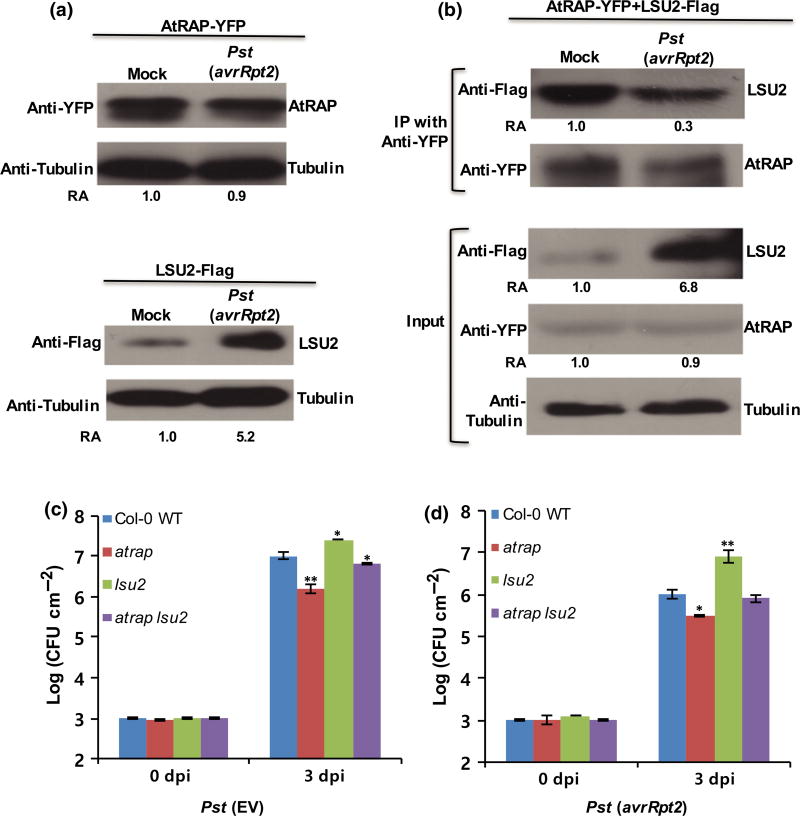
Since we found that LSU2 interacts with AtRAP (Fig. 3), we examined whether bacterial infection affects the interaction between LSU2 and AtRAP. Because the antibodies specific for LSU2 and AtRAP are not yet available, experiments were performed by transiently expressing LSU2-Flag and AtRAP-YFP in N. benthamiana as shown in the Co-IP assay (Fig. 3b). At 2 d after agroinfiltration of AtRAP-YFP and LSU2-Flag, Pst (avrRpt2) was inoculated into the same leaves. After 1 d, total proteins were extracted from the infiltrated leaves and subjected to Western blot analysis using anti-YFP or anti-Flag antibodies. As expected, expression of AtRAP in N. benthamiana did not change significantly upon Pst (avrRpt2) infection (Fig. 5a) because the 3′-untranslated region which contains the lsiRNA target site is not present in the expression construct. However, a higher accumulation of LSU2-Flag was observed in the leaves inoculated with Pst (avrRpt2) than in the leaves inoculated with inoculation buffer (control) (Fig. 5a), which is consistent with the RNA accumulation pattern of LUS2 after infection. To perform Co-IP, AtRAP-YFP and LSU2-Flag were coexpressed in N. benthamiana leaves by agroinfiltration. Two days later, Pst (avrRpt2) was inoculated into the N. benthamiana leaves. Total proteins were extracted from the inoculated leaves at 1 dpi and subjected to Co-IP using anti-YFP antibody-conjugated agarose beads. Western blot analyses using anti-YFP and anti-Flag antibodies were performed to examine the co-immunoprecipitated products. Interestingly, the amount of LSU2 which co-immunoprecipitated with AtRAP was significantly decreased in the leaf samples inoculated with Pst (avrRpt2) (Fig. 5b), although the entire accumulation level of LSU2 was significantly higher than that of the blank control. This result suggested that the physical interaction between AtRAP and LSU2 can be disrupted upon bacterial infection and that this results in release of LSU2, which positively regulates host defense. Conversely, this result suggests that AtRAP plays a regulatory role in suppressing defense responses by inactivating LSU2 through physical interaction.
Fig. 5.
Effects of bacterial infection on the interaction between AtRAP and LSU2. (a) AtRAP-YFP and LSU2-Flag were separately expressed in Nicotiana benthamiana leaves by agro-infiltration. After 2 d, Pseudomonas syringae pv. tomato (Pst) (avrRpt2) was inoculated into the leaves infiltrated with AtRAP-YFP or LSU2-Flag. After 1 d, total proteins were extracted from the infiltrated leaves and subjected to Western blot analysis using anti-YFP or anti-Flag antibodies. (b) Co-immunoprecipitation of AtRAP with LSU2. AtRAP-YFP and LSU2-Flag were coexpressed in N. benthamiana leaves by agro-infiltration. After 2 d, Pst (avrRpt2) was inoculated into the leaves. After 1 d, total proteins were extracted from the infiltrated leaves and subjected to co-immunoprecipitation using anti-YFP antibody-conjugated agarose beads. Western blot analysis using anti-YFP or anti-Flag antibodies was performed to analyze the resulting co-immunoprecipitated products. (c, d) Growth of Pst (EV) and Pst (avrRpt2) in the atrap lsu2 mutant plants at 0 and 3 dpi. The atrap lsu2 double mutant is less resistant than the atrap single mutant, and is less susceptible than the lsu2 single mutant to bacterial infection. Error bars represent ±SD of more than six replicates. Similar results were obtained from three independent experiments. **, P < 0.01; *, P < 0.05 (determined by Student’s t-test).
To validate the antagonistic relationship between AtRAP and LSU2 in vivo, we generated the double mutant of atrap lsu2. The double mutant still displayed the virescent phenotype as with the atrap single mutant (Fig. S4a,b). We then performed pathogen growth assays on atrap lsu2. The double mutant became less resistant than the single atrap mutant, but less susceptible than the single lsu2 mutant when challenged with both virulent and avirulent strains (Fig. 5c,d), indicating that AtRAP antagonistically regulates LSU2 by directing interacting with it.
Microarray analysis reveals altered gene expression in the atrap mutant
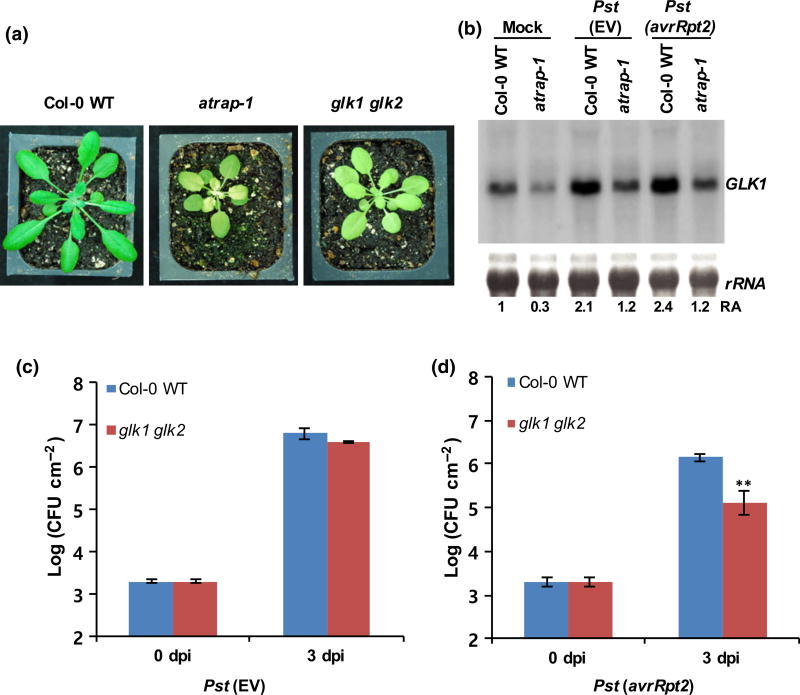
To identify genes that are regulated by AtRAP, we performed transcriptome analysis on the atrap mutant using Affymetrix ATH1 microarrays. Three biological replicates were analyzed using mRNAs extracted from rosette leaves of 3-wk-old Col-0 WT and the atrap mutant plants. The microarray results revealed that a total of 28 genes were expressed with significant differences between the atrap mutant and the Col-0 WT (i.e. 21 genes were downregulated and seven genes were upregulated in the atrap mutant; Tables S3, S4). Among them, the transcription factor GLK1 was downregulated in the atrap mutant (Table S3). GLK1 is a member of the GARP superfamily (Waters et al., 2008, 2009), and is closely related to another GARP family gene GLK2. GLK1 and GLK2 are differentially regulated, showing differential tissue expression patterns (Fitter et al., 2002). Interestingly, the double knockout mutant plant (glk1 glk2) shows a pale green leaf phenotype, somewhat similar to that of the atrap mutant (Fitter et al., 2002; Waters et al., 2009). Thus, we examined the functional significance of the GLK transcription factors in disease resistance to Pst. We first performed Northern blot analysis to confirm the downregulation of the GLK1 transcript level in the atrap mutant observed in the microarray analysis. To determine whether GLK1 is directly regulated by AtRAP or its downregulation in the atrap mutant is due to the retrograde signaling from the damaged plastids in the mutant, we examined the expression level of GLK1 in the AtRAP overexpression lines where there is no obvious plastid damage. We found that GLK1 expression was elevated in the AtRAP overexpression line, suggesting that AtRAP is likely to positively regulate GLK1 directly (Fig. S5), although we cannot rule out the possibility of retrograde signaling regulation after AtRAP expression change.
We also infiltrated Col-0 WT and the atrap mutant with either Pst (EV) or Pst (avrRpt2) to examine whether the expression level of GLK1 transcripts is altered by Pst infection. At 1 dpi, total RNAs were extracted from infected leaves and used for Northern blot analysis. Consistent with the microarray analysis, the Northern blot result showed that the GLK1 transcript level was down-regulated in the atrap mutant (Fig. 6b). Interestingly, the GLK1 transcript level was upregulated upon infection with Pst (EV) or Pst (avrRpt2) in both Col-0 WT and the atrap mutant. To assess the biological functions of the GLK1 and GLK2 transcription factors in disease resistance to Pst, we performed a pathogen growth assay using the double knockout mutant glk1 glk2, because GLK1 and GLK2 are functionally redundant. The analyses revealed that the glk1 glk2 mutant exhibits enhanced disease resistance to Pst (avrRpt2) when compared with Col-0 WT (Fig. 6c,d), suggesting that GLKs are negative regulators of plant defense against Pst (avrRpt2). Consistent results were obtained from three independent experiments.
Fig. 6.
GLK1 negatively regulates defense responses in Arabidopsis. (a) Phenotypes of the atrap-1 mutant and the glk1 glk2 mutant plants. (b) Detection of GLK1 mRNA in the Col-0 wild-type (WT) and the atrap mutant plants. Five-week-old Arabidopsis Col-0 WT and the atrap mutant plants were infiltrated with Pst (EV) or Pst (avrRpt2) at a concentration of 2 ×107 CFU ml−1. Total RNAs were extracted from the infiltrated leaves at 8 hpi after inoculation and subjected to Northern blot analysis. (c, d) Growth of Pst (EV) and Pst (avrRpt2) in the glk 1gkl2 mutant plants at 0 and 3 dpi. Error bars represent ±SD of more than six replicates. Similar results were obtained from three independent experiments. **, P < 0.01 (determined by Student’s t-test).
Since AtRAP has been shown to localize in the chloroplast (Fig. 3), it is noteworthy that, of 28 genes, eight have been annotated to be located in the chloroplast (TAIR; www.arabidopsis.org) (Tables S3, S4): ABC1 (a protein kinase superfamily protein; At5g05200), a dehydrin family protein (At1g54410), an aspartate-glutamate racemase family protein (At1g15410), FDH (an NAD-dependent formate dehydrogenase; At5g14780), MRL1 (a pentatricopeptide repeat protein; At4g34830), an RNA-binding (RRM motif) family protein (At1g70200), an FAD/NAD(P)-binding oxidoreductase family protein (At1g57770) and RNA polymerase beta′subunit-2 (Atcg00170). These genes have been shown to be intimately involved in chloroplast development and/or metabolic processes (Lange & Ghassemian, 2003; Dal Bosco et al., 2004; Friso et al., 2004; Ytterberg et al., 2006; Johnson et al., 2010; Steiner et al., 2011). Since the atrap mutant shows a virescent phenotype, further experiments may reveal how these genes are involved in contributing to its biological phenotype.
Discussion
Through loss- and gain-of-function studies using the knockout mutant and overexpression lines of AtRAP, respectively, we showed that AtRAP functions as a negative regulator of plant innate immunity because alteration of the AtRAP expression affects both compatible and incompatible interactions between Arabidopsis and Pst bacterial strains. Analyses of transcriptional levels of representative defense-related marker genes suggest that AtRAP is directly involved in suppression of plant immunity under normal growth conditions because the levels of PR1 and PR2 were constitutively upregulated in the atrap mutant (Fig. 1a,b). In addition, the atrap mutant induced more rapid and strong HR cell death upon pathogen infection than seen in the Col-0 WT (Fig. S1b). This seems to be associated with the constitutive accumulation, and sensitive production, of ROS in the atrap mutant (Fig. 1e), because ROS play a key role in both triggering HR and inducing defense-related genes (Torres et al., 2006). Both RBOHd and RBOHf are known to be involved in generating ROS in plant defense (Torres et al., 2002; Miller et al., 2009; Vellosillo et al., 2010). Our transcriptional analyses suggested that RBOHf, but not RBOHd, is responsible, at least in part, for sensitive production of ROS in the atrap mutant upon Pst infection (Fig. 1c,d). It has been shown that RBOHf is more important for the activation of HR cell death, although RBOHd contributes to the generation of total detected ROS in Arabidopsis (Torres et al., 2002). Transgenic plants that overexpress AtRAP, which have no obvious developmental defects, showed a more susceptible phenotype, further supporting that AtRAP is directly involved in regulating plant immunity.
Using a yeast two-hybrid screening with AtRAP as bait, we identified the AtRAP interacting protein, LSU2. A strong interaction between AtRAP and LSU2 was confirmed by Co-IP (Fig. 3b). LSU2 is known to be strongly expressed under sulfur deprivation (Zimmermann et al., 2004). A recent study showed that LSU2 is highly induced by a combination of light and plastid signaling, suggesting that LSU2 is involved in the optimization of chloroplast function (Ruckle et al., 2012). Chloroplast localization of LSU2 and its association with AtRAP may be associated with the function of LSU2 in chloroplast biogenesis. In this study, analysis of the lsu2 knockout mutant revealed that LSU2 is involved in disease resistance to both virulent and avirulent strains of Pst (EV) and Pst (avrRpt2), respectively (Fig. 4). While little information is available on the biological function of LSU2 in disease resistance, Mukhtar et al. (2011) have shown that LSU2 physically interacts with multiple effector proteins of Pst, and with Hyaloperonospora arabidopsidis (Hpa), and is required for full immune function during Hpa infection. Consistent with our results, their preliminary observations suggested that LSU2 is also required for Pst bacterial resistance mediated by the RPS2-dependent disease resistance pathway (Mukhtar et al., 2011). It is not clear how LSU2 mediates plant immunity against Pst (EV) and Pst (avrRpt2). Our observation that LSU2 was upregulated upon Pst infection at both the RNA and the protein levels (Figs 4c, 5a, S3) supports the role of LSU2 as a positive regulatory component in the surveillance system of plant defense. In addition, upon bacterial infection, the physical interaction between LSU2 and AtRAP was disrupted (Fig. 5b), indicating that an increase of the released LSU2 has a positive effect on plant defense. In this regard, it is likely that the activity of LSU2 as an immune activator may be suppressed by a physical interaction with AtRAP under normal growth conditions because these two proteins have antagonistic effects on plant immunity. This model was also supported by the results from functional characterization of the atrap lsu double mutant.
GLK1 and GLK2 are involved in chloroplast development and maintenance, and coordinate the expression of the photosynthetic apparatus (Waters et al., 2008, 2009). In the atrap mutant, the GLK1 transcript level was significantly downregulated (Figs 6b, S5), whereas in the AtRAP overexpression lines, GLK1 was highly induced (Fig. S5), suggesting that GLK1 expression is regulated by the AtRAP protein. Furthermore, our microarray results showed that expression levels of some nuclear genes involved in chloroplast development and/or metabolic processes were also significantly decreased in the atrap mutant (Table S3). Although it is unknown if GLK1 is a transcriptional activator that directly regulates these genes, the downregulation of those nuclear genes seems to be highly correlated with the biological phenotype of the atrap mutant, which is similar to the glk1 glk2 mutant. Interestingly, the overexpression of GLK1 resulted in significant down-regulation of PR1, but upregulation of PR10, suggesting that GLK1 may negatively modulate the defense signaling pathway (Qiu et al., 2007; Savitch et al., 2007). Our results are consistent with a study by Murmu et al. (2014), which indicates that GLK1 negatively regulates plant defense responses to the biotrophic oomycete pathogen H. arabidopsidis by activating a JA-dependent pathway. They and others also showed that GLK1 positively regulates plant defense responses to the necrotrophic fungal pathogens Botrytis cinerea and Fusarium graminearum (Savitch et al., 2007; Schreiber et al., 2011; Murmu et al., 2014). Thus, GLK1 is a negative regulator of plant immune responses against biotrophs, and a positive regulator of plant defense against necrotrophs.
Chloroplasts are important organelles for plant immunity. Retrograde signaling from chloroplasts regulates expression of various nuclear genes involved in the defense response (Nott et al., 2006). A recent study showed that expression of nuclear encoded chloroplast-targeted genes (NECGs) was significantly altered upon Pst infection (Zabala et al., 2015). Thus, some pathogen effectors, such as Hop1J, Hop U1 and HopR1 suppress plant immunity by targeting the chloroplast genes (Fu et al., 2007; Jelenska et al., 2007; Caplan et al., 2008; Zabala et al., 2015). The phenotypic characteristics of the atrap mutant, which are virescent and with retarded growth, further support the important role of AtRAP in the regulation of chloroplast functions under normal growth conditions. Accumulating evidence suggests a strong correlation between chloroplasts and the modulation of innate immunity (Wildermuth et al., 2001; Torres et al., 2006; Vlot et al., 2009). The discovery of photosynthesis inhibition by Pst infection suggests that comprehensive photosynthesis is essential for the plant defense system (Zabala et al., 2015). The chloroplast is not only a major source of ROS production, but also of antioxidants required to maintain ROS homeostasis (Mittler et al., 2004, 2011). The absence of functional AtRAP in chloroplasts might result in an imbalance of ROS homeostasis in the atrap mutant. Our results show that the atrap mutant exhibited sensitive induction of HR cell death and ROS accumulation upon pathogen inoculation (Figs 1, S1b). In addition, a recent study demonstrated that AtRAP functions in the maturation of 16S rRNA (Kleinknecht et al., 2014). Therefore, it is likely that AtRAP plays an essential role in the balancing and coupling of plant immunity and chloroplast functions.
A hypothetical working model for the AtRAP proteinmediated regulation in plant antibacterial immune responses is presented in Fig. S6. Under normal growth conditions in the absence of pathogen infection, the AtRAP protein directly interacts with LSU2 and suppresses LSU2-associated plant defense responses. At the same time, AtRAP is also required for the constitutive expression of GLK1, which is a transcription factor regulating the expression of various genes involved in chloroplast development and stress responses (Fig. S5a). Upon Pst infection, AtlsiRNA-1 is highly induced and silences its target gene AtRAP. Silencing of AtRAP results in the release and functional activation of LSU2, which initiates plant immune responses, as well as modulates chloroplast activities. Silencing of AtRAP also alters expression of the GLK1 transcription factor, resulting in the reprogramming of associated gene expression. Eventually, the AtlsiRNA1-induced silencing of AtRAP promptly activates defense responses upon pathogen attack. Further characterization of the biological functions of AtRAP and its associated proteins in plant defense and chloroplast function will expand our understanding of how plants fine-tune their signaling regulatory pathways to balance growth and defense.
Supplementary Material
Fig. S1 AtRAP negatively regulates plant defense to Pst.
Fig. S2 Phenotype of AtRAP-overexpressing plants.
Fig. S3 LSU2 is negatively regulated by AtRAP.
Fig. S4 Characterization of the atrap lsu2 double mutant.
Fig. S5 GLK1 is positively regulated by AtRAP.
Fig. S6 A hypothetical working model for the regulation of the AtRAP protein in plant antibacterial disease resistance.
Table S1 Quantitative-PCR primers used in this study
Table S2 DNA probes used in this study
Table S3 Genes down-regulated in the atrap mutant
Table S4 Genes up-regulated in the atrap mutant
Acknowledgments
We thank Jane A. Langdale for providing the glk1glk2 mutant seeds. The Arabidopsis yeast two-hybrid cDNA library was kindly provided by Thomas A. Eulgem. This work was supported by grants from the National Institute of Health (R01 GM093008), National Science Foundation (IOS-1557812) and an AES-CE Award (PPA-7517H) awarded to H.J.; and grants from the Agenda Program (PJ011306), and Rural Development Administration.
Footnotes
Author contributions
H.J. conceived the idea and designed the project. H.W., J-K.S. and S.G. performed the experiments. H.W., J-K.S. and H.J. analyzed the data and wrote the manuscript. X.C. analyzed microarray data. All authors have read and approved the manuscript.
Additional Supporting Information may be found online in the Supporting Information tab for this article:
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
References
- Anderson JP, Badruzsaufari E, Schenk PM, Manners JM, Desmond OJ, Ehlert C, Maclean DJ, Ebert PR, Kazan K. Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell. 2004;16:3460–3479. doi: 10.1105/tpc.104.025833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- Caplan JL, Mamillapalli P, Burch-Smith TM, Czymmek K, Dinesh-Kumar SP. Chloroplastic protein NRIP1 mediates innate immune receptor recognition of a viral effector. Cell. 2008;132:449–462. doi: 10.1016/j.cell.2007.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host–microbe interactions: shaping the evolution of the plant immune response. Cell. 2006;124:803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Dal Bosco C, Lezhneva L, Biehl A, Leister D, Strotmann H, Wanner G, Meurer J. Inactivation of the chloroplast ATP synthase gamma subunit results in high non-photochemical fluorescence quenching and altered nuclear gene expression in Arabidopsis thaliana. Journal of Biological Chemistry. 2004;279:1060–1069. doi: 10.1074/jbc.M308435200. [DOI] [PubMed] [Google Scholar]
- Dangl JL, Horvath DM, Staskawicz BJ. Pivoting the plant immune system from dissection to deployment. Science. 2013;341:746–751. doi: 10.1126/science.1236011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds PN, Rathjen JP. Plant immunity: towards an integrated view of plant–pathogen interactions. Nature Reviews Genetics. 2010;11:539–548. doi: 10.1038/nrg2812. [DOI] [PubMed] [Google Scholar]
- Dong X, Mindrinos M, Davis KR, Ausubel FM. Induction of Arabidopsis defense genes by virulent and avirulent Pseudomonas syringae strains and by a cloned avirulence gene. Plant Cell. 1991;3:61–72. doi: 10.1105/tpc.3.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys BJ, Parker JE. Interplay of signaling pathways in plant disease resistance. Trends in Genetics. 2000;16:449–455. doi: 10.1016/s0168-9525(00)02107-7. [DOI] [PubMed] [Google Scholar]
- Fitter DW, Martin DJ, Copley MJ, Scotland RW, Langdale JA. GLK gene pairs regulate chloroplast development in diverse plant species. Plant Journal. 2002;31:713–727. doi: 10.1046/j.1365-313x.2002.01390.x. [DOI] [PubMed] [Google Scholar]
- Friso G, Giacomelli L, Ytterberg AJ, Peltier JB, Rudella A, Sun Q, Wijk KJ. In-depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: new proteins, new functions, and a plastid proteome database. Plant Cell. 2004;16:478–499. doi: 10.1105/tpc.017814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu ZQ, Guo M, Jeong BR, Tian F, Elthon TE, Cerny RL, Staiger D, Alfano JR. A type III effector ADP-ribosylates RNA-binding proteins and quells plant immunity. Nature. 2007;447:284–288. doi: 10.1038/nature05737. [DOI] [PubMed] [Google Scholar]
- Grant MR, Jones JD. Hormone (dis)harmony moulds plant health and disease. Science. 2009;324:750–752. doi: 10.1126/science.1173771. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Research. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelenska J, Yao N, Vinatzer BA, Wright CM, Brodsky JL, Greenberg JT. A J domain virulence effector of Pseudomonas syringae remodels host chloroplasts and suppresses defenses. Current Biology. 2007;17:499–508. doi: 10.1016/j.cub.2007.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson X, Wostrikoff K, Finazzi G, Kuras R, Schwarz C, Bujaldon S, Nickelsen J, Stern DB, Wollman FA, Vallon O. MRL1, a conserved pentatricopeptide repeat protein, is required for stabilization of rbcL mRNA in Chlamydomonas and Arabidopsis. Plant Cell. 2010;22:234–248. doi: 10.1105/tpc.109.066266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Katagiri F. A global view of defense gene expression regulation – a highly interconnected signaling network. Current Opinion in Plant Biology. 2004;7:506–511. doi: 10.1016/j.pbi.2004.07.013. [DOI] [PubMed] [Google Scholar]
- Katiyar-Agarwal S, Gao S, Vivian-Smith A, Jin H. A novel class of bacteria-induced small RNAs in Arabidopsis. Genes & Development. 2007;21:3123–3134. doi: 10.1101/gad.1595107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar-Agarwal S, Jin H. Role of small RNAs in host-microbe interactions. Annual Review of Phytopathology. 2010;48:225–246. doi: 10.1146/annurev-phyto-073009-114457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar-Agarwal S, Morgan R, Dahlbeck D, Borsani O, Villegas A, Jr, Zhu JK, Staskawicz BJ, Jin H. A pathogen-inducible endogenous siRNA in plant immunity. Proceedings of the National Academy of Sciences, USA. 2006;103:18002–18007. doi: 10.1073/pnas.0608258103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinknecht L, Wang F, Stube R, Philippar K, Nickelsen J, Bohne AV. RAP, the sole octotricopeptide repeat protein in Arabidopsis is required for chloroplast 16S rRNA maturation. Plant Cell. 2014;26:777–787. doi: 10.1105/tpc.114.122853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel BN, Bent AF, Dahlbeck D, Innes RW, Staskawicz BJ. RPS2, an Arabidopsis disease resistance locus specifying recognition of Pseudomonas syringae strains expressing the avirulence gene avrRpt2. Plant Cell. 1993;5:865–875. doi: 10.1105/tpc.5.8.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel BN, Brooks DM. Cross talk between signaling pathways in pathogen defense. Current Opinion in Plant Biology. 2002;5:325–331. doi: 10.1016/s1369-5266(02)00275-3. [DOI] [PubMed] [Google Scholar]
- Lange BM, Ghassemian M. Genome organization in Arabidopsis thaliana: a survey for genes involved in isoprenoid and chlorophyll metabolism. Plant Molecular Biology. 2003;51:925–948. doi: 10.1023/a:1023005504702. [DOI] [PubMed] [Google Scholar]
- Mackey D, Belkhadir Y, Alonso JM, Ecker JR, Dangl JL. Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell. 2003;112:379–389. doi: 10.1016/s0092-8674(03)00040-0. [DOI] [PubMed] [Google Scholar]
- Miller G, Schlauch K, Tam R, Cortes D, Torres MA, Shulaev V, Dangl JL, Mittler R. The plant NADPH oxidase RBOHD mediates rapid systemic signaling in response to diverse stimuli. Science Signalling. 2009;2:ra45. doi: 10.1126/scisignal.2000448. [DOI] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Gollery M, Van Breusegem F. Reactive oxygen gene network of plants. Trends in Plant Science. 2004;9:490–498. doi: 10.1016/j.tplants.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Suzuki N, Miller G, Tognetti VB, Vandepoele K, Gollery M, Shulaev V, Van Breusegem F. ROS signaling: the new wave? Trends in Plant Science. 2011;16:300–309. doi: 10.1016/j.tplants.2011.03.007. [DOI] [PubMed] [Google Scholar]
- Mukhtar MS, Carvunis AR, Dreze M, Epple P, Steinbrenner J, Moore J, Tasan M, Galli M, Hao T, Nishimura MT, et al. Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science. 2011;333:596–601. doi: 10.1126/science.1203659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murmu J, Wilton M, Allard G, Pandeya R, Desveaux D, Singh J, Subramaniam R. Arabidopsis GOLDEN2-LIKE (GLK) transcription factors activate jasmonic acid (JA)-dependent disease susceptibility to the biotrophic pathogen Hyaloperonospora arabidopsidis, as well as JA-independent plant immunity against the necrotrophic pathogen Botrytis cinerea. Molecular Plant Pathology. 2014;15:174–184. doi: 10.1111/mpp.12077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu D, Lii YE, Chellappan P, Lei L, Peralta K, Jiang C, Guo J, Coaker G, Jin H. miRNA863-3p sequentially targets negative immune regulator ARLPKs and positive regulator SERRATE upon bacterial infection. Nature Communications. 2016;7:11324. doi: 10.1038/ncomms11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott A, Jung HS, Koussevitzky S, Chory J. Plastid-to-nucleus retrograde signaling. Annual Review of Plant Biology. 2006;57:739–759. doi: 10.1146/annurev.arplant.57.032905.105310. [DOI] [PubMed] [Google Scholar]
- Panstruga R, Parker JE, Schulze-Lefert P. SnapShot: plant immune response pathways. Cell. 2009;136:978.e971–978.e973. doi: 10.1016/j.cell.2009.02.020. [DOI] [PubMed] [Google Scholar]
- Pieterse CM, Leon-Reyes A, Van der Ent S, Van Wees SC. Networking by small-molecule hormones in plant immunity. Nature Chemical Biology. 2009;5:308–316. doi: 10.1038/nchembio.164. [DOI] [PubMed] [Google Scholar]
- Qiu D, Xiao J, Ding X, Xiong M, Cai M, Cao Y, Li X, Xu C, Wang S. OsWRKY13 mediates rice disease resistance by regulating defense-related genes in salicylate- and jasmonate-dependent signaling. Molecular Plant-Microbe Interactions. 2007;20:492–499. doi: 10.1094/MPMI-20-5-0492. [DOI] [PubMed] [Google Scholar]
- Ruckle ME, Burgoon LD, Lawrence LA, Sinkler CA, Larkin RM. Plastids are major regulators of light signaling in Arabidopsis. Plant Physiology. 2012;159:366–390. doi: 10.1104/pp.112.193599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Ferrer V, Voinnet O. Roles of plant small RNAs in biotic stress responses. Annual Review of Plant Biology. 2009;60:485–510. doi: 10.1146/annurev.arplant.043008.092111. [DOI] [PubMed] [Google Scholar]
- Savitch LV, Subramaniam R, Allard GC, Singh J. The GLK1 ‘regulon’ encodes disease defense related proteins and confers resistance to Fusarium graminearum in Arabidopsis. Biochemical and Biophysical Research Communications. 2007;359:234–238. doi: 10.1016/j.bbrc.2007.05.084. [DOI] [PubMed] [Google Scholar]
- Schreiber KJ, Nasmith CG, Allard G, Singh J, Subramaniam R, Desveaux D. Found in translation: high-throughput chemical screening in Arabidopsis thaliana identifies small molecules that reduce Fusarium head blight disease in wheat. Molecular Plant-Microbe Interactions. 2011;24:640–648. doi: 10.1094/MPMI-09-10-0210. [DOI] [PubMed] [Google Scholar]
- Schwessinger B, Ronald PC. Plant innate immunity: perception of conserved microbial signatures. Annual Review of Plant Biology. 2012;63:451–482. doi: 10.1146/annurev-arplant-042811-105518. [DOI] [PubMed] [Google Scholar]
- Seo JK, Kwon SJ, Choi HS, Kim KH. Evidence for alternate states of Cucumber mosaic virus replicase assembly in positive- and negative-strand RNA synthesis. Virology. 2009;383:248–260. doi: 10.1016/j.virol.2008.10.033. [DOI] [PubMed] [Google Scholar]
- Seo JK, Wu J, Lii Y, Li Y, Jin H. Contribution of small RNA pathway components in plant immunity. Molecular Plant-Microbe Interactions. 2013;26:617–625. doi: 10.1094/MPMI-10-12-0255-IA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner S, Schroter Y, Pfalz J, Pfannschmidt T. Identification of essential subunits in the plastid-encoded RNA polymerase complex reveals building blocks for proper plastid development. Plant Physiology. 2011;157:1043–1055. doi: 10.1104/pp.111.184515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ThordalChristensen H, Zhang ZG, Wei YD, Collinge DB. Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant Journal. 1997;11:1187–1194. [Google Scholar]
- Torres MA, Dangl JL, Jones JD. Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proceedings of the National Academy of Sciences, USA. 2002;99:517–522. doi: 10.1073/pnas.012452499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MA, Jones JD, Dangl JL. Reactive oxygen species signaling in response to pathogens. Plant Physiology. 2006;141:373–378. doi: 10.1104/pp.106.079467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellosillo T, Vicente J, Kulasekaran S, Hamberg M, Castresana C. Emerging complexity in reactive oxygen species production and signaling during the response of plants to pathogens. Plant Physiology. 2010;154:444–448. doi: 10.1104/pp.110.161273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlot AC, Dempsey DA, Klessig DF. Salicylic acid, a multifaceted hormone to combat disease. Annual Review of Phytopathology. 2009;47:177–206. doi: 10.1146/annurev.phyto.050908.135202. [DOI] [PubMed] [Google Scholar]
- Waters MT, Moylan EC, Langdale JA. GLK transcription factors regulate chloroplast development in a cell-autonomous manner. Plant Journal. 2008;56:432–444. doi: 10.1111/j.1365-313X.2008.03616.x. [DOI] [PubMed] [Google Scholar]
- Waters MT, Wang P, Korkaric M, Capper RG, Saunders NJ, Langdale JA. GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell. 2009;21:1109–1128. doi: 10.1105/tpc.108.065250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiberg A, Wang M, Bellinger M, Jin H. Small RNAs: a new paradigm in plant-microbe interactions. Annual Review of Phytopathology. 2014;52:495–516. doi: 10.1146/annurev-phyto-102313-045933. [DOI] [PubMed] [Google Scholar]
- Weiberg A, Wang M, Lin FM, Zhao H, Zhang Z, Kaloshian I, Huang HD, Jin H. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science. 2013;342:118–123. doi: 10.1126/science.1239705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature. 2001;414:562–565. doi: 10.1038/35107108. [DOI] [PubMed] [Google Scholar]
- Ytterberg AJ, Peltier JB, van Wijk KJ. Protein profiling of plastoglobules in chloroplasts and chromoplasts. A surprising site for differential accumulation of metabolic enzymes. Plant Physiology. 2006;140:984–997. doi: 10.1104/pp.105.076083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabala MDT, Littlejohn G, Jayaraman S, Studholme D, Bailey T, Lawson T, Tillich M, Licht D, Bolter B, Delfino L, et al. Chloroplasts play a central role in plant defence and are targeted by pathogen effectors. Nature Plants. 2015;1:15074. doi: 10.1038/nplants.2015.74. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhao H, Gao S, Wang WC, Katiyar-Agarwal S, Huang HD, Raikhel N, Jin H. Arabidopsis Argonaute 2 regulates innate immunity via miRNA393(*)-mediated silencing of a Golgi-localized SNARE gene, MEMB12. Molecular Cell. 2011;42:356–366. doi: 10.1016/j.molcel.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiology. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 AtRAP negatively regulates plant defense to Pst.
Fig. S2 Phenotype of AtRAP-overexpressing plants.
Fig. S3 LSU2 is negatively regulated by AtRAP.
Fig. S4 Characterization of the atrap lsu2 double mutant.
Fig. S5 GLK1 is positively regulated by AtRAP.
Fig. S6 A hypothetical working model for the regulation of the AtRAP protein in plant antibacterial disease resistance.
Table S1 Quantitative-PCR primers used in this study
Table S2 DNA probes used in this study
Table S3 Genes down-regulated in the atrap mutant
Table S4 Genes up-regulated in the atrap mutant