The synthesis and structures of three manganese(II) pyridine N-oxide complexes are presented.
Keywords: crystal structure, manganese, pyridine N-oxide ligand
Abstract
Three manganese(II) N-oxide complexes have been synthesized from the reaction of manganese(II) chloride with either pyridine N-oxide (PNO), 2-methylpyridine N-oxide (2MePNO) or 3-methylpyridine N-oxide (3MePNO). The compounds were synthesized from methanolic solutions of MnCl2·4H2O and the respective N-oxide, and subsequently characterized structurally by single-crystal X-ray diffraction. The compounds are catena-poly[[aquachloridomanganese(II)]-di-μ-chlorido-[aquachloridomanganese(II)]-bis(μ-pyridine N-oxide)], [MnCl2(C5H5NO)(H2O)]n or [MnCl2(PNO)(H2O)]n (I), catena-poly[[aquachloridomanganese(II)]-di-μ-chlorido-[aquachloridomanganese(II)]-bis(μ-2-methylpyridine N-oxide)], [MnCl2(C6H7NO)(H2O)]n or [MnCl2(2MePNO)(H2O)]n (II), and bis(μ-3-methylpyridine N-oxide)bis[diaquadichloridomanganese(II)], [Mn2Cl4(C6H7NO)2(H2O)4] or [MnCl2(3MePNO)(H2O)2]2 (III). The MnII atoms are found in pseudo-octahedral environments for each of the three complexes. Compound I forms a coordination polymer with alternating pairs of bridging N-oxide and chloride ligands. The coordination environment is defined by two PNO bridging O atoms, two chloride bridging atoms, a terminal chloride, and a terminal water. Compound II also forms a coordination polymer with a similar metal cation; however, the coordination polymer is bridged between MnII atoms by both a single chloride and 2MePNO. The distorted octahedrally coordinated metal cation is defined by two bridging 2MePNO trans to each other, two chlorides, also trans to one another in the equatorial (polymeric) plane, and a terminal chloride and terminal water. Finally, complex III forms a dimer with two bridging 3MePNOs, two terminal chlorides and two terminal waters forming the six-coordinate metal environment. All three compounds exhibit hydrogen bonding between the coordinating water(s) and terminal chlorides.
Chemical context
The utility of aromatic N-oxides to facilitate organic oxo-transfer reactions has been well documented over the years (see, for example, Eppenson, 2003 ▸). Many of these reactions are actually catalyzed by transition metal interactions with the N-oxide ligands (see, for example, Moustafa et al., 2014 ▸). Furthermore, N-oxide metal interactions have recently attracted much interest in a variety of other areas, including metal organic frameworks (MOFs) (Hu et al., 2014 ▸). These MOFs synthesized using N-oxide derivatives take advantage of the multiple binding modes of the sp 3 O atom and the ease of modification of the organic backbone of the N-oxide. The utility of the MOFs has been examined in areas such as catalysis (Liu et al., 2014 ▸) and sensors (Hu et al., 2014 ▸). The constructs extend to the supramolecular study of coordination polymers that have been found in this type of complex because of their incredible versatility as ligands (Sarma & Baruah, 2011 ▸).
In this context, we report the synthesis and solid-state structures of three pyridine N-oxide manganese(II) complexes. Notably, we used the ligands pyridine N-oxide, 2-methylpyridine N-oxide, and 3-methylpyridine N-oxide to study the impact of substitution of the pyridine on the two- and three-dimensional solid-state structures. The pyridine N-oxide (PNO) and 2-methylpyridine N-oxide (2MePNO) complexes form coordination polymers with subtle differences. The 3-methylpyridine N-oxide (3MePNO), however, forms a dimeric complex.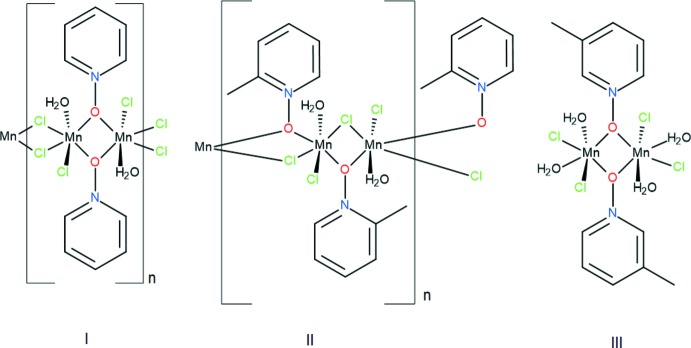
Structural commentary
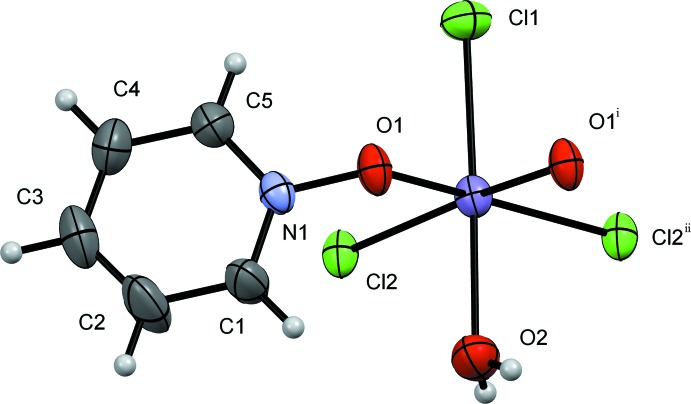
Complex I exhibits the repeating motif of [MnCl2(PNO)(H2O)]n and crystallizes in the triclinic space group P
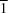 , containing two formula units per unit cell (Fig. 1 ▸). The coordination sphere around each MnII atom is a distorted octahedron, with the equatorial atoms being two bridging chlorides alternating with two bridging pyridine N-oxide (PNO) molecules (Fig. 2 ▸). In the equatorial plane, the bridging chlorides and the bridging pyridine N-oxides are cis to one another. The axial positions are a terminal chloride and a water molecule. The Mn1—O1 bond length is 2.177 (3) Å, whereas the Mn1—O1vii bond length is slightly longer at 2.182 (3) Å for the bridging PNO [symmetry code (vii) −x + 1, −y + 1, −z + 1]. The bridging chlorides are found to have Mn—Cl2 distances of 2.5240 (19) and 2.532 (19) Å, respectively. Axially, the water is located 2.250 (3) Å from the MnII cation and the terminal chloride is at 2.479 (2) Å. The bond angles around the equator are severely compressed at the two bridging N-oxides, with the O1—Mn1—O1i angle observed at 72.03 (10)°. The remaining three angles are found to all be similar at 95.58 (7) (Cl2—Mn1—Cl2i), 96.80 (8) (O1—Mn1—Cl2), and 94.69 (9)° (O1vii—Mn1—Cl2vii). Axially, the bond angle from the water through manganese(II) and the terminal chloride (O2—Mn1—Cl1) is nearly linear at 177.36 (8)°.
, containing two formula units per unit cell (Fig. 1 ▸). The coordination sphere around each MnII atom is a distorted octahedron, with the equatorial atoms being two bridging chlorides alternating with two bridging pyridine N-oxide (PNO) molecules (Fig. 2 ▸). In the equatorial plane, the bridging chlorides and the bridging pyridine N-oxides are cis to one another. The axial positions are a terminal chloride and a water molecule. The Mn1—O1 bond length is 2.177 (3) Å, whereas the Mn1—O1vii bond length is slightly longer at 2.182 (3) Å for the bridging PNO [symmetry code (vii) −x + 1, −y + 1, −z + 1]. The bridging chlorides are found to have Mn—Cl2 distances of 2.5240 (19) and 2.532 (19) Å, respectively. Axially, the water is located 2.250 (3) Å from the MnII cation and the terminal chloride is at 2.479 (2) Å. The bond angles around the equator are severely compressed at the two bridging N-oxides, with the O1—Mn1—O1i angle observed at 72.03 (10)°. The remaining three angles are found to all be similar at 95.58 (7) (Cl2—Mn1—Cl2i), 96.80 (8) (O1—Mn1—Cl2), and 94.69 (9)° (O1vii—Mn1—Cl2vii). Axially, the bond angle from the water through manganese(II) and the terminal chloride (O2—Mn1—Cl1) is nearly linear at 177.36 (8)°.
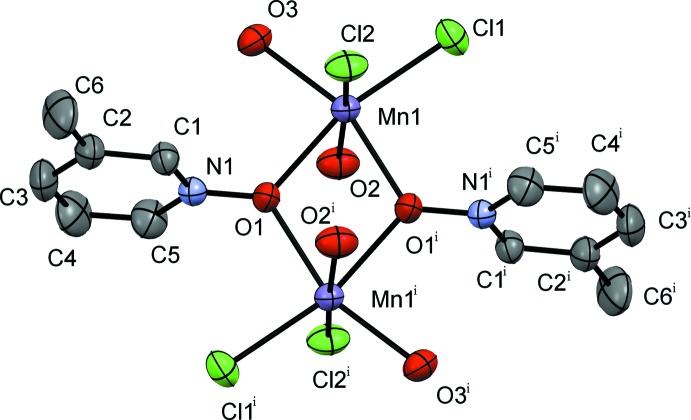
Figure 1.
A view of compound I, showing the atom labeling. Displacement ellipsoids are drawn at the 50% probability level. [Symmetry codes: (i) −x + 1, −y + 1, −z + 1; (ii) −x, -y+1, −z + 1]
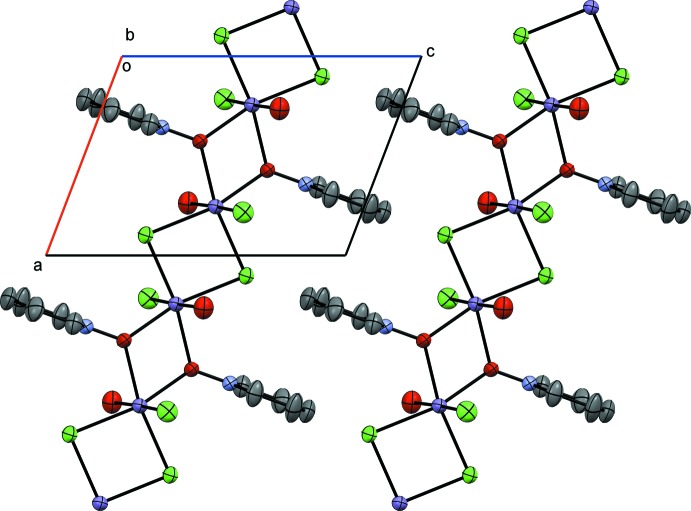
Figure 2.
Crystal packing diagram of compound I, viewed along the b axis. H atoms have been omitted for clarity.
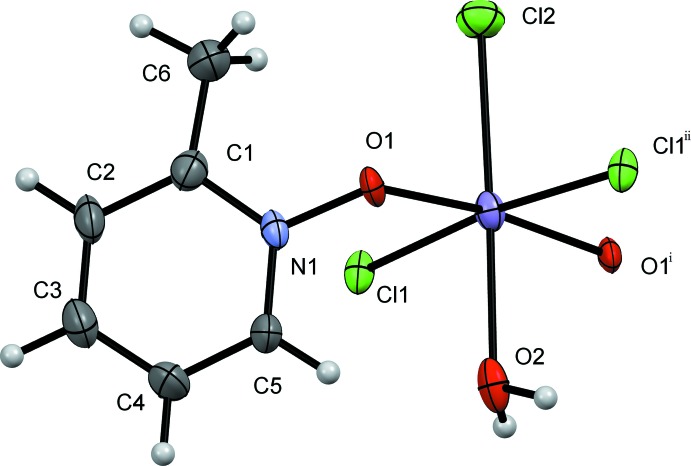
Complex II, [MnCl2(2MePNO)(H2O)]n, posseses a metal environment similar to complex I and crystallizes in the orthorhombic space group P212121. The major difference in structure II is in the bridging network, where the chlorides and N-oxides are trans to one another rather than cis as in I (Figs. 3 ▸ and 4 ▸). The pseudo-octahedral environment includes an Mn1—Cl1 bond length of 2.516 (4) Å and an Mn1—O1 (N-oxide) bond length of 2.170 (6) Å, with a Cl1—Mn1—O1 bond angle of 84.37 (19)°. The bond angle across the Cl atoms, Cl1—Mn1—Cl1viii, is 174.02 (5)° and across the O atoms of 2MePNO, O1—Mn1—O1ix, is 173.12 (6)°; a slight compression is observed across the bridges [symmetry codes: (viii) −x − 1, y +  , −z +
, −z +  ; (ix) −x, y +
; (ix) −x, y +  , −z +
, −z +  ]. The axial (non-bridging) Mn1—Cl2 bond length is 2.503 (4) Å, while the axial water is found at a distance of 2.268 (6) Å from the metal center.
]. The axial (non-bridging) Mn1—Cl2 bond length is 2.503 (4) Å, while the axial water is found at a distance of 2.268 (6) Å from the metal center.
Figure 3.
A view of compound II, showing the atom labeling. Displacement ellipsoids are drawn at the 50% probability level. [Symmetry codes: (i) −x − 1, y +  , −z +
, −z +  ; (ii) −x, y +
; (ii) −x, y +  , −z +
, −z +  .]
.]
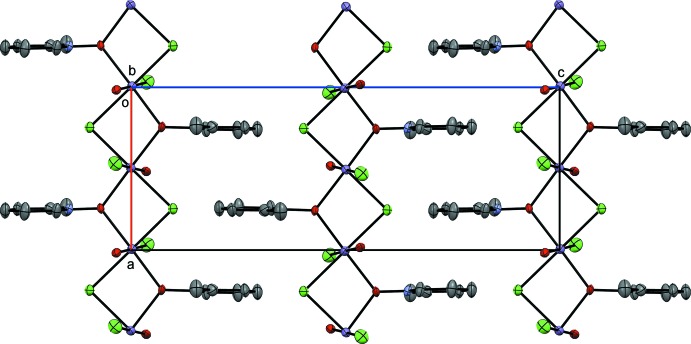
Figure 4.
Crystal packing diagram of compound II, viewed along the b axis. H atoms have been omitted for clarity.
The dimeric complex III, [MnCl2(3MePNO)(OH2)2]2, crystallizes in the triclinic P
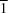 space group, with the inversion center sitting in the center of the dimer (Fig. 5 ▸). The 3-methyl derivative does not form a coordination polymer but discrete dimeric molecules. The structure possesses two bridging 3MePNO ligands, four terminal chlorides, and four terminal waters. Two waters and two chlorides are in the equatorial plane coincident with the N-oxide bridge, and the other equivalents are axial in the pseudo-octahedral geometry around the MnII atoms. The Mn1—Cl1 and Mn1—Cl2 bond lengths are 2.4601 (5) and 2.4903 (19) Å, respectively, with a Cl1—Mn1—Cl2 bond angle of 98.32 (4)°. The bridging N-oxide is at a distance of 2.1791 (18) Å from Mn1—O1, with an O1—Mn1—O1vii bond angle of 71.86 (7)° [symmetry code: (vii) −x + 1, −y + 1, −z + 1]. The Mn1—O2(water) and Mn1—O3(water) bond lengths are 2.245 (2) and 2.1696 (17) Å, respectively, with an O2—Mn1—O3 bond angle of 85.83 (7)°.
space group, with the inversion center sitting in the center of the dimer (Fig. 5 ▸). The 3-methyl derivative does not form a coordination polymer but discrete dimeric molecules. The structure possesses two bridging 3MePNO ligands, four terminal chlorides, and four terminal waters. Two waters and two chlorides are in the equatorial plane coincident with the N-oxide bridge, and the other equivalents are axial in the pseudo-octahedral geometry around the MnII atoms. The Mn1—Cl1 and Mn1—Cl2 bond lengths are 2.4601 (5) and 2.4903 (19) Å, respectively, with a Cl1—Mn1—Cl2 bond angle of 98.32 (4)°. The bridging N-oxide is at a distance of 2.1791 (18) Å from Mn1—O1, with an O1—Mn1—O1vii bond angle of 71.86 (7)° [symmetry code: (vii) −x + 1, −y + 1, −z + 1]. The Mn1—O2(water) and Mn1—O3(water) bond lengths are 2.245 (2) and 2.1696 (17) Å, respectively, with an O2—Mn1—O3 bond angle of 85.83 (7)°.
Figure 5.
A view of the molecular structure of compound III, showing the atom labeling. Displacement ellipsoids are drawn at the 50% probability level. H atoms have been omitted for clarity. [Symmetry code: (i) −x − 1, −y + 1, −z + 1.]
The formation of the polymeric structure in I and II versus the dimer in III is likely due to the steric influence of the methyl group in the 3-position in 3MePNO and the core constituents. One can define the Mn2 ‘N-oxide diamond core’ in each of the structures as follows: I is alternating Mn2Cl2 and Mn2O2 (oxygen bridges via PNO) cores, II is Mn2ClO (oxygen bridge via 2MePNO) and III Mn2O2 (oxygen bridges via 3MePNO). In I, the unsubstituted pyridine N-oxide group does not generate as much steric strain, allowing for polymer formation. In II, the core is formed to permit alternating up and down pyridine N-oxides with the 2-methyl substituents also facing in opposite directions. This limits the steric interactions and the N-oxide slightly tilts out of the polymeric core line to allow the methyl group to effect less steric interactions. In III, the methyl group appears to inhibit polymer formation due to the position of this bulky substituent. Subsequently, when the polymer is not formed, an extra water molecule is required to fill the sixth coordination site on the metal cation occupied by a bridging atom in I and II.
Supramolecular features
The packing of I forms a coordination polymer of alternating bis-bridges of two chlorides and two pyridine N-oxides in the a-axis direction (Fig. 2 ▸). The aromatic rings stack at 6.860 (7) Å, outside of π-stacking distance due to the alternating chloride and pyridine N-oxide bridges. The single water molecule is locked into weak hydrogen-bonding interactions in two different modes. One hydrogen-bond interaction (H2A) is located down the bridge to the terminal chloride (Cl1), on the adjacent MnII atom, and the O2—H2A⋯Cl1i distance is 2.53 (2) Å. The other hydrogen-bond interaction (H2B) is across to the next polymeric chain with Cl1; the O2—H2B⋯Cl1ii distance is 2.52 (3) Å (see Table 1 ▸ for hydrogen-bond details and symmetry codes).
Table 1. Hydrogen-bond geometry (Å, °) for I .
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O2—H2A⋯Cl1i | 0.83 (2) | 2.53 (2) | 3.348 (4) | 168 (4) |
| O2—H2B⋯Cl1ii | 0.82 (2) | 2.52 (3) | 3.232 (4) | 147 (4) |
Symmetry codes: (i) 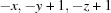 ; (ii)
; (ii)  .
.
Complex II packs as a coordination polymer in the a direction similar to I (Fig. 4 ▸). However, as I has alternating pyridine N-oxide and chloride bridges (placing these ligands cis to one another), II has a single 2-methylpyridine N-oxide and a single chloride in each bridge. Similar to I, the hydrogen-bonding interactions are to a terminal chloride (Cl2) on the adjacent MnII atom. There are two observed interactions, viz. O2—H2A⋯Cl2iii with a distance of 2.49 Å and O2—H2B⋯Cl2iv with a distance of 2.26 Å (see Table 2 ▸ for hydrogen-bond details and symmetry codes). The H2A⋯ Cl2 interaction is in the coordination polymer and the H2B⋯Cl2 interaction is across the polymeric chains. Similar to I, the aromatic rings stack too far apart to be interacting in the a direction, at a distance of 6.862 (11) Å.
Table 2. Hydrogen-bond geometry (Å, °) for II .
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O2—H2A⋯Cl2iii | 0.90 | 2.49 | 3.205 (7) | 137 |
| O2—H2B⋯Cl2iv | 0.89 | 2.26 | 3.145 (7) | 169 |
Symmetry codes: (iii) 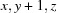 ; (iv)
; (iv) 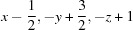 .
.
As noted above, compound III does not form a coordination polymer but is observed in the solid state as a dimer with two water molecules for each MnII atom (versus one aqua equivalent in I and II) (Fig. 5 ▸). The aromatic inter-centroid distance is longer than in the other two molecules, at 7.902 (7) Å. In compound III, a single water molecule hydrogen bonds from the equatorial plane of one dimer to an axial chloride on another dimer. Conversely, the axial water hydrogen bonds to an equatorial chloride on a different dimer. These interactions are found to be O2—H2B⋯Cl1v [distance 2.38 (2) Å] and O3—H3A⋯Cl2vi [distance 2.28 (2) Å] (see Table 3 ▸ for hydrogen-bond details and symmetry codes).
Table 3. Hydrogen-bond geometry (Å, °) for III .
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O2—H2B⋯Cl1v | 0.80 (2) | 2.38 (2) | 3.147 (3) | 161 (2) |
| O3—H3A⋯Cl2vi | 0.86 (2) | 2.28 (2) | 3.120 (2) | 167 (2) |
Symmetry codes: (v) 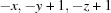 ; (vi)
; (vi) 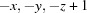 .
.
Database survey
A search in the Cambridge Structural Database (CSD; Groom et al., 2016 ▸) for aromatic N-oxides bound to manganese returned 87 entries. Similar N-oxides with simple counter-ions in the list include 4,4′-dipyridine N,N′-dioxide [FIVHAU (Ma et al., 2005 ▸) and XOHQUH (Jiu et al., 2008 ▸)], 1,2-bis(4-pyridyl)ethane N,N′-dioxide (TOJDAY and TOJDIG; Sun et al., 2008 ▸), and 1,3-bis(4-pyridyl)propane N,N′-dioxide (Zhang et al., 2003 ▸). Similarly, two derivatives of 3,5-dimethylpyridine N-oxide are found in the CSD (GIWQAF and GIWQEJ; Shi et al., 2007 ▸)
Synthesis and crystallization
The title compounds were all synthesized in a similar manner. 0.200 g of MnCl2·4H2O (1.01 mmol) was dissolved in a minimal amount of methanol, approximately 10 ml. Two stoichiometric equivalents of the appropriate N-oxide were also dissolved in approximately 20 ml of methanol (PNO: 0.191 g, 2.02 mmol; 2MePNO: 0.220 g, 2.02 mmol; 3MePNO: 0.220 g, 2.02 mmol). The solutions were stirred for approximately 10 min; during each reaction, a brown solution was observed upon mixing. The reaction solution was then allowed to sit and brown crystals were grown by slow evaporation in the near quantitative yields reported below based on the manganese(II) chloride starting material. The FT–IR spectra of the complexes all exhibit broad absorbances in the 3400–3000 cm−1 region due to the ν(O—H) of the coordinating water molecules, as well as the characteristic ν(N—O) of the N-oxide pyridyl moiety in the 1250–1150 cm−1 region noted previously (Mautner et al., 2017 ▸).
Compound I, Mn(PNO)Cl2·H2O, yield 0.215 g (90.3%). Selected IR bands (ATR, FT–IR, KBr composite, cm−1): 3364 (m, br), 3235 (m, br), 3068 (m, br), 1660 (w), 1471 (w), 1214 (m), 1205 (m), 1023 (s), 831 (s), 780 (s), 674 (s), 556 (s). Elemental analysis for MnCl2C5H7NO2, calculated (%): C 25.13, H 2.95, N 5.86; found (%): C 25.22, H 2.96, N 5.87.
Compound II, Mn(2MePNO)Cl2·H2O, yield 0.227 g (87.9%). Selected IR bands (ATR, FT–IR, KBr composite, cm−1): 3410 (m, br), 3247(m, br), 3073(m, br), 1716 (w), 1619 (w), 1578 (w), 1421 (m), 1264 (m), 1154 (m), 1029 (s), 831 (s), 799 (s), 684 (s), 584 (s). Elemental analysis for MnCl2C6H9NO2, calculated (%): C 28.48, H 3.59, N 5.53; found (%): C 28.75, H 3.53, N 5.28.
Compound III, Mn2(3MePNO)2Cl4·4H2O, yield 0.231 g (89.5%). Selected IR bands (ATR, FT–IR, KBr composite, cm−1): 3374 (m, br), 3251 (m, br), 3094 (m, br), 1663 (w), 1614(w), 1492 (m), 1261 (m), 1164 (m), 1019 (s), 946 (s), 750 (s), 672 (s). Elemental analysis for Mn2Cl4C12H22N2O6, calculated (%): C 26.59, H 4.09, N 5.16; found (%): C 26.53, H 4.04, N 5.21.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 4 ▸. All carbon-bound H atoms were positioned geometrically and refined as riding, with C—H = 0.95 or 0.98 Å and U iso(H) = 1.2U eq(C) or U iso(H) = 1.5U eq(C) for C(H) and CH3 groups, respectively. In order to ensure chemically meaningful O—H distances for the bound water molecules in compound I, the H2A—O2 and H2B—O2 distances were restrained to a target value of 0.84 (2) Å (using a DFIX command in SHELXL2017; Sheldrick, 2015b ▸). In compound II, water H atoms were refined as riding, with the O—H distance constrained to 0.892 Å and U iso(H) = 1.5U eq(O) using an AFIX 7 command, and in compound III, H2A—O2, H2B—O2, H3A—O3, and H3B—O3 were restrained using DFIX as for compound I. A rotating-group model was applied for the methyl groups. Structure refinement of II exhibits inversion twinning. Several crystals were tried and the centrosymmetric space group Pnma was tested. In all cases, there was a significant reduction in the R value for the inversion twinning P212121 solution.
Table 4. Experimental details.
| I | II | III | |
|---|---|---|---|
| Crystal data | |||
| Chemical formula | [MnCl2(C5H5NO)(H2O)] | [MnCl2(C6H7NO)(H2O)] | [Mn2Cl4(C6H7NO)2(H2O)4] |
| M r | 238.96 | 252.98 | 541.99 |
| Crystal system, space group | Triclinic, P
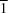
|
Orthorhombic, P212121 | Triclinic, P
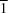
|
| Temperature (K) | 173 | 173 | 173 |
| a, b, c (Å) | 6.897 (2), 7.050 (1), 9.853 (3) | 6.862 (2), 7.491 (2), 18.047 (5) | 7.902 (7), 8.026 (7), 9.893 (8) |
| α, β, γ (°) | 101.042 (7), 109.559 (10), 94.196 (6) | 90, 90, 90 | 98.033 (1), 99.272 (7), 113.634 (11) |
| V (Å3) | 438.2 (2) | 927.7 (4) | 552.6 (8) |
| Z | 2 | 4 | 1 |
| Radiation type | Mo Kα | Mo Kα | Mo Kα |
| μ (mm−1) | 2.06 | 1.96 | 1.65 |
| Crystal size (mm) | 0.29 × 0.18 × 0.13 | 0.2 × 0.2 × 0.1 | 0.85 × 0.50 × 0.28 |
| Data collection | |||
| Diffractometer | Rigaku XtalLab mini CCD | Rigaku XtalLab mini CCD | Rigaku XtalLab mini CCD |
| Absorption correction | Multi-scan (REQAB; Rigaku, 1998 ▸) | Multi-scan (REQAB; Rigaku, 1998 ▸) | Multi-scan (REQAB; Rigaku, 1998 ▸) |
| T min, T max | 0.613, 0.765 | 0.563, 0.737 | 0.482, 0.630 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 4655, 2004, 1770 | 8438, 2109, 1800 | 5837, 2553, 2375 |
| R int | 0.040 | 0.051 | 0.072 |
| (sin θ/λ)max (Å−1) | 0.651 | 0.649 | 0.652 |
| Refinement | |||
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.031, 0.080, 1.14 | 0.051, 0.100, 1.12 | 0.031, 0.087, 1.07 |
| No. of reflections | 2004 | 2109 | 2553 |
| No. of parameters | 108 | 112 | 135 |
| No. of restraints | 2 | 0 | 4 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement | H-atom parameters constrained | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.40, −0.44 | 0.95, −0.73 | 0.56, −0.41 |
| Absolute structure | – | Refined as an inversion twin | – |
| Absolute structure parameter | – | 0.44 (8) | – |
Supplementary Material
Crystal structure: contains datablock(s) I, II, III. DOI: 10.1107/S2056989017012038/zl2709sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989017012038/zl2709Isup2.hkl
Structure factors: contains datablock(s) II. DOI: 10.1107/S2056989017012038/zl2709IIsup3.hkl
Structure factors: contains datablock(s) III. DOI: 10.1107/S2056989017012038/zl2709IIIsup4.hkl
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
The authors would like to thank Armstrong State University, Department of Chemistry and Physics, for financial support of this work.
supplementary crystallographic information
catena-poly[[aquachloridomanganese(II)]-di-µ-chlorido-[aquachloridomanganese(II)]-bis(µ-pyridine N-oxide)] (I) . Crystal data
| [MnCl2(C5H5NO)(H2O)] | Z = 2 |
| Mr = 238.96 | F(000) = 238 |
| Triclinic, P1 | Dx = 1.811 Mg m−3 |
| a = 6.897 (2) Å | Mo Kα radiation, λ = 0.71073 Å |
| b = 7.050 (1) Å | Cell parameters from 1189 reflections |
| c = 9.853 (3) Å | θ = 2.3–27.5° |
| α = 101.042 (7)° | µ = 2.06 mm−1 |
| β = 109.559 (10)° | T = 173 K |
| γ = 94.196 (6)° | Prism, clear brown |
| V = 438.2 (2) Å3 | 0.29 × 0.18 × 0.13 mm |
catena-poly[[aquachloridomanganese(II)]-di-µ-chlorido-[aquachloridomanganese(II)]-bis(µ-pyridine N-oxide)] (I) . Data collection
| Rigaku XtalLab mini CCD diffractometer | 1770 reflections with I > 2σ(I) |
| ω scans | Rint = 0.040 |
| Absorption correction: multi-scan (REQAB; Rigaku, 1998) | θmax = 27.6°, θmin = 2.3° |
| Tmin = 0.613, Tmax = 0.765 | h = −8→8 |
| 4655 measured reflections | k = −9→9 |
| 2004 independent reflections | l = −12→12 |
catena-poly[[aquachloridomanganese(II)]-di-µ-chlorido-[aquachloridomanganese(II)]-bis(µ-pyridine N-oxide)] (I) . Refinement
| Refinement on F2 | Primary atom site location: dual |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.031 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.080 | w = 1/[σ2(Fo2) + 0.444P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.14 | (Δ/σ)max = 0.001 |
| 2004 reflections | Δρmax = 0.40 e Å−3 |
| 108 parameters | Δρmin = −0.44 e Å−3 |
| 2 restraints |
catena-poly[[aquachloridomanganese(II)]-di-µ-chlorido-[aquachloridomanganese(II)]-bis(µ-pyridine N-oxide)] (I) . Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
catena-poly[[aquachloridomanganese(II)]-di-µ-chlorido-[aquachloridomanganese(II)]-bis(µ-pyridine N-oxide)] (I) . Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Mn1 | 0.24478 (6) | 0.51944 (6) | 0.49588 (4) | 0.02748 (12) | |
| Cl1 | 0.21608 (12) | 0.81797 (10) | 0.39668 (9) | 0.04150 (19) | |
| O1 | 0.5727 (3) | 0.6094 (3) | 0.63106 (19) | 0.0327 (4) | |
| N1 | 0.6425 (3) | 0.6894 (3) | 0.7767 (2) | 0.0283 (5) | |
| C1 | 0.6940 (6) | 0.5762 (5) | 0.8731 (3) | 0.0493 (8) | |
| H1 | 0.681940 | 0.441595 | 0.840110 | 0.059* | |
| Cl2 | 0.10246 (10) | 0.65268 (10) | 0.69397 (7) | 0.03317 (16) | |
| O2 | 0.2634 (4) | 0.2541 (3) | 0.5927 (3) | 0.0447 (5) | |
| H2A | 0.146 (4) | 0.218 (6) | 0.594 (5) | 0.081 (16)* | |
| H2B | 0.305 (7) | 0.162 (5) | 0.551 (5) | 0.087 (16)* | |
| C2 | 0.7655 (7) | 0.6608 (6) | 1.0222 (4) | 0.0655 (11) | |
| H2 | 0.802254 | 0.582800 | 1.090313 | 0.079* | |
| C3 | 0.7826 (6) | 0.8564 (6) | 1.0704 (3) | 0.0543 (9) | |
| H3 | 0.829678 | 0.913307 | 1.171227 | 0.065* | |
| C4 | 0.7299 (6) | 0.9692 (5) | 0.9693 (4) | 0.0532 (9) | |
| H4 | 0.742760 | 1.104037 | 1.001068 | 0.064* | |
| C5 | 0.6577 (5) | 0.8836 (5) | 0.8203 (3) | 0.0421 (7) | |
| H5 | 0.619974 | 0.959566 | 0.750808 | 0.051* |
catena-poly[[aquachloridomanganese(II)]-di-µ-chlorido-[aquachloridomanganese(II)]-bis(µ-pyridine N-oxide)] (I) . Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Mn1 | 0.0228 (2) | 0.0328 (2) | 0.0259 (2) | 0.00479 (16) | 0.00858 (16) | 0.00467 (16) |
| Cl1 | 0.0478 (4) | 0.0329 (4) | 0.0481 (4) | 0.0073 (3) | 0.0188 (4) | 0.0156 (3) |
| O1 | 0.0230 (9) | 0.0460 (12) | 0.0238 (9) | 0.0052 (8) | 0.0062 (8) | −0.0006 (8) |
| N1 | 0.0232 (11) | 0.0371 (12) | 0.0233 (10) | 0.0033 (9) | 0.0078 (9) | 0.0052 (9) |
| C1 | 0.076 (2) | 0.0386 (17) | 0.0325 (15) | 0.0152 (16) | 0.0152 (16) | 0.0124 (13) |
| Cl2 | 0.0307 (3) | 0.0377 (4) | 0.0267 (3) | 0.0016 (3) | 0.0102 (3) | −0.0016 (3) |
| O2 | 0.0553 (16) | 0.0394 (13) | 0.0420 (13) | 0.0117 (11) | 0.0186 (12) | 0.0115 (10) |
| C2 | 0.094 (3) | 0.073 (3) | 0.0290 (16) | 0.031 (2) | 0.0135 (19) | 0.0204 (17) |
| C3 | 0.057 (2) | 0.071 (3) | 0.0249 (15) | 0.0074 (18) | 0.0091 (15) | −0.0009 (15) |
| C4 | 0.067 (2) | 0.0425 (19) | 0.0391 (17) | −0.0022 (17) | 0.0151 (17) | −0.0064 (15) |
| C5 | 0.054 (2) | 0.0388 (17) | 0.0325 (15) | 0.0027 (14) | 0.0149 (14) | 0.0075 (13) |
catena-poly[[aquachloridomanganese(II)]-di-µ-chlorido-[aquachloridomanganese(II)]-bis(µ-pyridine N-oxide)] (I) . Geometric parameters (Å, º)
| Mn1—Cl1 | 2.479 (2) | C1—C2 | 1.376 (5) |
| Mn1—O1 | 2.177 (3) | O2—H2A | 0.833 (19) |
| Mn1—O1i | 2.182 (2) | O2—H2B | 0.819 (19) |
| Mn1—Cl2ii | 2.5324 (19) | C2—H2 | 0.9300 |
| Mn1—Cl2 | 2.5240 (19) | C2—C3 | 1.353 (5) |
| Mn1—O2 | 2.250 (3) | C3—H3 | 0.9300 |
| O1—N1 | 1.341 (3) | C3—C4 | 1.364 (5) |
| N1—C1 | 1.331 (4) | C4—H4 | 0.9300 |
| N1—C5 | 1.339 (4) | C4—C5 | 1.377 (4) |
| C1—H1 | 0.9300 | C5—H5 | 0.9300 |
| Cl1—Mn1—Cl2 | 93.43 (6) | C5—N1—O1 | 118.0 (2) |
| Cl1—Mn1—Cl2ii | 92.57 (6) | N1—C1—H1 | 120.4 |
| O1i—Mn1—Cl1 | 95.12 (8) | N1—C1—C2 | 119.1 (3) |
| O1—Mn1—Cl1 | 93.65 (7) | C2—C1—H1 | 120.4 |
| O1—Mn1—O1i | 72.02 (10) | Mn1—Cl2—Mn1ii | 84.42 (7) |
| O1—Mn1—Cl2ii | 165.77 (6) | Mn1—O2—H2A | 108 (3) |
| O1i—Mn1—Cl2ii | 94.69 (9) | Mn1—O2—H2B | 116 (3) |
| O1i—Mn1—Cl2 | 166.32 (5) | H2A—O2—H2B | 110 (4) |
| O1—Mn1—Cl2 | 96.81 (8) | C1—C2—H2 | 119.7 |
| O1—Mn1—O2 | 86.96 (9) | C3—C2—C1 | 120.5 (3) |
| O1i—Mn1—O2 | 87.46 (10) | C3—C2—H2 | 119.7 |
| Cl2—Mn1—Cl2ii | 95.58 (7) | C2—C3—H3 | 120.4 |
| O2—Mn1—Cl1 | 177.42 (7) | C2—C3—C4 | 119.1 (3) |
| O2—Mn1—Cl2ii | 87.40 (8) | C4—C3—H3 | 120.4 |
| O2—Mn1—Cl2 | 84.01 (9) | C3—C4—H4 | 120.0 |
| Mn1—O1—Mn1i | 107.98 (10) | C3—C4—C5 | 120.1 (3) |
| N1—O1—Mn1 | 123.78 (14) | C5—C4—H4 | 120.0 |
| N1—O1—Mn1i | 126.50 (16) | N1—C5—C4 | 119.1 (3) |
| C1—N1—O1 | 119.9 (2) | N1—C5—H5 | 120.5 |
| C1—N1—C5 | 122.0 (3) | C4—C5—H5 | 120.5 |
Symmetry codes: (i) −x+1, −y+1, −z+1; (ii) −x, −y+1, −z+1.
catena-poly[[aquachloridomanganese(II)]-di-µ-chlorido-[aquachloridomanganese(II)]-bis(µ-pyridine N-oxide)] (I) . Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O2—H2A···Cl1ii | 0.83 (2) | 2.53 (2) | 3.348 (4) | 168 (4) |
| O2—H2B···Cl1iii | 0.82 (2) | 2.52 (3) | 3.232 (4) | 147 (4) |
Symmetry codes: (ii) −x, −y+1, −z+1; (iii) x, y−1, z.
catena-Poly[[aquachloridomanganese(II)]-di-µ-chlorido-[aquachloridomanganese(II)]-bis(µ-2-methylpyridine N-oxide)] (II) . Crystal data
| [MnCl2(C6H7NO)(H2O)] | Dx = 1.811 Mg m−3 |
| Mr = 252.98 | Mo Kα radiation, λ = 0.71075 Å |
| Orthorhombic, P212121 | Cell parameters from 2686 reflections |
| a = 6.862 (2) Å | θ = 2.7–27.5° |
| b = 7.491 (2) Å | µ = 1.96 mm−1 |
| c = 18.047 (5) Å | T = 173 K |
| V = 927.7 (4) Å3 | Prism, colorless |
| Z = 4 | 0.2 × 0.2 × 0.1 mm |
| F(000) = 508 |
catena-Poly[[aquachloridomanganese(II)]-di-µ-chlorido-[aquachloridomanganese(II)]-bis(µ-2-methylpyridine N-oxide)] (II) . Data collection
| Rigaku XtalLab mini CCD diffractometer | 1800 reflections with I > 2σ(I) |
| ω scans | Rint = 0.051 |
| Absorption correction: multi-scan (REQAB; Rigaku, 1998) | θmax = 27.5°, θmin = 2.3° |
| Tmin = 0.563, Tmax = 0.737 | h = −8→8 |
| 8438 measured reflections | k = −9→9 |
| 2109 independent reflections | l = −23→23 |
catena-Poly[[aquachloridomanganese(II)]-di-µ-chlorido-[aquachloridomanganese(II)]-bis(µ-2-methylpyridine N-oxide)] (II) . Refinement
| Refinement on F2 | Hydrogen site location: mixed |
| Least-squares matrix: full | H-atom parameters constrained |
| R[F2 > 2σ(F2)] = 0.051 | w = 1/[σ2(Fo2) + (0.004P)2 + 2.3909P] where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.100 | (Δ/σ)max < 0.001 |
| S = 1.12 | Δρmax = 0.95 e Å−3 |
| 2109 reflections | Δρmin = −0.73 e Å−3 |
| 112 parameters | Absolute structure: Refined as an inversion twin. |
| 0 restraints | Absolute structure parameter: 0.44 (8) |
| Primary atom site location: dual |
catena-Poly[[aquachloridomanganese(II)]-di-µ-chlorido-[aquachloridomanganese(II)]-bis(µ-2-methylpyridine N-oxide)] (II) . Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refined as a 2-component inversion twin. |
catena-Poly[[aquachloridomanganese(II)]-di-µ-chlorido-[aquachloridomanganese(II)]-bis(µ-2-methylpyridine N-oxide)] (II) . Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Mn1 | 0.5059 (2) | 0.73290 (15) | 0.50137 (7) | 0.0197 (2) | |
| Cl1 | 0.2537 (3) | 0.6800 (2) | 0.40260 (8) | 0.0226 (3) | |
| O1 | 0.7561 (8) | 0.7085 (6) | 0.42855 (19) | 0.0179 (9) | |
| N1 | 0.7503 (11) | 0.7249 (7) | 0.3544 (2) | 0.0199 (11) | |
| C1 | 0.7553 (14) | 0.5782 (9) | 0.3112 (3) | 0.0276 (15) | |
| Cl2 | 0.5309 (3) | 0.4110 (3) | 0.53742 (11) | 0.0325 (5) | |
| O2 | 0.4833 (8) | 1.0223 (7) | 0.4655 (3) | 0.0265 (12) | |
| H2A | 0.546895 | 1.093811 | 0.497004 | 0.040* | |
| H2B | 0.358601 | 1.056880 | 0.464095 | 0.040* | |
| C2 | 0.7532 (15) | 0.6041 (10) | 0.2345 (3) | 0.0321 (16) | |
| H2 | 0.758660 | 0.503891 | 0.202342 | 0.038* | |
| C3 | 0.7436 (16) | 0.7719 (11) | 0.2056 (4) | 0.0370 (18) | |
| H3 | 0.740766 | 0.787849 | 0.153377 | 0.044* | |
| C4 | 0.7378 (16) | 0.9180 (10) | 0.2515 (4) | 0.0371 (19) | |
| H4 | 0.731159 | 1.035141 | 0.231495 | 0.044* | |
| C5 | 0.7418 (15) | 0.8925 (9) | 0.3261 (3) | 0.0283 (16) | |
| H5 | 0.738625 | 0.992359 | 0.358479 | 0.034* | |
| C6 | 0.7629 (17) | 0.4059 (10) | 0.3484 (4) | 0.040 (2) | |
| H6A | 0.757534 | 0.310158 | 0.311520 | 0.048* | |
| H6B | 0.651845 | 0.395059 | 0.382265 | 0.048* | |
| H6C | 0.884583 | 0.396660 | 0.376629 | 0.048* |
catena-Poly[[aquachloridomanganese(II)]-di-µ-chlorido-[aquachloridomanganese(II)]-bis(µ-2-methylpyridine N-oxide)] (II) . Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Mn1 | 0.0171 (4) | 0.0266 (5) | 0.0154 (4) | −0.0028 (5) | 0.0017 (4) | −0.0008 (4) |
| Cl1 | 0.0212 (7) | 0.0300 (9) | 0.0167 (6) | 0.0003 (10) | 0.0004 (8) | −0.0069 (6) |
| O1 | 0.020 (2) | 0.025 (3) | 0.0089 (16) | −0.005 (3) | −0.002 (2) | −0.0013 (16) |
| N1 | 0.019 (2) | 0.029 (3) | 0.012 (2) | 0.002 (3) | 0.002 (3) | −0.001 (2) |
| C1 | 0.028 (4) | 0.028 (4) | 0.026 (3) | 0.003 (5) | −0.001 (4) | −0.005 (3) |
| Cl2 | 0.0377 (12) | 0.0234 (10) | 0.0365 (11) | −0.0021 (10) | 0.0037 (9) | 0.0050 (8) |
| O2 | 0.015 (3) | 0.048 (3) | 0.016 (2) | −0.009 (3) | 0.002 (2) | −0.003 (2) |
| C2 | 0.036 (4) | 0.046 (5) | 0.015 (3) | 0.007 (6) | 0.002 (4) | −0.004 (3) |
| C3 | 0.048 (5) | 0.046 (5) | 0.017 (3) | 0.004 (6) | 0.001 (4) | 0.001 (3) |
| C4 | 0.060 (5) | 0.027 (4) | 0.024 (4) | 0.002 (6) | 0.003 (5) | 0.005 (3) |
| C5 | 0.042 (4) | 0.026 (4) | 0.017 (3) | 0.007 (5) | 0.004 (4) | −0.002 (3) |
| C6 | 0.061 (6) | 0.029 (4) | 0.029 (4) | 0.007 (6) | 0.001 (5) | 0.000 (3) |
catena-Poly[[aquachloridomanganese(II)]-di-µ-chlorido-[aquachloridomanganese(II)]-bis(µ-2-methylpyridine N-oxide)] (II) . Geometric parameters (Å, º)
| Mn1—Cl1i | 2.514 (3) | O2—H2B | 0.8947 |
| Mn1—Cl1 | 2.516 (4) | C2—H2 | 0.9500 |
| Mn1—O1ii | 2.174 (5) | C2—C3 | 1.363 (10) |
| Mn1—O1 | 2.171 (6) | C3—H3 | 0.9500 |
| Mn1—Cl2 | 2.503 (4) | C3—C4 | 1.374 (10) |
| Mn1—O2 | 2.268 (6) | C4—H4 | 0.9500 |
| O1—N1 | 1.345 (6) | C4—C5 | 1.360 (9) |
| N1—C1 | 1.348 (8) | C5—H5 | 0.9500 |
| N1—C5 | 1.357 (8) | C6—H6A | 0.9800 |
| C1—C2 | 1.397 (9) | C6—H6B | 0.9800 |
| C1—C6 | 1.456 (10) | C6—H6C | 0.9800 |
| O2—H2A | 0.8951 | ||
| Cl1i—Mn1—Cl1 | 174.01 (5) | N1—C1—C6 | 117.1 (6) |
| O1ii—Mn1—Cl1 | 84.38 (18) | C2—C1—C6 | 125.5 (7) |
| O1—Mn1—Cl1i | 84.49 (18) | Mn1—O2—H2A | 110.9 |
| O1ii—Mn1—Cl1i | 94.57 (18) | Mn1—O2—H2B | 110.5 |
| O1—Mn1—Cl1 | 95.84 (18) | H2A—O2—H2B | 108.1 |
| O1—Mn1—O1ii | 173.11 (6) | C1—C2—H2 | 119.7 |
| O1—Mn1—Cl2 | 91.26 (14) | C3—C2—C1 | 120.5 (7) |
| O1ii—Mn1—Cl2 | 95.58 (14) | C3—C2—H2 | 119.7 |
| O1ii—Mn1—O2 | 85.39 (19) | C2—C3—H3 | 119.8 |
| O1—Mn1—O2 | 87.78 (19) | C2—C3—C4 | 120.3 (6) |
| Cl2—Mn1—Cl1i | 91.40 (9) | C4—C3—H3 | 119.8 |
| Cl2—Mn1—Cl1 | 94.57 (9) | C3—C4—H4 | 120.5 |
| O2—Mn1—Cl1 | 84.34 (16) | C5—C4—C3 | 119.0 (7) |
| O2—Mn1—Cl1i | 89.71 (16) | C5—C4—H4 | 120.5 |
| O2—Mn1—Cl2 | 178.46 (15) | N1—C5—C4 | 120.3 (6) |
| Mn1ii—Cl1—Mn1 | 86.32 (13) | N1—C5—H5 | 119.9 |
| Mn1—O1—Mn1i | 104.73 (19) | C4—C5—H5 | 119.9 |
| N1—O1—Mn1i | 125.7 (5) | C1—C6—H6A | 109.5 |
| N1—O1—Mn1 | 124.9 (5) | C1—C6—H6B | 109.5 |
| O1—N1—C1 | 120.1 (5) | C1—C6—H6C | 109.5 |
| O1—N1—C5 | 117.4 (5) | H6A—C6—H6B | 109.5 |
| C1—N1—C5 | 122.5 (5) | H6A—C6—H6C | 109.5 |
| N1—C1—C2 | 117.4 (7) | H6B—C6—H6C | 109.5 |
| Mn1—O1—N1—C1 | −102.4 (8) | C1—N1—C5—C4 | 0.0 (15) |
| Mn1i—O1—N1—C1 | 105.6 (8) | C1—C2—C3—C4 | 0.7 (17) |
| Mn1—O1—N1—C5 | 78.4 (9) | C2—C3—C4—C5 | 0.0 (17) |
| Mn1i—O1—N1—C5 | −73.7 (9) | C3—C4—C5—N1 | −0.3 (17) |
| O1—N1—C1—C2 | −178.6 (8) | C5—N1—C1—C2 | 0.6 (14) |
| O1—N1—C1—C6 | 1.2 (13) | C5—N1—C1—C6 | −179.6 (10) |
| O1—N1—C5—C4 | 179.3 (9) | C6—C1—C2—C3 | 179.2 (11) |
| N1—C1—C2—C3 | −1.0 (16) |
Symmetry codes: (i) x+1/2, −y+3/2, −z+1; (ii) x−1/2, −y+3/2, −z+1.
catena-Poly[[aquachloridomanganese(II)]-di-µ-chlorido-[aquachloridomanganese(II)]-bis(µ-2-methylpyridine N-oxide)] (II) . Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O2—H2A···Cl2iii | 0.90 | 2.49 | 3.205 (7) | 137 |
| O2—H2B···Cl2ii | 0.89 | 2.26 | 3.145 (7) | 169 |
Symmetry codes: (ii) x−1/2, −y+3/2, −z+1; (iii) x, y+1, z.
Bis(µ-3-methylpyridine N-oxide)bis[diaquadichloridomanganese(II)] (III) . Crystal data
| [Mn2Cl4(C6H7NO)2(H2O)4] | Z = 1 |
| Mr = 541.99 | F(000) = 274 |
| Triclinic, P1 | Dx = 1.629 Mg m−3 |
| a = 7.902 (7) Å | Mo Kα radiation, λ = 0.71073 Å |
| b = 8.026 (7) Å | Cell parameters from 5886 reflections |
| c = 9.893 (8) Å | θ = 2.8–27.5° |
| α = 98.033 (1)° | µ = 1.65 mm−1 |
| β = 99.272 (7)° | T = 173 K |
| γ = 113.634 (11)° | Prism, clear brown |
| V = 552.6 (8) Å3 | 0.85 × 0.5 × 0.28 mm |
Bis(µ-3-methylpyridine N-oxide)bis[diaquadichloridomanganese(II)] (III) . Data collection
| Rigaku XtalLab mini CCD diffractometer | 2375 reflections with I > 2σ(I) |
| ω scans | Rint = 0.072 |
| Absorption correction: multi-scan (REQAB; Rigaku, 1998) | θmax = 27.6°, θmin = 2.8° |
| Tmin = 0.482, Tmax = 0.630 | h = −10→10 |
| 5837 measured reflections | k = −10→10 |
| 2553 independent reflections | l = −12→12 |
Bis(µ-3-methylpyridine N-oxide)bis[diaquadichloridomanganese(II)] (III) . Refinement
| Refinement on F2 | Primary atom site location: dual |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.031 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.087 | w = 1/[σ2(Fo2) + (0.031P)2 + 0.023P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.07 | (Δ/σ)max < 0.001 |
| 2553 reflections | Δρmax = 0.56 e Å−3 |
| 135 parameters | Δρmin = −0.41 e Å−3 |
| 4 restraints |
Bis(µ-3-methylpyridine N-oxide)bis[diaquadichloridomanganese(II)] (III) . Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Bis(µ-3-methylpyridine N-oxide)bis[diaquadichloridomanganese(II)] (III) . Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Mn1 | 0.27031 (3) | 0.40104 (3) | 0.53553 (2) | 0.02936 (11) | |
| Cl1 | 0.14922 (6) | 0.48607 (7) | 0.73553 (4) | 0.04342 (14) | |
| Cl2 | 0.32299 (6) | 0.13672 (6) | 0.60668 (6) | 0.04544 (15) | |
| O1 | 0.56562 (15) | 0.60090 (16) | 0.62683 (12) | 0.0345 (3) | |
| O2 | 0.2605 (2) | 0.6467 (2) | 0.45620 (16) | 0.0443 (3) | |
| H2A | 0.355 (3) | 0.710 (3) | 0.436 (2) | 0.055 (7)* | |
| H2B | 0.167 (3) | 0.639 (3) | 0.404 (2) | 0.062 (8)* | |
| O3 | 0.00694 (17) | 0.23434 (18) | 0.37696 (14) | 0.0395 (3) | |
| H3A | −0.069 (3) | 0.128 (2) | 0.390 (2) | 0.062 (7)* | |
| H3B | −0.054 (4) | 0.288 (4) | 0.352 (3) | 0.081 (10)* | |
| N1 | 0.62886 (17) | 0.71302 (18) | 0.75570 (13) | 0.0294 (3) | |
| C1 | 0.6561 (2) | 0.8903 (2) | 0.77007 (17) | 0.0342 (4) | |
| H1 | 0.631326 | 0.934009 | 0.690425 | 0.041* | |
| C2 | 0.7204 (3) | 1.0102 (3) | 0.90084 (19) | 0.0415 (4) | |
| C3 | 0.7538 (3) | 0.9400 (3) | 1.0169 (2) | 0.0568 (6) | |
| H3 | 0.795802 | 1.016517 | 1.106619 | 0.068* | |
| C4 | 0.7250 (4) | 0.7571 (4) | 0.9997 (2) | 0.0611 (6) | |
| H4 | 0.748138 | 0.710090 | 1.077885 | 0.073* | |
| C5 | 0.6620 (3) | 0.6431 (3) | 0.8670 (2) | 0.0472 (5) | |
| H5 | 0.642729 | 0.519251 | 0.854911 | 0.057* | |
| C6 | 0.7494 (4) | 1.2087 (3) | 0.9131 (3) | 0.0701 (7) | |
| H6A | 0.873126 | 1.282752 | 0.900126 | 0.105* | |
| H6B | 0.739622 | 1.256760 | 1.004510 | 0.105* | |
| H6C | 0.653880 | 1.213706 | 0.842403 | 0.105* |
Bis(µ-3-methylpyridine N-oxide)bis[diaquadichloridomanganese(II)] (III) . Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Mn1 | 0.02821 (16) | 0.02780 (17) | 0.03004 (17) | 0.00959 (12) | 0.00843 (11) | 0.00651 (12) |
| Cl1 | 0.0471 (3) | 0.0617 (3) | 0.0309 (2) | 0.0299 (2) | 0.01431 (18) | 0.0142 (2) |
| Cl2 | 0.0436 (3) | 0.0349 (3) | 0.0596 (3) | 0.0137 (2) | 0.0151 (2) | 0.0236 (2) |
| O1 | 0.0326 (6) | 0.0310 (6) | 0.0313 (6) | 0.0074 (5) | 0.0091 (5) | −0.0010 (5) |
| O2 | 0.0389 (7) | 0.0414 (7) | 0.0584 (9) | 0.0185 (6) | 0.0137 (6) | 0.0231 (7) |
| O3 | 0.0339 (6) | 0.0322 (7) | 0.0438 (7) | 0.0079 (5) | 0.0039 (5) | 0.0080 (6) |
| N1 | 0.0295 (6) | 0.0293 (7) | 0.0271 (6) | 0.0107 (5) | 0.0061 (5) | 0.0062 (5) |
| C1 | 0.0443 (9) | 0.0307 (8) | 0.0273 (7) | 0.0159 (7) | 0.0082 (6) | 0.0067 (6) |
| C2 | 0.0484 (10) | 0.0363 (9) | 0.0336 (8) | 0.0138 (8) | 0.0112 (7) | 0.0016 (7) |
| C3 | 0.0674 (13) | 0.0598 (13) | 0.0262 (9) | 0.0158 (11) | 0.0049 (9) | 0.0014 (9) |
| C4 | 0.0766 (14) | 0.0687 (15) | 0.0355 (10) | 0.0289 (12) | 0.0028 (10) | 0.0246 (10) |
| C5 | 0.0591 (11) | 0.0424 (10) | 0.0425 (10) | 0.0236 (9) | 0.0064 (9) | 0.0186 (9) |
| C6 | 0.105 (2) | 0.0393 (11) | 0.0571 (14) | 0.0257 (13) | 0.0237 (14) | −0.0048 (10) |
Bis(µ-3-methylpyridine N-oxide)bis[diaquadichloridomanganese(II)] (III) . Geometric parameters (Å, º)
| Mn1—Cl1 | 2.4602 (15) | C1—H1 | 0.9300 |
| Mn1—Cl2 | 2.4900 (19) | C1—C2 | 1.381 (2) |
| Mn1—O1i | 2.2228 (17) | C2—C3 | 1.381 (3) |
| Mn1—O1 | 2.1792 (18) | C2—C6 | 1.500 (3) |
| Mn1—O2 | 2.246 (2) | C3—H3 | 0.9300 |
| Mn1—O3 | 2.1704 (17) | C3—C4 | 1.373 (4) |
| O1—N1 | 1.3411 (19) | C4—H4 | 0.9300 |
| O2—H2A | 0.791 (15) | C4—C5 | 1.377 (3) |
| O2—H2B | 0.802 (16) | C5—H5 | 0.9300 |
| O3—H3A | 0.863 (16) | C6—H6A | 0.9600 |
| O3—H3B | 0.795 (17) | C6—H6B | 0.9600 |
| N1—C1 | 1.334 (2) | C6—H6C | 0.9600 |
| N1—C5 | 1.340 (2) | ||
| Cl1—Mn1—Cl2 | 98.31 (4) | C1—N1—O1 | 119.45 (13) |
| O1—Mn1—Cl1 | 95.44 (6) | C1—N1—C5 | 121.69 (16) |
| O1i—Mn1—Cl1 | 165.45 (4) | C5—N1—O1 | 118.86 (16) |
| O1i—Mn1—Cl2 | 89.66 (5) | N1—C1—H1 | 119.3 |
| O1—Mn1—Cl2 | 93.11 (7) | N1—C1—C2 | 121.31 (16) |
| O1—Mn1—O1i | 71.87 (7) | C2—C1—H1 | 119.3 |
| O1i—Mn1—O2 | 82.07 (6) | C1—C2—C3 | 117.8 (2) |
| O1—Mn1—O2 | 81.64 (7) | C1—C2—C6 | 119.87 (19) |
| O2—Mn1—Cl1 | 89.20 (6) | C3—C2—C6 | 122.4 (2) |
| O2—Mn1—Cl2 | 171.24 (4) | C2—C3—H3 | 120.0 |
| O3—Mn1—Cl1 | 101.10 (7) | C4—C3—C2 | 119.99 (19) |
| O3—Mn1—Cl2 | 97.13 (6) | C4—C3—H3 | 120.0 |
| O3—Mn1—O1i | 89.88 (8) | C3—C4—H4 | 119.9 |
| O3—Mn1—O1 | 159.02 (5) | C3—C4—C5 | 120.16 (18) |
| O3—Mn1—O2 | 85.76 (7) | C5—C4—H4 | 119.9 |
| Mn1—O1—Mn1i | 108.13 (7) | N1—C5—C4 | 119.10 (19) |
| N1—O1—Mn1 | 124.29 (9) | N1—C5—H5 | 120.5 |
| N1—O1—Mn1i | 127.41 (10) | C4—C5—H5 | 120.5 |
| Mn1—O2—H2A | 114.4 (17) | C2—C6—H6A | 109.5 |
| Mn1—O2—H2B | 121.9 (18) | C2—C6—H6B | 109.5 |
| H2A—O2—H2B | 112 (2) | C2—C6—H6C | 109.5 |
| Mn1—O3—H3A | 118.3 (16) | H6A—C6—H6B | 109.5 |
| Mn1—O3—H3B | 116 (2) | H6A—C6—H6C | 109.5 |
| H3A—O3—H3B | 109 (3) | H6B—C6—H6C | 109.5 |
Symmetry code: (i) −x+1, −y+1, −z+1.
Bis(µ-3-methylpyridine N-oxide)bis[diaquadichloridomanganese(II)] (III) . Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O2—H2B···Cl1ii | 0.80 (2) | 2.38 (2) | 3.147 (3) | 161 (2) |
| O3—H3A···Cl2iii | 0.86 (2) | 2.28 (2) | 3.120 (2) | 167 (2) |
Symmetry codes: (ii) −x, −y+1, −z+1; (iii) −x, −y, −z+1.
References
- Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K. & Puschmann, H. (2009). J. Appl. Cryst. 42, 339–341.
- Eppenson, J. H. (2003). Adv. Inorg. Chem. 54, 157–202.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Hu, Z. C., Deibert, B. J. & Li, J. (2014). Chem. Soc. Rev. 43, 5815–5840. [DOI] [PubMed]
- Jiu, J., Blake, A. J., Champness, N. R., Hubberstey, P., Wilson, C. & Schröder, M. (2008). Inorg. Chem. 47, 8652–8664. [DOI] [PubMed]
- Liu, J., Chen, L., Cui, H., Zhang, Y., Zhang, L. & Su, C.-Y. (2014). Chem. Soc. Rev. 43, 6011–6061. [DOI] [PubMed]
- Ma, B.-Q., Sun, H.-L. & Gao, S. (2005). Inorg. Chem. 44, 837–839. [DOI] [PubMed]
- Mautner, F. A., Berger, C., Fischer, R. C., Massoud, S. S. & Vicente, R. (2017). Polyhedron, 134, 126–134.
- Moustafa, M. E., Boyle, P. D. & Puddephatt, R. J. (2014). Organometallics, 33, 5402–5413.
- Rigaku (1998). REQAB. Rigaku Corporation, Tokyo, Japan.
- Rigaku (2011). CrystalClearSM Expert. Rigaku Corporation, Tokyo, Japan.
- Sarma, R. & Baruah, J. B. (2011). Solid State Sci. 13, 1692–1700.
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Shi, J.-M., Liu, Z., Li, W.-N., Zhao, H. Y. & Liu, L.-D. (2007). J. Coord. Chem. 60, 1077–1082.
- Sun, H.-L., Wang, Z.-M., Gao, S. & Batten, S. R. (2008). CrystEngComm, 10, 1796–1802.
- Zhang, L.-P., Lu, W.-J. & Mak, T. C. (2003). Chem. Commun. 22, 2830–2831. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, II, III. DOI: 10.1107/S2056989017012038/zl2709sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989017012038/zl2709Isup2.hkl
Structure factors: contains datablock(s) II. DOI: 10.1107/S2056989017012038/zl2709IIsup3.hkl
Structure factors: contains datablock(s) III. DOI: 10.1107/S2056989017012038/zl2709IIIsup4.hkl
Additional supporting information: crystallographic information; 3D view; checkCIF report