Tranexamic acid is an antifibrinolytic amino acid that exists as a zwitterion [trans-4-(ammoniomethyl)cyclohexane-1-carboxylate] in the solid state. Its reaction with copper chloride lead to the formation of a copper(II) paddle-wheel structure.
Keywords: crystal structure, tranexamic acid, antifibrinolytic amino acid, fibrinolytic inhibitor, copper(II), paddle-wheel, hydrogen bonding
Abstract
Tranexamic acid [systematic name: trans-4-(aminomethyl)cyclohexane-1-carboxylic acid], is an antifibrinolytic amino acid that exists as a zwitterion [trans-4-(ammoniomethyl)cyclohexane-1-carboxylate] in the solid state. Its reaction with copper chloride leads to the formation of a compound with a copper(II) paddle-wheel structure that crystallizes as a hexahydrate, [Cu2Cl2(C8H15NO2)4]2+·2Cl−·6H2O. The asymmetric unit is composed of a copper(II) cation, two zwitterionic tranexamic acid units, a coordinating Cl− anion and a free Cl− anion, together with three water molecules of crystallization. The whole structure is generated by inversion symmetry, with the Cu⋯Cu axle of the paddle-wheel dication being located about a center of symmetry. The cyclohexane rings of the zwitterionic tranexamic acid units have chair conformations. The carboxylate groups that bridge the two copper(II) cations are inclined to one another by 88.4 (8)°. The copper(II) cation is ligated by four carboxylate O atoms in the equatorial plane and by a Cl− ion in the axial position. Hence, it has a fivefold O4Cl coordination sphere with a perfect square-pyramidal geometry and a τ 5 index of zero. In the crystal, the paddle-wheel dications are linked by a series of N—H⋯Cl hydrogen bonds, involving the coordinating and free Cl− ions, forming a three-dimensional network. This network is strengthened by a series of N—H⋯Owater, Owater—H⋯Cl and Owater—H⋯O hydrogen bonds.
Chemical context
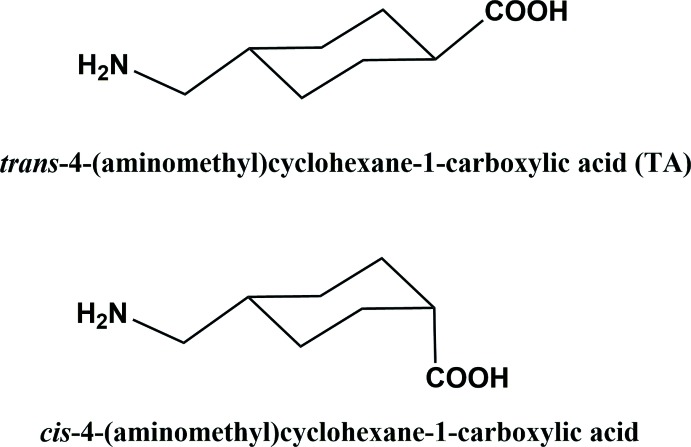
Tranexamic acid (TA) is a derivative of the amino acid lysine. It has important antifibrinolytic activity and is used extensively in both trauma and normal surgery to prevent excessive blood loss (Napolitano et al., 2013 ▸; Melvin et al., 2015 ▸). It was first synthesized in the early 1960s by the Japanese husband and wife team Shosuke and Utako Okamato (1962 ▸). They showed amino-methyl-cyclohexane-carboxylic acid (AMCHA) to be a new inhibitor of fibrinolysis. Almost simultaneously with a Swedish group (Melander et al., 1965 ▸), they were able to show that the antifibrinolytic active isomer (Okamoto et al., 1964 ▸) has a trans-conformation (TA; Fig. 1 ▸) with the aminomethyl and carboxylic acid substituents on the cyclohexane ring occupying equatorial positions (Fig. 1 ▸). The cis-isomer (Fig. 1 ▸), in which the carboxylic acid moiety is axial, is almost inactive. The latter was shown to exist as the free acid in the solid state (Yamazaki et al., 1981 ▸), in contrast to the trans-isomer, which exists as a zwitterion in the solid state (Groth, 1968 ▸; Shahzadi et al., 2007 ▸). Recently, Tengborn et al. (2015 ▸) published an excellent review article, entitled ‘Tranexamic acid – an old drug still going strong and making a revival’, in which they recount the history of the development of TA and its mechanism of action, pharmokinetics and other details, including clinical uses. Herein, we report on the first crystal structure of a metal complex of tranexamic acid. The reaction of TA with copper(II) chloride leads to the formation of the title compound with a copper(II) paddle-wheel structure, that crystallizes as a hexahydrate. The reaction of TA with copper(II) bromide leads to the formation of an isotypical compound; however, the crystals were twinned and the subsequent X-ray analysis was of insufficient quality to be submitted or deposited.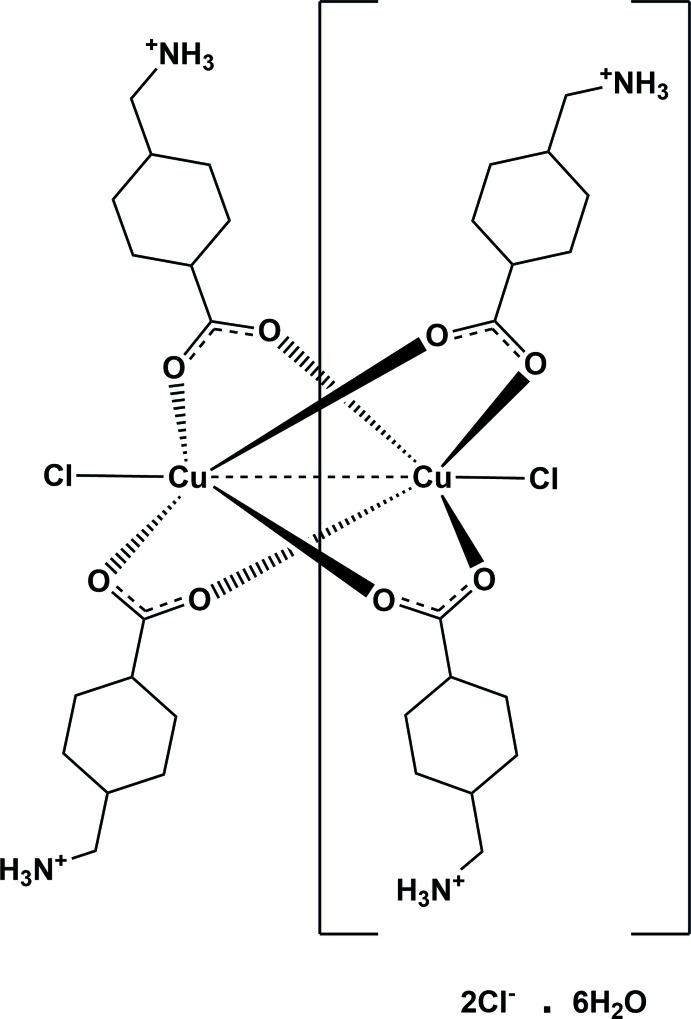
Figure 1.
The trans- and cis-isomers of 4-(aminomethyl)cyclohexane-1-carboxylic acid.
Structural commentary
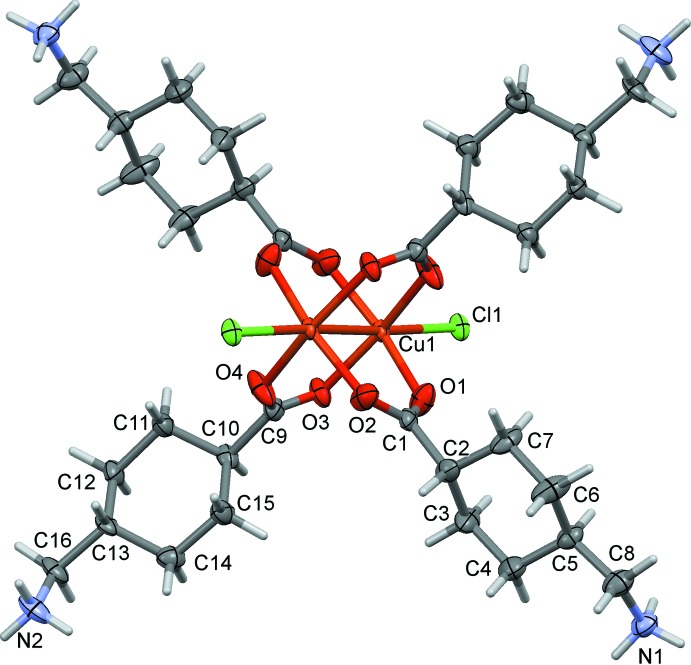
The molecular structure of the dication of the title compound is illustrated in Fig. 2 ▸. The asymmetric unit is composed of a copper(II) cation coordinated by the carboxylate O atoms (O1–O4) of two zwitterionic tranexamic acid units and a Cl− anion, Cl1, together with a free Cl− anion, Cl2, and three water molecules of crystallization. The whole structure is generated by inversion symmetry, with the Cu1⋯Cu1i axle [2.6649 (11) Å; symmetry code (i): −x + 1, −y + 1, −z + 1] of the paddle-wheel being located about a center of symmetry. Selected bond lengths and angles in the paddle-wheel dication are given in Table 1 ▸. Atom Cu1 is coordinated by four carboxylate O atoms (O1–O4) in the equatorial plane and a Cl− ion, Cl1, in the axial position. The Cu—O distances vary from 1.950 (4) to 1.991 (3) Å, with a longer Cu1—Cl1 axial distance of 2.499 (1) Å (Table 1 ▸). The copper(II) cation, Cu1 (Cu1i), has a perfect square-pyramidal coordination sphere with a τ 5 index of 0.0 (τ 5 = 0 for an ideal square-pyramidal coordination sphere, and = 1 for an ideal trigonal–pyramidal coordination sphere; Addison et al., 1984 ▸).
Figure 2.
A view of the molecular structure of the title dication, with atom labelling. Displacement ellipsoids are drawn at the 50% probability level. Unlabelled atoms are related to the labelled atoms by inversion symmetry (symmetry operation: −x + 1, −y + 1, −z + 1).
Table 1. Selected geometric parameters (Å, °).
| Cu1—Cu1i | 2.6649 (11) | Cu1—O4i | 1.965 (4) |
| Cu1—O1 | 1.950 (4) | Cu1—O3 | 1.991 (3) |
| Cu1—O2i | 1.955 (4) | Cu1—Cl1 | 2.4990 (12) |
| O1—Cu1—O2i | 167.02 (15) | O4i—Cu1—Cl1 | 92.57 (11) |
| O1—Cu1—O4i | 89.3 (2) | O3—Cu1—Cl1 | 100.08 (10) |
| O2i—Cu1—O4i | 89.9 (2) | O1—Cu1—Cu1i | 83.04 (11) |
| O1—Cu1—O3 | 88.33 (18) | O2i—Cu1—Cu1i | 84.05 (11) |
| O2i—Cu1—O3 | 89.60 (18) | O4i—Cu1—Cu1i | 80.24 (11) |
| O4i—Cu1—O3 | 167.27 (15) | O3—Cu1—Cu1i | 87.06 (10) |
| O1—Cu1—Cl1 | 94.31 (11) | Cl1—Cu1—Cu1i | 172.34 (4) |
| O2i—Cu1—Cl1 | 98.68 (11) |
Symmetry code: (i) 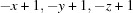 .
.
The conformations of the two zwitterionic tranexamic acid units differ slightly. The cyclohexane rings (C2–C7 and C10–C15) have chair conformations; puckering parameters for ring C2–C7 are Q = 0.569 (7) Å, θ = 176.3 (6)°, φ = 358 (13)°, and for ring C10–C15 are Q = 0.568 (6) Å, θ = 6.0 (6)°, φ = 137 (6)°. The carboxylate groups (C1/O1/O2 and C9/O3/O4) are inclined to the mean planes of the four planar atoms of the respective cyclohexane rings (C3/C4/C6/C7 and C11/C12/C14/C15) by 67.5 (6) and 85.8 (7)°, while they are inclined to one another by 88.4 (8)°. The ammoniomethyl units, C5/C8/N1 and C13/C16/N2, are inclined to the mean planes of the four planar atoms of the respective cyclohexane rings (C3/C4/C6/C7 and C11/C12/C14/C15) by 34.9 (6) and 47.5 (6)°.
Supramolecular features
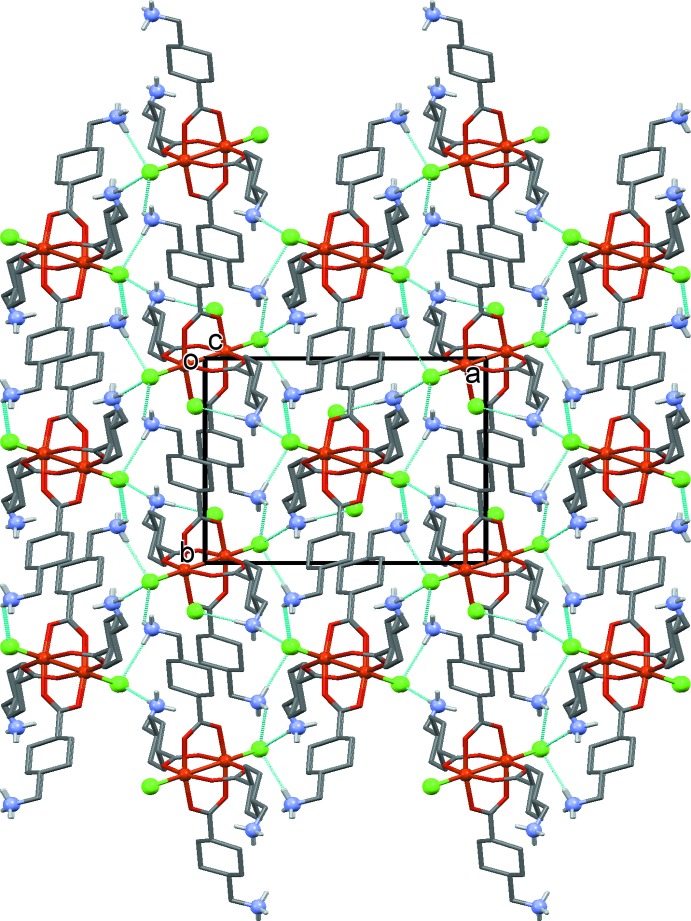
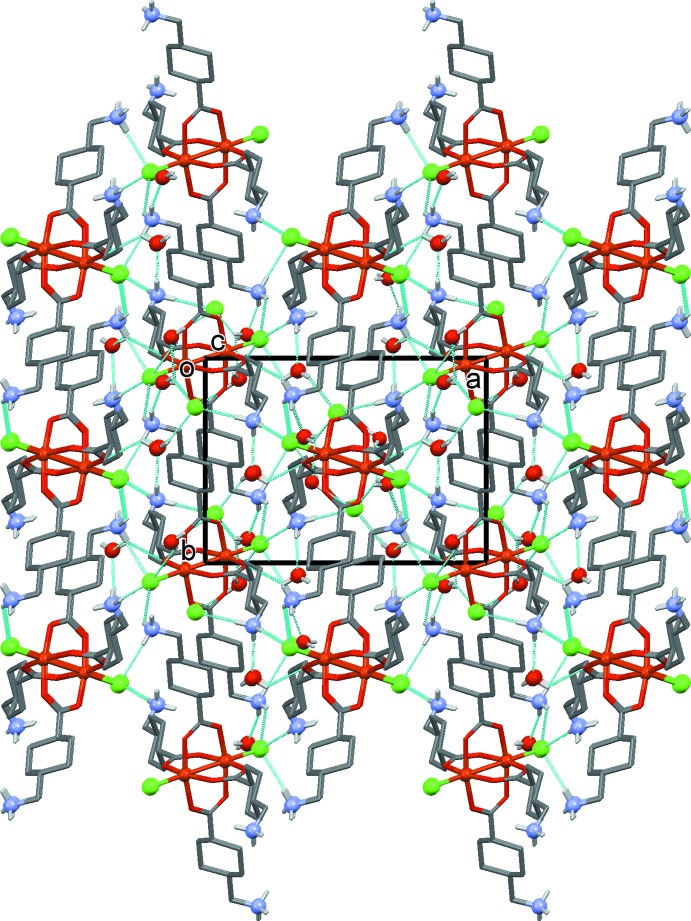
In the crystal, the NH3 + groups of the zwitterionic tranexamic acid units and the coordinating and free Cl– ions are linked by a series of N—H⋯Cl hydrogen bonds forming a three-dimensional framework (Table 2 ▸ and Fig. 3 ▸). This framework is strengthened by a series of N—H⋯Owater, Owater—H⋯Cl and Owater—H⋯O hydrogen bonds (Table 2 ▸ and Fig. 4 ▸). The packing index, or percentage of filled space, is 67.1 (Kitajgorodskij, 1973 ▸) and the unit cell contains no residual solvent-accessible voids.
Table 2. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1A⋯Cl1ii | 0.91 | 2.63 | 3.316 (5) | 133 |
| N1—H1C⋯Cl1iii | 0.91 | 2.52 | 3.394 (5) | 160 |
| N2—H2B⋯Cl1iv | 0.91 | 2.25 | 3.123 (5) | 160 |
| N2—H2C⋯Cl2v | 0.91 | 2.24 | 3.151 (6) | 179 |
| N1—H1B⋯O1W vi | 0.91 | 1.90 | 2.760 (7) | 156 |
| N2—H2A⋯O2W vii | 0.91 | 2.06 | 2.789 (9) | 137 |
| O1W—H1WA⋯Cl2v | 0.89 (2) | 2.57 (8) | 3.218 (6) | 130 (8) |
| O2W—H2WA⋯Cl2viii | 0.90 (2) | 2.46 (7) | 3.176 (6) | 137 (9) |
| O2W—H2WB⋯O3i | 0.90 (2) | 2.22 (8) | 2.820 (6) | 124 (8) |
| O3W—H3WA⋯Cl1ix | 0.88 (2) | 2.66 (6) | 3.258 (5) | 127 (6) |
| O3W—H3WB⋯Cl2x | 0.88 (2) | 2.35 (3) | 3.182 (5) | 158 (7) |
Symmetry codes: (i) 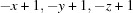 ; (ii)
; (ii)  ; (iii)
; (iii) 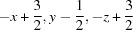 ; (iv)
; (iv) 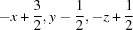 ; (v)
; (v) 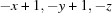 ; (vi)
; (vi) 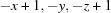 ; (vii)
; (vii) 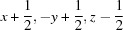 ; (viii)
; (viii) 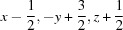 ; (ix)
; (ix) 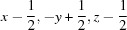 ; (x)
; (x) 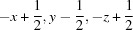 .
.
Figure 3.
A view along the c axis of the crystal structure of the title compound with the water solvent molecules omitted. The N—H⋯Cl hydrogen bonds are shown as dashed lines (see Table 2 ▸), and the C-bound H atoms have been omitted for clarity.
Figure 4.
A view along the c axis of the crystal structure of the title compound, with the hydrogen bonds shown as dashed lines (see Table 2 ▸). The C-bound H atoms have been omitted for clarity.
Database survey
A search of the Cambridge Structural Database (CSD, Version 5.38, update May 2017; Groom et al., 2016 ▸) for the skeleton of amino-methyl-cyclohexane-carboxylic acid gave 13 hits, of which six structures concern the cis- and trans-isomers. The crystal structures of the hydrobromide of the trans-isomer (CSD refcode: CHCAHB) and the hydrochloride of the cis-isomer (CHCAHC) were reported in 1966 (Kadoya et al., 1966 ▸). The crystal structure of the hydrobromide of the cis-isomer has also been reported (AMHCAC; Groth & Hassel, 1965 ▸), and that of the free cis-isomer (AMCHCA; Yamazaki et al., 1981 ▸), which does not exist as a zwitterion in the solid state. The room temperature analysis of the trans-isomer (TA), viz. tranexamic acid (AMMCHC10; Groth, 1968 ▸), and a low-temperature analysis at 173 K (AMMCHC11; Shahzadi et al., 2007 ▸) showed that it crystallizes in the chiral orthorhombic space group P212121 and exists as a zwitterion in the solid state. Interestingly, in the low-temperature structure it can be seen that the carboxylate group (COO−) is inclined to the mean plane of the four planar atoms of the cyclohexane ring by 48.9 (2)°, compared to 67.5 (6) and 85.8 (7)° in the title compound. The plane of the ammoniomethyl unit (Car—C—N) is inclined to the same mean plane of the four planar atoms of the cyclohexane ring by 37.4 (2)°, compared to 34.9 (6) and 47.5 (6)° in the title compound. Hence, on complexation the cyclohexane rings are rotated about the Ccarboxylate—Ccyclohexane bonds (C1—C2 and C9—C10), most probably to minimize steric hindrance.
In the CSD over 1500 copper(II) paddle-wheel structures have been deposited. There are only 13 compounds in which the axial position is occupied by a Cl− anion (see Supporting information). The Cu⋯Cu distances vary from ca 2.63 to 2.84 Å, with the carboxylate groups being inclined to one another by ca 84.65–90°, and the Cu—Cl distances varying from ca 2.41 to 2.49 Å. The values observed for the title compound fall within these limits (see Section 2, Structural commentary). In all 13 compounds the copper atoms have perfect square-pyramidal geometry, with τ 5 = 0.0.
Synthesis and crystallization
Tranexamic acid (0.785 g, 0.5 mmol) dissolved in 10 ml of deionized water was added dropwise to a transparent blue solution of CuCl2·2H2O (0.426 g, 0.25 mmol) in 20 ml of acetonitrile at ambient temperature and the mixture was stirred for 30 min. The green solution obtained was filtered and the filtrate kept undisturbed at room temperature for slow evaporation. After five days green plate-like crystals of the title compound were obtained.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 3 ▸. The H atoms of the water molecules were located in difference-Fourier maps and refined with distance restraints: O—H = 0.88 (2) Å with U iso(H) = 1.5U eq(O). The ammonium H atoms and the C-bound H atoms were included in calculated positions and treated as riding: N—H = 0.91 Å, C-H = 0.99–1.00 Å with U iso(H) = 1.5U eq(N-ammonium) and 1.2U eq(C) for other H atoms. In the final difference-Fourier map the residual density peaks [Δρmax, Δρmin 1.80, −0.93 e Å−3] are located at a distance of 1.2 and 0.9 Å, respectively, from the copper atoms.
Table 3. Experimental details.
| Crystal data | |
| Chemical formula | [Cu2Cl2(C8H15NO2)4](Cl2)·6H2O |
| M r | 1005.81 |
| Crystal system, space group | Monoclinic, P21/n |
| Temperature (K) | 153 |
| a, b, c (Å) | 14.7100 (11), 10.7163 (6), 14.9312 (12) |
| β (°) | 91.828 (10) |
| V (Å3) | 2352.5 (3) |
| Z | 2 |
| Radiation type | Mo Kα |
| μ (mm−1) | 1.19 |
| Crystal size (mm) | 0.34 × 0.30 × 0.20 |
| Data collection | |
| Diffractometer | STOE IPDS 1 |
| Absorption correction | Multi-scan (MULABS; Spek, 2009 ▸) |
| T min, T max | 0.712, 1.000 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 18018, 4545, 3172 |
| R int | 0.091 |
| (sin θ/λ)max (Å−1) | 0.615 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.067, 0.196, 1.02 |
| No. of reflections | 4545 |
| No. of parameters | 273 |
| No. of restraints | 9 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 1.80, −0.93 |
Supplementary Material
Crystal structure: contains datablock(s) I, Global. DOI: 10.1107/S2056989017012543/wm5416sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989017012543/wm5416Isup2.hkl
CSD search of axially Cl- ligated Cu-Cu paddle-wheel structures. DOI: 10.1107/S2056989017012543/wm5416sup3.pdf
CCDC reference: 1571897
Additional supporting information: crystallographic information; 3D view; checkCIF report
supplementary crystallographic information
Crystal data
| [Cu2Cl2(C8H15NO2)4](Cl2)·6H2O | F(000) = 1060 |
| Mr = 1005.81 | Dx = 1.420 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| a = 14.7100 (11) Å | Cell parameters from 8000 reflections |
| b = 10.7163 (6) Å | θ = 2.3–25.9° |
| c = 14.9312 (12) Å | µ = 1.19 mm−1 |
| β = 91.828 (10)° | T = 153 K |
| V = 2352.5 (3) Å3 | Plate, green |
| Z = 2 | 0.34 × 0.30 × 0.20 mm |
Data collection
| STOE IPDS 1 diffractometer | 4545 independent reflections |
| Radiation source: fine-focus sealed tube | 3172 reflections with I > 2σ(I) |
| Plane graphite monochromator | Rint = 0.091 |
| φ rotation scans | θmax = 25.9°, θmin = 2.3° |
| Absorption correction: multi-scan (MULABS; Spek, 2009) | h = −18→18 |
| Tmin = 0.712, Tmax = 1.000 | k = −13→13 |
| 18018 measured reflections | l = −18→18 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.067 | Hydrogen site location: mixed |
| wR(F2) = 0.196 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.02 | w = 1/[σ2(Fo2) + (0.1331P)2] where P = (Fo2 + 2Fc2)/3 |
| 4545 reflections | (Δ/σ)max < 0.001 |
| 273 parameters | Δρmax = 1.80 e Å−3 |
| 9 restraints | Δρmin = −0.93 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Cu1 | 0.57200 (3) | 0.53341 (6) | 0.55087 (4) | 0.0195 (2) | |
| Cl1 | 0.69386 (7) | 0.59126 (13) | 0.66374 (7) | 0.0253 (3) | |
| O1 | 0.5727 (3) | 0.3572 (4) | 0.5833 (3) | 0.0384 (10) | |
| O2 | 0.4505 (3) | 0.3014 (4) | 0.5009 (3) | 0.0369 (10) | |
| O3 | 0.6477 (2) | 0.4905 (4) | 0.4471 (2) | 0.0297 (9) | |
| O4 | 0.5250 (2) | 0.4381 (5) | 0.3640 (3) | 0.0445 (12) | |
| N1 | 0.6876 (3) | −0.1791 (5) | 0.8123 (3) | 0.0339 (11) | |
| H1A | 0.715646 | −0.207468 | 0.762784 | 0.051* | |
| H1B | 0.688742 | −0.239545 | 0.855069 | 0.051* | |
| H1C | 0.717108 | −0.110116 | 0.833690 | 0.051* | |
| N2 | 0.6727 (3) | 0.1961 (6) | −0.0237 (3) | 0.0451 (14) | |
| H2A | 0.674668 | 0.130093 | 0.014673 | 0.068* | |
| H2B | 0.702133 | 0.175997 | −0.074399 | 0.068* | |
| H2C | 0.613684 | 0.215079 | −0.038025 | 0.068* | |
| C1 | 0.5154 (3) | 0.2771 (5) | 0.5551 (3) | 0.0244 (11) | |
| C2 | 0.5225 (4) | 0.1471 (5) | 0.5916 (4) | 0.0299 (12) | |
| H2 | 0.490146 | 0.089283 | 0.548741 | 0.036* | |
| C3 | 0.6224 (4) | 0.1033 (6) | 0.6034 (4) | 0.0377 (14) | |
| H3A | 0.650187 | 0.098871 | 0.543974 | 0.045* | |
| H3B | 0.657016 | 0.165255 | 0.640008 | 0.045* | |
| C4 | 0.6295 (4) | −0.0230 (6) | 0.6482 (4) | 0.0388 (14) | |
| H4A | 0.694444 | −0.045681 | 0.657197 | 0.047* | |
| H4B | 0.600371 | −0.086743 | 0.608849 | 0.047* | |
| C5 | 0.5841 (3) | −0.0227 (5) | 0.7379 (4) | 0.0288 (12) | |
| H5 | 0.614942 | 0.042295 | 0.776158 | 0.035* | |
| C6 | 0.4837 (4) | 0.0149 (6) | 0.7258 (6) | 0.0472 (18) | |
| H6A | 0.451049 | −0.048267 | 0.688621 | 0.057* | |
| H6B | 0.455311 | 0.017746 | 0.784995 | 0.057* | |
| C7 | 0.4754 (3) | 0.1418 (6) | 0.6811 (4) | 0.0372 (14) | |
| H7A | 0.502404 | 0.205796 | 0.721666 | 0.045* | |
| H7B | 0.410239 | 0.162109 | 0.671203 | 0.045* | |
| C8 | 0.5915 (4) | −0.1465 (6) | 0.7881 (4) | 0.0397 (14) | |
| H8A | 0.556328 | −0.141231 | 0.843406 | 0.048* | |
| H8B | 0.564509 | −0.213527 | 0.750187 | 0.048* | |
| C9 | 0.6099 (3) | 0.4514 (5) | 0.3760 (3) | 0.0244 (11) | |
| C10 | 0.6679 (3) | 0.4182 (5) | 0.2966 (3) | 0.0244 (11) | |
| H10 | 0.733515 | 0.427604 | 0.315003 | 0.029* | |
| C11 | 0.6461 (4) | 0.5081 (6) | 0.2187 (4) | 0.0288 (12) | |
| H11A | 0.579838 | 0.507245 | 0.204844 | 0.035* | |
| H11B | 0.663545 | 0.593965 | 0.236560 | 0.035* | |
| C12 | 0.6970 (4) | 0.4710 (6) | 0.1351 (3) | 0.0299 (12) | |
| H12A | 0.681611 | 0.530032 | 0.085930 | 0.036* | |
| H12B | 0.763338 | 0.475985 | 0.147985 | 0.036* | |
| C13 | 0.6722 (3) | 0.3405 (5) | 0.1063 (3) | 0.0262 (12) | |
| H13 | 0.604822 | 0.337427 | 0.095228 | 0.031* | |
| C14 | 0.6965 (4) | 0.2495 (6) | 0.1830 (4) | 0.0332 (13) | |
| H14A | 0.677871 | 0.164112 | 0.164965 | 0.040* | |
| H14B | 0.763281 | 0.249287 | 0.193720 | 0.040* | |
| C15 | 0.6507 (4) | 0.2838 (6) | 0.2693 (4) | 0.0319 (12) | |
| H15A | 0.584401 | 0.270120 | 0.261501 | 0.038* | |
| H15B | 0.673527 | 0.227990 | 0.317868 | 0.038* | |
| C16 | 0.7174 (4) | 0.3051 (6) | 0.0196 (4) | 0.0347 (14) | |
| H16A | 0.714618 | 0.376924 | −0.022083 | 0.042* | |
| H16B | 0.782239 | 0.285487 | 0.032601 | 0.042* | |
| Cl2 | 0.53120 (11) | 0.73439 (19) | 0.07190 (12) | 0.0537 (5) | |
| O1W | 0.3515 (3) | 0.3850 (6) | 0.0849 (3) | 0.0600 (14) | |
| H1WA | 0.385 (4) | 0.405 (9) | 0.038 (3) | 0.090* | |
| H1WB | 0.393 (4) | 0.383 (9) | 0.130 (3) | 0.090* | |
| O2W | 0.1694 (4) | 0.5612 (6) | 0.5040 (4) | 0.0651 (15) | |
| H2WA | 0.134 (5) | 0.589 (9) | 0.548 (5) | 0.098* | |
| H2WB | 0.222 (3) | 0.602 (8) | 0.514 (6) | 0.098* | |
| O3W | 0.1194 (3) | 0.1039 (6) | 0.3118 (3) | 0.0619 (15) | |
| H3WA | 0.104 (5) | 0.075 (8) | 0.259 (3) | 0.093* | |
| H3WB | 0.073 (4) | 0.148 (8) | 0.329 (5) | 0.093* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cu1 | 0.0152 (3) | 0.0291 (4) | 0.0144 (3) | 0.0005 (2) | 0.0036 (2) | 0.0010 (2) |
| Cl1 | 0.0213 (6) | 0.0352 (8) | 0.0193 (6) | −0.0045 (5) | 0.0001 (4) | 0.0005 (5) |
| O1 | 0.035 (2) | 0.039 (3) | 0.041 (2) | −0.0092 (18) | −0.0129 (17) | 0.0117 (19) |
| O2 | 0.035 (2) | 0.033 (2) | 0.041 (2) | 0.0018 (17) | −0.0158 (17) | −0.0035 (19) |
| O3 | 0.0195 (16) | 0.050 (3) | 0.0195 (17) | −0.0029 (16) | 0.0022 (13) | −0.0049 (17) |
| O4 | 0.0187 (18) | 0.088 (4) | 0.027 (2) | −0.0061 (19) | 0.0057 (14) | −0.018 (2) |
| N1 | 0.031 (2) | 0.032 (3) | 0.038 (3) | 0.007 (2) | −0.0064 (19) | 0.001 (2) |
| N2 | 0.041 (3) | 0.056 (4) | 0.039 (3) | 0.001 (3) | 0.018 (2) | −0.019 (3) |
| C1 | 0.024 (2) | 0.031 (3) | 0.018 (2) | 0.004 (2) | 0.0067 (19) | 0.004 (2) |
| C2 | 0.032 (3) | 0.025 (3) | 0.032 (3) | 0.001 (2) | −0.008 (2) | −0.003 (2) |
| C3 | 0.032 (3) | 0.048 (4) | 0.034 (3) | 0.015 (3) | 0.016 (2) | 0.011 (3) |
| C4 | 0.039 (3) | 0.040 (4) | 0.038 (3) | 0.015 (3) | −0.001 (2) | 0.003 (3) |
| C5 | 0.020 (2) | 0.029 (3) | 0.037 (3) | 0.001 (2) | −0.002 (2) | 0.004 (2) |
| C6 | 0.022 (3) | 0.036 (4) | 0.084 (5) | 0.002 (2) | 0.010 (3) | 0.022 (4) |
| C7 | 0.019 (2) | 0.037 (4) | 0.056 (4) | 0.007 (2) | 0.010 (2) | 0.019 (3) |
| C8 | 0.028 (3) | 0.040 (4) | 0.051 (4) | 0.005 (3) | 0.004 (2) | 0.009 (3) |
| C9 | 0.022 (2) | 0.030 (3) | 0.022 (2) | 0.001 (2) | 0.0045 (19) | 0.004 (2) |
| C10 | 0.014 (2) | 0.039 (3) | 0.021 (2) | 0.001 (2) | 0.0046 (17) | 0.000 (2) |
| C11 | 0.034 (3) | 0.029 (3) | 0.024 (3) | 0.002 (2) | 0.008 (2) | −0.001 (2) |
| C12 | 0.032 (3) | 0.037 (4) | 0.021 (3) | 0.004 (2) | 0.010 (2) | 0.004 (2) |
| C13 | 0.020 (2) | 0.035 (3) | 0.024 (2) | 0.004 (2) | 0.0101 (18) | −0.006 (2) |
| C14 | 0.031 (3) | 0.030 (3) | 0.039 (3) | 0.002 (2) | 0.011 (2) | 0.000 (3) |
| C15 | 0.036 (3) | 0.031 (3) | 0.030 (3) | 0.003 (2) | 0.011 (2) | 0.002 (2) |
| C16 | 0.029 (3) | 0.047 (4) | 0.028 (3) | 0.003 (2) | 0.013 (2) | −0.012 (3) |
| Cl2 | 0.0453 (9) | 0.0647 (13) | 0.0518 (10) | −0.0012 (8) | 0.0122 (7) | 0.0128 (9) |
| O1W | 0.062 (3) | 0.068 (4) | 0.051 (3) | 0.008 (3) | 0.014 (2) | 0.021 (3) |
| O2W | 0.048 (3) | 0.075 (4) | 0.071 (4) | 0.012 (3) | −0.008 (3) | −0.019 (3) |
| O3W | 0.052 (3) | 0.081 (4) | 0.053 (3) | 0.018 (3) | 0.000 (2) | −0.034 (3) |
Geometric parameters (Å, º)
| Cu1—Cu1i | 2.6649 (11) | C6—H6A | 0.9900 |
| Cu1—O1 | 1.950 (4) | C6—H6B | 0.9900 |
| Cu1—O2i | 1.955 (4) | C7—H7A | 0.9900 |
| Cu1—O4i | 1.965 (4) | C7—H7B | 0.9900 |
| Cu1—O3 | 1.991 (3) | C8—H8A | 0.9900 |
| Cu1—Cl1 | 2.4990 (12) | C8—H8B | 0.9900 |
| O1—C1 | 1.265 (7) | C9—C10 | 1.526 (6) |
| O2—C1 | 1.258 (6) | C10—C15 | 1.516 (8) |
| O3—C9 | 1.255 (6) | C10—C11 | 1.536 (7) |
| O4—C9 | 1.264 (6) | C10—H10 | 1.0000 |
| N1—C8 | 1.489 (7) | C11—C12 | 1.527 (7) |
| N1—H1A | 0.9100 | C11—H11A | 0.9900 |
| N1—H1B | 0.9100 | C11—H11B | 0.9900 |
| N1—H1C | 0.9100 | C12—C13 | 1.505 (8) |
| N2—C16 | 1.479 (8) | C12—H12A | 0.9900 |
| N2—H2A | 0.9100 | C12—H12B | 0.9900 |
| N2—H2B | 0.9100 | C13—C16 | 1.522 (7) |
| N2—H2C | 0.9100 | C13—C14 | 1.538 (8) |
| C1—C2 | 1.499 (8) | C13—H13 | 1.0000 |
| C2—C7 | 1.526 (8) | C14—C15 | 1.518 (7) |
| C2—C3 | 1.547 (7) | C14—H14A | 0.9900 |
| C2—H2 | 1.0000 | C14—H14B | 0.9900 |
| C3—C4 | 1.512 (9) | C15—H15A | 0.9900 |
| C3—H3A | 0.9900 | C15—H15B | 0.9900 |
| C3—H3B | 0.9900 | C16—H16A | 0.9900 |
| C4—C5 | 1.515 (8) | C16—H16B | 0.9900 |
| C4—H4A | 0.9900 | O1W—H1WA | 0.89 (2) |
| C4—H4B | 0.9900 | O1W—H1WB | 0.90 (2) |
| C5—C8 | 1.526 (8) | O2W—H2WA | 0.90 (2) |
| C5—C6 | 1.536 (7) | O2W—H2WB | 0.90 (2) |
| C5—H5 | 1.0000 | O3W—H3WA | 0.88 (2) |
| C6—C7 | 1.518 (9) | O3W—H3WB | 0.88 (2) |
| O1—Cu1—O2i | 167.02 (15) | C5—C6—H6B | 109.6 |
| O1—Cu1—O4i | 89.3 (2) | H6A—C6—H6B | 108.1 |
| O2i—Cu1—O4i | 89.9 (2) | C6—C7—C2 | 112.7 (5) |
| O1—Cu1—O3 | 88.33 (18) | C6—C7—H7A | 109.0 |
| O2i—Cu1—O3 | 89.60 (18) | C2—C7—H7A | 109.0 |
| O4i—Cu1—O3 | 167.27 (15) | C6—C7—H7B | 109.0 |
| O1—Cu1—Cl1 | 94.31 (11) | C2—C7—H7B | 109.0 |
| O2i—Cu1—Cl1 | 98.68 (11) | H7A—C7—H7B | 107.8 |
| O4i—Cu1—Cl1 | 92.57 (11) | N1—C8—C5 | 112.0 (5) |
| O3—Cu1—Cl1 | 100.08 (10) | N1—C8—H8A | 109.2 |
| O1—Cu1—Cu1i | 83.04 (11) | C5—C8—H8A | 109.2 |
| O2i—Cu1—Cu1i | 84.05 (11) | N1—C8—H8B | 109.2 |
| O4i—Cu1—Cu1i | 80.24 (11) | C5—C8—H8B | 109.2 |
| O3—Cu1—Cu1i | 87.06 (10) | H8A—C8—H8B | 107.9 |
| Cl1—Cu1—Cu1i | 172.34 (4) | O3—C9—O4 | 124.6 (5) |
| C1—O1—Cu1 | 125.2 (3) | O3—C9—C10 | 119.4 (4) |
| C1—O2—Cu1i | 123.8 (4) | O4—C9—C10 | 116.0 (4) |
| C9—O3—Cu1 | 119.4 (3) | C15—C10—C9 | 109.8 (4) |
| C9—O4—Cu1i | 128.7 (3) | C15—C10—C11 | 111.3 (4) |
| C8—N1—H1A | 109.5 | C9—C10—C11 | 109.5 (4) |
| C8—N1—H1B | 109.5 | C15—C10—H10 | 108.7 |
| H1A—N1—H1B | 109.5 | C9—C10—H10 | 108.7 |
| C8—N1—H1C | 109.5 | C11—C10—H10 | 108.7 |
| H1A—N1—H1C | 109.5 | C12—C11—C10 | 111.1 (4) |
| H1B—N1—H1C | 109.5 | C12—C11—H11A | 109.4 |
| C16—N2—H2A | 109.5 | C10—C11—H11A | 109.4 |
| C16—N2—H2B | 109.5 | C12—C11—H11B | 109.4 |
| H2A—N2—H2B | 109.5 | C10—C11—H11B | 109.4 |
| C16—N2—H2C | 109.5 | H11A—C11—H11B | 108.0 |
| H2A—N2—H2C | 109.5 | C13—C12—C11 | 110.8 (4) |
| H2B—N2—H2C | 109.5 | C13—C12—H12A | 109.5 |
| O2—C1—O1 | 123.9 (5) | C11—C12—H12A | 109.5 |
| O2—C1—C2 | 118.0 (5) | C13—C12—H12B | 109.5 |
| O1—C1—C2 | 118.1 (4) | C11—C12—H12B | 109.5 |
| C1—C2—C7 | 108.9 (5) | H12A—C12—H12B | 108.1 |
| C1—C2—C3 | 112.2 (5) | C12—C13—C16 | 111.5 (5) |
| C7—C2—C3 | 110.2 (4) | C12—C13—C14 | 109.2 (4) |
| C1—C2—H2 | 108.5 | C16—C13—C14 | 112.2 (5) |
| C7—C2—H2 | 108.5 | C12—C13—H13 | 107.9 |
| C3—C2—H2 | 108.5 | C16—C13—H13 | 107.9 |
| C4—C3—C2 | 112.0 (5) | C14—C13—H13 | 107.9 |
| C4—C3—H3A | 109.2 | C15—C14—C13 | 112.3 (5) |
| C2—C3—H3A | 109.2 | C15—C14—H14A | 109.1 |
| C4—C3—H3B | 109.2 | C13—C14—H14A | 109.1 |
| C2—C3—H3B | 109.2 | C15—C14—H14B | 109.1 |
| H3A—C3—H3B | 107.9 | C13—C14—H14B | 109.1 |
| C3—C4—C5 | 111.3 (5) | H14A—C14—H14B | 107.9 |
| C3—C4—H4A | 109.4 | C10—C15—C14 | 112.5 (5) |
| C5—C4—H4A | 109.4 | C10—C15—H15A | 109.1 |
| C3—C4—H4B | 109.4 | C14—C15—H15A | 109.1 |
| C5—C4—H4B | 109.4 | C10—C15—H15B | 109.1 |
| H4A—C4—H4B | 108.0 | C14—C15—H15B | 109.1 |
| C4—C5—C8 | 113.9 (5) | H15A—C15—H15B | 107.8 |
| C4—C5—C6 | 110.2 (5) | N2—C16—C13 | 111.6 (5) |
| C8—C5—C6 | 109.8 (5) | N2—C16—H16A | 109.3 |
| C4—C5—H5 | 107.5 | C13—C16—H16A | 109.3 |
| C8—C5—H5 | 107.5 | N2—C16—H16B | 109.3 |
| C6—C5—H5 | 107.5 | C13—C16—H16B | 109.3 |
| C7—C6—C5 | 110.5 (5) | H16A—C16—H16B | 108.0 |
| C7—C6—H6A | 109.6 | H1WA—O1W—H1WB | 102 (3) |
| C5—C6—H6A | 109.6 | H2WA—O2W—H2WB | 103 (3) |
| C7—C6—H6B | 109.6 | H3WA—O3W—H3WB | 106 (3) |
| Cu1i—O2—C1—O1 | 1.1 (7) | Cu1—O3—C9—O4 | 1.0 (8) |
| Cu1i—O2—C1—C2 | 178.1 (4) | Cu1—O3—C9—C10 | −179.8 (4) |
| Cu1—O1—C1—O2 | 0.9 (7) | Cu1i—O4—C9—O3 | −2.3 (9) |
| Cu1—O1—C1—C2 | −176.1 (4) | Cu1i—O4—C9—C10 | 178.5 (4) |
| O2—C1—C2—C7 | −94.2 (6) | O3—C9—C10—C15 | 122.8 (5) |
| O1—C1—C2—C7 | 82.9 (6) | O4—C9—C10—C15 | −57.9 (6) |
| O2—C1—C2—C3 | 143.5 (5) | O3—C9—C10—C11 | −114.7 (5) |
| O1—C1—C2—C3 | −39.4 (7) | O4—C9—C10—C11 | 64.5 (6) |
| C1—C2—C3—C4 | 174.7 (5) | C15—C10—C11—C12 | −53.7 (6) |
| C7—C2—C3—C4 | 53.1 (7) | C9—C10—C11—C12 | −175.2 (4) |
| C2—C3—C4—C5 | −56.3 (7) | C10—C11—C12—C13 | 58.9 (6) |
| C3—C4—C5—C8 | −178.1 (5) | C11—C12—C13—C16 | 176.2 (4) |
| C3—C4—C5—C6 | 57.9 (7) | C11—C12—C13—C14 | −59.3 (5) |
| C4—C5—C6—C7 | −57.2 (8) | C12—C13—C14—C15 | 56.5 (6) |
| C8—C5—C6—C7 | 176.5 (6) | C16—C13—C14—C15 | −179.4 (5) |
| C5—C6—C7—C2 | 55.9 (8) | C9—C10—C15—C14 | 172.3 (4) |
| C1—C2—C7—C6 | −176.8 (5) | C11—C10—C15—C14 | 50.9 (6) |
| C3—C2—C7—C6 | −53.3 (7) | C13—C14—C15—C10 | −52.9 (6) |
| C4—C5—C8—N1 | 63.3 (7) | C12—C13—C16—N2 | −162.4 (5) |
| C6—C5—C8—N1 | −172.5 (5) | C14—C13—C16—N2 | 74.9 (6) |
Symmetry code: (i) −x+1, −y+1, −z+1.
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1A···Cl1ii | 0.91 | 2.63 | 3.316 (5) | 133 |
| N1—H1C···Cl1iii | 0.91 | 2.52 | 3.394 (5) | 160 |
| N2—H2B···Cl1iv | 0.91 | 2.25 | 3.123 (5) | 160 |
| N2—H2C···Cl2v | 0.91 | 2.24 | 3.151 (6) | 179 |
| N1—H1B···O1Wvi | 0.91 | 1.90 | 2.760 (7) | 156 |
| N2—H2A···O2Wvii | 0.91 | 2.06 | 2.789 (9) | 137 |
| O1W—H1WA···Cl2v | 0.89 (2) | 2.57 (8) | 3.218 (6) | 130 (8) |
| O2W—H2WA···Cl2viii | 0.90 (2) | 2.46 (7) | 3.176 (6) | 137 (9) |
| O2W—H2WB···O3i | 0.90 (2) | 2.22 (8) | 2.820 (6) | 124 (8) |
| O3W—H3WA···Cl1ix | 0.88 (2) | 2.66 (6) | 3.258 (5) | 127 (6) |
| O3W—H3WB···Cl2x | 0.88 (2) | 2.35 (3) | 3.182 (5) | 158 (7) |
Symmetry codes: (i) −x+1, −y+1, −z+1; (ii) x, y−1, z; (iii) −x+3/2, y−1/2, −z+3/2; (iv) −x+3/2, y−1/2, −z+1/2; (v) −x+1, −y+1, −z; (vi) −x+1, −y, −z+1; (vii) x+1/2, −y+1/2, z−1/2; (viii) x−1/2, −y+3/2, z+1/2; (ix) x−1/2, −y+1/2, z−1/2; (x) −x+1/2, y−1/2, −z+1/2.
Funding Statement
This work was funded by Swiss National Science Foundation grant . the University of Neuchâtel grant .
References
- Addison, A. W., Rao, T. N., Reedijk, J., van Rijn, J. & Verschoor, G. C. (1984). J. Chem. Soc. Dalton Trans. pp. 1349–1356.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Groth, P., Hassel, O., Klaeboe, P. & Enari, T.-M. (1965). Acta Chem. Scand. 19, 1709–1714.
- Groth, P., Rasmussen, S. E., Taylor, D. B., Haug, A., Enzell, C. & Francis, G. (1968). Acta Chem. Scand. 22, 143–158.
- Kadoya, S., Hanazaki, F. & Iitaka, Y. (1966). Acta Cryst. 21, 38–49.
- Kitajgorodskij, A. I. (1973). Molecular Crystals and Molecules. New York: Academic Press, USA.
- Macrae, C. F., Bruno, I. J., Chisholm, J. A., Edgington, P. R., McCabe, P., Pidcock, E., Rodriguez-Monge, L., Taylor, R., van de Streek, J. & Wood, P. A. (2008). J. Appl. Cryst. 41, 466–470.
- Melander, B., Gliniecki, G., Granstrand, B. & Hanshoff, G. (1965). Acta Pharmacol. Toxicol. 22, 340–352. [DOI] [PubMed]
- Melvin, J. S., Stryker, L. S. & Sierra, R. J. (2015). J. Am. Acad. Orthop. Surg. 23, 732–740. [DOI] [PubMed]
- Napolitano, L. M., Cohen, M. J., Cotton, B. A., Schreiber, M. A. & Moore, E. E. (2013). J. Trauma Acute Care Surg. 74, 1575–1786. [DOI] [PubMed]
- Okamoto, S. & Okamoto, U. (1962). Keio j. Med. 11, 105–115.
- Okamoto, S., Sato, S., Takada, Y. & Okamoto, U. (1964). Keio J. Med. 13, 177–185. [DOI] [PubMed]
- Shahzadi, S., Ali, S. M., Parvez, M., Badshah, A., Ahmed, E. & Malik, A. (2007). Russ. J. Inorg. Chem. 52, 386–393.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2015). Acta Cryst. C71, 3–8.
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Stoe & Cie (2004). IPDSI Bedienungshandbuch. Stoe & Cie GmbH, Darmstadt, Germany.
- Tengborn, L., Blombäck, M. & Berntorp, E. (2015). Thromb. Res. 135, 231–242. [DOI] [PubMed]
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
- Yamazaki, K., Watanabe, A., Moroi, R. & Sano, M. (1981). Acta Cryst. B37, 1447–1449.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, Global. DOI: 10.1107/S2056989017012543/wm5416sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989017012543/wm5416Isup2.hkl
CSD search of axially Cl- ligated Cu-Cu paddle-wheel structures. DOI: 10.1107/S2056989017012543/wm5416sup3.pdf
CCDC reference: 1571897
Additional supporting information: crystallographic information; 3D view; checkCIF report