Fig. 2.
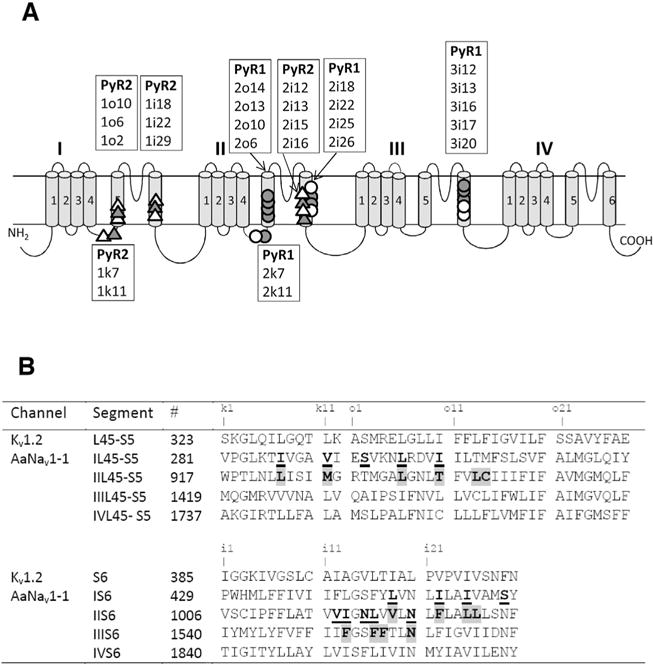
(A) Topology of sodium channel indicating residues within/around PyR1 and PyR2. The open circles and open triangles indicate, respectively, positions of mutations within/around PyR1 and PyR2, which affect action of pyrethroids. The filled circles and filled triangles indicate, respectively, positions of mutations within/around PyR1 and PyR2 that affect action of both pyrethroids and DDT, see (Du et al., 2015; Du et al., 2016) and references therein. Labels of respective positions are shown in boxes above transmembrane helices S5, S6 or below L45 linker-helices. To describe sequential positions of residues within the channel we use a residue-labeling scheme (Du et al., 2013; Zhorov and Tikhonov, 2004) where a label includes the domain number (1–4), segment type (k, the linker-helix L45; i, the inner helix S6; and o, the outer helix S5), and relative number of the residue in the segment. This provides the same labels to residues in the matching positions of the sequence alignment of sodium channels from different organisms and highlights symmetric location of residues in different channel domains. See Table 1 for more details. (B) The aligned sequences of Kv1.2 and AaNav1-1 channels. Residues predicted to contribute to PyR1 or control ligand access to PyR1 are highlighted. Residues predicted to contribute to PyR2 or control ligand access to PyR1 are underlined. Substitutions of these residues have been tested experimentally (Du et al., 2013, 2015, 2016; O’Reilly et al., 2006; Usherwood et al., 2007). Characters “k1”, “o1”, “i1”, etc. mark relative positions of residues, respectively, in the linker-helices L45, outer helices S5 and inner helices S6.
