The crystal structure of the triterpene lactone ochraceolide A (3-oxolup-20 (29)-en-30,21α-olide) isolated from Elaeodendron trichotomum (Turcz.) Lundell is reported.
Keywords: crystal structure, ochraceolide A, triterpene lactone, Elaeodendron trichotomum
Abstract
The title compound, C30H44O3 [systematic name: 6aR,6 bR,8aS,9aR,12aR,14bR)-4,4,6a,6;b,8a,14b-hexamethyl-12-methyleneicosahydro-3H-phenanthro[1′,2′:6,7]indeno[2,1-b]furan-3,11(2H)-dione], is a triterpene lactone, which was isolated from dichloromethane extract of Elaeodendron trichotomum (Turcz.) Lundell (celastraceae) stem bark. The compound has a lupane skeleton and consists of four fused six-membered rings and two five-membered rings. In the crystal, molecules are linked by weak C—H⋯O hydrogen bonds into a three-dimensional network. The configuration of ochraceolide A was proposed based on analogue compounds which belong to the lupane type.
Chemical context
Ochraceolides A–E are a group of cytotoxic lupane γ-lactones isolated from the Celastraceae family. Ochraceolide A was firstly isolated from Kokoona ochracea (Elm.) Merril stem bark (Ngassapa et al., 1991 ▸) and afterwards from Lophopetalum wallichii (Sturm et al., 1996 ▸) and Cassine xylocarpa (Callies et al., 2015 ▸). The title compound has shown significant cytotoxic activity against murine lymphocytic leukemia cells (P-388) with an ED50 of 0.6 µM; human oral epidermoid carcinoma (KB-3) with an ED50 of 6.0 µM; and hormone-dependent breast cancer with an ED50 of 9.9 µM (Ngassapa et al., 1991 ▸; Sturm et al., 1996 ▸). In the same way, this compound has exhibited significant inhibitory activity in the FPTase assay with an IC50 of 2.2 µM (Sturm et al., 1996 ▸) and inhibitory effects of human immunodeficiency virus type 1 replication with an IC50 of 39.0 µM (Callies et al., 2015 ▸). Ochraceolide A is part of the structure of the Diels–Alder adduct (i.e. celastroidine A or volubilide) isolated from Hippocratea celastroides K. (Jiménez-Estrada et al., 2000 ▸) and Hyppocratea volubilis L. (Alvarenga et al., 2000 ▸). In these publications, the crystal structure of the adduct was reported as a solvate of dichloromethane and toluene, respectively. The X-ray analysis showed that the Diels–Alder adduct was integrated by the triterpene ochraceolide A and a theoretical diterpene, in which the former seems to have acted as dienophile and the latter as diene in the biosynthesis. Herein the first isolation of ochraceolide A from Elaeodendron trichotomum (Turcz.) Lundell stem bark is reported and the crystal structure described.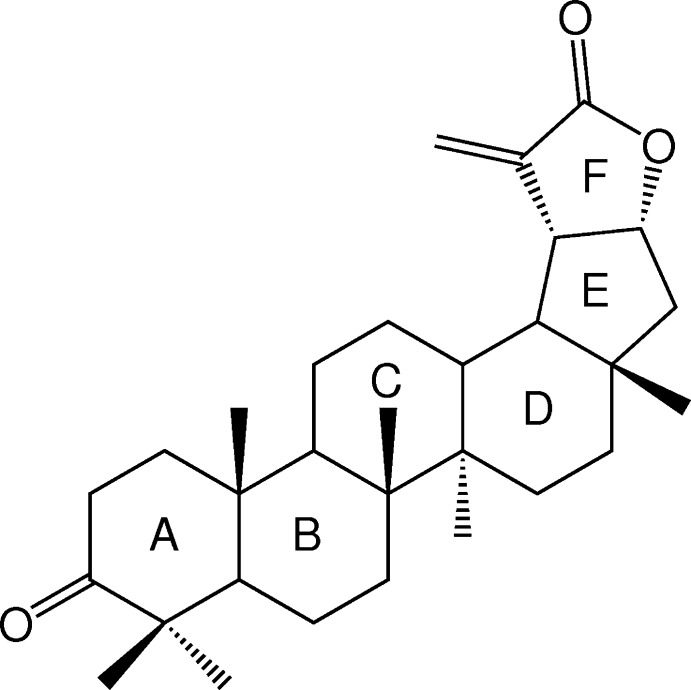
Structural commentary
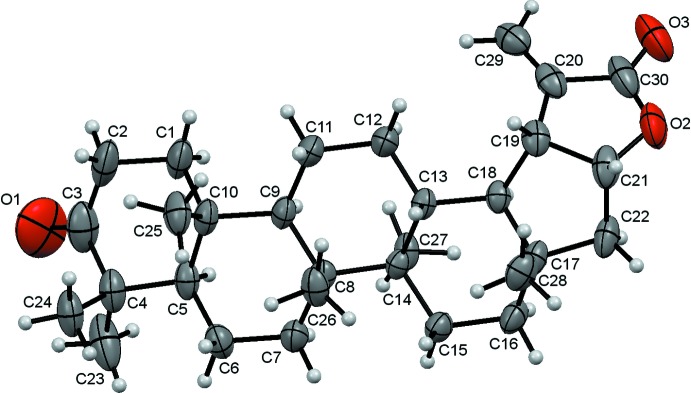
The title compound has a lupane skeleton and crystallizes in the orthorhombic space group P212121 with one molecule in the asymmetric unit (Fig. 1 ▸). The triterpene skeleton consists of four fused six-membered rings (A–D) and two five-membered rings (E and F). The cyclohexane rings are trans-fused and in standard chair conformations. The cyclopentane (C17–C19/C21/C22) ring is trans-fused to the triterpene D ring and exhibits an envelope conformation [Q = 0.451 (4) Å and θ = 356.7 (5)°] with the puckered C17 atom having the maximum deviation of 0.285 (4) Å. The α-methylene γ-lactone is cis-fused at C19–C21 to the cyclopentane E ring and is essentially planar with a maximum deviation of 0.006 (4) Å for atom C19. The torsion angle C20—C19—C21—O2 is 0.8 (4)° and the weighted average absolute internal torsion angle for the lactone ring is 0.7 (2)°
Figure 1.
The molecular structure of the title compound with the atom labelling. Displacement ellipsoids are drawn at the 50% probability level and H atoms are shown as spheres of arbitrary radius.
Supramolecular features
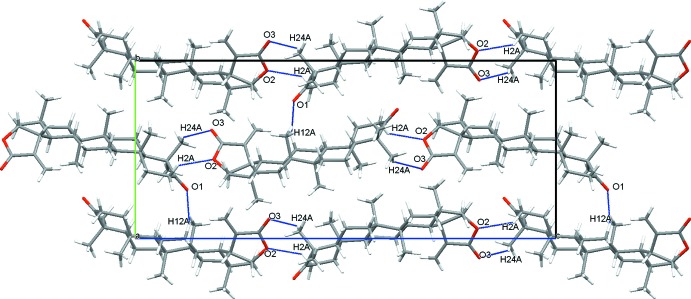
In the crystal, molecules are linked by weak C—H⋯O hydrogen bonds (Table 1 ▸, Fig. 2 ▸). The lactone and A rings of adjacent molecules interact through two hydrogen bonds (C2—H2A⋯O2 and C24—H24A⋯O3) in a head-to-tail arrangement, forming chains along [001]. These chains are further connected through a weak hydrogen bond between the oxygen of the ketone group (O1) and a methylene group on the C ring (C12), forming an overall three-dimensional network.
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C2—H2A⋯O2i | 0.99 | 2.57 | 3.395 (5) | 141 |
| C12—H12A⋯O1ii | 0.99 | 2.45 | 3.310 (6) | 146 |
| C24—H24A⋯O3i | 0.98 | 2.58 | 3.357 (6) | 137 |
Symmetry codes: (i) 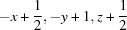 ; (ii)
; (ii) 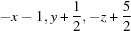 .
.
Figure 2.
Part of the crystal structure showing hydrogen bonds as blue lines.
Database survey
A search of the Cambridge Structural Database (CSD Version 5.38, update November 2016; Groom et al., 2016 ▸) for α-methylene γ-lactone fused to a cyclopentane ring gave only one entry for 6,6-dimethyl-3-methylenetetrahydro-2H-cyclopenta[b]furan-2,5(3H)-dione (CCDC 658922; Edwards et al., 2008 ▸). In both compounds, the principal supramolecular interactions are C—H⋯O hydrogen bonds and the α-methylene γ-lactones are cis-fused to the corresponding cyclopentane ring. However, unlike the title compound, the γ-lactone of the synthetic compound presents a twisted conformation.
Isolation and crystallization
Elaeodendron trichotomum (Turcz.) Lundell was collected from Chunchucmil, Yucatán, México (20o 51.032′ N, 90o 11.488′ W). A voucher specimen (JTun2328) was deposited at the Herbarium Alfredo Barrera Marín, Universidad Autónoma de Yucatán, México. Dried and milled stem bark (2100 g) was exhaustively extracted by dichloromethane using a Soxhlet extraction apparatus to yield 184.2 g of crude extract. A portion of the extract (100 g) was chromatographed on silica gel (40–60 µm) using a gradient elution with n-hexane–ethyl acetate (10–100% ethyl acetate), to obtain 44 fractions. Single crystals suitable for X-ray structure analysis were obtained by slow evaporation of the mixture of solvents present in fractions 7–10 at room temperature.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. Hydrogen atoms bonded to C atoms were positioned geometrically and refined using a riding model with C—H = 0.95–1.00 Å with U iso(H) = 1.2U eq(C) or 1.5U eq(methyl C).
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C30H44O3 |
| M r | 452.65 |
| Crystal system, space group | Orthorhombic, P212121 |
| Temperature (K) | 150 |
| a, b, c (Å) | 7.6131 (5), 11.7216 (7), 27.7076 (17) |
| V (Å3) | 2472.6 (3) |
| Z | 4 |
| Radiation type | Cu Kα |
| μ (mm−1) | 0.59 |
| Crystal size (mm) | 0.36 × 0.27 × 0.25 |
| Data collection | |
| Diffractometer | Bruker D8 Venture |
| Absorption correction | Multi-scan (SADABS; Krause et al., 2015 ▸) |
| T min, T max | 0.783, 0.864 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 14632, 4513, 4057 |
| R int | 0.061 |
| (sin θ/λ)max (Å−1) | 0.603 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.061, 0.164, 1.09 |
| No. of reflections | 4513 |
| No. of parameters | 304 |
| H-atom treatment | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.28, −0.19 |
| Absolute structure | Flack x determined using 1515 quotients [(I +)−(I −)]/[(I +)+(I −)] (Parsons et al., 2013 ▸). |
| Absolute structure parameter | 0.2 (3) |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989017012816/lh4023sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989017012816/lh4023Isup2.hkl
CCDC reference: 1573017
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
ADHE, GJMR and GML are grateful to Dra Reyna Reyes-Martínez for assistance in preparing the manuscript. ADHE thanks the Consejo Nacional de Ciencias y Tecnología–CONACYT for a postdoctoral fellowship.
supplementary crystallographic information
Crystal data
| C30H44O3 | Dx = 1.216 Mg m−3 |
| Mr = 452.65 | Cu Kα radiation, λ = 1.54178 Å |
| Orthorhombic, P212121 | Cell parameters from 9889 reflections |
| a = 7.6131 (5) Å | θ = 3.2–68.3° |
| b = 11.7216 (7) Å | µ = 0.59 mm−1 |
| c = 27.7076 (17) Å | T = 150 K |
| V = 2472.6 (3) Å3 | Prism, colourless |
| Z = 4 | 0.36 × 0.27 × 0.25 mm |
| F(000) = 992 |
Data collection
| Bruker D8 Venture diffractometer | 4513 independent reflections |
| Radiation source: micro-focus X-ray source | 4057 reflections with I > 2σ(I) |
| Detector resolution: 52.0833 pixels mm-1 | Rint = 0.061 |
| ω–scans | θmax = 68.3°, θmin = 3.2° |
| Absorption correction: multi-scan (SADABS; Krause et al., 2015) | h = −9→8 |
| Tmin = 0.783, Tmax = 0.864 | k = −13→14 |
| 14632 measured reflections | l = −33→33 |
Refinement
| Refinement on F2 | Hydrogen site location: inferred from neighbouring sites |
| Least-squares matrix: full | H-atom parameters constrained |
| R[F2 > 2σ(F2)] = 0.061 | w = 1/[σ2(Fo2) + (0.0789P)2 + 0.8039P] where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.164 | (Δ/σ)max < 0.001 |
| S = 1.09 | Δρmax = 0.28 e Å−3 |
| 4513 reflections | Δρmin = −0.19 e Å−3 |
| 304 parameters | Absolute structure: Flack x determined using 1515 quotients [(I+)-(I-)]/[(I+)+(I-)] (Parsons et al., 2013). |
| 0 restraints | Absolute structure parameter: 0.2 (3) |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.7000 (9) | 0.2747 (4) | 1.1228 (2) | 0.127 (2) | |
| O2 | 0.2109 (5) | 0.5554 (3) | 0.68677 (10) | 0.0712 (10) | |
| O3 | 0.0663 (6) | 0.3910 (4) | 0.68208 (13) | 0.0865 (12) | |
| C1 | 0.4287 (7) | 0.3773 (4) | 1.02722 (13) | 0.0568 (11) | |
| H1A | 0.3012 | 0.3872 | 1.0216 | 0.068* | |
| H1B | 0.4687 | 0.3118 | 1.0075 | 0.068* | |
| C2 | 0.4602 (8) | 0.3508 (4) | 1.08081 (14) | 0.0692 (14) | |
| H2A | 0.3999 | 0.4094 | 1.1005 | 0.083* | |
| H2B | 0.4059 | 0.2762 | 1.0884 | 0.083* | |
| C3 | 0.6490 (8) | 0.3471 (4) | 1.09523 (16) | 0.0674 (14) | |
| C4 | 0.7745 (6) | 0.4361 (3) | 1.07509 (13) | 0.0498 (10) | |
| C5 | 0.7231 (6) | 0.4658 (3) | 1.02182 (11) | 0.0415 (8) | |
| H5 | 0.7540 | 0.3957 | 1.0031 | 0.050* | |
| C6 | 0.8379 (6) | 0.5589 (3) | 0.99999 (13) | 0.0482 (9) | |
| H6A | 0.9620 | 0.5455 | 1.0091 | 0.058* | |
| H6B | 0.8022 | 0.6339 | 1.0131 | 0.058* | |
| C7 | 0.8213 (5) | 0.5604 (3) | 0.94485 (12) | 0.0443 (8) | |
| H7A | 0.8673 | 0.4876 | 0.9319 | 0.053* | |
| H7B | 0.8953 | 0.6228 | 0.9319 | 0.053* | |
| C8 | 0.6322 (5) | 0.5770 (3) | 0.92684 (11) | 0.0347 (7) | |
| C9 | 0.5071 (5) | 0.4933 (3) | 0.95387 (10) | 0.0372 (8) | |
| H9 | 0.5416 | 0.4159 | 0.9420 | 0.045* | |
| C10 | 0.5260 (5) | 0.4859 (3) | 1.01054 (11) | 0.0420 (8) | |
| C11 | 0.3177 (5) | 0.5085 (4) | 0.93680 (12) | 0.0460 (9) | |
| H11A | 0.2746 | 0.5839 | 0.9478 | 0.055* | |
| H11B | 0.2436 | 0.4494 | 0.9522 | 0.055* | |
| C12 | 0.2954 (5) | 0.5006 (3) | 0.88168 (12) | 0.0424 (8) | |
| H12A | 0.3119 | 0.4204 | 0.8715 | 0.051* | |
| H12B | 0.1743 | 0.5235 | 0.8730 | 0.051* | |
| C13 | 0.4255 (5) | 0.5761 (3) | 0.85450 (11) | 0.0326 (7) | |
| H13 | 0.3992 | 0.6569 | 0.8634 | 0.039* | |
| C14 | 0.6178 (4) | 0.5509 (3) | 0.87041 (11) | 0.0321 (7) | |
| C15 | 0.7510 (5) | 0.6247 (3) | 0.84160 (12) | 0.0410 (8) | |
| H15A | 0.7459 | 0.7039 | 0.8539 | 0.049* | |
| H15B | 0.8708 | 0.5953 | 0.8478 | 0.049* | |
| C16 | 0.7197 (5) | 0.6269 (4) | 0.78673 (12) | 0.0457 (9) | |
| H16A | 0.7982 | 0.6842 | 0.7718 | 0.055* | |
| H16B | 0.7496 | 0.5514 | 0.7730 | 0.055* | |
| C17 | 0.5299 (5) | 0.6555 (3) | 0.77439 (12) | 0.0405 (8) | |
| C18 | 0.4109 (4) | 0.5670 (3) | 0.79938 (11) | 0.0336 (7) | |
| H18 | 0.4567 | 0.4900 | 0.7903 | 0.040* | |
| C19 | 0.2307 (5) | 0.5807 (3) | 0.77408 (12) | 0.0415 (8) | |
| H19 | 0.1522 | 0.6334 | 0.7925 | 0.050* | |
| C20 | 0.1359 (5) | 0.4732 (3) | 0.76020 (13) | 0.0460 (9) | |
| C21 | 0.2797 (6) | 0.6321 (4) | 0.72350 (13) | 0.0522 (10) | |
| H21 | 0.2275 | 0.7099 | 0.7198 | 0.063* | |
| C22 | 0.4785 (6) | 0.6384 (4) | 0.72141 (12) | 0.0504 (10) | |
| H22A | 0.5178 | 0.7034 | 0.7013 | 0.060* | |
| H22B | 0.5291 | 0.5670 | 0.7083 | 0.060* | |
| C30 | 0.1290 (6) | 0.4657 (5) | 0.70646 (16) | 0.0625 (13) | |
| C29 | 0.0717 (5) | 0.3894 (4) | 0.78608 (15) | 0.0525 (10) | |
| H29A | 0.0215 | 0.3248 | 0.7706 | 0.063* | |
| H29B | 0.0754 | 0.3932 | 0.8203 | 0.063* | |
| C26 | 0.5821 (6) | 0.7027 (3) | 0.93685 (12) | 0.0470 (9) | |
| H26A | 0.6189 | 0.7235 | 0.9696 | 0.071* | |
| H26B | 0.4546 | 0.7118 | 0.9339 | 0.071* | |
| H26C | 0.6411 | 0.7523 | 0.9134 | 0.071* | |
| C28 | 0.4863 (6) | 0.7796 (3) | 0.78893 (14) | 0.0491 (9) | |
| H28A | 0.5315 | 0.7947 | 0.8214 | 0.074* | |
| H28B | 0.3587 | 0.7905 | 0.7886 | 0.074* | |
| H28C | 0.5410 | 0.8324 | 0.7660 | 0.074* | |
| C27 | 0.6651 (5) | 0.4251 (3) | 0.85928 (11) | 0.0382 (8) | |
| H27A | 0.6767 | 0.4150 | 0.8243 | 0.057* | |
| H27B | 0.5721 | 0.3750 | 0.8715 | 0.057* | |
| H27C | 0.7765 | 0.4057 | 0.8750 | 0.057* | |
| C25 | 0.4458 (7) | 0.5894 (4) | 1.03594 (13) | 0.0546 (11) | |
| H25A | 0.3345 | 0.6096 | 1.0203 | 0.082* | |
| H25B | 0.5271 | 0.6540 | 1.0338 | 0.082* | |
| H25C | 0.4243 | 0.5709 | 1.0699 | 0.082* | |
| C24 | 0.7708 (7) | 0.5395 (4) | 1.10971 (13) | 0.0584 (12) | |
| H24A | 0.6501 | 0.5677 | 1.1127 | 0.088* | |
| H24B | 0.8460 | 0.6002 | 1.0969 | 0.088* | |
| H24C | 0.8140 | 0.5161 | 1.1415 | 0.088* | |
| C23 | 0.9581 (8) | 0.3866 (5) | 1.07703 (17) | 0.0815 (18) | |
| H23A | 0.9834 | 0.3605 | 1.1099 | 0.122* | |
| H23B | 1.0434 | 0.4453 | 1.0677 | 0.122* | |
| H23C | 0.9665 | 0.3220 | 1.0547 | 0.122* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.166 (5) | 0.085 (3) | 0.131 (4) | 0.003 (3) | −0.022 (4) | 0.066 (3) |
| O2 | 0.079 (2) | 0.100 (3) | 0.0340 (13) | 0.015 (2) | −0.0172 (15) | 0.0057 (15) |
| O3 | 0.090 (3) | 0.108 (3) | 0.062 (2) | 0.011 (2) | −0.0361 (19) | −0.023 (2) |
| C1 | 0.084 (3) | 0.058 (2) | 0.0289 (17) | −0.010 (2) | 0.0078 (19) | 0.0033 (16) |
| C2 | 0.116 (4) | 0.061 (3) | 0.0311 (19) | −0.014 (3) | 0.013 (2) | 0.0049 (18) |
| C3 | 0.115 (4) | 0.041 (2) | 0.046 (2) | 0.011 (2) | −0.007 (3) | 0.0053 (18) |
| C4 | 0.076 (3) | 0.0434 (19) | 0.0296 (16) | 0.0128 (19) | −0.0052 (17) | 0.0002 (14) |
| C5 | 0.064 (2) | 0.0337 (16) | 0.0270 (15) | 0.0099 (16) | −0.0007 (15) | −0.0032 (13) |
| C6 | 0.057 (2) | 0.050 (2) | 0.0371 (18) | −0.0026 (18) | −0.0093 (17) | 0.0000 (15) |
| C7 | 0.050 (2) | 0.049 (2) | 0.0336 (16) | −0.0053 (17) | −0.0016 (16) | 0.0048 (15) |
| C8 | 0.0472 (19) | 0.0259 (14) | 0.0312 (15) | −0.0022 (13) | 0.0003 (14) | −0.0001 (12) |
| C9 | 0.050 (2) | 0.0376 (17) | 0.0242 (14) | −0.0016 (15) | 0.0050 (14) | −0.0011 (12) |
| C10 | 0.060 (2) | 0.0402 (18) | 0.0253 (14) | 0.0005 (17) | 0.0048 (15) | −0.0029 (13) |
| C11 | 0.045 (2) | 0.061 (2) | 0.0320 (16) | −0.0069 (17) | 0.0054 (15) | 0.0050 (16) |
| C12 | 0.0439 (19) | 0.050 (2) | 0.0329 (16) | −0.0072 (16) | 0.0017 (15) | 0.0037 (15) |
| C13 | 0.0406 (17) | 0.0297 (15) | 0.0274 (14) | 0.0047 (13) | 0.0028 (13) | 0.0006 (12) |
| C14 | 0.0400 (17) | 0.0275 (15) | 0.0288 (14) | 0.0006 (13) | 0.0038 (13) | 0.0023 (11) |
| C15 | 0.0418 (19) | 0.0447 (18) | 0.0364 (17) | −0.0028 (15) | 0.0029 (14) | 0.0093 (14) |
| C16 | 0.047 (2) | 0.054 (2) | 0.0360 (18) | 0.0031 (17) | 0.0089 (15) | 0.0156 (16) |
| C17 | 0.049 (2) | 0.0415 (18) | 0.0305 (16) | 0.0104 (15) | 0.0075 (15) | 0.0103 (14) |
| C18 | 0.0394 (18) | 0.0333 (16) | 0.0280 (15) | 0.0087 (14) | −0.0002 (13) | 0.0013 (12) |
| C19 | 0.0452 (19) | 0.0445 (18) | 0.0348 (16) | 0.0176 (15) | −0.0005 (15) | 0.0002 (14) |
| C20 | 0.0397 (19) | 0.055 (2) | 0.0432 (19) | 0.0165 (17) | −0.0098 (16) | −0.0102 (17) |
| C21 | 0.063 (2) | 0.058 (2) | 0.0356 (18) | 0.024 (2) | −0.0016 (17) | 0.0059 (16) |
| C22 | 0.062 (2) | 0.061 (2) | 0.0288 (17) | 0.0212 (19) | 0.0064 (16) | 0.0122 (16) |
| C30 | 0.057 (3) | 0.087 (3) | 0.044 (2) | 0.022 (3) | −0.021 (2) | −0.014 (2) |
| C29 | 0.043 (2) | 0.060 (2) | 0.055 (2) | 0.0029 (18) | −0.0046 (18) | −0.0162 (19) |
| C26 | 0.075 (3) | 0.0307 (17) | 0.0350 (17) | −0.0006 (17) | −0.0062 (18) | −0.0047 (13) |
| C28 | 0.063 (3) | 0.0358 (18) | 0.049 (2) | 0.0035 (17) | 0.0063 (19) | 0.0128 (16) |
| C27 | 0.052 (2) | 0.0354 (17) | 0.0271 (14) | 0.0116 (15) | −0.0014 (14) | 0.0004 (13) |
| C25 | 0.070 (3) | 0.062 (2) | 0.0316 (17) | 0.020 (2) | 0.0030 (18) | −0.0088 (17) |
| C24 | 0.093 (3) | 0.052 (2) | 0.0298 (16) | 0.011 (2) | −0.0087 (19) | −0.0049 (16) |
| C23 | 0.106 (4) | 0.095 (4) | 0.044 (2) | 0.049 (4) | −0.019 (3) | −0.010 (2) |
Geometric parameters (Å, º)
| O1—C3 | 1.206 (6) | C14—C15 | 1.553 (5) |
| O2—C30 | 1.338 (7) | C15—C16 | 1.539 (5) |
| O2—C21 | 1.455 (6) | C15—H15A | 0.9900 |
| O3—C30 | 1.205 (6) | C15—H15B | 0.9900 |
| C1—C2 | 1.536 (5) | C16—C17 | 1.522 (5) |
| C1—C10 | 1.544 (6) | C16—H16A | 0.9900 |
| C1—H1A | 0.9900 | C16—H16B | 0.9900 |
| C1—H1B | 0.9900 | C17—C22 | 1.532 (5) |
| C2—C3 | 1.492 (8) | C17—C18 | 1.542 (5) |
| C2—H2A | 0.9900 | C17—C28 | 1.545 (5) |
| C2—H2B | 0.9900 | C18—C19 | 1.549 (5) |
| C3—C4 | 1.521 (7) | C18—H18 | 1.0000 |
| C4—C23 | 1.515 (7) | C19—C20 | 1.502 (6) |
| C4—C24 | 1.546 (5) | C19—C21 | 1.570 (5) |
| C4—C5 | 1.566 (4) | C19—H19 | 1.0000 |
| C5—C6 | 1.523 (6) | C20—C29 | 1.311 (6) |
| C5—C10 | 1.550 (6) | C20—C30 | 1.492 (5) |
| C5—H5 | 1.0000 | C21—C22 | 1.517 (6) |
| C6—C7 | 1.533 (5) | C21—H21 | 1.0000 |
| C6—H6A | 0.9900 | C22—H22A | 0.9900 |
| C6—H6B | 0.9900 | C22—H22B | 0.9900 |
| C7—C8 | 1.536 (5) | C29—H29A | 0.9500 |
| C7—H7A | 0.9900 | C29—H29B | 0.9500 |
| C7—H7B | 0.9900 | C26—H26A | 0.9800 |
| C8—C26 | 1.546 (5) | C26—H26B | 0.9800 |
| C8—C9 | 1.559 (5) | C26—H26C | 0.9800 |
| C8—C14 | 1.597 (4) | C28—H28A | 0.9800 |
| C9—C11 | 1.528 (5) | C28—H28B | 0.9800 |
| C9—C10 | 1.579 (4) | C28—H28C | 0.9800 |
| C9—H9 | 1.0000 | C27—H27A | 0.9800 |
| C10—C25 | 1.530 (5) | C27—H27B | 0.9800 |
| C11—C12 | 1.539 (4) | C27—H27C | 0.9800 |
| C11—H11A | 0.9900 | C25—H25A | 0.9800 |
| C11—H11B | 0.9900 | C25—H25B | 0.9800 |
| C12—C13 | 1.527 (5) | C25—H25C | 0.9800 |
| C12—H12A | 0.9900 | C24—H24A | 0.9800 |
| C12—H12B | 0.9900 | C24—H24B | 0.9800 |
| C13—C18 | 1.535 (4) | C24—H24C | 0.9800 |
| C13—C14 | 1.557 (5) | C23—H23A | 0.9800 |
| C13—H13 | 1.0000 | C23—H23B | 0.9800 |
| C14—C27 | 1.549 (4) | C23—H23C | 0.9800 |
| C30—O2—C21 | 111.6 (3) | C16—C15—H15B | 108.6 |
| C2—C1—C10 | 112.4 (4) | C14—C15—H15B | 108.6 |
| C2—C1—H1A | 109.1 | H15A—C15—H15B | 107.6 |
| C10—C1—H1A | 109.1 | C17—C16—C15 | 111.9 (3) |
| C2—C1—H1B | 109.1 | C17—C16—H16A | 109.2 |
| C10—C1—H1B | 109.1 | C15—C16—H16A | 109.2 |
| H1A—C1—H1B | 107.8 | C17—C16—H16B | 109.2 |
| C3—C2—C1 | 114.5 (4) | C15—C16—H16B | 109.2 |
| C3—C2—H2A | 108.6 | H16A—C16—H16B | 107.9 |
| C1—C2—H2A | 108.6 | C16—C17—C22 | 115.4 (3) |
| C3—C2—H2B | 108.6 | C16—C17—C18 | 108.0 (3) |
| C1—C2—H2B | 108.6 | C22—C17—C18 | 101.1 (3) |
| H2A—C2—H2B | 107.6 | C16—C17—C28 | 110.7 (4) |
| O1—C3—C2 | 120.0 (6) | C22—C17—C28 | 108.5 (3) |
| O1—C3—C4 | 120.9 (6) | C18—C17—C28 | 112.9 (3) |
| C2—C3—C4 | 119.1 (4) | C13—C18—C17 | 111.0 (3) |
| C23—C4—C3 | 107.7 (4) | C13—C18—C19 | 120.5 (3) |
| C23—C4—C24 | 107.2 (4) | C17—C18—C19 | 104.3 (3) |
| C3—C4—C24 | 107.4 (4) | C13—C18—H18 | 106.8 |
| C23—C4—C5 | 110.4 (3) | C17—C18—H18 | 106.8 |
| C3—C4—C5 | 110.0 (4) | C19—C18—H18 | 106.8 |
| C24—C4—C5 | 113.9 (3) | C20—C19—C18 | 117.0 (3) |
| C6—C5—C10 | 111.5 (3) | C20—C19—C21 | 101.9 (3) |
| C6—C5—C4 | 113.0 (3) | C18—C19—C21 | 103.5 (3) |
| C10—C5—C4 | 117.7 (3) | C20—C19—H19 | 111.2 |
| C6—C5—H5 | 104.3 | C18—C19—H19 | 111.2 |
| C10—C5—H5 | 104.3 | C21—C19—H19 | 111.2 |
| C4—C5—H5 | 104.3 | C29—C20—C30 | 119.2 (4) |
| C5—C6—C7 | 110.9 (3) | C29—C20—C19 | 131.9 (3) |
| C5—C6—H6A | 109.5 | C30—C20—C19 | 108.8 (4) |
| C7—C6—H6A | 109.5 | O2—C21—C22 | 111.3 (3) |
| C5—C6—H6B | 109.5 | O2—C21—C19 | 107.6 (4) |
| C7—C6—H6B | 109.5 | C22—C21—C19 | 106.9 (3) |
| H6A—C6—H6B | 108.0 | O2—C21—H21 | 110.3 |
| C6—C7—C8 | 113.7 (3) | C22—C21—H21 | 110.3 |
| C6—C7—H7A | 108.8 | C19—C21—H21 | 110.3 |
| C8—C7—H7A | 108.8 | C21—C22—C17 | 103.0 (3) |
| C6—C7—H7B | 108.8 | C21—C22—H22A | 111.2 |
| C8—C7—H7B | 108.8 | C17—C22—H22A | 111.2 |
| H7A—C7—H7B | 107.7 | C21—C22—H22B | 111.2 |
| C7—C8—C26 | 107.0 (3) | C17—C22—H22B | 111.2 |
| C7—C8—C9 | 109.7 (3) | H22A—C22—H22B | 109.1 |
| C26—C8—C9 | 111.3 (3) | O3—C30—O2 | 121.8 (4) |
| C7—C8—C14 | 111.0 (3) | O3—C30—C20 | 128.1 (5) |
| C26—C8—C14 | 110.0 (3) | O2—C30—C20 | 110.1 (4) |
| C9—C8—C14 | 107.9 (2) | C20—C29—H29A | 120.0 |
| C11—C9—C8 | 110.7 (3) | C20—C29—H29B | 120.0 |
| C11—C9—C10 | 113.6 (3) | H29A—C29—H29B | 120.0 |
| C8—C9—C10 | 117.1 (3) | C8—C26—H26A | 109.5 |
| C11—C9—H9 | 104.6 | C8—C26—H26B | 109.5 |
| C8—C9—H9 | 104.6 | H26A—C26—H26B | 109.5 |
| C10—C9—H9 | 104.6 | C8—C26—H26C | 109.5 |
| C25—C10—C1 | 108.9 (3) | H26A—C26—H26C | 109.5 |
| C25—C10—C5 | 114.5 (3) | H26B—C26—H26C | 109.5 |
| C1—C10—C5 | 106.2 (3) | C17—C28—H28A | 109.5 |
| C25—C10—C9 | 112.2 (3) | C17—C28—H28B | 109.5 |
| C1—C10—C9 | 107.4 (3) | H28A—C28—H28B | 109.5 |
| C5—C10—C9 | 107.3 (3) | C17—C28—H28C | 109.5 |
| C9—C11—C12 | 113.8 (3) | H28A—C28—H28C | 109.5 |
| C9—C11—H11A | 108.8 | H28B—C28—H28C | 109.5 |
| C12—C11—H11A | 108.8 | C14—C27—H27A | 109.5 |
| C9—C11—H11B | 108.8 | C14—C27—H27B | 109.5 |
| C12—C11—H11B | 108.8 | H27A—C27—H27B | 109.5 |
| H11A—C11—H11B | 107.7 | C14—C27—H27C | 109.5 |
| C13—C12—C11 | 112.5 (3) | H27A—C27—H27C | 109.5 |
| C13—C12—H12A | 109.1 | H27B—C27—H27C | 109.5 |
| C11—C12—H12A | 109.1 | C10—C25—H25A | 109.5 |
| C13—C12—H12B | 109.1 | C10—C25—H25B | 109.5 |
| C11—C12—H12B | 109.1 | H25A—C25—H25B | 109.5 |
| H12A—C12—H12B | 107.8 | C10—C25—H25C | 109.5 |
| C12—C13—C18 | 113.8 (3) | H25A—C25—H25C | 109.5 |
| C12—C13—C14 | 111.1 (3) | H25B—C25—H25C | 109.5 |
| C18—C13—C14 | 109.7 (3) | C4—C24—H24A | 109.5 |
| C12—C13—H13 | 107.3 | C4—C24—H24B | 109.5 |
| C18—C13—H13 | 107.3 | H24A—C24—H24B | 109.5 |
| C14—C13—H13 | 107.3 | C4—C24—H24C | 109.5 |
| C27—C14—C15 | 106.0 (3) | H24A—C24—H24C | 109.5 |
| C27—C14—C13 | 110.0 (3) | H24B—C24—H24C | 109.5 |
| C15—C14—C13 | 111.3 (3) | C4—C23—H23A | 109.5 |
| C27—C14—C8 | 111.2 (2) | C4—C23—H23B | 109.5 |
| C15—C14—C8 | 110.6 (3) | H23A—C23—H23B | 109.5 |
| C13—C14—C8 | 107.8 (2) | C4—C23—H23C | 109.5 |
| C16—C15—C14 | 114.6 (3) | H23A—C23—H23C | 109.5 |
| C16—C15—H15A | 108.6 | H23B—C23—H23C | 109.5 |
| C14—C15—H15A | 108.6 | ||
| C10—C1—C2—C3 | −52.1 (6) | C18—C13—C14—C8 | 172.8 (2) |
| C1—C2—C3—O1 | −139.3 (5) | C7—C8—C14—C27 | 62.4 (4) |
| C1—C2—C3—C4 | 41.2 (6) | C26—C8—C14—C27 | −179.3 (3) |
| O1—C3—C4—C23 | 24.3 (6) | C9—C8—C14—C27 | −57.8 (4) |
| C2—C3—C4—C23 | −156.2 (4) | C7—C8—C14—C15 | −55.1 (3) |
| O1—C3—C4—C24 | −90.8 (6) | C26—C8—C14—C15 | 63.2 (4) |
| C2—C3—C4—C24 | 88.7 (5) | C9—C8—C14—C15 | −175.3 (3) |
| O1—C3—C4—C5 | 144.7 (5) | C7—C8—C14—C13 | −176.9 (3) |
| C2—C3—C4—C5 | −35.8 (5) | C26—C8—C14—C13 | −58.7 (4) |
| C23—C4—C5—C6 | −64.0 (5) | C9—C8—C14—C13 | 62.9 (3) |
| C3—C4—C5—C6 | 177.2 (3) | C27—C14—C15—C16 | 72.7 (4) |
| C24—C4—C5—C6 | 56.6 (5) | C13—C14—C15—C16 | −46.9 (4) |
| C23—C4—C5—C10 | 163.7 (4) | C8—C14—C15—C16 | −166.6 (3) |
| C3—C4—C5—C10 | 44.9 (4) | C14—C15—C16—C17 | 50.6 (4) |
| C24—C4—C5—C10 | −75.7 (5) | C15—C16—C17—C22 | −169.3 (3) |
| C10—C5—C6—C7 | −61.6 (4) | C15—C16—C17—C18 | −57.2 (4) |
| C4—C5—C6—C7 | 163.1 (3) | C15—C16—C17—C28 | 66.9 (4) |
| C5—C6—C7—C8 | 57.3 (4) | C12—C13—C18—C17 | 173.2 (3) |
| C6—C7—C8—C26 | 72.4 (4) | C14—C13—C18—C17 | −61.6 (3) |
| C6—C7—C8—C9 | −48.5 (4) | C12—C13—C18—C19 | 51.0 (4) |
| C6—C7—C8—C14 | −167.6 (3) | C14—C13—C18—C19 | 176.2 (3) |
| C7—C8—C9—C11 | 179.7 (3) | C16—C17—C18—C13 | 64.1 (4) |
| C26—C8—C9—C11 | 61.4 (4) | C22—C17—C18—C13 | −174.3 (3) |
| C14—C8—C9—C11 | −59.3 (3) | C28—C17—C18—C13 | −58.6 (4) |
| C7—C8—C9—C10 | 47.2 (4) | C16—C17—C18—C19 | −164.7 (3) |
| C26—C8—C9—C10 | −71.1 (4) | C22—C17—C18—C19 | −43.1 (3) |
| C14—C8—C9—C10 | 168.2 (3) | C28—C17—C18—C19 | 72.6 (4) |
| C2—C1—C10—C25 | −66.7 (5) | C13—C18—C19—C20 | −98.8 (4) |
| C2—C1—C10—C5 | 57.0 (5) | C17—C18—C19—C20 | 135.8 (3) |
| C2—C1—C10—C9 | 171.6 (4) | C13—C18—C19—C21 | 150.0 (3) |
| C6—C5—C10—C25 | −68.7 (4) | C17—C18—C19—C21 | 24.7 (3) |
| C4—C5—C10—C25 | 64.3 (4) | C18—C19—C20—C29 | 63.3 (5) |
| C6—C5—C10—C1 | 171.1 (3) | C21—C19—C20—C29 | 175.3 (4) |
| C4—C5—C10—C1 | −56.0 (4) | C18—C19—C20—C30 | −113.1 (3) |
| C6—C5—C10—C9 | 56.5 (4) | C21—C19—C20—C30 | −1.0 (4) |
| C4—C5—C10—C9 | −170.6 (3) | C30—O2—C21—C22 | 116.4 (4) |
| C11—C9—C10—C25 | −55.7 (4) | C30—O2—C21—C19 | −0.3 (4) |
| C8—C9—C10—C25 | 75.5 (4) | C20—C19—C21—O2 | 0.8 (4) |
| C11—C9—C10—C1 | 64.0 (4) | C18—C19—C21—O2 | 122.7 (3) |
| C8—C9—C10—C1 | −164.8 (3) | C20—C19—C21—C22 | −118.8 (4) |
| C11—C9—C10—C5 | 177.8 (3) | C18—C19—C21—C22 | 3.1 (4) |
| C8—C9—C10—C5 | −51.0 (4) | O2—C21—C22—C17 | −146.9 (3) |
| C8—C9—C11—C12 | 53.1 (4) | C19—C21—C22—C17 | −29.8 (4) |
| C10—C9—C11—C12 | −172.7 (3) | C16—C17—C22—C21 | 160.8 (3) |
| C9—C11—C12—C13 | −49.7 (4) | C18—C17—C22—C21 | 44.7 (4) |
| C11—C12—C13—C18 | 178.0 (3) | C28—C17—C22—C21 | −74.3 (4) |
| C11—C12—C13—C14 | 53.6 (4) | C21—O2—C30—O3 | −178.3 (4) |
| C12—C13—C14—C27 | 60.9 (3) | C21—O2—C30—C20 | −0.4 (5) |
| C18—C13—C14—C27 | −65.8 (3) | C29—C20—C30—O3 | 1.8 (7) |
| C12—C13—C14—C15 | 178.1 (3) | C19—C20—C30—O3 | 178.7 (4) |
| C18—C13—C14—C15 | 51.4 (3) | C29—C20—C30—O2 | −176.0 (4) |
| C12—C13—C14—C8 | −60.5 (3) | C19—C20—C30—O2 | 0.9 (5) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C2—H2A···O2i | 0.99 | 2.57 | 3.395 (5) | 141 |
| C12—H12A···O1ii | 0.99 | 2.45 | 3.310 (6) | 146 |
| C24—H24A···O3i | 0.98 | 2.58 | 3.357 (6) | 137 |
Symmetry codes: (i) −x+1/2, −y+1, z+1/2; (ii) −x−1, y+1/2, −z+5/2.
Funding Statement
This work was funded by Facultad de Química, Universidad Autonoma de Yucatan grant SISTPROY FQUI-2016-0006.
References
- Alvarenga, N. L., Ferro, E. A., Ravelo, A. G., Kennedy, M. L., Maestro, M. A. & González, A. G. (2000). Tetrahedron, 56, 3771–3774.
- Bruker (2014). APEX2, SAINT and SADABS. Bruker AXS Inc., Madison, Wisconsin, USA.
- Callies, O., Bedoya, L. M., Beltrán, M., Muñoz, A., Calderón, P. O., Osorio, A. A., Jiménez, I. A., Alcamí, J. & Bazzocchi, I. L. (2015). J. Nat. Prod. 78, 1045–1055. [DOI] [PubMed]
- Edwards, M. G., Kenworthy, M. N., Kitson, R. A., Scott, M. S. & Taylor, R. K. (2008). Angew. Chem. Int. Ed. 47, 1935–1937. [DOI] [PubMed]
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Jiménez-Estrada, M., Reyes-Chilpa, R., Hernández-Ortega, S., Cristobal-Telésforo, E., Torres-Colín, L., Jankowski, C. K., Aumelas, A. & Van Calsteren, M. R. (2000). Can. J. Chem. 78, 248–254.
- Krause, L., Herbst-Irmer, R., Sheldrick, G. M. & Stalke, D. (2015). J. Appl. Cryst. 48, 3–10. [DOI] [PMC free article] [PubMed]
- Macrae, C. F., Edgington, P. R., McCabe, P., Pidcock, E., Shields, G. P., Taylor, R., Towler, M. & van de Streek, J. (2006). J. Appl. Cryst. 39, 453–457.
- Ngassapa, O. D., Soejarto, D. D., Che, C., Pezzuto, J. M. & Farnsworth, N. R. (1991). J. Nat. Prod. 54, 1353–1359. [DOI] [PubMed]
- Parsons, S., Flack, H. D. & Wagner, T. (2013). Acta Cryst. B69, 249–259. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2015). Acta Cryst. A71, 3–8.
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Sturm, S., Gil, R. R., Chai, H., Ngassapa, O. D., Santisuk, T., Reutrakul, V., Howe, A., Moss, M., Besterman, J. M., Yang, S., Farthing, J. E., Tait, R. M., Lewis, J. A., O’Neill, M. J., Farnsworth, N. R., Cordell, G. A., Pezzuto, J. M. & Kinghorn, A. D. (1996). J. Nat. Prod. 59, 658–663. [DOI] [PubMed]
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989017012816/lh4023sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989017012816/lh4023Isup2.hkl
CCDC reference: 1573017
Additional supporting information: crystallographic information; 3D view; checkCIF report