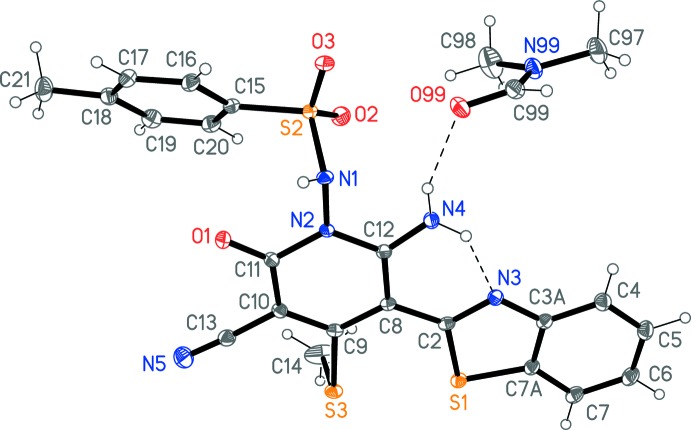
In the title compound, the toluenesulfonamide ring and the combined ring system involving the pyridone and benzothiazole rings subtend an interplanar angle of 39.86 (4)°. The pyridone and benzothiazyl rings are linked by an intramolecular N—Hamine⋯Nthiazole hydrogen bond. The molecules are linked by hydrogen bonds and an S⋯O contact to form layers parallel to the bc plane.
Keywords: crystal structure, 2-pyridone, benzothiazole, dimethylformamide
Abstract
In the title compound, C21H17N5O3S3·C3H7NO, the toluenesulfonamide ring and the combined ring system involving the pyridone and benzothiazole rings subtend an interplanar angle of 39.86 (4)°. The pyridone and benzothiazyl rings are linked by the intramolecular hydrogen bond N—Hamine⋯Nthiazole. The DMF O atom accepts two classical hydrogen bonds. The molecules are linked by hydrogen bonds and an S⋯O contact to form layers parallel to the bc plane.
Chemical context
Cyanoketene dithioacetals are versatile synthetic intermediates (Elgemeie et al., 2003a
▸, 2015 ▸) that have been utilized as building blocks for the synthesis of a wide range of heterocyclic compounds (Elgemeie et al., 2009 ▸, 2017a
▸); they are also of general interest in pharmaceutical chemistry (Elgemeie & Abou-Zeid, 2015 ▸; Elgemeie et al., 2016 ▸). Recently, we have described the synthesis of various antimetabolites starting from cyanoketene dithioacetals and related compounds, viz. cyanoketene S,S-acetals (Elgemeie, Mohamed, 2006 ▸), cyanoketene N,S-acetals (Elgemeie et al. 2017b
▸), and cyanoketene N,N-acetals (Elgemeie et al., 2003b
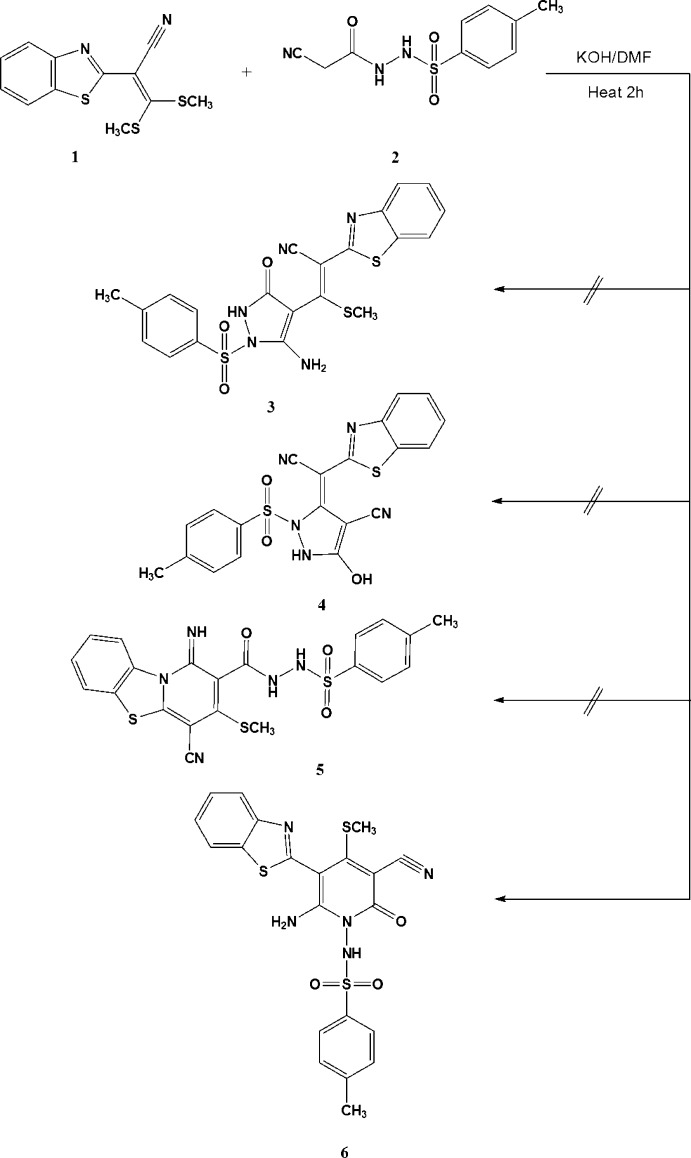
▸). As a part of this programme, the reaction of 2-(benzo[d]thiazol-2-yl)-3,3-bis(methylthio)acrylonitrile (1) with N-(2-cyanoacetyl)-4-methylbenzenesulfonohydrazide (2) was investigated. The reaction between 1 and 2 in KOH–DMF gives an adduct for which four possible isomeric structures were considered (structures 3–6). Spectroscopic methods did not allow us to identify the product unambiguously and therefore the X-ray crystal structure was determined, confirming the exclusive presence of structure 6 in the solid state. The formation of 6 from the reaction of 1 and 2 is assumed to proceed via initial addition of the active methylene carbon atom of 2 to the double bond of 1, followed by elimination of CH3SH and cyclization via addition of the NH group to the cyano group of benzothiazole to give the favoured, kinetically and thermodynamically controlled product 6. The 1H NMR spectra of the product revealed the presence of an amino group at δ = 8.84 p.p.m. and a pyridine methylthio group at δ = 2.45 p.p.m. in solution. Compound 6 and its derivatives showed interesting preclinical biological results and are currently being patented (Elgemeie et al., 2017c
▸).
Structural commentary
The solid-state structure of 6 is shown in Fig. 1 ▸, the structure analysis thereby confirming the nature of the product. The molecule essentially consists of two planes; the toluenesulfonamide ring and the combined ring system involving the pyridone and benzothiazole rings. The former has a r.m.s. deviation of 0.04 Å and the latter of 0.01 Å (including all direct substituents), and the interplanar angle is 39.86 (4)°. The pyridone and benzothiazyl rings are held coplanar by the intramolecular hydrogen bond N4—H03⋯N3 (Table 1 ▸). The contact N4—H02⋯N1 might also be classified as a hydrogen bond, with H⋯N 2.24 (2) Å, but its angle is only 105.7 (15)°. The nitrogen N4 is planar (angle sum 359.7°) but N1 is pyramidalized (343.9°).
Figure 1.
The structure of the title compound in the crystal. Displacement ellipsoids represent 50% probability levels.
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H01⋯O99i | 0.888 (18) | 1.872 (18) | 2.7583 (13) | 175.7 (16) |
| N4—H02⋯O99 | 0.84 (2) | 2.05 (2) | 2.8334 (14) | 154.6 (18) |
| N4—H03⋯N3 | 0.86 (2) | 1.86 (2) | 2.5760 (15) | 139.9 (17) |
| N4—H02⋯N1 | 0.84 (2) | 2.237 (19) | 2.5932 (14) | 105.7 (15) |
| C7—H7⋯O3ii | 0.95 | 2.54 | 3.3161 (16) | 139 |
| C20—H20⋯O2iii | 0.95 | 2.64 | 3.5605 (16) | 164 |
| C97—H97C⋯N5iv | 0.98 | 2.59 | 3.504 (2) | 155 |
Symmetry codes: (i) 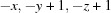 ; (ii)
; (ii) 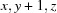 ; (iii)
; (iii) 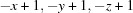 ; (iv)
; (iv) 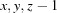 .
.
Supramolecular features
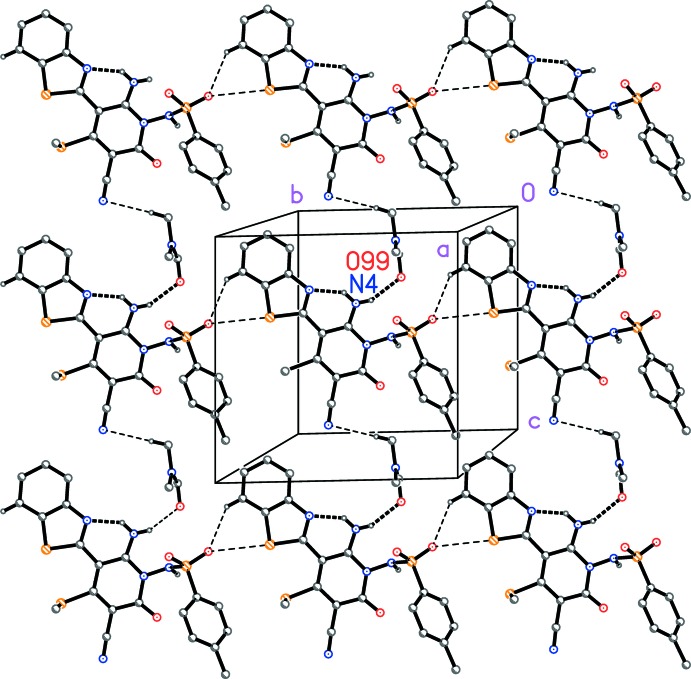
The oyxgen atom of the dimethylformamide accepts two classical hydrogen bonds. The clearest packing feature is the formation of layers parallel to the bc plane (Fig. 2 ▸), in which the hydrogen bonds H02⋯O99, H7⋯O3ii and H97C⋯N5iv are involved (Table 1 ▸), together with the short contact S1⋯O3(x, 1 + y, z) 3.2662 (10) Å. The hydrogen bond H01⋯O99i connects the layers in the third dimension.
Figure 2.
Packing diagram of the title compound viewed perpendicular to the bc plane. Dashed lines indicate classical hydrogen bonds (thick) or C—H⋯X and S⋯O interactions (thin).
Database survey
The 2-pyridone ring displays the usual features of a narrow angle at nitrogen and a wide angle at the carbonyl carbon (Table 2 ▸). A database search gave 555 hits (745 values) for the 2-pyridone ring, with average angles of 123.9° at nitrogen and 115.3° at C=O. No other structures could be found in which a 2-pyridone ring is attached at the 5-position to the C2 atom of a thiazol ring.
Table 2. Selected bond angles (°).
| N2—C11—C10 | 113.44 (10) | C12—N2—C11 | 125.63 (10) |
Synthesis and crystallization
2-(Benzo[d]thiazol-2-yl)-3,3-bis(methylthio)acrylonitrile (1) (2.78 g, 0.01 mol) was added to a solution of N-(2-cyanoacetyl)-4-methylbenzenesulfonohydrazide (2) (2.53 g., 0.01 mol) in dry DMF (30 ml) containing pulverized potassium hydroxide (0.56 g, 0.01 mol). The reaction mixture was refluxed with stirring for 2 h (TLC monitoring). After cooling, the reaction mixture was poured into ice-cold water and neutralized with HCl. The solid product was filtered off, washed with water, and dried. It was further purified from hot ethyl acetate: petroleum ether (1:1). The precipitated solid was crystallized from DMF to give yellow crystals, m.p. = 494 K, yield 78%.
IR (KBr, cm−1): ν 3393, 3208 (NH, NH2), 3072 (ArCH), 2922 (CH3), 2210 (CN), 1677 (CO), 1594 (C=N), 1350, 1170 (O=S=O); 1H NMR (400 MHz, DMSO-d 6): δ 2.42 (s, 3H, CH3), 2.45 (s, 3H, SCH3), 7.42 (d, J = 8 Hz, 2H, C6H4), 7.49 (t, J = 8 Hz, 1H, benzothiazole H), 7.56 (t, J = 8 Hz, 1H, benzothiazole H), 7.71 (d, J = 8 Hz, 2H, C6H4), 8.06 (d, J = 8 Hz, 1H, benzothiazole H), 8.13 (d, J = 8 Hz, 1H, benzothiazole H), 8.84 (br, 2H, NH2), 11.44 (s, 1H, NH). Analysis calculated for C21H17N5O3S3 (483.59): C 52.16, H 3.54, N 14.48%; found: C 52.11; H 3.48; N 14.50%; MS m/z (%): 484 (M+1, 1.03%), 384 (84%), 356 (100%), 283 (60%), 117 (77%).
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 3 ▸. NH hydrogen atoms were refined freely. Methyl hydrogen atoms were refined as idealized rigid groups allowed to rotate but not tip (AFIX 137), with C—H 0.98 Å and H—C—H 109.5°. Other hydrogen atoms were included using a riding model starting from calculated positions (C—Haromatic 0.95, C—Hmethine 1.00 Å) with U iso(H) = 1.5U eq(C) for methyl H atoms and 1.2U eq(C) for all others.
Table 3. Experimental details.
| Crystal data | |
| Chemical formula | C21H17N5O3S3·C3H7NO |
| M r | 556.67 |
| Crystal system, space group | Triclinic, P
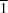
|
| Temperature (K) | 100 |
| a, b, c (Å) | 9.9916 (5), 11.7805 (6), 11.9776 (6) |
| α, β, γ (°) | 88.809 (4), 79.159 (4), 67.245 (5) |
| V (Å3) | 1274.80 (12) |
| Z | 2 |
| Radiation type | Mo Kα |
| μ (mm−1) | 0.34 |
| Crystal size (mm) | 0.5 × 0.4 × 0.2 |
| Data collection | |
| Diffractometer | Oxford Diffraction Xcalibur Eos |
| Absorption correction | Multi-scan (CrysAlis PRO; Rigaku OD, 2015 ▸) |
| T min, T max | 0.972, 1.000 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 68326, 7630, 6682 |
| R int | 0.036 |
| (sin θ/λ)max (Å−1) | 0.726 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.033, 0.082, 1.04 |
| No. of reflections | 7630 |
| No. of parameters | 350 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.61, −0.36 |
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S2056989017015778/hg5500sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989017015778/hg5500Isup2.hkl
CCDC reference: 1582798
Additional supporting information: crystallographic information; 3D view; checkCIF report
supplementary crystallographic information
Crystal data
| C21H17N5O3S3·C3H7NO | Z = 2 |
| Mr = 556.67 | F(000) = 580 |
| Triclinic, P1 | Dx = 1.450 Mg m−3 |
| a = 9.9916 (5) Å | Mo Kα radiation, λ = 0.71073 Å |
| b = 11.7805 (6) Å | Cell parameters from 19857 reflections |
| c = 11.9776 (6) Å | θ = 2.3–30.6° |
| α = 88.809 (4)° | µ = 0.34 mm−1 |
| β = 79.159 (4)° | T = 100 K |
| γ = 67.245 (5)° | Tablet, yellow |
| V = 1274.80 (12) Å3 | 0.5 × 0.4 × 0.2 mm |
Data collection
| Oxford Diffraction Xcalibur Eos diffractometer | 7630 independent reflections |
| Radiation source: fine-focus sealed X-ray tube | 6682 reflections with I > 2σ(I) |
| Detector resolution: 16.1419 pixels mm-1 | Rint = 0.036 |
| ω–scan | θmax = 31.1°, θmin = 2.3° |
| Absorption correction: multi-scan (CrysAlis PRO; Rigaku Oxford Diffraction, 2015) | h = −14→14 |
| Tmin = 0.972, Tmax = 1.000 | k = −16→16 |
| 68326 measured reflections | l = −17→17 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.033 | Hydrogen site location: mixed |
| wR(F2) = 0.082 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.04 | w = 1/[σ2(Fo2) + (0.0337P)2 + 0.772P] where P = (Fo2 + 2Fc2)/3 |
| 7630 reflections | (Δ/σ)max = 0.001 |
| 350 parameters | Δρmax = 0.61 e Å−3 |
| 0 restraints | Δρmin = −0.36 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| S1 | 0.20917 (3) | 1.05419 (3) | 0.44384 (2) | 0.01311 (6) | |
| C2 | 0.17424 (12) | 0.91906 (10) | 0.43367 (10) | 0.0117 (2) | |
| N3 | 0.13892 (11) | 0.90316 (9) | 0.33646 (8) | 0.01298 (18) | |
| C3A | 0.13978 (12) | 0.99549 (11) | 0.26319 (10) | 0.0130 (2) | |
| C4 | 0.10933 (14) | 1.00118 (12) | 0.15352 (10) | 0.0165 (2) | |
| H4 | 0.086143 | 0.938809 | 0.122941 | 0.020* | |
| C5 | 0.11382 (14) | 1.10006 (12) | 0.09052 (11) | 0.0183 (2) | |
| H5 | 0.094063 | 1.105310 | 0.015622 | 0.022* | |
| C6 | 0.14719 (14) | 1.19255 (12) | 0.13597 (11) | 0.0185 (2) | |
| H6 | 0.148137 | 1.260042 | 0.091474 | 0.022* | |
| C7 | 0.17885 (14) | 1.18784 (11) | 0.24436 (11) | 0.0167 (2) | |
| H7 | 0.201612 | 1.250566 | 0.274740 | 0.020* | |
| C7A | 0.17590 (13) | 1.08692 (11) | 0.30715 (10) | 0.0134 (2) | |
| C8 | 0.18455 (12) | 0.82947 (10) | 0.52233 (10) | 0.0113 (2) | |
| C9 | 0.21641 (12) | 0.84103 (10) | 0.63037 (10) | 0.0119 (2) | |
| C10 | 0.22112 (13) | 0.75420 (11) | 0.71150 (10) | 0.0130 (2) | |
| C11 | 0.19296 (13) | 0.64662 (11) | 0.69151 (10) | 0.0126 (2) | |
| C12 | 0.16105 (12) | 0.72035 (10) | 0.49822 (9) | 0.0112 (2) | |
| C13 | 0.25168 (14) | 0.76681 (11) | 0.82143 (11) | 0.0161 (2) | |
| C14 | 0.44732 (16) | 0.90452 (15) | 0.64337 (18) | 0.0375 (4) | |
| H14A | 0.482909 | 0.832100 | 0.688175 | 0.056* | |
| H14B | 0.484276 | 0.965711 | 0.663453 | 0.056* | |
| H14C | 0.483227 | 0.879848 | 0.562098 | 0.056* | |
| S2 | 0.28523 (3) | 0.39958 (3) | 0.51130 (2) | 0.01363 (7) | |
| S3 | 0.24867 (3) | 0.97028 (3) | 0.67335 (3) | 0.01445 (7) | |
| O1 | 0.18556 (10) | 0.56948 (8) | 0.75942 (7) | 0.01650 (17) | |
| O2 | 0.39814 (10) | 0.43053 (8) | 0.44172 (8) | 0.01993 (19) | |
| O3 | 0.22015 (11) | 0.32617 (8) | 0.46616 (8) | 0.01989 (19) | |
| N1 | 0.14142 (11) | 0.53246 (9) | 0.55209 (8) | 0.01215 (18) | |
| H01 | 0.0686 (19) | 0.5236 (16) | 0.6018 (15) | 0.024 (4)* | |
| N2 | 0.17243 (11) | 0.63362 (9) | 0.57965 (8) | 0.01120 (18) | |
| N4 | 0.13014 (12) | 0.69607 (10) | 0.40197 (9) | 0.01497 (19) | |
| H02 | 0.116 (2) | 0.6318 (18) | 0.3914 (16) | 0.031 (5)* | |
| H03 | 0.118 (2) | 0.7536 (18) | 0.3545 (16) | 0.030 (5)* | |
| N5 | 0.27764 (14) | 0.77140 (11) | 0.91037 (10) | 0.0249 (2) | |
| C15 | 0.35093 (13) | 0.32693 (11) | 0.63125 (10) | 0.0142 (2) | |
| C16 | 0.27838 (14) | 0.25780 (11) | 0.69169 (11) | 0.0162 (2) | |
| H16 | 0.196647 | 0.250018 | 0.667958 | 0.019* | |
| C17 | 0.32777 (14) | 0.20066 (11) | 0.78708 (11) | 0.0174 (2) | |
| H17 | 0.278268 | 0.154275 | 0.829314 | 0.021* | |
| C18 | 0.44894 (14) | 0.21006 (11) | 0.82216 (11) | 0.0173 (2) | |
| C19 | 0.51825 (14) | 0.28051 (12) | 0.76017 (11) | 0.0184 (2) | |
| H19 | 0.600293 | 0.288172 | 0.783499 | 0.022* | |
| C20 | 0.46997 (13) | 0.33974 (11) | 0.66507 (11) | 0.0168 (2) | |
| H20 | 0.517566 | 0.388092 | 0.623997 | 0.020* | |
| C21 | 0.50385 (16) | 0.14328 (14) | 0.92318 (12) | 0.0247 (3) | |
| H21A | 0.419338 | 0.152364 | 0.984240 | 0.037* | |
| H21B | 0.567230 | 0.178318 | 0.950152 | 0.037* | |
| H21C | 0.560779 | 0.055639 | 0.901004 | 0.037* | |
| C97 | 0.20835 (19) | 0.51005 (15) | −0.00331 (12) | 0.0303 (3) | |
| H97A | 0.105803 | 0.547512 | −0.014305 | 0.045* | |
| H97B | 0.258431 | 0.429728 | −0.045575 | 0.045* | |
| H97C | 0.259893 | 0.563981 | −0.031259 | 0.045* | |
| C98 | 0.35300 (17) | 0.44892 (18) | 0.15100 (14) | 0.0356 (4) | |
| H98A | 0.339323 | 0.441322 | 0.233601 | 0.053* | |
| H98B | 0.400502 | 0.507200 | 0.129860 | 0.053* | |
| H98C | 0.415656 | 0.368141 | 0.112343 | 0.053* | |
| C99 | 0.08612 (15) | 0.51648 (12) | 0.19300 (11) | 0.0186 (2) | |
| H99 | −0.003677 | 0.541604 | 0.165524 | 0.022* | |
| N99 | 0.20970 (13) | 0.49363 (11) | 0.11715 (9) | 0.0198 (2) | |
| O99 | 0.07862 (10) | 0.50763 (9) | 0.29665 (7) | 0.01853 (18) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1 | 0.01528 (13) | 0.01090 (13) | 0.01511 (13) | −0.00706 (10) | −0.00348 (10) | 0.00176 (10) |
| C2 | 0.0104 (5) | 0.0097 (5) | 0.0145 (5) | −0.0042 (4) | −0.0008 (4) | 0.0002 (4) |
| N3 | 0.0147 (4) | 0.0121 (4) | 0.0126 (4) | −0.0060 (4) | −0.0022 (3) | 0.0015 (3) |
| C3A | 0.0114 (5) | 0.0121 (5) | 0.0137 (5) | −0.0038 (4) | −0.0004 (4) | 0.0013 (4) |
| C4 | 0.0173 (5) | 0.0168 (5) | 0.0156 (5) | −0.0069 (4) | −0.0032 (4) | 0.0016 (4) |
| C5 | 0.0184 (6) | 0.0200 (6) | 0.0148 (5) | −0.0059 (5) | −0.0029 (4) | 0.0046 (4) |
| C6 | 0.0188 (6) | 0.0156 (6) | 0.0195 (6) | −0.0062 (5) | −0.0020 (5) | 0.0059 (4) |
| C7 | 0.0174 (5) | 0.0130 (5) | 0.0198 (6) | −0.0067 (4) | −0.0021 (4) | 0.0038 (4) |
| C7A | 0.0124 (5) | 0.0122 (5) | 0.0143 (5) | −0.0042 (4) | −0.0013 (4) | 0.0020 (4) |
| C8 | 0.0114 (5) | 0.0092 (5) | 0.0130 (5) | −0.0041 (4) | −0.0016 (4) | −0.0003 (4) |
| C9 | 0.0108 (5) | 0.0108 (5) | 0.0139 (5) | −0.0041 (4) | −0.0017 (4) | −0.0015 (4) |
| C10 | 0.0140 (5) | 0.0127 (5) | 0.0123 (5) | −0.0049 (4) | −0.0032 (4) | −0.0008 (4) |
| C11 | 0.0133 (5) | 0.0126 (5) | 0.0108 (5) | −0.0039 (4) | −0.0023 (4) | −0.0005 (4) |
| C12 | 0.0115 (5) | 0.0105 (5) | 0.0113 (5) | −0.0044 (4) | −0.0007 (4) | 0.0004 (4) |
| C13 | 0.0178 (5) | 0.0139 (5) | 0.0172 (6) | −0.0060 (4) | −0.0053 (4) | 0.0001 (4) |
| C14 | 0.0149 (6) | 0.0276 (8) | 0.0685 (12) | −0.0089 (6) | −0.0016 (7) | −0.0143 (8) |
| S2 | 0.01854 (14) | 0.00982 (12) | 0.01155 (13) | −0.00509 (10) | −0.00151 (10) | −0.00011 (9) |
| S3 | 0.01584 (13) | 0.01206 (13) | 0.01707 (14) | −0.00662 (10) | −0.00418 (10) | −0.00170 (10) |
| O1 | 0.0234 (4) | 0.0140 (4) | 0.0126 (4) | −0.0076 (3) | −0.0040 (3) | 0.0024 (3) |
| O2 | 0.0211 (4) | 0.0179 (4) | 0.0164 (4) | −0.0061 (4) | 0.0033 (3) | 0.0016 (3) |
| O3 | 0.0315 (5) | 0.0121 (4) | 0.0181 (4) | −0.0088 (4) | −0.0089 (4) | −0.0005 (3) |
| N1 | 0.0155 (5) | 0.0092 (4) | 0.0131 (4) | −0.0067 (4) | −0.0018 (4) | 0.0001 (3) |
| N2 | 0.0153 (4) | 0.0090 (4) | 0.0108 (4) | −0.0063 (3) | −0.0028 (3) | 0.0000 (3) |
| N4 | 0.0237 (5) | 0.0127 (5) | 0.0129 (5) | −0.0107 (4) | −0.0062 (4) | 0.0025 (4) |
| N5 | 0.0317 (6) | 0.0252 (6) | 0.0212 (6) | −0.0117 (5) | −0.0121 (5) | 0.0009 (5) |
| C15 | 0.0168 (5) | 0.0100 (5) | 0.0139 (5) | −0.0038 (4) | −0.0017 (4) | 0.0002 (4) |
| C16 | 0.0190 (6) | 0.0137 (5) | 0.0177 (6) | −0.0080 (4) | −0.0046 (4) | 0.0019 (4) |
| C17 | 0.0210 (6) | 0.0139 (5) | 0.0172 (6) | −0.0073 (5) | −0.0030 (4) | 0.0025 (4) |
| C18 | 0.0171 (5) | 0.0147 (5) | 0.0152 (5) | −0.0013 (4) | −0.0026 (4) | −0.0005 (4) |
| C19 | 0.0130 (5) | 0.0194 (6) | 0.0208 (6) | −0.0040 (4) | −0.0030 (4) | −0.0009 (5) |
| C20 | 0.0145 (5) | 0.0151 (5) | 0.0190 (6) | −0.0054 (4) | 0.0001 (4) | −0.0003 (4) |
| C21 | 0.0214 (6) | 0.0284 (7) | 0.0198 (6) | −0.0041 (5) | −0.0063 (5) | 0.0065 (5) |
| C97 | 0.0417 (9) | 0.0361 (8) | 0.0130 (6) | −0.0163 (7) | −0.0027 (6) | 0.0032 (5) |
| C98 | 0.0233 (7) | 0.0561 (11) | 0.0247 (7) | −0.0136 (7) | −0.0018 (6) | −0.0001 (7) |
| C99 | 0.0228 (6) | 0.0182 (6) | 0.0164 (6) | −0.0094 (5) | −0.0045 (5) | 0.0006 (4) |
| N99 | 0.0238 (5) | 0.0229 (5) | 0.0126 (5) | −0.0098 (4) | −0.0021 (4) | 0.0011 (4) |
| O99 | 0.0248 (5) | 0.0228 (5) | 0.0127 (4) | −0.0153 (4) | −0.0016 (3) | −0.0004 (3) |
Geometric parameters (Å, º)
| S1—C7A | 1.7375 (12) | S2—N1 | 1.6678 (10) |
| S1—C2 | 1.7677 (12) | S2—C15 | 1.7597 (12) |
| C2—N3 | 1.3153 (15) | N1—N2 | 1.4020 (13) |
| C2—C8 | 1.4706 (15) | N1—H01 | 0.888 (18) |
| N3—C3A | 1.3848 (14) | N4—H02 | 0.84 (2) |
| C3A—C4 | 1.3977 (17) | N4—H03 | 0.86 (2) |
| C3A—C7A | 1.4013 (17) | C15—C20 | 1.3872 (17) |
| C4—C5 | 1.3855 (17) | C15—C16 | 1.3955 (17) |
| C4—H4 | 0.9500 | C16—C17 | 1.3875 (17) |
| C5—C6 | 1.4031 (19) | C16—H16 | 0.9500 |
| C5—H5 | 0.9500 | C17—C18 | 1.3970 (18) |
| C6—C7 | 1.3880 (18) | C17—H17 | 0.9500 |
| C6—H6 | 0.9500 | C18—C19 | 1.3948 (18) |
| C7—C7A | 1.4005 (16) | C18—C21 | 1.5034 (18) |
| C7—H7 | 0.9500 | C19—C20 | 1.3894 (18) |
| C8—C9 | 1.4108 (16) | C19—H19 | 0.9500 |
| C8—C12 | 1.4372 (15) | C20—H20 | 0.9500 |
| C9—C10 | 1.3897 (16) | C21—H21A | 0.9800 |
| C9—S3 | 1.7781 (12) | C21—H21B | 0.9800 |
| C10—C13 | 1.4295 (16) | C21—H21C | 0.9800 |
| C10—C11 | 1.4340 (16) | C97—N99 | 1.4536 (17) |
| C11—O1 | 1.2213 (14) | C97—H97A | 0.9800 |
| C11—N2 | 1.4132 (14) | C97—H97B | 0.9800 |
| C12—N4 | 1.3124 (15) | C97—H97C | 0.9800 |
| C12—N2 | 1.3851 (14) | C98—N99 | 1.4554 (19) |
| C13—N5 | 1.1499 (17) | C98—H98A | 0.9800 |
| C14—S3 | 1.7952 (15) | C98—H98B | 0.9800 |
| C14—H14A | 0.9800 | C98—H98C | 0.9800 |
| C14—H14B | 0.9800 | C99—O99 | 1.2343 (15) |
| C14—H14C | 0.9800 | C99—N99 | 1.3242 (17) |
| S2—O3 | 1.4317 (10) | C99—H99 | 0.9500 |
| S2—O2 | 1.4326 (9) | ||
| C7A—S1—C2 | 89.58 (6) | N2—N1—S2 | 117.20 (8) |
| N3—C2—C8 | 121.49 (10) | N2—N1—H01 | 113.6 (11) |
| N3—C2—S1 | 113.55 (8) | S2—N1—H01 | 113.1 (11) |
| C8—C2—S1 | 124.95 (9) | C12—N2—N1 | 115.94 (9) |
| C2—N3—C3A | 112.58 (10) | C12—N2—C11 | 125.63 (10) |
| N3—C3A—C4 | 125.08 (11) | N1—N2—C11 | 117.88 (9) |
| N3—C3A—C7A | 114.40 (10) | C12—N4—H02 | 121.2 (13) |
| C4—C3A—C7A | 120.52 (11) | C12—N4—H03 | 114.9 (13) |
| C5—C4—C3A | 118.33 (12) | H02—N4—H03 | 123.6 (18) |
| C5—C4—H4 | 120.8 | C20—C15—C16 | 121.33 (11) |
| C3A—C4—H4 | 120.8 | C20—C15—S2 | 120.72 (9) |
| C4—C5—C6 | 120.82 (12) | C16—C15—S2 | 117.94 (9) |
| C4—C5—H5 | 119.6 | C17—C16—C15 | 118.80 (12) |
| C6—C5—H5 | 119.6 | C17—C16—H16 | 120.6 |
| C7—C6—C5 | 121.60 (11) | C15—C16—H16 | 120.6 |
| C7—C6—H6 | 119.2 | C16—C17—C18 | 121.22 (12) |
| C5—C6—H6 | 119.2 | C16—C17—H17 | 119.4 |
| C6—C7—C7A | 117.34 (12) | C18—C17—H17 | 119.4 |
| C6—C7—H7 | 121.3 | C19—C18—C17 | 118.44 (11) |
| C7A—C7—H7 | 121.3 | C19—C18—C21 | 121.38 (12) |
| C7—C7A—C3A | 121.37 (11) | C17—C18—C21 | 120.17 (12) |
| C7—C7A—S1 | 128.74 (10) | C20—C19—C18 | 121.46 (12) |
| C3A—C7A—S1 | 109.88 (8) | C20—C19—H19 | 119.3 |
| C9—C8—C12 | 116.48 (10) | C18—C19—H19 | 119.3 |
| C9—C8—C2 | 125.41 (10) | C15—C20—C19 | 118.73 (11) |
| C12—C8—C2 | 118.11 (10) | C15—C20—H20 | 120.6 |
| C10—C9—C8 | 122.53 (10) | C19—C20—H20 | 120.6 |
| C10—C9—S3 | 115.37 (9) | C18—C21—H21A | 109.5 |
| C8—C9—S3 | 122.08 (9) | C18—C21—H21B | 109.5 |
| C9—C10—C13 | 122.46 (11) | H21A—C21—H21B | 109.5 |
| C9—C10—C11 | 122.30 (10) | C18—C21—H21C | 109.5 |
| C13—C10—C11 | 115.24 (10) | H21A—C21—H21C | 109.5 |
| O1—C11—N2 | 119.46 (11) | H21B—C21—H21C | 109.5 |
| O1—C11—C10 | 127.10 (11) | N99—C97—H97A | 109.5 |
| N2—C11—C10 | 113.44 (10) | N99—C97—H97B | 109.5 |
| N4—C12—N2 | 116.83 (10) | H97A—C97—H97B | 109.5 |
| N4—C12—C8 | 123.94 (11) | N99—C97—H97C | 109.5 |
| N2—C12—C8 | 119.23 (10) | H97A—C97—H97C | 109.5 |
| N5—C13—C10 | 176.92 (13) | H97B—C97—H97C | 109.5 |
| S3—C14—H14A | 109.5 | N99—C98—H98A | 109.5 |
| S3—C14—H14B | 109.5 | N99—C98—H98B | 109.5 |
| H14A—C14—H14B | 109.5 | H98A—C98—H98B | 109.5 |
| S3—C14—H14C | 109.5 | N99—C98—H98C | 109.5 |
| H14A—C14—H14C | 109.5 | H98A—C98—H98C | 109.5 |
| H14B—C14—H14C | 109.5 | H98B—C98—H98C | 109.5 |
| O3—S2—O2 | 121.42 (6) | O99—C99—N99 | 125.03 (13) |
| O3—S2—N1 | 102.99 (5) | O99—C99—H99 | 117.5 |
| O2—S2—N1 | 106.32 (5) | N99—C99—H99 | 117.5 |
| O3—S2—C15 | 106.76 (6) | C99—N99—C97 | 121.56 (12) |
| O2—S2—C15 | 109.03 (6) | C99—N99—C98 | 121.18 (12) |
| N1—S2—C15 | 109.88 (5) | C97—N99—C98 | 117.25 (12) |
| C9—S3—C14 | 98.98 (6) | ||
| C7A—S1—C2—N3 | −0.89 (9) | C2—C8—C12—N4 | −0.37 (17) |
| C7A—S1—C2—C8 | 178.19 (10) | C9—C8—C12—N2 | −0.79 (15) |
| C8—C2—N3—C3A | −178.06 (10) | C2—C8—C12—N2 | 179.01 (10) |
| S1—C2—N3—C3A | 1.06 (13) | C10—C9—S3—C14 | 83.40 (11) |
| C2—N3—C3A—C4 | 178.78 (11) | C8—C9—S3—C14 | −98.15 (12) |
| C2—N3—C3A—C7A | −0.70 (14) | O3—S2—N1—N2 | −167.59 (8) |
| N3—C3A—C4—C5 | 179.67 (11) | O2—S2—N1—N2 | −38.90 (9) |
| C7A—C3A—C4—C5 | −0.89 (17) | C15—S2—N1—N2 | 78.95 (9) |
| C3A—C4—C5—C6 | −0.39 (18) | N4—C12—N2—N1 | −3.39 (15) |
| C4—C5—C6—C7 | 0.93 (19) | C8—C12—N2—N1 | 177.19 (10) |
| C5—C6—C7—C7A | −0.15 (18) | N4—C12—N2—C11 | −174.61 (10) |
| C6—C7—C7A—C3A | −1.15 (17) | C8—C12—N2—C11 | 5.97 (17) |
| C6—C7—C7A—S1 | −179.72 (9) | S2—N1—N2—C12 | 103.12 (10) |
| N3—C3A—C7A—C7 | −178.80 (11) | S2—N1—N2—C11 | −84.95 (11) |
| C4—C3A—C7A—C7 | 1.70 (17) | O1—C11—N2—C12 | 172.21 (11) |
| N3—C3A—C7A—S1 | 0.01 (13) | C10—C11—N2—C12 | −7.76 (16) |
| C4—C3A—C7A—S1 | −179.49 (9) | O1—C11—N2—N1 | 1.15 (16) |
| C2—S1—C7A—C7 | 179.17 (12) | C10—C11—N2—N1 | −178.82 (9) |
| C2—S1—C7A—C3A | 0.47 (9) | O3—S2—C15—C20 | 152.17 (10) |
| N3—C2—C8—C9 | −177.62 (11) | O2—S2—C15—C20 | 19.34 (12) |
| S1—C2—C8—C9 | 3.36 (16) | N1—S2—C15—C20 | −96.82 (10) |
| N3—C2—C8—C12 | 2.61 (16) | O3—S2—C15—C16 | −28.68 (11) |
| S1—C2—C8—C12 | −176.41 (8) | O2—S2—C15—C16 | −161.51 (9) |
| C12—C8—C9—C10 | −1.79 (16) | N1—S2—C15—C16 | 82.33 (10) |
| C2—C8—C9—C10 | 178.44 (11) | C20—C15—C16—C17 | −0.33 (18) |
| C12—C8—C9—S3 | 179.88 (8) | S2—C15—C16—C17 | −179.47 (9) |
| C2—C8—C9—S3 | 0.10 (16) | C15—C16—C17—C18 | −0.79 (19) |
| C8—C9—C10—C13 | −179.23 (11) | C16—C17—C18—C19 | 1.19 (18) |
| S3—C9—C10—C13 | −0.79 (15) | C16—C17—C18—C21 | −177.66 (12) |
| C8—C9—C10—C11 | −0.40 (18) | C17—C18—C19—C20 | −0.49 (19) |
| S3—C9—C10—C11 | 178.04 (9) | C21—C18—C19—C20 | 178.35 (12) |
| C9—C10—C11—O1 | −175.15 (12) | C16—C15—C20—C19 | 1.01 (18) |
| C13—C10—C11—O1 | 3.76 (18) | S2—C15—C20—C19 | −179.87 (9) |
| C9—C10—C11—N2 | 4.82 (16) | C18—C19—C20—C15 | −0.59 (19) |
| C13—C10—C11—N2 | −176.27 (10) | O99—C99—N99—C97 | −178.03 (13) |
| C9—C8—C12—N4 | 179.84 (11) | O99—C99—N99—C98 | 3.0 (2) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H01···O99i | 0.888 (18) | 1.872 (18) | 2.7583 (13) | 175.7 (16) |
| N4—H02···O99 | 0.84 (2) | 2.05 (2) | 2.8334 (14) | 154.6 (18) |
| N4—H03···N3 | 0.86 (2) | 1.86 (2) | 2.5760 (15) | 139.9 (17) |
| N4—H02···N1 | 0.84 (2) | 2.237 (19) | 2.5932 (14) | 105.7 (15) |
| C7—H7···O3ii | 0.95 | 2.54 | 3.3161 (16) | 139 |
| C20—H20···O2iii | 0.95 | 2.64 | 3.5605 (16) | 164 |
| C97—H97C···N5iv | 0.98 | 2.59 | 3.504 (2) | 155 |
Symmetry codes: (i) −x, −y+1, −z+1; (ii) x, y+1, z; (iii) −x+1, −y+1, −z+1; (iv) x, y, z−1.
References
- Elgemeie, G. H. & Abou-Zeid, M. (2015). Nucleosides Nucleotides Nucleic Acids, 34, 834–847. [DOI] [PubMed]
- Elgemeie, G. H., Abou-Zeid, M., Alsaid, S., Hebishy, A. & Essa, H. (2015). Nucleosides Nucleotides Nucleic Acids, 34, 659–673. [DOI] [PubMed]
- Elgemeie, G. H., Abou-Zeid, M. & Azzam, R. (2016). Nucleosides Nucleotides Nucleic Acids, 35, 211–222. [DOI] [PubMed]
- Elgemeie, G. H., Azzam, R. A. & Elsayed, R. E. (2017c). Patent No. 1554/2017. Egyptian Academy of Scientific Research.
- Elgemeie, G. H., El-Ezbawy, S. R. & Sood, S. A. (2003a). Synth. Commun. 33, 2095–2101.
- Elgemeie, G. H., Elghandour, A. H. & Abd Elaziz, G. W. (2003b). Synth. Commun. 33, 1659–1664.
- Elgemeie, G. H., Elsayed, S. H. & Hassan, A. S. (2009). Synth. Commun. 39, 1781–1792.
- Elgemeie, G. H., Fathy, N., Zaghary, W. & Farag, A. (2017b). Nucleosides Nucleotides Nucleic Acids, 36, 198–212. [DOI] [PubMed]
- Elgemeie, G. H. & Mohamed, M. A. (2006). Synth. Commun. 36, 1025–1038.
- Elgemeie, G. H., Salah, A. M., Abbas, N. S., Hussein, H. A. & Mohamed, R. A. (2017a). Nucleosides Nucleotides Nucleic Acids, 36, 213–223. [DOI] [PubMed]
- Rigaku OD (2015). CrysAlis PRO. Rigaku Oxford Diffraction, Abingdon, UK.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2015). Acta Cryst. C71, 3–8.
- Siemens (1994). XP. Siemens Analytical X–Ray Instruments, Madison, Wisconsin, Wisconsin USA.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S2056989017015778/hg5500sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989017015778/hg5500Isup2.hkl
CCDC reference: 1582798
Additional supporting information: crystallographic information; 3D view; checkCIF report