Abstract
The genomic revolution promises to transform our approach to treating patients by individualizing treatments, reducing adverse events and decreasing healthcare costs. The early advances using this have been realized primarily by optimizing preventive and therapeutic approaches in cancer using human genome sequencing. The ability to characterize the microbiome, which includes all the microbes that reside within and upon us and all their genetic elements, using next-generation sequencing, allows us to now incorporate this important contributor to human disease in developing new preventive and therapeutic strategies. In this review we highlight the importance of the microbiome in all aspects of human disease including pathogenesis, phenotype, prognosis and response to treatment as well their role as diagnostic and therapeutic biomarkers. We provide a role for next-generation sequencing in both precise microbial identification for infectious diseases as well as characterization of microbial communities and their function. Taken together, the microbiome is emerging as an integral part of precision medicine approach as it not only contributes to inter-individual variability in all aspects of a disease but also represents a potentially modifiable factor that is amenable to targeting by therapeutics.
Introduction
The focus of biomedical research for the majority of its existence has been the ability to identify and target specific disease-associated pathways, leading to therapeutic strategies targeting a pathway. This approach remains mostly naïve to inter-individual variability in development of disease and response to therapy especially relevant in multi-factorial diseases. However, the genomic revolution has provided a window into individual-specific information and its impact on human physiology paving the way for personalized or precision medicine.1 Over the past decade, efforts in oncology have allowed human genomic screening to identify spectrum of germ-line encoded mutations allowing individual-specific application of preventive and therapeutic strategies. In addition to personalization of treatment based on genetic contribution to disease pathogenesis, precision medicine efforts have also allowed stratification of patients based on response to treatment and development of adverse events.
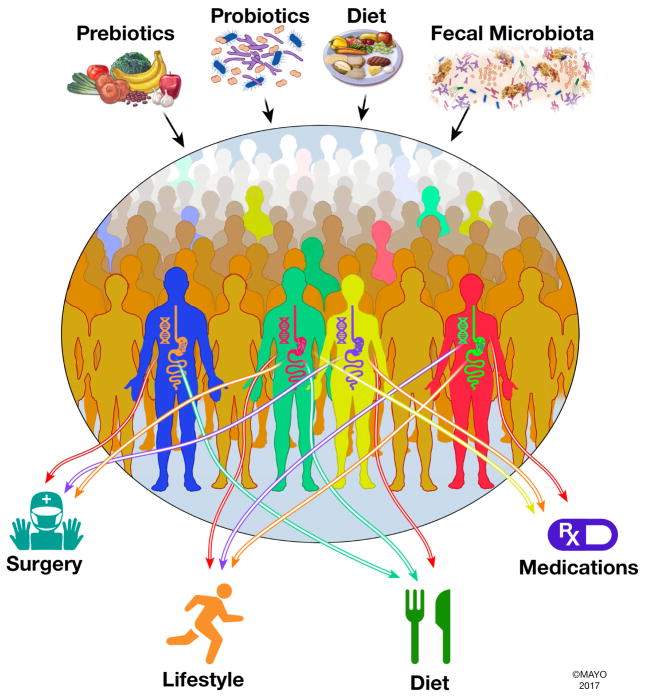
The advent of microbiome research has identified microbiome as a significant contributor to human health and in this review we highlight why microbiome is an integral component of the precision medicine initiative (Figure 1). Microbiome represents the complex collection of microorganisms both within and upon us, their genomes and collective functions.2 The field has benefited vastly from the genomic revolution allowing DNA-based identification of non-culturable bacteria inhabiting various body sites. Alteration in microbial communities (often referred to as dysbiosis) has been shown to be associated with diseases ranging from infectious (Clostridium difficile infection) to inflammatory (inflammatory bowel disease, rheumatoid arthritis) and metabolic (diabetes, obesity) diseases suggesting an important role for them in the pathogenesis of multi-factorial conditions.3 An important aspect about the microbiome is its resilience as well as its plasticity making it more mutable than human cells. While on first impression these appear opposing concepts, the resilience of the microbiome is evident in health, where in spite of temporary insults, (travel, diet, antibiotics etc.) the microbiome maintains a relatively stable steady state. On the other hand, it represents a malleable organ and can be modified by dietary and other directed therapies (Figure 1). Further the inter-individual variability in composition and metabolic capacity of the microbiome play an important role in interactions with the environment resulting in development of disease, as well as response to treatment and development of adverse events. The microbiome has been shown to be determined in part by the host genome, but this contribution seems small when compared with the vast environmental microbiome modulation. Hence, the important role of microbiome in human health, the inter-individual variability and contribution to host function in health, and its plasticity making it a targetable factor, all point towards the importance of incorporating the microbiome in precision medicine (Figure 1).
Figure 1.
Gut microbiome as a determinant of human health and response to therapeutic intervention. Gut microbiome plays an important role in an individual’s response to interventions raging from dietary and lifestyle changes to medications and surgical interventions; hence in addition to host genetics it is important to consider the role of gut microbiome when selecting appropriate therapy. Gut microbiome unlike host genes represents a modifiable factor that can be targeted by probiotics, prebiotics, diet as well as community replacement approaches such as fecal microbiota transplantation.
The current methods employ a spectrum of strategies to characterize the microbiome. The simplest being the marker gene using variable regions within the highly conserved 16S rRNA gene. This approach, while valuable in assessing alterations in microbial community structure, fails to provide resolution at species or strain level and does not provide sufficient functional insight into the community. Complimentary approaches including metagenomics (study of all genomes in an ecosystem), metatranscriptomics (characterization of gene expression from all microbes in an ecosystem), metabolomics (characterization of all small molecule metabolites in an ecosystem) and metaproteomics (characterization of all proteins in an ecosystem), provide greater insight into functional potential, as well as the expression of microbiome-derived bioactive molecules necessary to understand the therapeutic implications of the microbiome. While microbiome represents an attractive target for development of personalized treatment approaches, standardization of methods to develop reliable and reproducible microbiome based diagnostic and therapeutic strategies remains a challenge. The strong effort from the scientific community, as well as collaboration with rapidly emerging biotech companies, provide an optimistic outlook for developing microbiome-dependent and microbiome targeted diagnostics and therapeutics.
Sequencing Revolution Allows Development of Precise Microbial Diagnostics
Bacterial Typing
Awareness of the role of the microbiome in health has both benefited from, and been spurred by sequencing technology. Once considered milestone achievements requiring the resources of dedicated genomics centers, the sequencing of a complete bacterial genome can now be performed on a lab bench for about a hundred dollars per sample. Rapidly declining costs and continuing development of software and algorithms for assembling genomes, either from existing reference databases or de novo, promise to fundamentally alter the clinical paradigm by improving our ability to track, understand, and identify disease-causing agents.4
Here we will describe some of the applications of bacterial genome sequencing, and attempt to summarize some of the many efforts going on worldwide to bring genomic data to various problems from bacterial typing5,6 to anti-terrorism.7 While these might seem like very disparate use cases, what unites them is the data contained within the genome; which contains mutations that reflect evolutionary relationships, 8 and genes that underlie important phenotypes such as antibiotic resistance.9, 10, 11
Infectious disease tracking involves the ability to detect and trace outbreaks of disease. This assists hospitals in preventing the spread of nosocomial infections, food distributors in tracing back contaminated food sources, and governments in protecting people from biological agents. The most common of these uses is in the hospital where an indication of nosocomial disease spread can be used to improve the practice of medicine. However, these efforts have largely relied on event count and statistics—i.e., they are more reactive than proactive.
The most prominent bacterial typing technique is pulse-field gel electrophoresis (PFGE),12 which relies on restriction enzymes and gels in order to obtain a rough distribution of genome fragment sizes, and in essence provides very little detailed information and must generally be used with care and attention to detail.13 Great impact and effort has been made through the standardization of PFGE techniques to enhance the comparability of results between different gels run at different labs.14,15,16 However, PFGE retains its difficulties in detecting infectious outbreaks across multiple centers.17
Where PFGE falls short on finer resolution and reproducibility, genome sequencing excels. Genomic data provides a base-by-base genomic ‘fingerprint’ that enhances the resolution with which monitoring becomes possible. The fact that this may one day enable us to identify potential outbreaks sooner and act to prevent them before they become larger has prompted numerous studies on the efficacy of different comparison methodologies.18,19,20 These methods have been tested across various species and range from SNP-based tests to use of whole genome comparisons.21,22 This has also given rise to a large number of publically-available phylogenetic reconstruction algorithms that analyze genomic data for mutational signatures of relatedness in order to track relationships between different pathogens.23,24,25 These algorithms use the evolutionary principle of descent with modification to assess which strains descended from a very recent common ancestor.
In addition, sequencing provides a great deal of information about the characteristics of an infection. One can query for antibiotic resistance genes, identifying susceptibilities in antibiotic resistant pathogens. This can be done using PCR amplicon sequencing26,27 or whole-genome sequencing.9, 10 While PCR-based approaches currently have an advantage in turnaround time and cost, whole-genome approaches provide more information and a common platform for evaluating multiple species. Both methods have the potential to directly assess antibiotic resistance without culture, a feature that becomes especially important in the case of slow-growing bacteria, such as tuberculosis, where culture-based tests can take weeks to complete.
The utility of sequencing bacteria goes beyond pathogen or pathogen-complex evaluation. They can be used to directly assess more complex specimens revealing microbial ecosystems with multiple species present and represent a potential tool for diagnosis infections of unknown origin.28,29,30 Such broad searches require even greater bioinformatics and database support. This need has spurred a rapid growth in the number of publically available resources for identifying potential infectious agents from complex microbiome data31,32
Microbiome Sequencing
The revolutionary change in our ability to understand the role of the microbiome came with the advent of next- generation sequencing has allowed in depth characterization of gut microbiota using multi-omics approaches without the need to culture individual microbes, which in some instances can be quite challenging. The most popular method to characterizing microbial communities is using the marker gene approach using 16S rRNA gene which is highly conserved among bacteria with little evidence of horizontal gene transfer. However this approach lacks species and strain level resolution which often requires metagenomic sequencing and de novo assembly of genomes, providing better compositional as well as functional resolution of the microbiome.2 Metatranscriptomics compliments metagenomics by allowing identification of microbial genes that are expressed under different conditions. Metabolomics and Metaproteomics help identify metabolites and proteins resulting from microbe-host co-metabolism which can serve as reliable biomarkers given that they represent end products of metabolic interactions among the microbe and host. The combination of multi-omic technologies increases confidence in identified diagnostic and therapeutic biomarkers as well as provides testable hypotheses. In order to test emergent hypotheses generated as a result of these technologies and delineate mechanisms by which microbes influence the host, germ free animal models provide a highly controllable experimental system with reduced complexity to study interactions between the host and its resident microbiota.
Microbiome as a Tool for Precision Diagnosis and Personalized Treatment Strategies
There is an emerging role of gut microbiome as a biomarker for disease phenotype, prognosis and response to treatment; in addition, to the well described associations of alterations in microbial community structure in different disease states. Inflammatory bowel disease is one of the best studied conditions associated with dysbiosis, with the microbiome serving as an important marker of disease phenotype and response to treatment. Inflammatory bowel disease (IBD) is heterogeneous with three major subtypes: ulcerative colitis (UC), Crohns disease (CD), and indeterminate colitis, which not only differ in their presentation and location but also have different therapeutic strategies making it important to obtain a precise diagnosis. The microbial populations are quite distinct even within CD with a decrease in Faecalibacterium prauznitzii, and increase in Escherichia coli as well as antibodies against E.coli outer membrane protein C seen in ileal CD compared to colonic CD,33, 34 as well as extra-intestinal manifestations such as peripheral spondyloarthritis.35 Gut microbiome signatures have also been associated with surgical outcomes in CD with an increase in F. prausnitzii in the ileal mucosa associated with decreased disease recurrence at 6 months. Inspite of several studies highlighting changes in the microbiome in IBD, there is lack of agreement among studies, making it imperative to have large cohorts from different geographic locations to overcome the effect of disease subtype, antibiotic use, diet and other factors affecting the gut microbiome. This was highlighted in a study of treatment naïve Crohn’s disease patients where a large patient cohort was needed to identify discriminatory taxa.36 The study further demonstrated the need to study mucosa associated bacteria which may be more relevant in inflammatory diseases like IBD. In addition to IBD, microbiome signatures have been described in several other gastrointestinal diseases. Fusobacterium nucleatum has been implicated in colorectal cancer through its FadA adhesion serving as both a diagnostic and therapeutic marker.37 C. difficile infection has been associated with decreased microbial diversity and a decrease in secondary bile acid production.38 In addition, recently two studies have identified microbiome signatures that allow prediction of disease outcome allowing therapeutic stratification.39, 40 An expansion of Proteobacteria in the setting of dysbiotic microbiota was described in patients with celiac disease with gastrointestinal symptoms compared to those with extra-intestinal manifestations of celiac disease.41 In addition to diseases within gastrointestinal tract, it is interesting that several studies have described gut microbiome signatures in systemic disorders such as rheumatoid arthritis. An expansion of Prevotella copri has been described in new onset rheumatoid arthritis (RA).42 Another recent study identified enrichment of Collinsella, Eggerthella and Faecalibacterium in patients with RA and a strong association of Collinsella with high levels of alpha-aminoadipic acid, and asparagine as well as production of the alpha-aminoadipic cytokine IL-17A, and experimental arthritis.43 These few examples are just a window to accumulating experimental evidence for the role of microbiome in human disease and the future of microbiome based diagnostic and therapeutic biomarkers. While these studies are helpful in identifying biomarkers, much work still needs to be done in validating these signatures in large multicenter cohorts as well identifying potential causative role using a combination of in vitro and in vivo models.
Microbiome as a Determinant of Human Therapeutics
The ecology of a microbial population, as in any ecosystem, involves a lot of cross talk between different species. Microbial survival and growth is governed strongly by their chemical environment, and unsurprisingly, they have evolved gene cassettes for chemical warfare.44,45 Indeed, the discovery of antibiotics first occurred in microbial culture as a unique characteristic of colonies46 and since then broader surveys of the soil microbiota have revealed an even greater array of antibiotic compounds.47,48 Recently, this has been extended to the human microbiome as well across multiple sites along the human body,49 which means the source of compounds we need to harness control over our microbiome might already be within us.
In addition to antibiotics and signaling agents, the discovery of the so-called bacterial immune system, i.e., the CRISPR-Cas system, allows bacteria to resist and exclude bacteriophages from the population by targeting specific sequences for cleavage.50 While providing adaptive immunity to viruses, the industrial uses of this biological system have been widely recognized leading to the implementation of synthetic CRISPR-Cas systems51 that have led to the implementation of species specific antimicrobial agents,52 that may be able to preserve the bulk of the microbiome while still making key alterations.
In addition to being a source of therapeutics with implication for human disease, the microbiome serves as both as a modulator of traditional therapies as well as target for therapies. The inter-individual variability in response to therapy and development of adverse events has been attributed to individual specific disease phenotype and host genetics, but gut microbiota is often overlooked. However gut microbiota plays an important role in drug transformation affecting their efficacy. Acetaminophen, a commonly used analgesic may compete with bacteria generated p-cresol for O-sulfonation resulting in acetaminophen glucuronidation, which can explain in part inter-individual variability in analgesic response53 as well as differences in adverse events due to accumulation of it toxic metabolite NAPQI(N-acetyl-p-benzoquinone imine). Microbiome markers of drug efficacy ranging from chemotherapeutic agents to statins have been widely described. Bifidobacterium has been found to augment tumor control in mouse models of melanoma treated with anti-PD L1 (Programmed Death-Ligand 1).54 Similarly in humans, Bacteroides have been suggested to be responsible for antitumor effects of CTLA-4 blockade, commonly used for cancer immunotherapy.55 Irinotecan (CPT-11), a chemotherapeutic agent commonly used for colorectal cancer can undergo beta-glucuronidation by gut bacteria resulting in an active metabolite that causes severe diarrhea.56 It is important to note that host genetic variation also plays an important role in shaping the gut microbiome. 57 Bacteria-derived coprostanol levels have been associated with clinical response to statins, which are commonly used as LDL (low density lipoprotein) cholesterol lowering agents. Digoxin, a cardiac glycoside with narrow therapeutic window can be inactivated by an Eggerthella lenta in the gut. Finally, a recent study highlights the role of gut microbiota in mediating the antidiabetic effects of metformin.58 These examples clearly highlight the importance of considering the gut microbiota when determining drug responses akin to pharmacogenomics (Figure 1). The combination of the two approaches will allow us to impart more precise and effective therapeutics while decreasing overall adverse events.
Targeting the Microbiome to Improve Health
In addition to serving as diagnostic and therapeutic biomarkers and modulating therapeutic responses to drugs, the appealing aspect of the microbiome is its plasticity and the ability to modify components of the microbiome. The traditional approach to targeting microbial populations has been with the use of antibiotics, which are both essential and effective for treating systemic infections typically resulting from pathogen invasion. However, the unintended off target effects on microbial community structure as well as side effects in humans makes it less appealing as precise therapies to target the microbiome.59,60,61 There is a continued role for developing pathogen targeted antibiotics by identifying specific targets which narrow the spectrum of the antibiotic. A novel approach includes mining the microbiota for therapeutic targets by identifying specific functions that impact the host allowing us to modify microbial community functionality without harming the community itself.62 An example is the role tri-methyl amine oxidase (TMAO) in atherosclerosis and the inhibition of bacterial TMA lyases by 3, 3-dimethyl-1-butanol (DMB) decreasing bacterial TMA production in a high choline diet-fed murine model.63 Even with precise targeting of a single pathway, there were still alterations in the microbiome by DMB, highlighting the complexity of microbial interactions within these ecosystems. There are several other approaches to targeting the microbiome including the use of pro-, pre-biotics as well as dietary interventions. The early probiotics (live microorganisms that when given in sufficient amounts confer a health benefit on the host) were dominated by members of the genus Lactobacillus and Bifidobacteria, but lacked precision in terms of targeting a biological function. A recent systematic review of medium to high quality randomized controlled trials using probiotics found that there was no significant effect on gut microbiota compared to placebo. The clinical efficacy of currently available probiotics is difficult to assess given the small sample sizes limiting the power, heterogeneity in strains of bacteria used, end points, duration of treatment and molecular methods of studying the gut microbiota, recording of baseline measurements such as diet and often a lack of good preclinical mechanistic data.64 However recent work highlights the promise of next-generation probiotics which will be developed using targeted approaches to alter microbial metabolism in a diseases specific manner. A precision approach by utilizing Clostridium scindens to augment resistance to C. difficile infection by targeting secondary bile acid pathway38 is one such example. Similarly a multicomponent probiotic was shown to modulate the gut microbiome with resultant suppression of hepatocellular carcinoma in a mouse model.65 The advent of genetic engineering and synthetic biology approaches also hold promise for development of precision probiotics.66 An example is the engineering of a common gut commensal to secrete the molecular signal cholera autoinducer (CAI-1), inhibiting V. cholerae virulence in a mouse model.67 Furthermore, tunable expression tools in robust colonizers of the human gut provides us the ability further calibrate delivery of bioactive compounds by these precision probiotics.68 Prebiotic (ingredients that are selectively fermented by gut microbes and confer a health benefit) approaches aim to modulate the microbial community in a way that is beneficial to human health. While the early prebiotics have focused on promoting the growth of a single or group of beneficial bacteria, they fail to account for the downstream effects on other microbial members. Similar to probiotics, the prebiotics which are mainly comprised of fermentable oligosaccharides such as inulin and fructooligosaccharides have focused on increasing growth of potentially beneficial bacteria such as Bifidobacteria. The lack of ecosystem approach is reflected in the modest clinical efficacy of available prebiotics. The development of next generation prebiotics will require careful modeling of the metabolic interactions among the members of the ecosystem to better understand the overall effects on the community and host physiology. Fecal microbiota transplant (FMT) which entails transfer of healthy gut microbiota from a donor either orally via capsules or endoscopically has been highly successful as an ecosystem approach in treating recurrent C. difficile infection.69 A similar approach with FMT has been tested in multiple diseases associated with the microbiome but has failed to show clinical efficacy. However the use of FMT in disease like IBD has provided insight into donor specificity70 in terms of response, suggesting a role for individualizing FMT approaches for multifactorial diseases like IBD, in contrast to the approach in C. difficile infection.
Finally, diet has major implications for the microbiome as it is the primary nutrient source for microbes. Dietary manipulations fall with three distinct approaches. The use of microbiome markers in optimizing dietary interventions, modulating the diet based on the microbiome and using diet to alter the microbiome. Dietary interventions limiting fermentable oligo-, di- mono- saccharides and polyols (FODMAP) have shown to be beneficial in ameliorating symptoms in patients with irritable bowel syndrome (IBS).71 However, long term use of such an intervention can decrease microbial short chain fatty acid production which in turn may have negative implications for human health. A recent study identified microbiome markers which predict a positive response to FODMAP72 with the potential to allow optimization of therapy and minimizing undesirable adverse effects in individuals less likely to respond. An important aspect of the gut microbiome is its role in determining host responses to dietary components given that the microbiome plays an important role in metabolism of dietary nutrients. Zeevi et al. found large interpersonal differences in post-prandial glycemic responses to dietary components in an elegant study of 800 subjects. The prediction engine used to make the predictions incorporated multiple host and microbial parameters, and they found incorporation of microbiome-derived features significantly improved the accuracy of prediction of glycemic responses.73 In a follow up study the authors found significant inter-personal variability in the glycemic response to different bread types, and the glycemic response to different types of bread could be predicted solely on microbiome data prior to the intervention.74 These studies highlight the ability to personalize nutritional intervention to improve host physiology based on an individual’s microbiome. It is important to note that both short term and long term dietary patterns have a significant impact on shaping the microbiome. A diet high in protein and fat in the long term has been associated with enrichment of Bacteroides while a carbohydrate rich diet has been associated with Prevotella.75 Sonnenburg et al. demonstrated that a western diet low in microbiota accessible carbohydrates leads to decreased diversity in the microbiota of humanized mice, which are largely reversible within a single generation but over several generations this leads to a progressive loss of diversity which cannot be recovered by diet alone and needs replacement of the microbiota.76 This has significant implications for populations consuming a western diet, which has been associated with decreased diversity and an increase in autoimmune disease. The study suggests that even long term dietary effects may be reversible within a certain timeframe. Interestingly short term dietary effects on the microbiome seem to be easily reversible even when using extreme dietary interventions.77 Moreover, short term dietary interventions have also shown to have beneficial effects on the host and gut microbiome. In the study by Zeevi et al. mentioned previously, introduction of meals associated with low post-prandial glucose response led to an increase in bacteria thought to be protective against type 2 diabetes mellitus such as Roseburia inulinivorans, Eubacterium eligens and Bacteroides vulgatus.73 Similarly, a 3 day dietary intervention with barley-based bread was associated with higher Prevotella/Bacteroides ratio and improved glucose metabolism.78 It is interesting to note that changes in gut microbiota to a similar dietary intervention can vary based on an individual’s microbiome.79 Taken together it is apparent the while relationship of diet and gut microbiome is complex, it is highly relevant in determining host responses to diet as well as predicting changes in the microbiome in response to diet.
Conclusion
In this review we highlight the importance of incorporating microbiome as a component of personalized or precision medicine to improve diagnosis, reduce disease risk and optimize early detection and treatment. Microbial fingerprints could serve as precise, non-invasive, accessible and economic tools that could be used for personalized disease diagnosis including phenotypes, severity and prognosis. The role of microbiome in the metabolism of many chemical compounds makes it a key player in determining drug availability, efficacy, and toxicity making in indispensable for developing personalized drug therapies. Finally, the ability to manipulate the microbiome makes it appealing in developing personalized treatment approaches by using precision microbiome targeting approaches. The use of approaches targeting specific microbial pathways tailored to an individual’s microbiota may enable the development of treatment for multi-factorial disorders such as inflammatory bowel disease, obesity and diabetes mellitus. The development of precision probiotics using genetic engineering approaches, next generation prebiotics resulting from a better understanding of metabolic interactions among members of the microbial ecosystem, and personalized dietary therapies tailored to an individual’s microbiota will form the new frontier in the field of personalized medicine.
Overall, the outlook is very optimistic, but there are also substantial challenges in the field. In order to implement microbiome-based diagnostics and therapeutics, we need to develop uniform collection, sequencing, and analysis standards that would enhance reproducibility of results across centers and reduce biases in their interpretation. The majority of current studies are based on disease association but we need to better define the mechanisms by which microbiota influence aspects of human disease in order to develop more reliable biomarkers. Further we are only starting to appreciate the contribution of other microorganisms such as fungi, bacteriophages and parasites as well as the inter-kingdom signaling among the microorganisms and the host. As we unravel aspects of these complex interactions, we will begin to develop more robust strategies to address the impact of microbiome on the host.
The plasticity of microbiome, while being advantageous in terms of making it amenable to intervention, also poses a challenge in terms of stability of changes. This was highlighted above, wherein dietary interventions can be developed based on an individual’s microbiome; however, it has the potential to change the microbiome itself. Hence, a systems approach to better understanding the diet-microbiome interaction will allow identification of dependencies between dietary compounds and bacterial taxa and prediction of trends in their variation resulting from dietary intervention.
These challenges apart, the incorporation of microbiome based diagnostics and therapeutics with other components of personalized medicine such as pharmacogenomics and epigenomics will be an integral part of the new era in patient care. This integration will further enhance our ability to find the right treatment for the right patient while, at the same time, reducing adverse events and health care cost.
Acknowledgments
Funding
This work was supported in part by NIH R01 DK114007 and R03 DK DK111850 (PCK)
We would like to thank Lyndsay Busby for her administrative assistance.
Abbreviations
- CD
Crohns Disease
- FMT
Fecal microbiota transplant
- IBD
Inflammatory bowel disease
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jameson JL, Longo DL. Precision medicine--personalized, problematic, and promising. N Engl J Med. 2015;372(23):2229–2234. doi: 10.1056/NEJMsb1503104. [DOI] [PubMed] [Google Scholar]
- 2.Marchesi JR, Ravel J. The vocabulary of microbiome research: a proposal. Microbiome. 2015;3:31. doi: 10.1186/s40168-015-0094-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marchesi JR, Adams DH, Fava F, et al. The gut microbiota and host health: a new clinical frontier. Gut. 2016;65(2):330–339. doi: 10.1136/gutjnl-2015-309990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Didelot X, Bowden R, Wilson DJ, Peto TE, Crook DW. Transforming clinical microbiology with bacterial genome sequencing. Nat Rev Genet. 2012;13(9):601–612. doi: 10.1038/nrg3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koser CU, Ellington MJ, Cartwright EJP, et al. Routine Use of Microbial Whole Genome Sequencing in Diagnostic and Public Health Microbiology. Plos Pathogens. 2012;8(8) doi: 10.1371/journal.ppat.1002824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Joensen KG, Scheutz F, Lund O, et al. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J Clin Microbiol. 2014;52(5):1501–1510. doi: 10.1128/JCM.03617-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keim P, Van Ert MN, Pearson T, Vogler AJ, Huynh LY, Wagner DM. Anthrax molecular epidemiology and forensics: using the appropriate marker for different evolutionary scales. Infect Genet Evol. 2004;4(3):205–213. doi: 10.1016/j.meegid.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 8.Harris SR, Feil EJ, Holden MT, et al. Evolution of MRSA during hospital transmission and intercontinental spread. Science. 2010;327(5964):469–474. doi: 10.1126/science.1182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leopold SR, Goering RV, Witten A, Harmsen D, Mellmann A. Bacterial whole-genome sequencing revisited: portable, scalable, and standardized analysis for typing and detection of virulence and antibiotic resistance genes. J Clin Microbiol. 2014;52(7):2365–2370. doi: 10.1128/JCM.00262-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Veenemans J, Overdevest IT, Snelders E, et al. Next-generation sequencing for typing and detection of resistance genes: performance of a new commercial method during an outbreak of extended-spectrum-beta-lactamase-producing Escherichia coli. J Clin Microbiol. 2014;52(7):2454–2460. doi: 10.1128/JCM.00313-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Machado MP, Ribeiro-Goncalves B, Silva M, Ramirez M, Carrico JA. Epidemiological Surveillance and Typing Methods to Track Antibiotic Resistant Strains Using High Throughput Sequencing. Methods Mol Biol. 2017;1520:331–356. doi: 10.1007/978-1-4939-6634-9_20. [DOI] [PubMed] [Google Scholar]
- 12.Schwartz DC, Cantor CR. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell. 1984;37(1):67–75. doi: 10.1016/0092-8674(84)90301-5. [DOI] [PubMed] [Google Scholar]
- 13.Foxman B, Zhang L, Koopman JS, Manning SD, Marrs CF. Choosing an appropriate bacterial typing technique for epidemiologic studies. Epidemiol Perspect Innov. 2005;2:10. doi: 10.1186/1742-5573-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Swaminathan B, Barrett TJ, Hunter SB, Tauxe RV Force CDCPT. PulseNet: the molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg Infect Dis. 2001;7(3):382–389. doi: 10.3201/eid0703.010303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kam KM, Luey CK, Parsons MB, et al. Evaluation and validation of a PulseNet standardized pulsed-field gel electrophoresis protocol for subtyping Vibrio parahaemolyticus: an international multicenter collaborative study. J Clin Microbiol. 2008;46(8):2766–2773. doi: 10.1128/JCM.00424-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scharff RL, Besser J, Sharp DJ, Jones TF, Peter GS, Hedberg CW. An Economic Evaluation of PulseNet: A Network for Foodborne Disease Surveillance. Am J Prev Med. 2016;50(5 Suppl 1):S66–73. doi: 10.1016/j.amepre.2015.09.018. [DOI] [PubMed] [Google Scholar]
- 17.Price JR, Didelot X, Crook DW, Llewelyn MJ, Paul J. Whole genome sequencing in the prevention and control of Staphylococcus aureus infection. J Hosp Infect. 2013;83(1):14–21. doi: 10.1016/j.jhin.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Viau RA, Kiedrowski LM, Kreiswirth BN, et al. A Comparison of Molecular Typing Methods Applied to Enterobacter cloacae complex: hsp60 Sequencing, Rep- PCR, and MLST. Pathog Immun. 2017;2(1):23–33. doi: 10.20411/pai.v2i1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salipante SJ, SenGupta DJ, Cummings LA, Land TA, Hoogestraat DR, Cookson BT. Application of whole-genome sequencing for bacterial strain typing in molecular epidemiology. J Clin Microbiol. 2015;53(4):1072–1079. doi: 10.1128/JCM.03385-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jolley KA, Maiden MC. Automated extraction of typing information for bacterial pathogens from whole genome sequence data: Neisseria meningitidis as an exemplar. Euro Surveill. 2013;18(4):20379. doi: 10.2807/ese.18.04.20379-en. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coll F, McNerney R, Guerra-Assuncao JA, et al. A robust SNP barcode for typing Mycobacterium tuberculosis complex strains. Nat Commun. 2014;5:4812. doi: 10.1038/ncomms5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cunningham SA, Chia N, Jeraldo PR, et al. Comparison of Whole-Genome Sequencing Methods for Analysis of Three Methicillin-Resistant Staphylococcus aureus Outbreaks. J Clin Microbiol. 2017;55(6):1946–1953. doi: 10.1128/JCM.00029-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol. 2004;186(5):1518–1530. doi: 10.1128/JB.186.5.1518-1530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vaz C, Francisco AP, Silva M, et al. TypOn: the microbial typing ontology. J Biomed Semantics. 2014;5(1):43. doi: 10.1186/2041-1480-5-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nascimento M, Sousa A, Ramirez M, Francisco AP, Carrico JA, Vaz C. PHYLOViZ 2.0: providing scalable data integration and visualization for multiple phylogenetic inference methods. Bioinformatics. 2017;33(1):128–129. doi: 10.1093/bioinformatics/btw582. [DOI] [PubMed] [Google Scholar]
- 26.Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38(3):1008–1015. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ng LK, Martin I, Alfa M, Mulvey M. Multiplex PCR for the detection of tetracycline resistant genes. Mol Cell Probes. 2001;15(4):209–215. doi: 10.1006/mcpr.2001.0363. [DOI] [PubMed] [Google Scholar]
- 28.Wilson MR, Naccache SN, Samayoa E, et al. Actionable diagnosis of neuroleptospirosis by next-generation sequencing. N Engl J Med. 2014;370(25):2408–2417. doi: 10.1056/NEJMoa1401268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scholz CF, Jensen A. Development of a Single Locus Sequence Typing (SLST) Scheme for Typing Bacterial Species Directly from Complex Communities. Methods Mol Biol. 2017;1535:97–107. doi: 10.1007/978-1-4939-6673-8_7. [DOI] [PubMed] [Google Scholar]
- 30.Sahl JW, Schupp JM, Rasko DA, Colman RE, Foster JT, Keim P. Phylogenetically typing bacterial strains from partial SNP genotypes observed from direct sequencing of clinical specimen metagenomic data. Genome Med. 2015;7(1):52. doi: 10.1186/s13073-015-0176-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wattam AR, Abraham D, Dalay O, et al. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res. 2014;42(Database issue):D581–591. doi: 10.1093/nar/gkt1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zolfo M, Tett A, Jousson O, Donati C, Segata N. MetaMLST: multi-locus strain- level bacterial typing from metagenomic samples. Nucleic Acids Res. 2017;45(2):e7. doi: 10.1093/nar/gkw837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Willing B, Halfvarson J, Dicksved J, et al. Twin studies reveal specific imbalances in the mucosa-associated microbiota of patients with ileal Crohn’s disease. Inflamm Bowel Dis. 2009;15(5):653–660. doi: 10.1002/ibd.20783. [DOI] [PubMed] [Google Scholar]
- 34.Willing BP, Dicksved J, Halfvarson J, et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology. 2010;139(6):1844–1854. e1841. doi: 10.1053/j.gastro.2010.08.049. [DOI] [PubMed] [Google Scholar]
- 35.Viladomiu M, Kivolowitz C, Abdulhamid A, et al. IgA-coated E. coli enriched in Crohn’s disease spondyloarthritis promote TH17-dependent inflammation. Sci Transl Med. 2017;9(376) doi: 10.1126/scitranslmed.aaf9655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gevers D, Kugathasan S, Denson LA, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15(3):382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14(2):195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buffie CG, Bucci V, Stein RR, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2015;517(7533):205–208. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khanna S, Montassier E, Schmidt B, et al. Gut microbiome predictors of treatment response and recurrence in primary Clostridium difficile infection. Aliment Pharmacol Ther. 2016;44(7):715–727. doi: 10.1111/apt.13750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seekatz AM, Rao K, Santhosh K, Young VB. Dynamics of the fecal microbiome in patients with recurrent and nonrecurrent Clostridium difficile infection. Genome Med. 2016;8(1):47. doi: 10.1186/s13073-016-0298-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wacklin P, Kaukinen K, Tuovinen E, et al. The duodenal microbiota composition of adult celiac disease patients is associated with the clinical manifestation of the disease. Inflamm Bowel Dis. 2013;19(5):934–941. doi: 10.1097/MIB.0b013e31828029a9. [DOI] [PubMed] [Google Scholar]
- 42.Scher JU, Sczesnak A, Longman RS, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife. 2013;2:e01202. doi: 10.7554/eLife.01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen J, Wright K, Davis JM, et al. An expansion of rare lineage intestinal microbes characterizes rheumatoid arthritis. Genome Med. 2016;8(1):43. doi: 10.1186/s13073-016-0299-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shank EA, Kolter R. New developments in microbial interspecies signaling. Curr Opin Microbiol. 2009;12(2):205–214. doi: 10.1016/j.mib.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cornforth DM, Foster KR. Antibiotics and the art of bacterial war. Proc Natl Acad Sci U S A. 2015;112(35):10827–10828. doi: 10.1073/pnas.1513608112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jones D, Metzger HJ, Schatz A, Waksman SA. Control of Gram-Negative Bacteria in Experimental Animals by Streptomycin. Science. 1944;100(2588):103–105. doi: 10.1126/science.100.2588.103. [DOI] [PubMed] [Google Scholar]
- 47.Aigle B, Lautru S, Spiteller D, et al. Genome mining of Streptomyces ambofaciens. J Ind Microbiol Biotechnol. 2014;41(2):251–263. doi: 10.1007/s10295-013-1379-y. [DOI] [PubMed] [Google Scholar]
- 48.Bachmann BO, Van Lanen SG, Baltz RH. Microbial genome mining for accelerated natural products discovery: is a renaissance in the making? J Ind Microbiol Biotechnol. 2014;41(2):175–184. doi: 10.1007/s10295-013-1389-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Donia MS, Cimermancic P, Schulze CJ, et al. A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell. 2014;158(6):1402–1414. doi: 10.1016/j.cell.2014.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barrangou R, Fremaux C, Deveau H, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315(5819):1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 51.Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bikard D, Euler CW, Jiang W, et al. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat Biotechnol. 2014;32(11):1146–1150. doi: 10.1038/nbt.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Clayton TA, Baker D, Lindon JC, Everett JR, Nicholson JK. Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc Natl Acad Sci U S A. 2009;106(34):14728–14733. doi: 10.1073/pnas.0904489106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sivan A, Corrales L, Hubert N, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350(6264):1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vetizou M, Pitt JM, Daillere R, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350(6264):1079–1084. doi: 10.1126/science.aad1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wallace BD, Wang H, Lane KT, et al. Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science. 2010;330(6005):831–835. doi: 10.1126/science.1191175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blekhman R, Goodrich JK, Huang K, et al. Host genetic variation impacts microbiome composition across human body sites. Genome Biol. 2015;16:191. doi: 10.1186/s13059-015-0759-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu H, Esteve E, Tremaroli V, et al. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat Med. 2017 doi: 10.1038/nm.4345. [DOI] [PubMed] [Google Scholar]
- 59.Rubinstein E, Camm J. Cardiotoxicity of fluoroquinolones. J Antimicrob Chemother. 2002;49(4):593–596. doi: 10.1093/jac/49.4.593. [DOI] [PubMed] [Google Scholar]
- 60.Galatti L, Giustini SE, Sessa A, et al. Neuropsychiatric reactions to drugs: an analysis of spontaneous reports from general practitioners in Italy. Pharmacol Res. 2005;51(3):211–216. doi: 10.1016/j.phrs.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 61.Rubin BK, Tamaoki J. Antibiotics as anti-inflammatory and immunomodulatory agents. Basel; Boston: Birkhäuser; 2005. [Google Scholar]
- 62.Wallace BD, Redinbo MR. The human microbiome is a source of therapeutic drug targets. Curr Opin Chem Biol. 2013;17(3):379–384. doi: 10.1016/j.cbpa.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang Z, Roberts AB, Buffa JA, et al. Non-lethal Inhibition of Gut Microbial Trimethylamine Production for the Treatment of Atherosclerosis. Cell. 2015;163(7):1585–1595. doi: 10.1016/j.cell.2015.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kristensen NB, Bryrup T, Allin KH, Nielsen T, Hansen TH, Pedersen O. Alterations in fecal microbiota composition by probiotic supplementation in healthy adults: a systematic review of randomized controlled trials. Genome Med. 2016;8(1):52. doi: 10.1186/s13073-016-0300-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li J, Sung CY, Lee N, et al. Probiotics modulated gut microbiota suppresses hepatocellular carcinoma growth in mice. Proc Natl Acad Sci U S A. 2016;113(9):E1306–1315. doi: 10.1073/pnas.1518189113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Amalaradjou MA, Bhunia AK. Bioengineered probiotics, a strategic approach to control enteric infections. Bioengineered. 2013;4(6):379–387. doi: 10.4161/bioe.23574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ruder WC, Lu T, Collins JJ. Synthetic biology moving into the clinic. Science. 2011;333(6047):1248–1252. doi: 10.1126/science.1206843. [DOI] [PubMed] [Google Scholar]
- 68.Whitaker WR, Shepherd ES, Sonnenburg JL. Tunable Expression Tools Enable Single-Cell Strain Distinction in the Gut Microbiome. Cell. 2017;169(3):538–546. e512. doi: 10.1016/j.cell.2017.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kelly CR, Kahn S, Kashyap P, et al. Update on Fecal Microbiota Transplantation 2015: Indications, Methodologies, Mechanisms, and Outlook. Gastroenterology. 2015;149(1):223–237. doi: 10.1053/j.gastro.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Moayyedi P, Surette MG, Kim PT, et al. Fecal Microbiota Transplantation Induces Remission in Patients With Active Ulcerative Colitis in a Randomized Controlled Trial. Gastroenterology. 2015;149(1):102–109. e106. doi: 10.1053/j.gastro.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 71.Halmos EP, Power VA, Shepherd SJ, Gibson PR, Muir JG. A diet low in FODMAPs reduces symptoms of irritable bowel syndrome. Gastroenterology. 2014;146(1):67–75. e65. doi: 10.1053/j.gastro.2013.09.046. [DOI] [PubMed] [Google Scholar]
- 72.Bennet SMP, Bohn L, Storsrud S, et al. Multivariate modelling of faecal bacterial profiles of patients with IBS predicts responsiveness to a diet low in FODMAPs. Gut. 2017 doi: 10.1136/gutjnl-2016-313128. [DOI] [PubMed] [Google Scholar]
- 73.Zeevi D, Korem T, Zmora N, et al. Personalized Nutrition by Prediction of Glycemic Responses. Cell. 2015;163(5):1079–1094. doi: 10.1016/j.cell.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 74.Korem T, Zeevi D, Zmora N, et al. Bread Affects Clinical Parameters and Induces Gut Microbiome-Associated Personal Glycemic Responses. Cell Metab. 2017;25(6):1243–1253. e1245. doi: 10.1016/j.cmet.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 75.Wu GD, Chen J, Hoffmann C, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334(6052):105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sonnenburg ED, Smits SA, Tikhonov M, Higginbottom SK, Wingreen NS, Sonnenburg JL. Diet-induced extinctions in the gut microbiota compound over generations. Nature. 2016;529(7585):212–215. doi: 10.1038/nature16504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505(7484):559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kovatcheva-Datchary P, Nilsson A, Akrami R, et al. Dietary Fiber-Induced Improvement in Glucose Metabolism Is Associated with Increased Abundance of Prevotella. Cell Metab. 2015;22(6):971–982. doi: 10.1016/j.cmet.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 79.Smits SA, Marcobal A, Higginbottom S, Sonnenburg JL, Kashyap PC. Individualized Responses of Gut Microbiota to Dietary Intervention Modeled in Humanized Mice. mSystems. 2016;1(5) doi: 10.1128/mSystems.00098-16. [DOI] [PMC free article] [PubMed] [Google Scholar]