SUMMARY
Evasion of apoptosis is a hallmark of cancer. Bcl-2 and p53 represent two important nodes in apoptosis signaling pathways. We find that concomitant p53 activation and Bcl-2 inhibition overcome apoptosis resistance and markedly prolong survival in three mouse models of resistant acute myeloid leukemia (AML). Mechanistically, p53 activation negatively regulates the Ras/Raf/MEK/ERK pathway and activates GSK3 to modulate Mcl-1 phosphorylation and promote its degradation, thus overcoming AML resistance to Bcl-2 inhibition. Moreover, Bcl-2 inhibition reciprocally overcomes apoptosis resistance to p53 activation by switching cellular response from G1 arrest to apoptosis. The efficacy, together with the mechanistic findings, reveals the potential of simultaneously targeting these two apoptosis regulators and provides a rational basis for clinical testing of this therapeutic approach.
Keywords: Bcl-2, p53, Mcl-1, apoptosis resistance, synthetic lethality, p21, Ras, G1 arrest, leukemia
Graphical abstract
Pan et al. show that p53 activation promotes Mcl-1 degradation, and Bcl-2 inhibition shifts the p53 activation response from G1 arrest to apoptosis. Combining p53 activation and Bcl-2 inhibition overcomes resistance to either alone and provides better therapeutic efficacy in mouse models of acute myeloid leukemia.
INTRODUCTION
Evasion of apoptosis is integral to tumorigenesis and drug resistance (Hanahan and Weinberg, 2011; Juin et al., 2013). Upregulation of anti-apoptotic Bcl-2 family members and inactivation of p53 functions are two canonical approaches exploited by cancer cells to escape apoptosis (Delbridge et al., 2016; Hanahan and Weinberg, 2011; Hoe et al., 2014; Juin et al., 2013). Both approaches shift the balance of anti- and pro-apoptotic proteins toward survival and render cancer cells resistant to various therapies. Tremendous efforts to develop Bcl-2 inhibitors culminated in ABT-199 (ABT, venetoclax), a selective and potent Bcl-2 inhibitor that showed promising efficacy in several cancers (Delbridge et al., 2016; Hata et al., 2015; Souers et al., 2013). The advent of ABT allows scientists to specifically study the effects of Bcl-2 inhibition. We have previously reported that targeting Bcl-2 by ABT effectively induced apoptosis in acute myeloid leukemia (AML). However, the expression of Mcl-1, another important anti-apoptotic protein, renders leukemia cells resistant to both ABT and its predecessor ABT-737 (Konopleva et al., 2006; Pan et al., 2014; Pan et al., 2015).
TP53, arguably the most important tumor suppressor gene, is mutated in ~50% of human solid tumors but rarely mutated in hematological malignancies (Cerami et al., 2012; Weinberg, 2014). However, wild-type p53 functions are frequently suppressed by MDM2, an E3 ubiquitin ligase that targets p53 for proteasomal degradation (Haupt et al., 1997; Kubbutat et al., 1997). Nutlins represented a breakthrough in the development of p53-reactivating compounds (Vassilev et al., 2004). The recently developed RG7388 (RG, idasanutlin) is a second-generation MDM2 inhibitor with greater potency, selectivity, and bioavailability compared to its predecessors Nutlin-3a and RG7112 and able to effectively activate p53 (Ding et al., 2013). Nevertheless, some AML cell lines and primary patient samples are resistant to p53-induced apoptosis following RG treatment. In the current study, we utilized RG, ABT, and various genetic and functional assays to investigate whether p53 activation and Bcl-2 inhibition could reciprocally overcome apoptosis resistance to either approach alone and studied the underlying mechanism.
RESULTS
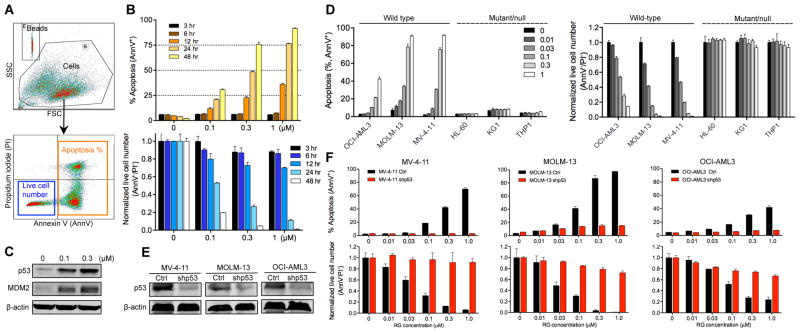
RG Activates p53 and Induces Apoptosis in a p53-Dependent Manner
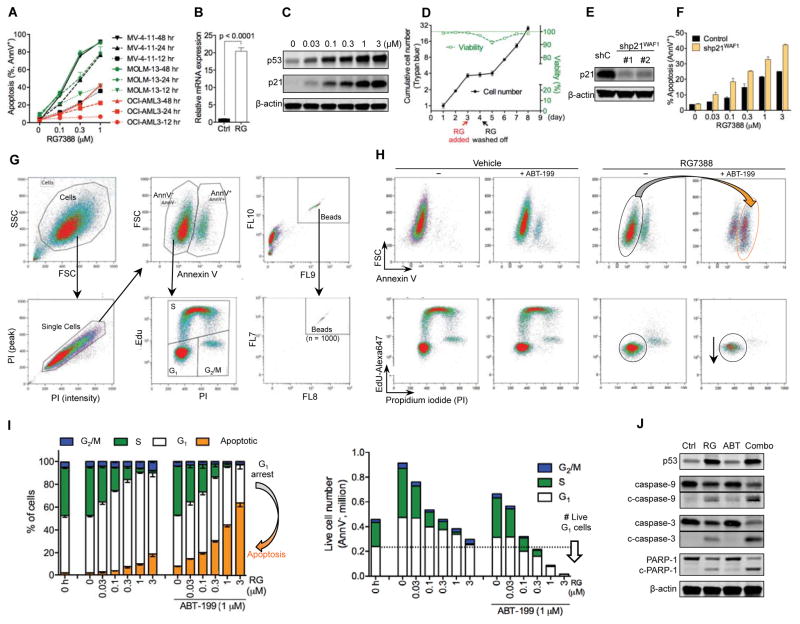
We first tested whether the MDM2 inhibitor RG depends on wild-type p53 for its activity using annexin V (AnnV) and propidium iodide (PI) co-staining followed by flow cytometry analysis (Figure 1A). In three p53 wild-type (p53WT) AML cell lines (MV-4-11, OCI-AML3, and MOLM-13), RG induced a rapid accumulation of p53 as well as time- and dose-dependent apoptotic cell death and growth inhibition (Figures 1B, 1C, S1A, and S1B). In contrast, RG did not trigger apoptosis in p53null HL-60 cells or p53mutant KG1 and THP1 cells (Figure 1D). Furthermore, stable p53 knockdown substantially attenuated RG efficacy in the three p53WT cell lines (Figures 1E and 1F). These results demonstrate that RG can effectively activate p53 and induce apoptosis in a p53-dependent manner, indicating that RG can serve as a superior compound to study p53 activation in cancers.
Figure 1. MDM2 Inhibitor RG Activates p53 and Induces Apoptosis in a p53-Dependent Manner.
(A) Flow cytometry plots showing the gating strategy to determine apoptosis (AnnV+) and the numbers of live cells (AnnV−/PI−).
(B) Time- and dose-dependent induction of apoptosis and reduction of live cell numbers by RG in MV-4-11 cells.
(C) Immunoblots of p53 and MDM2 in MV-4-11 cells after RG treatment for 8 hr. β-actin served as the loading control.
(D) Sensitivity comparison of p53WT and p53 deficient AML cells. AML cells were treated with indicated concentrations of RG for 48 hr.
(E) Immunoblot showing p53 knockdown by lentiviral shRNA in three AML cell lines. The cells were treated with 1 μM RG for 12 hr to induce p53 prior to immunoblotting.
(F) Sensitivity comparison of control and p53 knockdown AML cells. Cells were treated with indicated concentrations of RG for 48 hr.
Cell numbers were enumerated by flow analysis using CountBright counting beads and normalized to untreated controls (B, D, F). Data in the bar graphs (B, D, F) represent the means of triplicate experiments. Error bars, mean ± SD. See also Figure S1.
We also tested whether there is synergism between p53 activation and Bcl-2 inhibition in sensitive AML cells. MV-4-11 and MOLM-13 are two AML cell lines sensitive to the Bcl-2–specific inhibitor ABT (IC50 values < 20 nM). Combining the p53 activator RG with ABT at a 5:1 or 1:1 ratio augmented apoptosis and reduced live cell numbers to a significantly greater extent than either agent alone (Figures S1C and S1D). The combination index (CI) values indicate a strong synergy between the two compounds in the two ABT-sensitive cell lines (Figure S1E).
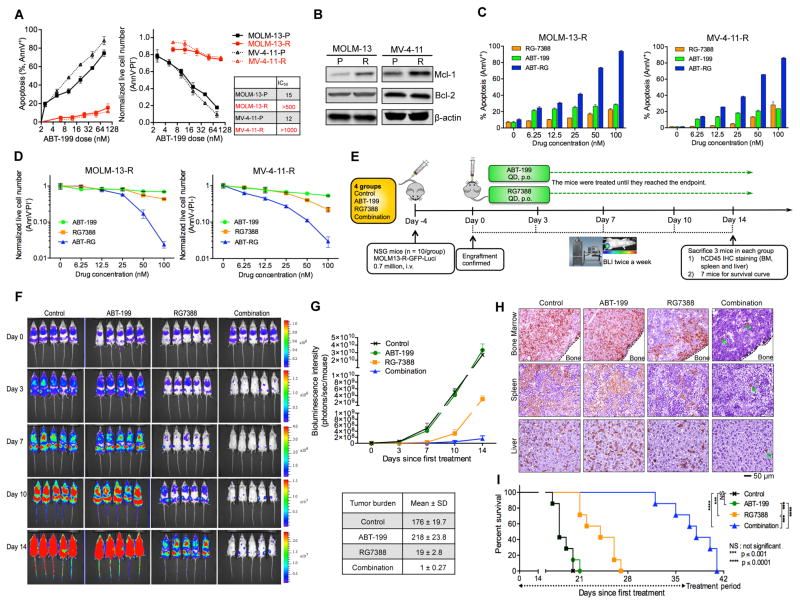
p53 Activation Overcomes Resistance to Bcl-2 Inhibition in Surviving AML Cells
Although initially sensitive to Bcl-2 inhibitors, a fraction of MOLM-13 and MV-4-11 cells survived prolonged exposure to ABT. While Bcl-2 inhibition readily induced apoptosis in parental cells, only marginal apoptosis occurred in the surviving resistant cells (MOLM-13-R and MV-4-11-R, Figure 2A). The ABT IC50 increased by over 30- and 80-fold for MOLM-13-R and MV-4-11-R cells, respectively. Compared to their parental counterparts, the resistant cells expressed higher levels of Mcl-1 (Figure 2B). Ectopic Mcl-1 expression also conferred resistance to ABT in the parental MOLM-13 and MV-4-11 cells (Figures S2A–S2D), indicating that the ABT resistance of the surviving cells could be attributed, at least in part, to the elevated expression of Mcl-1.
Figure 2. p53 Activator RG Overcomes the Resistance of AML Cells Which Survived ABT Treatment.
(A) Sensitivity comparison of parental (P) and generated resistant (R) cells. The cells were treated with ABT for 48 hr. The IC50 values were calculated with Calcusyn software based on live cell numbers.
(B) Immunoblots showing Mcl-1 and Bcl-2 levels in ABT-resistant MOLM-13R and MV-4-11R cells.
(C, D) Apoptosis induction (C) and decrease of live cell numbers (D) in the MOLM-13-R and MV-4-11R resistant cells after treatment with ABT, RG, or the combination (1:1 ratio) for 48 hr.
(E) Schematic outline of the mouse model of ABT resistance (MOLM-13-R). Treatment was started after confirmation of AML engraftment on day 0 (4 days after injection).
(F) Serial bioluminescence images of mice bearing MOLM-13-R cells after treatment with vehicle, ABT, RG, or the combination.
(G) Quantification of bioluminescence emitted from the whole body of each mouse in panel F. Data represent the mean ± SD of 5 examined mice in each group.
(H) Representative immunohistochemical (IHC) staining of murine bone marrow, spleen, and liver for human CD45 antigen on day 14. Three mice from each group were sacrificed. Rare leukemic cells in tissue sections from combination-treated mice are marked by arrows.
(I) Kaplan-Meier survival curves of mice injected with MOLM-13-R cells (n = 7 per group; statistical significance was evaluated by the log-rank test).
Data in the bar/line graphs (A, C, D) represent the means of triplicate experiments. Error bars, mean ± SD. See also Figure S2.
Next we examined if the resistance to Bcl-2 inhibition could be overcome by p53 activation. Bcl-2 inhibition by ABT triggered little apoptosis in MOLM-13-R or MV-4-11-R cells. However, the combination of p53 activator RG with ABT markedly induced apoptosis and decreased live cell numbers (Figures 2C and 2D), implying that the ABT resistance of the surviving cells could be abrogated by p53 activation (Similarly, RG overcame ABT resistance in engineered Mcl-1–overexpressing MOLM-13/MV-4-11 cells; Figures S2E and S2F). To test this in vivo, we labeled the resistant MOLM-13-R cells with luciferase (Luci) and injected them into NOD SCID gamma (NSG) mice (Figure 2E). After confirmation of engraftment by bioluminescence imaging (BLI), the mice were randomly assigned to four groups (n = 10/group) and treated with vehicle, ABT, RG, or both ABT and RG. After 2 weeks of treatment, the leukemic burden in the combination group was ~1/200 of that in the ABT and control groups and ~1/19 of that in the RG group (Figures 2F and 2G). Meanwhile, the combination dramatically reduced leukemic infiltration in spleen, liver, and bone marrow (BM, Figure 2H). MOLM-13-R cells rapidly killed control and ABT-treated mice at about the same time (Figure 2I). RG prolonged survival by 6 days, while the combination significantly extended mouse survival by 20 days (Figure 2I). These results suggest that p53 activation by RG could overcome the ABT resistance of MOLM-13-R cells both in vitro and in vivo.
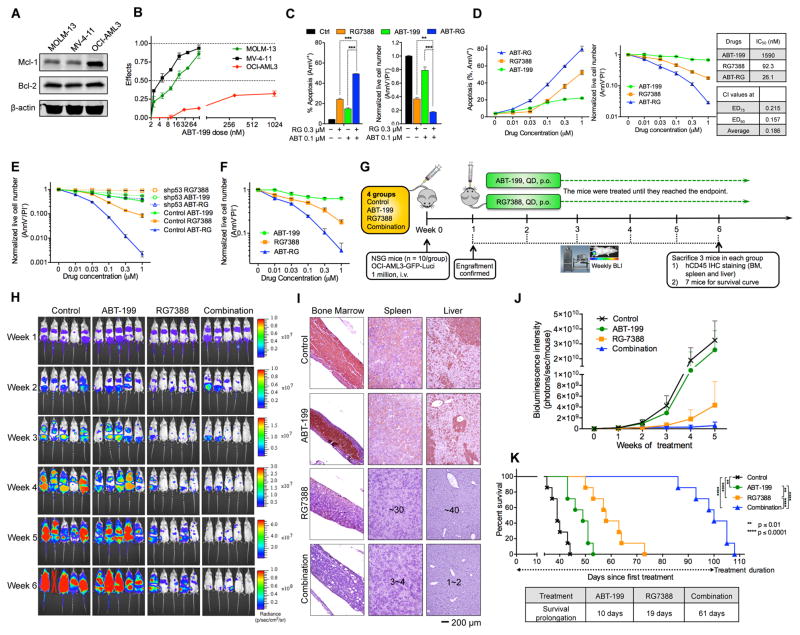
p53 Activation Abrogates Inherent Resistance to Bcl-2 Inhibition in OCI-AML3 cells
OCI-AML3 cells express high levels of Mcl-1 and are inherently resistant to ABT (Figures 3A and 3B). Both Mcl-1 knockdown (Pan et al., 2014) and a selective Mcl-1 inhibitor A-1210477 (Figure S3) abolished ABT resistance, further validating Mcl-1 as an inherent resistance factor to ABT. We then asked whether the inherent resistance could be abrogated by p53 activation. Combining RG with ABT at a 3:1 or 1:1 ratio increased apoptosis and decreased live cell numbers to a significantly greater extent than either agent alone (Figures 3C and 3D). The p53 activator RG reduced ABT IC50 from 1,590 nM to 26 nM. These results indicate that p53 activation could overcome the inherent resistance to ABT. Importantly, RG failed to overcome the inherent resistance in p53 knockdown OCI-AML3 cells (Figure 3E), suggesting a pivotal role of p53 in this process.
Figure 3. p53 Activation by RG Abrogates Inherent Resistance to Bcl-2 Inhibitor ABT.
(A) Basal expression of Mcl-1 and Bcl-2 in MOLM-13, MV-4-11, and OCI-AML3 cells.
(B) AML cells were treated with ABT for 48 hr. Effects = 1 – live cell numbertreated/live cell numbercontrol.
(C, D) OCI-AML3 cells were treated with indicated compounds at a 3:1 ratio (C) or a 1:1 ratio (D) for 48 hr. The combination index (CI) and IC50 values were calculated using Calcusyn software based on live cell numbers. ED75, 75% effective dose; ED90, 90% effective dose.
(E) Comparison of live cell numbers in control and p53 knockdown OCI-AML3 cells after treatment with ABT, RG, or ABT/RG combination for 48 hr.
(F) Reduction of live cell numbers of luciferase-labeled OCI-AML3 cells after 48 hr treatment with ABT, RG, or ABT/RG in vitro.
(G) Schematic outline of the mouse model of inherent ABT resistance (OCI-AML3).
(H) Serial bioluminescence images of OCI-AML3–engrafted NSG mice. Treatment was initiated on day 7 after confirmation of engraftment.
(I) Representative IHC staining of murine BM, spleen, and liver for human CD45 antigen at week 6. Three mice from each group were sacrificed. Countable numbers of hCD45+ cells were marked on each slide.
(J) Quantitation of bioluminescence emitted from the whole body of each mouse. Data represent the mean ± SD of 5 examined mice in each group as shown in (H).
(K) Kaplan-Meier survival curves of mice injected with luciferase-labeled OCI-AML3 cells (n = 7 per group; statistical significance and median survival were evaluated using the log-rank test).
Data in the bar/line graphs B–F represent the means of triplicate experiments with error bars indicating SD. **p < 0.01, ***p < 0.001 as determined by two-tailed unpaired Student’s t-test. See also Figure S3.
To determine if p53 activation could overcome the inherent resistance in vivo, we labeled OCI-AML3 cells with luciferase and injected them into NSG mice (Figures 3F and 3G). One week after injection, OCI-AML3 engraftment was confirmed by BLI (Figure 3H). The mice were then randomly divided into four groups and treated with vehicle, ABT, RG, or the combination. Bcl-2 inhibition by ABT slightly reduced leukemic burden, as indicated by BLI and immunohistochemical (IHC) analysis (Figures 3H–3J). RG significantly decreased tumor burden, while the combination further slowed leukemia progression and reduced tumor infiltration in BM, spleen, and liver (Figures 3H–3J). The ABT/RG combination markedly extended mouse survival by 61 days (3.2- and 6.1-fold prolongation relative to RG or ABT monotherapy, respectively; Figure 3K). These data suggest that p53 activation by RG can overcome the Mcl-1–mediated inherent resistance to ABT both in vitro and in vivo.
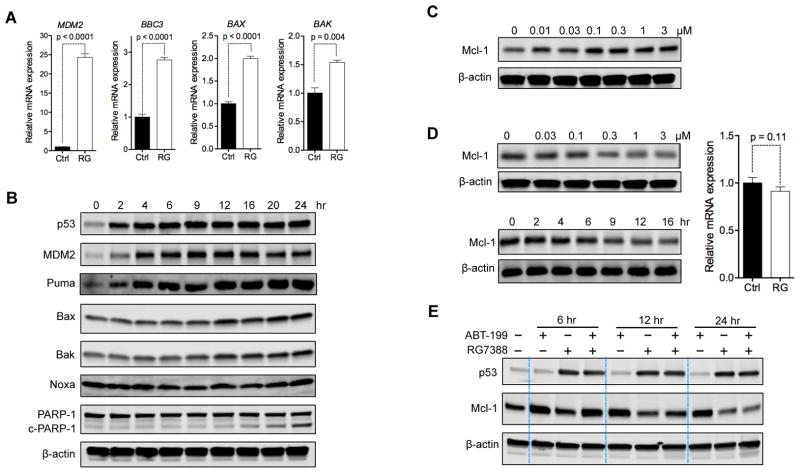
p53 Activation Reverses ABT–Induced Upregulation of Mcl-1
We next sought to address the mechanism by which p53 activation overcame Mcl-1–mediated resistance. We first used OCI-AML3 for the mechanistic studies because this cell line is not only resistant to ABT but also relatively resistant to RG, when compared to MV-4-11 and MOLM-13 cells (Figures S1A and 1B). We found that RG treatment significantly increased mRNA levels of p53 target genes MDM2, BBC3, BAX, and BAK (Figure 4A), indicating the activation of p53. Similarly, kinetic study using immunoblotting revealed a rapid and strong upregulation of MDM2 and Puma (Figure 4B) and a delayed upregulation of Bax and Bak. These observations correspond with previous reports that p53 has lower affinity for BAX and BAK promoters than for MDM2 and BBC3 promoters (Kracikova et al., 2013; Kruiswijk et al., 2015). Since Puma, Bax, and Bak each binds to and counteracts Mcl-1 (Czabotar et al., 2014; Hata et al., 2015), their upregulation could potentially contribute to overcoming Mcl-1–mediated resistance.
Figure 4. p53 Activation Upregulates Pro-Apoptotic Proteins and Reduces Anti-Apoptotic Mcl-1.
(A) Relative mRNA expression of MDM2, BAX, BAK, and BBC3 (Puma-encoding gene) in OCI-AML3 cells after 12 hr treatment with 1 μM RG.
(B) Immunoblots showing the levels of indicated proteins in OCI-AML3 cells after treatment with 2 μM RG for indicated time. c-PARP-1, cleaved PARP-1 protein.
(C) Immunoblot of Mcl-1 in OCI-AML3 cells treated with indicated concentrations of ABT for 24 hr.
(D) Immunoblots of Mcl-1 in OCI-AML3 cells treated with indicated concentrations of RG for 24 hr (upper left) or treated with 1 μM RG for the indicated durations (lower left) and the MCL1 mRNA levels in OCI-AML3 cells incubated without (Ctrl) or with 1 μM RG for 12 hr.
(E) Changes in Mcl-1 protein levels in OCI-AML3 cells treated with vehicle, 1 μM ABT, 1 μM RG, or the combination for 6, 12, or 24 hr.
Data in bar graphs (A, D) represent the means of triplicate experiments. Error bars, mean ± SD. p values were calculated using two-tailed unpaired Student’s t-test.
Although OCI-AML3 cells express high levels of Mcl-1, Mcl-1 was further upregulated after acute exposure to ABT (Figure 4C), which may further enhance their resistance to ABT. In contrast, RG induced a time- and dose-dependent downregulation of Mcl-1 (Figure 4D). The downregulation occurred within 9 hr, when no cleavage of PARP-1 could be detected and the cell viability was over 97%, excluding the possibility that the observed Mcl-1 degradation was a secondary consequence of apoptosis. ABT rapidly upregulated Mcl-1 (Figure 4E), even in the presence of p53 activator RG. Nevertheless, the Mcl-1 upregulation could be reversed later by RG (Figure 4E). These findings indicate that p53 activation by RG could decrease Mcl-1 levels and reverse the ABT–mediated upregulation of Mcl-1.
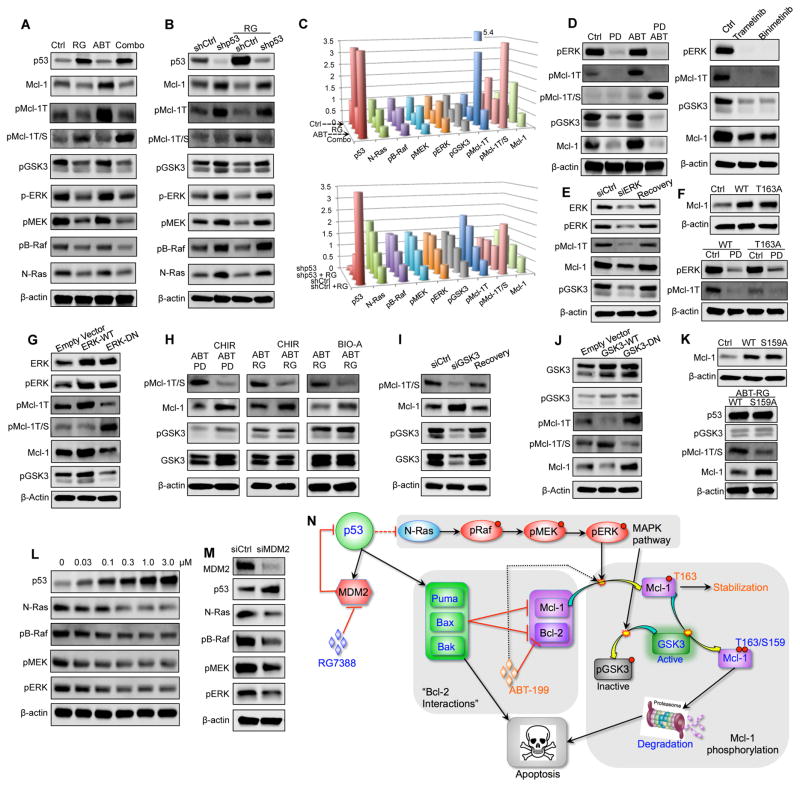
p53 Activation Inhibits pERK and Activates GSK3 to Modulate Mcl-1 Phosphorylation
To determine the mechanism underlying Mcl-1 reduction, we first conducted qRT-PCR analysis, which indicated that RG did not affect MCL1 mRNA levels significantly (Figure 4D, right). We postulated that Mcl-1 might be regulated by post-translational modifications (PTMs) that affect its stability. Indeed, p53 activation by RG decreased Mcl-1 mono-phosphorylation at threonine 163 (pMcl-1T) but increased its dual-phosphorylation at threonine 163 and serine 159 (pMcl-1T/S; Figure 5A, lane 2). It has been reported that T163 phosphorylation stabilizes Mcl-1 but also primes Mcl-1 to be further phosphorylated at S159, and that the dually-phosphorylated Mcl-1 is ubiquitinated and targeted for proteasomal degradation (Gores and Kaufmann, 2012; Opferman, 2006). To determine whether RG treatment increases Mcl-1 ubiquitination, we immunoprecipitated Mcl-1 from vehicle- or RG-treated cells and found that RG treatment indeed augmented Mcl-1 ubiquitination (Figure S4A). If proteasomes play critical roles in RG-induced Mcl-1 degradation, one would anticipate that proteasome inhibition could stabilize Mcl-1. Next, we co-treated the cells with RG and a proteasome inhibitor (bortezomib or ixazomib). As expected, both compounds stabilized Mcl-1 and protected it from RG–induced degradation (Figure S4B).
Figure 5. p53 Regulates MAPK/GSK3 Signaling to Modulate Mcl-1 Phosphorylation and Degradation.
(A) Immunoblots of the indicated proteins in OCI-AML3 cells treated with DMSO control, 1 μM RG, 1 μM ABT, or the combination for 24 hr.
(B) Immunoblots of the indicated proteins in control and p53 knockdown OCI-AML3 cells after treatment with vehicle DMSO or 1 μM RG for 24 hr.
(C) Three-dimensional cylinder charts showing protein levels in panels A (top) and B (bottom) as measured by quantitative immunoblot using the Odyssey Infrared Imaging System. Value = 1 for untreated control.
(D) Immunoblots of the indicated proteins in OCI-AML3 cells treated with DMSO control, 1 μM ABT, 100 nM PD0325901 (PD), or both compounds for 12 hr.
(E) Immunoblots of pMcl-1T and pGSK3 after transient ERK knockdown in OCI-AML3 cells.
(F) Immunoblot of Mcl-1 in control OCI-AML3 cells or OCI-AML3 cells overexpressing the wild-type or the T163A mutant Mcl-1 (upper) or immunoblots of pERK and pMcl-1T in Mcl-1WT and Mcl-1T163A overexpressing OCI-AML3 cells treated with 100 nM PD for 12 hr (lower).
(G) Immunoblots of the indicated proteins in OCI-AML3 cells overexpressing wild-type ERK or dominant-negative (DN) ERK.
(H) Immunoblots of the indicated proteins in OCI-AML3 cells treated with different combinations of 1 μM ABT, 100 nM PD, 1 μM RG, and GSK3 inhibitors CHIR-99021 (CHIR, 1 μM) or BIO-Acetoxime (BIO-A, 1 μM) for 24 hr.
(I) Immunoblots of the indicated proteins after transient GSK3 knockdown in OCI-AML3 cells. Cells were treated with both 1 μM ABT and 1 μM RG for 24 hr before immunoblotting.
(J) Immunoblots of the indicated proteins in OCI-AML3 cells stably overexpressing wild-type or dominant-negative (DN) GSK3. Cells were treated with ABT/RG (1 μM each) for 24 hr.
(K) Immunoblot of Mcl-1 in control MV-4-11 cells or MV-4-11 cells overexpressing the wild-type or S159A mutant Mcl-1 (upper) or immunoblots of the indicated proteins in Mcl-1WT and Mcl-1S159A overexpressing MV-4-11 cells after treatment with 1 μM ABT and 1 μM RG for 24 hr.
(L) Immunoblots of the indicated proteins in OCI-AML3 cells after treatment with escalating doses of RG for 24 hr.
(M) Immunoblots of the indicated proteins in control and MDM2 knockdown OCI-AML3 cells.
(N) Schematic of the proposed mechanism by which p53 activation overcomes Mcl-1–mediated resistance. See also Figure S4.
Bcl-2 inhibitor ABT by itself dramatically elevated pMcl-1T (Figure 5A, lane 3). Interestingly, RG converted the abundant stabilizing pMcl-1T to destabilizing pMcl-1T/S (Figure 5A, lanes 3,4), which may explain how p53 activation reversed the ABT–induced upregulation of Mcl-1. In agreement with these observations, p53 knockdown upregulated pMcl-1T, increased Mcl-1, and diminished RG’s ability to convert pMcl-1T to pMcl-1T/S (Figure 5B; see also Figure 5C for quantitative values of indicated proteins in Figures 5A and 5B), suggesting an essential role of p53 in this conversion. Of note, Bcl-2 protein levels remained largely unchanged after p53 activation or p53 knockdown (Figure S4C).
The next question was which kinase(s) phosphorylated Mcl-1 at T163 or S159 in this context. pJNK has been known to phosphorylate Mcl-1 at T163 (Morel et al., 2009; Opferman, 2006). It appeared that pJNK did not play a significant role here since neither pJNK nor total JNK decreased in response to p53 activation (Figure S4D). In contrast, p53 activation decreased pERK along with pMcl-1T (Figure 5A, lane 2). This finding agrees with previous reports that pERK phosphorylates Mcl-1 at T163 (Domina et al., 2004; Gores and Kaufmann, 2012). Consistently, p53 knockdown increased pERK and pMcl-1T (Figure 5B). To further examine ERK’s role in T163 phosphorylation, we tested whether ERK inhibition would lead to reduced levels of pMcl-1T. MEK inhibitor PD0325901 (PD) is highly selective and widely used to study ERK inhibition. As expected, PD treatment eliminated pERK and markedly reduced pMcl-1T (Figure 5D). Similarly, ERK inhibition by trametinib and binimetinib also drastically reduced pMcl-1T (Figure 5D, right).
Next, we transiently knocked down ERK expression and found it significantly decreased pMcl-1T (Figure 5E). In addition, after cells regained ERK expression, pMcl-1T reverted to its basal level (Figure 5E). Meanwhile, we stably overexpressed Mcl-1WT or the Mcl-1T163A mutant to similar levels in OCI-AML3 cells (Figure 5F). Presumably, Mcl-1T163A is no longer a substrate of pERK. Indeed, Mcl-1T163A overexpressors had lower levels of pMcl-1T than Mcl-1WT overexpressing cells (Figure 5F, lower panel). Nevertheless, ERK inhibition by PD was able to eliminate the residual pMcl-1T in Mcl-1T163A cells. As discussed above, dually-phosphorylated pMcl-1T/S could be ubiquitinated and then degraded. Since T163A point mutation abrogates the formation of pMcl-1T/S, T163A cells should have less ubiquitinated Mcl-1 after RG treatment or after ERK inhibition. Indeed, there was significantly less ubiquitinated Mcl-1 but more total Mcl-1 in T163A overexpressing cells, either after RG treatment or after ERK inhibition/knockdown (Figure S4E). Next we overexpressed wild-type and dominant-negative ERK to increase or inhibit ERK activity (Figure 5G). As expected, overexpression of wild-type ERK increased pMcl-1T levels while overexpressing dominant-negative ERK reduced pMcl-1T (Figure 5G). These results collectively indicate that pERK regulates Mcl-1 phosphorylation at T163.
GSK3 has been reported to associate with pMcl-1T and phosphorylate its S159 residue (Maurer et al., 2006; Opferman, 2006). GSK3 protein is constitutively active, but its activity is diminished by inhibitory phosphorylation at serine 9/21 (Cohen and Frame, 2001; Maurer et al., 2006). We found that p53 activation reduced inactive pGSK3 (Figure 5A, lanes 2 and 4) but not total GSK3 (Figure S4F), implying a larger fraction of active GSK3, which could conceivably be responsible for transforming ABT–induced pMcl-1T to pMcl-1T/S (Figure 5A, lane 4). Consistently, p53 knockdown did not affect total GSK3 levels (Figure S4G) but increased GSK3 inhibitory phosphorylation, which likely restrained GSK3 from converting pMcl-1T to pMcl-1T/S (Figure 5B, lanes 2 and 4). To further investigate GSK3’s role in this conversion, we pretreated OCI-AML3 cells with GSK3 inhibitor CHIR-99021 (CHIR). Bcl-2 inhibitor ABT was also added to increase pMcl-1T (i.e., GSK3 substrate). We have shown that ABT alone induced abundant pMcl-1T but not pMcl-1T/S (Figure 5A, lane 3), implying that active GSK3 is probably present at low levels in OCI-AML3 cells. Therefore, p53 activator RG or MEK inhibitor PD was also added to release GSK3 activity. As expected, ABT/PD or ABT/RG combinations induced abundant pMcl-1T/S, while GSK3 inhibitor CHIR prevented the generation of destabilizing pMcl-1T/S and stabilized Mcl-1 (Figure 5H). Similarly, another GSK3 inhibitor, Bio-Acetoxime, also reduced S159 phosphorylation and stabilized Mcl-1. Notably, both GSK3 inhibitors attenuated apoptosis induction by ABT/RG co-treatment (Figure S4H).
We next transiently knocked down GSK3 using siRNAs. GSK3 knockdown significantly reduced pMcl-1T/S, stabilized Mcl-1 (Figure 5I), decreased Mcl-1 ubiquitination (Figure S4I), and protected AML cells from ABT/RG treatment (Figure S4J). In addition, after cells resumed GSK3 expression, pMcl-1T/S, total Mcl-1, and ubiquitinated Mcl-1 all reverted to their basal levels (Figures 5I and S4I). We next overexpressed wild-type GSK3 and its dominant-negative mutant to upregulate or downregulate GSK3 activity. As expected, cells overexpressing wild-type GSK3 converted more pMcl-1T into pMcl-1T/S (Figure 5J), increased Mcl-1 ubiquitination, and decreased total Mcl-1 after ABT/RG treatment (Figure S4K). In contrast, dominant negative GSK3 inhibited the conversion from pMcl-1T to pMcl-1T/S (Figure 5J), reduced Mcl-1 ubiquitination, and stabilized Mcl-1 (Figure S4K). All these results support that GSK3 phosphorylated Mcl-1 at S159 to promote its ubiquitination and degradation. We also stably overexpressed Mcl-1WT and Mcl-1S159A in MV-4-11 cells (Figure 5K) and treated them with ABT/RG. Since S159A mutation eliminates the GSK3 phosphorylation site, one would anticipate that S159A mutation could stabilize Mcl-1 and protect cells from ABT/RG treatment. Indeed, there was significantly more Mcl-1 in Mcl-1S159A overexpressors than in Mcl-1WT overexpressors (Figure 5K, lower panel), and more Mcl-1S159A overexpressors survived the ABT/RG treatment (Figure S4L). In addition, we have shown that ABT/RG reduced inhibitory phosphorylation of GSK3, increased Mcl-1 S159 phosphorylation, reduced Mcl-1, and effectively induced apoptosis in OCI-AML3 cells (Figures 5A and 3E). These findings collectively suggest that GSK3-dependent Mcl-1 phosphorylation and degradation are essential for the synergistic effect of the ABT/RG combination.
p53 Activation Downregulates the Ras/Raf/MEK/ERK Signaling Cascade
To determine the mechanism by which p53 activation regulates GSK3 activity, we first measured the effects of p53 expression on AKT, which is widely known to phosphorylate and inactivate GSK3 (Cohen and Frame, 2001; Maurer et al., 2006). However, p53 knockdown or p53 activation did not significantly affect AKT and pAKT levels (Figure S4M). Meanwhile, AKT inhibition by MK-2206 dramatically decreased pAKT but only marginally reduced pGSK3 (Figure S4N), suggesting that AKT did not play a significant role here. In contrast, pMEK and pERK, along with pGSK3, were decreased after RG–induced p53 activation (Figure 5A). This observation agreed with previous reports that MEK/ERK signaling could promote GSK3 phosphorylation (Cohen and Frame, 2001; Ding et al., 2005). Consistently, in p53 knockdown cells, pGSK3 was increased together with pMEK and pERK (Figure 5B, lane 2). In addition, in p53 knockdown cells, RG could not reduce pMEK/pERK. As a result, RG failed to reduce pGSK3 in these cells (Figure 5B, lane 4).
To further examine the role of MEK/ERK signaling in GSK3 phosphorylation, we blocked MEK/ERK signaling using MEK inhibitor PD and found that PD markedly reduced pGSK3 (Figure 5D). Similarly, another two MEK inhibitors trametinib and binimetinib decreased pGSK3 to very low levels (Figure 5D, right). We also found that transient ERK knockdown did not affect the levels of total GSK3 but significantly decreased GSK3 inhibitory phosphorylation (Figure 5E). After cells resumed ERK expression, pGSK3 reverted to its basal levels (Figure 5E). Importantly, overexpression of wild-type ERK increased pGSK3 while overexpressing dominant-negative ERK reduced pGSK3 levels (Figure 5G). These findings collectively implied that MEK/ERK signaling promoted the inhibitory phosphorylation of GSK3. We have shown that GSK3 promoted Mcl-1 ubiquitination. Consistently, ERK knockdown or overexpression of dominant-negative ERK, by virtue of inhibiting ERK and releasing GSK3 activity, increased Mcl-1 ubiquitination and reduced total Mcl-1 (Figures S4I and S4K).
Next, we tracked the Ras/Raf/MEK/ERK pathway bottom-up and found that p53 activation by RG led to the downregulation of N-Ras protein (Figure 5A). In contrast, RG treatment failed to downregulate N-Ras in p53 knockdown cells (Figure 5B, lane 4), suggesting an essential role of p53 in this process. Notably, p53 knockdown actually increased N-Ras protein (Figure 5B, lane 2). These data suggested that p53 negatively regulates N-Ras protein levels. Consistently, RG induced a dose-dependent upregulation of p53 and a corresponding downregulation of N-Ras (Figure 5L). We also transiently knocked down MDM2 to stabilize p53, which, as expected, reduced N-Ras and the downstream Ras/Raf/MEK/ERK signaling (Figure 5M). In addition, p53 activation by RG similarly reduced N-Ras and MAPK signaling in another cell line (Figure S4O). These results collectively suggested that p53 negatively modulated N-Ras expression, though the detailed mechanism through which this is accomplished needs to be defined in future studies.
As mentioned above, we also quantitated and normalized the levels of proteins involved in this regulatory cascade and generated two cylinder charts corresponding to Figures 5A and 5B (Figure 5C). The cylinder charts show similar trends between the Ras/Raf/MEK/ERK signaling cascade and Mcl-1 protein levels but opposite trends between p53 and Mcl-1 levels and between p53 levels and the MAPK signaling cascade. These correlations are consistent with the proposed mechanism. We also used ABT-resistant MV-4-11_Mcl-1 cells to further examine our mechanistic findings. As in OCI-AML3 cells, p53 activation by RG reduced Mcl-1 levels (Figure S4O). Although MV-4-11_Mcl-1 cells express abundant Mcl-1 (comparable to OCI-AML3), ABT treatment further upregulated Mcl-1 levels. However, the upregulation could be reversed by RG co-treatment (Figure S4O, lane 4). These results are consistent with our findings in OCI-AML3 cells. Similarly, p53 activation downregulated N-Ras, pB-Raf, pMEK, and pERK and decreased the inhibitory phosphorylation of GSK3 (Figure S4O), implying an increase in active GSK3, which could conceivably convert stabilizing pMcl-1T to destabilizing pMcl-1T/S. Indeed, ABT/RG-treated cells had significantly less pMcl-1T but more pMcl-1T/S compared with ABT-treated cells (Figure S4O, lane 4). These results are virtually the same as what we found in OCI-AML3 cells and validated our mechanistic findings, which can be briefly summarized as follows: RG treatment activates p53, which negatively regulates the Ras/Raf/MEK/ERK pathway and releases GSK3 activity; active GSK3 then converts ABT–induced stabilizing pMcl-1T to destabilizing pMcl-1T/S to promote Mcl-1 degradation, which contributes significantly to the synergistic effects of the ABT/RG combination. Figure 5N schematically summarizes the proposed mechanism.
Bcl-2 Inhibition Reciprocally Overcomes Resistance to p53 Activation
RG treatment induced similar levels of p53 in MV-4-11, MOLM-13, and OCI-AML3 cells, but substantially less apoptosis occurred in OCI-AML3 cells (Figure 6A), suggesting that OCI-AML3 is relatively resistant to apoptosis induction by p53 and may have a higher apoptotic threshold. We postulated that if p53 level was not sufficient to push OCI-AML3 cells across the apoptotic threshold, abundant p21 protein induced by p53 might trigger pro-survival cell cycle arrest (CCA). Indeed, p53 activation by RG induced little apoptosis in OCI-AML3 cells (Figure 6A) but robustly elevated p21 expression at mRNA and protein levels (Figures 6B and 6C) and triggered CCA at G1 phase (Figure S5A). Notably, the induced G1 arrest was reversible as OCI-AML3 cells quickly resumed proliferation after removal of RG from the medium (Figure 6D). Moreover, p21 knockdown (Figure 6E) reduced RG–induced G1 arrest and decreased live cell numbers (Figures S5A and S5B) but significantly increased RG–induced apoptosis (Figure 6F). These findings suggest that p21 promotes pro-survival G1 arrest and may protect cancer cells from p53–induced apoptosis.
Figure 6. Bcl-2 Inhibition Reciprocally Overcomes Resistance to p53 Activation by Shifting Cellular Response from G1 Arrest to Apoptosis.
(A) Apoptosis percentage of MV-4-11, MOLM-13, and OCI-AML3 cells after treatment with indicated concentrations of RG for 12, 24, or 48 h.
(B) Relative mRNA expression of CDKN1A (p21-encoding gene) in OCI-AML3 cells after 12 hr treatment with 1 μM RG.
(C) Immunoblots of p53 and p21 in OCI-AML3 cells after 24 hr treatment with indicated concentrations of RG.
(D) The cell numbers and viability of OCI-AML3 cells treated with 1 μM RG as indicated. Cells were analyzed with Vi-CELL viability analyzer (Trypan blue exclusion assay).
(E) Immunoblots showing stable p21 knockdown in OCI-AML3 cells. Due to the low basal expression of p21, the control (shC) or knockdown cells were treated with 1 μM RG for 12 hr to induce p21 prior to immunoblot analysis.
(F) Apoptosis of the control and p21 knockdown OCI-AML3 cells after 48 hr treatment with indicated concentrations of RG.
(G) Flow cytometry plots showing the gating strategy to simultaneously analyze apoptosis and cell cycle distribution. The absolute cell numbers were enumerated using CountBright counting beads.
(H) Representative flow cytometry plots of OCI-AML3 cells after 24 hr treatment. The gating strategy was the same as in (G).
(I) Apoptosis and cell cycle analysis of OCI-AML3 cells treated with RG in the absence or presence of ABT for 24 hr. The gating strategy was the same as in (G).
(J) Cleavage of caspase-3, -9 and PARP-1 in response to 24 hr treatment with DMSO (vehicle), 1 μM RG, 1 μM ABT, or the ABT/RG combination. c-, cleaved.
Data in the line/bar graphs (A, B, D, F, I) represent the means of triplicate experiments. Error bars, mean ± SD. See also Figure S5.
The apoptotic threshold can be directly modulated up or down by Bcl-2 family proteins. OCI-AML3 expresses high levels of Bcl-2 and Mcl-1, both of which contribute to its high apoptotic threshold. As mentioned above, Bcl-2 protein levels were not affected by p53 activation or knockdown. Although Mcl-1 levels decreased upon p53 activation, the high levels of Bcl-2 may still be sufficient to keep most cells below the apoptotic threshold (We previously showed that knockdown of Mcl-1 by 85% did not significantly affect the viability or proliferation of OCI-AML3 cells (Pan et al., 2014; Pan et al., 2015)). Under this scenario, cells would preferentially enter G1 arrest in response to p53–induced p21 rather than undergo apoptosis. Based on these, we proposed that lowering the apoptotic threshold by Bcl-2 inhibition might be sufficient to switch the cellular response of p53 activation from pro-survival G1 arrest to apoptotic cell death. To test this hypothesis, we developed a protocol to simultaneously analyze apoptosis (AnnV+) and cell cycle distribution of AnnV− live cells (Figure 6G). Counting beads were also added to quantify cell numbers in different mitotic phases. As shown in Figures 6H and 6I, p53 activator RG triggered little apoptotic cell death but arrested most cells in G1 phase. Although ABT by itself did not affect cell cycle distribution, lowering apoptotic threshold by ABT substantially reduced RG-induced G1 arrest and augmented RG-induced apoptosis. Moreover, adding ABT to RG markedly increased the cleavage of caspase-9, caspase-3, and PARP-1 (Figure 6J), which further supported our hypothesis that Bcl-2 inhibition could overcome resistance to p53 activation by switching the cellular response from pro-survival G1 arrest to apoptosis. Consistently, in another two AML cell lines MV-4-11_Mcl-1 and MOLM-13_Mcl-1, p53 activator RG strongly induced p21 (Figures S5C) and arrested the majority of cells in G1 phase (Figure S5D). Co-treatment with ABT significantly shifted the cellular response from G1 arrest to apoptotic cell death (Figure S5D), despite the fact that ABT treatment alone did not induce much apoptosis in the two cell lines.
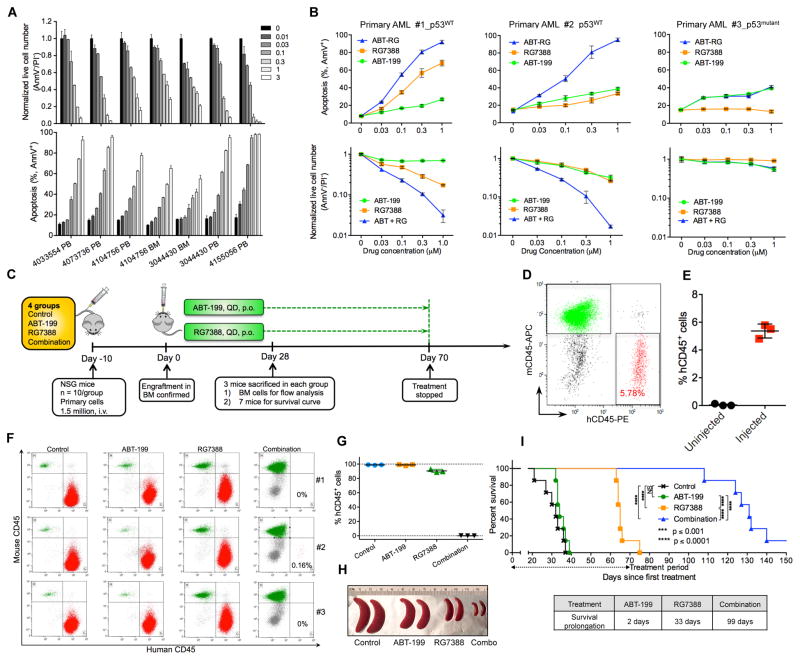
p53 Activation and Bcl-2 Inhibition Reciprocally Overcome Resistance of Primary AML Cells
p53 activator RG effectively induced apoptosis and reduced live cell numbers in p53WT patient samples (7/7 tested; Figure 7A). Consistent with our results in cell lines, RG could overcome ABT resistance in p53WT but not in p53-deficient primary cells (Figure 7B). Notably, one p53WT primary sample was highly resistant to both ABT and RG, as 48 hr treatment with 1 μM of either compound merely induced a marginal increase of apoptosis (Figure 7B, middle panels). Nonetheless, the combination could overcome the resistance and induced massive apoptotic cell death in this primary sample.
Figure 7. Bcl-2 Inhibition and p53 Activation Reciprocally Overcome Resistance of AML Primary Samples Both In Vitro and in a PDX Mouse Model.
(A) Apoptosis and live cell numbers of p53WT primary AML samples after treatment with indicated concentrations of RG (μM) for 48 hr. PB, peripheral blood. BM, bone marrow.
(B) Apoptosis and live cell numbers of three ABT-resistant primary samples after treatment with indicated concentrations of ABT, RG, or the combination for 48 hr.
(C) Schematic outline of the PDX mouse model of drug resistance.
(D) Representative flow cytometry plot showing engraftment of human AML cells (hCD45+) in murine femur BM on day 0 as in (C).
(E) Percentage of hCD45+ leukemic cells in murine BM on day 0 as in (C). The percentage of hCD45+ cells was calculated as: % hCD45+ cells = the number of hCD45+ cells/the sum of hCD45+ and mCD45+ cells.
(F) Flow cytometry analysis of hCD45+ leukemic cells in femur BM on day 28 as shown in (C). Three mice from each group were sacrificed and examined.
(G, H) Percentage of hCD45+ cells in femur BM (G) and images of representative spleens from sacrificed mice (H) on day 28 as in (C).
(I) Kaplan-Meier survival curves of mice engrafted with resistant primary AML cells (n = 7 per group). Statistical significance was evaluated using the log-rank test. NS, not significant.
Data in panels A, B, E, and G represent the means of triplicate experiments. Error bars, mean ± SD.
To test the combination in a patient-derived xenograft (PDX) model, we injected NSG mice with one resistant patient sample (p53WT) which could rapidly kill injected mice within few weeks (Figure 7C for experimental design). To gauge the level of engraftment, we sacrificed one extra group of three mice 10 days after injection and found that the BM (3/3) was uniformly engrafted with ~5% human CD45+ cells (Figures 7D and 7E). The treatment was then initiated as formerly described. After 4 weeks, three mice from each group were euthanized and examined. As shown in Figures 7F and 7G, BM from the control, ABT, or RG groups was heavily infiltrated with leukemic cells (>85% by flow cytometry analysis), indicating this patient sample was resistant to both ABT and RG. In contrast, the combinatorial treatment reduced leukemic cells to 0.16% in the BM of one mouse and to undetectable levels in the other two mice (Figure 7F). ABT or RG as monotherapy slightly or moderately decreased spleen size, whereas the combination dramatically reduced spleen size comparable to that of healthy mice (Figure 7H). Bcl-2 inhibitor ABT extended median survival by only 2 days, but combining ABT with RG substantially prolonged mouse survival by 99 days (Figure 7I). Collectively, these in vitro and in vivo data indicate that the combination of Bcl-2 inhibition (ABT) and p53 activation (RG) could cooperatively overcome apoptosis resistance to either treatment alone in primary AML cells.
In the three different types of ABT-resistant mouse models (Figures 2E, 3G, and 7C), p53 activator RG moderately prolonged mouse survival, suggesting that the three types of engrafted cells were also relatively resistant to p53 activation. Bcl-2 inhibitor ABT by itself prolonged mouse survival only marginally or not at all. Nonetheless, combining the two extended median survival at least 3-fold compared to RG alone in all three resistant mouse models. These in vivo data, together with our in vitro results and mechanistic studies, demonstrate that Bcl-2 inhibition and p53 activation can reciprocally overcome apoptosis resistance to either strategy alone.
DISCUSSION
Drug resistance is always a potential concern with targeted therapeutics. Most targeted compounds focus on upstream nodes of cancer signaling pathways. Although the initial response might be promising, resistance is often inevitable due to various compensatory mechanisms mediated by complex cancer signaling networks (Chandarlapaty, 2012; Pazarentzos and Bivona, 2015; Trusolino and Bertotti, 2012). For most cancer treatments, including chemotherapy, radiotherapy, and targeted therapies, induction of apoptosis represents the principal mechanism by which malignant cells are eliminated (Delbridge et al., 2016; Hata et al., 2015; Letai, 2008). Therefore, targeting the downstream apoptosis machinery appears to be a promising strategy since it directly elicits cell death and potentially minimizes the development of drug resistance. Bcl-2 family proteins and p53 are central regulators of apoptosis machinery. In this study, we found that p53 activation combined with Bcl-2 inhibition achieved impressive anti-leukemia efficacy in vitro and in vivo. The combination of RG and ABT was able to overcome resistance to either treatment alone, and significantly prolonged survival in three AML mouse models of resistance. These encouraging results provided the rationale for a multi-center phase II clinical trial in AML (NCT02670044). Actually, the proposed concept is not limited to AML. Other hematological malignancies such as myelodysplastic syndromes, multiple myeloma, chronic myeloid leukemia, acute lymphocytic leukemia, and chronic lymphocytic leukemia also have low p53 mutation rates and express high levels of Bcl-2. They might also respond well to the ABT/RG combination strategy. Additionally, several compounds have been reported to promote the proper folding of mutant p53 and restore its transcriptional function (Gurpinar and Vousden, 2015; Hoe et al., 2014). Combining these compounds with Bcl-2 inhibitors might represent a promising direction for p53 mutant cancers. We also found that low doses of ABT/RG synergized well with cytarabine and idarubicin (our unpublished data), revealing its potential to be combined with AML front-line therapeutics.
Anti-apoptotic Mcl-1 has been increasingly recognized as an important target in cancer therapy (Belmar and Fesik, 2015; Czabotar et al., 2014; Hata et al., 2015). Mcl-1 protein has a short half-life and can be cleaved by activated caspases during apoptotic cell death (Opferman, 2006). A number of studies observed Mcl-1 reduction after treatment with different anti-tumor therapeutics; but the Mcl-1 decrease was often accompanied by substantial levels of apoptosis. Therefore, it is ambiguous whether the observed Mcl-1 decrease was the cause of apoptosis or merely a secondary effect of apoptotic cell death. In the current study, we observed that p53 activation induced Mcl-1 reduction prior to apoptotic cell death and that p53 knockdown upregulated Mcl-1 while both the control and knockdown cells were highly viable. Moreover, we illustrated the underlying mechanism that p53 activation regulated MAPK/GSK3 signaling to modulate the phosphorylation, ubiquitination, and degradation of Mcl-1. These findings jointly support the negative regulation of Mcl-1 by p53 and exclude the possibility that the observed Mcl-1 reduction is merely a consequence of apoptosis.
Notably, ABT-sensitive cell lines such as MV-4-11 and MOLM-13 are not appropriate for studying Mcl-1 stability in response to ABT treatment, because presumably ABT can rapidly induce apoptosis of these cells, which leads to inevitable degradation of Mcl-1 by activated caspases. Indeed, in both MV-4-11 and MOLM-13 parental cells, 24 hr treatment with 1 μM ABT significantly reduced Mcl-1 levels (data not shown). To avoid this pitfall, we selected ABT-resistant OCI-AML3 cells for the mechanistic studies. To examine whether our mechanistic findings in OCI-AML3 cells are applicable to other cells, we also overexpressed Mcl-1 in ABT-sensitive MV-4-11 cells to generate ABT-resistant MV-4-11_Mcl-1 cells. In contrast to Mcl-1 reduction in parental cells, we observed further upregulation of Mcl-1 in MV-4-11_Mcl-1 cells after ABT treatment. This finding is consistent with the results in OCI-AML3 cells. Meanwhile, using the ABT-resistant MV-4-11_Mcl-1 cells, we also validated our mechanistic findings that p53 activation modulated the Ras/Raf/MEK/ERK/GSK3 signaling cascade, converted ABT–induced pMcl-1T to pMcl-1T/S, and abrogated ABT–induced upregulation of Mcl-1.
Biological outcomes of p53 activation can be determined by different cellular thresholds for CCA and apoptosis (Chen et al., 1996; Kracikova et al., 2013; Kruiswijk et al., 2015). For tumor cells with high apoptotic threshold, p53 activity might not be enough to push them through their apoptotic threshold but may induce sufficient cell cycle regulatory proteins to trigger reversible CCA, which may protect tumor cells from apoptotic cell death. To minimize CCA–mediated resistance, we proposed to convert the cellular response from CCA to apoptosis by reducing apoptotic threshold using Bcl-2 inhibitor ABT. Paradoxically, ABT induced an unwelcome increase of Mcl-1, which may further raise the apoptotic threshold. Given that a low dose of ABT (i.e., 10 nM) is sufficient to upregulate Mcl-1, special attention should be given to possible Mcl-1 elevation when using ABT in the laboratory or clinic. Nevertheless, in the current study, the Mcl-1 elevation could be reversed by p53 activator RG. In this scenario, Bcl-2 inhibition by ABT could effectively reduce the apoptotic threshold and switch the outcomes of p53 activation from pro-survival G1 arrest to pro-death apoptosis. In addition, we and others have shown that AML and other hematologic malignancies widely overexpress Bcl-2 (Coustan-Smith et al., 1996; Iqbal et al., 2006; Pan et al., 2014; Reed, 2008). This may explain why in this study, ABT almost always synergized with RG in p53WT cell lines, in p53WT primary samples, and in three mouse models of resistant AML.
In summary, we demonstrated that p53 activation overcomes the resistance to Bcl-2 inhibition by regulating the MAPK/GSK3 signaling to promote Mcl-1 degradation. Meanwhile, Bcl-2 inhibition reciprocally overcomes apoptosis resistance to p53 activation by shifting the cellular response from pro-survival G1 arrest to apoptosis. The mechanistic studies, together with the in vitro and in vivo efficacy, strongly support future research and clinical trials testing the combinations of Bcl-2 inhibitors with p53-reactivating compounds.
STAR METHODS
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-p53 (clone DO-1) | Santa Cruz Biotechnology | Cat#sc-126; RRID: AB_628082 |
| Mouse monoclonal anti-MDM2 (clone D-12) | Santa Cruz Biotechnology | Cat#sc-5304; RRID: AB_627918 |
| Mouse monoclonal anti-Bcl-2 (clone C-2) | Santa Cruz Biotechnology | Cat#sc-7382; RRID: AB_626736 |
| Rabbit polyclonal anti-Mcl-1 (clone S-19) | Santa Cruz Biotechnology | Cat#sc-819; RRID: AB_2144105 |
| Rabbit polyclonal anti-Puma (clone H-136) | Santa Cruz Biotechnology | Cat#sc-28226; RRID: AB_2064827 |
| Mouse monoclonal anti-ERK (cloned D-2) | Santa Cruz Biotechnology | Cat#sc-1647; RRID: AB_627547 |
| Mouse monoclonal anti-N-Ras (clone F-155) | Santa Cruz Biotechnology | Cat#sc-31; RRID: AB_628041 |
| Mouse monoclonal anti-GSK3 (clone 0011-A) | Santa Cruz Biotechnology | Cat#sc-7291; RRID: AB_2279451 |
| Rabbit polyclonal anti-PARP-1 | Cell Signaling Technology | Cat#9542; RRID: AB_2160739 |
| Rabbit polyclonal anti-Bim | Cell Signaling Technology | Cat#2819; RRID: AB_10692515 |
| Rabbit monoclonal anti-Bak (clone D4E4) | Cell Signaling Technology | Cat#12105 |
| Rabbit monoclonal anti-cleaved caspase-3 (D175) (clone 5A1E) | Cell Signaling Technology | Cat#9664; RRID: AB_2070042 |
| Mouse monoclonal anti-caspase-9 (clone C9) | Cell Signaling Technology | Cat#9508; RRID: AB_10695598 |
| Rabbit monoclonal anti-pMcl-1 (T163) (clone D5M9D) | Cell Signaling Technology | Cat#14765 |
| Rabbit monoclonal anti-B-Raf (clone 55C6) | Cell Signaling Technology | Cat#9433; RRID: AB_ 2259354 |
| Rabbit polyclonal anti-pB-Raf (S445) | Cell Signaling Technology | Cat#2696; RRID: AB_390721 |
| Rabbit polyclonal anti-JNK | Cell Signaling Technology | Cat#9252; RRID: AB_2250373 |
| Mouse monoclonal anti-pJNK (T183Y185) (clone G9) | Cell Signaling Technology | Cat#9255; RRID: AB_2307321 |
| Rabbit polyclonal anti-Ubiquitin | Cell Signaling Technology | Cat#3933; RRID: AB_2180538 |
| Rabbit monoclonal anti-pERK (T202/Y204) (clone D13.14.4E) | Cell Signaling Technology | Cat#4370; RRID: AB_2315112 |
| Rabbit polyclonal anti-pMEK (S217/S221) | Cell Signaling Technology | Cat#9121; RRID: AB_331648 |
| Mouse monoclonal anti-MEK (clone L38C12) | Cell Signaling Technology | Cat#4694; RRID: AB_10695868 |
| Mouse monoclonal anti-AKT (clone 40D4) | Cell Signaling Technology | Cat#2920; RRID: AB_1147620 |
| Rabbit polyclonal anti-pAKT(S473) | Cell Signaling Technology | Cat#9271; RRID: AB_329825 |
| Rabbit monoclonal anti-pGSK3 (S21/S9) (clone D17D2) | Cell Signaling Technology | Cat#8566; RRID: AB_10860069 |
| Mouse monoclonal anti-Bax (clone 2D2) | Sigma-Aldrich | Cat#B8554; RRID: AB_258645 |
| Mouse monoclonal anti-β-Actin (clone AC-74) | Sigma-Aldrich | Cat#A2228; RRID: AB_476697 |
| Mouse monoclonal anti-Noxa (clone 114C307) | Abcam | Cat#ab13654; RRID: AB_300536 |
| Rabbit polyclonal anti-pMcl-1 (S159) | Abcam | Cat#ab111574; RRID: AB_10865697 |
| Mouse monoclonal anti-p21WAF1 (clone EA10) | EMD Millipore | Cat#OP64; RRID: AB_2335868 |
| IRDye® 680RD Donkey anti-Rabbit IgG (H + L) | Li-COR Biosciences | Cat#925-68073 |
| IRDye® 800CW Donkey anti-Mouse IgG (H + L) | Li-COR Biosciences | Cat#925-32212 |
| Mouse monoclonal anti-human CD45 (clone PD7/26 + 2B11) | DAKO | Cat#M0701; RRID: AB_2314143 |
| Mouse monoclonal anti-human CD45-PE (clone HI30) | BD Biosciences | Cat#555483; RRID: AB_395875 |
| Rat monoclonal anti-mouse CD45-APC (clone 30-F11) | BD Biosciences | Cat#559864; RRID: AB_398672 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| RG7388 (idasanutlin) | Roche/Genentech | N/A |
| ABT-199 (venetoclax/GDC-0199) | AbbVie | N/A |
| A-1210477 | AbbVie | N/A |
| Annexin V, FITC conjugate | BD Biosciences | Cat#556419 |
| Annexin V, APC conjugate | BD Biosciences | Cat#550474 |
| Annexin V, Alexa Fluor 488 conjugate | Thermo Fisher | Cat#A13201 |
| D-Luciferin Firefly Potassium Salt | Gold Biotechnology | Cat#LUCK-1G CAS#115144-35-9 |
| Protease/Phosphatase Inhibitor Cocktail (100X) | Cell signaling | Cat#5872 |
| PD0325901 | Selleckchem | Cat#S1036 |
| Trametinib | Selleckchem | Cat#S2673 |
| Binimetinib | Selleckchem | Cat#S7007 |
| CHIR-99021 | Selleckchem | Cat#S1263 |
| Bortezomib | Selleckchem | Cat#S1013 |
| Ixazomib | Selleckchem | Cat#S2180 |
| BIO-Acetoxime | Selleckchem | Cat#S7915 |
| MK-2206 2HCl | Selleckchem | Cat#S1078 |
| Critical Commercial Assays | ||
| Click-iT EdU Alexa Fluor 647 Imaging Kit | Thermo Fisher | Cat#C10340 |
| FxCycle PI/RNase Staining Solution | Thermo Fisher | Cat#F10797 |
| Q5 Site-Directed Mutagenesis Kit | New England BioLabs | Cat#E0554S |
| Q5 Hot Start High-Fidelity 2X Master Mix | New England BioLabs | Cat#M0494S |
| jetPRIME Versatile Transfection Reagent | Polyplus | Cat#114-15 |
| Accell siRNA Delivery Media | Dharmacon | Cat#B-005000-500 |
| QIAprep Spin Miniprep Kit | QIAGEN | Cat#27104 |
| Wizard® SV Gel and PCR Clean-Up System | Promega | Cat#A9281 |
| Experimental Models: Cell Lines | ||
| Human: OCI-AML3 | DSMZ | Cat#ACC-582; RRID: CVCL_1844 |
| Human: MV-4-11 | ATCC | Cat#CRL-9591; RRID: CVCL_0064 |
| Human: MOLM-13 | DSMZ | Cat#ACC-554; RRID: CVCL_2119 |
| Human: HL-60 | ATCC | Cat#CCL-240; RRID: CVCL_0002 |
| Human: KG1 | ATCC | Cat#CCL-246; RRID: CVCL_0374 |
| Human: THP1 | ATCC | Cat#TIB-202; RRID: CVCL_0006 |
| Human: HEK293T | ATCC | Cat#CRL-3216; RRID:CVCL_0063 |
| Experimental Models: Organisms/Strains | ||
| Mice: NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ | The Jackson Laboratory | Strain: 005557; RRID: IMSR_JAX:005557 |
| Oligonucleotides | ||
| pLKO.1-TRC shRNA negative control: ccgcaggtatgcacgcgt | Moffat et al., 2006 A gift from David Root | Addgene Plasmid #10879 |
| pMKO.1 puro p53 shRNA 2: gactccagtggtaatctacttcaagagagtagattacactggagtcttttt | Masutomi et al., 2003 A gift from William Hahn | Addgene Plasmid #10672 |
| shRNA targeting human MCL1: ccggtgtgaattcatgggctcatcttcaagagagatgagcccatgaattcacttttttgg | This paper | N/A |
| TRC lentiviral human CDKN1A shRNA: ccagcactcttaggaacctct (#1) | Dharmacon | Cat#RHS3979-200795837 Clone ID: TRCN0000010401 |
| TRC lentiviral human CDKN1A shRNA: taaggcagaagatgtagagcg (#2) | Dharmacon | Cat#RHS3979-201789662 Clone ID: TRCN0000040123 |
| Accell siRNA non-targeting control pool | Dharmacon | Cat#D-001910-10 |
| Accell MAPK1 siRNA SMART pool | Dharmacon | Cat#E-003555-00 |
| Accell MAPK3 siRNA SMART pool | Dharmacon | Cat#E-003592-00 |
| Accell GSK3B siRNA SMART pool | Dharmacon | Cat#E-003010-00 |
| Accell GSK3A siRNA SMART pool | Dharmacon | Cat#E-003009-00 |
| Recombinant DNA | ||
| pCDH-EF1-MCS-BGH-PGK-GFP-T2A-Puro | System Biosciences | Cat#CD550A-1 |
| pLKO.1 Vector | Moffat et al., 2006 | Addgene Plasmid #10878 |
| psPAX2 Plasmid | A gift from Didier Trono | Addgene Plasmid #12260 |
| pMD2.G Plasmid | A gift from Didier Trono | Addgene Plasmid #12259 |
| Software and Algorithms | ||
| Image Studio Lite Software | LI-COR Biosciences | https://www.licor.com/bio/products/software/image_studio_lite/ |
| Prism Software v6.0 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| CalcuSyn 2.0 Software | Biosoft | http://www.biosoft.com/w/calcusyn.htm |
| Kaluza Analysis Software 1.3 | Beckman Coulter | https://www.beckman.com/coulter-flow-cytometers/software/kaluza |
| CellSens Software | OLYMPUS | https://www.olympus-lifescience.com/en/software/cellsens/ |
| Other | ||
| CountBright™ Absolute Counting Beads | Thermo Fisher Scientific | Cat#C36950 |
| Xenogen IVIS-200 In Vivo Imaging System | PerkinElmer | N/A |
| Odyssey Infrared Imaging System | LI-COR | N/A |
| BX43 Upright Microscope with DP72 Digital Color Camera | OLYMPUS | N/A |
| Vi-CELL XR Viability Analyzer | Beckman Coulter | Mode# 731050 |
| Gallios 10 Colors/3 Lasers Flow Cytometer (561 with Kaluza Acquisition Software) | Beckman Coulter | Mode# B43618 |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled according to institutional rules by the Lead Contact, Michael Andreeff (mandreef@mdanderson.org).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
All animal studies were performed in accordance with protocols approved by MD Anderson Cancer Center Institutional Animal Care and Use Committees. Six-week-old female NOD SCID gamma (NSG) mice (Jackson Laboratory, Bar Harbor, ME, USA) were intravenously injected with AML cell lines or primary patient cells as indicated in the schematic outlines for each mouse model (Figures 2E, 3G, and 7C). After confirmation of engraftment, the mice were randomly assigned to four groups (n = 10 per group) and treated with vehicle, ABT (100 mg/kg), RG (100 mg/kg) or the combination daily by oral gavage. ABT was prepared weekly in 10% ethanol, 30% polyethyleneglycol-400, and 60% phosal 50 propylene glycol (provided by AbbVie). RG was formulated daily in 4% DMA, 30% PEG400 and 66% Gelucire 44-14 (provided by Roche/Genentech). The leukemic burden of OCI-AML3 or MOLM-13-R cells was monitored by bioluminescence imaging at multiple time points. Briefly, mice were anesthetized and injected intraperitoneally with firefly luciferase substrate D-luciferin (Gold Biotechnology, St. Louis, MO, USA) and then imaged with the IVIS-200 in vivo imaging system (PerkinElmer, Waltham, MA, USA). After treatment for the indicated time, three mice were randomly selected from each group and euthanized by CO2 asphyxiation. Left femurs, spleens, and livers were harvested for IHC staining with human CD45 antibody. For the PDX mouse model of resistance (Figure 7C), BM cells from the right femurs were also collected and stained with human CD45-PE and murine CD45-APC antibodies (BD Biosciences) followed by flow cytometry analysis (Figure 7F). The remaining seven mice in each group were monitored for survival.
Patient Samples
Primary samples from AML patients at The University of Texas MD Anderson Cancer Center were obtained with informed consent from all subjects according to protocols approved by the Institutional Review Board. Mononuclear cells were isolated from peripheral blood or bone marrow samples by Ficoll density centrifugation. The primary cells were used immediately or resuspended in 90% FBS plus10% DMSO and viably cryopreserved in liquid nitrogen for future use. The isolated cells were treated with different compounds or combinations in RPMI-1640 medium supplemented with 10% FBS (final concentration of DMSO vehicle < 0.1%) at 37°C in a humidified atmosphere containing 5% CO2. After 48 hr, cell viability was determined by flow cytometry following concurrent AnnV and PI staining. Samples with a spontaneous apoptosis rate > 30% were excluded from the analysis.
Cell Lines
AML and other cell lines were purchased from the American Type Culture Collection (Manassas, VA, USA) or Deutsche Sammlung von Mikroorganismen und Zellkulturen (Braunschweig, Germany) and maintained according to the vendors’ instructions. OCI-AML3, MOLM-13, MV-4-11, KG1, HL-60 and THP1 were validated by short tandem repeat DNA fingerprinting using the Amp-FlSTR Identifier kit according to the manufacturer’s instructions (Thermo Fisher Scientific, Waltham, MA). Cells were cultured at 37°C in a humidified atmosphere containing 5% CO 2. Exponentially growing cells were used for all in vitro and in vivo studies.
METHOD DETAILS
Apoptosis, Live Cell Number, and IC50 Values
Exponentially growing cells were treated with different compounds or dimethyl sulfoxide (DMSO) as indicated in the figures and figure legends. DMSO served as the drug vehicle, and its final concentration was no more than 0.1%. After treatment, cells were collected, stained with annexin V (AnnV) and propidium iodide (PI; Sigma-Aldrich, St Louis, MO, USA), and analyzed by flow cytometry. Briefly, cells were washed twice by centrifugation (1,000 g, 5 min) with 2 mL AnnV binding buffer (ABB; 10 mM HEPES, 140 mM NaCl, and 5 mM CaCl2 at pH 7.4). After washing, cells were resuspended in 0.1 mL ABB containing fluorochrome-conjugated AnnV and incubated in darkness at room temperature for 15 min. The cells were washed again with 2 mL ABB and then resuspended in 0.3 mL ABB. PI was added at a final concentration of 1 μg/mL before analysis by a Gallios flow cytometer (Beckman Coulter, Indianapolis, IN, USA). To determine absolute cell number, CountBright Absolute Counting Beads (Life Technologies, Carlsbad, CA, USA) were also added. Data were analyzed by Kaluza software version 1.3 (Beckman Coulter). Calcusyn 2.0 software (Biosoft, Great Shelford, UK) was used to calculate IC50 values and combination index, based on the absolute number of live cells (i.e., AnnV−/PI−). AnnV-FITC and AnnV-APC were purchased from BD Biosciences (San Jose, CA, USA).
Immunoblotting and Antibodies
Cells were subjected to lysis at a density of 1 × 106/50 μL in cell lysis buffer (0.25 M Tris-HCl, 2% sodium dodecylsulfate, 4% β-mercaptoethanol, 10% glycerol, 0.02% bromophenol blue) supplemented with 1x protease/phosphatase inhibitor cocktail (#5872, Cell Signaling Technology, Beverly, MA, USA). Cell lysates were then loaded onto polyacrylamide gels with sodium dodecyl sulfate (Bio-Rad, Hercules, CA, USA). After electrophoresis, proteins were transferred to polyvinylidene difluoride membranes. The transblotted membranes were blocked for 1 hr and then probed with appropriate primary antibodies (dilution as recommended by manufacturers) overnight at 4°C. Next, the membranes were washed three times for a total of 30 min and then incubated with secondary antibodies in darkness at room temperature for 1 hr. After another three washes, Odyssey infrared imaging system and companion software (LI-COR Biosciences, Lincoln, NE, USA) were used to scan immunoblot membranes and to quantify band intensity according to the manufacturer’s manual. The ratios of proteins of interest/loading control in treated samples were normalized to their counterparts in untreated cells. Antibodies for immunoblotting were purchased from the following sources: p53 (sc-126), MDM2 (sc-5304), Bcl-2 (sc-7382), Mcl-1 (sc-819), Puma (sc-28226), ERK (sc-1647), N-Ras (sc-31) and GSK3 (sc-7291) from Santa Cruz Biotechnology (Dallas, TX, USA); pERK (T202/Y204, 4370), pMEK (S217/S221, 9121), MEK (4694), AKT (2920), pAKT(S473, 9271), pGSK3 (S21/S9, 8566), PARP-1 (9542), Bim (2819), Bak (12105), caspase-3 (9664), caspase-9 (9508), pMcl-1 (T163, 14765), B-Raf (9433), pB-Raf (S445, 2696), JNK (9252) and pJNK (T183Y185, 9255) from Cell Signaling Technology; Bax (B8554) and β-Actin (A2228) from Sigma-Aldrich; Noxa (ab13654) and pMcl-1 (T163S159, ab111574) from Abcam; p21 (OP64) from EMD Millipore (Darmstadt, Germany). IRDye 680 donkey anti-rabbit and IRDye 800 donkey anti-mouse secondary antibodies (#925-68073 and #925-32212) were purchased from Li-COR Biosciences.
Gene Knockdown by shRNA
p53, Mcl-1 and p21 were knocked down by lentivirus delivering well-validated shRNAs in pLKO.1 vector (Masutomi et al., 2003; Moffat et al., 2006). In parallel, control cells were transduced with lentivirus delivering a hairpin targeting GFP. Briefly, lentivirus was prepared by co-transfection of log-phase HEK293T cells with an equimolar mix of transfer vector psPAX2 and packaging plasmid pMD2.G (Addgene) using jetPrime transfection reagent according to the manufacturer’s instructions (Polyplus, Illkirch, France). Lentiviral supernatants were harvested 48 hr after transfection by centrifugation (800 g, 10 min) at room temperature. The supernatant was then filtered through 0.45 μM surfactant-free cellulose acetate membranes to remove lentivirus-producing cells. Polybrene (Chemicon, Temecula, CA, USA) was added to a final concentration of 8 μg/mL, and the virus stock was used immediately to infect AML cell lines. Exponentially growing cells were resuspended at a concentration of 0.5 × 106 cells per mL of virus stock, transferred to 12-well tissue plates, and centrifuged for 45 min (1,300 g, 30°C). After a dding one volume of fresh virus stock, the cells were centrifuged again and incubated in a tissue culture incubator (37°C, 5% CO 2) for 1 hr. The cells were then washed twice with growth medium to remove the polybrene and returned to the tissue culture incubator. After two doubling times, infected cells were subjected to selection with 0.5 μg/mL puromycin (Invitrogen, San Diego, CA, USA). Generated knockdown cells were assessed by immunoblot using Mcl-1, p53 and p21 antibodies. Because of the low basal expression of p53 and p21 in OCI-AML3, MOLM-13 and MV-4-11 cells, the control and knockdown cells were treated with 1 μM RG for 12 hr to induce p53 and p21 expression prior to immunoblot analysis.
Overexpression of Mcl-1, ERK, GSK3 and Their Mutants
Protein expression cassettes were delivered into AML cells by lentiviral transduction. Briefly, DNA sequence encoding human Mcl-1, ERK, GSK3, and their mutants were inserted into the MCS site of a commercially available lentiviral transfer vector pCDH-EF1-MCS-BGH-PGK-GFP-T2A-Puro (System Biosciences, Mountain View, CA, USA). After verification of the construction by Sanger sequencing, lentivirus was prepared and used to transduce AML cells as described above. In parallel, control cells were transduced with lentivirus delivering the empty vector. Infected cells were selected with puromycin up to 2 μg/mL. Increased expression of protein of interest was verified by immunoblot analysis.
Transient Gene Knockdown by Accell siRNAs
Transient gene knockdown was conducted using Accell siRNA transfection. Accell siRNAs, Accell siRNA delivery media and Accell non-targeting control were all purchased from GE Dharmacon. Transfection was performed according to the manufacturer’s protocol. Briefly, log-phase AML cells were collected by centrifugation at 1,000 g for 5 min. Because residual serum affects the knockdown efficiency of Accell siRNAs, growth medium should be removed as much as possible. The pelleted cells were then resuspended in appropriate volume of Accell siRNA Delivery Media (#B-005000). Next, siRNAs were added to a final concentration of 1 μM and gently mixed (Note: siRNAs targeting different genes can be multiplexed). The cells were then incubated with Accell siRNAs at 37°C in a humidified atmosphere containing 5% CO2. After 48 hr incubation, growth media (RPMI-1640 with 10% FBS) was added back to half of the transfection volume. After another 24 hr, AML cells were harvested and lysed as descried in Immunoblotting and Antibodies. Gene knockdown were validated by immunoblotting.
Concurrent Apoptosis and Cell Cycle Analysis
Exponentially growing cells were treated with vehicle, ABT, RG or ABT/RG combination for the indicated time. Then EdU (10 mM in phosphate-buffered saline solution [PBS]) was added to a final concentration of 10 μM. After incubation under optimal growth conditions for 1 hr, cells were washed with 3 mL ice-cold ABB buffer and pelleted by centrifugation at 4°C (1,000 g, 5 min). Supernatant was carefully removed by aspiration without disturbing the cell pellet. The cells were then placed on ice, mixed with 50 μL ABB containing 5 μL AnnV-Alexa488 (Life Technologies) and incubated in darkness for 15 min. After staining, cells were washed again with ice-cold ABB. The cells were fixed, permeabilized and then incubated with Click-iT Plus reaction cocktail (Life Technologies) according to the manufacturer’s instructions. After the Click-iT reaction, cells were mixed with 0.5 mL of FxCycle PI/RNase solution (Life Technologies) by gentle vortexing. After 30 min incubation in darkness at 37°C, CountBright Counting Beads were added. The samples were then analyzed by flow cytometry for apoptosis (AnnV+) and cell cycle distribution of AnnV− live cells (Figure 5G for gating strategy).
Generation of ABT-Resistant Cells
Parental MOLM-13 and MV-4-11 cells were cultured in RPMI-1640 medium (10% fetal bovine serum [FBS]) supplemented with escalating doses of ABT (starting at IC50 doses for each cell line). Every 2 days, the cells were pelleted by centrifugation (1,000 g, 5 min) and then resuspended in fresh medium supplemented with ABT. Cell viability was monitored by Vi-CELL viability analyzer (Beckman Coulter). Once the viability reached ≥ 90%, ABT concentration was doubled. The generated cells were at least 30-fold more resistant than their parental cells (Figure 2A).
Detection of ubiquitinated proteins
Mcl-1 protein was immunoprecipitated as described by Choo et al.(Choo and Zhang, 2009). Briefly, 5 million viable cells were processed as follows: wash cells with ice-cold PBS once; lyse cells with 100 μL cell lysis buffer (10 mM Tris-HCl, pH 8.0, 2% SDS, 150 mM NaCl, and 1X protease/phosphatase inhibitors); heat cell lysates immediately at 95°C for 10 min; shear cell lysates with a sonication device; add 1.5 mL of dilution buffer (10 mM Tris-HCl, pH 8.0, 150 mM NaCl, 2 mM EDTA, 1% Triton); incubate the diluted samples at 4°C for 30 min with gentle rotation; spin the samples at ≥10,000 g for 10 min at 4°C to get rid of cell debris; transfer the supernatant to a new 1.5 mL microfuge tube; add recommended amount of primary antibodies to the supernatant; incubate with gentle rocking overnight at 4°C; add 50 μL protein A/G plus-agarose beads (#sc-2003, Santa Cruz Biotechnology); incubate the mixture overnight at 4°C with gentle rotation; spin down the beads at 5, 000 g for 5 min at 4°C to get rid of supernatant; wash the beads with washing buffer (10 mM Tris-HCl, pH 8.0, 1 M NaCl, 1 mM EDTA, 1% NP-40) three times; aspirate washing buffer and resuspend the pellets in SDS gel-loading buffer; heat the mixture at 95°C for 5 min; spin for 5 min at 12,000 g; load the supernatant onto a SDS-PAGE gel for immunoblotting analysis. Immunoprecipitated Mcl-1 from 1 million viable cells was analyzed by standard immunoblotting using an ubiquitin-specific antibody (#3933 from Cell Signaling Technologies).
Immunohistochemical (IHC) Analysis
IHC analysis of human CD45 was performed as previously described (Pan et al., 2014). Briefly, spleen, and liver tissue was formalin-fixed and paraffin-embedded. Bone tissue was also decalcified before embedding in paraffin. Paraffin blocks were then sectioned into 5 μm thick slices and mounted onto microscope slides. Tissue sections were deparaffinized and rehydrated using 100% xylene and ethanol solution of decreasing concentrations. The rehydrated tissue sections were next incubated with monoclonal mouse anti-human CD45 antibody (M0701, DAKO, Carpinteria, CA, USA), followed by sequential incubation with biotinylated secondary antibody, peroxidase-labeled streptavidin, and 3,3′ diaminobenzidine tetrahydrochloride/H2O2, which resulted in a brown precipitate at the antigen site. Images were acquired by an OLYMPUS BX43 microscope with DP72 digital color camera and companion software CellSens (Olympus, Upper Saucon Township, PA, USA).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analyses were conducted using Prism software v6.0 (GraphPad Software, La Jolla, CA, USA). The log-rank test was used to compare mouse survival curves. For in vitro studies, statistical significance was determined by the two-tailed unpaired Student’s t-test. A p value < 0.05 was considered statistically significant. For all figures, NS, not significant, *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001. The statistical parameters (i.e., exact value of n, p values) have been noted in the figures and figure legends. Unless otherwise indicated, all data represent the mean ± standard deviation from at least three independent experiments.
Supplementary Material
SIGNIFICANCE.
Drug resistance is a potential concern with targeted therapeutics. Here, we demonstrate that concurrent Bcl-2 inhibition and p53 activation induce synthetic lethality and achieve remarkable efficacy in three types of resistant leukemia models, both in vitro and in vivo. We illustrate the underlying mechanisms by which Bcl-2 inhibition and p53 activation reciprocally overcome resistance to either strategy alone. Our results provide important mechanistic insights as well as critical preclinical evidence supporting the use of p53–reactivating compounds in combination with Bcl-2 inhibitors in cancer patients, and warrant clinical trials to assess the clinical benefit of such combined therapy.
HIGHLIGHTS.
p53 regulates Ras/Raf/MEK/ERK/GSK3 signaling cascade to promote Mcl-1 degradation
Bcl-2 inhibition shifts the outcomes of p53 activation from G1 arrest to apoptosis
p53 activation and Bcl-2 inhibition reciprocally overcome apoptosis resistance
The combination markedly prolongs survival in three mouse models of resistant AML
Acknowledgments
This work was supported in part by the National Institutes of Health through grant CA-55164 (to M.A.) and through MD Anderson’s Cancer Center Support Grant CA016672. This study was also supported by the Paul and Mary Haas Chair in Genetics (to M.A.) and the Presidents’ Research Scholarship from MD Anderson Cancer Center and UT Health Science Center (to R.P.).
Footnotes
POTENTIAL CONFLICTS OF INTEREST
M.A. and M.K. have collaborative research agreements from Roche, AbbVie, and Genentech. J.D.L. has ownership interest in AbbVie. G.N. and J.C.R. have ownership interest (including patents) in Roche. M.K. is a consultant/advisory board member of AbbVie Inc. The remaining authors declare no competing financial interests.
AUTHOR CONTRIBUTIONS
R.P. conceived the study, designed and conducted the experiments, analyzed and interpreted the data, and wrote the manuscript; V.R. and H.M. conducted the experiments and edited the manuscript; J.D.L. and M.K. contributed to study design, interpretation of data and editing of the manuscript; G.N. and J.C.R. provided thoughtful inputs and edited the manuscript; M.A. conceived and supervised the study, interpreted the data, and edited the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Belmar J, Fesik SW. Small molecule Mcl-1 inhibitors for the treatment of cancer. Pharmacol Ther. 2015;145:76–84. doi: 10.1016/j.pharmthera.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandarlapaty S. Negative feedback and adaptive resistance to the targeted therapy of cancer. Cancer Discov. 2012;2:311–319. doi: 10.1158/2159-8290.CD-12-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Ko LJ, Jayaraman L, Prives C. p53 levels, functional domains, and DNA damage determine the extent of the apoptotic response of tumor cells. Genes Dev. 1996;10:2438–2451. doi: 10.1101/gad.10.19.2438. [DOI] [PubMed] [Google Scholar]
- Choo YS, Zhang Z. Detection of protein ubiquitination. Journal of visualized experiments: JoVE. 2009 doi: 10.3791/1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P, Frame S. The renaissance of GSK3. Nat Rev Mol Cell Biol. 2001;2:769–776. doi: 10.1038/35096075. [DOI] [PubMed] [Google Scholar]
- Coustan-Smith E, Kitanaka A, Pui CH, McNinch L, Evans WE, Raimondi SC, Behm FG, Arico M, Campana D. Clinical relevance of BCL-2 overexpression in childhood acute lymphoblastic leukemia. Blood. 1996;87:1140–1146. [PubMed] [Google Scholar]
- Czabotar PE, Lessene G, Strasser A, Adams JM. Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat Rev Mol Cell Biol. 2014;15:49–63. doi: 10.1038/nrm3722. [DOI] [PubMed] [Google Scholar]
- Delbridge AR, Grabow S, Strasser A, Vaux DL. Thirty years of BCL-2: translating cell death discoveries into novel cancer therapies. Nature reviews Cancer. 2016;16:99–109. doi: 10.1038/nrc.2015.17. [DOI] [PubMed] [Google Scholar]
- Ding Q, Xia W, Liu JC, Yang JY, Lee DF, Xia J, Bartholomeusz G, Li Y, Pan Y, Li Z, et al. Erk associates with and primes GSK-3beta for its inactivation resulting in upregulation of beta-catenin. Mol Cell. 2005;19:159–170. doi: 10.1016/j.molcel.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Ding Q, Zhang Z, Liu JJ, Jiang N, Zhang J, Ross TM, Chu XJ, Bartkovitz D, Podlaski F, Janson C, et al. Discovery of RG7388, a potent and selective p53-MDM2 inhibitor in clinical development. J Med Chem. 2013 doi: 10.1021/jm400487c. [DOI] [PubMed] [Google Scholar]
- Domina AM, Vrana JA, Gregory MA, Hann SR, Craig RW. MCL1 is phosphorylated in the PEST region and stabilized upon ERK activation in viable cells, and at additional sites with cytotoxic okadaic acid or taxol. Oncogene. 2004;23:5301–5315. doi: 10.1038/sj.onc.1207692. [DOI] [PubMed] [Google Scholar]
- Gores GJ, Kaufmann SH. Selectively targeting Mcl-1 for the treatment of acute myelogenous leukemia and solid tumors. Genes Dev. 2012;26:305–311. doi: 10.1101/gad.186189.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurpinar E, Vousden KH. Hitting cancers’ weak spots: vulnerabilities imposed by p53 mutation. Trends Cell Biol. 2015;25:486–495. doi: 10.1016/j.tcb.2015.04.001. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hata AN, Engelman JA, Faber AC. The BCL2 Family: Key Mediators of the Apoptotic Response to Targeted Anticancer Therapeutics. Cancer discovery. 2015;5:475–487. doi: 10.1158/2159-8290.CD-15-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- Hoe KK, Verma CS, Lane DP. Drugging the p53 pathway: understanding the route to clinical efficacy. Nature reviews Drug discovery. 2014;13:217–236. doi: 10.1038/nrd4236. [DOI] [PubMed] [Google Scholar]
- Iqbal J, Neppalli VT, Wright G, Dave BJ, Horsman DE, Rosenwald A, Lynch J, Hans CP, Weisenburger DD, Greiner TC, et al. BCL2 expression is a prognostic marker for the activated B-cell-like type of diffuse large B-cell lymphoma. J Clin Oncol. 2006;24:961–968. doi: 10.1200/JCO.2005.03.4264. [DOI] [PubMed] [Google Scholar]
- Juin P, Geneste O, Gautier F, Depil S, Campone M. Decoding and unlocking the BCL-2 dependency of cancer cells. Nat Rev Cancer. 2013;13:455–465. doi: 10.1038/nrc3538. [DOI] [PubMed] [Google Scholar]
- Konopleva M, Contractor R, Tsao T, Samudio I, Ruvolo PP, Kitada S, Deng X, Zhai D, Shi YX, Sneed T, et al. Mechanisms of apoptosis sensitivity and resistance to the BH3 mimetic ABT-737 in acute myeloid leukemia. Cancer Cell. 2006;10:375–388. doi: 10.1016/j.ccr.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Kracikova M, Akiri G, George A, Sachidanandam R, Aaronson SA. A threshold mechanism mediates p53 cell fate decision between growth arrest and apoptosis. Cell Death Differ. 2013;20:576–588. doi: 10.1038/cdd.2012.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruiswijk F, Labuschagne CF, Vousden KH. p53 in survival, death and metabolic health: a lifeguard with a licence to kill. Nat Rev Mol Cell Biol. 2015;16:393–405. doi: 10.1038/nrm4007. [DOI] [PubMed] [Google Scholar]
- Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- Letai AG. Diagnosing and exploiting cancer’s addiction to blocks in apoptosis. Nature reviews Cancer. 2008;8:121–132. doi: 10.1038/nrc2297. [DOI] [PubMed] [Google Scholar]
- Masutomi K, Yu EY, Khurts S, Ben-Porath I, Currier JL, Metz GB, Brooks MW, Kaneko S, Murakami S, DeCaprio JA, et al. Telomerase maintains telomere structure in normal human cells. Cell. 2003;114:241–253. doi: 10.1016/s0092-8674(03)00550-6. [DOI] [PubMed] [Google Scholar]
- Maurer U, Charvet C, Wagman AS, Dejardin E, Green DR. Glycogen synthase kinase-3 regulates mitochondrial outer membrane permeabilization and apoptosis by destabilization of MCL-1. Mol Cell. 2006;21:749–760. doi: 10.1016/j.molcel.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- Morel C, Carlson SM, White FM, Davis RJ. Mcl-1 integrates the opposing actions of signaling pathways that mediate survival and apoptosis. Mol Cell Biol. 2009;29:3845–3852. doi: 10.1128/MCB.00279-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opferman JT. Unraveling MCL-1 degradation. Cell Death Differ. 2006;13:1260–1262. doi: 10.1038/sj.cdd.4401978. [DOI] [PubMed] [Google Scholar]
- Pan R, Hogdal LJ, Benito JM, Bucci D, Han L, Borthakur G, Cortes J, Deangelo DJ, Debose L, Mu H, et al. Selective BCL-2 inhibition by ABT-199 causes on-target cell death in acute myeloid leukemia. Cancer Discov. 2014;4:362–375. doi: 10.1158/2159-8290.CD-13-0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan R, Ruvolo VR, Wei J, Konopleva M, Reed JC, Pellecchia M, Andreeff M, Ruvolo PP. Inhibition of Mcl-1 with the pan-Bcl-2 family inhibitor (-)BI97D6 overcomes ABT-737 resistance in acute myeloid leukemia. Blood. 2015;126:363–372. doi: 10.1182/blood-2014-10-604975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazarentzos E, Bivona TG. Adaptive stress signaling in targeted cancer therapy resistance. Oncogene. 2015;34:5599–5606. doi: 10.1038/onc.2015.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JC. Bcl-2-family proteins and hematologic malignancies: history and future prospects. Blood. 2008;111:3322–3330. doi: 10.1182/blood-2007-09-078162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souers AJ, Leverson JD, Boghaert ER, Ackler SL, Catron ND, Chen J, Dayton BD, Ding H, Enschede SH, Fairbrother WJ, et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat Med. 2013;19:202–208. doi: 10.1038/nm.3048. [DOI] [PubMed] [Google Scholar]
- Trusolino L, Bertotti A. Compensatory pathways in oncogenic kinase signaling and resistance to targeted therapies: six degrees of separation. Cancer Discov. 2012;2:876–880. doi: 10.1158/2159-8290.CD-12-0400. [DOI] [PubMed] [Google Scholar]
- Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- Weinberg RA. The biology of cancer. New York, NY: Garland Science, Taylor & Francis Group LLC); 2014. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.