Abstract
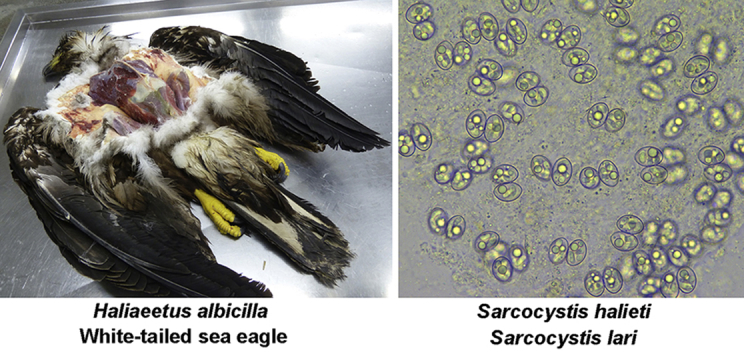
An emaciated white-tailed sea eagle (Haliaeetus albicilla) from Western Norway was found and nursed briefly before it died. The necropsy revealed that the principal cause of death was an inflammation and occlusion of the bile ducts. A secondary finding was the presence in the intestinal mucosa of numerous sporulated Sarcocystis oocysts measuring 21.8–22.8 × 16.0–17.0 μm. The aim of this study was to identify these oocysts to species level using molecular methods. Genomic DNA was extracted from 10 mucosal scrapings containing oocysts and subjected to PCR amplification and sequencing of four DNA regions: the 18S and 28S rRNA genes, the ITS1 region and the cox1 gene. DNA of three previously known Sarcocystis spp. was identified, but only two of these, Sarcocystis halieti n. sp. and Sarcocystis lari, both employing sea birds as intermediate hosts, were considered to have used the sea eagle as a definitive host and to have formed oocysts in its intestine. The third species found, Sarcocystis truncata, employs red deer as intermediate hosts and seems to use felids as definitive hosts based on its phylogenetic position and prevalence. The sea eagle had probably recently ingested portions of one of the latter hosts (red deer or cat/lynx) containing stages (sarcocysts/oocysts) and thus DNA of S. truncata. The species S. halieti and S. lari could only be unambiguously separated from their most closely related congeners on the basis of their ITS1 sequences. This is the first report of Sarcocystis oocysts in sea eagles and the first identification to species level of Sarcocystis oocysts in any type of eagle. The sea eagle also acted as intermediate host of an unidentified Sarcocystis spp. as evidenced by the finding of six thin-walled sarcocysts in a histological section of cardiac muscle.
Keywords: Sarcocystis, Haliaeetus albicilla, Oocysts, ITS1, Cox1, Phylogeny
Graphical abstract
Highlights
-
•
First record of the white-tailed sea eagle as definitive host of Sarcocystis spp.
-
•
Intestinal Sarcocystis oocysts molecularly characterized at four DNA markers.
-
•
Oocysts belonged to Sarcocystis lari and Sarcocystis halieti n. sp.
-
•
Sarcocysts of an unidentified species found in cardiac muscle of the sea eagle.
1. Introduction
Protozoans belonging to the genus Sarcocystis are intracellular parasites with an obligatory two-host life cycle comprising oocyst formation in the intestinal mucosa of their definitive hosts and a biphasic asexual multiplication in vascular endothelial cells and striated muscle cells, respectively, of their intermediate hosts. The latter hosts acquire a Sarcocystis infection through the accidental ingestion of oocysts/sporocysts in fecally contaminated feed or water, whereas definitive hosts get infected through ingestion of sarcocysts, mainly located in striated muscles, via predation or scavenging. Hence, mainly carnivorous animals (mammals, birds, reptiles), but also some omnivores, e.g., corvid birds (Gjerde and Dahlgren, 2010, Irie et al., 2017), act as definitive hosts, whereas herbivores, omnivores and carnivores may act as intermediate hosts.
More than 200 Sarcocystis spp. have been described and named on the basis of the morphology of the sarcocyst stage in the intermediate hosts, particularly in various domesticated mammals, but in recent years, an increasing number of Sarcocystis spp. have also been reported from various birds. The definitive hosts of most of the named Sarcocystis spp., including those with avian intermediate hosts, are, however, still unknown. Prior to the molecular era, the definitive host(s) of a given Sarcocystis sp. could only be determined through experimental infections, since the oocysts/sporocysts of various Sarcocystis spp. shed by their definitive hosts are morphologically indistinguishable. Through the use of molecular methods, comprising PCR amplification of the appropriate DNA regions (markers) followed by comparisons of the resulting nucleotide sequences, it has become possible to determine if the oocysts/sporocysts in the faeces or intestinal mucosa of naturally infected definitive hosts belong to particular Sarcocystis spp. that have already been similarly characterised from intermediate hosts or belong to species that have not yet been described or molecularly examined in the latter hosts. Hence, the appropriate use of molecular methods has largely obviated the need to perform expensive and ethically questionable transmission experiments in order to elucidate the complete life cycle of various Sarcocystis spp. Moreover, nucleotide sequences may be used to infer the phylogenetic relationship between different species and to predict the most likely definitive hosts of species for which this part of the life cycle is still unknown.
As regards Sarcocystis spp. with avian intermediate hosts, a few have been found to use mammalian carnivores as definitive hosts, like Sarcocystis falcatula using opossums (Box et al., 1984, Wünschmann et al., 2010), Sarcocystis rileyi using striped skunks, foxes and raccoon dogs (Cawthorn et al., 1981, Wicht, 1981, Prakas et al., 2015) and Sarcocystis albifronsi and possibly Sarcocystis anasi using foxes (Kutkienė et al., 2012a, Moré et al., 2016), whereas other Sarcocystis spp. have been found to use birds of prey (raptors) as definitive hosts. Černá and Kvašňovská (1986) were the first to report the latter type of bird–bird cycle after having observed sarcocysts in the muscle tissues of three canaries (Serinus canaria) fed sporocysts from the intestines of two goshawks (Accipiter gentilis). They named this species Sarcocystis accipitris after its definitive host. Similarly, Svobodová (1996) found sarcocysts in three great tits (Parus major) fed sporocysts from a goshawk, but, in the absence of molecular data at the time, they were unable to determine whether this species was identical to the aforementioned S. accipitris or not.
Experimental infections were also used by Olias et al. (2010a) to prove that Sarcocystis calchasi cycled between domestic pigeons (Columba livia) and goshawks, but they even employed molecular methods to verify that the sarcocysts and oocysts/sporocysts in the two host types belonged to the same species. In a subsequent study based solely on naturally infected birds, Olias et al. (2010b) showed through molecular methods that sparrow hawks (Accipiter nisus) acted as definitive hosts for Sarcocystis columbae in wood pigeons (Columba palumbus), as well as for another species with an unknown intermediate host, which in a later study was found to be near identical at the three genetic markers examined with Sarcocystis turdusi from the common blackbird (Turdus merula) (Kutkienė et al., 2012b). Using primer pairs specifically targeting S. calchasi, S. columbae and the still unnamed species (=S. turdusi), respectively, Olias et al. (2011) showed that oocysts of all three species were common in both goshawks and sparrow hawks in Germany, but they missed any other species that might have used these raptors as definitive hosts, since their primers were unable to detect them. In a similar survey of sparrow hawks and goshawks in Germany, Mayr et al. (2016) used both S. calchasi-specific and more general primers and confirmed that these hawks acted as definitive hosts for the aforementioned three species, but also for Sarcocystis cornixi from corvid birds (Kutkienė et al., 2009) and an unnamed Sarcocystis sp. that had been found in the great cormorant (Phalacrocorax carbo) in Lithuania (according to GenBank accession nos. JQ733511–JQ733513). Likewise, in a recent study from the United States, a Cooper's hawk (Accipiter cooperii) was found to act as definitive host of an unnamed species that was genetically most closely related to S. columbae (Lindsay et al., 2017). In an earlier study from the USA, Yabsley et al. (2009) obtained sequences of the partial 18S ribosomal (r) RNA gene from 10 intestinal samples from four species of hawks harbouring Sarcocystis oocysts/sporocysts, and found seven slightly different sequence variants (GenBank nos. EU810395–EU810402), which might represent separate species. However, subsequent studies have shown that various Sarcocystis spp. using birds as intermediate and/or definitive hosts differ very little, if at all, at this gene (Prakas et al., 2014), and hence cannot be reliably identified on the basis of this marker alone, and even less so when only partial sequences of the gene are available for comparison. It is therefore not possible to accurately identify the species found by Yabsley et al. (2009).
Thus, based primarily on sequences of the internal transcribed spacer 1 (ITS1) region of the nuclear ribosomal DNA unit, which seems to be the best marker for species delimitation within this group (Prakas et al., 2014), at least six Sarcocystis spp. with avian intermediate hosts have so far been identified in Accipiter hawks as outlined above. In addition, several other birds of prey are known to act as definitive hosts of Sarcocystis spp. with small rodents as natural intermediate hosts (Yabsley et al., 2009). The latter group comprises the two species initially assigned to the genus Frenkelia, that is, Frenkelia microti and Frenkelia glareoli, which are transmitted by buzzards of the genus Buteo within the family Accipitridae (Votýpka et al., 1998, Yabsley et al., 2009), as well as Sarcocystis spp., including Sarcocystis dispersa, using various owls (order: Strigiformes; families: Strigidae and Tytonidae) as definitive hosts (Černá et al., 1978, Kolárová, 1986).
The coincidental finding of Sarcocystis oocysts in the intestine of a diseased white-tailed sea eagle (Haliaeetus albicilla) submitted for necropsy provided the opportunity to identify these oocysts by molecular methods and thereby determine for the first time for which Sarcocystis spp. this large bird of prey within the family Accipitridae may act as a definitive host.
2. Materials and methods
2.1. Case history and major necropsy findings
On February 8, 2016, an emaciated young female white-tailed sea eagle was found in Oldedalen, Stryn municipality, Sogn og Fjordane county in Western Norway. The eagle was brought to a local ornithologist and environmental adviser, who had taken care of and rescued other injured wild birds previously. The eagle was force-fed a small amount of mutton (frozen and thawed), but died the following day. The carcass was then immediately shipped to the Norwegian Veterinary Institute, Oslo, Norway, for a post-mortem examination, which was performed on February 10, 2016, by one of the authors (T.V.) according to standard necropsy procedures (journal number: 2016–04–4079/V82). Prior to the necropsy, the carcass was X-rayed for the presence of bone fractures and shotgun pellets/bullet fragments. Tissue samples of the brain, heart, lungs, liver, kidneys, pancreas and a distended portion of the bile duct were fixed in 10% buffered formalin and processed routinely for histopathological examination, including staining of the sections (3 μm) with haematoxylin and eosin (HE). The contents of the distended bile duct were cultured for bacteria using calf blood agar plates, incubated aerobically at 37 °C for 24–48 h. A wet smear of the intestinal mucosa/contents was examined routinely for parasites under a light microscope.
The necropsy revealed that the eagle weighed 3.2 kg and was emaciated with severe muscle atrophy. There was only a small amount of ingested food in the gastrointestinal tract, including some remnants in the crop, proventriculus and gizzard of the mutton that had been fed to the eagle shortly before it died. A portion of the bile duct system was greatly distended, measuring about 4 cm in diameter, and contained a brownish semi-fluid material. Culturing of this material revealed a rich growth of Clostridium perfringens bacteria. In the adjacent bile ducts, the content was dry and the lumen was occluded. Histology revealed that the distended portion of the bile duct was heavily inflamed with necrosis, haemorrhages, infiltration of inflammatory cells and aggregations of large Gram-positive bacteria. In the pancreas, there was moderate fibrosis. In a histological section of cardiac muscle, six sarcocysts were found, but there was no inflammation associated with them. In the wet smear of the intestinal mucosa and contents, there were numerous Sarcocystis-like oocysts.
Based on the abovementioned findings, the major cause of the emaciation and death of the sea eagle was considered to have been inflammation and occlusion of the bile ducts with consequent blocking of the flow of bile to the intestine. It could not be determined if the bacterial colonization of the biliary ducts with C. perfringens was the primary cause of the cholangitis, or if it occurred secondary to this condition. The Sarcocystis infection in the intestinal mucosa was not believed to have contributed to the emaciation and death of the eagle, but this secondary finding will be the major topic of the remainder of this paper. Likewise, the presence of sarcocysts in cardiac muscle is not considered to have impacted the health of the eagle, but the finding is of importance from a parasitological point of view, and will therefore be further described in section 3.1.
2.2. Sampling and microscopic examination of the intestinal mucosa
Following the finding of numerous Sarcocystis-like oocysts in the wet smear examined routinely for parasites at the necropsy, the entire intestine was removed and transferred to the Parasitology lab. of the Veterinary Institute, where the intestinal content was examined in more detail under a light microscope the same day by one of the authors (I.S.H.). It was established that the mucosa contained numerous thin-walled sporulated Sarcocystis oocysts, as well as some free sporocysts, suggesting that the eagle had acted as a definitive host for one or more Sarcocystis spp. A few low-resolution digital photographs of the oocysts were taken and the intestine was then put in a freezer at −20 °C. About two weeks later, the first author (B.G.) was notified about the findings and it was decided to attempt to identify the oocysts by molecular methods. Hence, on February 24, 2016, the frozen intestine was transferred to the first author at the nearby Veterinary School, where, after thawing, 16 small portions of the mucosa were scraped off with a scalpel blade and transferred to separate 1.5-ml micro centrifuge tubes, each containing about 0.5 ml of distilled water. Most of the remaining mucosal lining was finally scraped off and transferred to a 50-ml screw cap tube containing 45 ml of distilled water. All the mucosal samples were kept frozen at −20 °C pending DNA extraction. During the sampling for molecular studies, a few small portions of the mucosa were also transferred to microscope slides and examined as wet mounts under a light microscope in order to take digital photographs of oocysts and sporocysts and measure their size. Moreover, a few small pieces of the mucosa were fixed in 10% neutral buffered formalin for histological examination, but by a mistake, the latter samples were discarded and not further processed. About eight months later, the pooled mucosal sample in the 50-ml tube was thawed, and small portions of the mucosa were transferred to two 1.5-ml tubes and washed several times in distilled water by repeated centrifugation. The sediment was finally transferred to microscope slides and examined as wet mounts under a light microscope and many additional digital photographs of the oocysts were recorded.
2.3. Molecular examination of mucosal samples
Genomic DNA was extracted on three separate occasions from a total of 10 samples of the intestinal mucosa containing Sarcocystis oocysts using the QIAmp DNA Mini Kit (Qiagen, Germany) according to the manufacturer's tissue protocol as previously described for sarcocysts (Gjerde, 2013). Two of the mucosal samples (Ha1.9, Ha1.10) were washed thoroughly by repeated centrifugation in several changes of distilled water before DNA extraction, whereas the other samples consisted of the mucosal scrapings as originally collected. The 10 resulting DNA samples (labelled Ha1.1–Ha1.10) were subsequently kept frozen at −20 °C in between the removal of aliquots to be used as templates for PCR amplifications.
Initially it was assumed that a single Sarcocystis sp. was present in the intestine of the eagle, and hence the complete 18S rRNA gene was amplified from two isolates (Ha1.1, Ha1.2) as two overlapping fragments using primer pairs ERIB1/S2r and S3f/Primer BSarc, whereas the ITS1 region and the partial 28S rRNA gene were amplified from the same two isolates using primer pairs SU1F/5.8SR2 and KL1/KL3, respectively, as previously described (Gjerde and Josefsen, 2015). The amplicons from all these reactions were sequenced directly, but the resulting sequence chromatograms turned out to be unintelligible or of poor quality, which seemed to be due to an overlap of sequences of two or more species. Surprisingly, the short identifiable sequence fragments matched previous sequences of Sarcocystis truncata from red deer (Dahlgren and Gjerde, 2010, Gjerde, 2014a). However, the concurrent sequencing of the partial mitochondrial cytochrome c oxidase subunit I gene (cox1) that had been PCR amplified from the same two isolates, resulted in two sequences that were 99.8% identical with each other, and which shared identities of 99.2–100% with published cox1 sequences (as of June 2016) of Sarcocystis lutrae from otters and an arctic fox, S. calchasi from pigeons, S. turdusi from the common blackbird and Sarcocystis arctica from an arctic fox. These findings suggested that one or two additional species were present in the eagle and that this/these species used carnivores or birds as intermediate hosts. In order to design primers selectively targeting the 18S rRNA gene of such species to the exclusion of S. truncata and other species using ruminant intermediate hosts, a multiple alignment comprising 18S rRNA gene sequences of both types of Sarcocystis spp. was generated using the ClustalW program integrated in MEGA7 (Kumar et al., 2016) as described for the phylogenetic analysis of that gene (section 2.4). The alignment was visually inspected for regions clearly separating the two assemblages, and three new primers targeting such regions were designed. Forward primer A1F targeted nucleotide positions 173–192 from the 5′ end of GenBank sequence KF601301 of S. arctica, whereas forward primer A2F and reverse primer A2R targeted positions 698–718 and 706–729, respectively of that sequence. Using two DNA samples from the eagle and one DNA sample of S. rileyi (Gjerde, 2014c) as templates, these primers were then tested with good results against each other (AF1/A2R; amplifying nucleotides 173–779) and singly against three of the abovementioned more general 18S rRNA gene primers. Finally, the complete 18S rRNA gene of two isolates (Ha1.6, Ha1.8) was amplified as three overlapping fragments with the following three primer pairs: ERIB1/A2R (nucleotides 1–729), A1F/S2r (nucleotides 173–1001), and A2F/Primer BSarc (nucleotides 698–1803/1804).
In order to amplify the partial 28S rRNA gene of isolates Ha1.1 and Ha1.8, a semi-nested PCR was used. In the first round of amplification, the new forward primer A2F was used together with reverse primer KL3 (amplifying an ∼5000-bp-long DNA fragment comprising a portion of the 18S rRNA gene, the ITS1 region, the 5.8S rRNA gene, the ITS2 region and the partial 28S rRNA gene), followed by amplification with primer pair KL1/KL3 in the second round. Similarly, in order to amplify the ITS1 region of isolates Ha1.6 and Ha1.8, the new forward primer A2F was used together with reverse primer 5.8SR2 (yielding an ∼2000-bp-long product) in the first round, followed by amplification with primer pair SU1F/5.8SR2 in the second round. The latter amplicons from each isolate were then cloned into a plasmid vector before sequencing as previously described (Gjerde, 2014b, Gjerde, 2016), since it was suspected that there might be some intra-isolate sequence divergence in this region.
The partial cox1 gene was amplified with primer pair SF1/SR5 from all 10 isolates and with primer pair SF4/SR8D from six of the isolates. Amplification with the latter primer pair was performed in order to confirm the presence of S. truncata DNA in the samples. Thus, reverse primer SR8D had a complete match against cox1 of S. truncata, but a fairly poor match against sequences of the other species found to be present. However, since primer SR8D tends to perform poorly with forward primer SF1 (probably due to primer-dimer formation), forward primer SF4 was used instead (Gjerde et al., 2017a). Moreover, primer SF4 had a better match against sequences of S. truncata than against those of the other species present.
The sequences of all primers used are given in Table S1 in Supplementary material. The PCR reactions were performed and the PCR products were evaluated and purified as described previously, as were the procedures and the reagents used for cloning and purification of plasmid DNA (Gjerde, 2014b, Gjerde, 2016). Purified amplicons and plasmid DNA (from cloning) were sent to Eurofins Genomics, Germany, for sequencing on both strands as previously described (Gjerde et al., 2017a, Gjerde et al., 2017b). Forward and reverse sequence reads, as well as overlapping sequences, were assembled into complete consensus sequences using the Alignment Explorer of the MEGA7 software (Kumar et al., 2016). Newly assembled sequences were compared with each other and with related sequences in GenBank using the Nucleotide BLAST program as previously described (Gjerde, 2013). The different sequences obtained by cloning of the ITS1 region of two isolates were also aligned in MEGA7 by ClustalW and carefully examined by eye for the purpose of describing the intraspecific variation.
2.4. Phylogenetic analyses
Phylogenetic analyses were conducted separately on nucleotide sequences of the four DNA regions examined (cox1, ITS1 region, 18S and 28S rRNA genes) by means of the MEGA7 software (Kumar et al., 2016). The intestinal coccidium Eimeria tenella (family: Eimeriidae) was used as outgroup species to root the trees in all analyses except that based on ITS1 sequences. In all analyses, the phylogeny was tested with the bootstrap method, using 1000 bootstrap replications. The accession numbers of all GenBank sequences used are given behind the taxon names in the various phylogenetic trees. In addition, a cox1 sequence of Sarcocystis neurona that had been artificially concatenated from expressed sequence tags (ESTs) retrieved from GenBank was included in the cox1 analysis (sequence available in Supplementary material of Gjerde and Schulze, 2014).
Concerning cox1, a total of 63 partial sequences from 61 taxa were included in the analysis, including four new sequences generated in the present study. A codon-based multiple alignment of all sequences was obtained by using the ClustalW program integrated in MEGA7 as described previously (Gjerde, 2013). Sequences longer than 1020 bp were truncated at their 3′ end, so that the final alignment comprised 1020 positions with no gaps except those caused by missing data in eight sequences that were shorter than 1020 bp (899–1011 bp). The phylogenetic tree was reconstructed using the neighbour joining method, in which evolutionary distances were computed according to the Kimura 2-parameter model. All codon positions were used. Gaps due to missing data were treated with the pairwise deletion option.
As regards the ITS1 region, a total of 60 sequences from 29 taxa were used in the analysis, including 21 new sequences from two Sarcocystis spp. in the present study. A multiple sequence alignment was generated with the MUSCLE program integrated in MEGA7, using the default settings. The resulting alignment was checked by eye and slightly edited for inconsistencies in the treatment of closely related taxa. Aligned sequences comprising portions of the flanking 18S and 5.8S rRNA genes were trimmed at both ends so that only the ITS1 region was included in the analysis. The final alignment comprised 1368 aligned positions, including gaps. The phylogenetic tree was reconstructed using the neighbour joining method, in which evolutionary distances were computed according to the Kimura 2-parameter model. Gaps were treated with the pairwise deletion option. Sequences of nine Sarcocystis spp. with ruminant intermediate hosts were used as outgroup species to root the tree.
Sequences of the 18S and 28S rRNA genes were aligned and truncated as previously described (Gjerde and Josefsen, 2015). The analysis based on near complete 18S rRNA gene sequences comprised 58 sequences from 56 taxa, including two new sequences from the present study. The final alignment comprised 1956 aligned positions, including gaps. All sites were used. The phylogenetic tree was reconstructed using the maximum likelihood method based on the Hasegawa–Kishino–Yano model with gamma distribution and invariable sites. Similarly, the analysis based on partial 28S rRNA gene sequences comprised 42 sequences from 40 taxa, including two new sequences from the present study. The final alignment comprised 1812 positions, including gaps. The phylogenetic tree was reconstructed using the maximum parsimony method with the Tree–Bisection–Regrafting algorithm. All sites were used.
3. Results
3.1. Sarcocysts in cardiac muscle
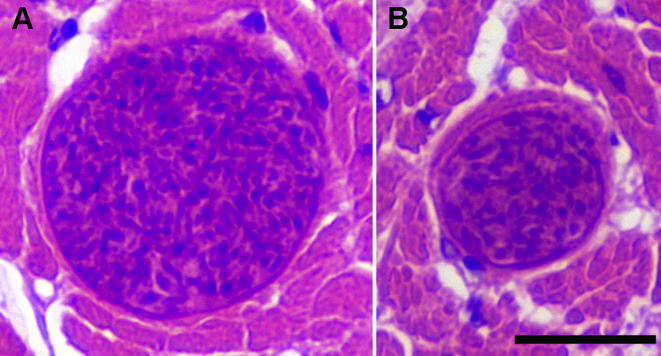
In the single HE-stained histological section of cardiac muscle examined, six small tissue cysts were found. Five of them had been cut nearly transversely in cross-sectioned muscle tissue and appeared as almost round profiles, whereas one cyst had been cut tangentially close to its surface, making only a very small portion of it visible. Hence, the length of the cysts could not be determined. The diameter of the largest cross-sectioned (portion of a) cyst was 40 μm (Fig. 1A). The cysts contained numerous small elongated cells and a few larger roundish cells (4–5 μm) with a pale stained cytoplasm. The larger cells were mainly located at the periphery (Fig. 1B). The outer wall of the cysts was about 0.5 μm thick and seemed to have a smooth surface with no visible protrusions (Fig. 1). Taken together, these features are consistent with those of sarcocysts of various Sarcocystis spp., which are known to contain round to oval metrocytes at their periphery and elongate cystozoites in the more central portions, but even at the periphery in older sarcocysts. Hence, we consider these cysts in the cardiac muscle of the sea eagle to represent sarcocysts of an as yet unidentified Sarcocystis sp. No attempts were made to identify the sarcocysts by molecular methods.
Fig. 1.
Cross-sections of two thin-walled sarcocysts in a HE-stained histological section of cardiac muscle from the white-tailed sea eagle (Bar = 20 μm). A – Fairly large profile of a sarcocyst. B – Smaller profile of a sarcocyst containing several roundish cells at the periphery.
3.2. Oocyst and sporocyst morphology
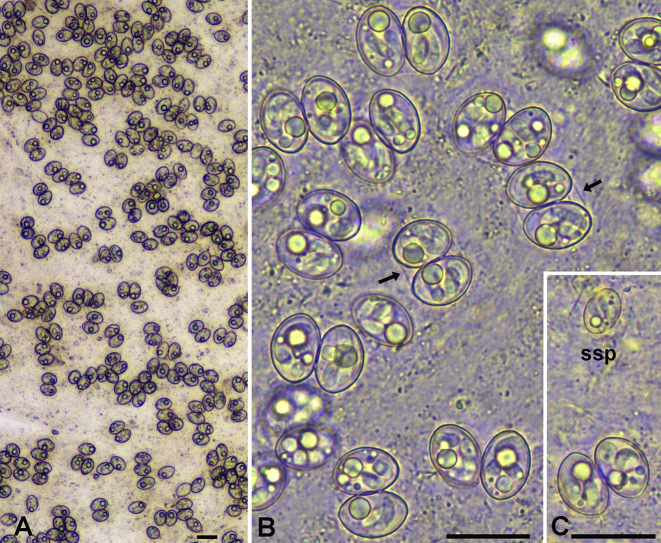
The intestinal mucosal samples contained numerous thin-walled sporulated Sarcocystis oocysts and a few free sporocysts (Fig. 2A). The vast majority of the oocysts/sporocysts were of about the same size; these oocysts measured 21.8–22.8 × 16.0–17.0 μm, whereas the sporocysts (free or within oocysts) measured 16.0–17.0 × 10.5–11.2 μm (Fig. 2B). Each sporocyst contained four sausage-shaped sporozoites, about 12.8 × 3.2 μm in size, and several sporocystic residual granules, which seemed to become larger and more prominent following freezing and thawing of the samples (Fig. 2B). After a careful examination of all the digital photographs recorded, two free sporocysts and a single intact oocysts with much smaller dimensions were detected among the larger oocysts/sporocysts (Fig. 2C). The single small oocyst measured 14.5 × 11.6 μm, whereas the four small sporocysts measured 11.5–11.7 × 7.6–8.6 μm.
Fig. 2.
Sporulated thin-walled oocysts of S. halieti and S. lari (based on molecular identification) in wet smears of the intestinal mucosa (frozen/thawed) of the white-tailed sea eagle (Bars = 20 μm). A – Low magnification of numerous oocysts in the mucosa. B – Higher magnification of sporulated oocysts with a thin wall (arrows). C – A fairly large oocyst of the predominant type and a much smaller free sporocyst (ssp), possibly of S. truncata.
3.3. Molecular identification and characterisation of oocysts
Comparisons of the new nucleotide sequences from this study with previous sequences in GenBank using BLAST, revealed the presence of DNA of three different Sarcocystis spp. in the intestine of the sea eagle. They included the two previously named species Sarcocystis lari (Prakas et al., 2014) and S. truncata (Dahlgren and Gjerde, 2010, Gjerde, 2014a), and the hitherto unnamed Sarcocystis sp. ex Phalacrocorax carbo from the great cormorant in Lithuania (GenBank sequences JQ733511–JQ733513). For the latter species, we have proposed the name Sarcocystis halieti n. sp., which will be used in the following. Identification of S. lari and S. halieti was based mainly on sequences of the ITS1 region, but supported by sequences of the 18S and 28S rRNA genes. There were no previous cox1 sequences of either species. In contrast, S. truncata was mainly identified on the basis of cox1 sequences, but the presence of this species was also suggested in the mixed sequences initially obtained from the three regions of the ribosomal DNA unit when using general primers as mentioned in section 2.3. No attempts were made to further characterize this species in these regions, since the sea eagle was not considered to have acted as a definitive host for this species (see Discussion). The DNA samples examined at each marker and the species identified in these isolates have been summarised in Table 1.
Table 1.
Overview of the molecular characterization at different markers of ten DNA isolates (Ha1.1–Ha1.10) from the intestinal mucosa of a white-tailed sea eagle and the Sarcocystis species identified in each isolate. The GenBank accession numbers of sequences obtained from particular isolates are given in parenthesis behind the isolates of origin.
| Species | Cox1a | 18S rRNA gene | ITS1 | 28S rRNA gene |
|---|---|---|---|---|
| S. halieti | Ha1.1 (MF946583)b, Ha1.2, Ha1.5, Ha1.6 | Ha1.6 (MF946587) |
Ha1.6 (MF946589–MF946596) | Ha1.1 (MF946610) |
| S. lari | Ha1.3 (MF946584)c, Ha1.4, Ha1.7, Ha1.8, Ha1.9, Ha1.10 | Ha1.8 (MF946588) | Ha1.8 (MF946597–MF946609) | Ha1.8 (MF946611) |
| S. truncata | Ha1.1 (MF946585)d, Ha1.3, Ha1.6; Ha1.8 (MF946586)d, Ha1.9, Ha1.10 |
(Ha1.1, Ha1.2) (mixed sequences)e |
(Ha1.1, Ha1.2) (mixed sequences)e |
(Ha1.1, Ha1.2) (mixed sequences)e |
DNA of S. truncata was identified with primer pair SF4/SR8D in isolates in which either S. halieti or S. lari were identified with primer pair SF1/SR5.
Identical sequences were obtained from all four isolates of S. halieti.
Identical sequences were obtained from all six isolates of S. lari.
Each sequence type was found in three isolates of S. truncata.
Short fragments of the mixed and largely unintelligible sequences were compatible with sequences of S. truncata.
3.3.1. Cox1 gene
Amplification of the partial cox1 gene from all 10 DNA isolates with primer pair SF1/SR5, resulted in two sequence types comprising six and four identical sequences, respectively, which differed from each other by substitutions at only two of 1053 nucleotide positions (99.8% identity). A careful inspection of the sequence chromatograms at these positions (nucleotides 424 and 642 of submitted sequences), revealed a single major peak (base) at each position and only occasionally a weak second peak, suggesting that DNA of one variant or species predominated in each DNA sample. This was confirmed in the subsequent amplification and sequencing of the three regions of the nuclear ribosomal DNA unit from selected isolates using the new primers, which yielded sequences of a single species from each isolate. Furthermore, the latter sequences, primarily those of the ITS1 region, revealed that the two cox1 sequence types represented two distinct Sarcocystis spp. The type comprising six identical sequences from six isolates were 100% identical with published cox1 sequences of S. lutrae (GenBank nos. KM657808–KM657809 and KF601326–KF601327) from otters and an arctic fox (Gjerde and Schulze, 2014, Gjerde and Josefsen, 2015) and thus seemed to belong to this species. However, sequences of the 18S and 28S rRNA genes and the ITS1 region from one of these isolates (Ha1.8; Table 1), all matched (see section 3.3.3) previous sequences of S. lari (GenBank nos. JQ733508–JQ733510) derived from sarcocysts in the great black-backed gull (Larus marinus) (Prakas et al., 2014), and hence these six cox1 sequences from the sea eagle were also attributed to S. lari in spite of their complete identity with those of S. lutrae. One of the six identical cox1 sequences of this type (from isolate Ha1.3) was submitted to GenBank and issued accession number MF946584.
From the four other mucosal samples examined, four identical cox1 sequences were obtained, which differed at two nucleotide positions (99.8% identity) from the abovementioned new sequences assigned to S. lari and the previous sequences of S. lutrae. They also differed at only two of 1053 positions from eight identical sequences (GenBank nos. KT588511–KT588518) of S. turdusi from the common blackbird. However, at the three regions of the ribosomal DNA unit, the sequences obtained from two of these mucosal samples (Ha1.1, Ha1.6; Table 1) were closely similar to sequences of an unnamed Sarcocystis sp. from the great cormorant (GenBank nos. JQ733511–JQ733513), and the former sequences, as well as those of the cox1 gene, were therefore considered to belong to this species, which we have named S. halieti. One of the four identical cox1 sequences was submitted to GenBank and issued accession number MF946583. The identities of the new cox1 sequences of S. halieti and S. lari with the most similar sequences available in GenBank have been summarized in Table S2 in Supplementary material. The high identity of both S. halieti and S. lari (100% and 98.8%, respectively) with the sequence of S. calchasi (GenBank no. KU220952) might be misleading, since the latter sequence is only 626 bp long, covering nucleotide positions 16–641 of the new sequences. Similarly, the new sequences of both species showed 99.9% identity to a 900-bp-long sequence (nucleotides 68–967) of an unnamed Sarcocystis sp. from A. cooperii (GenBank no. KY348756) (Lindsay et al., 2017).
Amplification and sequencing of cox1 with primer pair SF4/SR8D yielded fine 991-bp-long sequences of S. truncata from all six isolates examined, which included two and four isolates from which sequences of S. halieti and S. lari, respectively, had been obtained with primer pair SF1/SR5. The six sequences comprised two haplotypes (each from three isolates; Table 1), differing from each other by a single substitution (99.9% identity). One haplotype (GenBank no. MF946586) was identical in the overlapping region (nucleotides 39–1029) with three previous sequences of S. truncata (GenBank nos. KF241444, KF241447, KF241448) from red deer in Norway and differed at 1–5 of 991 positions from the remaining 18 previous cox1 sequences of this species. Likewise, the second haplotype (GenBank no. MF946585) differed at 1–5 positions (99.5–99.9% identity) from all 21 previous cox1 sequences of S. truncata from red deer in Norway (GenBank nos. KC209677–KC209683 and KF241439–KF241452) (Dahlgren and Gjerde, 2010, Gjerde, 2013, Gjerde, 2014a).
3.3.2. Ribosomal DNA unit of S. halieti
As stated above, one species from the sea eagle, which we have named S. halieti, showed a high identity with GenBank sequences JQ733511–JQ733513 of an unnamed species from the great cormorant in Lithuania (unpublished study by Prakas and co-workers), and is considered to be identical with that species. As regards the complete 18S rRNA gene, which was 1803 bp long, the new sequence (GenBank no. MF946587) was 100% identical with GenBank sequence JQ733511 from the great cormorant, but it was also completely identical with sequences of Sarcocystis wobeseri (GenBank nos. GQ922885, GQ922886, HM159419, EU502869). Moreover, the new sequence of S. halieti differed at only one nucleotide position (A rather than G in position 9 from the 3′ end) from GenBank sequence GQ245670 of S. calchasi, but this difference seems to be due to the inclusion of the terminal reverse primer (Primer B) in that sequence (Olias et al., 2010b). This primer does not match sequences of various Sarcocystis spp. in that position, and it was therefore modified to Primer BSarc (Gjerde, 2014b). Thus, S. calchasi actually has a G in this position as can be ascertained from GenBank sequence KC733718. Hence, at least two species, S. wobeseri and S. calchasi, were identical with S. halieti at the complete 18S rRNA gene. Moreover, sequences of this gene from several other Sarcocystis spp. with birds or carnivores as intermediate hosts (S. columbae, GenBank no. HM125054; Sarcocystis corvusi, GenBank no. JN256117; S. turdusi, GenBank no. JF975681; S. lutrae, GenBank no. KM657769; S. lari, GenBank no. JQ733508) differed by only 1–6 nucleotides (99.7–99.9% identity) from that of S. halieti.
The new 1504-bp-long sequence of the partial 28S rRNA gene that was assigned to S. halieti (GenBank no. MF946610) differed at only one of 1474 overlapping positions (T instead of A at position 445 of new sequence; 99.9% identity) from GenBank sequence JQ733513 of the unnamed species from the great cormorant. As regards other species, the most similar ones were S. columbae (GenBank nos. HM125053, GU253887; 99.5% identity), S. corvusi (GenBank no. JN256118; 99.5%), S. wobeseri (GenBank nos. GQ922887, GQ922888, HM159420, EF079886; 99.0%), and S. turdusi (GenBank no. JF975682; 99.0%), whereas many other species with birds or carnivores as intermediate hosts showed identities between 98 and 99%.
As regards the ITS1region, a total of eight clones of S. halieti from one isolate were processed and sequenced (GenBank nos. MF946589–MF946596; Table 1). The full-length sequences, including both primers, were 1083 or 1085 bp long, of which the ITS1 region comprised 828 or 830 bp, and the flanking portions of the 18S and 5.8S rRNA genes, 137 and 118 bp, respectively. The eight clones differed from each other at 1–9 nucleotide positions in the ITS1 region (98.9–99.9% identity within the ITS1 region; 99.2–99.9% identity between full-length sequences). These differences were mainly due to substitutions (single nucleotide polymorphisms, SNPs), but one clone (GenBank no. MF946596) differed from the seven others in having two deletions. Similarly, at the positions displaying substitutions, only one sequence deviated from the others. Hence, these variations might have been due to polymerase errors during the PCR amplification prior to cloning. The eight clones differed from GenBank sequence JQ733513 of the unnamed species from the great cormorant, which comprised the ITS1 region only, at 11–15 of 830 positions (98.2–98.7% identity). At 11 of these positions, all the eight new sequences differed from that sequence. With respect to other species, the ITS1 region of S. halieti showed the highest identity with this region of an unnamed Sarcocystis sp. from the intestine of A. cooperii (GenBank no. KY348755; 93%), S. columbae (GenBank nos. GU253885, HM125052; 92%) and S. corvusi (GenBank no. JN256119; 92%), followed by identities of about 84% with several sequences of S. calchasi and S. wobeseri.
3.3.3. Ribosomal DNA unit of S. lari
The new sequence of the complete 18S rRNA gene from isolate Ha1.8 (GenBank no. MF946588), which was 1804 bp long, was 100% identical with GenBank sequence JQ733508 of S. lari from sarcocysts in the great black-backed gull in Lithuania (Prakas et al., 2014). These sequences shared the highest identities with sequences of S. turdusi (99.8%), S. halieti, S. calchasi and S. wobeseri (99.7%); followed by those of S. corvusi and S. lutrae (99.6%).
The new 1506-bp-long sequence of the partial 28S rRNA gene from isolate Ha1.8 (GenBank no. MF946611) was 100% identical with GenBank sequence JQ733508 of S. lari from sarcocysts in the great black-backed gull, followed by an identity of 98.8% with sequences of S. calchasi (GenBank no. FJ232949) and S. wobeseri (GenBank nos. GQ922887, GQ922888, HM159420, EF079886), and 98.6% identity with a sequence of S. turdusi (GenBank no. JF975682). The new sequence of S. lari was 98.3% identical with the new sequence of S. halieti.
Concerning the ITS1region, a total of 14 clones from isolate Ha1.8 were processed and sequenced, but two clones were identical and therefore 13 clones were submitted to GenBank (accession nos. MF946597–MF946609; Table 1). These sequences were 1115–1117 bp long, of which the ITS1 region comprised 859–861 bp, and the portions of the flanking 18S and 5.8S rRNA genes, comprised 138 and 118 bp, respectively. The new sequences differed from each other at 1–11 nucleotide positions in the ITS1 region (98.7–99.9% identity). These differences were due to several substitutions (SNPs) and three 1-bp-long indels. In the variable positions caused by the substitutions and one of the indels, only one sequence deviated from the remaining 12 sequences, and thus they could have been due to polymerase errors during PCR amplification. As regards the two other indels, several clones possessed a deletion compared to the other sequences, but one of these indels occurred in a stretch of 9–10 consecutive Gs and the other in a stretch of 10–11 consecutive Ts, and thus could have been the result of polymerase slippage during amplification or sequencing. The 13 new sequences differed at 3–8 positions (99.1–99.8% identity) in the ITS1 region from GenBank sequence JQ733509 of S. lari, but there was only one position at which all the new sequences differed from the latter sequence. Hence, the new sequences from the sea eagle were considered to belong to S. lari. The identity with the ITS1 region of other Sarcocystis spp. was 80% or less, including an identity of about 74% with the new sequences of S. halieti and about 70% identity with sequences of S. lutrae (GenBank nos. KM657773–KM657805).
3.4. Phylogeny
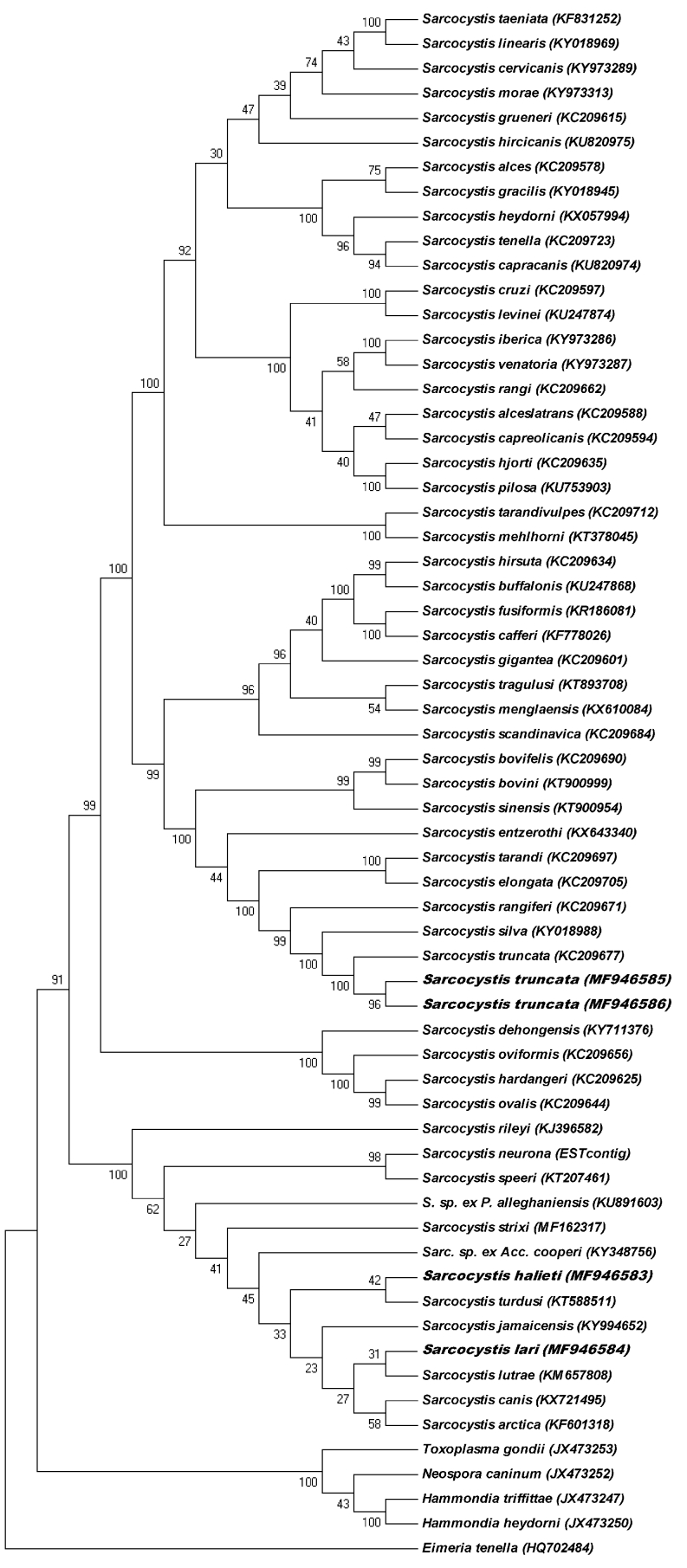
In the phylogenetic tree based on sequences of cox1 (Fig. 3), the new sequences of S. halieti and S. lari were sister to sequences of S. turdusi and S. lutrae, respectively, within a basal clade comprising mainly species with birds or carnivores as natural intermediate hosts. Some of these species are known to use either birds of prey or carnivores as definitive hosts, whereas the definitive hosts are still unknown for some of them. The relationships between the various species within this clade were poorly resolved due to the small differences between their nucleotide sequences. By comparison, the species with ruminant intermediate host formed three major clades according to their known or presumed definitive hosts (canids, felids/unknown, corvids), and the relationships between most of these species were well resolved. The two new sequences of S. truncata clustered with a previous sequence of this species.
Fig. 3.
Phylogenetic tree for members of the Sarcocystidae based on 63 sequences of the partial cox1 gene from 61 taxa and inferred using the neighbour-joining method. Evolutionary distances were computed using the Kimura 2-parameter method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches. The four new sequences from the present study are in boldface.
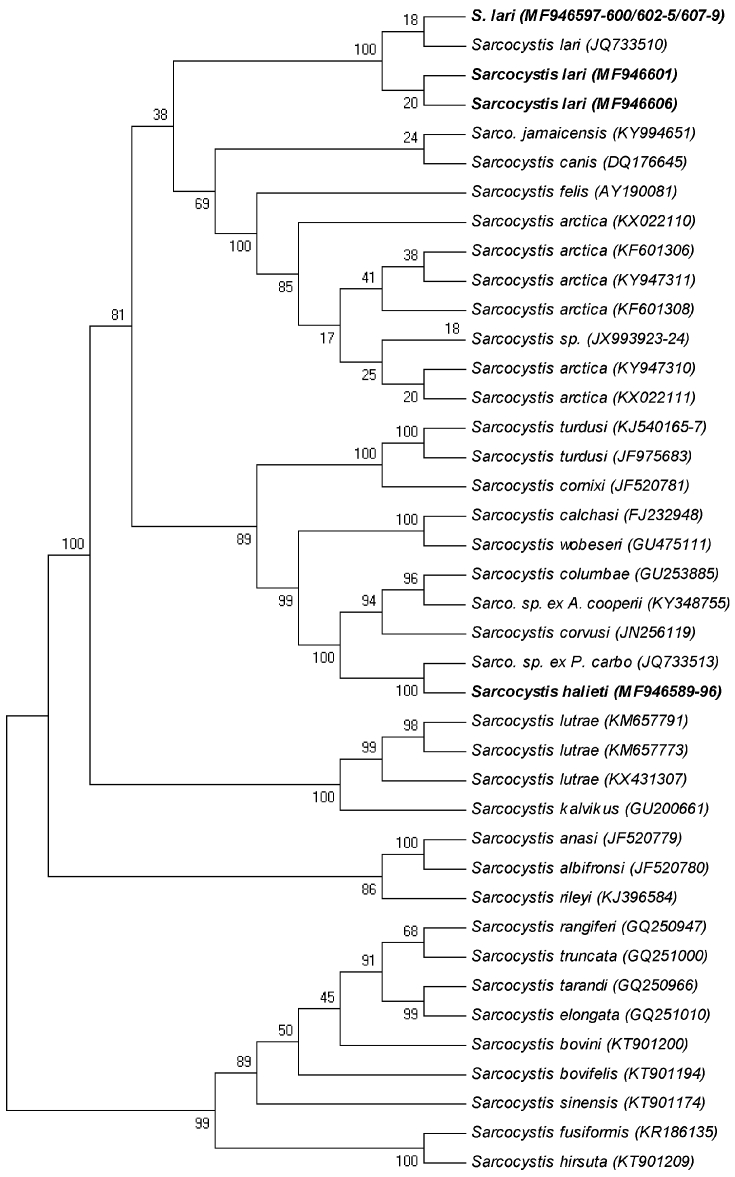
In the phylogenetic tree based on sequences of the ITS1 region (Fig. 4), the eight new sequences of S. halieti clustered with the single sequence of the unnamed species from the great cormorant. These sequences were separated with maximum support from a sister group comprising sequences of S. corvusi, S. columbae and an unnamed species using A. cooperii as definitive host. Within the same major clade were also sequences of S. calchasi, S. wobeseri, S. cornixi and S. turdusi. The 13 new sequences of S. lari, on the other hand, clustered with the previous sequence of that species in a sister clade, which also comprised sequences of S. arctica, Sarcocystis canis, Sarcocystis felis and Sarcocystis jamaicensis. Basal to both of these clades were a clade comprising sequences of S. lutrae and S. kalvikus, as well as a clade comprising sequences of S. rileyi, S. anasi and S. albifronsi.
Fig. 4.
Phylogenetic tree for members of the Sarcocystidae based on 60 sequences of the complete ITS1 region of 29 taxa and inferred using the neighbour-joining method. Evolutionary distances were computed using the Kimura 2-parameter method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches. The new sequences from the present study are in boldface. Some subtrees formed by two or more sequences of the same species have been collapsed.
In the analysis based on near complete 18S rRNA gene sequences (Fig. S1 in Supplementary material), the new sequences of S. halieti and S. lari were placed near previous sequences of the unnamed species from the great cormorant and S. lari, respectively, in a basal clade comprising mainly Sarcocystis spp. with birds or carnivores as intermediate hosts. The same was true in the analysis based on partial 28S rRNA sequences (Fig. S2 in Supplementary material), but in the latter analysis, the relationships between the species in this clade were better resolved than in the tree based on 18S rRNA gene sequences.
3.5. Taxonomic summary for Sarcocystis halieti
Definitive hosts: Haliaeetus albicilla, white-tailed see eagle (this study); Accipiter nisus, Eurasian (northern) sparrow hawk (Mayr et al., 2016).
Intermediate hosts: Phalacrocorax carbo, the great cormorant (according to GenBank sequences JQ733511–JQ733513); possibly other birds (Mayr et al., 2016; this study).
Distribution: Norway (this study), Lithuania, Germany (Mayr et al., 2016).
Oocyst and sporocyst morphology: Oocysts in mucosa sporulated and thin-walled, each containing two sporocysts with four sausage-shaped sporozoites. Oocysts 21.8–22.8 × 16.0–17.0 μm; sporocysts (free or within oocysts) 16.0–17.0 × 10.5–11.2 μm (Fig. 2).
Sarcocyst morphology: There are currently no published descriptions of the morphology of the sarcocysts in the great cormorant from which GenBank sequences JQ733511–JQ733513 were obtained by Prakas and co-workers, and which we consider to represent sarcocysts of S. halieti. However, it is likely that S. halieti has the same sarcocyst morphology as the species found to be most closely related in the phylogenetic analysis based on sequences of the ITS1 region (Fig. 4). These species include S. calchasi (Olias et al., 2010a), S. columbae (Olias et al., 2010b, Prakas et al., 2011a), S. corvusi (Prakas et al., 2013) and S. wobeseri (Kutkienė et al., 2010, Prakas et al., 2011b), all of which, according to these papers, have a smooth sarcocyst wall with no protrusions, but the cyst surface might be slightly wavy.
Molecular characteristics: GenBank sequences MF946583 (cox1), MF946587 (18S rRNA gene), MF946589–MF946596 (ITS1 region), MF946610 (28S rRNA gene). Previous GenBank sequences (JQ733511–JQ733513) derived from sarcocysts in the great cormorant in Lithuania might also be considered to represent this species.
Deposited material: Samples of oocysts/sporocysts from the intestinal mucosa of the sea eagle, as well as samples of genomic DNA extracted from such stages are stored at the Faculty of Veterinary Science, the Norwegian University of Life Sciences, Oslo, Norway.
Etymology: The specific epithet is derived from the genus name of its definitive host, Haliaeetus albicilla.
4. Discussion
The molecular methods used identified DNA of three Sarcocystis spp. in the intestine of the sea eagle, which contained large numbers of oocysts. All three species had previously been characterized molecularly on the basis of DNA from sarcocysts in their intermediate hosts, that is, S. lari in the great black-backed gull (Prakas et al., 2014), an unnamed species from the great cormorant (unpublished study by Prakas and co-workers), and S. truncata from red deer (Dahlgren and Gjerde, 2010, Gjerde, 2013, Gjerde, 2014a). However, we do not think that the sea eagle acted as a definitive host for the latter species. Thus, S. truncata is fairly common in red deer, which is also true for the closely related species Sarcocystis elongata in the same host, as well as for the related species Sarcocystis rangiferi and Sarcocystis tarandi in reindeer (Dahlgren and Gjerde, 2010) and Sarcocystis silva in roe deer (Gjerde, 2012, Gjerde et al., 2017a). It seems highly unlikely that the fairly low number of sea eagles (and golden eagles) that occasionally scavenge upon carcasses of these cervid hosts, should be able to contaminate the environment sufficiently with oocysts/sporocysts to cause the fairly high prevalence and infection intensity of these Sarcocystis spp. Moreover, the phylogenetic position of these species within a clade comprising several species known to be transmitted by felids (Fig. 3), suggests that they too use feline definitive hosts, which would be the domestic cat (Felis catus) and the Eurasian lynx (Lynx lynx) in Norway. This notion is also consistent with the geographic distribution and prevalence of these Sarcocystis spp. Hence, we believe that the DNA of S. truncata identified by PCR in this study originated from ingested material merely passing through the intestine of the sea eagle rather than from oocysts produced in its intestinal mucosa. It could not be determined if this material consisted of remnants of sarcocysts of S. truncata obtained by scavenging on a red deer carcass, or of oocysts/sporocysts of this species from the intestine of a definitive host (presumably a cat or a lynx) that had recently been preyed or scavenged upon by the sea eagle. However, the fact that DNA of S. truncata was also identified in the two mucosal samples that had been thoroughly washed by repeated centrifugation in several changes of water, suggests that this DNA originated from oocysts or free sporocysts in the intestinal contents, since cystozoites from digested sarcocysts might be expected to be removed by such washing, whereas the oocysts/sporocysts would be retained in the sediment after each centrifugation. Moreover, a few oocysts/sporocysts that were much smaller than the predominant type were indeed found, and these stages could have belonged to S. truncata. Interestingly, Kolárová (1986) encountered a similar problem when oocysts/sporocysts recovered from the intestinal scrapings of goshawks were fed to mice and induced the formation of thin-walled sarcocysts in the inoculated animals. This outcome suggested that the goshawks had acted as definitive hosts of a Sarcocystis sp. using mice as intermediate hosts, even though mice and other small rodents rarely got preyed upon by this raptor, whose diet mainly consisted of birds, including owls. Kolárová (1986) therefore believed that the oocysts/sporocysts in the intestines of the goshawks might have belonged to S. dispersa, a species cycling between mice and owls (Černá et al., 1978), and that the goshawks had acquired them by eating the intestines of infected owls.
As regards S. halieti and S. lari, on the other hand, we are confident that the sea eagle acted as a definitive host for both species, and that the DNA recognized by the molecular methods originated from the numerous oocysts found in the intestinal mucosa. Oocysts of the two species seemed to occupy separate regions of the mucosa, since DNA extracted from each of different portions belonged to one species only. In contrast, DNA of S. truncata were found in all samples examined, suggesting that it was present in the intestinal contents rather than the mucosa. The vast majority of the oocysts in the eagle were of about the same size and thus most likely represented oocysts of both S. halieti and S. lari. It could not be determined if the eagle had become infected by both species simultaneously by eating a single infected sea bird, or had acquired S. halieti and S. lari on different occasions from two separate birds. Sarcocystis spp. forming sarcocysts in birds have been found to be less intermediate host specific than most species with mammalian intermediate hosts, and it is therefore possible that a single bird harboured sarcocysts of both species.
Previously, S. lari has only been reported from its intermediate host, the great black-backed gull in Lithuania (Prakas et al., 2014). These authors mentioned that golden eagles, bald eagles and white-tailed sea eagles were the main predators of this seabird, but did not explicitly suggest that eagles might act as definitive hosts for this species, which we have shown to be the case as regards the sea eagle. The great black-backed gull breeds along the entire coast of Norway (https://artsdatabanken.no/Pages/186719), including in the area where the sea eagle was found, and hence the eagle might have acquired S. lari through consumption of this sea bird.
As regards the species that we named S. halieti, the nucleotide sequences derived from the oocysts in the sea eagle showed such a high identity with sequences derived from sarcocysts in the great cormorant in Lithuania by Prakas and co-workers (unpublished) that we consider them to be conspecific. Of particular importance for this conclusion was the high identity between sequences of the ITS1 region, which vary considerably more than sequences of the 18S and 28S rRNA genes among different species with avian intermediate hosts (Prakas et al., 2014). Using the ITS1 marker, Mayr et al. (2016) identified the same unnamed species in intestinal samples containing oocysts from three sparrow hawks in Germany, but did not deposit any sequences in GenBank. Anyway, S. halieti seems to use both sea eagles and sparrow hawks as definitive hosts, and might employ other members of the family Accipitridae as well in this capacity. The sea eagle examined might have acquired S. halieti through predation on great cormorants, which are present along the entire Norwegian coast (https://artsdatabanken.no/Pages/186784). However, the finding of this species in sparrow hawks in Germany, suggested to Mayr et al. (2016) that other (smaller) sea birds may act as additional intermediate hosts for this species.
The present molecular identification of oocysts of S. halieti and S. lari in the the white-tailed sea eagle is the first time that intestinal stages of particular Sarcocystis spp. have been identified in eagles. Moreover, to our knowledge, there have only been two previous reports on Sarcocystis-like oocysts in eagles, both from the USA. Mathey (1966) found such oocysts in a golden eagle (Aquila chrysaetos), but the paper only depicted morphologically similar oocysts found in an American kestrel (Falco sparverius). This was before the two-host life cycle of Sarcocystis spp. had been elucidated, and Mathey (1966) therefore assigned the sporulated oocysts in both raptors to the ‘species’ Isospora buteonis, which was a designation used about such oocysts in several birds. In a later study, Tuggle and Schmeling (1982) found typical Sarcocystis oocysts in the intestine of five bald eagles (Haliaeetus leucocephalus) from Alaska, California and Colorado. In the absence of transmission experiments (and molecular identification), they could not determine if the oocysts belonged to a Sarcocystis sp. or to a species of the then newly erected genus Frenkelia. Hence, they referred to the oocysts as Sarcocystis-like or as sarcosporidian oocysts.
The finding of six small sarcocysts in the histological section of cardiac muscle from the sea eagle, established for the first time that this raptor may itself become infected by Sarcocystis oocysts/sporocysts and act as an intermediate host. Previously, sarcocysts have been reported from the bald eagle and the golden eagle in the USA (Crawley et al., 1982, Olson et al., 2007, Wünschmann et al., 2010), and in the latter study they were identified as sarcocysts of S. falcatula, which uses opossums as definitive hosts. The thin-walled sarcocysts found in the sea eagle are morphologically similar to those reported from the bald eagle by Crawley et al. (1982). We made no attempts to identify the sarcocysts in the sea eagle by molecular methods, but consider it unlikely that they belong to either S. halieti or S. lari, which formed oocysts in the intestine of the same bird.
With respect to the three regions of the nuclear ribosomal DNA unit examined, this study again showed that Sarcocystis spp. with avian intermediate hosts can only be unambiguously delimited on the basis of sequences of the ITS1 region, whereas they are identical or nearly identical at the complete 18S rRNA gene, and almost identical in the portion of the 28S rRNA gene (D2/D3 domain) that is usually examined (Prakas et al., 2014, Lindsay et al., 2017). This study provided for the first time partial cox1 sequences of S. halieti and S. lari. Surprisingly, those of S. lari were identical with sequences of S. lutrae from otters and an arctic fox (Gjerde and Josefsen, 2015), whereas these species differed clearly from each other at the three other markers, and particularly in the ITS1 region. Likewise, a cox1 sequence of S. neurona that was artificially concatenated from expressed sequence tags (ESTs) retrieved from GenBank (Gjerde and Schulze, 2014), is completely identical with a cox1 sequence of Sarcocystis speeri (GenBank no. KT207461), but the latter two species are also almost identical in the ITS1 region, and might possibly represent a single species. Several other Sarcocystis spp. using birds or mammalian carnivores as intermediate hosts also differ very little (usually less than 1%) at the cox1 marker as shown in Table S2 in Supplementary material. On the other hand, these species also seem to possess little or no intraspecific variation at cox1 (Gjerde, 2014c, Gjerde and Schulze, 2014, Gjerde and Josefsen, 2015). In this study, all isolates of S. halieti were identical, as were all those of S. lari. By contrast, Sarcocystis spp. using ruminant intermediate hosts show greater intraspecific as well as interspecific sequence variation at cox1. This marker has therefore become useful for separating closely related Sarcocystis spp. in ruminants, which cannot be reliably separated on the basis of sequences of either the 18S or 28S rRNA genes or the ITS1 region (Gjerde, 2013, Gjerde, 2014a, Gjerde, 2014b, Gjerde, 2016, Gjerde et al., 2017a, Gjerde et al., 2017b). However, with respect to Sarcocystis spp. using birds and carnivores as intermediate hosts, the ITS1 marker seems to better suited than cox1 to separate closely related species.
In conclusion, the present study has shown that the white-tailed sea eagle may act as a definitive host of S. halieti and S. lari, forming sarcocysts in sea birds, but it may also act as an intermediate host of an unidentified Sarcocystis sp. with an unknown definitive host. Further studies are necessary to determine whether additional Sarcocystis spp. are transmitted by sea eagles and to identify the species forming sarcocysts in this large bird of prey.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgements
The authors would like to thank Alv Ottar Folkestad for taking care of the emaciated sea eagle and for submitting the carcass for necropsy. This research did not receive any specific grants from funding agencies in the public, commercial, or not-for-profit sectors.
Footnotes
Nucleotide sequence data reported in this paper are available in the GenBank™, EMBL and DDBJ databases under the accession numbers MF946583–MF946611.
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ijppaw.2017.12.001.
Contributor Information
Bjørn Gjerde, Email: bjorn.gjerde@nmbu.no.
Turid Vikøren, Email: turid.vikoren@vetinst.no.
Inger Sofie Hamnes, Email: inger.hamnes@vetinst.no.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- Box E.D., Meier J.L., Smith J.H. Description of Sarcocystis falcatula Stiles, 1893, a parasite of birds and opossums. J. Protozool. 1984;31:521–524. doi: 10.1111/j.1550-7408.1984.tb05495.x. https://doi.org/10.1111/j.1550-7408.1984.tb05495.x [DOI] [PubMed] [Google Scholar]
- Cawthorn R.J., Rainnie D., Wobeser G. Experimental transmission of Sarcocystis sp. (Protozoa: Sarcocystidae) between the shoveler (Anas clypeata) duck and the striped skunk (Mephitis mephitis) J. Wildl. Dis. 1981;17:389–394. doi: 10.7589/0090-3558-17.3.389. https://doi.org/10.7589/0090-3558-17.3.389 [DOI] [PubMed] [Google Scholar]
- Černá Z., Kolárová I., Šulc P. Contribution to the problem of cyst-producing coccidians. Folia Parasitol. 1978;25:9–16. [PubMed] [Google Scholar]
- Černá Z., Kvašňovská Z. Life-cycle involving bird-bird relation in Sarcocystis-coccidia with the description of Sarcocystis accipitris sp. n. Folia Parasitol. 1986;33:305–309. [Google Scholar]
- Crawley R.R., Ernst J.V., Milton J.L. Sarcocystis in a bald eagle (Haliaeetus leucocephalus) J. Wildl. Dis. 1982;18:253–255. doi: 10.7589/0090-3558-18.2.253. https://doi.org/10.7589/0090-3558-18.2.253 [DOI] [PubMed] [Google Scholar]
- Dahlgren S.S., Gjerde B. Molecular characterization of five Sarcocystis species in red deer (Cervus elaphus), including Sarcocystis hjorti n. sp., reveals that these species are not intermediate host specific. Parasitology. 2010;137:815–840. doi: 10.1017/S0031182009991569. https://doi.org/10.1017/S0031182009991569 [DOI] [PubMed] [Google Scholar]
- Gjerde B. Morphological and molecular characterization and phylogenetic placement of Sarcocystis capreolicanis and Sarcocystis silva n. sp. from roe deer (Capreolus capreolus) in Norway. Parasitol. Res. 2012;110:1225–1237. doi: 10.1007/s00436-011-2619-6. https://doi.org/10.1007/s00436-011-2619-6 [DOI] [PubMed] [Google Scholar]
- Gjerde B. Phylogenetic relationships among Sarcocystis species in cervids, cattle and sheep inferred from the mitochondrial cytochrome c oxidase subunit I gene. Int. J. Parasitol. 2013;43:579–591. doi: 10.1016/j.ijpara.2013.02.004. https://doi.org/10.1016/j.ijpara.2013.02.004 [DOI] [PubMed] [Google Scholar]
- Gjerde B. Sarcocystis species in red deer revisited: with a re-description of two known species as Sarcocystis elongata n. sp. and Sarcocystis truncata n. sp. based on mitochondrial cox1 sequences. Parasitology. 2014;141:441–452. doi: 10.1017/S0031182013001819. https://doi.org/10.1017/S0031182013001819 [DOI] [PubMed] [Google Scholar]
- Gjerde B. Morphological and molecular characteristics of four Sarcocystis spp. in Canadian moose (Alces alces), including Sarcocystis taeniata n. sp. Parasitol. Res. 2014;113:1591–1604. doi: 10.1007/s00436-014-3806-z. https://doi.org/10.1007/s00436-014-3806-z [DOI] [PubMed] [Google Scholar]
- Gjerde B. Molecular characterisation of Sarcocystis rileyi from a common eider (Somateria mollissima) in Norway. Parasitol. Res. 2014;113:3501–3509. doi: 10.1007/s00436-014-4062-y. https://doi.org/10.1007/s00436-014-4062-y [DOI] [PubMed] [Google Scholar]
- Gjerde B. Molecular characterisation of Sarcocystis bovifelis, Sarcocystis bovini n. sp., Sarcocystis hirsuta and Sarcocystis cruzi from cattle (Bos taurus) and Sarcocystis sinensis from water buffaloes (Bubalus bubalis) Parasitol. Res. 2016;115:1473–1492. doi: 10.1007/s00436-015-4881-5. https://doi.org/10.1007/s00436-015-4881-5 [DOI] [PubMed] [Google Scholar]
- Gjerde B., Dahlgren S.S. Corvid birds (Corvidae) act as definitive hosts for Sarcocystis ovalis in moose (Alces alces) Parasitol. Res. 2010;107:1445–1453. doi: 10.1007/s00436-010-2017-5. https://doi.org/10.1007/s00436-010-2017-5 [DOI] [PubMed] [Google Scholar]
- Gjerde B., Josefsen T.D. Molecular characterisation of Sarcocystis lutrae n. sp. and Toxoplasma gondii from the musculature of two Eurasian otters (Lutra lutra) in Norway. Parasitol. Res. 2015;114:873–886. doi: 10.1007/s00436-014-4251-8. https://doi.org/10.1007/s00436-014-4251-8 [DOI] [PubMed] [Google Scholar]
- Gjerde B., Schulze J. Muscular sarcocystosis in two arctic foxes (Vulpes lagopus) due to Sarcocystis arctica n. sp.: sarcocyst morphology, molecular characteristics and phylogeny. Parasitol. Res. 2014;113:811–821. doi: 10.1007/s00436-013-3711-x. https://doi.org/10.1007/s00436-013-3711-x [DOI] [PubMed] [Google Scholar]
- Gjerde B., Giacomelli S., Bianchi A., Bertoletti I., Mondani H., Gibelli L.R. Morphological and molecular characterization of four Sarcocystis spp., including Sarcocystis linearis n. sp., from roe deer (Capreolus capreolus) in Italy. Parasitol. Res. 2017;116:1317–1338. doi: 10.1007/s00436-017-5410-5. https://doi.org/10.1007/s00436-017-5410-5 [DOI] [PubMed] [Google Scholar]
- Gjerde B., Luzón M., Alunda J.M., de la Fuente C. Morphological and molecular characteristics of six Sarcocystis spp. from red deer (Cervus elaphus) in Spain, including Sarcocystis cervicanis and three new species. Parasitol. Res. 2017;116:2795–2811. doi: 10.1007/s00436-017-5590-z. https://doi.org/10.1007/s00436-017-5590-z [DOI] [PubMed] [Google Scholar]
- Irie T., Ikeda T., Nakamura T., Ichii O., Yamada N., Ito T., Yamazaki A., Takai S., Yagi K. First molecular detection of Sarcocystis ovalis in the intestinal mucosa of a Japanese jungle crow (Corvus macrorhynchos) in Hokkaido. Jpn. Vet. Parasitol. Reg. Stud. Rep. 2017;10:54–57. doi: 10.1016/j.vprsr.2017.08.005. https://doi.org/10.1016/j.vprsr.2017.08.005 [DOI] [PubMed] [Google Scholar]
- Kolárová L. Mouse (Mus musculus) as intermediate host of Sarcocystis sp. from the goshawk (Accipiter gentilis) Folia Parasitol. 1986;33:15–19. [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. http://dx.doi.org/10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutkienė L., Prakas P., Sruoga A., Butkauskas D. Sarcocystis in the birds family Corvidae with description of Sarcocystis cornixi sp. nov. from the hooded crow (Corvus cornix) Parasitol. Res. 2009;104:329–336. doi: 10.1007/s00436-008-1196-9. https://doi.org/10.1007/s00436-008-1196-9 [DOI] [PubMed] [Google Scholar]
- Kutkienė L., Prakas P., Sruoga A., Butkauskas D. The mallard duck (Anas platyrhynchos) as intermediate host for Sarcocystis wobeseri sp. nov. from the barnacle goose (Branta leucopsis) Parasitol. Res. 2010;107:879–888. doi: 10.1007/s00436-010-1945-4. https://doi.org/10.1007/s00436-010-1945-4 [DOI] [PubMed] [Google Scholar]
- Kutkienė L., Prakas P., Sruoga A., Butkauskas D. Description of Sarcocystis anasi sp. nov. and Sarcocystis albifronsi sp. nov. in birds of the order Anseriformes. Parasitol. Res. 2012;110:1043–1946. doi: 10.1007/s00436-011-2588-9. https://doi.org/10.1007/s00436-011-2588-9 [DOI] [PubMed] [Google Scholar]
- Kutkienė L., Prakas P., Butkauskas D., Sruoga A. Description of Sarcocystis turdusi sp. nov. from the common blackbird (Turdus merula) Parasitology. 2012;139:1438–1443. doi: 10.1017/S0031182012000819. https://doi.org/10.1017/S0031182012000819 [DOI] [PubMed] [Google Scholar]
- Lindsay D.S., Verma S.K., Scott D., Dubey J.P., von Dohlen A.R. Isolation, molecular characterization, and in vitro schizogonic development of Sarcocystis sp. ex Accipiter cooperii from a naturally infected Cooper's hawk (Accipiter cooperii) Parasitol. Int. 2017;66:106–111. doi: 10.1016/j.parint.2016.12.002. https://doi.org/10.1016/j.parint.2016.12.002 [DOI] [PubMed] [Google Scholar]
- Mathey W.J. Isospora buteonis Henry 1932 in an American kestrel (Falco sparverius) and a golden eagle (Aquila chrysaetos) Bull. Wildl. Dis. Assoc. 1966;2:20–22. https://doi.org/10.7589/0090-3558-2.2.20 [Google Scholar]
- Mayr S.L., Maier K., Müller J., Enderlein D., Gruber A.D., Lierz M. Accipiter hawks (Accipitridae) confirmed as definitive hosts of Sarcocystis turdusi, Sarcocystis cornixi and Sarcocystis sp. ex Phalacrocorax carbo. Parasitol. Res. 2016;115:3041–3047. doi: 10.1007/s00436-016-5059-5. https://doi.org/10.1007/s00436-016-5059-5 [DOI] [PubMed] [Google Scholar]
- Moré G., Maksimov A., Conraths F.J., Schares G. Molecular identification of Sarcocystis spp. in foxes (Vulpes vulpes) and raccoon dogs (Nyctereutes procyonoides) from Germany. Vet. Parasitol. 2016;220:9–14. doi: 10.1016/j.vetpar.2016.02.011. https://doi.org/10.1016/j.vetpar.2016.02.011 [DOI] [PubMed] [Google Scholar]
- Olias P., Gruber A.D., Hafez H.M., Heydorn A.O., Mehlhorn H., Lierz M. Sarcocystis calchasi sp. nov. of the domestic pigeon (Columba livia f. domestica) and the Northern goshawk (Accipiter gentilis): light and electron microscopical characteristics. Parasitol. Res. 2010;106:577–585. doi: 10.1007/s00436-009-1701-9. https://doi.org/10.1007/s00436-009-1701-9 [DOI] [PubMed] [Google Scholar]
- Olias P., Olias L., Lierz M., Mehlhorn H., Gruber A.D. Sarcocystis calchasi is distinct to Sarcocystis columbae sp. nov. from the wood pigeon (Columba palumbus) and Sarcocystis sp. from the sparrowhawk (Accipiter nisus) Vet. Parasitol. 2010;171:7–14. doi: 10.1016/j.vetpar.2010.03.021. https://doi.org/10.1016/j.vetpar.2010.03.021 [DOI] [PubMed] [Google Scholar]
- Olias P., Olias L., Krücken J., Lierz M., Gruber A.D. High prevalence of Sarcocystis calchasi sporocysts in European Accipiter hawks. Vet. Parasitol. 2011;175:230–236. doi: 10.1016/j.vetpar.2010.10.025. https://doi.org/10.1016/j.vetpar.2010.10.025 [DOI] [PubMed] [Google Scholar]
- Olson E.J., Wünschmann A., Dubey J.P. Sarcocystis sp.-associated meningoencephalitis in a bald eagle (Haliaeetus leucocephalus) J. Vet. Diagn. Invest. 2007;19:564–568. doi: 10.1177/104063870701900519. [DOI] [PubMed] [Google Scholar]
- Prakas P., Butkauskas D., Sruoga A., Švažas S., Kutkienė L. Identification of Sarcocystis columbae in wood pigeons (Columba palumbus) in Lithuania. Vet. Med. Zoot. 2011;55:33–39. [Google Scholar]
- Prakas P., Kutkienė L., Sruoga A., Butkauskas D. Sarcocystis sp. from the herring gull (Larus argentatus) identity to Sarcocystis wobeseri based on cyst morphology and DNA results. Parasitol. Res. 2011;109:1603–1608. doi: 10.1007/s00436-011-2421-5. https://doi.org/10.1007/s00436-011-2421-5 [DOI] [PubMed] [Google Scholar]
- Prakas P., Kutkienė L., Butkauskas D., Sruoga A., Zalakevičius M. Molecular and morphological investigations of Sarcocystis corvusi sp. nov. from the jackdaw (Corvus monedula) Parasitol. Res. 2013;112:1163–1167. doi: 10.1007/s00436-012-3247-5. http://dx.doi.org/10.1007/s00436-012-3247-5 [DOI] [PubMed] [Google Scholar]
- Prakas P., Kutkienė L., Butkauskas D., Sruoga A., Žalakevičius M. Description of Sarcocystis lari sp. n. (Apicomplexa: Sarcocystidae) from the great black-backed gull, Larus marinus (Charadriiformes: Laridae), on the basis of cyst morphology and molecular data. Folia Parasitol. 2014;61:11–17. https://doi.org/10.14411/fp.2014.002 [PubMed] [Google Scholar]
- Prakas P., Liaugaudaitė S., Kutkienė L., Sruoga A., Švažas S. Molecular identification of Sarcocystis rileyi sporocysts in red foxes (Vulpes vulpes) and raccoon dogs (Nyctereutes procyonoides) in Lithuania. Parasitol. Res. 2015;114:1671–1676. doi: 10.1007/s00436-015-4348-8. https://doi.org/10.1007/s00436-015-4348-8 [DOI] [PubMed] [Google Scholar]
- Svobodová M. A Sarcocystis species from goshawk (Accipiter gentilis) with great tit (Parus major) as intermediate host. Acta Protozool. 1996;35:223–226. [Google Scholar]
- Tuggle B.N., Schmeling S.K. Parasites of the bald eagle (Haliaeetus leucocephalus) of North America. J. Wildl. Dis. 1982;18:501–506. doi: 10.7589/0090-3558-18.4.501. https://doi.org/10.7589/0090-3558-18.4.501 [DOI] [PubMed] [Google Scholar]
- Votýpka J., Hypsa V., Jirků M., Flegr J., Vávra J., Lukes J. Molecular phylogenetic relatedness of Frenkelia spp. (Protozoa, Apicomplexa) to Sarcocystis falcatula Stiles 1893: is the genus Sarcocystis paraphyletic? J. Eukaryot. Microbiol. 1998;45:137–141. doi: 10.1111/j.1550-7408.1998.tb05081.x. [DOI] [PubMed] [Google Scholar]
- Wicht R.J. Transmission of Sarcocystis rileyi to the striped skunk (Mephitis mephitis) J. Wildl. Dis. 1981;17:387–388. doi: 10.7589/0090-3558-17.3.387. https://doi.org/10.7589/0090-3558-17.3.387 [DOI] [PubMed] [Google Scholar]
- Wünschmann A., Rejmanek D., Conrad P.A., Hall N., Cruz-Martinez L., Vaughn S.B., Barr B.C. Natural fatal Sarcocystis falcatula infections in free-ranging eagles in North America. J. Vet. Diagn. Invest. 2010;22:282–289. doi: 10.1177/104063871002200222. https://doi.org/10.1177/104063871002200222 [DOI] [PubMed] [Google Scholar]
- Yabsley M.J., Ellis A.E., Stallknecht D.E., Howerth E.W. Characterization of Sarcocystis from four species of hawks from Georgia, USA. J. Parasitol. 2009;95:256–259. doi: 10.1645/GE-1567.1. https://doi.org/10.1645/GE-1567.1 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.