Abstract
Introduction
Metabolomics is a valuable tool for biomarker screening of colorectal cancer (CRC). In this study, we profiled the urinary metabolomes of patients enrolled in a prospective patient cohort (ColoCare). We aimed to describe changes in the metabolome in the longer clinical follow-up and describe initial predictors as candidate markers with possibly prognostic significance
Methods
In total, 199 urine samples from CRC patients pre-surgery (n=97), 1–8 days post-surgery (n=12) and then after 6 and 12 months (n=52 and 38, respectively) were analyzed using both GC-MS and 1H-NMR. Both datasets were analyzed separately with built in uni- and multivariate analyses of Metaboanalyst 2.0. Furthermore, adjusted linear mixed effects regression models were constructed.
Results
Many concentrations of the metabolites derived from the gut microbiome were affected by CRC surgery, presumably indicating a tumor-induced shift in bacterial species. Associations of the microbial metabolites with disease stage indicate an important role of the gut microbiome in CRC.
We were able to differentiate the metabolite profiles of CRC patients prior to surgery from those at any post-surgery timepoint using a multivariate model containing 20 marker metabolites (AUCROC=0.89; 95% CI:0.84–0.95).
Conclusion
To the best of our knowledge, this is one of the first metabolomic studies to follow CRC patients in a prospective setting with repeated urine sampling over time. We were able to confirm markers initially identified in case-control studies and pin point metabolites which may serve as candidates for prognostic biomarkers of CRC.
Keywords: Colorectal cancer, Biomarkers, Patient cohort, Prospective design, Gut microbiome, Gas chromatography-mass spectrometry (GC-MS), Nuclear magnetic resonance (NMR), Metabolomics
1. Introduction
In 2012, colorectal cancer (CRC) was the second most common cancer in Europe, and the second most common cause of death from cancer with almost 215,000 deaths each year (Ferlay et al. 2013). In the US, CRC was the third most common cancer for both genders with more than 50,000 deaths in 2013 (ACS 2013). While improvements in the early detection of disease occurrence are crucial, biomarkers for tertiary prevention, to improve the prediction of disease recurrence, are urgently needed. To date, treatment and clinical follow-up of CRC patients largely follows standardized guidelines. Only a few markers for personalized therapies exist, such as KRAS mutation as a predictive biomarker for anti-EGFR therapy in metastasized patients (Normanno et al. 2013). Moreover, the detection of early stage metastasis is limited by the sensitivity of currently available tumor markers, CA19-9 and CEA, and imaging technologies such as abdominal sonography, computed tomography (CT) and magnetic resonance imaging (MRI) (Bellomi et al. 2005).
Metabolomics, the systematic investigation of metabolites in a given biological system (Fiehn 2002), has been more extensively used in cancer research in recent years (Spratlin et al. 2009). To date, many mass spectrometry (MS)-based metabolomic studies investigating CRC have been published using different hyphenated techniques such as liquid chromatography (LC-MS), gas chromatography (GC-MS) and capillary electrophoresis (CE-MS). Among these methods, GC-MS has the longest history. Extensive electron-impact mass spectral databases (NIST, GOLM, WILEY (Hummel et al. 2007)) on biochemicals are available, making it a robust method for metabolic profiling (Dunn et al. 2005). Nuclear magnetic resonance (NMR)-based techniques are known to have less sensitivity in comparison to MS-based methods, but compensate for this disadvantage by being highly reproducible and, furthermore, quantitative without the need for individual calibration of each detected analyte (Pan et al. 2007).
Metabolomic studies on CRC to date focus either on profiling of tumor tissue (Chan et al. 2009; Denkert et al. 2008; Mal et al. 2009; Mal et al. 2012) to gain insight into cancer biology, or use serum (Farshidfar et al. 2012; Kondo et al. 2011; Leichtle et al. 2012; Ma et al. 2010; Nishiumi et al. 2012; Qiu et al. 2009; Ritchie et al. 2010) and/or urine samples (Cheng et al. 2012; Ma et al. 2009; Qiu et al. 2010; Wang et al. 2010) with the aim to detect novel markers for early disease detection. As previously reviewed by our own group (Liesenfeld et al. 2013), most studies have used a simple case-control study design with only a few investigations conducting follow-up studies with repeated sampling of biospecimens over time (Farshidfar et al. 2012; Ma et al. 2010). Studies with a prospective design have the advantage of allowing the assessment of the predictive value of metabolomics with regard to patients’ outcomes, such as recurrence rates, metastasis or survival. Additionally, effects of therapy (primarily surgical intervention, but also radio- and chemotherapy) can also be monitored and assessed.
In this GC-MS- and 1H-NMR-based metabolomics study, we profiled 199 urine samples at different study timepoints in a prospective CRC patient cohort, the ColoCare study. We aimed to compare urinary metabolomes of CRC patients prior to surgery with those a few days and at 6 and 12 months post-surgery, assessing potential effects related to the primary therapeutic intervention. Additionally, we aimed to describe changes in the metabolome in the longer follow-up and describe initial predictors as candidate markers with possibly prognostic significance. To the best of our knowledge, this study is one of the first CRC metabolomic approaches embedded in a prospective study design.
2. Materials and methods
2.1 Study population
This study is part of the international prospective cohort study, ColoCare, recruiting newly-diagnosed CRC patients prior to surgery. ColoCare investigates predictors of cancer recurrence, survival, treatment toxicities and health-related quality of life. Within the study, extensive follow-up observations over the duration of 5 years are being performed: three follow-up timepoints in year one, followed by one each year thereafter, including a comprehensive collection of biospecimens and data. The ColoCare study has been approved by the ethics committee of the medical faculty at the University of Heidelberg and study participants provided their written informed consent. Patients were categorized according to the American Joint Committee on Cancer (AJCC) staging system based on histopathologic findings. Both patients with colon carcinoma (ICD-18) and patients with rectal or rectosigmoidal cancer (ICD-19/20) were included. Urine, generally from morning fasting, was collected from patients at baseline, prior to surgery, and in some cases from patients 1–8 days post-surgery. As part of the ColoCare follow-up regime, additional biosepecimens were obtained approximately 6 and 12 months post enrollment at the National Center of Tumor Disease, Heidelberg (NCT). Spot urine samples from these respective follow-up visits were included (see Table 1 for study population and sample availability). If patients underwent adjuvant chemotherapy, follow-up visits were scheduled at least 2 weeks after their last chemotherapy cycle had been completed. Urine samples were aliquoted immediately and stored at −80 °C until analysis. Self-reported food and fluid intake before sample collection was monitored during follow-up visits. Patients who reported alcohol or tobacco use 24 h prior to urine collection were excluded from this dataset.
Table 1.
Study population and sample availability for the urinary metabolomics analysis
| CRC patients (pre-surgery) |
CRC patients (post-surgery) |
CRC patients (6 months follow-up) |
CRC patients (12 months follow-up) |
Total | |||||
|---|---|---|---|---|---|---|---|---|---|
| Analytical data | |||||||||
| Total n | 97 | 12 | 52 | 38 | 199 | ||||
| GC-MS (n) | 79 | 9 | 46 | 36 | 170 | ||||
| 1H-NMR (n) | 88 | 10 | 48 | 37 | 183 | ||||
|
| |||||||||
| Biomedical data | |||||||||
| Male (n (%)) | 59 (60.8 %) | 10 (83.3 %) | 38 (73.0 %) | 24 (63.2 %) | 131 (65.8 %) | ||||
| Age (mean ± SD) | 64.8 ± 12.9 | 63.9 ± 12.5 | 60.1 ± 11.0 | 61.5 ± 11.6 | 62.9 ± 12.2 | ||||
| BMI (mean ± SD) | 26.3 ± 4.0 | 27.3 ± 3.3 | 25.4 ± 4.1 | 26.8 ± 3.9 | 26.2 ± 4.0 | ||||
|
| |||||||||
| Tumor data | |||||||||
| Site | Colon | Rectum | Colon | Rectum | Colon | Rectum | Colon | Rectum | Subtotal |
| Stage 0 (n) | 2 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 5 |
| Stage I (n) | 3 | 9 | 3 | 1 | 6 | 6 | 3 | 4 | 35 |
| Stage II (n) | 20 | 20 | 1 | 3 | 5 | 12 | 5 | 8 | 74 |
| Stage III (n) | 12 | 10 | 1 | 1 | 8 | 7 | 9 | 5 | 53 |
| Stage IV (n) | 6 | 12 | 2 | 0 | 4 | 4 | 1 | 3 | 32 |
|
| |||||||||
| Total (n) | 43 | 54 | 7 | 5 | 23 | 29 | 18 | 20 | 199 |
2.2 Chemicals and reagents
HPLC-grade methanol and acetonitrile were purchased from VWR International (Fontenay-sous-Bois, France). Sodium 3-(trimethylsilyl)-propionate-2,2,3,3-d4 99 % D (TSP) was purchased from Euriso-top (Saint-Aubin Cedex, France). Bis(trimethylsilyl)trifluoroacetamide (BSTFA) containing 1 % trimethylchlorosilane was purchased from Supelco (Bellefonte, USA). Deuterium oxide (D2O), pyridine, methoxyamine.HCl, l-4-chlorophenylalanine, uridine 5′-diphosphoglucuronic acid trisodium salt (UGA), saturated alkane mixture (C7–C40) and all analytical reference standards were purchased from Sigma Aldrich (Steinheim, Germany) at the highest purity available.
2.3 Sample preparation
For GC-MS analysis, the method for urine sample preparation was adapted from literature (Cheng et al. 2012), but without urease treatment because low enzyme specificity resulting in distorted metabolite levels have been reported by several research groups (Kind et al. 2007; Perroud et al. 2006). Pooled quality control (QC) samples and Kovat’s alkane mixtures were injected with each analytical batch.
For 1H-NMR, 540 µL urine were spiked with 60 µL K2HPO4/NaH2PO4 buffer in D2O (pH 6.5, 1.5 M) (Xiao et al. 2009) containing 10 mM TSP to a final D2O concentration of 10 % (v/v) for lock and shim purposes and TSP concentration of 1 mM. QC samples were run at the beginning and end of each analytical batch.
2.4 GC-MS data acquisition
Analysis was carried out on an Agilent 6890 GC/ 5973 MS single quadrupole system. Aliquots of 1 µL were injected onto a HP-5 MS fused silica column (30 m × 0.25 mm; 0.25 µm film thickness of the 5 % phenyl 95 % dimethylpolysiloxane stationary phase; Agilent J&W Scientific) in splitless mode. Chromatographic and mass spectrometric conditions are described in the Supplementary Data.
2.5 1H-NMR data acquisition
1H-NMR spectra were acquired using a Bruker Avance II NMR spectrometer equipped with a 5-mm, inverse-configuration probe with triple-axis gradient capability at a field strength of 14.1 T operating at 600.1 MHz for 1H. Spectra were acquired at 25 °C using the 1D NOESY with pre-saturation pulse program resident in the spectrometer software. Detailed 1H-NMR acquisition parameters can be found in the Supplementary Data.
2.6 Data pre-processing
GC-MS raw files were converted to netCDF format and processed with MZMine 2.0 (Pluskal et al. 2010) (see Supplementary Data for parameters). Chromatographic peaks were annotated based on four different levels following literature recommendations (Sumner et al. 2007). Level 1: authentic reference standard available, level 2: electron impact (EI) spectral match (>75 %) and Kovat’s retention indices (RI) match (±20) to the NIST 2011 database, level 3: (EI) spectral match (>75 %) to multiple components of a chemical class (i.e. sugars), level 4: unknown metabolite. A data matrix was constructed using the peak area of the base peak of each metabolite divided by the peak area of the internal standard (l-4-chlorophenylalanine). Artefact peaks resulting from the derivatization process and interferences due to urea were removed from the dataset. Urea interferences have been noted before in urinary GC-MS studies, leading to effects such as ion suppression, peak distortion and/or column overload (Kind et al. 2007). We excluded metabolites with >30 % coefficient of variance (CV) in QC samples from the final data analysis (Dunn et al. 2011).
For 1H-NMR, a targeted profiling approach using spectral fitting was applied. Bruker NMR files were imported into Chenomx NMR suite software (Chenomx, Edmonton, Canada), spline baseline and automatic shim correction were performed prior to manual fitting of spectra against the built in library version 7.7.
2.7 Data analysis
Data analysis was conducted using the Metaboanalyst 2.0 toolbox program (Xia et al. 2012) separately for GC-MS and 1H-NMR data. GC-MS data was normalized by sum, log-transformed and auto-scaled prior to analysis, whereas urinary concentrations determined by 1H-NMR were normalized to creatinine (µmol/mmol creatinine), log transformed and auto-scaled. If >40 % missing values were present for a given metabolite, the metabolite was not used for data analysis. For model building we tested and compared three different methods for missing value imputation (minimum, mean and median). Non-parametric univariate analyses (Wilcoxon–Mann–Whitney tests) were used for the comparison of two groups and one-way ANOVA for multiple group comparison. Furthermore, partial least squares discriminant analysis (PLS-DA) models were constructed with Metaboanalyst. Finally, to account for possible confounding variables, linear mixed effects regression models, not yet implemented in Metaboanalyst, were built into the statistical software R 3.0.1 (www.r-project.org) adjusting raw p-values for sex, disease stage, BMI, adjuvant and neoadjuvant chemo- and radiotherapy, NSAID use, smoking and fasting status. The linear mixed effects models were built with the "mer" function of the package lme4 (Bates et al. 2013) using patient ID as random-effect term, while the p-values for the fixed-effects terms were calculated using an approximate F-test based on the Kenward–Roger approach as implemented in the package pbkrtest (Højsgaard 2013). As a last step, adjusted p-values (p-adjust) were corrected for multiple testing using the Benjamini–Hochberg procedure (FDR) (Benjamini et al. 1995).
Binary class discrimination abilities (i.e. comparing pre- vs. post-surgery patients) were assessed with ROCCET toolbox to construct receiver operator characteristic curves (ROC) (Xia et al. 2013).
2.8 Pathway visualization
To visualize the overall effect of therapeutic intervention on CRC patients’ metabolomes, we used the approach of chemical similarity (PubChem ID) and biochemical pathway mapping (KEGG ID) with the MetaMapp tool and Cytoscape as previously described (Barupal et al. 2012). In the case of compound classes (i.e. level 3 annotations), we mapped metabolites to their closest annotated chemical entity (i.e. gluconic acid for sugar acids, sucrose for disaccharides). Unknowns and glucuronides were mapped as a separate group. 3.
3. Results
From the GC-MS data, a total of 198 metabolites were annotated after removal of all artifacts arising from derivatization. More than 85 % (169 metabolites) passed the initial QC criteria of <30 % CV in the QC samples. The median % CV in QC samples across all 198 metabolites was 12.3 % indicating a robust analytical method. For metabolites that showed multiple peaks during derivatization, i.e. from isomers or partial silylation of amino groups, only the most abundant peak was used for analysis.
A total of 161 metabolites were annotated by 1H-NMR spectral fitting. The reproducibility was generally higher than for GC-MS (median % CV: 7.3%; see also Supplementary Figure 4). Without chromatographic separation prior to 1H-NMR analysis, the fitting of signals in crowded regions of the spectra (e.g. 3–5 ppm) is problematic due to spectral overlap. We therefore validated both methodologies against each other by applying a conservative approach: metabolites with a Spearman rank correlation coefficient <0.5 over the whole dataset (GC-MS peak area vs. 1H-NMR concentrations) were subject to manual inspection and deleted from either the 1H-NMR or GC-MS dataset (see Supplementary Table S1 for the exclusion criteria). The remaining cleaned datasets, containing 168 metabolites for GC-MS and 152 metabolites for 1H-NMR, produced very consistent results throughout the analyses (vide infra).
3.1 Patient urine metabolomes change after surgery
A primary aim of this study was to monitor patient urine metabolomes on a prospective basis. We therefore compared two groups:
patients pre-surgery, still bearing a tumor, and
patients after therapeutic surgery (n = 12 at 1–8 days post-surgery; n = 52 at 6 months follow-up, n = 38 at 12 months follow-up)
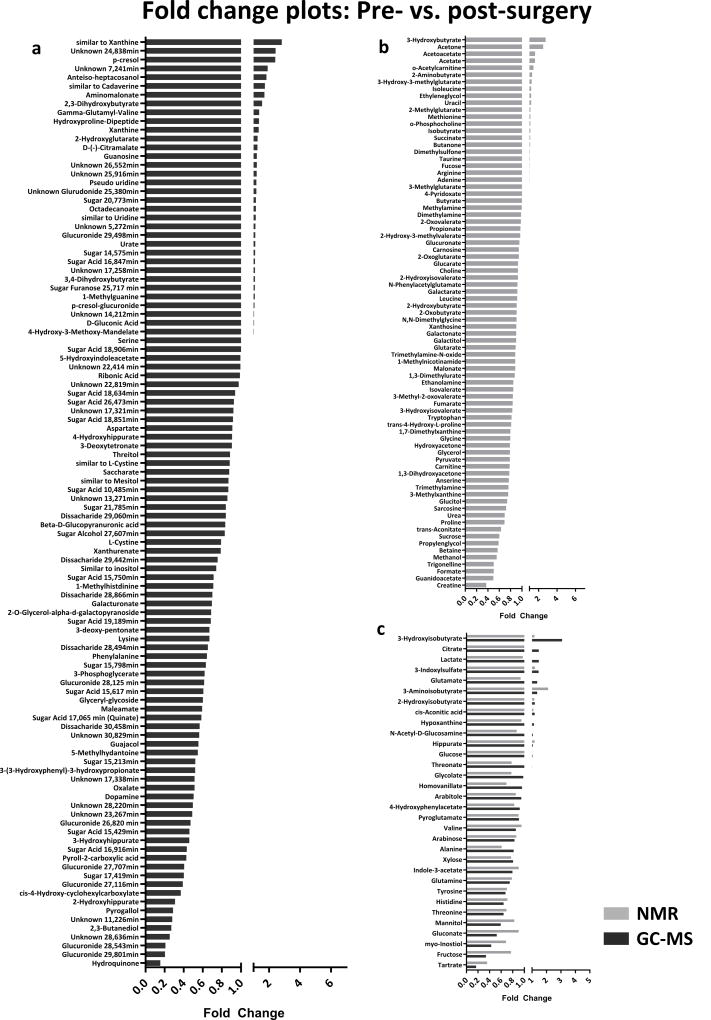
Significantly altered metabolites between the groups are listed in Table 2. Corresponding fold-change plots for GC-MS and 1H-NMR can be found in Fig. 1. Altogether, 39 metabolites detected by GC-MS remained significant after adjustment for covariables and multiple testing (FDR <0.05), whereas only 2-aminobutyrate was found to be significant in our 1H-NMR analysis. Many unknown metabolites (level 3 or 4 identification) were of high significance in the GC-MS dataset, however, they were absent from the 1H-NMR data due to its use as a targeted profiling technique in our study.
Table 2.
Significantly altered metabolites distinguishing pre- from post-surgery CRC patients
| Metabolite | fold changea | p-value (raw)b | p-adjustedc | FDR adjustedd | Identification |
|---|---|---|---|---|---|
| GC-MS | |||||
| Sugar, Rt 15.798 min | 0.632 | 4.5E-10 | 3.7E-08 | 5.3E-06 | Level 3 |
| Glucuronide, Rt 29.801 min | 0.204 | 8.7E-09 | 3.3E-07 | 1.6E-05 | Level 3 |
| Unknown, Rt 28.636 min | 0.254 | 1.6E-12 | 2.5E-07 | 1.8E-05 | Level 4 |
| 2,3-Butanediol | 0.271 | 5.9E-11 | 4.3E-06 | 2.0E-04 | Level 2 |
| 3-Hydroxyisobutyrate | 3.083 | 1.7E-07 | 8.0E-06 | 2.9E-04 | Level 1 |
| Guajacol | 0.555 | 1.5E-05 | 3.6E-05 | 0.001 | Level 1 |
| Hydroquinone | 0.155 | 8.2E-09 | 4.3E-05 | 0.001 | Level 1 |
| Glucuronide, Rt 27.116 min | 0.390 | 1.3E-08 | 5.0E-05 | 0.001 | Level 3 |
| Unknown, Rt 28.220 min | 0.496 | 2.5E-08 | 5.8E-05 | 0.001 | Level 4 |
| Glyceryl glycoside | 0.601 | 1.9E-07 | 7.3E-05 | 0.001 | Level 2 |
| Maleamate | 0.593 | 4.5E-11 | 7.9E-05 | 0.001 | Level 2 |
| γ-Glutamyl valine | 1.368 | 2.9E-08 | 8.3E-05 | 0.001 | Level 2 |
| Tartrate | 0.167 | 9.8E-08 | 1.3E-04 | 0.002 | Level 1 |
| 3-Phosphoglycerate | 0.619 | 1.4E-04 | 1.7E-04 | 0.002 | Level 2 |
| Glucuronide, Rt 27.707 min | 0.405 | 4.2E-06 | 1.8E-04 | 0.002 | Level 3 |
| 3-Hydroxyhippurate | 0.458 | 0.003 | 1.6E-04 | 0.002 | Level 2 |
| Sugar acid, Rt 15.617 min | 0.607 | 2.8E-07 | 2.6E-04 | 0.002 | Level 3 |
| Sugar acid, Rt 15.750 min | 0.713 | 3.3E-04 | 3.1E-04 | 0.002 | Level 3 |
| Citrate | 1.471 | 4.7E-05 | 3.2E-04 | 0.002 | Level 1 |
| Glucuronide, Rt 28.543 min | 0.208 | 8.5E-12 | 3.5E-04 | 0.003 | Level 3 |
| Indole-3-acetate | 0.796 | 3.2E-04 | 4.7E-04 | 0.003 | Level 1 |
| 3-Deoxypentonate | 0.669 | 1.8E-05 | 4.7E-04 | 0.003 | Level 2 |
| Sugar acid, Rt 15.429 min | 0.460 | 9.7E-09 | 0.001 | 0.004 | Level 3 |
| Sugar, Rt 17.419 min | 0.402 | 1.2E-05 | 0.001 | 0.004 | Level 3 |
| Glucuronide, Rt 26.820 min | 0.471 | 0.001 | 0.002 | 0.009 | Level 3 |
| Glucuronide, Rt 28.125 min | 0.614 | 4.3E-05 | 0.002 | 0.009 | Level 3 |
| 5-Methylhydantoine | 0.548 | 5.7E-06 | 0.003 | 0.015 | Level 2 |
| Phenylalanine | 0.644 | 8.8E-07 | 0.003 | 0.016 | Level 1 |
| Unknown, Rt 17.338 min | 0.514 | 0.020 | 0.003 | 0.018 | Level 4 |
| Unknown, Rt 24.838 min | 2.420 | 1.6E-12 | 0.004 | 0.018 | Level 4 |
| Sugar, Rt 15.213 min | 0.522 | 6.5E-06 | 0.004 | 0.018 | Level 3 |
| 4-Hydroxyhippurate | 0.906 | 0.027 | 0.004 | 0.019 | Level 2 |
| myo-Inositol | 0.429 | 3.9E-07 | 0.005 | 0.021 | Level 1 |
| Dissacharide, Rt 28.866 min | 0.700 | 0.014 | 0.005 | 0.022 | Level 3 |
| Mannitol | 0.593 | 1.5E-06 | 0.007 | 0.029 | Level 1 |
| Oxalate | 0.513 | 2.2E-06 | 0.007 | 0.029 | Level 1 |
| Arabinose | 0.831 | 0.031 | 0.010 | 0.039 | Level 1 |
| Dissacharide, Rt 28.494 min | 0.653 | 0.003 | 0.012 | 0.045 | Level 3 |
| Pyrogallol | 0.288 | 2.1E-09 | 0.012 | 0.045 | Level 1 |
| Similar to quinic acid | 0.586 | 0.001 | 0.017 | 0.058 | Level 3 |
| Pyrrole-2-carboxylic acid | 0.428 | 0.002 | 0.017 | 0.058 | Level 2 |
| Gluconate | 0.524 | 0.006 | 0.018 | 0.059 | Level 2 |
| Unknown, Rt 11.226 min | 0.280 | 1.3E-06 | 0.020 | 0.061 | Level 4 |
| p-Cresol-β-O-glucuronide | 1.064 | 0.052 | 0.020 | 0.061 | Level 1 |
| 2-Hydroxyglutarate | 1.270 | 0.035 | 0.019 | 0.061 | Level 2 |
| Threonine | 0.642 | 7.0E-07 | 0.022 | 0.065 | Level 1 |
| 3-Indoxylsulfate | 1.456 | 4.1E-04 | 0.023 | 0.065 | Level 1 |
| 2-Deoxytetronate | 0.905 | 8.4E-06 | 0.023 | 0.067 | Level 2 |
| 2-Hydroxyhippurate | 0.310 | 0.329 | 0.024 | 0.068 | Level 2 |
| Dopamine | 0.504 | 1.5E-05 | 0.031 | 0.080 | Level 1 |
| Tyrosine | 0.675 | 1.1E-04 | 0.031 | 0.080 | Level 1 |
| 2,3-Dihydroxybutyrate | 1.556 | 4.4E-05 | 0.034 | 0.086 | Level 2 |
| cis-4-Hydroxycyclohexylcarboxylate | 0.369 | 2.6E-05 | 0.050 | 0.119 | Level 2 |
| Similar to cadaverine | 1.737 | 2.2E-08 | 0.072 | 0.160 | Level 3 |
| Xanthine | 1.337 | 0.001 | 0.076 | 0.165 | Level 1 |
| Glutamine | 0.749 | 0.004 | 0.090 | 0.191 | Level 1 |
| Pseudo uridine | 1.187 | 0.001 | 0.098 | 0.206 | Level 2 |
| Histidine | 0.643 | 1.1E-04 | 0.110 | 0.227 | Level 1 |
| 2-Hydroxyisobutyrate | 1.191 | 0.002 | 0.116 | 0.235 | Level 1 |
| p-Cresol | 2.405 | 2.8E-05 | 0.148 | 0.284 | Level 1 |
| Similar to inositol | 0.741 | 0.010 | 0.165 | 0.308 | Level 3 |
| 2-O-Glycerol-α-d-galactopyranoside | 0.688 | 0.006 | 0.191 | 0.339 | Level 2 |
| Unknown, Rt 26.552 min | 1.217 | 1.3E-05 | 0.189 | 0.339 | Level 4 |
| Lysine | 0.669 | 0.002 | 0.209 | 0.363 | Level 1 |
| Galacturonate | 0.695 | 0.001 | 0.303 | 0.463 | Level 2 |
| Glucuronide, Rt 25.380 min | 1.182 | 0.010 | 0.332 | 0.489 | Level 3 |
| 5-Hydroxyindole acetate | 0.995 | 0.020 | 0.334 | 0.489 | Level 2 |
| 1-Methylhistidine | 0.710 | 0.001 | 0.375 | 0.527 | Level 2 |
| Xanthurenate | 0.788 | 4.6E-05 | 0.387 | 0.534 | Level 2 |
| Similar to xanthine | 2.827 | 0.001 | 0.413 | 0.564 | Level 3 |
| Sugar acid, Rt 16.916 min | 0.433 | 0.001 | 0.496 | 0.640 | Level 3 |
| Sugar acid, Rt 10.485 min | 0.868 | 0.002 | 0.490 | 0.640 | Level 3 |
| Fructose | 0.338 | 0.002 | 0.496 | 0.640 | Level 1 |
| Unknown, Rt 25.916 min | 1.207 | 0.026 | 0.530 | 0.678 | Level 4 |
| Unknown, Rt 30.829 min | 0.562 | 0.008 | 0.6 | 0.765 | Level 4 |
| Alanine | 0.817 | 0.001 | 0.635 | 0.784 | Level 1 |
| Lactate | 1.471 | 0.012 | 0.722 | 0.826 | Level 1 |
| Glutamate | 1.361 | 0.024 | 0.733 | 0.826 | Level 1 |
| Sugar furanose, Rt 25.717 min | 1.087 | 0.044 | 0.752 | 0.831 | Level 3 |
| Sugar, Rt 14.575 min | 1.104 | 0.043 | 0.792 | 0.853 | Level 3 |
| Unknown, Rt 7.241 min | 1.914 | 1.9E-05 | 0.783 | 0.853 | Level 4 |
| Sugar, Rt 20.773 min | 1.157 | 0.001 | 0.996 | 0.996 | Level 3 |
|
| |||||
| NMR | |||||
|
| |||||
| 2-Aminobutyrate | 1.295 | 0.001 | 8.07E-11 | 9.04E-09 | NMR-Lib |
| 2-Oxobutyrate | 0.907 | 0.279 | 0.003 | 0.147 | NMR-Lib |
| 3-Hydroxyisobutyrate | 1.170 | 0.038 | 0.02 | 0.202 | NMR-Lib |
| Tryptophan | 0.818 | 0.009 | 0.04 | 0.317 | NMR-Lib |
| Glycine | 0.794 | 0.001 | 0.061 | 0.371 | NMR-Lib |
| Ethanolamine | 0.848 | 1.2E-04 | 0.078 | 0.398 | NMR-Lib |
| Lactate | 0.975 | 0.002 | 0.084 | 0.41 | NMR-Lib |
| Asparagine | 0.788 | 0.022 | 0.133 | 0.484 | NMR-Lib |
| Trimethylamine | 0.762 | 0.001 | 0.131 | 0.484 | NMR-Lib |
| Fructose | 0.770 | 0.004 | 0.141 | 0.494 | NMR-Lib |
| Pyruvate | 0.786 | 0.001 | 0.158 | 0.499 | NMR-Lib |
| Glutamine | 0.785 | 0.001 | 0.174 | 0.499 | NMR-Lib |
| Glucitol | 0.739 | 0.005 | 0.147 | 0.499 | NMR-Lib |
| trans-Aconitate | 0.631 | 1.8E-07 | 0.169 | 0.499 | NMR-Lib |
| Arabinitol | 0.863 | 0.009 | 0.185 | 0.504 | NMR-Lib |
| Threonate | 0.781 | 1.0E-04 | 0.182 | 0.504 | NMR-Lib |
| Trigonelline | 0.503 | 0.029 | 0.189 | 0.505 | NMR-Lib |
| 3-Methylxanthine | 0.758 | 0.006 | 0.237 | 0.585 | NMR-Lib |
| 2-Hydroxyisobutyrate | 1.161 | 0.023 | 0.24 | 0.585 | NMR-Lib |
| Xylose | 0.773 | 0.037 | 0.249 | 0.591 | NMR-Lib |
| Glycerol | 0.788 | 0.001 | 0.258 | 0.591 | NMR-Lib |
| Arabinose | 0.852 | 0.050 | 0.292 | 0.641 | NMR-Lib |
| Acetone | 2.495 | 6.1E-10 | 0.344 | 0.697 | NMR-Lib |
| Threonine | 0.691 | 7.1E-05 | 0.397 | 0.765 | NMR-Lib |
| myo-Inositol | 0.684 | 1.7E-06 | 0.467 | 0.799 | NMR-Lib |
| Creatine | 0.371 | 0.008 | 0.436 | 0.799 | NMR-Lib |
| Leucine | 0.917 | 0.002 | 0.449 | 0.799 | NMR-Lib |
| Isoleucine | 1.199 | 0.008 | 0.525 | 0.841 | NMR-Lib |
| Urea | 0.695 | 0.005 | 0.567 | 0.872 | NMR-Lib |
| Proline | 0.692 | 1.5E-05 | 0.576 | 0.872 | NMR-Lib |
| Propylenglycol | 0.589 | 4.7E-07 | 0.597 | 0.885 | NMR-Lib |
| Methanol | 0.555 | 4.6E-04 | 0.603 | 0.885 | NMR-Lib |
| Tartrate | 0.358 | 2.5E-07 | 0.693 | 0.906 | NMR-Lib |
| 3-Aminoisobutyrate | 2.114 | 1.6E-04 | 0.67 | 0.906 | NMR-Lib |
| Acetoacetate | 1.618 | 0.009 | 0.736 | 0.906 | NMR-Lib |
| Glycolate | 0.777 | 0.045 | 0.674 | 0.906 | NMR-Lib |
| Mannitol | 0.826 | 0.002 | 0.683 | 0.906 | NMR-Lib |
| 1,7-Dimethylxanthine | 0.797 | 0.011 | 0.719 | 0.906 | NMR-Lib |
| O-Acetylcarnitine | 1.449 | 0.001 | 0.725 | 0.906 | NMR-Lib |
| Hydroxyproline | 0.811 | 0.037 | 0.748 | 0.908 | NMR-Lib |
| Histidine | 0.708 | 0.020 | 0.817 | 0.93 | NMR-Lib |
| 1,3-Dihydroxyacetone | 0.779 | 0.039 | 0.873 | 0.964 | NMR-Lib |
| 1-Methylnicotinamide | 0.882 | 0.032 | 0.878 | 0.964 | NMR-Lib |
| Homovanillate | 0.692 | 0.010 | 0.918 | 0.984 | NMR-Lib |
| Tyrosine | 0.700 | 8.9E-07 | 0.94 | 0.984 | NMR-Lib |
| N-Acetylglucosamine | 0.870 | 0.036 | 0.946 | 0.984 | NMR-Lib |
| Guanidoacetate | 0.496 | 9.2E-07 | 0.968 | 0.984 | NMR-Lib |
| Alanine | 0.609 | 6.7E-13 | 0.98 | 0.984 | NMR-Lib |
| Anserine | 0.768 | 0.002 | 0.936 | 0.984 | NMR-Lib |
Fold change; values <1 indicate lower concentrations in pre-surgery CRC patients.
Raw unadjusted p-values based on a non-parametric Wilcoxon test.
Adjusted p-values were based on our mixed regression model and were adjusted for sex, disease stage, BMI, adjuvant and neoadjuvant chemo- and radiotherapy, NSAID use, smoking and fasting status.
Adjusted p-values were corrected for multiple testing (FDR).
Fig. 1.
a) Fold-change plot comparing pre- vs. post-surgery CRC patients; fold changes >1 indicate higher urinary concentrations pre-surgery, fold changes <1 indicate lower urinary concentrations pre-surgery. a) Metabolites present in GC-MS dataset. b) Metabolites present in NMR dataset. C) Metabolites present in both GC-MS and NMR datasets.
Many significant metabolites, not present in human biochemical pathways, were presumably of microbial origin, for example 2,3-butanediol, maleamate, hydroquinone, guajacol, pyrogallol and the hippuric acid derivatives 2-, 3- and 4-hydroxyhippurate. All of them were diminished in pre-surgery CRC patients’ urine compared to post-surgery urine (fold change <1, Fig. 1a). In contrast, two microbial metabolites, p-cresol and its glucuronide (p-cresol-β-O-glucuronide), were found to be excreted in higher concentrations in patients pre-surgery (p-adjusted < 0.05 < FDR).
Several amino acids, phenylalanine, threonine, tyrosine, lysine, tryptophan, asparagine and glycine, were detected at significantly lower concentration in the urine of patients pre-surgery compared to post-surgical timepoints by both GC-MS and 1H-NMR analysis. Alanine, histidine, hydroxyproline, glutamine, leucine, isoleucine and lysine had highly significant raw p-values, but became non-significant after adjusting for covariates. Some metabolites of valine (3-hydroxyisobutyrate and γ-glutamyl-valine) were present in higher concentrations pre-surgery. Additionally, concentrations of some tryptophan catabolites, e.g. 3-indoxylsulfate, xanthurenate and indole-3-acetate, were found to be altered. We also observed significant decreased excretions of myo-inositol, 5-methylhydantoine, oxalate and tartrate in pre-surgery patients’ urine.
Several metabolites were significant after adjustment for covariates, but did not sustain multiple testing (p-adjusted < 0.05 < FDR): the oncometabolite 2-hydroxyglutarate was elevated in pre-surgery patients’ urine, whereas pyrrole-2-carboxylic acid, a metabolite of collagen and a product of the d-hydroxyproline isomer (Yamanishi et al. 1972), was found at higher levels in urine of patients after surgical intervention. A metabolite similar to cadaverine (putatively a polyamine derivative) and xanthine were elevated in pre-surgery patients’ urine at borderline significance.
We observed a combined modulating effect of adjuvant chemo- and radiotherapy on the described changes in metabolite levels when pre- and post-surgery individuals were compared (see Supplementary Table S3). The differences between pre- and post-surgical samples were more pronounced among patients undergoing additional adjuvant chemotherapy. However, the same direction of change in metabolite levels was already observed in patients who only underwent surgery with no adjuvant treatment. In contrast, neoadjuvant treatment, prior to surgery, did not have a distinct effect on patients’ metabolomes.
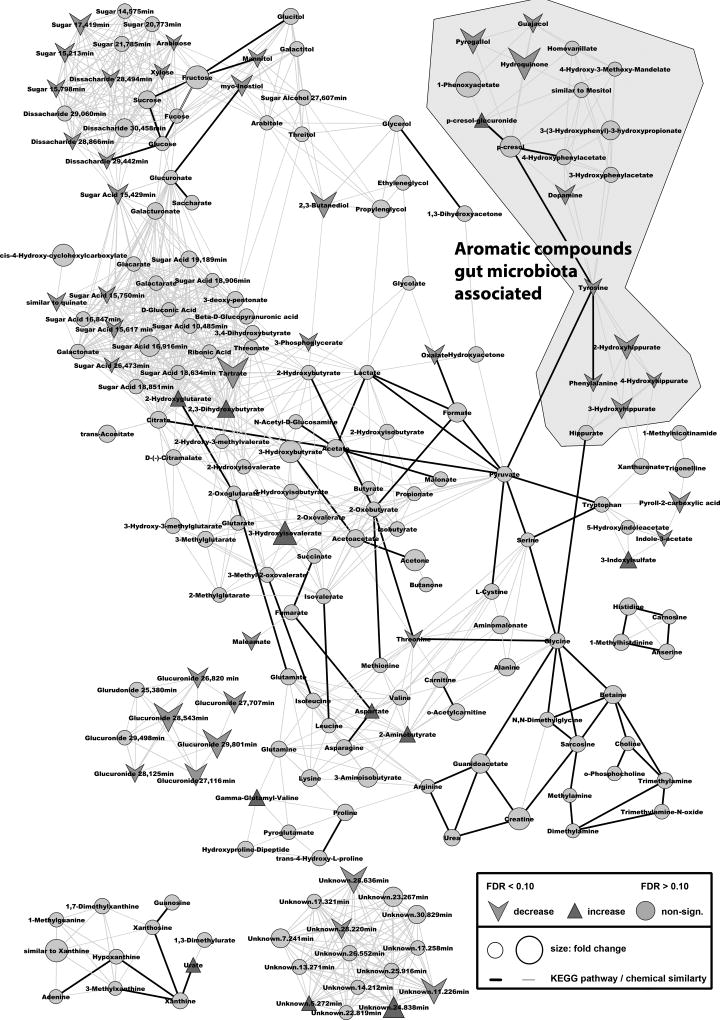
Overall effects of the therapeutic intervention were visualized with MetaMapp and cytoscape where groups of altered metabolites were clustered together according to their chemical similarity (see Fig. 2).
Fig. 2.
Mapped pathway for the therapeutic effects on colorectal cancer patients’ metabolomes. Metabolites were mapped according to their biochemical pathway (KEGG – bold line) and chemical similarity (PubChem ID – light gray line). Fold changes and adjusted/FDR-corrected p-values were abstracted by comparing patients pre- and post surgery. Only metabolites with an adjusted/ FDR-corrected p-value <0.1 were labeled as increased/decreased.
3.2 Distinguishing early and late stage disease
To investigate whether metabolites in the urine could discriminate CRC patients by disease stage, patients prior to surgery were compared based on staging of their excised tumors according to the AJCC staging system. CRC patients were grouped as follows:
stage 0 (carcinoma in situ) and stage I (localized) as "early"
stage II (locally advanced) and stage III (locally advanced with lymphnodes affected) as "intermediate"
stage IV (metastasized) as "late".
A PLS-DA score plot of pre-surgery CRC patients by stage is illustrated in Supplementary Fig. S3. Additionally, discriminating metabolites were selected using one-way ANOVA. Sixteen metabolites in both GC-MS and 1H-NMR datasets were found to significantly discriminate CRC stages (p-values <0.05), but only two of them remained significant after adjusting for multiple testing, a dipeptide of hydroxyproline (Hyp-Hyp) and p-cresol-β-O-glucuronide.
p-Cresol and its glucuronide were present at higher concentrations both in pre-surgery patients compared to post-surgery counterparts and in later stages of the disease (Fig. 1a and Table 3). p-Cresol-β-O-glucuronide was determined to be a level 2 metabolite but showed a very high correlation with p-cresol (data not shown). A peak at 375 m/z in the electron-impact spectrum indicated the presence of a glucuronide group for the latter compound (Thevis et al. 2001). We therefore chose to synthesize the glucuronide enzymatically (Supplementary Material) to unequivocally identify this phase II metabolite of p-cresol as the β-O-glucuronide.
Table 3.
Metabolites distinguishing early-, intermediate- and late-stage CRC patients pre-surgery
| Metabolite | Origin/Pathway | Early (Stage 0–I)a |
Intermediate (Stage II–III)a |
Late (Stage IV)a |
p-value (raw)b |
FDR | Identification |
|---|---|---|---|---|---|---|---|
| GC-MS | |||||||
|
| |||||||
| Hyp-Hyp | extracellular matrix | ↓ | ↑↑ | ↑ | 0.0002 | 0.0346 | Level 2 |
| p-Cresol-β-O-glucuronide | microbial metabolism | ↓ | ↑↑ | ↑ | 0.0004 | 0.0364 | Level 1 |
| Hippurate | mammalian-microbial cometabolite | ↓ | ↑ | ↑ | 0.0046 | 0.2341 | Level 1 |
| p-Cresol | microbial metabolism | ↓ | ↑ | ↑ | 0.0056 | 0.2341 | Level 1 |
| Sugar alcohol, Rt 13.507 min | - | ↑ | ↓ | ↓ | 0.0185 | 0.6217 | Level 3 |
| Unknown, Rt 22.819 min | - | ↓ | ↑ | ↑↑ | 0.0235 | 0.6579 | Level 4 |
| Unknown, Rt 7.241 min | - | ↓ | ↑ | ↑ | 0.0319 | 0.6610 | Level 4 |
| Glucuronide, Rt 27.688 min | - | ↓ | ↑ | ↑ | 0.0386 | 0.6610 | Level 3 |
| Glucuronide, Rt 26.658 min | - | ↓ | ↑ | ↑ | 0.0435 | 0.6610 | Level 3 |
| Unknown, Rt 23.073 min | - | ↓ | ↑ | ↑↑ | 0.0458 | 0.6610 | Level 4 |
|
| |||||||
| 1H-NMR | |||||||
|
| |||||||
| Urea | urea cycle | ↑ | ↓ | ↓ | 0.0015 | 0.1580 | NMR library |
| Hippurate | mammalian-microbial cometabolite | ↑ | ↑↑ | ↑ | 0.0113 | 0.3052 | NMR library |
| Glycerol | triacylglycerol/phospholipid precursor | ↓ | ↓ | ↑ | 0.0211 | 0.3600 | NMR library |
| Butyrate | microbial metabolism | ↓ | ↑ | ↑↑ | 0.0233 | 0.3600 | NMR library |
| Carnitine | fatty acid metabolism | ↑ | ↑ | ↓ | 0.0326 | 0.4403 | NMR library |
| Galactarate | sugar acid | ↓ | ↓ | ↑ | 0.0486 | 0.5550 | NMR library |
Arrows indicate relative high or low concentrations in the respective groups.
Raw p-values based on one-way ANOVA were corrected for multiple testing (FDR).
We observed that Hyp-Hyp was increased in intermediate- and late-stage pre-surgery patients when compared to early-stage patients. Though it was increased in pre-surgery patients, the increase was not considered significant. We also found hippurate to be excreted in higher concentrations in intermediate- and late-stage patients compared to early-stage patients and detected the same trend by GC-MS and 1H-NMR analysis. Urea levels were diminished in late-stage patients compared to early- or intermediate-stage patients. Glycerol, galactarate and butyrate levels were increased only in late stage CRCs, whereas the opposite trend was seen for carnitine. Moreover, we detected 3 unknowns, 2 glucuronides and one sugar alcohol that enabled discrimination between the different stages in our GC-MS dataset.
Generally, early-stage patients were easier to distinguish from more advanced stages of the disease, whereas intermediate stages were only poorly differentiated from either of these groups (see Supplementary Fig. S3).
3.3 Colon vs. rectal cancer patients
Among pre-surgery CRC patients enrolled in our study, 54 had rectal cancer and 43 had colon cancer. Seven metabolites were significantly different after adjusting for all covariates and multiple testing if colon and rectal cancer patients prior to surgery were compared. Results are shown in Supplementary Table S2. Hippurate and 3-(3-hydroxyphenyl)-3-hydroxypropionate were excreted in higher concentrations in colon cancer patients, whereas the opposite was true for cis-aconitic acid, pyroglutamate, 4-hydroxy-3-methoxy-mandelate and two unidentified metabolites. For many of the previously mentioned gut microbiota-associated metabolites, e.g. 3- and 4-hydroxyhippurate, guajacol, p-cresol and p-cresol-β-O-glucuronide, they were excreted in higher concentrations from colon cancer patients, although the differences were not statistically significant after multiple testing. Interestingly, Hyp-Hyp and p-cresol, which were present in higher concentrations in late-stage patients were elevated in colon carcinoma patients compared to rectal cancer patients (p-adjusted = 0.056). We furthermore compared pre-surgery patients who received neoadjuvant therapy (n = 36) against pre-surgery patients without any therapy prior to surgery (n = 50). No metabolites differed significantly after adjusting for multiple testing.
3.4 Metabolite markers for CRC
We chose to build two binary classification models for the differentiation of tumor bearing individuals (pre-surgery CRC patients) from patients after therapeutic intervention:
Model 1 – Manual metabolite selection
We manually selected those metabolites that were significant after adjusting for covariates (p-adjusted), with fewer than 10% missing values, where a direct effect of surgery was seen in patients 1–8 days post-surgery (labeled as "post") and maintained over a period of up to one year (6 month and 12 month follow-up). Box plots of 20 metabolites fulfilling these requirements are illustrated in Fig. 2. We selected 4 amino acids (tyrosine, threonine, tryptophan and glycine), 10 metabolites with high confidence in identification (level 1 or 2) and 6 unidentified, but highly significant species (see Supplementary Table S4 for list of selected metabolites; all p-adjust < 0.10). These 20 metabolites were used to construct a multivariate model (based on partial least squares-discriminat analysis) with the ROCCET toolbox (Xia et al. 2013).
Model 2 – Automatic metabolite selection
A second model was constructed by selecting the ten best differentiating metabolites from both GC-MS and 1H-NMR based on their univariate ROC. Metabolite selection was performed with fasted individuals only (n = 108) to avoid marker selection due to differences in fasting state. This second model, based on these 20 automatically selected metabolites, was constructed using the same ROCCET parameters as in Model 1.
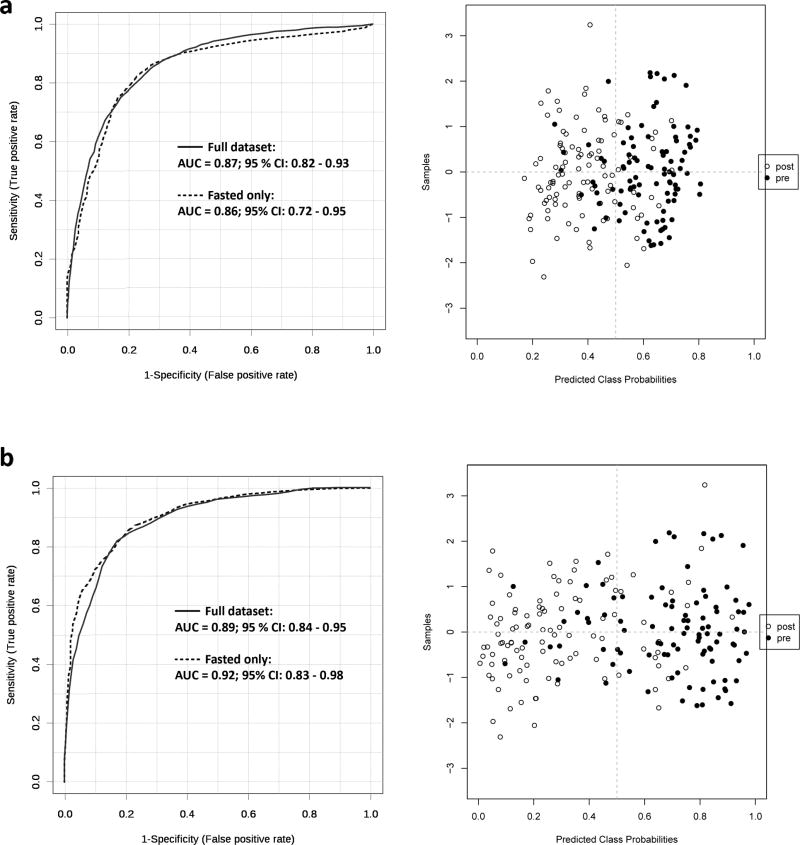
Both metabolite sets were used to discriminate pre- from post-surgery individuals in the full dataset and a subset of fasted individuals. The results are illustrated in Fig. 3 with metabolites and univariate AUCs provided in the Supplementary Data.
Fig. 3.
Receiver operator characteristic (ROC) curves for the differentiation between pre- and post-surgery CRC patients. a) Model 1 using manual variable selection; ROC curve and score plot from a partial least squares-discriminant analysis (PLS-DA) model containing 20 metabolites for all patients in our dataset (bold line) and a subset of fasted patients (dahed line). b) Model 2 using automatic variable selection; ROC curve and score plot from a partial least squares-discriminant analysis (PLS-DA) model containing 20 metabolites for all patients in our dataset (bold line) and a subset of fasted patients (dashed line).
We were able to differentiate pre- from post-surgery patients with high accuracy in both models. The method for imputation of missing values only had a marginal effect on the overall model performance (see Supplementary Data). The automatically selected set of metabolites performed slightly better than the manually selected set (cf. Fig. 3a vs. 3b). Overall, only small differences in the model performance were observed between a full dataset and a dataset consisting of fasted patients (AUCs in the range of 0.85–0.92), indicating that both metabolite sets were independent of a patients’ fasting state.
4. Discussion
Our results show that urine metabolomes of CRC patients prior to surgery can be distinguished from those of patients after surgery. This indicates that patients’ metabolomes are primarily affected by the disease or disease-associated removal of the tumor and substantial parts of the intestine, although an exacerbating effect of adjuvant chemotherapy was also observed. Furthermore, we were able to identify several metabolites associated with particular disease stages as candidates for prognostic markers. Generally, we observed many metabolites with unknown identity (level 3 or 4) to be highly and significantly altered. Unfortunately, due to a targeted 1H-NMR profiling approach and a low mass accuracy in single-quadrupole MS, we were not able to fully identify these unknowns; a process that remains one of the main challenges of metabolomics studies (Sumner et al. 2007).
Most studies published thus far have employed simple case-control designs, comparing CRC patients with healthy individuals (Liesenfeld et al. 2013). Many of the metabolites, which were discovered as potential biomarkers in these studies, were also significant in our prospective study.
4.1 Differences by stage: degradation products of collagen as candidates for prognostic biomarkers for CRC
We were able to identify several urinary metabolites that distinguished early- from late-stage CRC patients. We identified Hyp-Hyp at higher levels in late-stage cancer patients’ urine. The metabolite was also significantly altered in the urine of hepatocellular cancer patients (Wu et al. 2009). Hydroxyproline (Hyp) itself, a major component of collagen, showed no difference in excretion levels by stage. It has long been known that cancer cells need to degrade extracellular matrix components in order to disseminate through connective tissue (Deryugina et al. 2006). Dipeptides of hydroxyproline might be derived from the process of disseminating tumor cells, degrading collagen during the process of early metastasis, although the definitive origin of the metabolite remains to be determined. Interestingly, CRC patients prior to surgical intervention and post-surgery patients did not show a significant difference in their urinary excretion of Hyp-Hyp. However, colon cancer patients excreted higher levels of Hyp-Hyp compared to rectal cancer patients–an observation that cannot be explained by neoadjuvant therapy. Future mechanistic studies are needed to determine whether or not dipeptides of Hyp are formed during early metastasis and if the marker has a prognostic value for CRC patients. Other metabolites differentiating early- from intermediate- or late-stage CRC patients are discussed below.
4.2 Post-surgery urine metabolome changes indicative of the importance of the gut microbiome
We found many small aromatic components, such as guajacol, three derivatives of hydroxyhippuric acids, pyrogallol and hydroquinone lower in CRC patients prior to surgery compared to post-surgery timepoints. In contrast, p-cresol and its glucuronide tended to be present at higher concentrations in pre-surgical patients. Moreover, p-cresol and its glucuronide were of value for discerning the different stages of the disease (Table 3). All of the above metabolites can be classified as being mainly derived from the gut microbiota, due to their absence of human biochemical pathways. Some of these metabolites have previously been found to discriminate healthy controls from CRC patients. Qui et al. and Cheng et al. both reported altered p-cresol levels in the urine of CRC patients compared to healthy controls, but with an opposing direction (Cheng et al. 2012; Qiu et al. 2010). We not only measured elevated concentrations of p-cresol and its glucuronide in the late stages of CRC, but, moreover, surgical intervention markedly reduced the levels in patients after surgery by approximately 50 %. Interestingly, Zhang et al. found the glucuronide of p-cresol, identified using characteristic precursor ions and neutral loss fragments in LC-MS/MS, to be up-regulated in rats with 2,4,6 trinitrobenzene induced colitis. p-Cresol and its glucuronide might therefore reflect an inflammatory response of the gut microbiota (Zhang et al. 2012).
Our study and the results of several other authors (Cheng et al. 2012; Qiu et al. 2010; Zhu et al. 2013) indicate that metabolites derived from the gut microbiome show marked differences either over the course of the illness or in case-control designs pointing towards an important role of the gut microbiota in the pathogenesis of CRC (Cheng et al. 2012; Qiu et al. 2010) (Zhu et al. 2013). The origin of small aromatic compounds is mainly from microbial metabolism of aromatic amino acids, though eukaryotic pathways may also contribute. Qui et al. discussed their findings of altered p-cresol metabolites in the context of perturbed populations of Clostridium sp. (Qiu et al. 2010). Studies led by Bone et al. showed that p-cresol species arise primarily from tyrosine conversion of anaerobic bacteria in the left colon (Bone et al. 1976). The tumor promoting activities of p-cresol and related compounds have been known for decades (Boutwell et al. 1959). In addition to p-cresol, we observed 3-(3-hydroxyphenyl)-3-hydroxypropionate, a metabolite that has been associated with members of Clostridiae (Shaw 2010), to be elevated in colon cancer patients’ urine compared to rectal cancer patients’ urine. Moreover, urinary butyrate levels were increased in late-stage CRC patients prior to surgery. Butyrate shows growth promoting effects in normal colon cells but also tumor inhibitory effects (hence it is known as the "butyrate paradox") (Vanhoutvin et al. 2009). Its effects on secondary prevention of CRC by its induction of glutathione S-transferases might, though, be a double edged sword, by enhancing detoxification in both normal and cancer cells (Scharlau et al. 2009).
Our findings suggest that surgical removal of the tumor and parts of the intestine may affect microbial metabolites and possibly the microbiota itself, with adjuvant chemotherapy exacerbating this effect. The microbial activity in the gut may lead to the production of toxins (Heavey et al. 2004), mutagens (Carman et al. 1988) or the production of reactive oxygen intermediates as previous demonstrated (Owen et al. 2000). On the other hand, the tumor microenvironment can also influence and select bacterial communities, for example by providing a niche for certain bacterial species (Marchesi et al. 2011). Therapeutical intervention and/or removal of parts of the intestine might therefore change the bacterial population, which in turn leads to an altered metabolism in the gut.
Interestingly, our results support the hypothesis that the gut microbiota play a more important role for colon cancer than for rectal or rectosigmoidal cancer patients. Most of the microbiome associated metabolites were found to be altered in colon carcinoma patients, irrespective of prior neoadjuvant treatment (see Supplementary Table S2), which is in line with the findings that these compounds arise directly in the colon (Bone et al. 1976).
Many of these bacterial metabolites are absorbed by the intestine and further metabolized by the host and/or excreted in the urine. The identified glucuronide of p-cresol is an example of these detoxification mechanisms. Many of the partially identified metabolites (i.e. labeled with the level 3 tag "glucuronide") might serve as exposure markers for such exogenous or endogenous substances. However, future studies will have to establish how the composition of the microbiome (assessed, for example, by genotyping analyses) is associated with the metabolites excreted in urine. Moreover, it remains to be determined whether a decreased absorption area after removal of parts of the intestine, or a change in the composition of the microbiome itself, might account for the different excretion rates of microbial metabolites.
4.3 Long-known hallmarks of cancer monitored by the metabolomic profile of CRC patients’ urine
4.3.1 Amino acid metabolism and protein synthesis
Although the concentrations of some amino acids were primarily affected by co-variables such as fasting status, many other amino acids, such as phenylalanine, threonine, tyrosine, lysine, tryptophan, asparagine and glycine for example, were excreted at lower concentrations in pre-surgery CRC patients’ urine. Alterations in the amino acid profiles of cancer patients have been described multiple times (Leichtle et al. 2012; Maeda et al. 2010; Miyagi et al. 2011). Miyagi et al. compared targeted plasma amino acid profiles of various cancer entities against controls and also reported decreased levels for most amino acids in CRC, except for isoleucine, proline and glycine (Miyagi et al. 2011).
Decreased levels of 2-oxobutyrate in combination with an increase in 2-aminobutyrate might favor the hypothesis of increased amino acid catabolism of threonine (Paxton et al. 1986; Steele 1982). On the other hand, lower urinary excretion might also reflect an increased uptake of amino acids by the tumor for protein biosynthesis, accompanied by cellular turnover and tumor growth. Interestingly, we observed lower urinary urea levels among late-stage patients. Urinary urea, as the prime end product of amino acid catabolism, is an indicator of nitrogen excretion. The utilization of nitrogen is of special interest for the cancer cachexia syndrome, which accompanies about half of cancer patients (Kern et al. 1988). Lower levels of urinary urea in late-stage cancers might indicate less nitrogen flux into catabolic reactions, and a likely higher amino acid flux into protein biosynthesis. This is in line with the theory that tumors act as "nitrogen sinks" on a systemic level (Kern et al. 1988). This theory is further supported by animal studies led by Beck and Tisdale who observed an initial increase in urinary urea excretion after transplantation of colon adenocarinomas in mice, but a decrease below the baseline level at late stages of tumor growth (Beck et al. 1989).
Additionally, phenylalanine and tyrosine, precursors for catecholamine biosynthesis, were significantly altered in post-surgery patients. These aromatic amino acids serve as precursors for aromatic microbial products, such as p-cresol, as indicated by our chemical similarity network (see Fig. 2). Furthermore, urinary dopamine, a downstream metabolite of tyrosine, was found to be diminished in patients still bearing a tumor. Dopamine may have anti-angiogenic effects, inhibiting vascular endothelial growth factor (VEGF) in vitro (Chakroborty et al. 2009). Moreover, it reduced tumor growth in tumor bearing mice by blocking the effects of daily restraint stress (immobilization) (Moreno-Smith et al. 2013).
In addition, four members of tryptophan metabolism were affected by the surgical intervention: tryptophan itself (0.82-fold) as well as xanthurenate (0.79-fold) and indole-3-acetate (0.80-fold) were diminished in pre-surgery compared to post-surgery patients, whereas 3-indoxylsulfate levels were increased 1.46-fold (Fig. 1). Cheng et al. also reported diminished levels of tryptophan metabolism (5-hydroxy-Trp, indoleacetate, indole and others) in CRC patients compared to healthy individuals (Cheng et al. 2012). Tryptophan is the precursor of several neurotransmitters and hormones such as serotonin and melatonin, but also of kynurenine and its downstream catabolites. Recently, kynurenine was described as an endogenous ligand of the arylhydrocarbon receptor, promoting a tumor progressive mechanism (Opitz et al. 2011). We did not detect kynurenine in our urinary samples but noted a catabolite, xanthurenate, to be lowered in CRC patients prior to surgery compared to post-surgery patients. Moreover Cheng et al. reported diminished levels of another catabolites of kynurenine (kynurenate) in the urine of CRC patients compared to urine of healthy controls (Cheng et al. 2012).
4.3.2 Oncometabolites
We observed moderately higher (1.27-fold, FDR = 0.068) urinary excretion of the oncometabolite 2-hydroxyglutarate in patients at pre- compared to post-surgery. Oncometabolic activities of 2-hydroxyglutarate have recently been discovered (Losman et al. 2013): 2-hydroxyglutarate can promote epigenetically driven carcinogenesis by directly inhibiting histone lysine demethylases (Højsgaard 2013) and 5-methylcytosine hydroxylases (Pollard et al. 2005). The metabolite itself has been described as being derived from the neomorphic activity of mutated isocitrate dehydrogenase (IDH). While IDH mutations are most common in glioblastomas and acute myelogenous leukemia (Bates et al. 2013), they were initially discovered in metastatic colon cancer (Rakheja et al. 2013).
myo-Inositol was diminished by 0.43 and 0.68-fold, respectively, according to GC-MS and 1H-NMR analysis in pre-surgery CRC patients compared to post-surgery. myo-Inositol and its derivatives have previously been associated with different cancer-types/biological matrices and varying direction of change (Aa et al. 2012; Perroud et al. 2006; Thysell et al. 2010). Denkert et al. reported lower levels of an inositol stereoisomer in colon cancer tissue compared to normal colon mucosa (Denkert et al. 2008), in concordance with the observed lower urinary myo-inositol levels in pre-surgery, tumor bearing patients in our study. Another group compared CRC patients at different stages and observed that inositol was less abundant in tissues of CRC liver metastasis (Farshidfar et al. 2012). Inositol might therefore play a role in cellular signaling, via its link to the phosphoinositol-3-kinase PI3K pathway, impacting CRC pathogenesis.
Interestingly, 1.74-fold higher levels (borderline significance; p-adjusted 0.07) of a metabolite similar to cadaverine was also observed in pre-surgery CRC patients. We hypothesize the metabolite to be a polyamine derivative, but further investigations will have to confirm the exact chemical identity of the metabolite. Increased levels of polyamines have been reported in the plasma and urine of cancer patients (Liu et al. 2013).
5. Concluding remarks
To the best of our knowledge, this study is one of the first reporting urine metabolomic data with repeated sampling from a prospective CRC patient cohort. While most studies published so far have compared CRC patients to healthy controls, we were able to monitor patient metabolomes over time. Moreover, we used two complimentary methods, GC-MS and 1H-NMR, enabling us to independently validate our findings. A main limitation of our study was the low sensitivity of NMR and single-quadrupole MS as well as the low unit mass accuracy for the latter technique. Subsequently, we were unable to identify a number of metabolites of high significance by not being able to conduct more sophisticated library searches or sum formula calculations.
Surgical intervention had an impact on several metabolites derived from the gut microbiota. Moreover, amino acids and downstream metabolites of aromatic amino acids were altered. Nevertheless, we were able to formulate a discrimination model consisting of thirteen marker metabolites to differentiate pre- from post-surgery CRC patients with good accuracy. Additionally, we observed a dipeptide of hydroxyproline, together with p-cresol and its glucuronide, to be present in higher concentrations in the urine of late stage CRC patients. To further evaluate whether these candidate metabolites are of prognostic value for patients’ clinical outcome, a validation study with continued follow-up over a longer time period will be needed.
Supplementary Material
Acknowledgments
Acknowledgements and Funding
The ColoCare study has been funded by the German Consortium for Translational Cancer Research (DKTK), Matthias Lackas Foundation and the Helmholtz International Graduate School for Cancer Research, Heidelberg. We thank our collaborators, in particular Prof. Hermann Brenner, Prof. Jenny Chang-Claude and Dr. Michael Hoffmeister. We also thank all the study staff who through their energy and effort have made this study possible, particularly Dr. Clare Abbenhardt, Dr. Stephanie Zschäbitz, Thorsten Kölsch, Judith Kammer, Stephanie Tosic, Verena Widmer, Manja Ghajar Rahimi, Biljana Gigic, Werner Diehl, Rifraz Farook and many others. Last, we are truly grateful for the time and cooperation of our study participants.
Footnotes
Conflict of interest: The authors declare that there are no conflicts of interest
References
- Aa J, et al. Metabolic features of the tumor microenvironment of gastric cancer and the link to the systemic macroenvironment. Metabolomics. 2012;8:164–173. [Google Scholar]
- ACS, A. C. S. Cancer Facts & Figures 2013. Atlanta: 2013. [Google Scholar]
- Barupal DK, et al. MetaMapp: mapping and visualizing metabolomic data by integrating information from biochemical pathways and chemical and mass spectral similarity. BMC Bioinformatics. 2012;13:99. doi: 10.1186/1471-2105-13-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D, Maechler M, Bolker B, Walker S. lme4: Linear mixed-effects models using Eigen and S4. 2013 1.0–5 edn. [Google Scholar]
- Beck SA, Tisdale MJ. Nitrogen excretion in cancer cachexia and its modification by a high fat diet in mice. Cancer Res. 1989;49:3800–4. [PubMed] [Google Scholar]
- Bellomi M, et al. Accuracy of computed tomography and magnetic resonance imaging in the detection of lymph node involvement in cervix carcinoma. Eur Radiol. 2005;15:2469–74. doi: 10.1007/s00330-005-2847-1. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57:289–300. [Google Scholar]
- Bone E, Tamm A, Hill M. The production of urinary phenols by gut bacteria and their possible role in the causation of large bowel cancer. Am J Clin Nutr. 1976;29:1448–54. doi: 10.1093/ajcn/29.12.1448. [DOI] [PubMed] [Google Scholar]
- Boutwell RK, Bosch DK. The tumor-promoting action of phenol and related compounds for mouse skin. Cancer Res. 1959;19:413–24. [PubMed] [Google Scholar]
- Carman RJ, Van Tassell RL, Kingston DG, Bashir M, Wilkins TD. Conversion of IQ, a dietary pyrolysis carcinogen to a direct-acting mutagen by normal intestinal bacteria of humans. Mutat Res. 1988;206:335–42. doi: 10.1016/0165-1218(88)90119-x. [DOI] [PubMed] [Google Scholar]
- Chakroborty D, Sarkar C, Basu B, Dasgupta PS, Basu S. Catecholamines regulate tumor angiogenesis. Cancer Res. 2009;69:3727–30. doi: 10.1158/0008-5472.CAN-08-4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EC, et al. Metabolic profiling of human colorectal cancer using high-resolution magic angle spinning nuclear magnetic resonance (HR-MAS NMR) spectroscopy and gas chromatography mass spectrometry (GC/MS) J Proteome Res. 2009;8:352–61. doi: 10.1021/pr8006232. [DOI] [PubMed] [Google Scholar]
- Cheng Y, et al. Distinct urinary metabolic profile of human colorectal cancer. J Proteome Res. 2012;11:1354–1363. doi: 10.1021/pr201001a. [DOI] [PubMed] [Google Scholar]
- Denkert C, et al. Metabolite profiling of human colon carcinoma - Deregulation of TCA cycle and amino acid turnover. Molecular Cancer. 2008;7 doi: 10.1186/1476-4598-7-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deryugina EI, Quigley JP. Matrix metalloproteinases and tumor metastasis. Cancer Metastasis Rev. 2006;25:9–34. doi: 10.1007/s10555-006-7886-9. [DOI] [PubMed] [Google Scholar]
- Dunn WB, et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat Protoc. 2011;6:1060–83. doi: 10.1038/nprot.2011.335. [DOI] [PubMed] [Google Scholar]
- Dunn WB, Ellis DI. Metabolomics: Current analytical platforms and methodologies. Trends in Analytical Chemistry. 2005;24:285–294. [Google Scholar]
- Farshidfar F, et al. Serum metabolomic profile as a means to distinguish stage of colorectal cancer. Genome Medicine. 2012;4:2–13. doi: 10.1186/gm341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferlay J, et al. Cancer incidence and mortality patterns in Europe: estimates for 40 countries in 2012. Eur J Cancer. 2013;49:1374–403. doi: 10.1016/j.ejca.2012.12.027. [DOI] [PubMed] [Google Scholar]
- Fiehn O. Metabolomics-the link between genotypes and phenotypes. Plant molecular biology. 2002;48:155–71. [PubMed] [Google Scholar]
- Heavey PM, Rowland IR. Microbial-gut interactions in health and disease. Gastrointestinal cancer. Best Pract Res Clin Gastroenterol. 2004;18:323–36. doi: 10.1016/j.bpg.2003.10.003. [DOI] [PubMed] [Google Scholar]
- Højsgaard UHS. pbkrtest: Parametric bootstrap and Kenward Roger based methods for mixed model comparison. 2013 0.3–8 edn. [Google Scholar]
- Hummel J, Selbig J, Walther D, Kopka J. The Golm Metabolome Database: a database for GC-MS based metabolite profiling. In: Nielsen J, Jewett M, editors. Metabolomics. Topics in Current Genetics. Vol. 18. Springer; Berlin Heidelberg: 2007. pp. 75–95. [Google Scholar]
- Kern KA, Norton JA. Cancer cachexia. JPEN J Parenter Enteral Nutr. 1988;12:286–98. doi: 10.1177/0148607188012003286. [DOI] [PubMed] [Google Scholar]
- Kind T, Tolstikov V, Fiehn O, Weiss RH. A comprehensive urinary metabolomic approach for identifying kidney cancer. Analytical Biochemistry. 2007;363:185–195. doi: 10.1016/j.ab.2007.01.028. [DOI] [PubMed] [Google Scholar]
- Kondo Y, et al. Serum fatty acid profiling of colorectal cancer by gas chromatography/mass spectrometry. Biomarkers in Medicine. 2011;5:451–460. doi: 10.2217/bmm.11.41. [DOI] [PubMed] [Google Scholar]
- Leichtle AB, et al. Serum amino acid profiles and their alterations in colorectal cancer. Metabolomics. 2012;8:643–653. doi: 10.1007/s11306-011-0357-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liesenfeld DB, Habermann N, Owen RW, Scalbert A, Ulrich CM. Review of mass spectrometry-based metabolomics in cancer research. Cancer Epidemiol Biomarkers Prev. 2013;22:2182–201. doi: 10.1158/1055-9965.EPI-13-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Li Q, Ma R, Lin X, Xu H, Bi K. Determination of polyamine metabolome in plasma and urine by ultrahigh performance liquid chromatography-tandem mass spectrometry method: application to identify potential markers for human hepatic cancer. Anal Chim Acta. 2013;791:36–45. doi: 10.1016/j.aca.2013.06.044. [DOI] [PubMed] [Google Scholar]
- Losman JA, Kaelin WG., Jr What a difference a hydroxyl makes: mutant IDH, (R)-2-hydroxyglutarate, and cancer. Genes Dev. 2013;27:836–52. doi: 10.1101/gad.217406.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma YL, et al. A pilot study of gas chromatograph/mass spectrometry-based serum metabolic profiling of colorectal cancer after operation. Molecular Biology Reports. 2010;37:1403–1411. doi: 10.1007/s11033-009-9524-4. [DOI] [PubMed] [Google Scholar]
- Ma YL, et al. Ultra-High Performance Liquid Chromatography-Mass Spectrometry for the Metabolomic Analysis of Urine in Colorectal Cancer. Digestive Diseases and Sciences. 2009;54:2655–2662. doi: 10.1007/s10620-008-0665-4. [DOI] [PubMed] [Google Scholar]
- Maeda J, et al. Possibility of multivariate function composed of plasma amino acid profiles as a novel screening index for non-small cell lung cancer: a case control study. BMC Cancer. 2010;10:690–698. doi: 10.1186/1471-2407-10-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mal M, Koh PK, Cheah PY, Chan ECY. Development and validation of a gas chromatography/mass spectrometry method for the metabolic profiling of human colon tissue. Rapid Communications in Mass Spectrometry. 2009;23:487–494. doi: 10.1002/rcm.3898. [DOI] [PubMed] [Google Scholar]
- Mal M, Koh PK, Cheah PY, Chan ECY. Metabotyping of human colorectal cancer using two-dimensional gas chromatography mass spectrometry. Anal Bioanal Chem. 2012;403:483–493. doi: 10.1007/s00216-012-5870-5. [DOI] [PubMed] [Google Scholar]
- Marchesi JR, et al. Towards the Human Colorectal Cancer Microbiome. Plos One. 2011;6:e20447. doi: 10.1371/journal.pone.0020447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagi Y, et al. Plasma free amino acid profiling of five types of cancer patients and its application for early detection. Plos One. 2011;6:e24143. doi: 10.1371/journal.pone.0024143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Smith M, et al. Biologic effects of dopamine on tumor vasculature in ovarian carcinoma. Neoplasia. 2013;15:502–10. doi: 10.1593/neo.121412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiumi S, et al. A Novel Serum Metabolomics-Based Diagnostic Approach for Colorectal Cancer. Plos One. 2012;7 doi: 10.1371/journal.pone.0040459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normanno N, et al. Molecular diagnostics and personalized medicine in oncology: challenges and opportunities. J Cell Biochem. 2013;114:514–24. doi: 10.1002/jcb.24401. [DOI] [PubMed] [Google Scholar]
- Opitz CA, et al. An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature. 2011;478:197–203. doi: 10.1038/nature10491. [DOI] [PubMed] [Google Scholar]
- Owen RW, Spiegelhalder B, Bartsch H. Generation of reactive oxygen species by the faecal matrix. Gut. 2000;46:225–32. doi: 10.1136/gut.46.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Z, Raftery D. Comparing and combining NMR spectroscopy and mass spectrometry in metabolomics. Anal Bioanal Chem. 2007;387:525–7. doi: 10.1007/s00216-006-0687-8. [DOI] [PubMed] [Google Scholar]
- Paxton R, Scislowski PW, Davis EJ, Harris RA. Role of branched-chain 2-oxo acid dehydrogenase and pyruvate dehydrogenase in 2-oxobutyrate metabolism. Biochem J. 1986;234:295–303. doi: 10.1042/bj2340295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perroud B, et al. Pathway analysis of kidney cancer using proteomics and metabolic profiling. Molecular Cancer. 2006;5:1–17. doi: 10.1186/1476-4598-5-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluskal T, Castillo S, Villar-Briones A, Oresic M. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics. 2010;11:395–406. doi: 10.1186/1471-2105-11-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard PJ, et al. Accumulation of Krebs cycle intermediates and over-expression of HIF1alpha in tumours which result from germline FH and SDH mutations. Hum Mol Genet. 2005;14:2231–9. doi: 10.1093/hmg/ddi227. [DOI] [PubMed] [Google Scholar]
- Qiu YP, et al. Urinary Metabonomic Study on Colorectal Cancer. J Proteome Res. 2010;9:1627–1634. doi: 10.1021/pr901081y. [DOI] [PubMed] [Google Scholar]
- Qiu YP, et al. Serum Metabolite Profiling of Human Colorectal Cancer Using GC-TOFMS and UPLC-QTOFMS. J Proteome Res. 2009;8:4844–4850. doi: 10.1021/pr9004162. [DOI] [PubMed] [Google Scholar]
- Computing, R. F. f. S, editor. R-Development-Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: 2013. [Google Scholar]
- Rakheja D, Medeiros LJ, Bevan S, Chen W. The emerging role of d-2-hydroxyglutarate as an oncometabolite in hematolymphoid and central nervous system neoplasms. Front Oncol. 2013;3:169. doi: 10.3389/fonc.2013.00169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie SA, et al. Reduced levels of hydroxylated, polyunsaturated ultra long-chain fatty acids in the serum of colorectal cancer patients: implications for early screening and detection. Bmc Medicine. 2010;8:1–20. doi: 10.1186/1741-7015-8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharlau D, et al. Mechanisms of primary cancer prevention by butyrate and other products formed during gut flora-mediated fermentation of dietary fibre. Mutat Res. 2009;682:39–53. doi: 10.1016/j.mrrev.2009.04.001. [DOI] [PubMed] [Google Scholar]
- Shaw W. Increased urinary excretion of a 3-(3-hydroxyphenyl)-3-hydroxypropionic acid (HPHPA), an abnormal phenylalanine metabolite of Clostridia spp. in the gastrointestinal tract, in urine samples from patients with autism and schizophrenia. Nutr Neurosci. 2010;13:135–43. doi: 10.1179/147683010X12611460763968. [DOI] [PubMed] [Google Scholar]
- Spratlin JL, Serkova NJ, Eckhardt SG. Clinical applications of metabolomics in oncology: a review. Clin Cancer Res. 2009;15:431–40. doi: 10.1158/1078-0432.CCR-08-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele RD. Transaminative metabolism of alpha-amino-n-butyrate in rats. Metabolism. 1982;31:318–25. doi: 10.1016/0026-0495(82)90106-8. [DOI] [PubMed] [Google Scholar]
- Sumner L, et al. Proposed minimum reporting standards for chemical analysis. Metabolomics. 2007;3:211–221. doi: 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevis M, Opfermann G, Schmickler H, Schänzer W. Mass spectrometry of steroid glucuronide conjugates. I. Electron impact fragmentation of 5α-/5β-androstan-3α-ol-17-one glucuronides, 5α-estran-3α-ol-17-one glucuronide and deuterium-labelled analogues. Journal of Mass Spectrometry. 2001;36:159–168. doi: 10.1002/jms.117. [DOI] [PubMed] [Google Scholar]
- Thysell E, et al. Metabolomic characterization of human prostate cancer bone metastases reveals increased levels of cholesterol. Plos One. 2010;5:e14175. doi: 10.1371/journal.pone.0014175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhoutvin SA, et al. Butyrate-induced transcriptional changes in human colonic mucosa. Plos One. 2009;4:e6759. doi: 10.1371/journal.pone.0006759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WZ, et al. Urinary metabolic profiling of colorectal carcinoma based on online affinity solid phase extraction-high performance liquid chromatography and ultra performance liquid chromatography-mass spectrometry. Molecular Biosystems. 2010;6:1947–1955. doi: 10.1039/c004994h. [DOI] [PubMed] [Google Scholar]
- Wu H, et al. Metabolomic profiling of human urine in hepatocellular carcinoma patients using gas chromatography/mass spectrometry. Analytica Chimica Acta. 2009;648:98–104. doi: 10.1016/j.aca.2009.06.033. [DOI] [PubMed] [Google Scholar]
- Xia J, Broadhurst D, Wilson M, Wishart DS. Translational biomarker discovery in clinical metabolomics: an introductory tutorial. Metabolomics. 2013;9:280–299. doi: 10.1007/s11306-012-0482-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Mandal R, Sinelnikov IV, Broadhurst D, Wishart DS. MetaboAnalyst 2.0--a comprehensive server for metabolomic data analysis. Nucleic Acids Res. 2012;40:2. doi: 10.1093/nar/gks374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C, Hao F, Qin X, Wang Y, Tang H. An optimized buffer system for NMR-based urinary metabonomics with effective pH control, chemical shift consistency and dilution minimization. Analyst. 2009;134:916–25. doi: 10.1039/b818802e. [DOI] [PubMed] [Google Scholar]
- Yamanishi Y, Iguchi M, Oyama H, Matsumura Y. Collagen metabolism. I. Significance of urinary pyrrole-2-carboxylic acid. J Clin Endocrinol Metab. 1972;35:55–8. doi: 10.1210/jcem-35-1-55. [DOI] [PubMed] [Google Scholar]
- Zhang X, et al. Metabolite profiling of plasma and urine from rats with TNBS-induced acute colitis using UPLC-ESI-QTOF-MS-based metabonomics--a pilot study. FEBS J. 2012;279:2322–38. doi: 10.1111/j.1742-4658.2012.08612.x. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Gao R, Wu W, Qin H. The role of gut microbiota in the pathogenesis of colorectal cancer. Tumor Biology. 2013;34:1285–1300. doi: 10.1007/s13277-013-0684-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.