In the cation of the title molecular salt, one of the non-H substituents on the piperidine ring occupies an equatorial site and the other an axial site. The ions are linked into sheets by a combination of one N—H⋯O and two C—H⋯O hydrogen bonds.
Keywords: molecular structure, disorder, conformation, hydrogen bonding, supramolecular assembly, crystal structure
Abstract
Ebastine, 4-(benzhydryloxy)-1-[4-(4-tert-butylphenyl)-4-oxobutyl]piperidine, reacts with 3,5-dinitrobenzoic acid in methanol solution to give the title 1:1 salt, ebastinium 3,5-dinitrobenzoate, C32H40NO2 +·C7H3N2O6 −. In the cation, the disubstituted aryl ring exhibits orientational disorder over two sets of atomic sites having occupancies 0.706 (4) and 0.294 (6), with a dihedral angle of 41.2 (5)° between the two orientations: the bulky Ph2CH—O– substituent occupies an axial site on the piperidine ring. The two ions in the selected asymmetric unit are linked by a nearly linear N—H⋯O hydrogen bond and this, in combination with two C—H⋯O hydrogen bonds, links the ions into complex sheets.
Chemical context
Ebastine, or 4-(benzhydryloxy)-1-[4-(4-tert-butylphenyl)-4-oxobutyl]piperidine, is a non-sedating second generation H1 receptor antagonist, which is effective in the treatment of both allergic rhinitis, whether seasonal or perennial, and chronic idiopathic urticaria (Wiseman & Faulds, 1996 ▸; Van Cauwenberge et al., 2004 ▸). The structure of ebastine has been the subject of two recent reports (Cheng et al., 2005 ▸: Sharma et al., 2015 ▸). Herein, we report the molecular and supramolecular structure of the 1:1 salt ebastinium 3,5-dinitrobenzoate (I), formed in the reaction between ebastine and 3,5-dinitrobenzoic acid.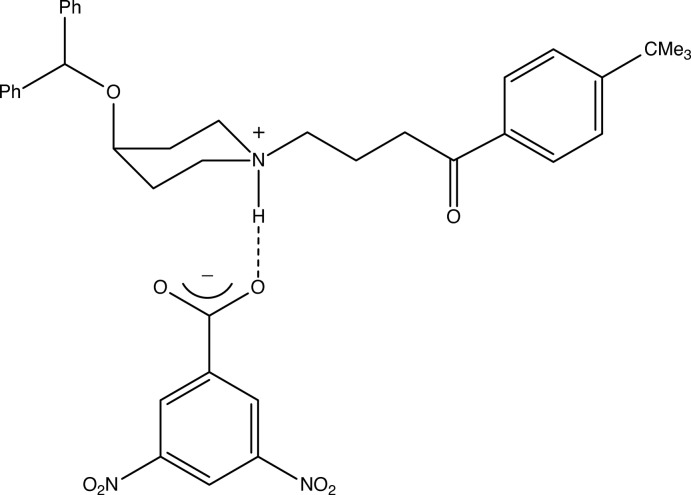
Structural commentary
The title compound (I), consists of an N-protonated ebastinium cation and a 3,5-dinitrobenzoate anion (Fig. 1 ▸), which are linked within the selected asymmetric unit a by a fairly short and nearly linear N—H⋯O hydrogen bond (Fig. 1 ▸, Table 1 ▸). The disubstituted aryl ring in the cation is disordered over two sets of atomic sites having occupancies 0.706 (4) for the major ring orientation, labelled C161–C166, and 0.294 (4) for the minor orientation, labeled C171–C176: the dihedral angle between these two ring planes is 41.2 (5)° (Fig. 1 ▸). The piperidine ring adopts an almost perfect chair conformation, with a ring-puckering angle, calculated for the atom sequence (N1,C2,C3,C4,C5,C6) of θ = 0.0 (3)°, identical within experimental uncertainty to the idealized value for a perfect chair form of θ = 0.0° (Boeyens, 1978 ▸). However, although the non-H substituent at atom N1 in the ring occupies an equatorial site, as expected, the bulky Ph2CHO substituent at atom C4 unexpectedly occupies an axial site. This observation is the more surprising since in ebastine itself, both non-H substituents on the piperidine ring occupy equatorial sites (Cheng et al., 2005 ▸: Sharma et al., 2015 ▸). The 3,5-dinitrobenzoate anion in compound (I) is nearly planar: the dihedral angles between the aryl ring and the substituents at atoms C21, C23 and C25 are 1.4 (2), 4.2 (2) and 10.7 (2)°, respectively: only the O atoms of the 5-nitro group are significantly displaced from the mean plane of the anion as a whole, 0.219 (2) Å for atom O25 and 0.187 (2) Å for atom O26: the r.m.s. deviation from the mean plane for the entire anion is only 0.082 Å.
Figure 1.
The molecular structure of the ionic components of compound (I), showing the atom-labelling scheme, the N—H⋯O hydrogen bond within the selected asymmetric unit, and the orientational disorder of the disubstituted aryl ring (the major component is drawn with full lines and the minor component with broken lines). Displacement ellipsoids are drawn at the 30% probability level and, for clarity, a few of the atom labels have been omitted.
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1⋯O22 | 0.99 (3) | 1.66 (2) | 2.634 (3) | 167 (2) |
| C2—H2A⋯O25i | 0.97 | 2.50 | 3.444 (3) | 163 |
| C11—H11A⋯O14ii | 0.97 | 2.49 | 3.358 (4) | 150 |
Symmetry codes: (i)  ; (ii)
; (ii)  .
.
Supramolecular features
In addition to the N—H⋯O hydrogen bond within the selected asymmetric unit, already noted (cf. Fig. 1 ▸ and Table 1 ▸), there are two C—H⋯O hydrogen bonds in the crystal of compound (I), which link the components into complex sheets, whose formation can, however, be readily analysed in terms of two simple, one-dimensional sub-structures (Ferguson et al., 1998a
▸,b
▸; Gregson et al., 2000 ▸). In the simpler of the two sub-structures, cations related by translation are linked by a single C—H⋯O hydrogen bond to form a C(6) chain running parallel to the [100] direction (Fig. 2 ▸, Table 1 ▸). The second sub-structure involves the cations and the anions, and a combination of the N—H⋯O hydrogen bond and a second C—H⋯O hydrogen bond links ions related by a c-glide plane into a  (11) chain, running parallel to the [20
(11) chain, running parallel to the [20 ] direction, in which cations and anions alternate (Fig. 3 ▸, Table 1 ▸). The combination of these two chain motifs generates a sheet lying parallel to (010) in the domain 0.5 < y < 1.0, and a second such sheet, related to the first by inversion, lies in the domain 0.0 < y < 0.5, but there are no direction-specific interactions between adjacent sheets. It is interesting to note that none of the hydrogen bonds in compound (I) involves the Ph2CHO substituent, so that direction-specific interactions cannot be held responsible for the location of this substituent at an axial site on the piperidine ring.
] direction, in which cations and anions alternate (Fig. 3 ▸, Table 1 ▸). The combination of these two chain motifs generates a sheet lying parallel to (010) in the domain 0.5 < y < 1.0, and a second such sheet, related to the first by inversion, lies in the domain 0.0 < y < 0.5, but there are no direction-specific interactions between adjacent sheets. It is interesting to note that none of the hydrogen bonds in compound (I) involves the Ph2CHO substituent, so that direction-specific interactions cannot be held responsible for the location of this substituent at an axial site on the piperidine ring.
Figure 2.
Part of the crystal structure of compound (I), showing a hydrogen-bonded C(6) chain of cations running parallel to [100]. For clarity, the anions, the minor disorder component of the cation, and the H atoms bonded to carrier atoms not involved in the motif shown have been omitted. The atoms marked with an asterisk (*) or a hash (#) are at the symmetry positions (−1 + x, y, z) and (1 + x, y, z) respectively.
Figure 3.
Part of the crystal structure of compound (I), showing a hydrogen-bonded  (11) chain running parallel to [20
(11) chain running parallel to [20 ]. For clarity, the minor disorder component of the cation, and the H atoms bonded to C atoms not involved in the motif shown have been omitted.
]. For clarity, the minor disorder component of the cation, and the H atoms bonded to C atoms not involved in the motif shown have been omitted.
Database survey
The molecular structure of neutral ebastine (Cheng et al., 2005 ▸; Sharma et al., 2015 ▸) differs from that of the ebastinium cation in compound (I) in two significant respects. Firstly, there is no disorder in the neutral compound as opposed to the orientation disorder of the disubstituted aryl ring in (I) and secondly, both of the non-H substituents on the piperidine ring occupy equatorial sites in the neutral compound as opposed to the presence of one axial and one equatorial substituent in (I). Neither of the two reports on the structure of ebastine gave any description of the supramolecular assembly: one (Cheng et al., 2005 ▸) noted the presence of hydrogen bonds, but the second (Sharma et al., 2015 ▸) did not record these. Accordingly, we have now examined the supramolecular assembly of ebastine using the most recently reported atomic coordinates (Sharma et al., 2015 ▸): a combination of one C—H⋯N hydrogen bond and one C—H⋯O hydrogen bond links the molecules into sheets lying parallel to (100) and containing  (20) and
(20) and  (48) rings, both centrosymmetric, arranges in chess board fashion (Fig. 4 ▸). Structures have also been reported recently for some structurally related compounds with pharmacological activity, including the picrate salt of the anticholinergic drug propiverine, 4-(2,2-diphenyl-2-propoxyacetoxy)-1-methylpiperidin-1-ium picrate (Jasinski et al., 2009 ▸), and the anti-spasmodic drug pargeverine, N,N-dimethyl-[2-(2,2-diphenyl)-2-prop-2-ynyloxy)acetoxy]ethylamine and its picrate and (2R,3R)-(hydrogentartrate) salts (Shaibah et al., 2017 ▸).
(48) rings, both centrosymmetric, arranges in chess board fashion (Fig. 4 ▸). Structures have also been reported recently for some structurally related compounds with pharmacological activity, including the picrate salt of the anticholinergic drug propiverine, 4-(2,2-diphenyl-2-propoxyacetoxy)-1-methylpiperidin-1-ium picrate (Jasinski et al., 2009 ▸), and the anti-spasmodic drug pargeverine, N,N-dimethyl-[2-(2,2-diphenyl)-2-prop-2-ynyloxy)acetoxy]ethylamine and its picrate and (2R,3R)-(hydrogentartrate) salts (Shaibah et al., 2017 ▸).
Figure 4.
Part of the crystal structure of ebastine showing the formation of a hydrogen-bonded sheet of  (20) and
(20) and  (48) rings. The original atomic coordinates (Sharma et al., 2015 ▸) have been used and, for the sake of clarity, the H atoms not involved in the motifs shown have been omitted.
(48) rings. The original atomic coordinates (Sharma et al., 2015 ▸) have been used and, for the sake of clarity, the H atoms not involved in the motifs shown have been omitted.
Synthesis and crystallization
A sample of ebastine was a gift from RL Fine Chem, Pvt. Ltd., Bengaluru, India. For the synthesis of compound (I), ebastine (100 mg, 0.20 mmol) and 3,5-dinitrobenzoic acid (45 mg, 0.20 mmol) were dissolved in hot methanol and held at 333 K for 30 min, with magnetic stirring throughout. The resulting solution was then allowed to cool slowly to room temperature, giving colourless block-like crystals (m.p. 424–428 K).
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. Three low-angle reflections (021), (002) and (012), which had been attenuated by the beam stop, were omitted from the refinements. It was apparent from an early stage in the refinement that the disubstituted aryl ring was disordered over two sets of atomic sights having unequal occupancies, and corresponding to different orientations of this ring relative to its substituents. For the minor orientation, the bonded distances and the 1,3-non-bonded distances were restrained to be the same as the corresponding distances in the major orientation, subject to s.u.s of 0.01 and 0.02 Å, respectively: in addition, the anisotropic displacement parameters for corresponding pairs of atomic sites were constrained to be equal. All H atoms, other than those in the minor disorder components, were located in difference-Fourier maps. The C-bound H atoms were all treated as riding atoms in geometrically idealized positions: C—H 0.93 Å (aromatic), 0.96 Å (CH3), 0.97 Å (CH2) or 0.98 Å (aliphatic C—H), with U iso(H) = 1.5U eq(C-methyl) and 1.2U eq(C) for other H atoms. The methyl groups were permitted to rotate but not to tilt. For the H atom bonded to the N atom, the atomic coordinates were refined with U iso(H) = 1.2U eq(N), giving an N—H distance of 0.99 (3) Å. Subject to these conditions, the occupancies of the two disordered components refined to 0.706 (4) and 0.294 (4). In the final analysis of variance there was a large value, 15.256, of K = [mean(F o 2)/mean(F c 2)] for the group of 867 very weak reflections having F c/F c(max) in the range 0.000 < F c/F c(max) < 0.005.
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C32H40NO2·C7H3N2O6 |
| M r | 681.76 |
| Crystal system, space group | Monoclinic, P21/c |
| Temperature (K) | 293 |
| a, b, c (Å) | 5.9168 (3), 28.3733 (12), 21.0782 (11) |
| β (°) | 97.836 (5) |
| V (Å3) | 3505.6 (3) |
| Z | 4 |
| Radiation type | Mo Kα |
| μ (mm−1) | 0.09 |
| Crystal size (mm) | 0.23 × 0.21 × 0.18 |
| Data collection | |
| Diffractometer | Rigaku Saturn724 |
| Absorption correction | Multi-scan (SADABS; Krause et al., 2015 ▸) |
| T min, T max | 0.956, 0.984 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 40112, 7331, 4388 |
| R int | 0.061 |
| (sin θ/λ)max (Å−1) | 0.629 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.065, 0.179, 1.05 |
| No. of reflections | 7331 |
| No. of parameters | 470 |
| No. of restraints | 22 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.20, −0.25 |
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S205698901701324X/su5391sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S205698901701324X/su5391Isup2.hkl
Supporting information file. DOI: 10.1107/S205698901701324X/su5391Isup3.cml
CCDC reference: 1574718
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
The authors thank the DST–PURSE Lab. (Mangalore University) for the diffractometer and other facilities. MAES thanks the University of Mysore for research facilities and BKS thanks the UGC for the award of a Rajiv Gandhi National Fellowship.
supplementary crystallographic information
Crystal data
| C32H40NO2+·C7H3N2O6− | F(000) = 1448 |
| Mr = 681.76 | Dx = 1.292 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| a = 5.9168 (3) Å | Cell parameters from 10431 reflections |
| b = 28.3733 (12) Å | θ = 2.4–31.2° |
| c = 21.0782 (11) Å | µ = 0.09 mm−1 |
| β = 97.836 (5)° | T = 293 K |
| V = 3505.6 (3) Å3 | Block, colourless |
| Z = 4 | 0.23 × 0.21 × 0.18 mm |
Data collection
| Rigaku Saturn724 diffractometer | 4388 reflections with I > 2σ(I) |
| Radiation source: fine focus sealed tube | Rint = 0.061 |
| φ and ω scans | θmax = 26.6°, θmin = 2.4° |
| Absorption correction: multi-scan (SADABS; Krause et al., 2015) | h = −7→7 |
| Tmin = 0.956, Tmax = 0.984 | k = −35→35 |
| 40112 measured reflections | l = −26→25 |
| 7331 independent reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.065 | Hydrogen site location: mixed |
| wR(F2) = 0.179 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.05 | w = 1/[σ2(Fo2) + (0.0661P)2 + 1.0412P] where P = (Fo2 + 2Fc2)/3 |
| 7331 reflections | (Δ/σ)max = 0.001 |
| 470 parameters | Δρmax = 0.20 e Å−3 |
| 22 restraints | Δρmin = −0.25 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| N1 | 0.3135 (4) | 0.72898 (6) | 0.37455 (11) | 0.0510 (5) | |
| H1 | 0.374 (4) | 0.7192 (8) | 0.3349 (12) | 0.061* | |
| C2 | 0.4460 (4) | 0.77114 (8) | 0.40021 (11) | 0.0479 (6) | |
| H2B | 0.6062 | 0.7629 | 0.4097 | 0.057* | |
| H2A | 0.3938 | 0.7811 | 0.4398 | 0.057* | |
| C3 | 0.4185 (4) | 0.81101 (8) | 0.35285 (11) | 0.0457 (6) | |
| H3A | 0.4826 | 0.8018 | 0.3147 | 0.055* | |
| H3B | 0.5027 | 0.8382 | 0.3713 | 0.055* | |
| C4 | 0.1701 (4) | 0.82448 (8) | 0.33437 (11) | 0.0457 (6) | |
| H4 | 0.1579 | 0.8488 | 0.3011 | 0.055* | |
| C5 | 0.0386 (4) | 0.78137 (9) | 0.30906 (12) | 0.0553 (7) | |
| H5A | −0.1219 | 0.7892 | 0.2994 | 0.066* | |
| H5B | 0.0916 | 0.7713 | 0.2697 | 0.066* | |
| C6 | 0.0676 (4) | 0.74172 (9) | 0.35682 (13) | 0.0575 (7) | |
| H6A | 0.0043 | 0.7510 | 0.3950 | 0.069* | |
| H6B | −0.0160 | 0.7144 | 0.3388 | 0.069* | |
| O4 | 0.0623 (3) | 0.84055 (5) | 0.38735 (7) | 0.0484 (4) | |
| C41 | 0.1566 (4) | 0.88157 (8) | 0.41962 (11) | 0.0460 (6) | |
| H41 | 0.3107 | 0.8739 | 0.4403 | 0.055* | |
| C141 | 0.0098 (4) | 0.89282 (8) | 0.47183 (11) | 0.0453 (6) | |
| C142 | −0.1660 (4) | 0.86412 (9) | 0.48464 (11) | 0.0511 (6) | |
| H142 | −0.1975 | 0.8367 | 0.4610 | 0.061* | |
| C143 | −0.2974 (5) | 0.87546 (10) | 0.53237 (12) | 0.0613 (7) | |
| H143 | −0.4159 | 0.8557 | 0.5403 | 0.074* | |
| C144 | −0.2527 (6) | 0.91565 (10) | 0.56772 (13) | 0.0686 (8) | |
| H144 | −0.3422 | 0.9236 | 0.5991 | 0.082* | |
| C145 | −0.0755 (6) | 0.94408 (10) | 0.55665 (15) | 0.0798 (10) | |
| H145 | −0.0426 | 0.9711 | 0.5812 | 0.096* | |
| C146 | 0.0550 (6) | 0.93287 (9) | 0.50904 (14) | 0.0719 (8) | |
| H146 | 0.1749 | 0.9525 | 0.5019 | 0.086* | |
| C151 | 0.1735 (4) | 0.92244 (8) | 0.37464 (11) | 0.0469 (6) | |
| C152 | 0.3761 (5) | 0.94689 (9) | 0.37596 (14) | 0.0612 (7) | |
| H152 | 0.5016 | 0.9380 | 0.4050 | 0.073* | |
| C153 | 0.3962 (6) | 0.98416 (10) | 0.33510 (18) | 0.0770 (9) | |
| H153 | 0.5338 | 1.0003 | 0.3368 | 0.092* | |
| C154 | 0.2134 (7) | 0.99730 (11) | 0.29230 (17) | 0.0808 (10) | |
| H154 | 0.2264 | 1.0222 | 0.2644 | 0.097* | |
| C155 | 0.0086 (6) | 0.97356 (11) | 0.29040 (15) | 0.0754 (9) | |
| H155 | −0.1161 | 0.9827 | 0.2613 | 0.091* | |
| C156 | −0.0118 (5) | 0.93637 (9) | 0.33152 (13) | 0.0603 (7) | |
| H156 | −0.1504 | 0.9207 | 0.3302 | 0.072* | |
| C11 | 0.3376 (6) | 0.68892 (10) | 0.42095 (17) | 0.0865 (11) | |
| H11A | 0.2213 | 0.6656 | 0.4071 | 0.104* | |
| H11B | 0.3101 | 0.7005 | 0.4625 | 0.104* | |
| C12 | 0.5687 (5) | 0.66527 (9) | 0.42801 (16) | 0.0724 (9) | |
| H12A | 0.6523 | 0.6664 | 0.4737 | 0.087* | |
| H12B | 0.6523 | 0.6776 | 0.4014 | 0.087* | |
| C13 | 0.5462 (5) | 0.61430 (10) | 0.41343 (17) | 0.0833 (10) | |
| H13A | 0.4697 | 0.6108 | 0.3700 | 0.100* | |
| H13B | 0.4481 | 0.6005 | 0.4417 | 0.100* | |
| C14 | 0.7637 (6) | 0.58644 (10) | 0.41954 (15) | 0.0709 (8) | |
| O14 | 0.9451 (4) | 0.60527 (8) | 0.43408 (16) | 0.1170 (10) | |
| C161 | 0.7488 (5) | 0.53513 (9) | 0.40590 (14) | 0.0659 (8) | 0.706 (4) |
| C162 | 0.5497 (7) | 0.51590 (14) | 0.3727 (3) | 0.0902 (17) | 0.706 (4) |
| H162 | 0.4245 | 0.5351 | 0.3599 | 0.108* | 0.706 (4) |
| C163 | 0.5386 (7) | 0.46828 (14) | 0.3589 (3) | 0.0907 (18) | 0.706 (4) |
| H163 | 0.4054 | 0.4561 | 0.3364 | 0.109* | 0.706 (4) |
| C164 | 0.7209 (5) | 0.43792 (9) | 0.37782 (13) | 0.0587 (7) | 0.706 (4) |
| C165 | 0.9157 (9) | 0.45915 (18) | 0.4048 (5) | 0.103 (4) | 0.706 (4) |
| H165 | 1.0458 | 0.4407 | 0.4147 | 0.124* | 0.706 (4) |
| C166 | 0.9308 (9) | 0.50664 (17) | 0.4185 (4) | 0.099 (3) | 0.706 (4) |
| H166 | 1.0697 | 0.5192 | 0.4367 | 0.119* | 0.706 (4) |
| C171 | 0.7488 (5) | 0.53513 (9) | 0.40590 (14) | 0.0659 (8) | 0.294 (4) |
| C172 | 0.5662 (15) | 0.5073 (3) | 0.4211 (5) | 0.0902 (17) | 0.294 (4) |
| H172 | 0.4497 | 0.5216 | 0.4396 | 0.108* | 0.294 (4) |
| C173 | 0.5554 (15) | 0.4594 (3) | 0.4093 (5) | 0.0907 (18) | 0.294 (4) |
| H173 | 0.4382 | 0.4415 | 0.4223 | 0.109* | 0.294 (4) |
| C174 | 0.7209 (5) | 0.43792 (9) | 0.37782 (13) | 0.0587 (7) | 0.294 (4) |
| C175 | 0.9160 (17) | 0.4626 (4) | 0.3793 (15) | 0.103 (4) | 0.294 (4) |
| H175 | 1.0483 | 0.4468 | 0.3726 | 0.124* | 0.294 (4) |
| C176 | 0.9267 (18) | 0.5104 (4) | 0.3905 (12) | 0.099 (3) | 0.294 (4) |
| H176 | 1.0625 | 0.5262 | 0.3872 | 0.119* | 0.294 (4) |
| C181 | 0.7009 (5) | 0.38550 (9) | 0.36135 (13) | 0.0591 (7) | |
| C182 | 0.6091 (6) | 0.35909 (12) | 0.41549 (16) | 0.0925 (11) | |
| H18A | 0.5908 | 0.3264 | 0.4044 | 0.139* | |
| H18B | 0.4643 | 0.3721 | 0.4220 | 0.139* | |
| H18C | 0.7145 | 0.3622 | 0.4541 | 0.139* | |
| C183 | 0.9278 (5) | 0.36413 (11) | 0.35081 (18) | 0.0903 (11) | |
| H18D | 1.0326 | 0.3661 | 0.3897 | 0.135* | |
| H18E | 0.9883 | 0.3811 | 0.3175 | 0.135* | |
| H18F | 0.9059 | 0.3317 | 0.3386 | 0.135* | |
| C184 | 0.5349 (5) | 0.37825 (11) | 0.29982 (14) | 0.0753 (8) | |
| H18G | 0.5895 | 0.3949 | 0.2653 | 0.113* | |
| H18H | 0.3872 | 0.3900 | 0.3058 | 0.113* | |
| H18I | 0.5240 | 0.3453 | 0.2898 | 0.113* | |
| C21 | 0.7522 (4) | 0.69940 (8) | 0.19856 (11) | 0.0431 (5) | |
| C22 | 0.8763 (4) | 0.66079 (8) | 0.22368 (11) | 0.0458 (6) | |
| H22 | 0.8338 | 0.6448 | 0.2587 | 0.055* | |
| C23 | 1.0631 (4) | 0.64614 (8) | 0.19644 (12) | 0.0486 (6) | |
| C24 | 1.1306 (4) | 0.66812 (9) | 0.14419 (12) | 0.0525 (6) | |
| H24 | 1.2571 | 0.6579 | 0.1263 | 0.063* | |
| C25 | 1.0032 (4) | 0.70587 (9) | 0.11964 (11) | 0.0505 (6) | |
| C26 | 0.8156 (4) | 0.72192 (8) | 0.14567 (11) | 0.0493 (6) | |
| H26 | 0.7327 | 0.7476 | 0.1278 | 0.059* | |
| C27 | 0.5535 (4) | 0.71822 (9) | 0.23013 (13) | 0.0504 (6) | |
| O21 | 0.4514 (3) | 0.75292 (7) | 0.20644 (9) | 0.0689 (5) | |
| O22 | 0.5158 (3) | 0.69618 (6) | 0.27979 (9) | 0.0649 (5) | |
| N23 | 1.1983 (4) | 0.60593 (8) | 0.22480 (13) | 0.0672 (6) | |
| O23 | 1.1460 (4) | 0.58894 (8) | 0.27369 (12) | 0.0920 (7) | |
| O24 | 1.3561 (4) | 0.59235 (8) | 0.19822 (12) | 0.1010 (8) | |
| N25 | 1.0724 (5) | 0.72964 (9) | 0.06325 (12) | 0.0700 (7) | |
| O25 | 1.2546 (4) | 0.71894 (9) | 0.04679 (11) | 0.0974 (8) | |
| O26 | 0.9409 (5) | 0.75808 (9) | 0.03539 (11) | 0.1034 (8) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| N1 | 0.0585 (13) | 0.0405 (10) | 0.0606 (13) | 0.0002 (9) | 0.0321 (11) | −0.0031 (10) |
| C2 | 0.0490 (14) | 0.0496 (13) | 0.0474 (14) | 0.0035 (11) | 0.0147 (11) | −0.0072 (11) |
| C3 | 0.0470 (14) | 0.0421 (12) | 0.0508 (14) | −0.0017 (10) | 0.0168 (11) | −0.0075 (11) |
| C4 | 0.0480 (14) | 0.0472 (13) | 0.0444 (13) | 0.0039 (11) | 0.0150 (11) | −0.0043 (11) |
| C5 | 0.0430 (14) | 0.0664 (16) | 0.0576 (16) | 0.0005 (12) | 0.0108 (12) | −0.0223 (13) |
| C6 | 0.0514 (16) | 0.0545 (15) | 0.0722 (18) | −0.0128 (12) | 0.0285 (14) | −0.0229 (14) |
| O4 | 0.0520 (10) | 0.0442 (9) | 0.0527 (10) | 0.0011 (7) | 0.0203 (8) | −0.0131 (7) |
| C41 | 0.0446 (13) | 0.0415 (12) | 0.0521 (14) | 0.0040 (10) | 0.0076 (11) | −0.0054 (11) |
| C141 | 0.0513 (14) | 0.0397 (12) | 0.0453 (13) | 0.0080 (11) | 0.0082 (11) | −0.0019 (10) |
| C142 | 0.0510 (15) | 0.0579 (15) | 0.0441 (14) | 0.0011 (12) | 0.0058 (12) | −0.0045 (12) |
| C143 | 0.0581 (17) | 0.0772 (19) | 0.0501 (15) | 0.0059 (14) | 0.0130 (13) | 0.0065 (14) |
| C144 | 0.088 (2) | 0.0690 (18) | 0.0540 (17) | 0.0236 (17) | 0.0277 (16) | 0.0054 (15) |
| C145 | 0.122 (3) | 0.0537 (17) | 0.070 (2) | 0.0072 (18) | 0.040 (2) | −0.0174 (15) |
| C146 | 0.100 (2) | 0.0518 (16) | 0.0696 (19) | −0.0091 (15) | 0.0334 (18) | −0.0129 (14) |
| C151 | 0.0498 (14) | 0.0416 (12) | 0.0521 (14) | 0.0045 (11) | 0.0171 (12) | −0.0075 (11) |
| C152 | 0.0595 (17) | 0.0526 (15) | 0.0755 (19) | 0.0007 (13) | 0.0234 (15) | −0.0081 (14) |
| C153 | 0.079 (2) | 0.0557 (17) | 0.106 (3) | 0.0001 (16) | 0.047 (2) | −0.0010 (18) |
| C154 | 0.117 (3) | 0.0544 (17) | 0.083 (2) | 0.0105 (19) | 0.056 (2) | 0.0096 (16) |
| C155 | 0.092 (2) | 0.0709 (19) | 0.0646 (19) | 0.0251 (18) | 0.0133 (17) | 0.0088 (16) |
| C156 | 0.0614 (17) | 0.0567 (16) | 0.0640 (17) | 0.0072 (13) | 0.0131 (14) | 0.0013 (14) |
| C11 | 0.109 (3) | 0.0547 (16) | 0.110 (3) | 0.0158 (17) | 0.069 (2) | 0.0279 (17) |
| C12 | 0.089 (2) | 0.0500 (15) | 0.086 (2) | 0.0124 (15) | 0.0381 (18) | 0.0181 (15) |
| C13 | 0.087 (2) | 0.0635 (18) | 0.095 (2) | 0.0207 (16) | −0.0026 (19) | −0.0269 (17) |
| C14 | 0.073 (2) | 0.0543 (16) | 0.083 (2) | 0.0038 (15) | 0.0015 (17) | −0.0062 (15) |
| O14 | 0.0782 (17) | 0.0650 (14) | 0.200 (3) | −0.0039 (12) | −0.0078 (17) | −0.0152 (16) |
| C161 | 0.0586 (17) | 0.0523 (15) | 0.085 (2) | 0.0033 (14) | 0.0038 (15) | −0.0064 (14) |
| C162 | 0.071 (3) | 0.063 (2) | 0.125 (5) | 0.028 (2) | −0.031 (3) | −0.023 (3) |
| C163 | 0.062 (2) | 0.065 (2) | 0.136 (5) | 0.0107 (19) | −0.023 (3) | −0.029 (3) |
| C164 | 0.0531 (16) | 0.0537 (15) | 0.0714 (18) | 0.0057 (13) | 0.0157 (14) | −0.0063 (13) |
| C165 | 0.064 (2) | 0.056 (2) | 0.181 (11) | 0.0166 (17) | −0.013 (3) | −0.014 (4) |
| C166 | 0.061 (2) | 0.057 (2) | 0.170 (10) | 0.0066 (17) | −0.017 (3) | −0.011 (3) |
| C171 | 0.0586 (17) | 0.0523 (15) | 0.085 (2) | 0.0033 (14) | 0.0038 (15) | −0.0064 (14) |
| C172 | 0.071 (3) | 0.063 (2) | 0.125 (5) | 0.028 (2) | −0.031 (3) | −0.023 (3) |
| C173 | 0.062 (2) | 0.065 (2) | 0.136 (5) | 0.0107 (19) | −0.023 (3) | −0.029 (3) |
| C174 | 0.0531 (16) | 0.0537 (15) | 0.0714 (18) | 0.0057 (13) | 0.0157 (14) | −0.0063 (13) |
| C175 | 0.064 (2) | 0.056 (2) | 0.181 (11) | 0.0166 (17) | −0.013 (3) | −0.014 (4) |
| C176 | 0.061 (2) | 0.057 (2) | 0.170 (10) | 0.0066 (17) | −0.017 (3) | −0.011 (3) |
| C181 | 0.0610 (17) | 0.0529 (15) | 0.0659 (17) | 0.0014 (13) | 0.0175 (14) | 0.0020 (13) |
| C182 | 0.121 (3) | 0.087 (2) | 0.073 (2) | −0.007 (2) | 0.028 (2) | 0.0090 (18) |
| C183 | 0.079 (2) | 0.069 (2) | 0.124 (3) | 0.0164 (17) | 0.019 (2) | −0.014 (2) |
| C184 | 0.085 (2) | 0.0685 (18) | 0.074 (2) | −0.0021 (16) | 0.0180 (17) | −0.0076 (16) |
| C21 | 0.0455 (13) | 0.0423 (12) | 0.0432 (13) | −0.0045 (10) | 0.0127 (11) | −0.0105 (10) |
| C22 | 0.0518 (14) | 0.0467 (13) | 0.0406 (13) | −0.0047 (11) | 0.0124 (11) | −0.0067 (10) |
| C23 | 0.0479 (14) | 0.0481 (13) | 0.0496 (14) | 0.0050 (11) | 0.0062 (12) | −0.0117 (11) |
| C24 | 0.0476 (14) | 0.0601 (15) | 0.0527 (15) | −0.0056 (12) | 0.0173 (12) | −0.0186 (13) |
| C25 | 0.0535 (15) | 0.0581 (15) | 0.0428 (14) | −0.0097 (12) | 0.0169 (12) | −0.0076 (12) |
| C26 | 0.0536 (15) | 0.0480 (13) | 0.0472 (14) | −0.0034 (11) | 0.0106 (12) | −0.0072 (11) |
| C27 | 0.0486 (15) | 0.0505 (14) | 0.0548 (16) | −0.0031 (12) | 0.0166 (12) | −0.0165 (13) |
| O21 | 0.0677 (13) | 0.0620 (12) | 0.0798 (13) | 0.0171 (10) | 0.0207 (10) | −0.0063 (10) |
| O22 | 0.0747 (13) | 0.0646 (11) | 0.0634 (12) | 0.0063 (9) | 0.0386 (10) | −0.0071 (10) |
| N23 | 0.0668 (16) | 0.0624 (15) | 0.0715 (16) | 0.0143 (12) | 0.0064 (14) | −0.0117 (13) |
| O23 | 0.1038 (18) | 0.0849 (15) | 0.0873 (16) | 0.0259 (13) | 0.0133 (14) | 0.0247 (13) |
| O24 | 0.0920 (17) | 0.1013 (17) | 0.1138 (19) | 0.0466 (14) | 0.0293 (15) | −0.0112 (15) |
| N25 | 0.0849 (19) | 0.0748 (17) | 0.0557 (15) | −0.0129 (14) | 0.0292 (15) | −0.0015 (13) |
| O25 | 0.0960 (18) | 0.127 (2) | 0.0806 (16) | −0.0088 (15) | 0.0543 (14) | −0.0002 (14) |
| O26 | 0.132 (2) | 0.1008 (18) | 0.0844 (17) | 0.0167 (16) | 0.0413 (16) | 0.0340 (15) |
Geometric parameters (Å, º)
| N1—C2 | 1.491 (3) | C13—H13A | 0.9700 |
| N1—C11 | 1.493 (3) | C13—H13B | 0.9700 |
| N1—C6 | 1.496 (3) | C14—O14 | 1.201 (3) |
| N1—H1 | 0.99 (3) | C14—C161 | 1.484 (4) |
| C2—C3 | 1.503 (3) | C161—C166 | 1.344 (5) |
| C2—H2B | 0.9700 | C161—C162 | 1.397 (5) |
| C2—H2A | 0.9700 | C162—C163 | 1.382 (5) |
| C3—C4 | 1.517 (3) | C162—H162 | 0.9300 |
| C3—H3A | 0.9700 | C163—C164 | 1.396 (4) |
| C3—H3B | 0.9700 | C163—H163 | 0.9300 |
| C4—O4 | 1.434 (3) | C164—C165 | 1.355 (6) |
| C4—C5 | 1.507 (3) | C164—C181 | 1.528 (4) |
| C4—H4 | 0.9800 | C165—C166 | 1.378 (5) |
| C5—C6 | 1.504 (4) | C165—H165 | 0.9300 |
| C5—H5A | 0.9700 | C166—H166 | 0.9300 |
| C5—H5B | 0.9700 | C172—C173 | 1.381 (8) |
| C6—H6A | 0.9700 | C172—H172 | 0.9300 |
| C6—H6B | 0.9700 | C173—H173 | 0.9300 |
| O4—C41 | 1.423 (3) | C175—C176 | 1.376 (8) |
| C41—C151 | 1.510 (3) | C175—H175 | 0.9300 |
| C41—C141 | 1.526 (3) | C176—H176 | 0.9300 |
| C41—H41 | 0.9800 | C181—C183 | 1.517 (4) |
| C141—C142 | 1.377 (3) | C181—C182 | 1.526 (4) |
| C141—C146 | 1.386 (3) | C181—C184 | 1.530 (4) |
| C142—C143 | 1.390 (3) | C182—H18A | 0.9600 |
| C142—H142 | 0.9300 | C182—H18B | 0.9600 |
| C143—C144 | 1.368 (4) | C182—H18C | 0.9600 |
| C143—H143 | 0.9300 | C183—H18D | 0.9600 |
| C144—C145 | 1.368 (4) | C183—H18E | 0.9600 |
| C144—H144 | 0.9300 | C183—H18F | 0.9600 |
| C145—C146 | 1.384 (4) | C184—H18G | 0.9600 |
| C145—H145 | 0.9300 | C184—H18H | 0.9600 |
| C146—H146 | 0.9300 | C184—H18I | 0.9600 |
| C151—C152 | 1.382 (3) | C21—C26 | 1.380 (3) |
| C151—C156 | 1.383 (4) | C21—C22 | 1.383 (3) |
| C152—C153 | 1.379 (4) | C21—C27 | 1.524 (3) |
| C152—H152 | 0.9300 | C22—C23 | 1.378 (3) |
| C153—C154 | 1.363 (5) | C22—H22 | 0.9300 |
| C153—H153 | 0.9300 | C23—C24 | 1.371 (3) |
| C154—C155 | 1.382 (5) | C23—N23 | 1.473 (3) |
| C154—H154 | 0.9300 | C24—C25 | 1.370 (3) |
| C155—C156 | 1.381 (4) | C24—H24 | 0.9300 |
| C155—H155 | 0.9300 | C25—C26 | 1.381 (3) |
| C156—H156 | 0.9300 | C25—N25 | 1.472 (3) |
| C11—C12 | 1.512 (4) | C26—H26 | 0.9300 |
| C11—H11A | 0.9700 | C27—O21 | 1.226 (3) |
| C11—H11B | 0.9700 | C27—O22 | 1.265 (3) |
| C12—C13 | 1.481 (4) | N23—O24 | 1.215 (3) |
| C12—H12A | 1.0216 | N23—O23 | 1.215 (3) |
| C12—H12B | 0.8703 | N25—O26 | 1.215 (3) |
| C13—C14 | 1.501 (4) | N25—O25 | 1.215 (3) |
| C2—N1—C11 | 112.0 (2) | C11—C12—H12A | 113.2 |
| C2—N1—C6 | 110.00 (18) | C13—C12—H12B | 107.6 |
| C11—N1—C6 | 110.5 (2) | C11—C12—H12B | 110.1 |
| C2—N1—H1 | 107.3 (14) | H12A—C12—H12B | 110.5 |
| C11—N1—H1 | 109.0 (14) | C12—C13—C14 | 116.4 (3) |
| C6—N1—H1 | 107.9 (15) | C12—C13—H13A | 108.2 |
| N1—C2—C3 | 111.04 (19) | C14—C13—H13A | 108.2 |
| N1—C2—H2B | 109.4 | C12—C13—H13B | 108.2 |
| C3—C2—H2B | 109.4 | C14—C13—H13B | 108.2 |
| N1—C2—H2A | 109.4 | H13A—C13—H13B | 107.3 |
| C3—C2—H2A | 109.4 | O14—C14—C161 | 120.9 (3) |
| H2B—C2—H2A | 108.0 | O14—C14—C13 | 120.9 (3) |
| C2—C3—C4 | 111.94 (19) | C161—C14—C13 | 118.2 (3) |
| C2—C3—H3A | 109.2 | C166—C161—C162 | 117.5 (3) |
| C4—C3—H3A | 109.2 | C166—C161—C14 | 121.8 (3) |
| C2—C3—H3B | 109.2 | C162—C161—C14 | 120.3 (3) |
| C4—C3—H3B | 109.2 | C163—C162—C161 | 120.0 (4) |
| H3A—C3—H3B | 107.9 | C163—C162—H162 | 120.0 |
| O4—C4—C5 | 105.72 (18) | C161—C162—H162 | 120.0 |
| O4—C4—C3 | 113.48 (19) | C162—C163—C164 | 122.1 (4) |
| C5—C4—C3 | 108.75 (19) | C162—C163—H163 | 119.0 |
| O4—C4—H4 | 109.6 | C164—C163—H163 | 119.0 |
| C5—C4—H4 | 109.6 | C165—C164—C163 | 115.2 (3) |
| C3—C4—H4 | 109.6 | C165—C164—C181 | 124.3 (3) |
| C6—C5—C4 | 111.3 (2) | C163—C164—C181 | 120.2 (3) |
| C6—C5—H5A | 109.4 | C164—C165—C166 | 123.3 (5) |
| C4—C5—H5A | 109.4 | C164—C165—H165 | 118.3 |
| C6—C5—H5B | 109.4 | C166—C165—H165 | 118.3 |
| C4—C5—H5B | 109.4 | C161—C166—C165 | 121.3 (5) |
| H5A—C5—H5B | 108.0 | C161—C166—H166 | 119.3 |
| N1—C6—C5 | 111.49 (19) | C165—C166—H166 | 119.3 |
| N1—C6—H6A | 109.3 | C173—C172—H172 | 119.0 |
| C5—C6—H6A | 109.3 | C172—C173—H173 | 120.1 |
| N1—C6—H6B | 109.3 | C176—C175—H175 | 118.8 |
| C5—C6—H6B | 109.3 | C175—C176—H176 | 118.7 |
| H6A—C6—H6B | 108.0 | C183—C181—C182 | 109.0 (3) |
| C41—O4—C4 | 116.44 (17) | C183—C181—C164 | 112.3 (2) |
| O4—C41—C151 | 112.52 (19) | C182—C181—C164 | 109.4 (2) |
| O4—C41—C141 | 106.93 (18) | C183—C181—C184 | 107.5 (2) |
| C151—C41—C141 | 112.66 (18) | C182—C181—C184 | 108.4 (2) |
| O4—C41—H41 | 108.2 | C164—C181—C184 | 110.1 (2) |
| C151—C41—H41 | 108.2 | C181—C182—H18A | 109.5 |
| C141—C41—H41 | 108.2 | C181—C182—H18B | 109.5 |
| C142—C141—C146 | 117.8 (2) | H18A—C182—H18B | 109.5 |
| C142—C141—C41 | 122.5 (2) | C181—C182—H18C | 109.5 |
| C146—C141—C41 | 119.7 (2) | H18A—C182—H18C | 109.5 |
| C141—C142—C143 | 121.1 (2) | H18B—C182—H18C | 109.5 |
| C141—C142—H142 | 119.4 | C181—C183—H18D | 109.5 |
| C143—C142—H142 | 119.4 | C181—C183—H18E | 109.5 |
| C144—C143—C142 | 120.1 (3) | H18D—C183—H18E | 109.5 |
| C144—C143—H143 | 120.0 | C181—C183—H18F | 109.5 |
| C142—C143—H143 | 120.0 | H18D—C183—H18F | 109.5 |
| C145—C144—C143 | 119.7 (3) | H18E—C183—H18F | 109.5 |
| C145—C144—H144 | 120.2 | C181—C184—H18G | 109.5 |
| C143—C144—H144 | 120.2 | C181—C184—H18H | 109.5 |
| C144—C145—C146 | 120.3 (3) | H18G—C184—H18H | 109.5 |
| C144—C145—H145 | 119.9 | C181—C184—H18I | 109.5 |
| C146—C145—H145 | 119.9 | H18G—C184—H18I | 109.5 |
| C145—C146—C141 | 121.0 (3) | H18H—C184—H18I | 109.5 |
| C145—C146—H146 | 119.5 | C26—C21—C22 | 119.2 (2) |
| C141—C146—H146 | 119.5 | C26—C21—C27 | 120.1 (2) |
| C152—C151—C156 | 118.5 (2) | C22—C21—C27 | 120.7 (2) |
| C152—C151—C41 | 120.3 (2) | C23—C22—C21 | 119.6 (2) |
| C156—C151—C41 | 121.2 (2) | C23—C22—H22 | 120.2 |
| C153—C152—C151 | 121.4 (3) | C21—C22—H22 | 120.2 |
| C153—C152—H152 | 119.3 | C24—C23—C22 | 122.4 (2) |
| C151—C152—H152 | 119.3 | C24—C23—N23 | 118.5 (2) |
| C154—C153—C152 | 119.7 (3) | C22—C23—N23 | 119.2 (2) |
| C154—C153—H153 | 120.2 | C25—C24—C23 | 117.0 (2) |
| C152—C153—H153 | 120.2 | C25—C24—H24 | 121.5 |
| C153—C154—C155 | 120.0 (3) | C23—C24—H24 | 121.5 |
| C153—C154—H154 | 120.0 | C24—C25—C26 | 122.6 (2) |
| C155—C154—H154 | 120.0 | C24—C25—N25 | 117.6 (2) |
| C156—C155—C154 | 120.4 (3) | C26—C25—N25 | 119.8 (2) |
| C156—C155—H155 | 119.8 | C21—C26—C25 | 119.3 (2) |
| C154—C155—H155 | 119.8 | C21—C26—H26 | 120.3 |
| C155—C156—C151 | 120.1 (3) | C25—C26—H26 | 120.3 |
| C155—C156—H156 | 120.0 | O21—C27—O22 | 127.0 (2) |
| C151—C156—H156 | 120.0 | O21—C27—C21 | 118.1 (2) |
| N1—C11—C12 | 114.0 (2) | O22—C27—C21 | 115.0 (2) |
| N1—C11—H11A | 108.8 | O24—N23—O23 | 124.5 (3) |
| C12—C11—H11A | 108.8 | O24—N23—C23 | 117.9 (3) |
| N1—C11—H11B | 108.8 | O23—N23—C23 | 117.7 (2) |
| C12—C11—H11B | 108.8 | O26—N25—O25 | 124.2 (3) |
| H11A—C11—H11B | 107.7 | O26—N25—C25 | 117.6 (3) |
| C13—C12—C11 | 111.0 (3) | O25—N25—C25 | 118.1 (3) |
| C13—C12—H12A | 104.2 | ||
| C11—N1—C2—C3 | −179.77 (19) | O14—C14—C161—C166 | 10.3 (6) |
| C6—N1—C2—C3 | −56.5 (2) | C13—C14—C161—C166 | −170.9 (5) |
| N1—C2—C3—C4 | 57.2 (2) | O14—C14—C161—C162 | −162.5 (4) |
| C2—C3—C4—O4 | 61.4 (2) | C13—C14—C161—C162 | 16.4 (5) |
| C2—C3—C4—C5 | −55.9 (2) | C166—C161—C162—C163 | 5.6 (7) |
| O4—C4—C5—C6 | −66.3 (2) | C14—C161—C162—C163 | 178.6 (4) |
| C3—C4—C5—C6 | 55.8 (3) | C161—C162—C163—C164 | 0.8 (7) |
| C2—N1—C6—C5 | 57.0 (2) | C162—C163—C164—C165 | −6.2 (8) |
| C11—N1—C6—C5 | −178.8 (2) | C162—C163—C164—C181 | −179.9 (4) |
| C4—C5—C6—N1 | −57.7 (3) | C163—C164—C165—C166 | 5.7 (11) |
| C5—C4—O4—C41 | −179.83 (19) | C181—C164—C165—C166 | 179.0 (6) |
| C3—C4—O4—C41 | 61.1 (3) | C162—C161—C166—C165 | −6.3 (10) |
| C4—O4—C41—C151 | 53.7 (3) | C14—C161—C166—C165 | −179.2 (6) |
| C4—O4—C41—C141 | 177.88 (18) | C164—C165—C166—C161 | 0.5 (13) |
| O4—C41—C141—C142 | 4.6 (3) | C165—C164—C181—C183 | −24.9 (6) |
| C151—C41—C141—C142 | 128.7 (2) | C163—C164—C181—C183 | 148.2 (4) |
| O4—C41—C141—C146 | −176.5 (2) | C165—C164—C181—C182 | 96.3 (6) |
| C151—C41—C141—C146 | −52.4 (3) | C163—C164—C181—C182 | −90.6 (4) |
| C146—C141—C142—C143 | 1.4 (4) | C165—C164—C181—C184 | −144.6 (6) |
| C41—C141—C142—C143 | −179.7 (2) | C163—C164—C181—C184 | 28.4 (4) |
| C141—C142—C143—C144 | −0.2 (4) | C26—C21—C22—C23 | 1.4 (3) |
| C142—C143—C144—C145 | −1.2 (4) | C27—C21—C22—C23 | −176.3 (2) |
| C143—C144—C145—C146 | 1.4 (5) | C21—C22—C23—C24 | −0.9 (4) |
| C144—C145—C146—C141 | −0.1 (5) | C21—C22—C23—N23 | 178.1 (2) |
| C142—C141—C146—C145 | −1.3 (4) | C22—C23—C24—C25 | 0.0 (4) |
| C41—C141—C146—C145 | 179.8 (3) | N23—C23—C24—C25 | −179.0 (2) |
| O4—C41—C151—C152 | −129.7 (2) | C23—C24—C25—C26 | 0.4 (4) |
| C141—C41—C151—C152 | 109.3 (2) | C23—C24—C25—N25 | −179.6 (2) |
| O4—C41—C151—C156 | 50.5 (3) | C22—C21—C26—C25 | −1.0 (3) |
| C141—C41—C151—C156 | −70.5 (3) | C27—C21—C26—C25 | 176.7 (2) |
| C156—C151—C152—C153 | −0.6 (4) | C24—C25—C26—C21 | 0.1 (4) |
| C41—C151—C152—C153 | 179.6 (2) | N25—C25—C26—C21 | −180.0 (2) |
| C151—C152—C153—C154 | −0.2 (4) | C26—C21—C27—O21 | 1.3 (3) |
| C152—C153—C154—C155 | 0.6 (5) | C22—C21—C27—O21 | 179.0 (2) |
| C153—C154—C155—C156 | −0.3 (5) | C26—C21—C27—O22 | −177.2 (2) |
| C154—C155—C156—C151 | −0.5 (4) | C22—C21—C27—O22 | 0.5 (3) |
| C152—C151—C156—C155 | 0.9 (4) | C24—C23—N23—O24 | −4.2 (4) |
| C41—C151—C156—C155 | −179.3 (2) | C22—C23—N23—O24 | 176.8 (2) |
| C2—N1—C11—C12 | −72.4 (3) | C24—C23—N23—O23 | 174.9 (2) |
| C6—N1—C11—C12 | 164.5 (3) | C22—C23—N23—O23 | −4.1 (4) |
| N1—C11—C12—C13 | −122.9 (3) | C24—C25—N25—O26 | 168.7 (3) |
| C11—C12—C13—C14 | −179.2 (3) | C26—C25—N25—O26 | −11.3 (4) |
| C12—C13—C14—O14 | −1.7 (5) | C24—C25—N25—O25 | −9.9 (4) |
| C12—C13—C14—C161 | 179.5 (3) | C26—C25—N25—O25 | 170.2 (2) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1···O22 | 0.99 (3) | 1.66 (2) | 2.634 (3) | 167 (2) |
| C2—H2A···O25i | 0.97 | 2.50 | 3.444 (3) | 163 |
| C11—H11A···O14ii | 0.97 | 2.49 | 3.358 (4) | 150 |
Symmetry codes: (i) x−1, −y+3/2, z+1/2; (ii) x−1, y, z.
References
- Boeyens, J. C. A. (1978). J. Cryst. Mol. Struct. 8, 317–320.
- Cheng, J., Zhou, Z. & Yang, G. (2005). Acta Cryst. E61, o2932–o2933.
- Ferguson, G., Glidewell, C., Gregson, R. M. & Meehan, P. R. (1998a). Acta Cryst. B54, 129–138.
- Ferguson, G., Glidewell, C., Gregson, R. M. & Meehan, P. R. (1998b). Acta Cryst. B54, 139–150.
- Gregson, R. M., Glidewell, C., Ferguson, G. & Lough, A. J. (2000). Acta Cryst. B56, 39–57. [DOI] [PubMed]
- Jasinski, J. P., Butcher, R. J., Hakim Al-Arique, Q. N. M., Yathirajan, H. S. & Narayana, B. (2009). Acta Cryst. E65, o1738–o1739. [DOI] [PMC free article] [PubMed]
- Krause, L., Herbst-Irmer, R., Sheldrick, G. M. & Stalke, D. (2015). J. Appl. Cryst. 48, 3–10. [DOI] [PMC free article] [PubMed]
- Rigaku (2011). CrystalClear. Rigaku Corporation, Tokyo, Japan.
- Shaibah, M. A. E., Yathirajan, H. S., Kumar, S. M., Byrappa, K. & Glidewell, C. (2017). E73, 1488–1493. [DOI] [PMC free article] [PubMed]
- Sharma, R., Prasher, D. & Tiwari, R. K. (2015). J. Appl. Cryst. 48, 1299–1301.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2015). Acta Cryst. C71, 3–8.
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Van Cauwenberge, P., De Belder, T. & Sys, L. (2004). Expert Opin. Pharmacother. 5, 1807–1813. [DOI] [PubMed]
- Wiseman, L. R. & Faulds, D. (1996). Drugs, 51, 260–277. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S205698901701324X/su5391sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S205698901701324X/su5391Isup2.hkl
Supporting information file. DOI: 10.1107/S205698901701324X/su5391Isup3.cml
CCDC reference: 1574718
Additional supporting information: crystallographic information; 3D view; checkCIF report






