Abstract
Effective antiviral protection in multicellular organisms relies on both cell autonomous and systemic immunity. Systemic immunity mediates the spread of antiviral signals from infection sites to distant uninfected tissues. In arthropods, RNA interference (RNAi) is responsible for antiviral defense. Here we show that flies have a sophisticated systemic RNAi-based immunity mediated by macrophage-like haemocytes. Haemocytes take up dsRNA from infected cells and, through endogenous transposon reverse transcriptases, produce virus-derived complementary DNAs (vDNA). These vDNAs template de novo synthesis of secondary viral siRNAs (vsRNA) which are secreted in exosome-like vesicles. Strikingly, exosomes containing vsRNAs, purified from haemolymph of infected flies, confers passive protection against virus challenge in naïve animals. Thus, similar to vertebrates, insects use immune cells to generate immunological memory, in the form of stable vDNAs, that generate systemic immunity, which is mediated by the vsRNA-containing exosomes.
In brief
Drosophila, thought to lack an adaptive immune system, have dedicated immune cells that can alter their genetic repertoire to amplify and spread systemically in combat viral infection, providing protection to naive cells.

Introduction
Viruses are stealth invaders that hijack the cellular machinery of their host to replicate, leading to host dysfunction and often disease. To combat viral infection, multicellular organisms evolved mechanisms to limit replication of viral pathogens. In jawed vertebrates, a protein-based antiviral response produced by circulating immune cells spreads systemically through the organism to protect uninfected tissues. Once activated, immune cells can sustain production of specific antibodies and other antiviral effectors(Banchereau and Steinman, 1998). In addition, these immune cells provide a reservoir of immunological memory to target specific viral pathogens.
In plants and invertebrates, antiviral immunity involves a nucleic-acid based, sequence-specific mechanism, known as RNA interference (RNAi)(Harvey et al., 2011; Nayak et al., 2013). Immune specificity is mediated by small interfering RNAs (siRNAs) that guide an endonucleolytic RNA-Silencing Complex (RISC) to cognate viral transcripts. There are three types of siRNA: microRNA (miRNA), siRNA and piwi-interacting RNA (piRNAs)(Carthew and Sontheimer, 2009). Of these, miRNAs are endogenously expressed and mediate mostly host gene regulation, while piRNAs suppress transposons in the germline. In Drosophila, the siRNA pathway constitutes the prime antiviral defense mechanism (van Rij et al., 2006). RNAi is initiated upon recognition and cleavage of long double-stranded RNA by Dicer-2, an RNAse III family dsRNA endonuclease, into ~21-nucleotide (nt) long siRNA duplexes with characteristic 2 nt 3′ overhangs. These siRNAs are then loaded into the Argonaute-2 (Ago2)-containing RNA-induced silencing complex (RISC). Upon loading into RISC, one of the siRNA strands remains associated with Ago2. Base-pairing of the guide strand to a complementary target single stranded RNA leads to Ago2-mediated cleavage of the target. All these processes occur within infected cell, conferring cell-autonomous immunity. Whether antiviral RNAi signals produced in infected cells can also spread systemically has been a matter of debate. In nematodes and plants, organism-specific channels for systemic RNAi, known as plasmodesmata in plants (Cilia and Jackson, 2004), and SID in nematodes (Winston et al., 2002), allow siRNA intercellular trafficking. However, as they spread through the organism, RNAi molecules are expected to get diluted, thereby greatly limiting their range of action. To maintain their efficiency, systemic immune responses must rely on amplification of siRNAs (Carroll and Isenman, 2012; Sijen et al., 2001). Accordingly, plants and nematodes have RNA-dependent RNA polymerases (RdRp) that are recruited to RISC-targeted transcripts to synthesize secondary siRNAs (Himber et al., 2003; Sijen et al., 2001), thereby amplifying the RNAi signal and sustaining its systemic spread.
Unlike nematodes and plants, flies do not have endogenous RdRps, raising the question of whether there is antiviral RNAi amplification and spread in arthropods. This led to a proposed model where dsRNA diffusion mediates a short-range systemic RNAi response. Long dsRNA intermediates produced during viral replication and released by infected cells would be taken up by neighboring cells and processed into virus-derived siRNAs (vsRNAs), thereby priming their RNAi machinery against the invading virus. Supporting this model, effective antiviral immunity in Drosophila requires exogenous dsRNA uptake (Saleh et al., 2009; 2006). However, another feature of this model, namely that dsRNA diffusion limits systemic RNAi spread to local sites of infection, is inconsistent with observations that RNAi signals efficiently spread to distal tissues(Saleh et al., 2009). We thus considered whether distal spread of an RNAi antiviral response in Drosophila involves hitherto unrecognized amplification and distribution mechanisms.
Here we investigate this hypothesis and uncover that macrophage-like circulating immune cells in Drosophila, known as haemocytes, are key to the amplification and spread of an antiviral signal, even when they themselves are not infected by the virus. Haemocytes take up viral RNA from infected cells and copy them into viral cDNA templates through the action of endogenous transposon-encoded reverse transcriptases. These viral cDNA serve as a template for the synthesis of vsRNAs de novo, characterized by the production of a hitherto undescribed class of 5′-triphosphorylated vsRNAs. These vsRNAs are then incorporated and secreted into exosome-like vesicles (ELVs). The ELVs have antiviral activity, and can confer virus-specific immunity when injected into a naïve animal, which last several weeks after clearance of the virus. Indeed, the stable incorporation of virus-derived cDNAs into haemocytes provides the means for immunological memory. Thus, Drosophila evolved a sophisticated adaptive immune system with remarkable parallels to the protein-based antibody responses in higher vertebrates revealing general design principles of multicellular adaptive immunity.
Results
Circulating haemocytes are required for a systemic antiviral RNAi response
Systemic antiviral immunity may involve specialized immune tissues. The fly immune system is comprised of two tissues: the fat body and the haemocytes (Fauvarque and Williams, 2011). Haemocytes, circulating macrophage-like immune cells, have recently been shown to clear virally-infected cells by phagocytosis (Nainu et al., 2015). To assess their role in antiviral responses, we first examined the effect of haemocyte depletion on virus replication. To this end, we expressed the pro-apoptotic factor reaper (White et al., 1996) under the control of the haemocyte-specific enhancer of hemolectin(Sinenko and Mathey-Prevot, 2004), which resulted in a near complete haemocyte depletion (hml> reaper, Fig. 1A and S1A). Following infection with Sindbis virus (SINV), viral loads in hml>reaper flies were significantly higher compared to control flies (hml>dsCtr) (Fig. 1B, p < 0.05 Mann-Whitney-Wilcoxon test and Fig. S1B), suggesting that haemocytes are indeed involved in antiviral immunity.
Figure 1. Viral infection in Drosophila leads to an haemocyte-dependent immune response and virus-derived siRNAs (vsRNAs) de novo synthesis.
(A) Upper panel: Schematic of the Gal4 UAS system used to induce haemocyte-specific cell ablation (UAS-reaper) or knock-down of Ago2 (UAS-dsAgo2). Lower panel: eGFP expressing haemocytes are no longer detected in hml>GFP/reaper larvae.
(B) Hypersensitivity of haemocyte-deficient flies to Sindbis virus (SINV) infection. Normalized SINV titers measured by RT-qPCR in adult flies at 4 days post infection (d.p.i.) show increase viral replication in flies depleted of haemocytes (hml>reaper) or haemocyte-specific knock-down of Ago2 (hml>dsAgo2) compared to control line (hml>dsCtr) (n=4 independent experiments, Mann Whitney test p<0.05 compared to control flies).
(C) Haemocytes do not support detectable viral replication. Absolute SINV titers measured by RT-qPCR in carcasses versus haemocyte-enriched fraction show high level of viral replication in carcasses by no detectable level in haemocytes.
(D) Identification of 5′-triphosphorylated virus-derived siRNAs (vsRNA) in SINV-infected Drosophila by tripRNA-seq. The 100% stack bar graph shows the relative amounts of 5′-monophosphorylated (by pRNA-seq) and 5′-triphosphorylated (by tripRNA-seq) ends for the four main classes of siRNAs (miRNAs, piRNAs, esiRNAs and vsRNAs) measured at 4 d.p.i. Only vsRNAs present a high proportion of 5′-triphosphorylated ends. Above, the total number of reads for each class is given in reads per million (reads/106).
(E) 5′-triphosphorylated vsRNAs have similar size and strand orientation than classical vsRNAs. The size distribution plot shows the number of vsRNAs, in read per million, based on their size (from 18 to 30 nucleotides), orientation to the viral genome (sense or antisense) and 5′ mono (pRNA-seq) or tri-phosphorylated (tripRNA-seq) end.
We next examined whether the RNAi pathway within haemocytes is required for effective antiviral defense. We expressed a dsRNA hairpin directed against Ago2 under the control of the hemolectin enhancer to specifically deplete Ago2 in haemocytes (hml>dsAgo2). Surprisingly, downregulation of Ago2 (hml>dsAgo2) in haemocytes, which should not block phagocytosis of infected cells, also resulted in a significant increase of viral load (Fi. 1B, p < 0.05, Mann-Whitney-Wilcoxon test), suggesting that the Ago2 pathway of haemocytes is required for their antiviral activity. To test whether this related to an increase in viral replication within the Ago2-defficient haemocytes, we next determined viral RNA levels in isolated haemocytes. While the virus was abundantly detected in the carcasses, there was no detectable SINV RNA in haemocytes (Fig. 1C). We concluded that haemocytes, while not infected with SINV, are required for antiviral protection of the organism through a mechanism that involves the Ago2 pathway of haemocytes.
An amplification step involving de novo production of virus-derived small RNAs
Since haemocyte Ago2 was important for antiviral immunity, we asked whether haemocytes amplify or spread an antiviral RNAi response. To address this question, we next examined the small RNAs accumulating in SINV-infected flies. Our analysis distinguished between two types of virus-derived small RNAs (vsRNAs). Dicer processing of double stranded viral RNA replication intermediates produces small vsRNAs with a monophosphate group at their 5′ end (herein primary vsRNAs, gray in Fig. 1D, E and Fig. 2B). Primary vsRNAs are then used by Ago2-RNA Interference Silencing Complex (Ago2-RISC) to specifically target cognate viral RNA genomes. In plants and nematodes, RISC activation triggers the recruitment of RNA-dependent RNA polymerases to the RNA target for de novo synthesis of siRNAs. This amplification step of nematodes and plants, leads to production of secondary vsRNAs required for effective systemic immunity(Seo et al., 2013). Since secondary vsRNAs are produced by de novo synthesis, they have a 5′-triphosphate moiety (herein secondary vsRNAs, red in Fig. 1D, E and Fig. 2B)(Pak and Fire, 2007).
Figure 2. vsRNAs de novo synthesis occurs in haemocytes and is Ago2-dependent.
(A) Viral genome-wide production of secondary (5′-triphosphorylated) vsRNAs requires RNAi-competent haemocytes. Coverage of SINV genome by secondary vsRNAs (tripRNA-seq) in control (hml>dsCtr, solid lines), haemocyte-depleted flies (hml>rpr, thick dash lines) or flies with RNAi-deficient haemocytes (hml>dsAgo2, thin dash lines).
(B) Production of 5′-triphosphorylated vsRNAs is concentrated in haemocytes. 100% stack bar graphs show the relative amounts of 5′-monophosphorylated (pRNA-seq) and 5′-triphosphorylated (tripRNA-seq) miRNAs, piRNAs, esiRNAs and vsRNAs detected in carcasses versus haemocyte-enriched fractions. Of note, endosiRNAs (esiRNAs) and germline-specific piRNAs were not detected (n.d.) in the haemocyte fraction.
(C) siRNA de novo synthesis is specific to virally derived siRNAs and Ago2-dependent. 5′-mono and -triphosphorylated siRNAs were measured in SINV infected control flies (w1118, in black), homozygous ago2 mutant flies (ago2414, in grey), or in non-infected flies ubiquitously expressing eGFP and injected with dsRNA against eGFP (act5C>eGFP + dseGFP, in white). Relative abundance of SINV derived or eGFP-derived siRNA isolated by tripRNA-seq versus pRNA-seq is given as the log2 ratio normalized to the relative abundance of miRNAs in both sequencing approaches for each experiment (dash line indicates no enrichment relative to miRNA).
To assess if the RNAi response to virus infection in flies entails an amplification step, we tested the production of secondary vsRNAs, which should have a 5′-triphosphate moiety, in addition to the expected presence of primary vsRNAs, which should have a monophosphate 5′ moiety. We examined whether SINV infected flies accumulate secondary vsRNAs. Because 5′-triphosphate groups prevent linker ligation, conventional siRNA sequencing approaches (herein pRNA-seq) cannot detect 5′-triphosphorylated RNA species(Pak et al., 2012). To overcome this limitation, we created an alternative sequencing protocol to enable identification of secondary 5′-triphosphorylated vsRNA (herein tripRNA-seq). Briefly, small RNAs (approximately 18–30 nucleotides in length) were purified by polyacrylamide gel electrophoresis and treated with shrimp alkaline phosphatase (SAP), to remove 5′-monophosphates from small RNAs produced by endonuclease activity. The RNA preparation was then treated with Tobacco Acid Pyrophosphatase treatment (TAP) or RNA 5′ Pyrophosphohydrolase (RppH) to convert 5′-triphosphorylated RNAs into monophosphorylated RNAs that can be ligated to oligonucleotides for sequencing (STAR Methods and Fig. S2A). By comparing the spectrum of small vsRNAs identified by pRNA-seq and tripRNA-seq, we determined the presence and relative abundance of primary vs secondary vsRNAs. As expected for products of endonucleolytic processing (Carthew and Sontheimer, 2009; Han et al., 2015), the majority (95 to 99%) of miRNAs, piRNAs and esiRNAs, were detected only by pRNA-seq, but not by tripRNA-seq (Fig. 1D, compare gray with red bars and Table S1, p < 2.2e-16, Fisher exact test). Strikingly, approximately 50% of all vsRNAs accumulating in SINV-infected adult flies at 4 days post-infection (d.p.i.) were secondary vsRNAs (Fig. 1D, vsRNA, red bars). Specific enrichment of vsRNAs by tripRNA-seq was observed in 3 independent experiments with hml>eGFP flies (Table S2) and 3 experiments with wild type flies (w1118) (Table S1 and S4). We next analyzed the strand and size distribution of secondary and primary vsRNAs. Secondary 5′-triphosphorylated vsRNAs were similar to 5′-monophosphorylated vsRNAs in size (21 nt long) and corresponded to both sense and antisense viral RNA (Fig. 1E, compare gray and red bars). Of note, Calf Intestine Phosphatase (CIP) treatment prior to tripRNA-seq (CIP-smRNA-seq, Fig. S2B), which removes 5′ monophosphate and triphosphates but not 5′cap, completely abolished the detection of vsRNAs, indicating that vsRNAs are not capped. The finding that 5′-triphosphorylated secondary vsRNAs accumulate during viral infection is indicative of an amplification step that includes de novo RNA synthesis.
Secondary vsRNAs are synthesized in circulating haemocytes
We next examined whether haemocytes are required for the amplification step involving de novo production of secondary vsRNAs. To this end, we repeated the pRNA-seq and tripRNA-seq analyses in flies lacking haemocytes (hml>reaper) and in flies where the haemocytes lack Ago2 (hml>dsAgo2). Analysis of vsRNA production in SINV-infected flies showed that 5′-triphosphorylated secondary vsRNAs were significantly reduced in haemocyte-depleted flies (hml>reaper) compared to control flies (hml>dsCtr) (Fig. 2A and Table S2, p < 3.38 e-15). Down-regulation of Ago2 in haemocytes (hml>dsAgo2) also prevented accumulation of secondary vsRNAs (Fig. 2A and Table S2 p = 0.4901). These data suggested that secondary vsRNAs are produced in haemocytes in an Ago2-dependent manner. It thus appears that haemocytes participate in the amplification of the antiviral RNAi response through a de novo synthesis mechanism, that involves a RISC-associated processing event.
To further substantiate this conclusion, we extracted the haemolymph from wild type (w1118) flies and isolated haemocytes by low speed centrifugation (Fig S3A). Comparing small RNA species in the haemocyte fraction and the carcasses of infected flies showed that haemocytes contain the majority (>75%) of secondary vsRNAs produced during infection (Fig. 2B, and Table S3, Fisher exact test p < 2.2e-16, gray vs red bars in carcasses and haemocytes). Indeed, 5′-triphosphorylated secondary vsRNA constitute the most abundant siRNA species identified by tripRNA-seq in haemocytes. These observations support the idea that secondary vsRNAs are specifically produced in circulating haemocytes.
The role of Ago-2 in amplification of vsRNAs was further tested by examining production of secondary vsRNAs in Ago2 null mutant flies (ago2414)(Okamura et al., 2004). Following infection, ago2414 flies produced high amounts of primary siRNAs, as expected from the higher level of viral RNA replication (Table S4). In contrast, Ago2 null flies had undetectable amounts of secondary vsRNAs (Fig. 2C and Table S4, p = 0.1937 and p = 0,081 after miRNA normalization, Fisher exact test). This confirms that Ago2 is required for the amplification of the RNAi signal, which involves de novo vsRNA synthesis.
Since haemocytes are immune cells, we considered whether the amplification pathway is stimulated by viral infection. We examined whether de novo production of secondary siRNAs can be triggered without infection, by inoculation with exogenous dsRNA. We injected dsRNA directed against eGFP into non-infected flies expressing eGFP (act5C>eGFP). These dsRNAs gave rise to functional primary siRNAs against GFP, but not to secondary siRNAs, since no eGFP-derived 5′-triphosphorylated siRNAs were detected (Fig. 2C and Table S4, p = 0.8892 and p = 0,07635 after miRNA normalization, Fisher exact test). Thus, production of secondary siRNAs does not take place in the absence of infection. We conclude that haemocytes mediate an antiviral response that amplifies the RNAi signal through de novo synthesis of secondary vsRNAs in an Ago2-dependent mechanism.
Reverse transcription produces viral cDNAs as a template for secondary vsRNA synthesis
The finding that flies produce de novo secondary vsRNAs is surprising since endogenous RNA-dependent-RNA polymerases (RdRp), which mediate amplification in plants and nematodes, are absent in flies (Duan et al., 2010). We wondered whether the reported reverse transcription of viral RNA into complementary DNAs (vDNA) (Goic et al., 2013) could provide a template for de novo synthesis of secondary vsRNAs in haemocytes. To test this idea, we initially determined whether vDNA is produced within the haemocytes of SINV-infected flies (Fig. 3A). Indeed, we found that vDNA accumulates in the haemocytes of infected flies while being undetectable in the carcass (Fig. 3B, lanes 1–4). Inhibition of endogenous reverse-transcriptases by injection of azidothymidine (AZT) (Malki et al., 2014), abolished the production of SINV vDNA (Fig. 3B, lanes 5–8). Strikingly, genetic ablation of haemocytes in hml>reaper flies also eliminated accumulation of viral cDNA in the hemolymph fraction (Fig. 3B, lanes 9–10). This suggests that the process of vDNA synthesis occurs preferentially in haemocytes. To further demonstrate that haemocytes exhibit reverse transcriptase (RT) activity, we developed an assay derived from a previously described qPCR-based product-enhancement RT assay (Vermeire et al., 2012). Using haemocyte fraction lysate from hml>eGFP flies, we found that haemocytes harbor AZT-sensitive RT activity, specifically upon SINV infection (Fig. S3C). Thus, the hameocytes’ ability to produce vDNA appears to be directly linked to the presence of virus and further supports our observation that siRNA amplification is specific to the antiviral response (Figure 2C).
Figure 3. Stable viral cDNA (vDNA) production in haemocytes is an Ago2 and reverse transcriptase-dependent process, involved in vsRNA de novo synthesis.
(A) Experimental design to identify vDNA production in haemocytes and its role in vsRNA de novo synthesis.
(B) Production of vDNA in haemocytes. DNA was extracted from haemocyte-enriched fraction (Hem) or carcasses (Carc) of naïve (lanes 1, 2) or SINV-infected flies (lanes 3 to 11), treated with RNase A then assessed for SINV DNA product by end-point PCR. Crq gene was used as loading control. w1118 flies were used in lanes 1 to 8.
(C) Inhibition of vDNA production by AZT injection in flies reduces the production of primary (pRNA-seq) and secondary (tripRNA-seq) vsRNAs. Flies were injected with 2.5 nmoles AZT (red) or PBS only (grey) 1 day before and 1 day after SINV injection. Small RNAs were extracted at 4 d.p.i and subjected in parallel to pRNA-seq and tripRNA-seq. vsRNA counts given per 104 SINV genomes.
(D) AZT treatment increases SINV replication in an Ago2-dependent manner. Absolute SINV titers measured by RT-qPCR in wild type (w1118) or ago2 mutant (ago2414) flies treated with AZT (as in C) (n=3 injection experiments).
(E) vDNAs are retained specifically in haemocytes as a form of long lasting immunological memory. 3 weeks after SINV infection (+SINV), the haemocyte-enriched fraction (Hem) was isolated from flies expressing eGFP in their haemocytes (hml>eGFP). Half of the fraction was further processed by FACS sorting to obtain pure haemocytes samples (Hem FACS). Both preparations were treated with RNase A then assessed for SINV DNA product by end-point PCR. Crq gene was used as loading control.
(F) Model for the interactions between Ago2, reverse-transcriptase (endoRT), vDNA and vsRNA synthesis in haemocytes.
We next tested whether vDNAs produced in haemocytes serves as a template for de novo synthesis of secondary vsRNAs. If this were the case, inhibition of vDNA formation by AZT treatment should lead to a loss of secondary vsRNAs. Indeed, we found that AZT treated flies were defective for the production of secondary vsRNAs (Fig. 3C, 19.39 and 29.15 fold decrease compared to control for both primary and secondary vsRNAs respectively). Of note, the amount of vsRNAs cloned by pRNA-seq was also affected, albeit to a lesser extent, by AZT treatment. However, we could not detect any effect of AZT treatment on dsRNA-induced RNAi in cell culture (not shown, see Discussion). In addition, miRNA levels were not affected in either carcass or haemocyte fractions (Fig S3B). We conclude that, upon SINV infection, haemocytes reverse-transcribe the viral RNA to produce vDNA, which in turn is used as a template for transcription of secondary vsRNAs (Fig. 3F).
We next examined the role of Ago2 in vsRNA de novo synthesis in haemocytes. Since haemocytes do not support detectable Sindbis replication, one possibility is that these macrophage-like cells depend on Ago2 function to process viral RNA and facilitate vDNA synthesis. In principle, Ago2 could be required either prior to the vDNA synthesis step, or afterwards, to process the new vsRNA transcript. We found that haemocyte-specific knock-down (hml>dsAgo2) as well as organismal loss of function of Ago2 (ago2414) both prevented vDNA synthesis (Fig. 3B, lane 11–13). This result suggested that Ago2 is required for vDNA synthesis. The conclusion that vDNA synthesis requires Ago2 predicts that AZT treatment should increase viral replication in wild type flies but not in Ago2-deficient flies. Consistent with this prediction, AZT injection led to 3-fold higher SINV titers in SINV-infected wild type flies (w1118), but did not further increase virus replication in Ago2 mutant flies (ago2414) (Fig. 3D). Thus, AZT treatment per se does not enhance virus replication, but acts downstream of Ago2, through its inhibitory effect on vDNA synthesis, which is required for de novo vsRNA production.
The finding that production of virus-derived cDNA molecules within haemocytes serves to control viral infection raises the question of the persistence of this antiviral response. We thus considered whether vDNA is retained in haemocytes after the acute phase of infection. Haemocytes from SINV-infected hml>eGFP flies were isolated 3 weeks after infection and purified through FACS sorting of GFP positive cells. End-point PCR analysis showed that, even after this lengthy post-infection period, equivalent to a third of the lifetime of a fly, SINV-derived cDNA was detected in the haemocyte-enriched fraction and even more enriched in purified haemocytes (Fig. 3E). This suggests that vDNAs are not only a mechanism for amplification of RNAi signal, but also provide the means for bona fide immunological memory that lasts for a significant part of the fly’s lifespan.
Exogenous viral dsRNA stimulates vDNA and secondary vsRNA production
Since haemocytes are not efficiently infected by SINV, we were puzzled by what act as a template for synthesis of viral cDNA in haemocytes. Previous results indicate that inoculation of naked long RNA can induce systemic immunity and protect flies from infection (Saleh et al., 2009). However, not all Drosophila cell lines can efficiently take up dsRNA in vitro (Zhou et al., 2013; 2014). To identify what cells are able to take up dsRNA in vivo in the intact animal, we injected 50 ng of anti-eGFP dsRNA (dseGFP) into flies ubiquitously expressing eGFP (hsp70>eGFP). This treatment was ineffective at silencing eGFP in the majority of tissues (Fig. S4A), suggesting that only some cell type can take up dsRNA to initiate RNAi. In contrast, eGFP expression in the haemocytes of (hml>eGFP) was efficiently inhibited by injection of just 1 ng dseGFP (Fig. 4A). This inhibition was long lasting and was observed to last for up to 10 days (Fig. 4A, lower panel). Thus, unlike most tissues, the phagocytic haemocytes are specialized to efficiently take up dsRNA to initiate RNAi.
Figure 4. Uptake of exogenous dsRNA by haemocytes stimulates vDNA production.
(A) Haemocytes can efficiently take up dsRNA to silence an endogenously expressed reporter gene (eGFP) in vivo. 50 ng of control dsRNA (dsCTR) or anti-eGFP dsRNA (dsGFP) were injected in flies expressing eGFP in the haemocytes (hml>eGFP). eGFP expression was monitored directly by fluorescence microscopy at 5 days post injection (d.p.i.) (i. top panel) or by western blot at 0, 5, 10 and 20 d.p.i.(ii. lower panel).
(B) Loss of RNAi-competent haemocytes inhibits antiviral protection induced by dsRNA immunization. Control (dsCTR) or anti-SINV (dsSIN) dsRNA were co-injected with 500 pfu SINV in control flies (hml>dsCtr), haemocyte-depleted flies (hml>reaper) or flies with RNAi-deficient haemocytes (hml>dsAgo2). dsSIN protection efficiency was measured by RT-qPCR as the ratio of SINV titers in dsSIN versus dsCTR treated flies at 3 and 4 d.p.i (n = 3 injection experiments).
(C) anti-SINV dsRNA injection increases production of SINV vDNA in haemocytes. SINV vDNA production was assessed by end point PCR against SINV NSP1 gene (insert) and by qPCR in haemocytes of SINV infected flies co-injected with control (dsCtr) or anti-SINV (dsSIN) dsRNA. qPCR analysis shows that vDNA production from genes in both the viral genomic region (NSP1, in red) or the viral subgenomic region (capsid, in grey) was equally stimulated by dsSIN injection.
(D) Model for stimulation of vDNA formation in haemocytes by exogenous dsRNA. Uptake of viral genomic RNA (single stranded and/or double-stranded RNA) in haemocytes stimulates vDNA production which allows increased de novo synthesis of vsRNAs and efficient systemic antiviral RNAi response.
In vertebrates, phagocytic immune cells take up antigens from infected cells to produce antibodies and prime T-cells. The above results led us to hypothesize that haemocytes take up viral dsRNA to produce viral cDNA and secondary vsRNAs to initiate systemic RNAi response. To test this idea, we examined the effect of exogenous dsRNA inoculation in flies depleted of haemocytes (hml>reaper) or with haemocytes deficient in Ago2 (hml>dsAgo2). Injection of dsRNA directed against SINV (dsSIN) resulted in almost 6 orders of magnitude viral load reduction compare to control non-specific dsRNA (dsCtr). However, the antiviral effect of exogenous dsSIN was significantly reduced in flies depleted of haemocytes (hml>reaper) or if the expression of Ago2 in haemocytes was down regulated by dsRNA (hml>dsAgo2) (Fig 4B, p < 0.001). Importantly, flies injected with dsSIN produced significantly more SINV vDNA compared to control dsRNA inoculation (Fig. 4C, inset: end point PCR, bar graphic: qPCR). This indicates that Drosophila haemocytes take up viral dsRNA to stimulate synthesis of vDNA in an Ago2-dependent manner (Fig. 3B, 4B and D). To further assess the role of dsRNA uptake in antiviral RNAi amplification, we injected SINV-infected flies with a control dsRNA (dsCtr) or anti-SINV dsRNA (dsSIN) containing several mismatches in order to identify vsRNA derived from the injected dsRNA from vsRNA derived from the viral genome. Although the mutated dsSIN was less efficient at silencing SINV replication (Fig. S4B), it did lead to an increase number of secondary vsRNAs compare to dsCtr treated flies (Fig. S4C, p < 5.37e-6, Fisher exact test). We concluded that during infection, haemocytes can convert viral dsRNA into vDNAs that serves as a template for secondary vsRNA production, which mediate systemic antiviral activity.
Haemocytes secrete secondary vsRNAs in exosome-like vesicles
Once secondary vsRNAs are produced in haemocytes, how are they delivered to uninfected tissues to control infection? Simple secretion of siRNAs is unlikely to be effective, as naked siRNAs are labile and not efficiently taken up by Drosophila cells (Saleh et al., 2006) or by tissues in intact flies (Fig. S5A). Since extracellular vesicles known as exosomes have been shown to contain miRNAs (Turturici et al., 2014), we considered whether exosomes could facilitate the systemic spread of secondary vsRNAs produced in haemocytes. Exosomes originate from multivesicular bodies (MVB), where their formation requires the small GTPase, Rab11, and their secretion involves the membrane fusion component Syntaxin 1A (Syx1A) (Beckett et al., 2013; Koles et al., 2012). We reasoned that if exosomes produced by haemocytes participate in antiviral immunity, hameocyte-specific down-regulation of Rab11 or Syx1A (hml>dsRab11 or hml>dsSyn1A) should disrupt antiviral immunity and increase SINV replication. This was indeed the case, as SINV viral titers were significantly higher in hml>dsRab11 and hml>dsSyn1A compared to control flies (hml>dsCtr) (Fig. 5A, p < 0.05 Mann-Whitney-Wilcoxon test). Of note, a previous genome-wide RNAi screen did not find Rab11 and Syx1A deficiencies to prevent or enhance SINV-infection (Panda et al., 2013). Furthermore, haemocytes do not support robust SINV replication. We thus concluded that the effect of Rab11 and Syx1A haemocyte-specific knock-down on SINV titers results from a defect in the immune response.
Figure 5. Haemocytes secrete vsRNAs in exosome-like vesicles.
(A) Tissue-specific knockdown of Rab11 (hml>dsRab11) or Syntaxin 1A (hml>dsSyx1A) in haemocytes leads to higher organismal viral titers compared to control flies (hml>dsCtr). Flies were injected with 500 pfu SINV and virus production in whole flies was measured after 4 days by RT-qPCR (n=4 independent experiments, Mann Whitney test, p<0.05).
(B) Loss of exosome secretion in haemocytes inhibits antiviral protection induced by dsRNA immunization. Control (dsCTR) or anti-SINV (dsSIN) dsRNA were co-injected with 500 pfu SINV in control flies (hml>dsCtr), or flies with haemocytes deficient for exosome formation (hml>dsRab11) or secretion (hml>dsSyx1A). dsSIN immunization efficiency was measured by RT-qPCR as the ratio of SINV titers in dsSIN versus dsCTR treated flies at 3 and 4 d.p.i.
(C) Strand and size distribution of vsRNAs found in exosomes of SINV-infected control flies (hml>eGFP) or flies with rab11 knockdown haemocytes (hml>dsRab11). Exosomes from SINV-infected flies were treated with RNase A (RNase A) prior to RNA extraction and cloning for pRNA-seq analysis.
(D) vsRNA secretion into exosomes requires functional RNAi machinery, viral DNA production and exosome secretion in haemocytes. Exosomal vsRNA content was measured by pRNA-seq at 4 d.p.i from SINV infected hml>eGFP, hml>dsRab11, hml>dsSyx1A, hml>dsAgo2 flies, and in wild-type flies co-treated with PBS (w1118 – AZT) or 2.5 nmoles AZT (w1118 +AZT, red). Single asterisks represent p<0.0001 compared to hml>dsCtr. Double asterisks represent p<0.001 compared to -AZT control.
(E) Model for vsRNA secretion by haemocytes into Exosome-Like Vesicles (ELVs).
We further tested this idea by direct inoculation of dsRNA. Anti-SINV dsRNA led to a strong antiviral response in control flies (hml>Ctrl). However, its antiviral effect was abrogated in flies where Rab11 was specifically down-regulated in haemocytes (Fig. 5B, compare hml>Rab11 with control hml>Ctrl). This experiment demonstrated that the organismal antiviral protection observed upon systemic injection of exogenous dsRNA is dependent on the ability of haemocytes to secrete exosomes.
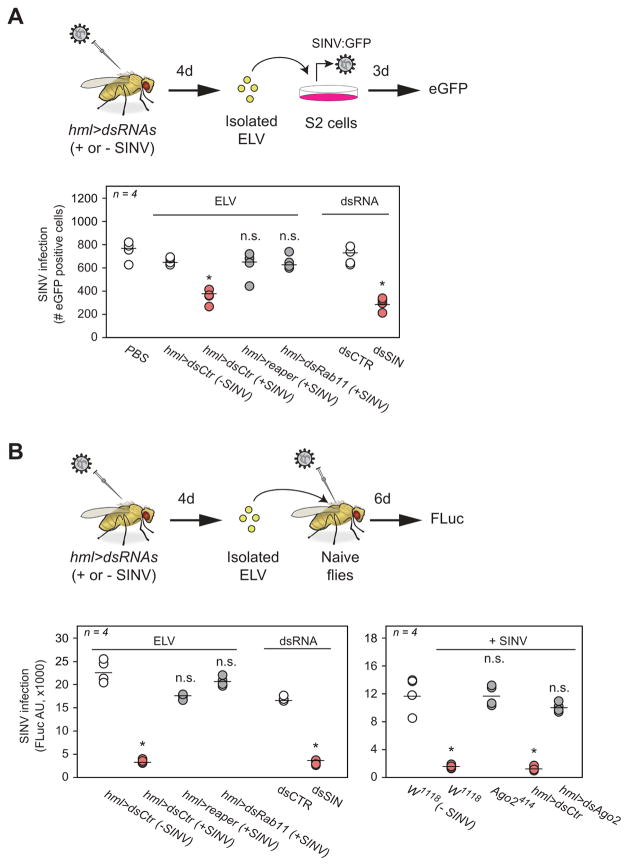
A prediction from this conclusion is that virus-infected flies should have circulating ELVs containing vsRNAs. We isolated exosomes from the haemolymph of SINV-infected flies by two independent methods (ultracentrifugation and exosome precipitation, see Methods). These purification protocols yield exosome-like vesicles (ELV) based on their size and protein content (Fig S5 B and C). ELVs should protect their RNA content from RNase digestion, but detergent treatment should disrupt their membrane and release the RNAs to be digested. We thus examined the RNA content of these ELVs after RNAse A treatment in the presence or absence of detergent. These experiments demonstrated that ELVs contained within them 21 nt long small RNAs (Fig. S5D), the size expected for vsRNAs. We then examined whether the ELVs secreted by haemocytes of virally infected flies contained SINV-specific vsRNAs. Isolation and sequencing of the small RNA content of ELVs isolated from the haemolymph of SINV-infected control flies (hml>dsCtr) showed that these ELVs contained a high proportion of SINV-specific vsRNAs (Fig. 5C). In contrast, haemocyte-specific knock-down of Rab11 and Syx1A (hml>dsRab11, hml>dsSyx1A) caused a significant reduction in the amount of vsRNAs found in ELVs (Fig. 5C and D, Fisher Exact Test, p < 0.0001 and Fig. S5E). Although the vsRNAs contained in ELVs are detected by pRNA-seq and not tripRNA-seq, they present similar characteristics to secondary vsRNAs. Firstly, they are 21 nt in length; secondly, they are depleted upon downregulation of Ago2 in haemocytes (hml>dsAgo2)(Fig. 5D, Fisher Exact Test, p < 0.0001) and thirdly, AZT treatment, which prevents formation of vDNA, significantly reduces exosomal vsRNA content compared to controls (Fig. 5D, Fisher Exact Test p < 0.001 for AZT treatment). Of note, loading of siRNAs into haemocyte-derived ELVs appeared to be specific to vsRNA, as we could not detect any small RNAs derived from a hairpin RNA expressed in the haemocytes (Fig. S5F). We conclude that haemocyte-derived ELVs are loaded with virus-specific secondary vsRNAs produced in an Ago2 and RT dependent manner.
Haemocyte-derived exosome-like vesicles confer passive antiviral immunity
The antiviral pathway emerging from our data so far indicates that upon viral infection haemocytes take up exogenous dsRNA to produce vDNA. This vDNA is used as a template to synthesize secondary vsRNAs, which are secreted within exosomes into the haemolymph. Abrogation of Ago2 in haemocytes, vDNA synthesis or secretion of exosomes all lead to enhanced susceptibility to infection. We thus hypothesize that haemocyte-derived ELVs mediate the delivery of antiviral vsRNAs to uninfected tissues and protect flies from infection. This hypothesis predicts that ELVs secreted from the haemocytes of SINV-infected flies are themselves antiviral. We initially tested this prediction in cell culture. ELVs were purified from uninfected (−SINV) or SINV-infected control flies (hml>dsCtr, +SINV) and added to cultured Drosophila S2 cells infected with SINV:eGFP. As a positive control, S2 cells were treated with control (dsCTR) or anti-SINV long dsRNA (dsSIN) previously shown to be efficiently taken up by S2 cells to initiate RNAi (Fig. 6A and (Saleh et al., 2006)). Strikingly, ELVs purified from SINV-infected flies reduced SINV replication to similar levels to those achieved with the anti-SINV dsRNA (Fig. 6A). ELVs from uninfected flies had no protective effect, suggesting the ELVs from infected flies contain an anti-SINV factor generated during infection. We next tested if the antiviral effect of ELVs from SINV-infected flies was dependent on the presence of haemocytes capable of exosome secretion. We thus purified ELVs from SINV-infected flies with either no haemocytes (hml>reaper, +SINV) or whose haemocytes were deficient for exosome secretion (hml>dsRab11, +SINV). Importantly, none of these flies produced ELVs with antiviral activity (Fig. 6A, p < 0.05). We concludeds that ELVs produced by the haemocytes of infected flies are antiviral, and can by themselves block viral replication in cultured Drosophila cells.
Figure 6. Exosome-Like Vesicles (ELVs) secreted by the haemocytes of SINV-infected flies can suppress SINV replication in cell culture and adult flies.
(A) ELVs from SINV-infected flies can suppress SINV replication in cell culture. PBS only or ELVs from naïve (hml>dsCtr −SINV) or SINV-infected control flies (hml>dsCtr +SINV), SINV-infected haemocyte-depleted flies (hml>reaper) or SINV-infected flies with rab11 knockdown haemocytes (hml>dsRab11) were added to the culture media of S2 cells transfected with a SINeGFP replicon. SINeGFP infection was monitored by the number of eGFP positive cells. Only ELVs from SINV-infected control flies (hml>dsCtr, red) and dsRNA against SINV (dsSIN, red) suppressed SINeGFP infection. Asterisks represent p<0.01 compared to SINV-infected control flies or control dsRNA for ELVs and dsRNA respectively.
(B) ELVs from SINV-infected flies can suppress SINV replication in flies.
Left panel shows SINV replication assessed by Fluciferase (FLuc) activity in control w1118 flies co-injected with 100 pfu SINV:Fluciferase (SINFluc) and ELVs from naïve (hml>eGFP Naive) or SINV-infected control flies (hml>eGFP), SINV-infected flies with rab11 knockdown haemocytes (hml>dsRab11) or SINV-infected haemocyte-depleted flies (hml>reaper) or with control (dsCTR) or anti-SINV (dsSIN) dsRNA. Right panel shows SINV replication assessed by FLuc activity in control w1118 flies co-injected with 100 pfu SINFluc and ELVs from naïve (w1118 −SINV) or SINV-infected control (w1118 ) or ago2 mutant (Ago2414) flies, or with ELVs from SINV-infected control flies (hml>eGFP), or SINV-infected flies with ago2 knockdown haemocytes (hml>dsAgo2). Asterisks represent p<0.05 (Mann Whitney test) compared to naive control flies or control dsRNA for ELVs and dsRNA respectively. Values not significantly different from control are labeled as n.s.
To further examine the systemic antiviral potential of haemocyte-derived ELVs, we next examined whether ELVs purified from SINV-infected flies could passively protect naïve flies from SINV infection. Adult female flies (w1118) were co-injected with a firefly luciferase expressing SINV-reporter virus and ELVs isolated from naïve or SINV-infected flies (hml>dsCtr). Co-injection of ELVs isolated from SINV-infected flies, hml>dsCtr (+SINV), significantly inhibited SINV replication compare to ELVs obtained from uninfected donor flies, hml>dsCtr (−SINV) (Fig. 6B, p < 0.05, Mann Whitney test). Inhibition of virus replication by exosome inoculation was comparable to injection of long naked dsRNA (Fig. 6B, dsSIN and dsCTR), which has been shown to efficiently protect flies from infection (Saleh et al., 2009). The protective effect was dependent on haemocytes having functional Ago2 and Rab11, since ELVs preparations from either hml>dsRab11, hml>reaper, hml>Ago2 or ago2414 mutant flies were unable to inhibit virus replication (Fig. 6B). To confirm that ELVs antiviral activity was mediated by their vsRNA content, we tested whether ELVs could induce passive immunity in RNAi deficient flies (ago2414). We found that ELVs isolated from SINV-infected wild type flies (w1118) did not protect ago2414 mutant flies (Fig. S6A), indicating that ELVs antiviral function is mediated by transmission of an antiviral RNAi signal. In addition, end point PCR and RT-qPCR analyses of ELVs from SINV-infected flies failed to detect any viral DNA nor viral RNA genome (Fig. S6B and C). These observations strongly suggest that transmission of antiviral RNAi signals by ELVs is mediated by small RNAs. Remarkably, the ELV antiviral protection of naïve flies was still effective when ELVs were isolated 2 weeks after SINV inoculation (Fig. 7A). Thus, the immunity mediated by ELVs persists long after the onset of viral infection.
Figure 7. ELV’s antiviral activity is long lasting and virus-specific.
(A) Immunological Memory in antiviral ELVs. ELVs from naïve (−SINV) or SINV-infected (+SINV) control flies (w1118 ) were extracted 14 days post infection and co-injected with 100 pfu SINV:Fluciferase (SINFluc) in new control (w1118) flies. SINV replication was assessed by FLuc activity 4 days later. Asterisk represents p<0.05 (Mann Whitney test) compared to naïve control flies.
(B) ELVs antiviral activity is virus-specific. ELVs from naïve (−SINV) or SINV-infected (+SINV) control flies (w1118 ) were extracted 4 days post infection and co-injected with 20 TCID50 Drosophila C virus (DCV) in new control (w1118) flies. Asterisk represents p<0.05 (Mann Whitney test) (n = 4 ). DCV replication was assessed by RT-qPCR 4 days later. Values not significantly different are labeled as n.s (n = 3).
The model supported by our data proposed that vsRNAs in ELVs mediate an adaptive antiviral immune response. If this were the case, exosomes purified from SINV-infected flies should have no antiviral activity against an unrelated virus. To test the specificity of the ELV-mediated immunity, we injected ELVs isolated from SINV-infected flies (hml>dsCtr) into flies infected with Drosophila C virus (DCV). We found that ELVs from SINV-infected flies did not protect against infection with another RNA virus, DCV (Fig. 7B), indicating that the ELV-mediated antiviral activity is virus-specific. This supports the model whereby haemocytes from SINV-infected flies secrete to the haemolymph circulating ELVs loaded with virus specific vsRNAs which confer systemic protection to uninfected tissues (Fig. 7C).
Discussion
This study describes an adaptive immune system in Drosophila that relies on amplification and systemic spread of RNAi-based immunity. This pathway involves uptake of viral RNA by haemocytes, followed by their reverse transcription to vDNAs that both amplify the signal by de novo synthesis of siRNAs and confer immunological memory that can last at least for a third of the lifetime of the flies. Antiviral RNAi amplification is characterized by the production of 5′-triphosphorylated secondary vsRNAs, that increases the pool of vsRNAs in haemocytes and fuels their loading into exosome-like vesicles (ELVs) which act as agents of systemic and virus-specific antiviral protection. In addition, the active haematopoeitic process recently discovered in Drosophila adults that generates new haemocytes (Ghosh et al., 2015), could further amplify the immune response and extend the immunological memory by producing new haemocytes carrying virus-derived cDNA.
We show that blocking RNAi amplification by AZT treatment or Ago2-depletion in haemocytes as well as impairment of ELV secretion in haemocytes, all enhance virus replication. Therefore, this pathway appears to be central to the Drosophila immune response against viruses. Our findings indicate that, unlike in plants and nematodes, Drosophila siRNA amplification depends on reverse-transcription of viral RNA sequences into DNA by endogenous retrotransposons, specifically in haemocytes. Such production of viral DNA is required for secondary vsRNA synthesis in vivo, suggesting that antiviral siRNA amplification relies on the transcription of viral DNA products into new vsRNAs. This may serve to create a sustained immune response, akin to the immunological memory observed in vertebrates. Future studies should clarify the mechanisms of vDNA synthesis and persistence in haemocytes, as well as the pathways that produce secondary vsRNAs. Because haemocytes contain most of the vDNA, support secondary vsRNA production and secrete most of the vsRNAs to protect the entire organism, these pathways must involve haemocyte-specific factors. Furthermore, we observed that haemocytes accumulate very low amount of primary viral dicing products compared to the rest of the organism (not shown). We also found that AZT treatment decreases the number of 5′-mono and triphosphorylated vsRNAs, cloned by pRNA-seq and tripRNA-seq respectively (Fig. 3C), while it does not affect dsRNA-induced RNAi (not shown). Taken together, these results suggest that viral DNA derived-transcripts may not be diced cis-natural antisense transcripts (cis-NATs), but rather processed as single stranded RNAs into a series of secondary vsRNAs. We speculate that they may be generated in a mechanism similar to anti-transposon piRNA biogenesis in the ovaries where long piRNA precursors are cleaved by endonuclease activity to generate a series of 5′-monophosphorylated small RNAs that are trimmed down to yield mature piRNAs of the correct length (Han et al., 2015). However, in Drosophila the well-defined length of 21 nucleotides observed for secondary vsRNA likely stems from a more precise trimming mechanism
Our data shows that that the core effector of RISC, Ago-2, is required for the production of vDNAs and secondary vsRNAs. Furthermore, priming the RNAi machinery with exogenous dsRNA enhances synthesis of vDNA. The most parsimonious interpretation of these results is that haemocytes use their siRNA machinery (Dicer2 and Ago2) to initiate vDNA synthesis and siRNA amplification following exogenous uptake of viral dsRNA. It is tempting to speculate that Ago2 functionally interacts with endogenous retrotransposon replication complexes and in this way, provides sequence specificity by siRNA target recognition of viral sequences, which may facilitate discrimination between host and viral RNA templates used for the production of cDNA. In addition, we found that viral infection triggers the reverse-transcriptase activity in haemocytes (Fig. S3C). We thus speculate the existence of two non-mutually exclusive mechanisms, where sensing of Ago2 activity itself, like viral infection, can trigger the RT activity in haemocytes. This observation highlights the complex relationship between virus sensors and antiviral response in insects.
A striking conclusion of our study is that circulating phagocytic haemocytes are the main tissue for systemic immunity through vsRNA de novo synthesis and spread. This link between viral DNA production and vsRNA spread points to the existence of a functional immunological memory in Drosophila, as evidenced by the persistence of viral DNA in haemocytes and the presence of circulating antiviral ELVs several weeks after infection (Fig. 3E and 7A).
From an evolutionary and organismal fitness context, haemocyte functional specialization in siRNA amplification could present a number of advantages. First, by confining de novo vsRNA synthesis in immune cells, the organism limits the cellular stress of producing virus-derived DNA and RNA to a subset of cells. In addition, whether the chimeric viral DNA elements exist as episomes or are integrated into the host genome, the ability to create a DNA-based template for an antiviral response may allow for sustained immunological memory without compromising the genome or function of other cells in the organism. The functional specialization of haemocyte in viral DNA production could restrict the potential deleterious effects (e.g.: mutagenic) of creating exogenously-derived DNA elements, to a non-essential tissue.
Our results establish for the first time that in infected Drosophila, immune cells use secretion of vsRNA-loaded ELVs as an adaptive antiviral response. Of note, siRNA loading into ELVs appears to be specific to vsRNAs, since siRNAs derived from a hairpin expressed in hameocytes were not detected in circulating ELVs (Fig. S4F). Remarkably, haemocytes are not efficiently infected with SINV, indicating they must have mechanisms to take up viral RNA from infected cells. Haemocytes’ role in scavenging pathogens and their ability to capture dsRNA must be key to amplify and spread antiviral RNAi signals essential for systemic antiviral RNAi immunity. Interestingly, in Drosophila, haemocytes have recently been shown to remove virally infected cells by phagocytosis (Nainu et al., 2015), making them perfectly suited for detecting circulating viruses and their cognate dsRNA. We suggest that haemocytes target and engulf virally infected cells, thereby facilitating their viral RNA acquisition and targeting to produce antiviral RNAi amplification and spread, in a process akin to cross-priming.
This study uncovers striking parallels between insect and mammalian adaptive immunity, albeit based on completely different molecular and cellular mechanisms. In Drosophila, a complex acquired antiviral immunity response relies on dedicated immune cells that alter their genetic repertoire to amplify and spread systemically a specific antiviral signal. In mammals, the ability to create an adaptive immune repertoire depends on endogenous B- and T-cell recombinase-activating gene (RAG-1 and -2)(Bednarski and Sleckman, 2012), while in insects reverse transcription and transposons could serve as acquired immunity driving forces.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Raul Andino (raul.andino@ucsf.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Fly Stocks
The following fly lines were used: w1118 (wild type), Ago-2414 (Okamura et al., 2004), w1118; P{w[+mC]=Hml-GAL4.Delta}2, P{w[+mC]=UAS-2xEGFP}AH2 (hml>eGFP), y1 v1; P{TRiP.JF02812}attP2 (UAS-dsRab11), y1 v1; P{TRiP.JF01355}attP2 (UAS-dsFluc), w1118; P{UAS-rpr.C}14 (UAS-reaper), y1 v1; P{TRiP.JF01829}attP2 (UAS-dsSyx1A), y1 sc* v1; P{TRiP.HMS00108}attP2 (dsAgo2). All UAS-RNAi lines and the UAS-reaper line were obtained from the Bloomington Drosophila Stock Center. hml>eGFP flies were crossed to the different UAS-RNAi lines and the heterozygous adult F1 progenies were used for experiments. hml>eGFP control flies used in the different experiments were heterozygous, obtained from crossing the above hml>eGFP parental line with w[1118] flies. hml>reaper were generated by crossing between hml>eGFP flies (BL#30140) and UAS-reaper flies (w[1118]; P{w[+mC]=UAS-rpr.C}14).
To be consistent with our previous studies of antiviral RNAi response (Saleh et al., 2009; van Rij et al., 2006) and to avoid heterogeneity in viral load due to natural size difference between adult fly males and females, only adult female flies were used in this study. For each experiment, 2 to 6 day old adult female flies were collected and pooled by strains before being subdivided into individual vials (40 flies per vial for large injection experiment or 10–15 flies per vial for dsRNA or exosome immunization experiments, see Method Details below). Only flies that were not alive at the time of collection after injection were not included and processed further.
Statistical analyses of the changes in viral titers between conditions were conducted with n = 4 independent experiments (e.g.: performed with different biological samples collected and treated on different days), which is the smaller sample size allowing two-tailed (e.g.: no assumption on the direction of change in titers) Mann-Whitney-Wilcoxon test (with a significance ∝ = 0.05).
All fly stocks were checked for wolbachia infection by PCR (Teixeira et al., 2008). Wolbachia positive stocks were treated with 0.05mg/mL tetracycline in instant drosophila medium (Carolina Biological Supply Company) for two generations, as described in (Teixeira et al., 2008), before being used in any experiment.
Viruses and Cells
Original Sindbis stock (Strauss et al., 1984) (Strauss et al., 1984)and recombinant Sindbis virus expressing GFP were obtained as described in (Saleh et al., 2009) identified by constructs generated by(Hahn et al., 1992). Briefly, pTE3’2J:SINV:eGFP was digested with XhoI. After DNA clean up (Nucleospin Gel and PCR clean-up, Macherey-Nagel), capped viral RNA was synthesized using mMESSAGE SP6 kit (Ambion) and transfected in BHK cells. P0 viral stock was collected at 24 hours post transfection and propagated in BHK cells to generated the P1 stock at 24 hours post infection. P1 stock used for fly injection, was tittered in BHK cells by plaque assay. Similarly, SIN:Fluc was generated by cloning Firefly Luciferase into the XbaI site of the double subgenomic Sindbis vector pTE3’2J.
DCV stocks were prepared on low-passage S2 cells and tittered by end-point dilution. Briefly, 25,000 S2 cells per well in a 96-well plate were inoculated with 10-fold dilutions of viral stocks. Cells were transferred to fresh medium at day 7, and cytopathic effect (CPE) was monitored visually over 14 d. Viral titers (in TCID50/mL, where tissue culture infectious dose is the virus dilution where 50% of the wells are infected) were calculated according to the Spearman-Karber formula: log10 50% end point dilution = −(x0 − d/2 + dΣri/ni), where x0 = log10 of the reciprocal of the highest dilution (lowest concentration) at which cells are dead (100% CPE) in all wells; d = log10 of the dilution factor; ni = number of wells used in each individual dilution; ri = number of wells with 100% CPE (out of ni); summation starts at dilution x0.
BHK cells were maintained in Dulbecco’s modified Eagle’s medium: Nutrient mixture F12 (DMEM/F12, UCSF Cell Culture Facility) supplemented with 10% fetal bovine serum (FBS) and 1X Penicillin/Streptomycin (Invitrogen) at 37°C with 5% CO2.
METHOD DETAILS
Injections and Preparation of Biological Samples
Fly Injection
Flies were reared on standard medium at 25°C. w1118 flies, hml>eGFP or hml>dsFluc, described above, were used as controls. Two to five day old female flies were injected with 50 nL of the appropriate virus dilution in 10 mM Tris (pH 7.5) using a nanoinjector (Nanoject II, Drummond Scientific).
For deep sequencing experiments, 180–200 flies per genotypic group were injected. For dsRNA or exosome immunization experiments, groups of 10–15 flies were injected.
At the indicated time points, flies were harvested and directly processed to extract total RNA using Trizol reagent (Ambion).
dsRNA Preparation and Injection
PCR products corresponding to SINV nucleotides 5851 to 6430, eGFP nucleotides 257 to 456, were used as template for in vitro transcription using T7 RNA-polymerase to generate the corresponding dsRNA. For dsRNA constructs used in deep sequencing experiments, gene fragments of SINV(5851–6430) and eGFP(257–456) regions containing point mutation every 40 bases (gBlock Gene Fragment, IDT) were used as in vitro transcription template. 1.5 nanogram dsRNA were injected per fly.
Exosome-Like Vesicles Injection
Exosome-like vesicles obtained by precipitation were quantified for protein content by Sypro Ruby (Invitrogen) on gel staining and confirmed by Nanodrop. For each experiment, ELVs were isolated from 50–80 flies, resuspended in PBS and filtered (see Haemocytes and Exosomes Isolation). ELV protein content was measured after filtration, and the relative amount of ELVs per fly was calculated as followed: (ELV prep protein content in ng/uL) x (Final volume after filtration in uL)/number of flies. Based on this calculation, the amount of ELV equivalent to 10 ng of ELV protein content (used for each injection) was found to represent about the quarter of the amount of ELV proteins extracted per fly.)
Exosomes were treated with 5 μg RNase A for 15 minutes at 37°C prior to injection, to eliminate potential dsRNA contamination. 200 ng/μL of exosomes were injected per fly (which corresponds to the exosomal protein content extracted from ~0.25 fly).
Haemocytes and Exosomes Isolation
Haemocyte samples were obtained from at least 80 adult female flies. For the entire procedure, samples were kept on ice and all centrifugations performed at 4°C. After a quick wash in cold 80% ethanol, flies were washed 3 times in cold PBS, transferred onto a sheet of parafilm in a drop of cold PBS (350 μL per 40 flies), and quickly wounded with a razor blade (avoiding slicing of the flies). Flies were then transferred to a 0.5 mL tube previously punctured with a 26G1/2 needle and placed into a 1.5 mL tube and spun at 800g 3 minutes. Centrifugation was performed twice, the ”bled” flies in the punctured 0.5 mL tubes were collected as the “Carcass” fraction. The flow through supernatant collected in the 1.5 mL was transferred to a new tube and spun 10 minutes at 100g to remove tissue contaminants. Haemocytes were isolated from the supernatants by two centrifugation cycles of 10 minutes at 800g. The pellets (P1) from the two 800g centrifugations were pooled and processed for RNA extraction with Trizol LS reagent (Ambion) or for DNA extraction (Nucleospin Tissue, Macherey-Nagel). Supernatants from the previous centrifugations (S1) were spun at 20,000g for 30 minutes. Supernatants (S20) were used to further isolate exosomes either by ultracentrifugation or by precipitation.
Exosome isolation by ultracentrifugation
S20 were transferred to thick wall tubes for TLA120 fixed-angle rotor (Beckman) and spun at 100,000g, 70 minutes. Pellets, which contain the exosomes, were resuspended in PBS, treated with 5 μg RNase A for 10 minutes at 37°C and then by 25 μg Proteinase K, 30 minutes at 37°C to eliminate RNA-protein complex contaminants. Samples were spun again at 100,000g, 70 minutes. Pellets were resuspended in 200 μL cold PBS, filtered through 0.2 μm membrane (Acrodisc, Pall Corporation) and stored at −80°C.
Exosome isolation by precipitation
S20 were mixed with 0.5 volume of Total Exosome Extraction from cell culture media (Invitrogen), incubated overnight at 4°C and spun at 10,000g for 1 hour. Exosome pellets were resuspended in 200 μL cold PBS, filtered through 0.2 μm membrane (Acrodisc, Pall Corporation), aliquoted and stored at −80°C.
FACS purification of haemocytes: haemocytes fraction were incubated with propidium iodide (1 μg/mL) in PBS on ice for 5 minutes then filtered through a 35μm nylon mesh and processed on an Aria II FACS machine (BD FACSDIVA software). FACS purified haemocytes were collected directly in Trizol for RNA extraction or DNAzol (Thermofisher) for DNA extraction.
Next Generation Sequencing library preparation and analysis
Small RNA Isolation and Cloning for NGS
Small RNA fractions were isolated from total RNA samples using mirVana kit (Ambion, Thermo Fisher Scientific) followed by gel purification of 17 to 30 nucleotide long RNAs on denaturing PAGE. Exosomal small RNAs were extracted from exosomes isolated by ultracentrifugation (Figure 5 and S5B, C and D) or by precipitation (Figure S5E and F) after RNase A and Proteinase K treatment, as described below.
Small RNA samples quality and quantity were assessed by Bioanalyzer small RNA chip (Agilent) before cloning.
pRNA-seq: small RNAs were ligated to a 10 fold excess of miRNA Cloning linker 1 (IDT) using T4 RNA ligase 2 truncated (NEB) in the presence of 40 U RNase Inhibitors, Murine (NEB). 3′ ligation products were gel purified. Before 5′ ligation, samples were pre-annealed to a 2S Block oligo (30 sec at 90°C; 5 min at 65°C; 15 min at 4°C) (Wickersheim and Blumenstiel, 2013), complementary to Drosophila 2S rRNA to prevent cloning of the corresponding rRNA. 5′ ligation was performed using T4 RNA ligase 1 (NEB). Initial libraries (Figure 1 and 2) were generated using the following 5′ DNA/RNA adapter (CCTTGrGrCrArCrCrCrGrArGrArArTrTrCrCrA). All other libraries were obtained using a modified version of the adapter (CCTTGrGrCrArCrCrCrGrArGrArArTrTrCrCrArNrNrNrNrN), with 5 random nucleotides at the 3′-end. This randomized region helped prevent ligation bias and improved reproducibility and quantitation in miRNA cloning. After reverse-transcription with Superscript III (Invitrogen), libraries were amplified and barcoded by PCR and size-selected for 150 bp products on 7% native acrylamide gels.
tripRNA-seq: after 3′ ligation and gel purification as above, ligation products were treated with Shrimp Alkaline Phosphatase (rSAP, Affymetrix), 1 unit per picomole of RNA, 1 hour at 37°C followed by 15 minutes at 65°C for enzyme heat inactivation. rSAP treated samples were extracted with acid phenol: chloroform, pH 4.5 (Ambion), precipitated with 15 micrograms Glycoblue (Ambion) at −80°C for 1 hour. After centrifugation (top speed, 15 minutes, 4°C) and 70% ethanol wash, RNA pellets were resuspended in TE0.1 (10mM Tris pH 7.4, 0.1mM EDTA) and treated with Tobacco Acid Pyrophosphatase, TAP (Epicenter), 1 unit per picomole of RNA, 90 minutes at 37°C. Alternatively, RNA pyrophosphoHydrolase (RppH, NEB) was used for 60 minute at 37°C. After acid phenol:chloroform extraction and precipitation as above, SAP and TAP/RppH treated samples were resuspended in TE0.1 and processed as in pRNA-seq, resuming at the 5′ ligation step.
CIP-smRNA-seq: same as the tripRNA-seq protocol, except that the rSAP treatment was replaced by Calf Intestinal Phosphatase (CIP, NEB).
Libraries quality was assessed by Bioanalyzer High Sensitivity DNA chip (Agilent) as 150 bp products and quantified by Library Quantification kit (KAPA Biosystems). Sequencing was performed on HiSeq platforms (Illumina) as single read 50 cycles rapid runs.
Small RNA Library Analysis
Only reads passing Q30 filters were kept for analysis. FASTXtool kit (Hannon lab) was used to trim off 3′ linker sequences and convert fastq files to fasta. Bowtie-1.1.2 (Langmead et al., 2009)was used to align reads to SINV and Drosophila genomes (Dmel Release 5.22) allowing for only 1 mismatch. miRNA contents were measured using the Drosophila melanogaster miRNA database from miRBase and an in-house python script. In-house python scripts were also used to determine siRNA size distributions and to remove the first 5 random bases from the modified adapter. piRNAs were identified as 24–26 nucleotide (nt) long reads uniquely mapping to the Drosophila genome (Dmel Release 5.22) and mapping to a Drosophila transposon database (D_mel_transposon_sequence_set.fav9.4.1) with Bowtie-1.1.2, allowing for 1 mismatch. Endo-siRNAs were identified as 21 nt long reads uniquely mapping to one of the 17 endo-siRNA loci previously identified (Brennecke et al., 2007).
Transcription Start Site (TSS)-derived small RNAs were identified as reads mapping to all 5′ UTR identified in the Drosophila genome (dmel_r6.06_FB2015_03, all-five_primeUTR) using Bowtie-1.1.2 (Langmead et al., 2009)and then further selected as mapping to the first 50 bases. Similarly, tRNA-derived small RNAs were identified as reads mapping to a Drosophila tRNA database (dmel_r6.06_FB2015_03, all-tRNA), while rRNA-derived small RNAs were mapped to rRNA database extracted with UCSC Genome Browser (all ribosomal genes from Dmel Release 5.22) allowing 1 mismatch.
3′ methylation of siRNAs is known to increase their stability, which could bias the ability of tripRNA-seq to capture only polyphosphorylated small RNAs. To avoid this potential bias, we compared vsRNAs and piRNAs (which are 3′ methylated) abundance after normalizing their read counts to miRNA populations (which are the most abundant small RNA species, mostly not 3′-methylated and unaffected by Ago2 defect) in each dataset to compare the specific capture of vsRNA by tripRNA-seq across treatments and genetic backgrounds (see Table S5). Only piRNAs uniquely mapping to Drosophila genome were considered for the statistical analysis to avoid the potential confounding effect of redundant piRNAs originating from different genomic regions.
Of note, tripRNA-seq dataset generated using TAP tended to yield more 5′-monophosphorylated small RNA, such as miRNAs, that RppH treated samples. On the other hand, tripRNA-seq dataset generated using RppH tended to more efficiently deplete miRNAs but also lack enrichment for TSS-derived small RNAs, due to the lack of conversion of capped RNAs. Overall, we found that RppH treatment gives a more robust depletion of 5′-monophosphorylated RNAs but with the caveat of ultimately yielding less cloning events and therefore requiring larger input material compared to TAP treated samples.
For the analysis of datasets with dsRNA containing single mutations every 40 bases (dseGFP in Figure 2C and dsSIN in Figure S4C), reads mapping perfectly (Bowtie-1.0.0) to their respective mutated dsRNA templates (that is, to the gene fragment synthesized by gBlock Gene Fragment) were subtracted from the datasets. Remaining reads were aligned to the respective target full length sequences (that is, eGFP coding region or the entire SINV genome) allowing for 1 mismatch. This allowed the removal of reads directly derived from the injected dsRNA while retaining reads derived from the cognate targets. Mutations were introduced every 40 bases to limit loss of efficiency in RNAi. However, because siRNAs are 21 nucleotide long, reads falling between mismatches are sorted as derived from the dsRNA trigger and not considered for further analysis. Therefore, this approach might underestimate the number of secondary siRNAs generated by dsRNA immunization.
For the dicing product analysis (Figure S7A), only SINV mapping reads were considered. Dicing products were identified as unique pairs of sense and antisense 21 bases long reads, overlapping each other from position 1 to 19.
Molecular and Cellular Assays
Luciferase Activity Assay
5 different exosome-like vesicles (ELVs) preps were used for injection (see “Exosome Injection” section) from each genetic background. 10 flies per group were homogenized in 100 μL Cell Culture Lysis Reagent (Promega), centrifuged at 10,000 g, 10 minutes at room temperature. 75 μL of supernatant were transfered to a new tube. Each sample was assessed in triplicate using 20 μL of lysate with 100 μL reconstituted Luciferase Assay reagent. Luciferase activity was measured on a Veritas luminometer (Turner BioSystems).
SIN:GFP Positive Cell Assay
50000 S2 cells expressing a SIN:eGFP replicon were plated in a 96 well plate. 6 different preps of exosome-like vesicles (ELVs) for each genetic background were tested. ELVs (500ng protein content in 100μL per well) were added 1 day after replicon transfection. GFP positive cell counts were measured at 3 days post treatment on a Cell Insight CX5 (Thermo Scientific).
Dynamic Light Scattering
The homogeneity and size of the vesicles found in exosomal fractions was assessed by Dynamic Light Scattering using a DynaPro Platereader II (Wyatt technology) and the Dynamics v7 software. Ten measurements were performed on three different exosome samples with an acquisition time of 2 seconds.
Fluorescence Microscopy
Hml>eGFP flies injected with 50ng control dsRNA or anti-eGFP dsRNA were anesthetized by CO2 on a black flypad, placed under fluorescence stereomicroscope and imaged for eGFP fluorescence (excitation wavelength: 485nm).
Western Blot Analysis
Equal amounts of protein were electrophoresed on 10% sodium dodecyl sulfate polyacrylamide gel (SDS-PAGE), and the bands were transferred to polyvinylidene fluoride membrane (Millipore, USA). The membrane was blocked and then incubated with the relevant primary antibodies (1/1000 dilution, overnight at 4°C). After washing, the membranes were incubated with horseradish peroxidase-conjugated secondary antibodies (GE Healthcare Life Sciences) for 1 h. Immunoreactive bands were detected with ECL Plus (Pierce, Thermo Fisher Scientific), and analyzed with BIO-RAD ChemiDoc MP Imaging System and Image Lab Software.
Antibodies
GFP antibody (rabbit polyclonal) was from Santa Cruz Biotechnology. Tsg101 (mouse monoclonal) antibody was from Fitzgerald Industries International. Beta Actin (AC-15) was from Pierce.
qPCR-based Reverse Transcriptase Activity Assay
Haemocytes-enriched fraction from naïve or SINV infected hml>eGFP flies were isolated (at 4 d.p.i.) as described above. 30 μL of haemocyte fraction in PBS w/o were mixed with 30μL 2x Lysis buffer (0.25% Triton X-100, 50 mM KCL, 100 mM TrisHCL pH 7.4, 40% glycerol) and supplemented with RNase Inhibitor (0.5U/μL) and proteinase inhibitors (Complete without EDTA, Sigma-Aldrich). Samples were pipetted up and down 10 times, kept on ice for 10 minutes, spun at top speed at 4°C 15 minutes. The protein concentration of supernatants was measured by A280 absorbance on Nanodrop. Samples were then diluted to 1μg/μL protein in 1x Lysis buffer before snap freezing in liquid nitrogen and stored at −80°C. Samples were thawed on ice before RT assay. For each qPCR-based RT activity reaction, 3 μL of sample (diluted 100 times in water) was mixed with 10 μL 2x SensiFast SYBR no-Rox mix (Bioline), 0.1 μL Firefly Luciferase (Fluc) RNA (100ng/μL), 500 nM of Fluc fw and rv primers with or without 1μL AZT (50mM stock) and water up to a final volume of 20 μL. Superscript II (Invitrogen)(10−4 unit/reaction) and water were used as positive and negative control respectively. Reactions were run in a 96 well qPCR plate with the following program: 42°C, 20 min (RT reaction); 95°C, 3 min; followed by 40 amplification cycles of 95°C, 5 sec; 60°C, 10 sec; 72°C, 15 sec (with acquisition during the elongation phase). Absolute copy number of FLuc DNA produced by each RT reaction was calculated from the serial dilution of Fluc plasmid standard run on each plate. Final RT activity was measured as the fold increase of (FLuc copies produced in absence of AZT)/(FLuc copies in presence of AZT).
PCR and qPCR
200 ng input genomic DNA were used to assess SINV DNA production by end point PCR using SIN414_fw and SIN913_rv primers. Loading of input DNA was controlled in parallel by end point PCR against Drosophila crq gene, using Crq_fw and Crq_rv primers. SINV DNA production was also assessed by qPCR using SINV_NSP1_fw and rv primers, and SINV_Capsid_fw and rv primers and normalized to detection of the upstream genomic region of the tube gene (tube_up fw and rv oligos, see Table 6).
To assess SINV titers and haemocyte-specific genes expression, 2 μg total RNA were treated with DNase I (NEB) according to manufacturer’s instruction. 700 ng of DNAse I-treated RNA were used for reverse transcription using iScript (Bio-Rad).
All qPCR experiments were performed using SensiFAST SYBR no ROX kit (Bioline).
Quantification and Statistical analysis
All statistical tests were performed using R statistics (R Core Team (2013), http://www.R-project.org/).
Analysis of Sindbis viral titers
Replicates and conditions are described in the figure legends and n corresponds to the number of independent experiments. Comparisons of SINV titers were performed using two-tailed Mann-Whitney-Wilcoxon (R statistics, Wilcox.test). The Mann-Whitney-Wilcoxon test is a non-parametric test and therefore does not assume normal distribution of the samples.
Analysis of small RNA capture by pRNA-seq versus tripRNA-seq
Comparisons of read counts from pRNA-seq and tripRNA-seq datasets were performed using Fisher exact test in 2×2 contingency tables (R statistics fisher.test), to test the significance of the association (e.g.: capture) of vsRNAs with tripRNA-seq compared to other small RNA species (e.g.: miRNAs and piRNAs). Enrichment for vsRNAs in exosomal fractions was calculated as the ratio of exosomal vsRNA counts divided by vsRNA counts from carcasses normalized by the ratio of exosomal miRNA counts divided by miRNA counts from carcasses (in reads per million). Similar to vsRNA enrichment by tripRNA-seq, statistical significance for exosomal vsRNA enrichment (Figure 5D) was measured as 2×2 contingency table using Fisher exact test (R statistics fisher.test).
DATA AVAILABILITY
Illumina sequencing raw data (FASTQ files) are available under the NCBI SRA Study number: SRP100925 (https://www.ncbi.nlm.nih.gov/bioproject/377533).
Supplementary Material
(A) Haemocyte-specific genes are undetectable or expressed at very low level after genetic cell ablation of haemocytes in hml>reaper flies. Bar graph shows relative gene expression of haemocyte-specific genes, hemolectin (hml, light grey) and croquemort (Crq, dark grey) compared to ubiquitously expressed gene rpL12 as measured by RT-qPCR in control (hml>dsCtr) or haemocyte-depleted (hml>reaper) flies.
(B) Hameocyte-depleted flies are hypersensitive to Sindbis virus (SINV) infection. Normalized SINV titers measured by RT-qPCR in adult flies at 4 days post infection (d.p.i.) show increase viral replication in flies depleted of haemocytes (hml>reaper) compared to control parental line (UAS:reaper) (n=4 independent experiments, Mann Whitney test p<0.05 compared to control flies).
(A) Schematic of pRNA-seq and tripRNA-seq protocols. pRNA-seq is the conventional cloning approach for miRNAs and other main siRNA species (e.g.: piRNAs, esiRNAs…) and relies on the direct cloning of oligonucleotide adapter to 5′-monophosphorylated small RNAs. Unlike pRNA-seq, tripRNA-seq involves pretreatment of small RNAs with Shrimp Alkaline Phosphatase to specifically remove 5′monophosphate groups, followed by a treatment with either Tobacco Acid Phosphatase (TAP) or RNA pyrophosphoHydrolase (RppH) to convert 5′-di-trihosphorylated or capped (in the case of TAP) RNA into 5′-monophosphorylated RNA amenable to 5′ ligation of an oligonucleotide adapter.
(B) CIP-smRNA-seq does not identify any capped virus-derived siRNAs. CIP-smRNA-seq approach is a modified version of tripRNA-seq (using TAP) where SAP is substituted for Calf Intestine Phosphatase that hydrolyses 5′mono, di, or triphosphate groups to specifically capture capped-small RNAs. The bar graph shows the number of virus-derived siRNAs in reads per million (rpm) captured from SINV-infected control flies (w1118) by tripRNA-seq versus CIP-smRNA-seq.
(A) Haemocyte-specific genes are enriched in haemocyte-enriched fraction. Bar graph shows relative gene expression of haemocyte-specific genes, hemolectin (hml, light grey) and croquemort (Crq, dark grey) compared to ubiquitously expressed gene rpL12 as measured by RT-qPCR in carcasses and haemocyte-enriched fraction (Haemocytes) from control flies (w1118).
(B) AZT treatment in adult flies does not affect global miRNA expression. Flies were injected with 2.5 nmoles AZT (+AZT) or PBS only (-AZT) 1 day before and 1 day after SINV injection. Small RNAs were extracted at 4 d.p.i from carcasses and haemocyte-enriched fraction and analyzed to pRNA-seq. miRNA counts in read per million (rpm) show no difference between control and AZT treatment in both carcasses (light grey squares) and haemocyte fractions (white diamonds).
(C) Haemocyte fractions from naïve or SINV-infected control flies (hml>eGFP) were isolated and processed from RT activity assay. RT activity in each sample was assessed as the amount of cDNA produced from an exogenous RNA (Firefly Luciferase) in the absence versus presence of AZT and measured by qPCR. Haemocytes from SINV-infected show significant AZT-sensitive RT activity compared to uninfected flies (p < 0.1, Mann-Whitney-Wilcoxon, n = 3). In parallel of each assay, 10μU of Superscript II (Invitrogen) and water were used as positive and negative control for RT activity respectively.
(A) dsRNA injection does not initiate RNAi ubiquitously in adult flies. 50 ng of anti-eGFP dsRNA (dseGFP) was injected in flies ubiquitously expressing low level of eGFP at 25°C (hps70>eGFP). eGFP expression was monitored by Western blot at 0, 1, 2, 3, 4 and 5 days post injection. Actin was used as a loading control.
(B and C) anti-SINV dsRNA injection leads to decrease viral titers and increase de novo production of vsRNAs. Flies were co-injected with 500 pfu SINV and control (dsCtr) or mutated anti-SINV(dsSIN) dsRNA. Of note, mutated dsSIN had 1 mismatch every 40 bases to allow sorting of dsRNA-derived versus SINV genome-derived siRNAs while maintaining efficient antiviral silencing. At 4 days post infection, large RNA were separated from small RNA fraction and used to measure SINV titers by RT-qPCR (B). Bar graph shows that mutated dsSIN efficiently decrease viral replication compared to dsCtr treatment. (C) Small RNAs were subjected to pRNA-seq and tripRNA-seq in parallel. Enrichment for vsRNAs in tripRNA-seq datasets was measured relative to miRNA abundance in both datasets. 5′-triphosphorylated vsRNA production was significantly higher in dsSIN versus dsCtr treated flies (p < 5.37 e-6, Fisher exact test).
(A) hml>eGFP flies were injected with 5′-mono and triphosphorylated control (lanes 1 and 2) or anti-eGFP siRNA (lanes 3 and 4) or anti-eGFP dsRNA (lane 5) or control dsRNA (lane 6). At 4 days post injection, only anti-eGFP dsRNA efficiently silence eGFP expression in haemocytes.
(B) ELV isolated from adult fly haemolymph are positive for the multivesicular body/exosome marker, Tsg101. Exosomal vesicles (Exo) were isolated from the haemolymph of 1 week old hml>dsCtr adult flies. 1:1000 of the carcass fraction (Carc) and 1:5 of the exosomal fraction were processed for anti-Tsg101 Western blot.
(C) ELVs isolated from adult fly haemolymph have the size of exosome-like vesicles (20–200nm). Size of the vesicles isolated in the exosome fraction was measured by Dynamic Light Scattering.
(D) A fraction of 21 nucleotide (nt) small RNAs associated with exosomes is protected from RNase A digestion. Bioanalyzer traces of small RNAs extracted from exosomes of SINV-infected flies and incubated 30 min at 37°C in PBS alone (no treatment), or with RNase A (RNase A) or RNase A in the presence of detergent Triton X-100 (RNaseA + Tx). Unlike other abundant RNAs (such as 24–26 nt-long RNAs, asterisk), 21 nt-long RNAs (black arrowhead) were protected from RNase A digestion in the absence of detergent.
(E and F) vsRNAs accumulate specifically in haemocyte-secreted ELVs of SINV-infected flies. Strand and size distribution of siRNAs found in circulating ELVs of SINV-infected control flies (hml>dsFluc, in grey) or flies with Syntaxin 1A knockdown haemocytes (hml>dsSyx1A, in white). ELVs were treated with RNase A (RNase A) prior to RNA extraction and siRNA counts were determined by pRNA-seq (in reads per million, rpm). (D) 21 nt long vsRNAs are detected in circulating ELVs from control flies (hml>dsFluc) but not from flies with haemocytes deficient for exosome secretion (hml>dsSyx1A). (E) siRNAs derived from hairpin (dsFluc) endogenously expressed in haemocytes (hml>dsFluc) were not detected in circulating ELVs.
(A) ELV immunization requires AGO2 in the receiving flies. ELVs from naïve or SINV-infected flies (w1118) were co-injected with 100 pfu SIN:Fluc in ago2414 mutant flies. Flies were collected at 4 d.p.i. and processed for Fluc activity to assess SIN:Fluc replication. Fluc activity was not significantly reduced by treatment with ELVs from SINV-infected flies (not significant, n.s., Mann Whitney test, n=4 independent experiments).
(B) ELV fractions from SINV-infected wild type flies do not contain detectable levels of SINV. RNA was extracted from either SINV stocks or from ELV fractions isolated from naive or SINV-infected control flies (w1118) and processed for RT-qPCR to assess SINV genome content. Note: the equivalent amount of ELV used for pRNA-seq was used for RT-qPCR (NTD indicates no detectable titers).
(C) ELV do not contain detectable levels of SINV vDNA. ELVs fractions from naïve (-SV) or SINV-infected (+SV) control flies (hml>dsCtr) or flies with Ago2-deficient haemocytes (hml>dsCtr) were purified at 4 d.p.i. and processed for DNA extraction. SINV-derived vDNA from two SINV genes (NSP1 and capsid) could not be detected by end-point PCR. Negative (Water) and positive (SINV replicon plasmid, SINV) control for the PCR reactions are shown.
(A) Viral dicing products are under-represented in haemocytes. The ratio of 5′-monophosphorylated dicing products (as defined in STAR Method section) over the total number of SINV mapping 21 mer reads (by pRNA-seq) were measured in carcass versus haemocyte fractions. Data from two independent experiments are presented (Exp1 and Exp2).
(B) Model for the de novo production of 21 nucleotide(nt)-long vsRNAs in hameocytes. Similarly to piRNA precursors biogenesis, transcripts produced from viral DNA template are transported to the cytoplasm where an endoribonuclease (i.e.: Zucchini in the case of piRNA precursors in Drosophila ovaries) cleaves them at relatively regular intervals to produce 5′ monophosphorylated products. Loading onto an Argonaute protein (Aubergine or PIWI in the case of piRNA) is followed by trimming of the protruding extra 3′ bases by an 3′–5′ exonuclease (PNLDC1 in silkworm1), thus producing mature small RNAs of 26 nt (piRNAs) or 21 nt (vsRNA). While mature piRNA act in the cytoplasm and the nucleus to silence transposable elements (TE) in the gonads, mature secondary 5′ monophosphorylated single stranded vsRNAs are loaded in haemocyte’s exosomes (ELV) to be secreted in the haemolymph.
Drosophila haemocytes can convert viral RNA genomes into viral DNA molecules
Viral DNA production allows de novo synthesis of secondary virus-derived siRNAs
RNAi amplification fuels virus-derived siRNA loading into exosome-like vesicles
Haemocyte-derived exosome-like vesicles confer systemic RNAi antiviral immunity
Acknowledgments
We thank Drs. Judith Frydman and members of Andino lab for critical comments on the manuscript. This work was supported by NIH (R01, AI36178, AI40085, P01 AI091575).
Footnotes
AUTHOR CONTRIBUTION
M.T. and R.A. conceived the idea and designed the experiments. M.T. performed most experiments, except for the microscopy data and eGFP Western Blot performed by M.K., M.T. and R.A. wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Banchereau J, Steinman RM. Dendritic cells and the control of immunity. Nature. 1998;392:245–252. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- Beckett K, Monier S, Palmer L, Alexandre C, Green H, Bonneil E, Raposo G, Thibault P, Le Borgne R, Vincent JP. Drosophila S2 cells secrete wingless on exosome-like vesicles but the wingless gradient forms independently of exosomes. Traffic. 2013;14:82–96. doi: 10.1111/tra.12016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Carroll MC, Isenman DE. Regulation of humoral immunity by complement. Immunity. 2012;37:199–207. doi: 10.1016/j.immuni.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cilia ML, Jackson D. Plasmodesmata form and function. Curr Opin Cell Biol. 2004;16:500–506. doi: 10.1016/j.ceb.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Fauvarque MO, Williams MJ. Drosophila cellular immunity: a story of migration and adhesion. J Cell Sci. 2011;124:1373–1382. doi: 10.1242/jcs.064592. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Singh A, Mandal S, Mandal L. Active hematopoietic hubs in Drosophila adults generate hemocytes and contribute to immune response. Dev Cell. 2015;33:478–488. doi: 10.1016/j.devcel.2015.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goic B, Vodovar N, Mondotte JA, Monot C, Frangeul L, Blanc H, Gausson V, Vera-Otarola J, Cristofari G, Saleh MC. RNA-mediated interference and reverse transcription control the persistence of RNA viruses in the insect model Drosophila. Nat Immunol. 2013;14:396–403. doi: 10.1038/ni.2542. [DOI] [PubMed] [Google Scholar]
- Hahn CS, Hahn YS, Braciale TJ, Rice CM. Infectious Sindbis virus transient expression vectors for studying antigen processing and presentation. Proc Natl Acad Sci U S A. 1992;89:2679–2683. doi: 10.1073/pnas.89.7.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han BW, Wang W, Li C, Weng Z, Zamore PD. Noncoding RNA. piRNA-guided transposon cleavage initiates Zucchini-dependent, phased piRNA production. Science. 2015;348:817–821. doi: 10.1126/science.aaa1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey JJW, Lewsey MG, Patel K, Westwood J, Heimstädt S, Carr JP, Baulcombe DC. An antiviral defense role of AGO2 in plants. PLoS ONE. 2011;6:e14639. doi: 10.1371/journal.pone.0014639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himber C, Dunoyer P, Moissiard G, Ritzenthaler C, Voinnet O. Transitivity-dependent and -independent cell-to-cell movement of RNA silencing. Embo J. 2003;22:4523–4533. doi: 10.1093/emboj/cdg431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koles K, Nunnari J, Korkut C, Barria R, Brewer C, Li Y, Leszyk J, Zhang B, Budnik V. Mechanism of evenness interrupted (Evi)-exosome release at synaptic boutons. J Biol Chem. 2012;287:16820–16834. doi: 10.1074/jbc.M112.342667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nainu F, Tanaka Y, Shiratsuchi A, Nakanishi Y. Protection of Insects against Viral Infection by Apoptosis-Dependent Phagocytosis. J Immunol. 2015;195:5696–5706. doi: 10.4049/jimmunol.1500613. [DOI] [PubMed] [Google Scholar]
- Nayak A, Tassetto M, Kunitomi M, Andino R. RNA interference-mediated intrinsic antiviral immunity in invertebrates. Curr Top Microbiol Immunol. 2013;371:183–200. doi: 10.1007/978-3-642-37765-5_7. [DOI] [PubMed] [Google Scholar]
- Okamura K, Ishizuka A, Siomi H, Siomi MC. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 2004;18:1655–1666. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak J, Fire A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science. 2007;315:241–244. doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
- Pak J, Maniar JM, Mello CC, Fire A. Protection from feed-forward amplification in an amplified RNAi mechanism. Cell. 2012;151:885–899. doi: 10.1016/j.cell.2012.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh MC, Tassetto M, van Rij RP, Goic B, Gausson V, Berry B, Jacquier C, Antoniewski C, Andino R. Antiviral immunity in Drosophila requires systemic RNA interference spread. Nature. 2009;458:346–350. doi: 10.1038/nature07712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh MC, van Rij RP, Hekele A, Gillis A, Foley E, O’Farrell PH, Andino R. The endocytic pathway mediates cell entry of dsRNA to induce RNAi silencing. Nat Cell Biol. 2006;8:793–802. doi: 10.1038/ncbl439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, Plasterk RH, Fire A. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- Sinenko SA, Mathey-Prevot B. Increased expression of Drosophila tetraspanin, Tsp68C, suppresses the abnormal proliferation of ytr-deficient and Ras/Raf-activated hemocytes. Oncogene. 2004;23:9120–9128. doi: 10.1038/sj.onc.1208156. [DOI] [PubMed] [Google Scholar]
- Strauss EG, Rice CM, Strauss JH. Complete nucleotide sequence of the genomic RNA of Sindbis virus. Virology. 1984;133:92–110. doi: 10.1016/0042-6822(84)90428-8. [DOI] [PubMed] [Google Scholar]
- Teixeira L, Ferreira A, Ashburner M. The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biology. 2008;6:e2. doi: 10.1371/journal.pbio.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turturici G, Tinnirello R, Sconzo G, Geraci F. Extracellular membrane vesicles as a mechanism of cell-to-cell communication: advantages and disadvantages. Am J Physiol, Cell Physiol. 2014;306:C621–C633. doi: 10.1152/ajpcell.00228.2013. [DOI] [PubMed] [Google Scholar]
- van Rij RP, Saleh MC, Berry B, Foo C, Houk A, Antoniewski C, Andino R. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev. 2006;20:2985–2995. doi: 10.1101/gad.1482006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeire J, Naessens E, Vanderstraeten H, Landi A, Iannucci V, Van Nuffel A, Taghon T, Pizzato M, Verhasselt B. Quantification of reverse transcriptase activity by real-time PCR as a fast and accurate method for titration of HIV, lenti- and retroviral vectors. PLoS ONE. 2012;7:e50859. doi: 10.1371/journal.pone.0050859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White K, Tahaoglu E, Steller H. Cell killing by the Drosophila gene reaper. Science. 1996;271:805–807. doi: 10.1126/science.271.5250.805. [DOI] [PubMed] [Google Scholar]
- Wickersheim ML, Blumenstiel JP. Terminator oligo blocking efficiently eliminates rRNA from Drosophila small RNA sequencing libraries. BioTechniques. 2013;55:269–272. doi: 10.2144/000114102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston WM, Molodowitch C, Hunter CP. Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science. 2002;295:2456–2459. doi: 10.1126/science.1068836. [DOI] [PubMed] [Google Scholar]
- Zhou R, Mohr S, Hannon GJ, Perrimon N. Inducing RNAi in Drosophila cells by transfection with dsRNA. Cold Spring Harb Protoc. 2013;2013:461–463. doi: 10.1101/pdb.prot074351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Mohr S, Hannon GJ, Perrimon N. Inducing RNAi in Drosophila cells by soaking with dsRNA. Cold Spring Harb Protoc. 2014;2014 doi: 10.1101/pdb.prot080747. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Haemocyte-specific genes are undetectable or expressed at very low level after genetic cell ablation of haemocytes in hml>reaper flies. Bar graph shows relative gene expression of haemocyte-specific genes, hemolectin (hml, light grey) and croquemort (Crq, dark grey) compared to ubiquitously expressed gene rpL12 as measured by RT-qPCR in control (hml>dsCtr) or haemocyte-depleted (hml>reaper) flies.
(B) Hameocyte-depleted flies are hypersensitive to Sindbis virus (SINV) infection. Normalized SINV titers measured by RT-qPCR in adult flies at 4 days post infection (d.p.i.) show increase viral replication in flies depleted of haemocytes (hml>reaper) compared to control parental line (UAS:reaper) (n=4 independent experiments, Mann Whitney test p<0.05 compared to control flies).
(A) Schematic of pRNA-seq and tripRNA-seq protocols. pRNA-seq is the conventional cloning approach for miRNAs and other main siRNA species (e.g.: piRNAs, esiRNAs…) and relies on the direct cloning of oligonucleotide adapter to 5′-monophosphorylated small RNAs. Unlike pRNA-seq, tripRNA-seq involves pretreatment of small RNAs with Shrimp Alkaline Phosphatase to specifically remove 5′monophosphate groups, followed by a treatment with either Tobacco Acid Phosphatase (TAP) or RNA pyrophosphoHydrolase (RppH) to convert 5′-di-trihosphorylated or capped (in the case of TAP) RNA into 5′-monophosphorylated RNA amenable to 5′ ligation of an oligonucleotide adapter.
(B) CIP-smRNA-seq does not identify any capped virus-derived siRNAs. CIP-smRNA-seq approach is a modified version of tripRNA-seq (using TAP) where SAP is substituted for Calf Intestine Phosphatase that hydrolyses 5′mono, di, or triphosphate groups to specifically capture capped-small RNAs. The bar graph shows the number of virus-derived siRNAs in reads per million (rpm) captured from SINV-infected control flies (w1118) by tripRNA-seq versus CIP-smRNA-seq.
(A) Haemocyte-specific genes are enriched in haemocyte-enriched fraction. Bar graph shows relative gene expression of haemocyte-specific genes, hemolectin (hml, light grey) and croquemort (Crq, dark grey) compared to ubiquitously expressed gene rpL12 as measured by RT-qPCR in carcasses and haemocyte-enriched fraction (Haemocytes) from control flies (w1118).
(B) AZT treatment in adult flies does not affect global miRNA expression. Flies were injected with 2.5 nmoles AZT (+AZT) or PBS only (-AZT) 1 day before and 1 day after SINV injection. Small RNAs were extracted at 4 d.p.i from carcasses and haemocyte-enriched fraction and analyzed to pRNA-seq. miRNA counts in read per million (rpm) show no difference between control and AZT treatment in both carcasses (light grey squares) and haemocyte fractions (white diamonds).
(C) Haemocyte fractions from naïve or SINV-infected control flies (hml>eGFP) were isolated and processed from RT activity assay. RT activity in each sample was assessed as the amount of cDNA produced from an exogenous RNA (Firefly Luciferase) in the absence versus presence of AZT and measured by qPCR. Haemocytes from SINV-infected show significant AZT-sensitive RT activity compared to uninfected flies (p < 0.1, Mann-Whitney-Wilcoxon, n = 3). In parallel of each assay, 10μU of Superscript II (Invitrogen) and water were used as positive and negative control for RT activity respectively.
(A) dsRNA injection does not initiate RNAi ubiquitously in adult flies. 50 ng of anti-eGFP dsRNA (dseGFP) was injected in flies ubiquitously expressing low level of eGFP at 25°C (hps70>eGFP). eGFP expression was monitored by Western blot at 0, 1, 2, 3, 4 and 5 days post injection. Actin was used as a loading control.
(B and C) anti-SINV dsRNA injection leads to decrease viral titers and increase de novo production of vsRNAs. Flies were co-injected with 500 pfu SINV and control (dsCtr) or mutated anti-SINV(dsSIN) dsRNA. Of note, mutated dsSIN had 1 mismatch every 40 bases to allow sorting of dsRNA-derived versus SINV genome-derived siRNAs while maintaining efficient antiviral silencing. At 4 days post infection, large RNA were separated from small RNA fraction and used to measure SINV titers by RT-qPCR (B). Bar graph shows that mutated dsSIN efficiently decrease viral replication compared to dsCtr treatment. (C) Small RNAs were subjected to pRNA-seq and tripRNA-seq in parallel. Enrichment for vsRNAs in tripRNA-seq datasets was measured relative to miRNA abundance in both datasets. 5′-triphosphorylated vsRNA production was significantly higher in dsSIN versus dsCtr treated flies (p < 5.37 e-6, Fisher exact test).
(A) hml>eGFP flies were injected with 5′-mono and triphosphorylated control (lanes 1 and 2) or anti-eGFP siRNA (lanes 3 and 4) or anti-eGFP dsRNA (lane 5) or control dsRNA (lane 6). At 4 days post injection, only anti-eGFP dsRNA efficiently silence eGFP expression in haemocytes.
(B) ELV isolated from adult fly haemolymph are positive for the multivesicular body/exosome marker, Tsg101. Exosomal vesicles (Exo) were isolated from the haemolymph of 1 week old hml>dsCtr adult flies. 1:1000 of the carcass fraction (Carc) and 1:5 of the exosomal fraction were processed for anti-Tsg101 Western blot.
(C) ELVs isolated from adult fly haemolymph have the size of exosome-like vesicles (20–200nm). Size of the vesicles isolated in the exosome fraction was measured by Dynamic Light Scattering.
(D) A fraction of 21 nucleotide (nt) small RNAs associated with exosomes is protected from RNase A digestion. Bioanalyzer traces of small RNAs extracted from exosomes of SINV-infected flies and incubated 30 min at 37°C in PBS alone (no treatment), or with RNase A (RNase A) or RNase A in the presence of detergent Triton X-100 (RNaseA + Tx). Unlike other abundant RNAs (such as 24–26 nt-long RNAs, asterisk), 21 nt-long RNAs (black arrowhead) were protected from RNase A digestion in the absence of detergent.
(E and F) vsRNAs accumulate specifically in haemocyte-secreted ELVs of SINV-infected flies. Strand and size distribution of siRNAs found in circulating ELVs of SINV-infected control flies (hml>dsFluc, in grey) or flies with Syntaxin 1A knockdown haemocytes (hml>dsSyx1A, in white). ELVs were treated with RNase A (RNase A) prior to RNA extraction and siRNA counts were determined by pRNA-seq (in reads per million, rpm). (D) 21 nt long vsRNAs are detected in circulating ELVs from control flies (hml>dsFluc) but not from flies with haemocytes deficient for exosome secretion (hml>dsSyx1A). (E) siRNAs derived from hairpin (dsFluc) endogenously expressed in haemocytes (hml>dsFluc) were not detected in circulating ELVs.
(A) ELV immunization requires AGO2 in the receiving flies. ELVs from naïve or SINV-infected flies (w1118) were co-injected with 100 pfu SIN:Fluc in ago2414 mutant flies. Flies were collected at 4 d.p.i. and processed for Fluc activity to assess SIN:Fluc replication. Fluc activity was not significantly reduced by treatment with ELVs from SINV-infected flies (not significant, n.s., Mann Whitney test, n=4 independent experiments).
(B) ELV fractions from SINV-infected wild type flies do not contain detectable levels of SINV. RNA was extracted from either SINV stocks or from ELV fractions isolated from naive or SINV-infected control flies (w1118) and processed for RT-qPCR to assess SINV genome content. Note: the equivalent amount of ELV used for pRNA-seq was used for RT-qPCR (NTD indicates no detectable titers).
(C) ELV do not contain detectable levels of SINV vDNA. ELVs fractions from naïve (-SV) or SINV-infected (+SV) control flies (hml>dsCtr) or flies with Ago2-deficient haemocytes (hml>dsCtr) were purified at 4 d.p.i. and processed for DNA extraction. SINV-derived vDNA from two SINV genes (NSP1 and capsid) could not be detected by end-point PCR. Negative (Water) and positive (SINV replicon plasmid, SINV) control for the PCR reactions are shown.
(A) Viral dicing products are under-represented in haemocytes. The ratio of 5′-monophosphorylated dicing products (as defined in STAR Method section) over the total number of SINV mapping 21 mer reads (by pRNA-seq) were measured in carcass versus haemocyte fractions. Data from two independent experiments are presented (Exp1 and Exp2).
(B) Model for the de novo production of 21 nucleotide(nt)-long vsRNAs in hameocytes. Similarly to piRNA precursors biogenesis, transcripts produced from viral DNA template are transported to the cytoplasm where an endoribonuclease (i.e.: Zucchini in the case of piRNA precursors in Drosophila ovaries) cleaves them at relatively regular intervals to produce 5′ monophosphorylated products. Loading onto an Argonaute protein (Aubergine or PIWI in the case of piRNA) is followed by trimming of the protruding extra 3′ bases by an 3′–5′ exonuclease (PNLDC1 in silkworm1), thus producing mature small RNAs of 26 nt (piRNAs) or 21 nt (vsRNA). While mature piRNA act in the cytoplasm and the nucleus to silence transposable elements (TE) in the gonads, mature secondary 5′ monophosphorylated single stranded vsRNAs are loaded in haemocyte’s exosomes (ELV) to be secreted in the haemolymph.
Data Availability Statement
Illumina sequencing raw data (FASTQ files) are available under the NCBI SRA Study number: SRP100925 (https://www.ncbi.nlm.nih.gov/bioproject/377533).