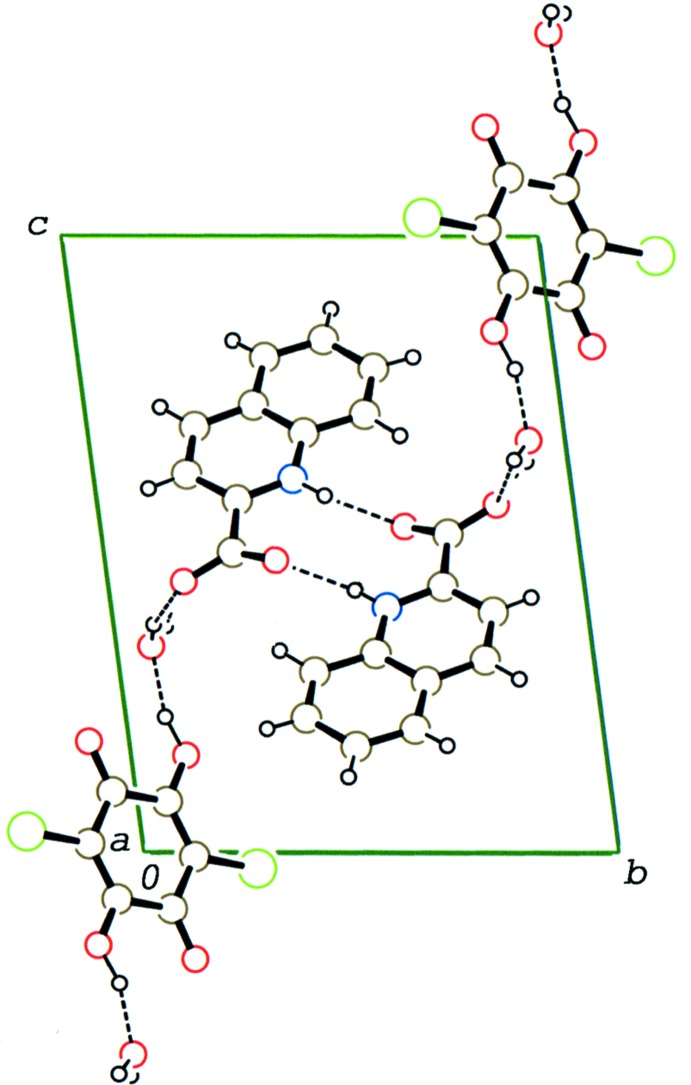
Crystal structures of hydrogen-bonded 1:2 dihydrate compounds of chloranilic acid with 2-carboxypyridine (I) and 2-carboxyquinoline (II) have been determined at 180 and 200 K, respectively. The base molecule in (I) is disordered over cationic and twitterionic states, while that in (II) is in a twitterionic form. In each crystal, the three components are linked by O—H⋯O and N—H⋯O hydrogen bonds, forming a layer structure.
Keywords: crystal structure, chloranilic acid, 2-carboxypyridine, 2-carboxyquinoline, twitterion, disorder, hydrogen bond
Abstract
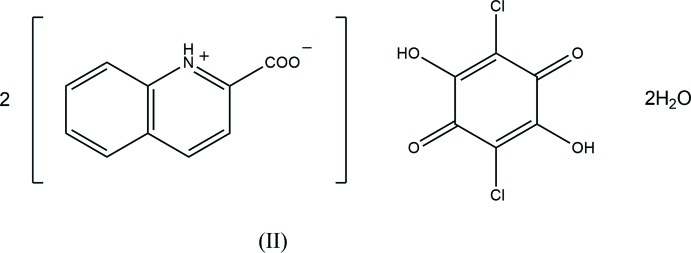
The crystal structure of the 1:2 dihydrate compound of chloranilic acid (systematic name: 2,5-dichloro-3,6-dihydroxy-1,4-benzoquinone) with 2-carboxypyridine (another common name: picolinic acid; systematic name: pyridine-2-carboxylic acid), namely, 2C6H5.5NO2 0.5+·C6HCl2O4 −·2H2O, (I), has been determined at 180 K, and the structure of the 1:2 dihydrate compound of chloranilic acid with 2-carboxyquinoline (another common name: quinaldic acid; systematic name: quinoline-2-carboxylic acid), namely, 2C10H7NO2·C6H2Cl2O4·2H2O, (II), has been redetermined at 200 K. This determination presents a higher precision crystal structure than the previously published structure [Marfo-Owusu & Thompson (2014 ▸). X-ray Struct. Anal. Online, 30, 55–56]. Compound (I) was analysed as a disordered structure over two states, viz. salt and co-crystal. The salt is bis(2-carboxypyridinium) chloranilate dihydrate, 2C6H6NO2 +·C6Cl2O4 2−·2H2O, and the co-crystal is bis(pyridinium-2-carboxylate) chloranilic acid dihydrate, 2C6H5NO2·C6H2Cl2O4·2H2O, including zwitterionic 2-carboxypyridine. In both salt and co-crystal, the water molecule links the chloranilic acid and 2-carboxypyridine molecules through O—H⋯O and N—H⋯O hydrogen bonds. The 2-carboxypyridine molecules are connected into a head-to-head inversion dimer by a short O—H⋯O hydrogen bond, in which the H atom is disordered over two positions. Compound (II) is a 1:2 dihydrate co-crystal of chloranilic acid and zwitterionic 2-carboxyquinoline. The water molecule links the chloranilic acid and 2-carboxyquinoline molecules through O—H⋯O hydrogen bonds. The 2-carboxyquinoline molecules are connected into a head-to-tail inversion dimer by a pair of N—H⋯O hydrogen bonds.
Chemical context
Chloranilic acid, a dibasic acid with hydrogen-bond donor as well as acceptor groups, appears particularly attractive as a template for generating tightly bound self-assemblies with various pyridine derivatives as well as being a model compound for investigating hydrogen-transfer motions in O—H⋯N and N—H⋯O hydrogen-bond systems (Zaman et al., 2004 ▸; Molčanov & Kojić-Prodić, 2010 ▸; Seliger et al., 2009 ▸; Asaji et al. 2010 ▸). Previously, we have prepared three 1:1 compounds of chloranilic acid with 2-, 3- and 4-carboxypyridine and analysed the crystal structures in order to extend our study on D—H⋯A hydrogen bonding (D = N, O or C; A = N, O or Cl) in chloranilic acid–substituted pyridine systems (Gotoh et al., 2006 ▸, 2009 ▸; Tabuchi et al., 2005 ▸). In the present study, we have prepared a 1:2 compound of chloranilic acid with 2-carboxypyridine and also redetermined the structure of a 1:2 compound of chloranilic acid with 2-carboxyquinoline with higher precision than previously reported structure [Marfo-Owusu & Thompson, 2014 ▸; although the title and text in this reference refer to the 1:1 adduct of chloranilic acid with 2-carboxyqulinone, the reported structure is the 1:2 compound, the same as the present compound (II)]. The crystal structure of the anhydrous 1:2 compound of chloranilic acid with 2-carboxyquinoline was also reported by Marfo-Owusu & Thompson (2016 ▸).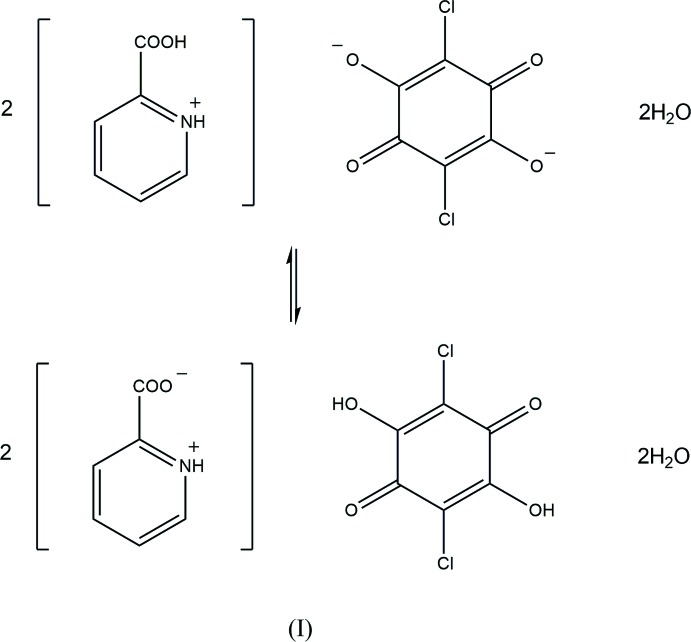
Structural commentary
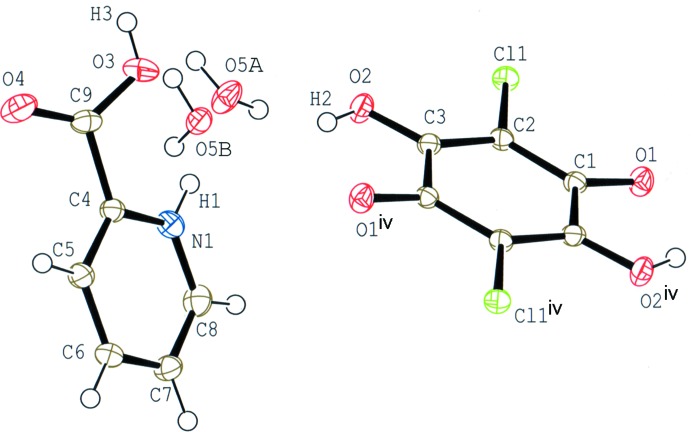
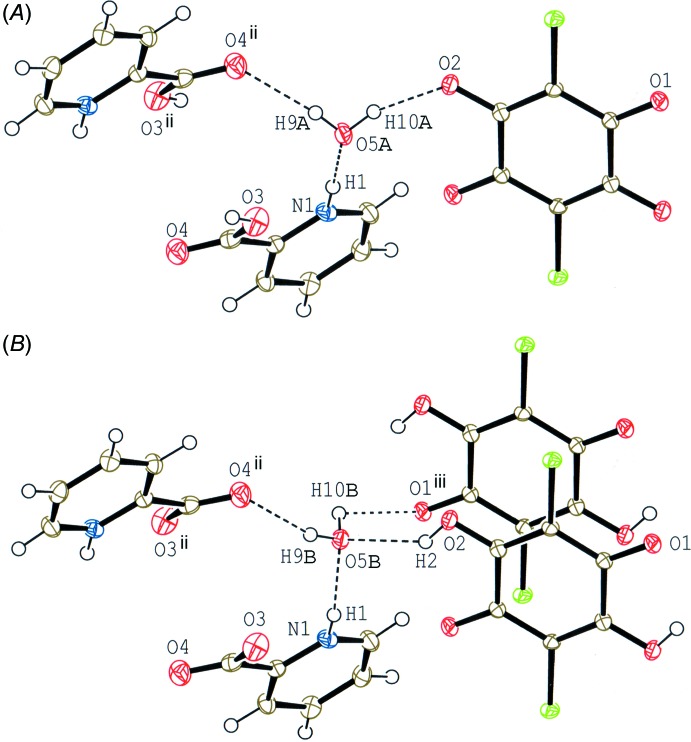
Compound (I) (Fig. 1 ▸) crystallizes with one-half of a chloranilic acid molecule, which is located on an inversion centre, one 2-carboxypyridine molecule and one water molecule in the asymmetric unit. In the crystal, the water molecule is disordered over two sites with equal occupancies of 0.5. The occupancies of the H atoms in the chloranilic acid molecule and the carboxy group of the 2-carboxypyridine molecule are also 0.5. The compound is, therefore, considered to be a disordered state over two forms, viz. bis(2-carboxypyridinium) chloranilate dihydrate, (A), and bis(pyridinium-2-carboxylate) chloranilic acid dihydrate, (B), as shown in the scheme and Fig. 2 ▸. In form (A), the water molecule acts as one N—H⋯O hydrogen-bond acceptor and two O—H⋯O hydrogen-bond donors (N1—H1⋯O5A, O5A—O9A⋯O4ii and O5A—H10A⋯O2; symmetry code as in Table 1 ▸), while in form (B), the water molecule acts as the acceptor of N—H⋯O and O—H⋯O hydrogen bonds, and as two O—H⋯O hydrogen-bond donors (N1—H1⋯O5B, O2—H2⋯O5B, O5B—H9B⋯O4ii and O5B—H10B⋯O1iii; Table 1 ▸). The dihedral angle between the pyridine ring and the carboxy plane in the base molecule is 23.32 (15)°.
Figure 1.
The molecular structure of compound (I), showing the atom-numbering scheme. Displacement ellipsoids of non-H atoms are drawn at the 50% probability level and H atoms are drawn as small spheres of arbitrary radii. The water molecule is disordered over two sites with equally occupancies. Atoms H2 and H3 have site-occupancy factors of 0.5. [Symmetry code: (iv) −x + 1, −y + 1, −z + 1.]
Figure 2.
A partial packing diagram of compound (I) around the disordered water molecule in bis(2-carboxypyridinium) chloranilate dihydrate (A) and bis(pyridinium-2-carboxylate) chloranilic acid dihydrate (B), showing O—H⋯O and N—H⋯O hydrogen bonds (dashed lines). [Symmetry codes: (ii) −x, −y, −z + 1; (iii) −x + 1, −y, −z + 1.]
Table 1. Hydrogen-bond geometry (Å, °) for (I) .
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1⋯O5A | 0.895 (17) | 1.843 (17) | 2.7380 (19) | 179 (2) |
| N1—H1⋯O5B | 0.895 (17) | 1.907 (17) | 2.7417 (18) | 154 (2) |
| O2—H2⋯O5B | 0.83 (3) | 2.12 (3) | 2.8111 (19) | 141 (3) |
| O3—H3⋯O3i | 0.82 (3) | 1.62 (4) | 2.4352 (14) | 174 (4) |
| O5A—H9A⋯O4ii | 0.82 (3) | 2.11 (3) | 2.927 (2) | 170 (3) |
| O5B—H9B⋯O4ii | 0.82 (3) | 2.02 (3) | 2.8159 (19) | 162 (3) |
| O5A—H10A⋯O2 | 0.84 (3) | 1.90 (3) | 2.6762 (19) | 154 (4) |
| O5B—H10B⋯O1iii | 0.85 (2) | 2.60 (3) | 3.095 (2) | 119 (2) |
| C8—H8⋯Cl1iii | 0.95 | 2.78 | 3.6524 (12) | 154 |
| C8—H8⋯O1iii | 0.95 | 2.47 | 3.1871 (14) | 132 |
Symmetry codes: (i) 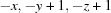 ; (ii)
; (ii) 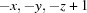 ; (iii)
; (iii) 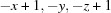 .
.
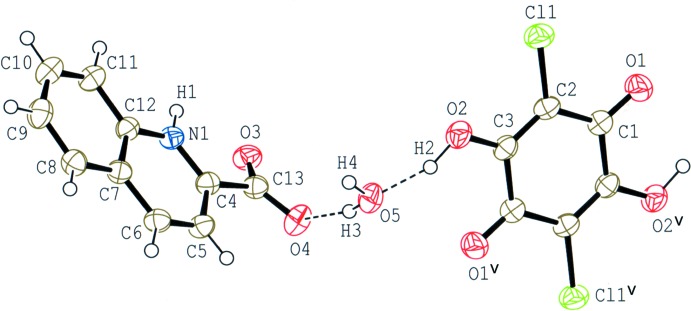
The asymmetric unit of compound (II) consists of one-half of a chloranilic acid molecule, which is located on an inversion centre, one 2-carboxyquinoline molecule and one water molecule. In the crystal, the 2-carboxyquinoline molecule is in a twitterionic form and no acid–base interaction involving H-atom transfer between chloranilic acid and 2-carboxyquinoline is observed (Fig. 3 ▸). The dihedral angle between the quinoline ring system and the carboxylate plane in the base molecule is 20.84 (19)°. The water molecule acts as an O—H⋯O hydrogen-bonding bridge between the chloranilic and 2-carboxyquinoline molecules (O2—H2⋯O5 and O5—H3⋯O4; Table 2 ▸).
Figure 3.
The molecular structure of compound (II), showing the atom-numbering scheme. Displacement ellipsoids of non-H atoms are drawn at the 50% probability level and H atoms are drawn as small spheres of arbitrary radii. O—H⋯O hydrogen bonds are shown as dashed lines. [Symmetry code: (v) −x, −y, −z.]
Table 2. Hydrogen-bond geometry (Å, °) for (II) .
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1⋯O3i | 0.88 (2) | 1.91 (2) | 2.7724 (17) | 167 (2) |
| O2—H2⋯O5 | 0.98 (3) | 1.59 (3) | 2.5092 (17) | 155 (3) |
| O5—H3⋯O4 | 0.82 (2) | 2.01 (2) | 2.8072 (19) | 164 (2) |
| O5—H4⋯O4ii | 0.85 (2) | 1.82 (2) | 2.6632 (19) | 171 (2) |
| C6—H6⋯O1iii | 0.95 | 2.54 | 3.392 (2) | 150 |
| C6—H6⋯O5iv | 0.95 | 2.47 | 3.211 (2) | 134 |
Symmetry codes: (i) 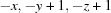 ; (ii)
; (ii) 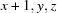 ; (iii)
; (iii) 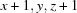 ; (iv)
; (iv) 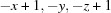 .
.
Supramolecular features
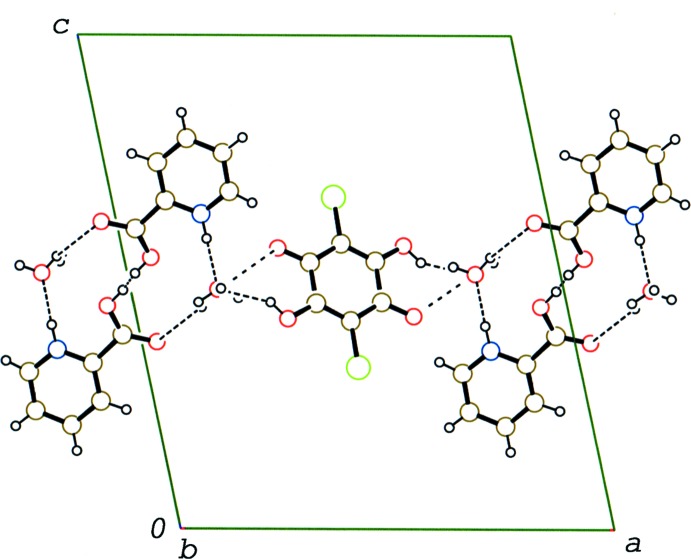
In the crystal of compound (I), the 2-carboxypyridine molecules, which are related by an inversion centre, are linked into a head-to-head dimer via a short O—H⋯O hydrogen bond, in which the H atom is disordered over two sites (O3—H3⋯O3i; Table 1 ▸), as observed in pyridinium-2-carboxylic acid pyridinium-2-carboxylate perchlorate (Wang et al., 2015 ▸). The three components are linked via the above-mentioned O—H⋯O and N—H⋯O hydrogen bonds together with weak C—H⋯Cl and C—H⋯O hydrogen bonds (C8—H8⋯Cl1iii and C8—H8⋯O1iii; Table 1 ▸), forming a layer parallel to the ab plane (Fig. 4 ▸). In the layer, the chloranilic acid rings are stacked along the b axis through a π–π interaction [centroid–centroid distance = 3.6851 (7) Å and interplanar spacing = 3.2118 (4) Å]. The pyridine rings are also stacked along the b axis through a π–π interaction [centroid–centroid distance = 3.6851 (7) Å and interplanar spacing = 3.4787 (5) Å]. Between the layers, a short Cl⋯Cl contact is observed [Cl1⋯Cl1v = 3.3717 (5) Å; symmetry code: (v) −x + 1, y −  , −z +
, −z +  ].
].
Figure 4.
A packing diagram of compound (I) viewed along the b axis, showing the layer structure. O—H⋯O and N—H⋯O hydrogen bonds are shown as dashed lines.
In the crystal of (II), two adjacent 2-carboxyquinoline molecules, which are related by an inversion centre, form a head-to-tail dimer via a pair of N—H⋯O hydrogen bonds (N—H1⋯O3i; symmetry code as in Table 2 ▸). The dimers are stacked in a column along the a axis through a weak π–π interaction between the N1/C4–C7/C12 and C7–C12 rings with a centroid–centroid distance of 3.9184 (10) Å. The water molecule links the stacked base molecules related by translation along a via O—H⋯O hydrogen bonds [O5—H3⋯O4 and O5—H4⋯O4ii; Table 2 ▸] and also links the acid molecule and the two base molecules via O—H⋯O hydrogen bonds, forming a layer structure parallel to (0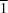 1) as shown in Fig. 5 ▸. No significant short contact between the acid molecules in the layer is observed. Between the layers, a bifurcated C—H⋯(O, O) hydrogen bond (C6—H6⋯O1iii and C6—H6⋯O5iv; Table 2 ▸) is observed, through which the 2-carboxyquinoline molecule is weakly linked with the chloranilic acid and water molecules.
1) as shown in Fig. 5 ▸. No significant short contact between the acid molecules in the layer is observed. Between the layers, a bifurcated C—H⋯(O, O) hydrogen bond (C6—H6⋯O1iii and C6—H6⋯O5iv; Table 2 ▸) is observed, through which the 2-carboxyquinoline molecule is weakly linked with the chloranilic acid and water molecules.
Figure 5.
A packing diagram of compound (II) viewed along the a axis, showing the layer structure formed by O—H⋯O and N—H⋯O hydrogen bonds (dashed lines).
Database survey
A search of the Cambridge Structural Database (Version 5.38, last update May 2017; Groom et al., 2016 ▸) for organic co-crystals of pyridinium-2-carboxylate (twitterionic form) gave six structures. For organic co-crystals of quinolinium-2-carboxylate (twitterionic form), eight structures were found.
Synthesis and crystallization
Single crystals of compound (I) were obtained by slow evaporation of an acetonitrile solution (200 ml) of chloranilic acid (250 mg) with 2-carboxypridine (310 mg) at room temperature. Single crystals of compound (II) were obtained by slow evaporation from a methanol solution (150 ml) of chloranilic acid (310 mg) with 2-carboxyquinoline (520 mg) at room temperature.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 3 ▸. The water molecule in compound (I) was found to be disordered over two sites in a difference-Fourier map. The occupancies were refined to 0.52 (2) and 0.48 (2) and then they were fixed at 0.5. The H atom in the carboxy group of the base molecule was also found in a difference-Fourier map to be disordered between the adjacent carboxy groups, which are related by an inversion centre, and the occupancy was set to be 0.5. Since the N-bound H atom refined reasonably with an occupancy of 1, the occupancy of the H atom of the acid molecule was set to be 0.5 to balance the total charge of the compound. All other H atoms were found in a difference-Fourier map. The N-bound H atom was refined freely, while the positions of O-bound H atoms were refined, with O—H = 0.84 (2) Å and U iso(H) = 1.5U eq(O). For the water H atoms, distant restraints of H⋯H = 1.37 (4) Å were also applied. C-bound H atoms were positioned geometrically (C—H = 0.95 Å) and were treated as riding with U iso(H) = 1.2U eq(C).
Table 3. Experimental details.
| (I) | (II) | |
|---|---|---|
| Crystal data | ||
| Chemical formula | 2C6H5.5NO2 0.5+·C6HCl2O4 −·2H2O | 2C10H7NO2·C6H2Cl2O4·2H2O |
| M r | 491.24 | 591.36 |
| Crystal system, space group | Monoclinic, P21/c | Triclinic, P
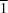
|
| Temperature (K) | 180 | 200 |
| a, b, c (Å) | 15.1028 (10), 3.6851 (3), 17.5689 (13) | 4.4745 (2), 10.5448 (8), 13.6111 (6) |
| α, β, γ (°) | 90, 101.871 (3), 90 | 96.652 (4), 94.109 (3), 99.009 (4) |
| V (Å3) | 956.89 (12) | 627.38 (6) |
| Z | 2 | 1 |
| Radiation type | Mo Kα | Mo Kα |
| μ (mm−1) | 0.40 | 0.32 |
| Crystal size (mm) | 0.35 × 0.18 × 0.13 | 0.41 × 0.21 × 0.03 |
| Data collection | ||
| Diffractometer | Rigaku R-AXIS RAPIDII | Rigaku R-AXIS RAPIDII |
| Absorption correction | Numerical (NUMABS; Higashi, 1999 ▸) | Numerical (NUMABS; Higashi, 1999 ▸) |
| T min, T max | 0.896, 0.949 | 0.925, 0.990 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 17752, 2789, 2537 | 12358, 3666, 2755 |
| R int | 0.021 | 0.122 |
| (sin θ/λ)max (Å−1) | 0.704 | 0.704 |
| Refinement | ||
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.030, 0.079, 1.11 | 0.052, 0.149, 1.01 |
| No. of reflections | 2789 | 3666 |
| No. of parameters | 176 | 197 |
| No. of restraints | 8 | 2 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.54, −0.24 | 0.52, −0.45 |
All H atoms in compound (II) were found in a difference-Fourier map. The O- and N-bound H atoms in the acid and base molecules were refined freely. The water H atoms were refined with O—H = 0.84 (2) Å. C-bound H atoms were positioned geometrically (C—H = 0.95 Å) and were treated as riding with U iso(H) = 1.2U eq(C).
Supplementary Material
Crystal structure: contains datablock(s) General, I, II. DOI: 10.1107/S2056989017015997/lh5860sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989017015997/lh5860Isup2.hkl
Structure factors: contains datablock(s) II. DOI: 10.1107/S2056989017015997/lh5860IIsup3.hkl
Supporting information file. DOI: 10.1107/S2056989017015997/lh5860IIsup4.cml
Additional supporting information: crystallographic information; 3D view; checkCIF report
supplementary crystallographic information
Bis(2-carboxypyridinium) chloranilate dihydrate–bis(pyridinium-2-carboxylate) chloranilic acid dihydrate (1/1) (I). Crystal data
| 2C6H5.5NO20.5+·C6HCl2O4−·2H2O | F(000) = 504.00 |
| Mr = 491.24 | Dx = 1.705 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71075 Å |
| a = 15.1028 (10) Å | Cell parameters from 15838 reflections |
| b = 3.6851 (3) Å | θ = 3.2–30.0° |
| c = 17.5689 (13) Å | µ = 0.40 mm−1 |
| β = 101.871 (3)° | T = 180 K |
| V = 956.89 (12) Å3 | Block, brown |
| Z = 2 | 0.35 × 0.18 × 0.13 mm |
Bis(2-carboxypyridinium) chloranilate dihydrate–bis(pyridinium-2-carboxylate) chloranilic acid dihydrate (1/1) (I). Data collection
| Rigaku R-AXIS RAPIDII diffractometer | 2537 reflections with I > 2σ(I) |
| Detector resolution: 10.000 pixels mm-1 | Rint = 0.021 |
| ω scans | θmax = 30.0°, θmin = 3.3° |
| Absorption correction: numerical (NUMABS; Higashi, 1999) | h = −21→21 |
| Tmin = 0.896, Tmax = 0.949 | k = −5→4 |
| 17752 measured reflections | l = −24→24 |
| 2789 independent reflections |
Bis(2-carboxypyridinium) chloranilate dihydrate–bis(pyridinium-2-carboxylate) chloranilic acid dihydrate (1/1) (I). Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.030 | Hydrogen site location: difference Fourier map |
| wR(F2) = 0.079 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.11 | w = 1/[σ2(Fo2) + (0.0445P)2 + 0.3065P] where P = (Fo2 + 2Fc2)/3 |
| 2789 reflections | (Δ/σ)max = 0.001 |
| 176 parameters | Δρmax = 0.54 e Å−3 |
| 8 restraints | Δρmin = −0.24 e Å−3 |
| Primary atom site location: structure-invariant direct methods |
Bis(2-carboxypyridinium) chloranilate dihydrate–bis(pyridinium-2-carboxylate) chloranilic acid dihydrate (1/1) (I). Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Bis(2-carboxypyridinium) chloranilate dihydrate–bis(pyridinium-2-carboxylate) chloranilic acid dihydrate (1/1) (I). Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| Cl1 | 0.48813 (2) | 0.21752 (7) | 0.32764 (2) | 0.01728 (8) | |
| O1 | 0.64061 (5) | 0.6016 (2) | 0.43110 (4) | 0.02004 (16) | |
| O2 | 0.34886 (5) | 0.1370 (2) | 0.42588 (4) | 0.02038 (17) | |
| H2 | 0.317 (2) | 0.137 (9) | 0.4593 (16) | 0.031* | 0.5 |
| O3 | 0.06134 (7) | 0.3759 (3) | 0.54909 (5) | 0.0313 (2) | |
| H3 | 0.018 (2) | 0.445 (12) | 0.516 (2) | 0.047* | 0.5 |
| O4 | −0.03494 (6) | 0.1653 (3) | 0.62162 (5) | 0.0303 (2) | |
| O5A | 0.19794 (11) | 0.1042 (7) | 0.48308 (10) | 0.0265 (4) | 0.5 |
| H9A | 0.1547 (18) | 0.038 (11) | 0.4493 (18) | 0.040* | 0.5 |
| H10A | 0.2453 (15) | 0.043 (11) | 0.4682 (18) | 0.040* | 0.5 |
| O5B | 0.20132 (11) | −0.1191 (6) | 0.48529 (9) | 0.0199 (3) | 0.5 |
| H9B | 0.1501 (15) | −0.090 (9) | 0.4587 (19) | 0.030* | 0.5 |
| H10B | 0.209 (2) | −0.347 (5) | 0.4873 (19) | 0.030* | 0.5 |
| N1 | 0.19857 (6) | 0.0209 (3) | 0.63805 (5) | 0.01977 (18) | |
| C1 | 0.57374 (6) | 0.5511 (3) | 0.45987 (5) | 0.01438 (18) | |
| C2 | 0.49294 (7) | 0.3688 (3) | 0.42158 (5) | 0.01476 (18) | |
| C3 | 0.42138 (6) | 0.3090 (3) | 0.45734 (6) | 0.01574 (19) | |
| C4 | 0.12231 (7) | 0.0793 (3) | 0.66495 (6) | 0.01658 (19) | |
| C5 | 0.12193 (7) | 0.0166 (3) | 0.74206 (6) | 0.0198 (2) | |
| H5 | 0.068573 | 0.056090 | 0.761605 | 0.024* | |
| C6 | 0.20078 (8) | −0.1057 (3) | 0.79117 (6) | 0.0226 (2) | |
| H6 | 0.201710 | −0.146932 | 0.844701 | 0.027* | |
| C7 | 0.27772 (7) | −0.1668 (3) | 0.76174 (7) | 0.0226 (2) | |
| H7 | 0.331694 | −0.251819 | 0.794609 | 0.027* | |
| C8 | 0.27478 (7) | −0.1023 (3) | 0.68384 (7) | 0.0236 (2) | |
| H8 | 0.326860 | −0.145258 | 0.662656 | 0.028* | |
| C9 | 0.04024 (8) | 0.2152 (3) | 0.60747 (6) | 0.0210 (2) | |
| H1 | 0.1993 (12) | 0.049 (5) | 0.5876 (10) | 0.039 (4)* |
Bis(2-carboxypyridinium) chloranilate dihydrate–bis(pyridinium-2-carboxylate) chloranilic acid dihydrate (1/1) (I). Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cl1 | 0.01956 (13) | 0.02040 (13) | 0.01184 (12) | −0.00159 (8) | 0.00314 (8) | −0.00202 (8) |
| O1 | 0.0170 (3) | 0.0273 (4) | 0.0174 (3) | −0.0029 (3) | 0.0071 (3) | −0.0028 (3) |
| O2 | 0.0159 (3) | 0.0286 (4) | 0.0168 (3) | −0.0068 (3) | 0.0037 (3) | −0.0038 (3) |
| O3 | 0.0340 (5) | 0.0363 (5) | 0.0200 (4) | −0.0003 (4) | −0.0028 (3) | 0.0107 (4) |
| O4 | 0.0212 (4) | 0.0433 (5) | 0.0243 (4) | 0.0042 (4) | −0.0003 (3) | 0.0001 (4) |
| O5A | 0.0152 (8) | 0.0467 (13) | 0.0185 (8) | −0.0033 (8) | 0.0052 (6) | −0.0080 (8) |
| O5B | 0.0160 (7) | 0.0243 (9) | 0.0188 (7) | −0.0013 (7) | 0.0028 (5) | −0.0010 (7) |
| N1 | 0.0231 (4) | 0.0215 (4) | 0.0160 (4) | 0.0002 (4) | 0.0071 (3) | −0.0002 (3) |
| C1 | 0.0148 (4) | 0.0154 (4) | 0.0132 (4) | 0.0008 (3) | 0.0035 (3) | 0.0012 (3) |
| C2 | 0.0171 (4) | 0.0174 (4) | 0.0100 (4) | −0.0015 (3) | 0.0033 (3) | −0.0007 (3) |
| C3 | 0.0147 (4) | 0.0186 (5) | 0.0137 (4) | 0.0012 (4) | 0.0025 (3) | 0.0021 (3) |
| C4 | 0.0177 (4) | 0.0163 (4) | 0.0151 (4) | −0.0002 (4) | 0.0022 (3) | 0.0005 (3) |
| C5 | 0.0183 (4) | 0.0251 (5) | 0.0166 (4) | 0.0003 (4) | 0.0052 (3) | 0.0026 (4) |
| C6 | 0.0236 (5) | 0.0271 (6) | 0.0163 (4) | −0.0015 (4) | 0.0018 (4) | 0.0051 (4) |
| C7 | 0.0185 (5) | 0.0209 (5) | 0.0261 (5) | 0.0010 (4) | −0.0009 (4) | 0.0032 (4) |
| C8 | 0.0197 (5) | 0.0245 (5) | 0.0281 (5) | 0.0025 (4) | 0.0083 (4) | −0.0013 (4) |
| C9 | 0.0241 (5) | 0.0207 (5) | 0.0156 (4) | 0.0022 (4) | −0.0017 (4) | −0.0013 (4) |
Bis(2-carboxypyridinium) chloranilate dihydrate–bis(pyridinium-2-carboxylate) chloranilic acid dihydrate (1/1) (I). Geometric parameters (Å, º)
| Cl1—C2 | 1.7293 (9) | N1—H1 | 0.895 (18) |
| O1—C1 | 1.2330 (11) | C1—C2 | 1.4335 (13) |
| O2—C3 | 1.2875 (12) | C1—C3i | 1.5307 (13) |
| O2—H2 | 0.830 (18) | C2—C3 | 1.3751 (13) |
| O3—C9 | 1.2801 (14) | C4—C5 | 1.3754 (13) |
| O3—H3 | 0.814 (17) | C4—C9 | 1.5139 (14) |
| O4—C9 | 1.2250 (15) | C5—C6 | 1.3939 (15) |
| O5A—H9A | 0.824 (18) | C5—H5 | 0.9500 |
| O5A—H10A | 0.841 (18) | C6—C7 | 1.3840 (16) |
| O5B—H9B | 0.824 (17) | C6—H6 | 0.9500 |
| O5B—H10B | 0.849 (17) | C7—C8 | 1.3808 (16) |
| N1—C8 | 1.3406 (14) | C7—H7 | 0.9500 |
| N1—C4 | 1.3492 (13) | C8—H8 | 0.9500 |
| C3—O2—H2 | 105 (2) | N1—C4—C9 | 117.38 (9) |
| C9—O3—H3 | 115 (3) | C5—C4—C9 | 122.95 (9) |
| H9A—O5A—H10A | 107 (3) | C4—C5—C6 | 119.20 (9) |
| H9B—O5B—H10B | 105 (3) | C4—C5—H5 | 120.4 |
| C8—N1—C4 | 122.23 (9) | C6—C5—H5 | 120.4 |
| C8—N1—H1 | 116.7 (11) | C7—C6—C5 | 119.83 (10) |
| C4—N1—H1 | 120.9 (11) | C7—C6—H6 | 120.1 |
| O1—C1—C2 | 124.63 (9) | C5—C6—H6 | 120.1 |
| O1—C1—C3i | 117.08 (9) | C8—C7—C6 | 118.95 (10) |
| C2—C1—C3i | 118.29 (8) | C8—C7—H7 | 120.5 |
| C3—C2—C1 | 122.26 (8) | C6—C7—H7 | 120.5 |
| C3—C2—Cl1 | 120.12 (8) | N1—C8—C7 | 120.11 (10) |
| C1—C2—Cl1 | 117.62 (7) | N1—C8—H8 | 119.9 |
| O2—C3—C2 | 124.19 (9) | C7—C8—H8 | 119.9 |
| O2—C3—C1i | 116.38 (8) | O4—C9—O3 | 128.74 (11) |
| C2—C3—C1i | 119.43 (8) | O4—C9—C4 | 118.74 (10) |
| N1—C4—C5 | 119.67 (9) | O3—C9—C4 | 112.51 (10) |
| O1—C1—C2—C3 | 177.79 (10) | N1—C4—C5—C6 | 0.18 (17) |
| C3i—C1—C2—C3 | −1.60 (16) | C9—C4—C5—C6 | −179.46 (10) |
| O1—C1—C2—Cl1 | −1.39 (14) | C4—C5—C6—C7 | −0.86 (17) |
| C3i—C1—C2—Cl1 | 179.22 (7) | C5—C6—C7—C8 | 0.46 (18) |
| C1—C2—C3—O2 | −177.81 (10) | C4—N1—C8—C7 | −1.34 (17) |
| Cl1—C2—C3—O2 | 1.35 (15) | C6—C7—C8—N1 | 0.62 (18) |
| C1—C2—C3—C1i | 1.61 (16) | N1—C4—C9—O4 | 157.55 (11) |
| Cl1—C2—C3—C1i | −179.22 (7) | C5—C4—C9—O4 | −22.81 (16) |
| C8—N1—C4—C5 | 0.93 (17) | N1—C4—C9—O3 | −23.17 (14) |
| C8—N1—C4—C9 | −179.41 (10) | C5—C4—C9—O3 | 156.48 (11) |
Symmetry code: (i) −x+1, −y+1, −z+1.
Bis(2-carboxypyridinium) chloranilate dihydrate–bis(pyridinium-2-carboxylate) chloranilic acid dihydrate (1/1) (I). Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1···O5A | 0.895 (17) | 1.843 (17) | 2.7380 (19) | 179 (2) |
| N1—H1···O5B | 0.895 (17) | 1.907 (17) | 2.7417 (18) | 154.4 (17) |
| O2—H2···O5B | 0.83 (3) | 2.12 (3) | 2.8111 (19) | 141 (3) |
| O3—H3···O3ii | 0.82 (3) | 1.62 (4) | 2.4352 (14) | 174 (4) |
| O5A—H9A···O4iii | 0.82 (3) | 2.11 (3) | 2.927 (2) | 170 (3) |
| O5B—H9B···O4iii | 0.82 (3) | 2.02 (3) | 2.8159 (19) | 162 (3) |
| O5A—H10A···O2 | 0.84 (3) | 1.90 (3) | 2.6762 (19) | 154 (4) |
| O5B—H10B···O1iv | 0.85 (2) | 2.60 (3) | 3.095 (2) | 119 (2) |
| C8—H8···Cl1iv | 0.95 | 2.78 | 3.6524 (12) | 154 |
| C8—H8···O1iv | 0.95 | 2.47 | 3.1871 (14) | 132 |
Symmetry codes: (ii) −x, −y+1, −z+1; (iii) −x, −y, −z+1; (iv) −x+1, −y, −z+1.
chloranilic acid–2-carboxyquinoline (1/2) dihydrate (II). Crystal data
| 2C10H7NO2·C6H2Cl2O4·2H2O | Z = 1 |
| Mr = 591.36 | F(000) = 304.00 |
| Triclinic, P1 | Dx = 1.565 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71075 Å |
| a = 4.4745 (2) Å | Cell parameters from 9714 reflections |
| b = 10.5448 (8) Å | θ = 3.0–30.1° |
| c = 13.6111 (6) Å | µ = 0.32 mm−1 |
| α = 96.652 (4)° | T = 200 K |
| β = 94.109 (3)° | Platelet, brown |
| γ = 99.009 (4)° | 0.41 × 0.21 × 0.03 mm |
| V = 627.38 (6) Å3 |
chloranilic acid–2-carboxyquinoline (1/2) dihydrate (II). Data collection
| Rigaku R-AXIS RAPIDII diffractometer | 2755 reflections with I > 2σ(I) |
| Detector resolution: 10.000 pixels mm-1 | Rint = 0.122 |
| ω scans | θmax = 30.0°, θmin = 3.0° |
| Absorption correction: numerical (NUMABS; Higashi, 1999) | h = −6→6 |
| Tmin = 0.925, Tmax = 0.990 | k = −14→14 |
| 12358 measured reflections | l = −19→19 |
| 3666 independent reflections |
chloranilic acid–2-carboxyquinoline (1/2) dihydrate (II). Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.052 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.149 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.00 | w = 1/[σ2(Fo2) + (0.0879P)2] where P = (Fo2 + 2Fc2)/3 |
| 3666 reflections | (Δ/σ)max < 0.001 |
| 197 parameters | Δρmax = 0.52 e Å−3 |
| 2 restraints | Δρmin = −0.45 e Å−3 |
| Primary atom site location: structure-invariant direct methods |
chloranilic acid–2-carboxyquinoline (1/2) dihydrate (II). Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
chloranilic acid–2-carboxyquinoline (1/2) dihydrate (II). Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Cl1 | 0.49390 (9) | 0.23664 (4) | −0.02842 (3) | 0.04300 (16) | |
| O1 | 0.0319 (3) | 0.08081 (12) | −0.17653 (8) | 0.0426 (3) | |
| O2 | 0.4150 (3) | 0.11710 (11) | 0.15510 (9) | 0.0391 (3) | |
| O3 | −0.1769 (3) | 0.36216 (11) | 0.47121 (9) | 0.0372 (3) | |
| O4 | −0.0712 (3) | 0.16160 (11) | 0.43604 (9) | 0.0425 (3) | |
| O5 | 0.3938 (3) | 0.07471 (12) | 0.33231 (9) | 0.0406 (3) | |
| N1 | 0.3233 (3) | 0.41970 (12) | 0.60390 (9) | 0.0298 (3) | |
| C1 | 0.0207 (3) | 0.04635 (14) | −0.09469 (11) | 0.0317 (3) | |
| C2 | 0.2276 (3) | 0.10762 (14) | −0.00985 (11) | 0.0320 (3) | |
| C3 | 0.2197 (3) | 0.06514 (14) | 0.07971 (11) | 0.0313 (3) | |
| C4 | 0.2151 (3) | 0.29703 (14) | 0.56909 (11) | 0.0305 (3) | |
| C5 | 0.3274 (3) | 0.19702 (15) | 0.61140 (11) | 0.0336 (3) | |
| H5 | 0.250293 | 0.109195 | 0.586437 | 0.040* | |
| C6 | 0.5493 (4) | 0.22680 (15) | 0.68906 (12) | 0.0342 (3) | |
| H6 | 0.622423 | 0.159293 | 0.719133 | 0.041* | |
| C7 | 0.6694 (3) | 0.35659 (14) | 0.72449 (11) | 0.0312 (3) | |
| C8 | 0.9050 (4) | 0.39332 (16) | 0.80217 (12) | 0.0363 (3) | |
| H8 | 0.985438 | 0.328869 | 0.834065 | 0.044* | |
| C9 | 1.0170 (4) | 0.52094 (18) | 0.83147 (13) | 0.0411 (4) | |
| H9 | 1.175836 | 0.545100 | 0.883608 | 0.049* | |
| C10 | 0.8980 (4) | 0.61764 (17) | 0.78463 (13) | 0.0418 (4) | |
| H10 | 0.980263 | 0.706066 | 0.805678 | 0.050* | |
| C11 | 0.6677 (4) | 0.58722 (15) | 0.70983 (13) | 0.0372 (3) | |
| H11 | 0.587483 | 0.653070 | 0.679646 | 0.045* | |
| C12 | 0.5532 (3) | 0.45558 (14) | 0.67897 (10) | 0.0294 (3) | |
| C13 | −0.0333 (3) | 0.27243 (14) | 0.48397 (11) | 0.0315 (3) | |
| H1 | 0.252 (5) | 0.481 (2) | 0.5760 (17) | 0.055 (6)* | |
| H2 | 0.359 (7) | 0.083 (3) | 0.216 (2) | 0.092 (10)* | |
| H3 | 0.255 (4) | 0.086 (2) | 0.3667 (15) | 0.054 (6)* | |
| H4 | 0.560 (4) | 0.111 (2) | 0.3651 (17) | 0.063 (7)* |
chloranilic acid–2-carboxyquinoline (1/2) dihydrate (II). Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cl1 | 0.0402 (2) | 0.0380 (2) | 0.0465 (2) | −0.00570 (16) | −0.00095 (18) | 0.00725 (17) |
| O1 | 0.0451 (7) | 0.0452 (7) | 0.0354 (6) | 0.0008 (5) | −0.0026 (5) | 0.0093 (5) |
| O2 | 0.0388 (6) | 0.0380 (6) | 0.0359 (6) | −0.0005 (5) | −0.0079 (5) | 0.0012 (4) |
| O3 | 0.0376 (6) | 0.0319 (6) | 0.0427 (6) | 0.0068 (4) | −0.0017 (5) | 0.0089 (4) |
| O4 | 0.0403 (6) | 0.0368 (6) | 0.0471 (6) | 0.0091 (5) | −0.0071 (5) | −0.0052 (5) |
| O5 | 0.0400 (7) | 0.0419 (7) | 0.0357 (6) | 0.0047 (5) | −0.0066 (5) | −0.0029 (5) |
| N1 | 0.0302 (6) | 0.0269 (6) | 0.0326 (6) | 0.0053 (5) | 0.0020 (5) | 0.0055 (5) |
| C1 | 0.0292 (7) | 0.0304 (7) | 0.0353 (7) | 0.0061 (5) | −0.0004 (6) | 0.0039 (6) |
| C2 | 0.0287 (7) | 0.0274 (7) | 0.0384 (7) | 0.0022 (5) | −0.0010 (6) | 0.0032 (5) |
| C3 | 0.0298 (7) | 0.0278 (7) | 0.0354 (7) | 0.0064 (5) | −0.0016 (6) | 0.0007 (5) |
| C4 | 0.0297 (7) | 0.0291 (7) | 0.0330 (7) | 0.0046 (5) | 0.0044 (6) | 0.0045 (5) |
| C5 | 0.0347 (7) | 0.0273 (7) | 0.0384 (7) | 0.0043 (5) | 0.0005 (6) | 0.0051 (5) |
| C6 | 0.0357 (7) | 0.0301 (7) | 0.0378 (7) | 0.0059 (6) | 0.0020 (6) | 0.0089 (6) |
| C7 | 0.0299 (7) | 0.0315 (7) | 0.0325 (7) | 0.0047 (5) | 0.0039 (6) | 0.0054 (5) |
| C8 | 0.0347 (7) | 0.0388 (8) | 0.0353 (7) | 0.0063 (6) | 0.0011 (6) | 0.0060 (6) |
| C9 | 0.0358 (8) | 0.0453 (9) | 0.0382 (8) | 0.0033 (7) | −0.0036 (7) | −0.0030 (7) |
| C10 | 0.0402 (8) | 0.0329 (8) | 0.0484 (9) | 0.0026 (6) | 0.0006 (7) | −0.0046 (7) |
| C11 | 0.0376 (8) | 0.0289 (7) | 0.0443 (8) | 0.0061 (6) | 0.0027 (7) | 0.0014 (6) |
| C12 | 0.0281 (6) | 0.0287 (7) | 0.0311 (6) | 0.0044 (5) | 0.0042 (6) | 0.0021 (5) |
| C13 | 0.0293 (7) | 0.0311 (7) | 0.0339 (7) | 0.0037 (5) | 0.0024 (6) | 0.0058 (5) |
chloranilic acid–2-carboxyquinoline (1/2) dihydrate (II). Geometric parameters (Å, º)
| Cl1—C2 | 1.7174 (15) | C4—C13 | 1.517 (2) |
| O1—C1 | 1.2130 (18) | C5—C6 | 1.370 (2) |
| O2—C3 | 1.3070 (17) | C5—H5 | 0.9500 |
| O2—H2 | 0.98 (3) | C6—C7 | 1.405 (2) |
| O3—C13 | 1.2453 (18) | C6—H6 | 0.9500 |
| O4—C13 | 1.2512 (18) | C7—C8 | 1.413 (2) |
| O5—H3 | 0.821 (16) | C7—C12 | 1.420 (2) |
| O5—H4 | 0.848 (16) | C8—C9 | 1.363 (2) |
| N1—C4 | 1.3270 (19) | C8—H8 | 0.9500 |
| N1—C12 | 1.3722 (19) | C9—C10 | 1.415 (3) |
| N1—H1 | 0.88 (2) | C9—H9 | 0.9500 |
| C1—C2 | 1.447 (2) | C10—C11 | 1.368 (2) |
| C1—C3i | 1.507 (2) | C10—H10 | 0.9500 |
| C2—C3 | 1.348 (2) | C11—C12 | 1.406 (2) |
| C4—C5 | 1.401 (2) | C11—H11 | 0.9500 |
| C3—O2—H2 | 111.3 (18) | C7—C6—H6 | 119.8 |
| H3—O5—H4 | 108 (2) | C6—C7—C8 | 123.01 (14) |
| C4—N1—C12 | 122.93 (13) | C6—C7—C12 | 118.61 (13) |
| C4—N1—H1 | 119.1 (16) | C8—C7—C12 | 118.37 (14) |
| C12—N1—H1 | 117.9 (16) | C9—C8—C7 | 120.18 (15) |
| O1—C1—C2 | 123.09 (14) | C9—C8—H8 | 119.9 |
| O1—C1—C3i | 118.94 (13) | C7—C8—H8 | 119.9 |
| C2—C1—C3i | 117.97 (13) | C8—C9—C10 | 120.32 (15) |
| C3—C2—C1 | 122.39 (13) | C8—C9—H9 | 119.8 |
| C3—C2—Cl1 | 120.62 (11) | C10—C9—H9 | 119.8 |
| C1—C2—Cl1 | 116.98 (11) | C11—C10—C9 | 121.78 (15) |
| O2—C3—C2 | 122.30 (14) | C11—C10—H10 | 119.1 |
| O2—C3—C1i | 118.13 (13) | C9—C10—H10 | 119.1 |
| C2—C3—C1i | 119.57 (12) | C10—C11—C12 | 118.00 (15) |
| N1—C4—C5 | 120.22 (13) | C10—C11—H11 | 121.0 |
| N1—C4—C13 | 116.94 (13) | C12—C11—H11 | 121.0 |
| C5—C4—C13 | 122.84 (13) | N1—C12—C11 | 120.39 (13) |
| C6—C5—C4 | 119.50 (14) | N1—C12—C7 | 118.27 (13) |
| C6—C5—H5 | 120.2 | C11—C12—C7 | 121.34 (14) |
| C4—C5—H5 | 120.2 | O3—C13—O4 | 128.04 (14) |
| C5—C6—C7 | 120.39 (14) | O3—C13—C4 | 117.19 (13) |
| C5—C6—H6 | 119.8 | O4—C13—C4 | 114.76 (13) |
| O1—C1—C2—C3 | −177.43 (15) | C12—C7—C8—C9 | −0.4 (2) |
| C3i—C1—C2—C3 | 2.8 (2) | C7—C8—C9—C10 | 0.2 (3) |
| O1—C1—C2—Cl1 | 1.2 (2) | C8—C9—C10—C11 | 0.5 (3) |
| C3i—C1—C2—Cl1 | −178.60 (10) | C9—C10—C11—C12 | −1.0 (3) |
| C1—C2—C3—O2 | 177.00 (13) | C4—N1—C12—C11 | 177.18 (14) |
| Cl1—C2—C3—O2 | −1.6 (2) | C4—N1—C12—C7 | −3.2 (2) |
| C1—C2—C3—C1i | −2.8 (2) | C10—C11—C12—N1 | −179.65 (14) |
| Cl1—C2—C3—C1i | 178.61 (10) | C10—C11—C12—C7 | 0.7 (2) |
| C12—N1—C4—C5 | 2.4 (2) | C6—C7—C12—N1 | 1.5 (2) |
| C12—N1—C4—C13 | −178.34 (12) | C8—C7—C12—N1 | −179.66 (13) |
| N1—C4—C5—C6 | 0.1 (2) | C6—C7—C12—C11 | −178.84 (14) |
| C13—C4—C5—C6 | −179.12 (14) | C8—C7—C12—C11 | 0.0 (2) |
| C4—C5—C6—C7 | −1.6 (2) | N1—C4—C13—O3 | −19.8 (2) |
| C5—C6—C7—C8 | −177.95 (14) | C5—C4—C13—O3 | 159.47 (14) |
| C5—C6—C7—C12 | 0.8 (2) | N1—C4—C13—O4 | 161.50 (14) |
| C6—C7—C8—C9 | 178.31 (16) | C5—C4—C13—O4 | −19.3 (2) |
Symmetry code: (i) −x, −y, −z.
chloranilic acid–2-carboxyquinoline (1/2) dihydrate (II). Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1···O3ii | 0.88 (2) | 1.91 (2) | 2.7724 (17) | 167 (2) |
| O2—H2···O5 | 0.98 (3) | 1.59 (3) | 2.5092 (17) | 155 (3) |
| O5—H3···O4 | 0.82 (2) | 2.01 (2) | 2.8072 (19) | 164 (2) |
| O5—H4···O4iii | 0.85 (2) | 1.82 (2) | 2.6632 (19) | 171 (2) |
| C6—H6···O1iv | 0.95 | 2.54 | 3.392 (2) | 150 |
| C6—H6···O5v | 0.95 | 2.47 | 3.211 (2) | 134 |
Symmetry codes: (ii) −x, −y+1, −z+1; (iii) x+1, y, z; (iv) x+1, y, z+1; (v) −x+1, −y, −z+1.
References
- Asaji, T., Seliger, J., Žagar, V. & Ishida, H. (2010). Magn. Reson. Chem. 48, 531–536. [DOI] [PubMed]
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Gotoh, K., Nagoshi, H. & Ishida, H. (2009). Acta Cryst. E65, o614. [DOI] [PMC free article] [PubMed]
- Gotoh, K., Tabuchi, Y., Akashi, H. & Ishida, H. (2006). Acta Cryst. E62, o4420–o4421.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Higashi, T. (1999). NUMABS. Rigaku Corporation, Tokyo, Japan.
- Marfo-Owusu, E. & Thompson, A. L. (2014). X-ray Struct. Anal. Online, 30, 55–56.
- Marfo-Owusu, E. & Thompson, A. L. (2016). X-ray Struct. Anal. Online, 32, 49–50.
- Molčanov, K. & Kojić-Prodić, B. (2010). CrystEngComm, 12, 925–939.
- Rigaku (2006). RAPID-AUTO. Rigaku Corporation, Tokyo, Japan.
- Rigaku (2010). CrystalStructure. Rigaku Corporation, Tokyo, Japan.
- Seliger, J., Žagar, V., Gotoh, K., Ishida, H., Konnai, A., Amino, D. & Asaji, T. (2009). Phys. Chem. Chem. Phys. 11, 2281–2286. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2015). Acta Cryst. C71, 3–8.
- Spek, A. L. (2015). Acta Cryst. C71, 9–18. [DOI] [PubMed]
- Tabuchi, Y., Takahashi, A., Gotoh, K., Akashi, H. & Ishida, H. (2005). Acta Cryst. E61, o4215–o4217.
- Wang, B.-Q., Yan, H.-B., Huang, Z.-Q., Zhang, Y.-H. & Sun, J. (2015). Acta Cryst. C71, 247–251. [DOI] [PubMed]
- Zaman, Md. B., Udachin, K. A. & Ripmeester, J. A. (2004). Cryst. Growth Des. 4, 585–589.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) General, I, II. DOI: 10.1107/S2056989017015997/lh5860sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989017015997/lh5860Isup2.hkl
Structure factors: contains datablock(s) II. DOI: 10.1107/S2056989017015997/lh5860IIsup3.hkl
Supporting information file. DOI: 10.1107/S2056989017015997/lh5860IIsup4.cml
Additional supporting information: crystallographic information; 3D view; checkCIF report