In the title compounds 1 and 2, the organic moieties adopt flattened conformations stabilized by an intramolecular N—H⋯N hydrogen bonds formed by the protonated amino group and the N atom of the pyridyl substituent. In 1, the organic moieties are linked with two N—H⋯Cl−-type hydrogen bonds, forming a C(4) graph-set. In its monohydrate, 2, the Cl− anion and a water molecule assemble the moieties into infinite bands showing hydrogen-bond patterns with graph sets C(6), 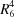 (12) and
(12) and 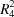 (8). Both crystals display π–π stacked supramolecular structures running along the b axis.
(8). Both crystals display π–π stacked supramolecular structures running along the b axis.
Keywords: crystal structure, tetrahydroindazole, tetrahydroindazolone, hydrochloride, hydrate
Abstract
The title compounds, C15H19N4O+·Cl− and C15H19N4O+·Cl−·H2O, obtained in attempts to synthesize metal complexes using tetrahydroindazole as a ligand, were characterized by NMR, IR and X-ray diffraction techniques. The partially saturated ring in the tetrahydroindazole core adopts a sofa conformation. An intramolecular N—H⋯N hydrogen bond formed by the protonated amino group and the N atom of the pyridyl substituent is found in the first structure. In the hydrochloride, the organic moieties are linked by two N—H⋯Cl− hydrogen bonds, forming a C(4) graph-set. In the hydrate crystal, a Cl− anion and a water molecule assemble the moieties into infinite bands showing hydrogen-bond patterns with graph sets C(6), R 6 4(12) and R 4 2(8). Organic moieties form π–π stacked supramolecular structures running along the b axis in both structures.
Chemical context
Tetrahydroindazoles can be regarded as annulated pyrazole analogs (Ansari et al., 2017 ▸) or as partially saturated indazoles (Gaikwad et al., 2015 ▸). In either of these categories they play an important role in medicinal chemistry. Tetrahydroindazoles are reported to be peripherally selective cannabinoid-1 receptor inverse agonists (Matthews et al., 2016 ▸), sigma-2 receptor ligands(Wu et al., 2015 ▸), and interleukin-2 inducible T-cell kinase inhibitors (Burch et al., 2015 ▸; Heifetz et al., 2016 ▸). Heterocyclic compounds containing a tetrahydroindazole core have been researched as antiviral agents (Bassyouni et al., 2016 ▸) and compounds with antioxidant properties (Polo et al., 2016 ▸). With appropriate side-chain decorations, they also possess COX-2 inhibitory activity (Abdel-Rahman et al., 2012 ▸) and can inhibit bacterial type II topoisomerases (Wiener et al., 2007 ▸). The latter has led to the development of compounds with both antitumor and antimicrobial activity (Faidallah et al., 2013 ▸), including novel antituberculosis agents (Guo et al., 2010 ▸).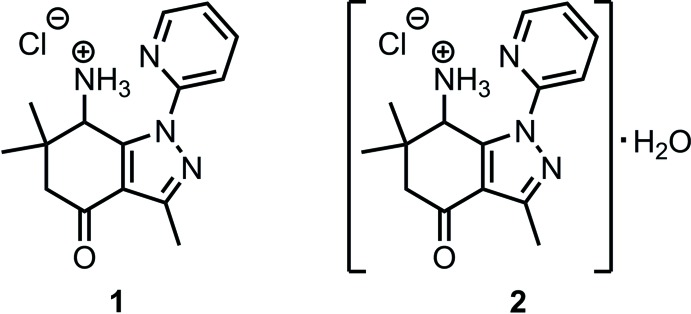
The broad application spectrum of tetrahydroindazoles has led to the development of synthetic methodologies. Thus, traditional approaches using a combination of either α,β-unsaturated ketones (Nakhai & Bergman, 2009 ▸) or dicarbonyl compounds (Murugavel et al., 2010 ▸), or tricarbonyl compounds (Kim et al., 2010 ▸; Scala et al., 2015 ▸) with hydrazines have been significantly updated and improved. In addition, the microwave-assisted synthesis of tetrahydroindazoles has been reported (Silva et al., 2006 ▸; Polo et al., 2016 ▸). It is interesting to note that compounds possessing free NH-functionality in the pyrazole ring have been studied thoroughly for their tautomeric equilibria (Claramunt et al., 2006 ▸). Additionally, tetrahydroindazolones substituted with 2-aminobenzamides have been studied as fluorescent probes (Jia et al., 2012 ▸). Other studies on side-chain modifications include the synthesis of polyfluoroalkyl-substituted analogs (Khlebnikova et al., 2012 ▸), triazole-functionalized tetrahydroindazolones (Strakova et al., 2009 ▸) and their conjugation with biologically active natural products such as lupane triterpenoids (Khlebnicova et al., 2017 ▸). Among other synthetic approaches, the Ritter reaction provides a fast entry into structural modifications and is applicable to obtain a combinatorial library of compounds (Turks et al., 2012 ▸). Combinatorial chemistry methodology has been reported for the construction of tetrahydroindazolones in enantiomerically pure pairs (Song et al., 2012 ▸). Also, enantiomerically pure 7-amino-tetrahydroindazolones (Strakova et al., 2011 ▸) have been obtained. For these reasons, we were interested in the synthesis of 7-amino-3,6,6-trimethyl-1-(pyridin-2-yl)-1,5,6,7-tetrahydro-4H-indazol-4-one for use as a starting material for further structural modifications. Herein, the structures of the corresponding hydrochloride 1 and its hydrate 2 are reported.
Structural commentary
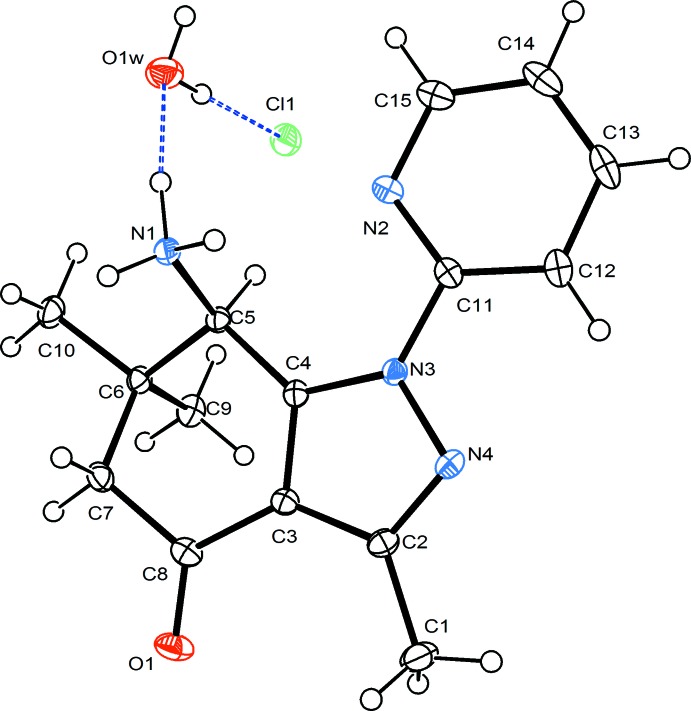
Figs. 1 ▸ and 2 ▸ show the asymmetric units of the hydrochloride (1) and its hydrate (2) with the symmetry-independent hydrogen bonds. The geometry and conformation of the organic cation in compounds 1 and 2 are substantially similar. The pyrazole ring is planar within an r.m.s. deviation of the fitted atoms of 0.0059 Å in 1 and 0.0092 Å in 2. In both structures, the partially saturated ring adopts a sofa conformation. The distance of atom C6 from the mean plane formed by atoms C3–C5/C7/C8 (r.m.s. deviation of fitted atoms = 0.0495 Å in 1 and 0.0558 Å in 2) is 0.639 (2) Å in 1 and 0.642 (2) Å in 2. The dihedral angle between the latter plane and pyrazole ring is 5.79 (6)° in 1 and 6.48 (4)° in 2. On the other hand, the dihedral angle between the pyrazole ring and its pyridyl substituent is 11.91 (6)° [torsion angle N4—N3—C11—C12 = 10.7 (2)°] in 1 and 7.22 (5)° [torsion angle N4—N3—C11—C12 = 4.6 (2)°] in 2. An intramolecular N—H⋯N hydrogen bond formed by the protonated amino group and nitrogen atom of pyridyl substituent is found in 1 (Table 1 ▸).
Figure 1.
ORTEP view of the asymmetric unit of 1 showing the atom-numbering scheme and 50% probability displacement ellipsoids. The intramolecular hydrogen bond is shown with dashed lines.
Figure 2.
ORTEP view of the asymmetric unit of 2 showing the atom-numbering scheme and 50% probability displacement ellipsoids. The intramolecular hydrogen bonds are shown with dashed lines.
Table 1. Hydrogen-bond geometry (Å, °) for (1) .
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1N1⋯N2 | 0.97 (2) | 2.42 (2) | 2.928 (2) | 112 (2) |
| N1—H2N1⋯Cl1i | 0.97 (2) | 2.08 (2) | 3.034 (2) | 168 (2) |
| N1—H3N1⋯Cl1ii | 0.93 (2) | 2.27 (2) | 3.188 (2) | 167 (2) |
Symmetry codes: (i) 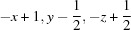 ; (ii)
; (ii)  .
.
Supramolecular features
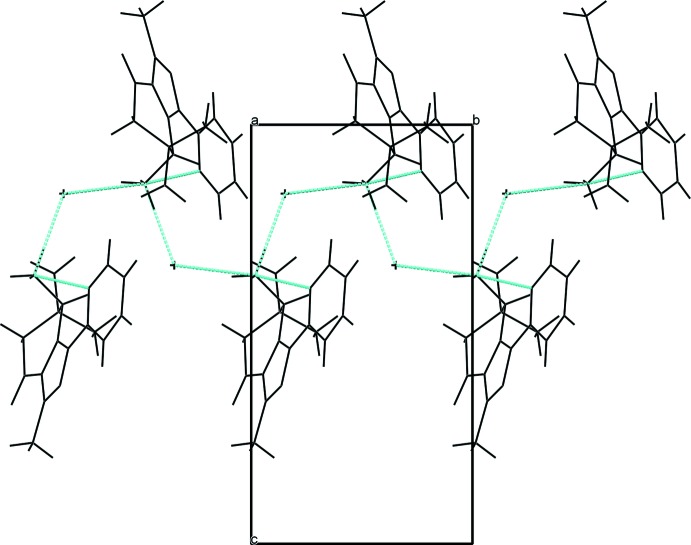
In the crystal of compound 1, the organic moieties are linked by two types of N—H⋯Cl− hydrogen bonds into infinite chains along the b-axis direction (Table 1 ▸). According to Etter (1990 ▸), the hydrogen-bond pattern in 1 can be described by a C(4) graph set. The packing of 1 is shown in Fig. 3 ▸. In the structure of 2, in addition to participating in an intramolecular hydrogen bond, the protonated amino group also forms two intermolecular hydrogen bonds with the Cl− anion and a water molecule (Table 2 ▸). Each Cl− anion and water molecule takes part in three intermolecular hydrogen bonds. The organic cations are bridged by a pair of Cl− anions and a water molecule, thus assembling the moieties into infinite bands running along the b-axis direction. The hydrogen-bond pattern can be described by graph sets C(6), 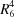 (12) and
(12) and 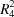 (8). The packing of 2 is shown in Fig. 4 ▸.
(8). The packing of 2 is shown in Fig. 4 ▸.
Figure 3.
The crystal packing of compound 1, viewed along the a axis. The hydrogen bonds are shown as dashed lines (see Table 1 ▸).
Table 2. Hydrogen-bond geometry (Å, °) for (2) .
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1W—H1W⋯Cl1i | 0.80 (3) | 2.39 (3) | 3.185 (2) | 176 (3) |
| O1W—H2W⋯Cl1 | 0.94 (3) | 2.31 (3) | 3.247 (2) | 179 (2) |
| N1—H1N1⋯Cl1ii | 0.85 (2) | 2.40 (2) | 3.228 (2) | 165 (2) |
| N1—H3N1⋯O1W | 0.95 (2) | 1.85 (3) | 2.775 (2) | 162 (2) |
Symmetry codes: (i) 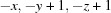 ; (ii)
; (ii)  .
.
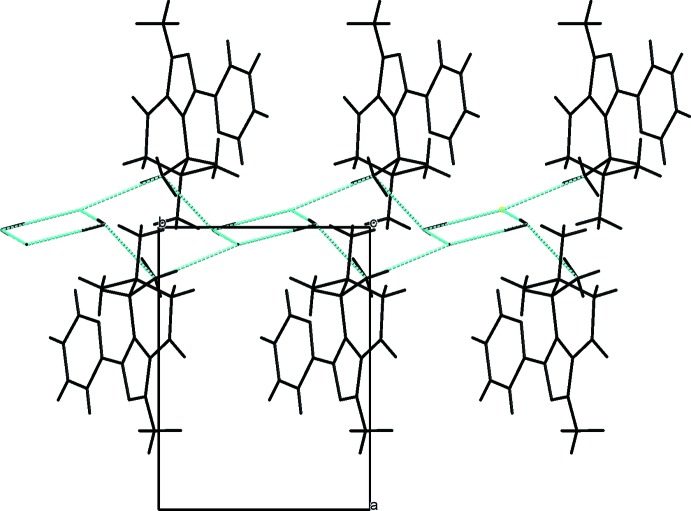
Figure 4.
The crystal packing of compound 2, viewed along the c axis. The hydrogen bonds are shown as dashed lines (see Table 2 ▸).
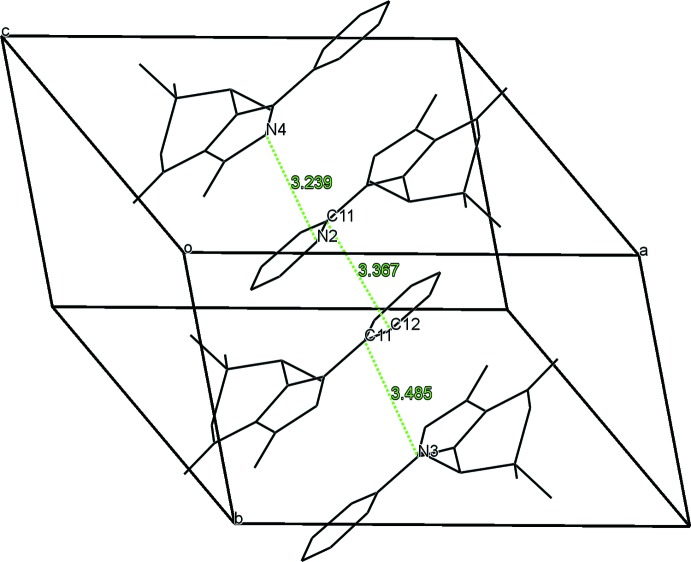
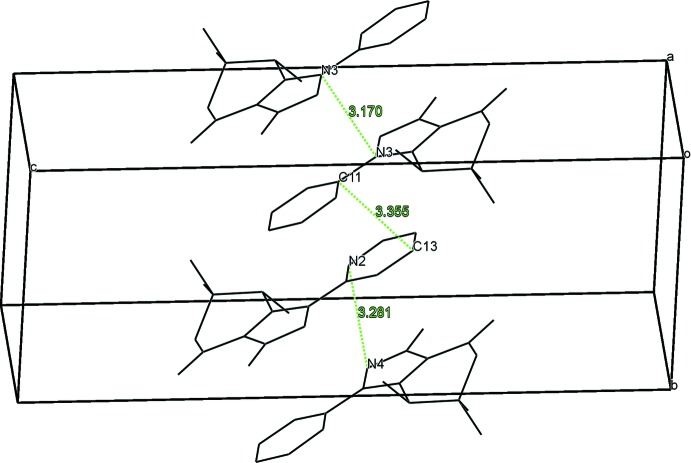
In the crystal of 1, the organic moieties form stacks running along the b axis which are stabilized by π–π interactions (Fig. 5 ▸). The distance between the centroids of the pyridine and pyrazole rings of adjacent molecules is 3.585 (2) Å. The shortest contact is 3.239 (2) Å between atoms N2 and N4 of two inversion-related molecules (Fig. 5 ▸). In the crystal of 2, the organic moieties also form π–π-stacked supramolecular structures running along the b-axis direction (Fig. 6 ▸). The distance between the centroids of the pyridine rings of adjacent molecules is 3.748 (2) Å. The shortest contact is 3.170 (2) Å between the N3 atoms of two inversion-related molecules (Fig. 6 ▸).
Figure 5.
View of stacks of organic moieties in the crystal structure of 1. H atoms and chloride anions are not shown for clarity.
Figure 6.
View of stacks of organic moieties in the crystal structure of 2. H atoms, chloride anions and water molecules are not shown for clarity.
Database survey
A search of the Cambridge Structural Database (Version 5.38; Groom et al., 2016 ▸) for the 3,6,6-trimethyl-4-oxo-4,5,6,7-tetrahydro-1H-indazole core revealed five structurally close compounds: UXAQUG, UXARAN, UXARER, UXARIV, UXAROB (Strakova et al., 2011 ▸). These compounds differ from compounds 1 and 2 by the substituents at the positions of atoms N3 and C5. In all examples, the partially saturated ring in the indazole fragment adopts a sofa conformation. However, the phenyl ring at the position N3 forms much larger dihedral angles with the pyrazole ring than with the pyridyl substituent in the structures reported here.
Synthesis and crystallization
The synthesis of the title compounds is depicted in the reaction scheme below. The 7-aminotetrahydroindazolone derivative 4 was prepared by an analogy of the procedure published by Strakova et al. (2011 ▸) from the known precursor 3 (Strakova et al., 2009 ▸). In our attempts to synthesize metal complexes with ligand 4, we obtained the hydrochloride salt 1 in its anhydrous form. It can be explained by the acidity of cobalt chloride hexahydrate, which was used in the selected experiment. This prompted us to develop a preparative synthesis of the hydrochloride salt. This was achieved by the formation and precipitation of crude hydrochloride in ethyl acetate solution. Its crystallization from water provided the hydrochloride hydrate 2.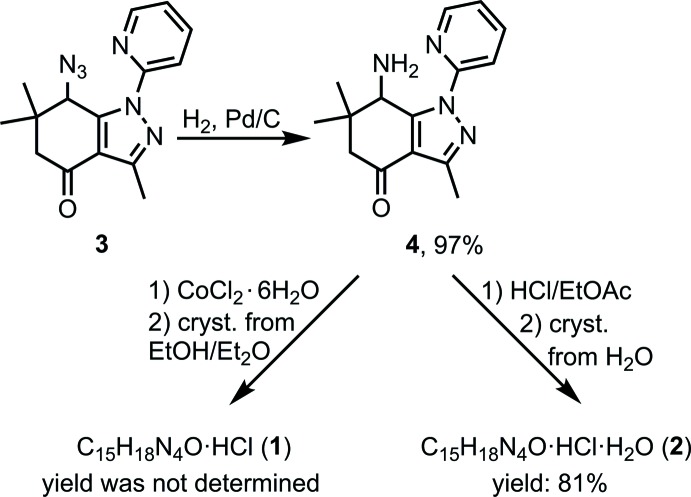
7-Amino-3,6,6-trimethyl-1-(pyridin-2-yl)-1,5,6,7-tetrahydro-4 H -indazol-4-one (4): Gaseous H2 was bubbled for 10 min. through a solution/suspension of compound 3 (0.80 g, 2.7 mmol) and 10% Pd/C (80 mg) in a mixture of EtOH (10 mL) and THF (2 mL). The resulting reaction mixture was stirred under an H2 atmosphere at standard temperature and pressure for 3 h (TLC control). The catalyst was filtered through a celite pad and the filtrate was evaporated to dryness. The resulting amorphous solid was dried under reduced pressure to yield amine 4 (0.71 g, 97%) as a colorless powder. M.p. 390–392 K; R f = 0.14 (Hex:EtOAc:Et3N = 8:1:0.5). IR (KBr), υ (cm−1): 3360, 3295, 3055, 2985, 2955, 2945, 2930, 2890, 2865, 1670, 1590, 1575, 1540, 1465, 1455, 1285, 1250, 1145, 1085, 1075, 1035, 995. 1H NMR (CDCl3, 300 MHz) δ (ppm): 8.48 [m, 1H, H-C(Py)], 7.99 [d, J = 8.3 Hz, 1H, H-C(Py)], 7.87 [m, 1H, H-C(Py)], 7.26 [m, 1H, H-C(Py)], 4.27 (s, 1H, H-C7), 2.82 (d, J = 16.8 Hz, 1H, Ha-C5), 2.54 (s, 3H, H3C-C3), 2.18 (d, J = 16.8 Hz, 1H, Hb-C5), 2.08 (bs, 2H, H2N-C7) 1.26, 1.02 (2s, 6H, H3C-C6).13C NMR (75.5 MHz, CDCl3), δ (ppm): 194.1, 153.9, 152.4, 150.4, 148.0, 139.1, 122.1, 116.5, 115.9, 53.8, 47.8, 38.4, 27.3, 26.6, 13.7. Analysis calculated: (C15H18N4O) C, 66.64; H, 6.71; N, 20.73. Found: C, 66.56; H, 6.68; N, 20.74.
3,6,6-Trimethyl-4-oxo-1-(pyridin-2-yl)-4,5,6,7-tetrahydro-1 H -indazol-7-aminium chloride (1): A solution of CoCl2·6H2O (24 mg, 0.1 mmol) in ethanol (2 mL) was added to a solution of amine 4 (27 mg, 0.1 mmol) in ethanol (2 mL). The resulting reaction mixture was maturated at ambient temperature for 24 h. Then a part of it (1.2 mL) was transferred into an NMR tube and Et2O (0.8 mL) was added carefully on the top of the ethanol solution. After two days, colorless crystals of 1 were collected form the wall of the NMR tube. The product was characterized spectroscopically in its hydrate form (see below).
3,6,6-Trimethyl-4-oxo-1-(pyridin-2-yl)-4,5,6,7-tetrahydro-1 H -indazol-7-aminium chloride hydrate (2): A solution of HCl in EtOAc (0.5 M, 1.48 mL, 0.74 mmol, 1.0 equiv.) was added to a solution of amine 4 (0.20 g, 0.74 mmol, 1.0 equiv.) in EtOAc (2 mL) at ambient temperature. The resulting precipitate was filtered and washed on the filter with DCM. The the crude product was crystallized from water to obtain colorless crystals of 2 (195 mg, 81%) suitable for X-ray analysis. M.p. 543 K (decomp.); IR (KBr), υ (cm−1): 3430 (br.s), 3145, 3100, 3035, 2965, 2880, 2750, 2575, 1955 (br.s), 1685, 1600, 1545, 1520, 1490, 1465, 1450, 1400, 1375, 1360, 1295, 1245, 1140, 1045, 1000, 955. 1H NMR (300MHz, D2O), δ (ppm): δ 8.55 [m, 1H, H-C(Py)], 8.12 [m,1H,H-C (Py)], 7.90 [d, J = 8.3 Hz, 1H, H-C(Py)], 7.53 [m, 1H, H-C(Py)], 4.84 (s, 1H, H-C7), 3.00 (d, J =17.8 Hz, 1H, Ha-C5), 2.54 (s, 3H, H3C-C3), 2.45 (d, J = 17.8Hz,1H,Hb-C5), 1.36, 1.10 (2s, 6H, H3C-C6). 13C NMR (75.5 MHz, DMSO-d 6), δ (ppm):192.3, 151.4, 149.0, 148.0, 144.4, 140.1, 122.8, 118.0, 114.6, 51.4, 47.1, 37.2, 26.8, 25.4, 13.2. Analysis calculated: (C15H18N4O·HCl·H2O) C, 55.47; H, 6.52; N, 17.25. Found: C, 55.78; H,6.40; N, 17.29.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 3 ▸. Hydrogen atoms bonded to heteroatoms were refined isotropically. Other H atoms were included in the refinement at geometrically calculated positions with C—H = 0.95–0.99Å and treated as riding with U iso(H) = 1.2U eq(C) or 1.5U eq(C-methyl).
Table 3. Experimental details.
| (1) | (2) | |
|---|---|---|
| Crystal data | ||
| Chemical formula | C15H19N4O+·Cl− | C15H19N4O+·Cl−·H2O |
| M r | 306.79 | 324.81 |
| Crystal system, space group | Monoclinic, P21/c | Monoclinic, P21/c |
| Temperature (K) | 190 | 190 |
| a, b, c (Å) | 13.5411 (4), 7.7421 (2), 19.2457 (5) | 10.1855 (2), 7.4951 (2), 20.7961 (4) |
| β (°) | 130.493 (2) | 100.545 (1) |
| V (Å3) | 1534.39 (8) | 1560.79 (6) |
| Z | 4 | 4 |
| Radiation type | Mo Kα | Mo Kα |
| μ (mm−1) | 0.25 | 0.26 |
| Crystal size (mm) | 0.38 × 0.32 × 0.15 | 0.42 × 0.25 × 0.14 |
| Data collection | ||
| Diffractometer | Nonius KappaCCD | Nonius KappaCCD |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 5860, 3486, 2715 | 5549, 3552, 2874 |
| R int | 0.027 | 0.023 |
| (sin θ/λ)max (Å−1) | 0.649 | 0.654 |
| Refinement | ||
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.043, 0.101, 1.03 | 0.039, 0.102, 1.06 |
| No. of reflections | 3486 | 3552 |
| No. of parameters | 205 | 222 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.29, −0.25 | 0.29, −0.28 |
Supplementary Material
Crystal structure: contains datablock(s) 1, 2, global. DOI: 10.1107/S205698901701667X/eb2002sup1.cif
Structure factors: contains datablock(s) 1. DOI: 10.1107/S205698901701667X/eb20021sup2.hkl
Supporting information file. DOI: 10.1107/S205698901701667X/eb20021sup4.mol
Structure factors: contains datablock(s) 2. DOI: 10.1107/S205698901701667X/eb20022sup3.hkl
Supporting information file. DOI: 10.1107/S205698901701667X/eb20022sup5.mol
Supporting information file. DOI: 10.1107/S205698901701667X/eb20021sup6.cml
Supporting information file. DOI: 10.1107/S205698901701667X/eb20022sup7.cml
Additional supporting information: crystallographic information; 3D view; checkCIF report
supplementary crystallographic information
3,6,6-Trimethyl-4-oxo-1-(pyridin-2-yl)-4,5,6,7-tetrahydro-1H-indazol-7-aminium chloride (1) . Crystal data
| C15H19N4O+·Cl− | F(000) = 648 |
| Mr = 306.79 | Dx = 1.328 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| a = 13.5411 (4) Å | Cell parameters from 6801 reflections |
| b = 7.7421 (2) Å | θ = 1.0–27.5° |
| c = 19.2457 (5) Å | µ = 0.25 mm−1 |
| β = 130.493 (2)° | T = 190 K |
| V = 1534.39 (8) Å3 | Block, colourless |
| Z = 4 | 0.38 × 0.32 × 0.15 mm |
3,6,6-Trimethyl-4-oxo-1-(pyridin-2-yl)-4,5,6,7-tetrahydro-1H-indazol-7-aminium chloride (1) . Data collection
| Nonius KappaCCD diffractometer | Rint = 0.027 |
| Radiation source: fine-focus sealed tube | θmax = 27.5°, θmin = 3.0° |
| CCD scans | h = −17→17 |
| 5860 measured reflections | k = −10→9 |
| 3486 independent reflections | l = −24→24 |
| 2715 reflections with I > 2σ(I) |
3,6,6-Trimethyl-4-oxo-1-(pyridin-2-yl)-4,5,6,7-tetrahydro-1H-indazol-7-aminium chloride (1) . Refinement
| Refinement on F2 | 0 restraints |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.043 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.101 | w = 1/[σ2(Fo2) + (0.0363P)2 + 0.7495P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.03 | (Δ/σ)max = 0.001 |
| 3486 reflections | Δρmax = 0.29 e Å−3 |
| 205 parameters | Δρmin = −0.25 e Å−3 |
3,6,6-Trimethyl-4-oxo-1-(pyridin-2-yl)-4,5,6,7-tetrahydro-1H-indazol-7-aminium chloride (1) . Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
3,6,6-Trimethyl-4-oxo-1-(pyridin-2-yl)-4,5,6,7-tetrahydro-1H-indazol-7-aminium chloride (1) . Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Cl1 | 0.63264 (4) | 0.65297 (6) | 0.33536 (3) | 0.03342 (14) | |
| O1 | 0.98736 (12) | −0.08281 (19) | 0.66919 (8) | 0.0361 (3) | |
| N3 | 0.59725 (12) | 0.17416 (17) | 0.53425 (8) | 0.0187 (3) | |
| N4 | 0.65309 (13) | 0.13670 (18) | 0.62325 (9) | 0.0219 (3) | |
| C4 | 0.67526 (15) | 0.1258 (2) | 0.51633 (10) | 0.0193 (3) | |
| N2 | 0.41873 (13) | 0.26763 (19) | 0.38981 (9) | 0.0230 (3) | |
| C12 | 0.42625 (16) | 0.3337 (2) | 0.51532 (11) | 0.0232 (4) | |
| H12 | 0.471148 | 0.327724 | 0.577688 | 0.028* | |
| C11 | 0.47588 (15) | 0.2621 (2) | 0.47773 (10) | 0.0190 (3) | |
| N1 | 0.55052 (14) | 0.0192 (2) | 0.35950 (9) | 0.0220 (3) | |
| C13 | 0.30686 (17) | 0.4145 (2) | 0.45593 (12) | 0.0282 (4) | |
| H13 | 0.267881 | 0.460408 | 0.477604 | 0.034* | |
| C3 | 0.78494 (15) | 0.0540 (2) | 0.59559 (10) | 0.0211 (3) | |
| C6 | 0.78026 (16) | 0.1295 (2) | 0.44709 (11) | 0.0246 (4) | |
| C15 | 0.30553 (16) | 0.3529 (2) | 0.33478 (12) | 0.0276 (4) | |
| H15 | 0.265092 | 0.362741 | 0.273282 | 0.033* | |
| C8 | 0.88681 (16) | −0.0231 (2) | 0.59948 (11) | 0.0244 (4) | |
| C5 | 0.65109 (15) | 0.1485 (2) | 0.42887 (10) | 0.0207 (3) | |
| H5 | 0.616867 | 0.265008 | 0.405413 | 0.025* | |
| C2 | 0.76718 (16) | 0.0664 (2) | 0.66070 (11) | 0.0224 (4) | |
| C10 | 0.75046 (18) | 0.1108 (3) | 0.35558 (12) | 0.0325 (4) | |
| H10A | 0.830139 | 0.113448 | 0.366104 | 0.049* | |
| H10B | 0.695686 | 0.204248 | 0.315864 | 0.049* | |
| H10C | 0.706960 | 0.002985 | 0.327613 | 0.049* | |
| C14 | 0.24569 (17) | 0.4268 (2) | 0.36430 (13) | 0.0297 (4) | |
| H14 | 0.166419 | 0.483251 | 0.323640 | 0.036* | |
| C7 | 0.85647 (16) | −0.0288 (2) | 0.50865 (11) | 0.0253 (4) | |
| H7A | 0.806434 | −0.132217 | 0.476113 | 0.030* | |
| H7B | 0.937384 | −0.037310 | 0.520035 | 0.030* | |
| C1 | 0.85803 (17) | 0.0141 (3) | 0.75907 (11) | 0.0312 (4) | |
| H1A | 0.809647 | −0.005846 | 0.778844 | 0.047* | |
| H1B | 0.920174 | 0.104542 | 0.795200 | 0.047* | |
| H1C | 0.902572 | −0.089806 | 0.766326 | 0.047* | |
| C9 | 0.86206 (17) | 0.2931 (2) | 0.49510 (13) | 0.0301 (4) | |
| H9A | 0.815252 | 0.391477 | 0.456570 | 0.045* | |
| H9B | 0.942494 | 0.281965 | 0.507027 | 0.045* | |
| H9C | 0.879617 | 0.308100 | 0.551734 | 0.045* | |
| H1N1 | 0.487 (2) | 0.000 (3) | 0.3670 (14) | 0.046 (6)* | |
| H2N1 | 0.503 (2) | 0.062 (3) | 0.2980 (16) | 0.054 (7)* | |
| H3N1 | 0.584 (2) | −0.088 (3) | 0.3627 (14) | 0.042 (6)* |
3,6,6-Trimethyl-4-oxo-1-(pyridin-2-yl)-4,5,6,7-tetrahydro-1H-indazol-7-aminium chloride (1) . Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cl1 | 0.0363 (3) | 0.0353 (3) | 0.0225 (2) | 0.0052 (2) | 0.01632 (19) | −0.00482 (19) |
| O1 | 0.0235 (7) | 0.0442 (8) | 0.0281 (6) | 0.0113 (6) | 0.0113 (6) | 0.0038 (6) |
| N3 | 0.0190 (7) | 0.0211 (7) | 0.0168 (6) | 0.0005 (6) | 0.0119 (5) | 0.0004 (6) |
| N4 | 0.0244 (7) | 0.0234 (7) | 0.0184 (6) | −0.0002 (6) | 0.0141 (6) | 0.0012 (6) |
| C4 | 0.0189 (8) | 0.0186 (8) | 0.0210 (7) | −0.0011 (6) | 0.0132 (7) | −0.0020 (7) |
| N2 | 0.0207 (7) | 0.0263 (7) | 0.0216 (7) | 0.0011 (6) | 0.0136 (6) | 0.0027 (6) |
| C12 | 0.0272 (9) | 0.0200 (8) | 0.0268 (8) | 0.0003 (7) | 0.0195 (7) | 0.0005 (7) |
| C11 | 0.0189 (8) | 0.0157 (8) | 0.0232 (8) | −0.0015 (6) | 0.0140 (7) | −0.0004 (7) |
| N1 | 0.0204 (7) | 0.0266 (8) | 0.0191 (7) | 0.0023 (6) | 0.0129 (6) | −0.0013 (6) |
| C13 | 0.0299 (9) | 0.0235 (9) | 0.0400 (10) | 0.0021 (8) | 0.0267 (8) | −0.0010 (8) |
| C3 | 0.0187 (8) | 0.0209 (8) | 0.0194 (7) | −0.0011 (7) | 0.0105 (6) | −0.0019 (7) |
| C6 | 0.0218 (8) | 0.0297 (9) | 0.0264 (8) | 0.0031 (7) | 0.0176 (7) | 0.0007 (8) |
| C15 | 0.0230 (8) | 0.0299 (9) | 0.0252 (8) | 0.0022 (8) | 0.0136 (7) | 0.0063 (8) |
| C8 | 0.0203 (8) | 0.0213 (8) | 0.0254 (8) | −0.0005 (7) | 0.0122 (7) | −0.0016 (7) |
| C5 | 0.0205 (8) | 0.0220 (8) | 0.0212 (7) | 0.0022 (7) | 0.0143 (7) | 0.0010 (7) |
| C2 | 0.0214 (8) | 0.0212 (8) | 0.0201 (8) | −0.0022 (7) | 0.0114 (7) | −0.0010 (7) |
| C10 | 0.0285 (9) | 0.0455 (12) | 0.0315 (9) | 0.0064 (9) | 0.0230 (8) | 0.0043 (9) |
| C14 | 0.0224 (9) | 0.0245 (9) | 0.0379 (10) | 0.0048 (7) | 0.0176 (8) | 0.0055 (8) |
| C7 | 0.0198 (8) | 0.0281 (9) | 0.0290 (9) | 0.0042 (7) | 0.0163 (7) | −0.0020 (8) |
| C1 | 0.0291 (9) | 0.0364 (11) | 0.0205 (8) | 0.0007 (8) | 0.0127 (8) | 0.0025 (8) |
| C9 | 0.0251 (9) | 0.0299 (10) | 0.0390 (10) | 0.0022 (8) | 0.0224 (8) | 0.0014 (8) |
3,6,6-Trimethyl-4-oxo-1-(pyridin-2-yl)-4,5,6,7-tetrahydro-1H-indazol-7-aminium chloride (1) . Geometric parameters (Å, º)
| O1—C8 | 1.223 (2) | C6—C10 | 1.537 (2) |
| N3—C4 | 1.360 (2) | C6—C7 | 1.544 (2) |
| N3—N4 | 1.3811 (18) | C6—C5 | 1.552 (2) |
| N3—C11 | 1.424 (2) | C15—C14 | 1.380 (3) |
| N4—C2 | 1.325 (2) | C15—H15 | 0.9300 |
| C4—C3 | 1.378 (2) | C8—C7 | 1.515 (2) |
| C4—C5 | 1.502 (2) | C5—H5 | 0.9800 |
| N2—C11 | 1.328 (2) | C2—C1 | 1.496 (2) |
| N2—C15 | 1.341 (2) | C10—H10A | 0.9600 |
| C12—C13 | 1.383 (2) | C10—H10B | 0.9600 |
| C12—C11 | 1.383 (2) | C10—H10C | 0.9600 |
| C12—H12 | 0.9300 | C14—H14 | 0.9300 |
| N1—C5 | 1.509 (2) | C7—H7A | 0.9700 |
| N1—H1N1 | 0.97 (2) | C7—H7B | 0.9700 |
| N1—H2N1 | 0.97 (2) | C1—H1A | 0.9600 |
| N1—H3N1 | 0.93 (2) | C1—H1B | 0.9600 |
| C13—C14 | 1.381 (3) | C1—H1C | 0.9600 |
| C13—H13 | 0.9300 | C9—H9A | 0.9600 |
| C3—C2 | 1.424 (2) | C9—H9B | 0.9600 |
| C3—C8 | 1.459 (2) | C9—H9C | 0.9600 |
| C6—C9 | 1.535 (3) | ||
| C4—N3—N4 | 111.53 (12) | C3—C8—C7 | 114.43 (14) |
| C4—N3—C11 | 129.76 (13) | C4—C5—N1 | 109.18 (13) |
| N4—N3—C11 | 118.61 (12) | C4—C5—C6 | 110.02 (13) |
| C2—N4—N3 | 105.47 (13) | N1—C5—C6 | 111.99 (13) |
| N3—C4—C3 | 106.66 (13) | C4—C5—H5 | 108.5 |
| N3—C4—C5 | 127.76 (14) | N1—C5—H5 | 108.5 |
| C3—C4—C5 | 125.56 (14) | C6—C5—H5 | 108.5 |
| C11—N2—C15 | 116.33 (14) | N4—C2—C3 | 110.63 (14) |
| C13—C12—C11 | 116.96 (15) | N4—C2—C1 | 120.64 (15) |
| C13—C12—H12 | 121.5 | C3—C2—C1 | 128.72 (16) |
| C11—C12—H12 | 121.5 | C6—C10—H10A | 109.5 |
| N2—C11—C12 | 125.07 (15) | C6—C10—H10B | 109.5 |
| N2—C11—N3 | 114.67 (13) | H10A—C10—H10B | 109.5 |
| C12—C11—N3 | 120.26 (14) | C6—C10—H10C | 109.5 |
| C5—N1—H1N1 | 110.3 (13) | H10A—C10—H10C | 109.5 |
| C5—N1—H2N1 | 110.9 (14) | H10B—C10—H10C | 109.5 |
| H1N1—N1—H2N1 | 106.3 (18) | C15—C14—C13 | 118.12 (16) |
| C5—N1—H3N1 | 114.2 (13) | C15—C14—H14 | 120.9 |
| H1N1—N1—H3N1 | 107.5 (19) | C13—C14—H14 | 120.9 |
| H2N1—N1—H3N1 | 107.3 (19) | C8—C7—C6 | 114.06 (14) |
| C14—C13—C12 | 119.76 (16) | C8—C7—H7A | 108.7 |
| C14—C13—H13 | 120.1 | C6—C7—H7A | 108.7 |
| C12—C13—H13 | 120.1 | C8—C7—H7B | 108.7 |
| C4—C3—C2 | 105.68 (14) | C6—C7—H7B | 108.7 |
| C4—C3—C8 | 121.68 (14) | H7A—C7—H7B | 107.6 |
| C2—C3—C8 | 132.55 (15) | C2—C1—H1A | 109.5 |
| C9—C6—C10 | 108.51 (15) | C2—C1—H1B | 109.5 |
| C9—C6—C7 | 109.37 (14) | H1A—C1—H1B | 109.5 |
| C10—C6—C7 | 110.59 (14) | C2—C1—H1C | 109.5 |
| C9—C6—C5 | 108.97 (14) | H1A—C1—H1C | 109.5 |
| C10—C6—C5 | 109.35 (13) | H1B—C1—H1C | 109.5 |
| C7—C6—C5 | 110.02 (14) | C6—C9—H9A | 109.5 |
| N2—C15—C14 | 123.68 (16) | C6—C9—H9B | 109.5 |
| N2—C15—H15 | 118.2 | H9A—C9—H9B | 109.5 |
| C14—C15—H15 | 118.2 | C6—C9—H9C | 109.5 |
| O1—C8—C3 | 123.57 (16) | H9A—C9—H9C | 109.5 |
| O1—C8—C7 | 121.97 (15) | H9B—C9—H9C | 109.5 |
| C4—N3—N4—C2 | 0.69 (18) | N3—C4—C5—N1 | −74.7 (2) |
| C11—N3—N4—C2 | −176.00 (14) | C3—C4—C5—N1 | 107.05 (18) |
| N4—N3—C4—C3 | 0.27 (18) | N3—C4—C5—C6 | 162.06 (16) |
| C11—N3—C4—C3 | 176.49 (15) | C3—C4—C5—C6 | −16.2 (2) |
| N4—N3—C4—C5 | −178.26 (15) | C9—C6—C5—C4 | −74.59 (17) |
| C11—N3—C4—C5 | −2.0 (3) | C10—C6—C5—C4 | 166.95 (14) |
| C15—N2—C11—C12 | 1.4 (2) | C7—C6—C5—C4 | 45.31 (18) |
| C15—N2—C11—N3 | −178.55 (14) | C9—C6—C5—N1 | 163.80 (14) |
| C13—C12—C11—N2 | 1.1 (3) | C10—C6—C5—N1 | 45.35 (19) |
| C13—C12—C11—N3 | −178.92 (15) | C7—C6—C5—N1 | −76.29 (16) |
| C4—N3—C11—N2 | 14.7 (2) | N3—N4—C2—C3 | −1.36 (18) |
| N4—N3—C11—N2 | −169.29 (14) | N3—N4—C2—C1 | 177.90 (15) |
| C4—N3—C11—C12 | −165.27 (16) | C4—C3—C2—N4 | 1.55 (19) |
| N4—N3—C11—C12 | 10.7 (2) | C8—C3—C2—N4 | −174.86 (17) |
| C11—C12—C13—C14 | −2.6 (3) | C4—C3—C2—C1 | −177.64 (17) |
| N3—C4—C3—C2 | −1.05 (18) | C8—C3—C2—C1 | 6.0 (3) |
| C5—C4—C3—C2 | 177.52 (15) | N2—C15—C14—C13 | 1.0 (3) |
| N3—C4—C3—C8 | 175.84 (15) | C12—C13—C14—C15 | 1.7 (3) |
| C5—C4—C3—C8 | −5.6 (3) | O1—C8—C7—C6 | −145.62 (17) |
| C11—N2—C15—C14 | −2.5 (3) | C3—C8—C7—C6 | 36.1 (2) |
| C4—C3—C8—O1 | 177.45 (17) | C9—C6—C7—C8 | 62.12 (18) |
| C2—C3—C8—O1 | −6.6 (3) | C10—C6—C7—C8 | −178.44 (14) |
| C4—C3—C8—C7 | −4.4 (2) | C5—C6—C7—C8 | −57.54 (19) |
| C2—C3—C8—C7 | 171.59 (17) |
3,6,6-Trimethyl-4-oxo-1-(pyridin-2-yl)-4,5,6,7-tetrahydro-1H-indazol-7-aminium chloride (1) . Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C12—H12···Cl1i | 0.93 | 2.80 | 3.4470 (17) | 127 |
| C14—H14···O1ii | 0.93 | 2.44 | 3.277 (2) | 150 |
| C7—H7A···Cl1iii | 0.97 | 2.71 | 3.6401 (18) | 160 |
| C1—H1B···O1iv | 0.96 | 2.60 | 3.503 (2) | 156 |
| N1—H1N1···N4v | 0.97 (2) | 2.29 (2) | 3.218 (2) | 161 (2) |
| N1—H1N1···N2 | 0.97 (2) | 2.42 (2) | 2.928 (2) | 112 (2) |
| N1—H2N1···Cl1vi | 0.97 (2) | 2.08 (2) | 3.034 (2) | 168 (2) |
| N1—H3N1···Cl1iii | 0.93 (2) | 2.27 (2) | 3.188 (2) | 167 (2) |
Symmetry codes: (i) −x+1, −y+1, −z+1; (ii) x−1, −y+1/2, z−1/2; (iii) x, y−1, z; (iv) −x+2, y+1/2, −z+3/2; (v) −x+1, −y, −z+1; (vi) −x+1, y−1/2, −z+1/2.
3,6,6-Trimethyl-4-oxo-1-(pyridin-2-yl)-4,5,6,7-tetrahydro-1H-indazol-7-aminium chloride monohydrate (2) . Crystal data
| C15H19N4O+·Cl−·H2O | F(000) = 688 |
| Mr = 324.81 | Dx = 1.382 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| a = 10.1855 (2) Å | Cell parameters from 8626 reflections |
| b = 7.4951 (2) Å | θ = 1.0–27.5° |
| c = 20.7961 (4) Å | µ = 0.26 mm−1 |
| β = 100.545 (1)° | T = 190 K |
| V = 1560.79 (6) Å3 | Block, colourless |
| Z = 4 | 0.42 × 0.25 × 0.14 mm |
3,6,6-Trimethyl-4-oxo-1-(pyridin-2-yl)-4,5,6,7-tetrahydro-1H-indazol-7-aminium chloride monohydrate (2) . Data collection
| Nonius KappaCCD diffractometer | Rint = 0.023 |
| Radiation source: fine-focus sealed tube | θmax = 27.7°, θmin = 3.6° |
| CCD scans | h = −13→13 |
| 5549 measured reflections | k = −9→9 |
| 3552 independent reflections | l = −27→26 |
| 2874 reflections with I > 2σ(I) |
3,6,6-Trimethyl-4-oxo-1-(pyridin-2-yl)-4,5,6,7-tetrahydro-1H-indazol-7-aminium chloride monohydrate (2) . Refinement
| Refinement on F2 | 0 restraints |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.039 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.102 | w = 1/[σ2(Fo2) + (0.0426P)2 + 0.794P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.06 | (Δ/σ)max < 0.001 |
| 3552 reflections | Δρmax = 0.29 e Å−3 |
| 222 parameters | Δρmin = −0.28 e Å−3 |
3,6,6-Trimethyl-4-oxo-1-(pyridin-2-yl)-4,5,6,7-tetrahydro-1H-indazol-7-aminium chloride monohydrate (2) . Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
3,6,6-Trimethyl-4-oxo-1-(pyridin-2-yl)-4,5,6,7-tetrahydro-1H-indazol-7-aminium chloride monohydrate (2) . Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Cl1 | 0.06154 (4) | 0.62526 (6) | 0.39636 (2) | 0.02975 (13) | |
| O1 | 0.45919 (12) | −0.12171 (17) | 0.26173 (5) | 0.0286 (3) | |
| N2 | 0.33632 (13) | 0.28400 (18) | 0.51823 (6) | 0.0210 (3) | |
| O1W | −0.00940 (13) | 0.25508 (19) | 0.46352 (7) | 0.0328 (3) | |
| N3 | 0.48296 (12) | 0.17260 (17) | 0.45521 (6) | 0.0159 (3) | |
| N1 | 0.18153 (14) | 0.01688 (19) | 0.43612 (7) | 0.0183 (3) | |
| N4 | 0.61172 (12) | 0.13967 (18) | 0.44629 (6) | 0.0196 (3) | |
| C5 | 0.24311 (14) | 0.1376 (2) | 0.39147 (7) | 0.0155 (3) | |
| H5 | 0.223932 | 0.261303 | 0.401963 | 0.019* | |
| C12 | 0.57141 (16) | 0.3394 (2) | 0.55440 (8) | 0.0219 (3) | |
| H12 | 0.658465 | 0.326736 | 0.547312 | 0.026* | |
| C11 | 0.46319 (15) | 0.2686 (2) | 0.51150 (7) | 0.0174 (3) | |
| C4 | 0.39179 (14) | 0.1150 (2) | 0.40322 (7) | 0.0153 (3) | |
| C6 | 0.18454 (15) | 0.1060 (2) | 0.31807 (7) | 0.0176 (3) | |
| C10 | 0.03220 (16) | 0.0870 (2) | 0.30801 (8) | 0.0246 (4) | |
| H10A | 0.009876 | −0.020414 | 0.328751 | 0.037* | |
| H10B | −0.003916 | 0.081667 | 0.262056 | 0.037* | |
| H10C | −0.004705 | 0.187831 | 0.326935 | 0.037* | |
| C9 | 0.21584 (16) | 0.2703 (2) | 0.27933 (8) | 0.0227 (3) | |
| H9A | 0.180810 | 0.375246 | 0.296617 | 0.034* | |
| H9B | 0.175456 | 0.256460 | 0.234144 | 0.034* | |
| H9C | 0.310786 | 0.282002 | 0.283039 | 0.034* | |
| C3 | 0.46350 (15) | 0.0404 (2) | 0.35966 (7) | 0.0166 (3) | |
| C8 | 0.39804 (16) | −0.0515 (2) | 0.30028 (7) | 0.0192 (3) | |
| C15 | 0.31230 (18) | 0.3783 (2) | 0.56960 (8) | 0.0270 (4) | |
| H15 | 0.224058 | 0.395222 | 0.574240 | 0.032* | |
| C7 | 0.24693 (16) | −0.0608 (2) | 0.29224 (7) | 0.0213 (3) | |
| H7A | 0.223032 | −0.164981 | 0.315275 | 0.026* | |
| H7B | 0.209284 | −0.075806 | 0.246229 | 0.026* | |
| C14 | 0.4120 (2) | 0.4514 (2) | 0.61590 (8) | 0.0299 (4) | |
| H14 | 0.391561 | 0.513786 | 0.651450 | 0.036* | |
| C13 | 0.54279 (19) | 0.4297 (2) | 0.60821 (8) | 0.0280 (4) | |
| H13 | 0.611825 | 0.475897 | 0.639260 | 0.034* | |
| C2 | 0.59994 (15) | 0.0631 (2) | 0.38810 (7) | 0.0196 (3) | |
| C1 | 0.72083 (17) | 0.0203 (3) | 0.36008 (9) | 0.0295 (4) | |
| H1A | 0.799516 | 0.046429 | 0.391767 | 0.044* | |
| H1B | 0.720784 | 0.090854 | 0.321550 | 0.044* | |
| H1C | 0.720059 | −0.104005 | 0.348878 | 0.044* | |
| H1W | −0.026 (3) | 0.288 (4) | 0.4977 (14) | 0.063 (9)* | |
| H2W | 0.012 (3) | 0.362 (4) | 0.4440 (13) | 0.066 (8)* | |
| H1N1 | 0.1557 (19) | −0.083 (3) | 0.4188 (9) | 0.024 (5)* | |
| H2N1 | 0.244 (2) | −0.009 (3) | 0.4766 (11) | 0.036 (5)* | |
| H3N1 | 0.112 (2) | 0.080 (3) | 0.4513 (11) | 0.049 (7)* |
3,6,6-Trimethyl-4-oxo-1-(pyridin-2-yl)-4,5,6,7-tetrahydro-1H-indazol-7-aminium chloride monohydrate (2) . Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cl1 | 0.0343 (2) | 0.0261 (2) | 0.0301 (2) | −0.00602 (18) | 0.00909 (17) | −0.00145 (17) |
| O1 | 0.0328 (7) | 0.0360 (7) | 0.0182 (6) | 0.0076 (5) | 0.0077 (5) | −0.0040 (5) |
| N2 | 0.0237 (7) | 0.0214 (7) | 0.0185 (6) | 0.0011 (6) | 0.0053 (5) | −0.0027 (6) |
| O1W | 0.0343 (7) | 0.0287 (7) | 0.0385 (8) | 0.0014 (6) | 0.0152 (6) | −0.0051 (6) |
| N3 | 0.0144 (6) | 0.0174 (6) | 0.0156 (6) | −0.0002 (5) | 0.0018 (5) | 0.0000 (5) |
| N1 | 0.0185 (7) | 0.0208 (7) | 0.0162 (6) | −0.0037 (6) | 0.0045 (5) | −0.0014 (6) |
| N4 | 0.0140 (6) | 0.0226 (7) | 0.0222 (7) | 0.0005 (5) | 0.0036 (5) | 0.0019 (6) |
| C5 | 0.0155 (7) | 0.0168 (7) | 0.0143 (7) | −0.0016 (6) | 0.0026 (5) | 0.0007 (6) |
| C12 | 0.0250 (8) | 0.0171 (7) | 0.0211 (8) | −0.0021 (6) | −0.0029 (6) | 0.0022 (6) |
| C11 | 0.0226 (8) | 0.0141 (7) | 0.0147 (7) | −0.0004 (6) | 0.0018 (6) | 0.0019 (6) |
| C4 | 0.0164 (7) | 0.0153 (7) | 0.0138 (7) | −0.0017 (6) | 0.0022 (5) | 0.0020 (6) |
| C6 | 0.0156 (7) | 0.0224 (8) | 0.0140 (7) | −0.0015 (6) | 0.0003 (5) | 0.0008 (6) |
| C10 | 0.0177 (8) | 0.0332 (9) | 0.0216 (8) | −0.0044 (7) | −0.0002 (6) | −0.0003 (7) |
| C9 | 0.0221 (8) | 0.0251 (9) | 0.0193 (7) | −0.0006 (7) | −0.0004 (6) | 0.0054 (7) |
| C3 | 0.0185 (7) | 0.0171 (7) | 0.0146 (7) | 0.0000 (6) | 0.0039 (6) | 0.0017 (6) |
| C8 | 0.0262 (8) | 0.0180 (7) | 0.0139 (7) | 0.0015 (6) | 0.0045 (6) | 0.0031 (6) |
| C15 | 0.0344 (9) | 0.0263 (9) | 0.0222 (8) | 0.0030 (7) | 0.0100 (7) | −0.0024 (7) |
| C7 | 0.0244 (8) | 0.0229 (8) | 0.0163 (7) | −0.0042 (7) | 0.0033 (6) | −0.0034 (6) |
| C14 | 0.0506 (12) | 0.0225 (9) | 0.0173 (7) | −0.0006 (8) | 0.0079 (7) | −0.0035 (7) |
| C13 | 0.0427 (11) | 0.0181 (8) | 0.0189 (8) | −0.0055 (7) | −0.0061 (7) | −0.0014 (7) |
| C2 | 0.0183 (8) | 0.0210 (8) | 0.0201 (7) | 0.0016 (6) | 0.0054 (6) | 0.0036 (6) |
| C1 | 0.0205 (8) | 0.0392 (11) | 0.0310 (9) | 0.0027 (8) | 0.0109 (7) | 0.0000 (8) |
3,6,6-Trimethyl-4-oxo-1-(pyridin-2-yl)-4,5,6,7-tetrahydro-1H-indazol-7-aminium chloride monohydrate (2) . Geometric parameters (Å, º)
| O1—C8 | 1.2207 (19) | C6—C7 | 1.543 (2) |
| N2—C11 | 1.330 (2) | C10—H10A | 0.9600 |
| N2—C15 | 1.340 (2) | C10—H10B | 0.9600 |
| O1W—H1W | 0.80 (3) | C10—H10C | 0.9600 |
| O1W—H2W | 0.94 (3) | C9—H9A | 0.9600 |
| N3—C4 | 1.3601 (18) | C9—H9B | 0.9600 |
| N3—N4 | 1.3800 (17) | C9—H9C | 0.9600 |
| N3—C11 | 1.4196 (19) | C3—C2 | 1.418 (2) |
| N1—C5 | 1.5127 (19) | C3—C8 | 1.464 (2) |
| N1—H1N1 | 0.85 (2) | C8—C7 | 1.519 (2) |
| N1—H2N1 | 0.98 (2) | C15—C14 | 1.378 (3) |
| N1—H3N1 | 0.95 (2) | C15—H15 | 0.9300 |
| N4—C2 | 1.325 (2) | C7—H7A | 0.9700 |
| C5—C4 | 1.499 (2) | C7—H7B | 0.9700 |
| C5—C6 | 1.5522 (19) | C14—C13 | 1.380 (3) |
| C5—H5 | 0.9800 | C14—H14 | 0.9300 |
| C12—C13 | 1.384 (2) | C13—H13 | 0.9300 |
| C12—C11 | 1.390 (2) | C2—C1 | 1.491 (2) |
| C12—H12 | 0.9300 | C1—H1A | 0.9600 |
| C4—C3 | 1.382 (2) | C1—H1B | 0.9600 |
| C6—C10 | 1.534 (2) | C1—H1C | 0.9600 |
| C6—C9 | 1.537 (2) | ||
| C11—N2—C15 | 116.89 (14) | H10B—C10—H10C | 109.5 |
| H1W—O1W—H2W | 103 (3) | C6—C9—H9A | 109.5 |
| C4—N3—N4 | 111.34 (12) | C6—C9—H9B | 109.5 |
| C4—N3—C11 | 129.57 (13) | H9A—C9—H9B | 109.5 |
| N4—N3—C11 | 118.90 (12) | C6—C9—H9C | 109.5 |
| C5—N1—H1N1 | 113.5 (13) | H9A—C9—H9C | 109.5 |
| C5—N1—H2N1 | 111.4 (12) | H9B—C9—H9C | 109.5 |
| H1N1—N1—H2N1 | 106.9 (18) | C4—C3—C2 | 105.85 (13) |
| C5—N1—H3N1 | 108.9 (15) | C4—C3—C8 | 122.00 (13) |
| H1N1—N1—H3N1 | 112.7 (19) | C2—C3—C8 | 132.00 (14) |
| H2N1—N1—H3N1 | 102.9 (18) | O1—C8—C3 | 123.26 (15) |
| C2—N4—N3 | 105.71 (12) | O1—C8—C7 | 122.43 (14) |
| C4—C5—N1 | 110.64 (12) | C3—C8—C7 | 114.23 (13) |
| C4—C5—C6 | 109.76 (12) | N2—C15—C14 | 123.21 (17) |
| N1—C5—C6 | 112.59 (12) | N2—C15—H15 | 118.4 |
| C4—C5—H5 | 107.9 | C14—C15—H15 | 118.4 |
| N1—C5—H5 | 107.9 | C8—C7—C6 | 113.45 (13) |
| C6—C5—H5 | 107.9 | C8—C7—H7A | 108.9 |
| C13—C12—C11 | 116.49 (16) | C6—C7—H7A | 108.9 |
| C13—C12—H12 | 121.8 | C8—C7—H7B | 108.9 |
| C11—C12—H12 | 121.8 | C6—C7—H7B | 108.9 |
| N2—C11—C12 | 124.81 (14) | H7A—C7—H7B | 107.7 |
| N2—C11—N3 | 114.70 (13) | C15—C14—C13 | 118.40 (16) |
| C12—C11—N3 | 120.48 (14) | C15—C14—H14 | 120.8 |
| N3—C4—C3 | 106.48 (13) | C13—C14—H14 | 120.8 |
| N3—C4—C5 | 128.05 (13) | C14—C13—C12 | 120.12 (16) |
| C3—C4—C5 | 125.34 (13) | C14—C13—H13 | 119.9 |
| C10—C6—C9 | 107.73 (13) | C12—C13—H13 | 119.9 |
| C10—C6—C7 | 110.38 (13) | N4—C2—C3 | 110.57 (13) |
| C9—C6—C7 | 109.20 (12) | N4—C2—C1 | 120.48 (14) |
| C10—C6—C5 | 110.21 (12) | C3—C2—C1 | 128.90 (15) |
| C9—C6—C5 | 108.27 (12) | C2—C1—H1A | 109.5 |
| C7—C6—C5 | 110.96 (12) | C2—C1—H1B | 109.5 |
| C6—C10—H10A | 109.5 | H1A—C1—H1B | 109.5 |
| C6—C10—H10B | 109.5 | C2—C1—H1C | 109.5 |
| H10A—C10—H10B | 109.5 | H1A—C1—H1C | 109.5 |
| C6—C10—H10C | 109.5 | H1B—C1—H1C | 109.5 |
| H10A—C10—H10C | 109.5 | ||
| C4—N3—N4—C2 | 0.70 (17) | N3—C4—C3—C2 | −1.90 (16) |
| C11—N3—N4—C2 | −174.73 (13) | C5—C4—C3—C2 | 174.12 (14) |
| C15—N2—C11—C12 | 1.5 (2) | N3—C4—C3—C8 | 174.09 (13) |
| C15—N2—C11—N3 | −178.02 (14) | C5—C4—C3—C8 | −9.9 (2) |
| C13—C12—C11—N2 | 1.0 (2) | C4—C3—C8—O1 | −178.45 (15) |
| C13—C12—C11—N3 | −179.50 (14) | C2—C3—C8—O1 | −3.6 (3) |
| C4—N3—C11—N2 | 9.7 (2) | C4—C3—C8—C7 | −1.5 (2) |
| N4—N3—C11—N2 | −175.82 (13) | C2—C3—C8—C7 | 173.30 (16) |
| C4—N3—C11—C12 | −169.86 (15) | C11—N2—C15—C14 | −2.8 (2) |
| N4—N3—C11—C12 | 4.6 (2) | O1—C8—C7—C6 | −148.01 (15) |
| N4—N3—C4—C3 | 0.81 (17) | C3—C8—C7—C6 | 35.01 (18) |
| C11—N3—C4—C3 | 175.62 (14) | C10—C6—C7—C8 | 179.76 (13) |
| N4—N3—C4—C5 | −175.06 (14) | C9—C6—C7—C8 | 61.51 (16) |
| C11—N3—C4—C5 | −0.3 (2) | C5—C6—C7—C8 | −57.76 (16) |
| N1—C5—C4—N3 | −72.98 (19) | N2—C15—C14—C13 | 1.5 (3) |
| C6—C5—C4—N3 | 162.17 (14) | C15—C14—C13—C12 | 1.2 (3) |
| N1—C5—C4—C3 | 111.88 (16) | C11—C12—C13—C14 | −2.3 (2) |
| C6—C5—C4—C3 | −13.0 (2) | N3—N4—C2—C3 | −1.93 (17) |
| C4—C5—C6—C10 | 167.33 (13) | N3—N4—C2—C1 | 175.62 (15) |
| N1—C5—C6—C10 | 43.61 (17) | C4—C3—C2—N4 | 2.45 (18) |
| C4—C5—C6—C9 | −75.07 (15) | C8—C3—C2—N4 | −172.97 (15) |
| N1—C5—C6—C9 | 161.21 (13) | C4—C3—C2—C1 | −174.84 (17) |
| C4—C5—C6—C7 | 44.75 (16) | C8—C3—C2—C1 | 9.7 (3) |
| N1—C5—C6—C7 | −78.96 (15) |
3,6,6-Trimethyl-4-oxo-1-(pyridin-2-yl)-4,5,6,7-tetrahydro-1H-indazol-7-aminium chloride monohydrate (2) . Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C12—H12···Cl1i | 0.93 | 2.90 | 3.7006 (17) | 145 |
| C14—H14···O1ii | 0.93 | 2.41 | 3.244 (2) | 149 |
| O1W—H1W···Cl1iii | 0.80 (3) | 2.39 (3) | 3.185 (2) | 176 (3) |
| O1W—H2W···Cl1 | 0.94 (3) | 2.31 (3) | 3.247 (2) | 179 (2) |
| N1—H1N1···Cl1iv | 0.85 (2) | 2.40 (2) | 3.228 (2) | 165 (2) |
| N1—H2N1···N4v | 0.98 (2) | 2.20 (2) | 3.1475 (19) | 164 (2) |
| N1—H3N1···O1W | 0.95 (2) | 1.85 (3) | 2.775 (2) | 162 (2) |
Symmetry codes: (i) −x+1, −y+1, −z+1; (ii) x, −y+1/2, z+1/2; (iii) −x, −y+1, −z+1; (iv) x, y−1, z; (v) −x+1, −y, −z+1.
Funding Statement
This work was funded by Latvian Council of Science grant 14.0593.
References
- Abdel-Rahman, H. M. & Ozadali, K. (2012). Arch. Pharm. Pharm. Med. Chem. 345, 878–883. [DOI] [PubMed]
- Ansari, A., Ali, A., Asif, M. & Shamsuzzaman (2017). New J. Chem. 41, 16–41.
- Bassyouni, F., El Din Gaffer, A., Roaiah, H., El-Senousy, W. M., El Nakkady, S. S. & &Rehim, M. A. (2016). Res. J. Pharm. Biol. Chem. Sci, 7, 24–37.
- Bruker (20014). COLLECT. Bruker AXS Inc., Madison, Wisconsin, USA.
- Burch, J. D., Barrett, K., Chen, Y., DeVoss, J., Eigenbrot, C., Goldsmith, R., Ismaili, M. H. A., Lau, K., Lin, Z., Ortwine, D. F., Zarrin, A. A., McEwan, P. A., Barker, J. J., Ellebrandt, C., Kordt, D., Stein, D. B., Wang, X., Chen, Y., Hu, B., Xu, X., Yuen, P.-W., Zhang, Y. & Pei, Z. (2015). J. Med. Chem. 58, 3806–3816. [DOI] [PubMed]
- Burla, M. C., Caliandro, R., Camalli, M., Carrozzini, B., Cascarano, G. L., De Caro, L., Giacovazzo, C., Polidori, G. & Spagna, R. (2005). J. Appl. Cryst. 38, 381–388.
- Claramunt, R. M., López, C., Pérez-Medina, C., Pinilla, E., Torres, M. R. & Elguero, J. (2006). Tetrahedron, 62, 11704–11713.
- Etter, M. C. (1990). Acc. Chem. Res. 23, 120–126.
- Faidallah, H. M., Khan, K. A., Rostom, S. A. F. & Asiri, A. M. (2013). J. Enzyme Inhib. Med. Chem. 28, 495–508. [DOI] [PubMed]
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Gaikwad, D. D., Chapolikar, A. D., Devkate, C. G., Warad, K. D., Tayade, A. P., Pawar, R. P. & Domb, A. J. (2015). Eur. J. Med. Chem. 90, 707–731. [DOI] [PubMed]
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Guo, S., Song, Y., Huang, Q., Yuan, H., Wan, B., Wang, Y., He, R., Beconi, M. G., Franzblau, S. G. & Kozikowski, A. P. (2010). J. Med. Chem. 53, 649–659. [DOI] [PubMed]
- Heifetz, A., Trani, G., Aldeghi, M., MacKinnon, C. H., McEwan, P. A., Brookfield, F. A., Chudyk, E. I., Bodkin, M., Pei, Z., Burch, J. D. & Ortwine, D. F. (2016). J. Med. Chem. 59, 4352–4363. [DOI] [PubMed]
- Jia, J., Xu, Q.-C., Li, R.-C., Tang, X., He, Y.-F., Zhang, M.-Y., Zhang, Y. & Xing, G.-W. (2012). Org. Biomol. Chem. 10, 6279–6286. [DOI] [PubMed]
- Khlebnicova, T. S., Piven, Y. A., Baranovsky, A. V., Lakhvich, F. A., Shishkina, S. V., Zicāne, D., Tetere, Z., Rāviņa, I., Kumpiņš, V., Rijkure, I., Mieriņa, I., Peipiņš, U. & Turks, M. (2017). Steroids, 117, 77–89. [DOI] [PubMed]
- Khlebnikova, T. S., Piven’, Y. A., Baranovskii, A. V. & Lakhvich, F. A. (2012). Russ. J. Org. Chem. 48, 411–418.
- Kim, J., Song, H. & Park, S. B. (2010). Eur. J. Org. Chem. pp. 3815–3822.
- Matthews, J. M., McNally, J. J., Connolly, P. J., Xia, M., Zhu, B., Black, S., Chen, C., Hou, C., Liang, Y., Tang, Y. & Macielag, M. J. (2016). Bioorg. Med. Chem. Lett. 26, 5346–5349. [DOI] [PubMed]
- Murugavel, K., Amirthaganesan, S. & R. T. S. (2010). Chem. Heterocycl. C. 46, 302–306.
- Nakhai, A. & Bergman, J. (2009). Tetrahedron, 65, 2298–2306.
- Otwinowski, Z. & Minor, W. (1997). Methods in Enzymology, Vol. 276, Macromolecular Crystallography, Part A, edited by C. W. Carter Jr & R. M. Sweet, pp. 307–326. New York: Academic Press.
- Polo, E., Trilleras, J., Ramos, J., Galdámez, A., Quiroga, J. & Gutierrez, M. (2016). Molecules, 21, article number 903. [DOI] [PMC free article] [PubMed]
- Scala, A., Piperno, A., Risitano, F., Cirmi, S., Navarra, M. & Grassi, G. (2015). Mol. Divers. 19, 473–480. [DOI] [PubMed]
- Sheldrick, G. M. (2015). Acta Cryst C71, 3–8.
- Silva, V. L. M., Silva, A. M. S., Pinto, D. C. G. A. & Cavaleiro, J. (2006). Synlett, pp. 1369–1373.
- Song, H., Lee, H., Kim, J. & Park, S. B. (2012). ACS Comb. Sci. 14, 66–74. [DOI] [PubMed]
- Strakova, I., Kumpiņa, I., Rjabovs, V., Lugiņina, J., Belyakov, S. & Turks, M. (2011). Tetrahedron Asymmetry, 22, 728–739.
- Strakova, I., Turks, M. & Strakovs, A. (2009). Tetrahedron Lett. 50, 3046–3049.
- Turks, M., Strakova, I., Gorovojs, K., Belyakov, S., Piven, Y. A., Khlebnicova, T. S. & Lakhvich, F. A. (2012). Tetrahedron, 68, 6131–6140.
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
- Wiener, J. J. M., Gomez, L., Venkatesan, H., Santillán, A. Jr, Allison, B. D., Schwarz, K. L., Shinde, S., Tang, L., Hack, M. D., Morrow, B. J., Motley, S. T., Goldschmidt, R. M., Shaw, K. J., Jones, T. K. & Grice, C. A. (2007). Bioorg. Med. Chem. Lett. 17, 2718–2722. [DOI] [PubMed]
- Wu, Z.-W., Song, S.-Y., Li, L., Lu, H.-L., Lieberman, B., Huang, Y.-S. & Mach, R. H. (2015). Bioorg. Med. Chem. 23, 1463–1471. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) 1, 2, global. DOI: 10.1107/S205698901701667X/eb2002sup1.cif
Structure factors: contains datablock(s) 1. DOI: 10.1107/S205698901701667X/eb20021sup2.hkl
Supporting information file. DOI: 10.1107/S205698901701667X/eb20021sup4.mol
Structure factors: contains datablock(s) 2. DOI: 10.1107/S205698901701667X/eb20022sup3.hkl
Supporting information file. DOI: 10.1107/S205698901701667X/eb20022sup5.mol
Supporting information file. DOI: 10.1107/S205698901701667X/eb20021sup6.cml
Supporting information file. DOI: 10.1107/S205698901701667X/eb20022sup7.cml
Additional supporting information: crystallographic information; 3D view; checkCIF report