Abstract.
The work explores the use of denoising autoencoders (DAEs) for brain lesion detection, segmentation, and false-positive reduction. Stacked denoising autoencoders (SDAEs) were pretrained using a large number of unlabeled patient volumes and fine-tuned with patches drawn from a limited number of patients (, 40, 65). The results show negligible loss in performance even when SDAE was fine-tuned using 20 labeled patients. Low grade glioma (LGG) segmentation was achieved using a transfer learning approach in which a network pretrained with high grade glioma data was fine-tuned using LGG image patches. The networks were also shown to generalize well and provide good segmentation on unseen BraTS 2013 and BraTS 2015 test data. The manuscript also includes the use of a single layer DAE, referred to as novelty detector (ND). ND was trained to accurately reconstruct nonlesion patches. The reconstruction error maps of test data were used to localize lesions. The error maps were shown to assign unique error distributions to various constituents of the glioma, enabling localization. The ND learns the nonlesion brain accurately as it was also shown to provide good segmentation performance on ischemic brain lesions in images from a different database.
Keywords: brain lesion, gliomas, magnetic resonance imaging, deep learning, stacked denoising autoencoder, denoising autoencoder
1. Introduction
Gliomas are a type of primary brain tumor that affects the glial cells in the brain. Based on severity, gliomas are further divided to high grade glioma (HGG) and low grade glioma (LGG). Automatic segmentation of gliomas from MRI, a preliminary step for treatment planning and determining disease progression, is a challenging task due to heterogeneity of tissue within the lesion, nonuniform intensity values of MR images and diffused borders of tumors. Furthermore, multiple MRI sequences, namely, T1, T2, fluid liquid attenuated inversion recovery (FLAIR), and T1 postcontrast (T1c), provide complementary information about the lesion and aids in the task of accurate segmentation. A fully automated image segmentation pipeline is thus necessary for evaluating large numbers of patients across multiple centers.
1.1. Literature Survey
In the recent past, various fully automated techniques have been proposed to segment gliomas, and they can be broadly classified as either generative or discriminative techniques.1 Generative techniques model the joint distribution of the voxel classes and voxel-specific features. A typical approach is to register the images onto a probabilistic atlas.2–5 An atlas represents a normal healthy brain and comprises white matter, gray matter, ventricles, brain stem, etc. Following registration, various techniques have been developed to classify the tumor as an outlier/additional class. For example, Prastawa et al.5 used a covariance determinant estimator to detect outliers followed by further segmentation using a K-means algorithm. Since the presence of large tumors or resectional cavities alters the structure of brain, the performance of generative models can be impacted by the registration technique that is used to align images and spatial priors.6 Overall, as stated in Refs. 1, 5–7, generative techniques perform well on unseen data. A recent work6 on a hybrid generative/discriminative model for glioma segmentation shows a boost in performance by combining a generative and discriminative approach.
Discriminative techniques model/determine the class conditional distribution given the image features, for example, voxel intensities. Discriminative techniques, such as random forest8–15 and support vector machines,16,17 have been applied to brain tumor segmentation. These techniques are suited for multiclass problems and use hand-coded features, such as mean, median, skewness, and symmetry of the brain, to name a few, to classify voxels. Discriminative techniques tend to misclassify certain voxels as lesion at anatomically and physiologically unlikely locations since each voxel is modeled to be independent from its neighboring voxels.18 However, conditional random fields and Markov random fields can be used to regularize the segmentation and could lead to improved results. The overall performance of discriminative techniques, in general, would depend on the quality of the computed features.
In the past decade, deep learning techniques, such as deep belief networks, convolutional neural networks (CNNs), and stacked denosing autoencoders (SDAEs), have been used in a variety of image classification and segmentation tasks.19–21 Deep learning techniques are capable of learning features, such as edges, textures, patterns, and various higher order features, from raw images. Recently, CNNs have been used for segmentation of gliomas from MR images18–27 and have outperformed other fully automatic techniques. CNNs can be considered discriminative models since they predict the posterior probability given the image features. Apart from segmentation of gliomas, deep neural networks, especially CNNs have performed well on a variety of medical image analysis tasks, such as focal pathology detection, segmentation of ischemic lesion, traumatic brain injuries, etc.28–31 Typically, a large number of labeled data are required to train discriminative models especially deep learning-based ones like CNNs.
Among deep learning models, restricted Boltzman machines (RBM) and deep Boltzman machines32 can be considered generative models. Convolutional RBMs33 have been used to extract features to aid in semiautomated segmentation of gliomas. The focus of this paper is on SDAEs that learn a compact encoding of the data, which can then be used as features for classification. Autoencoders and their variants, which include SDAEs and sparse stacked autoencoders, have been used in various medical image processing applications.34–36 Both RBMs and autoencoders can be pretrained using unlabeled data. RBMs being energy-based models are trained by optimizing the log-likelihood of the entire data or by minimizing contrastive divergence.37 Nonetheless, computing the log-likelihood of the entire data is an intractable quantity; thus, training of RBMs requires Monte Carlo Markov chain techniques like Gibbs sampling.38 Autoencoders, denoising autoencoders (DAEs), and stacked DAE (SDAE), on the other hand, are trained by minimizing the mean squared error or cross entropy, which enables the parameters to be learned using a gradient-based backpropagation algorithm. The absence of the sampling scheme during the training phase makes training of autoencoders relatively easier than RBMs.
One of the major issues that arises in training deep networks is class imbalance. Class imbalance is particularly acute in medical imaging problems since lesions constitute a minuscule percentage of image voxels. In such scenarios, a novelty/anomaly detection approach would be very effective. The principles of anomaly detection are well studied and typically involve detecting outliers or rare events by measuring a distance metric obtained from a parametric model of the data (excluding the anomalies). Autoencoders and other machine learning techniques39,40 have also found applications in novelty detection. In the next section, the original contribution is outlined based on the unsupervised training and novelty detection approach for glioma segmentation and ischemic lesion segmentation.
1.2. Contribution
This paper describes the application of DAEs for the detection and segmentation of brain lesions from multisequence MR images. Specifically, the contributions are:
-
•
False-positive (FP) reduction for gliomas and candidate detection for brain lesions (ischemic lesion) using a novelty detector (ND).
-
•
Variant of the ND, called cascaded ND (CND), which generates unique error distributions for various constituents of glioma.
-
•
Demonstrating semisupervised and weakly supervised learning by training SDAE using patches drawn from limited patient volumes ().
-
•
Transfer learning approach for LGG, which had limited number of labeled training data. The LGG network was obtained by fine-tuning the pretrained HGG network.
The manuscript is laid out as follows. Section 2 describes the dataset used, Sec. 3 describes the preprocessing of image data, training of SDAEs, and postprocessing using ND. Section 4 describes the results and discusses the performance of SDAEs on test data for the brain tumor segmentation task. The paper concludes with the summary and discussion of future direction in Sec. 5.
2. Training Data
The publicly available BraTS-2015 dataset1,41 was used for training the networks. The dataset comprises 220 HGG and 54 LGG patient data. The HGG dataset is composed of patients imaged only once [single-time (ST) point] and patients who are scanned multiple times (longitudinal data). The HGG dataset comprises 123 ST point patients and 97 longitudinal-time (LT) point patients, while no longitudinal studies were found in the LGG dataset. Each patient data comprises FLAIR, T2 weighted, T1 weighted, and T1c sequence. Each voxel in the image volumes is classified as one of the five classes, namely, normal, edema, nonenhancing tumor, necrotic region, and enhancing tumor. There exists a huge data imbalance among classes in both the HGG and LGG datasets (Table 1). Furthermore, in the aforementioned datasets, certain classes occur more frequently in one grade of glioma than the other, for example, enhancing tumor is more prominent in HGG while nonenhancing tumor is more prominent in LGG.
Table 1.
Amount of data imbalance in % in HGG and LGG (BraTS 2015 training data).
| Grade of glioma | Normal | Necrotic region | Edema | Nonenhancing tumor | Enhancing tumor |
|---|---|---|---|---|---|
| HGG | 98.76 | 0.06 | 0.80 | 0.13 | 0. 24 |
| LGG | 98.86 | 0.09 | 0.69 | 0.29 | 0.04 |
The ischemic lesion training database made available as part of the ISLES 2015 challenge42 was used for demonstrating candidate lesion detection using ND and CND. The database consists of 28 patient volumes comprising diffusion weighted images, FLAIR, T1- and T2-weighted sequences. Further details about the dataset are given in Appendix A.
3. Methods
3.1. Background
Autoencoders are neural networks that were originally used for dimensionality reduction. They are trained to reconstruct the input data, and dimensionality reduction is achieved using a fewer number of neurons in the hidden layer than in the input layer. A deep autoencoder is obtained by stacking multiple layers of encoders with each layer trained independently (pretraining) using an unsupervised learning criterion. A classification layer can be added to the pretrained encoder and further trained with labeled data (fine-tuning). Such an approach initially outlined in Ref. 20 was shown to be an effective way to train deep networks. DAE is a variant in which the hidden layer is pretrained with artificially corrupted data, and the reconstruction error is calculated against the uncorrupted data. DAEs provide robust features which in turn improves the classification accuracy.21 Autoencoders and their variants are explained in greater detail in Appendix B.
3.2. Overview
In this work, a single layer DAE was used as an anomaly/ND by training the network to reconstruct nonlesion patches. The reconstruction error corresponding to lesion and nonlesion patches would then be significantly different.
SDAEs were pretrained layer-by-layer using a large number of unlabeled patches. The network was fine-tuned using labeled patches drawn from a limited subset of patients after adding a classification layer. Voxelwise classification was done on test data volumes by selecting patches centered on every voxel to create a label image. The reconstruction error map was obtained for the entire volume using ND. A binary mask derived from the error map, indicating the lesion regions, was used to reject FPs in the label image. The entire pipeline is shown in Fig. 1 and is explained in the section below.
Fig. 1.
Flowchart of the pipeline used for segmentation of gliomas.
3.3. Preprocessing
3.3.1. Histogram matching
All the volumes in the database were histogram matched43 to an arbitrarily chosen reference image from the training data. This ensures the contrast and intensity range to be similar across image volumes [Figs. 2(a)–2(c)]. The same reference image was used for HGG, LGG, and ischemic dataset.
Fig. 2.
Histogram matching: (a) histogram of reference FLAIR sequence, (b) histogram of test FLAIR sequence, and (c) histogram of test data posthistogram matching.
3.3.2. z-score
Following histogram matching, all sequences corresponding to a patient volume were independently normalized to have zero mean and unit standard deviation.
3.4. Stacked Denoising Autoencoder
SDAE was trained to classify the center voxel of a patch of size as one of the five classes. There exists a notable difference in the expression of pathology in a HGG and a LGG subject, for example, the presence of an enhancing tumor would be fewer in LGG patients when compared with a HGG counterpart. Hence, separate networks with the same architecture were trained for LGG and HGG segmentation. The network architecture is given in Table 2.
Table 2.
SDAE architecture. , no. of neurons in the th hidden layer.
| Network | Input Layer | Output layer | ||||
|---|---|---|---|---|---|---|
| SDAE | 1764 | 3500 | 2000 | 1000 | 500 | 5 |
3.4.1. HGG network
The HGG network was pretrained using 130 HGG patients (70 ST and 60 LT data). The HGG network was fine-tuned using patches from 10 ST images and 10 LT images and validated on patches extracted from 11 ST and 10 LT.
Patch extraction
For pretraining, patches were sampled randomly using a sliding window of with a stride of 10 throughout the image volume, ignoring the voxel labels. The proposed scheme leads to the generation of a dataset skewed toward the most frequently occurring class; we refer to this as pretraining the network with “no class balance.”
Patches for fine-tuning were extracted from the tumor and its vicinity. This sampling scheme reduces the data imbalance between lesion and nonlesion patches to a great extent; however, this technique fails to eliminate class imbalance completely. The patch extraction scheme for the network is shown in Table 3.
Table 3.
Patch extraction scheme for DSSN.
| Pretraining | Fine-tuning | No. of patients | |
|---|---|---|---|
| DSSN | Systematic sampling, no class balance | Vicinity of tumor | , fine- |
To expose the network to the inherent class imbalance in gliomas, no attempts were made to maintain balance between various classes during training.
2-D patches were preferred over 3-D patches due to:
-
•
SDAEs being a fully connected neural network; the number of parameters in the 3-D patch network would be greater than a 2-D patch network. Further, as the number of parameters in the network increases, the network tends to overfit on the training data.
-
•
Most volumes in the BraTS dataset were acquired axially and hence had highest resolution along the axial plane.
Since the SDAEs were pretrained using unlabeled data and fine-tuned with limited labeled data, the networks are referred to as deep semisupervised network (DSSN).
Data augmentation
In the BraTS 2015 dataset, lesion classes constitute less than 2% of the image volume, which makes data augmentation unavoidable. Data augmentation was done by rotating image patches through various angles. The angles were chosen such that the fill in regions are minimized during the interpolation. Arbitrary angles are also possible but the impact of zero filling or zero padding would be difficult to determine. Augmentation is done on the fly, thus minimizing hard disk and RAM usage. Multithreaded training ensured that patches were augmented and loaded into GPU memory without slowing down training. It was observed that performing label preserving rotations during fine-tuning had a significant impact on classifying less prevalent classes like nonenhancing tumor and the necrotic region.
For the HGG network, data augmentation was carried out by rotation of patches (90 deg, and 180 deg). Rotation of patches by the aforementioned angles prevents introduction of zero padding. For a ST point patient patch in hand, an inbuilt random number generator was used to uniformly generate an integer between 1 and 3, where 1, 2, and 3 indicate rotation by 90 deg, , or 180 deg, respectively.
The presence of surgical cavities, radiation-induced necrosis, etc., in longitudinal patients makes segmentation of lesions from them more challenging than ST point patients. We observed that augmenting longitudinal patient data (HGG) improved segmentation on these volumes. To augment data, patches from longitudinal data were rotated by all three angles (90 deg, , and 180 deg). The original patch and its augmented version(s) were fed to the network.
Training of HGG network
The network was pretrained layer by layer using 941,716 patches with 25% masking noise for 50 epochs. The parameters of the network in each layer were learned by minimizing the reconstructional error with RmsProp44 as the optimizer. The networks used a sigmoid encoder and a linear decoder.
For fine-tuning, the weights and biases connecting the penultimate layer and decision layer were initialized with zeros. The network was trained using 3,304,035 patches (1,888,020-ST and 1,416,015-LT) and validated on 411,495 patches (235,140-ST; 176,355-LT). The weights of the network were learned by minimizing the negative log likelihood cost function using stochastic gradient descent with momentum equal to 0.9. The learning rate was initialized to 0.005 and was annealed as a function of number of epochs [Eq. (1)] with a learning rate decay of 0.001. To prevent overfitting, dropouts45 of 25% were used in all layers:
| (1) |
Hyperparameter optimization
Hyperparameters, such as patch size, optimizer, number of neurons in each layer, amount of data augmentation, etc., were set by random search. The hyperparameter of choice was found by retaining all other hyperparameters that were found to have positive impacts on performance.
3.4.2. LGG network
Due to limited amount of LGG volumes in the dataset, the network pretrained on the HGG data was fine-tuned using 20 patient volumes and validated on 11 patient volumes. The LGG network was fine-tuned by extracting patches in the same manner, as explained in Sec. 3.4.1.1.
Data augmentation
Due to a fewer number of training data, the extracted LGG patches were rotated by 90 deg, , 180 deg, 45 deg, and . During the training phase, LGG patches and their augmented versions were fed as input to the LGG network.
Training of LGG network
Due to the limited amount of LGG volumes, the network pretrained on the HGG data was fine-tuned with dropouts (35%) using LGG image patches (training—1,365,450 and validation—181,170). The LGG network was fine-tuned using the same learning rate, momentum, etc., as the ones used for fine-tuning the HGG network (Sec. 3.4.1.3).
The hyperparameters related to the LGG network were found in the similar fashion as that for the HGG network (Sec. 3.4.1.4).
3.4.3. Prediction
In the test phase for both networks (HGG and LGG), overlapping patches () of size were extracted from all four sequences and fed to the SDAE to classify the center voxel of the patch as one of the five classes and thereby to generate a label map for the entire volume.
3.5. Postprocessing Using ND and CND
ND and CND are one layer deep DAE (Table 4), trained on nonlesion patches to accurately reconstruct the input. The ND and its variant (CND) were used to reduce FPs in the predictions made by SDAE.
Table 4.
ND architecture. , no. of neurons in the th hidden layer.
| Network | Input layer | Output layer | |
|---|---|---|---|
| ND | 882 | 3500 | 882 |
3.5.1. Novelty detector
Patch extraction
In cases of gliomas, FLAIR and T2-weighted images are the sequences used by radiologists to evaluate patient’s response to therapy,46 so we used patches () from FLAIR and T2 images for training the ND. The nonlesion regions in the volume were sampled to obtain the patches.
Training of ND
ND, a one layer DAE, comprises a sigmoid encoding layer and a linear decoder. The ND was trained on 1,110,492 patches (576,636-ST; 533,856-LT) and validated on 438,275 patches (193,955-ST; 244,320-LT) extracted from the same subset of data that were used to fine-tune the HGG network. The training data were corrupted by 20% masking noise. The weights and biases of the network were randomly initialized. The network was trained by minimizing the mean squared error for 200 epochs with a learning rate of 0.001 and RmsProp as the optimizer. A L2 regularizer of 0.0001 was added to the cost function to prevent overfitting.
Error map generation using ND
Overlapping patches of size with stride set to 1 were extracted from T2 and FLAIR and were fed to the ND to generate a reconstruction error map. The reconstruction error map for a slice was constructed by assigning to every voxel in the image the mean reconstruction error, ND, of the patch centered at that voxel [Eq. (2)]. This led to a heat map like image with large error regions corresponding to the location of the glioma/lesion. In Eq. (2), () are the image coordinates of the center voxel of a patch; is the size of the patch, i.e., each patch was of size ; is the normalization term; and is the patch error. Patch error [Eq. (3)] is defined as the squared difference between the FLAIR () and () patches centered around and their respective reconstruction i.e. the reconstructed FLAIR patch (RF) and reconstructed T2 patch (RT). The error map was then binarized using Otsu’s thresholding47 technique:
| (2) |
| (3) |
Following the generation of binary mask, connected component analysis was carried on the image predicted by the HGG and LGG networks. Connected components that had a nonempty intersection with the binary error mask were retained while the rest were discarded.
Appendix C elaborates on the relationship between input patch size and performance of ND.
3.5.2. CND
The CND differs from ND by the error value assigned to the voxel of interest. Thus, CND does not involve an exclusive training phase and thereby makes use of the same one layer DAE network trained based on Sec. 3.5.1.2.
Error map generation using CND
Similar to ND, overlapping patches of size with stride set to 1 were extracted from T2 and FLAIR and were fed to the CND to generate the error map. In CND, the final resultant error value corresponding to a voxel was calculated by maintaining a cumulative error sum over all the image patches containing the voxel. The calculation of the error for a voxel with and as its coordinates is given in Eq. (4), where and are offsets of the patch centers that contribute to the reconstruction error at (, ) and is the patch error, as defined in Eq. (3). The error map generated by CND was binarized by setting a threshold one standard deviation away from the mean:
| (4) |
Following the generation of binary mask, connected component analysis was conducted on the image predicted by the HGG and LGG networks. Connected components that had a nonempty intersection with the binary error mask were retained while the rest were discarded.
3.6. Segmentation of Ischemic Lesion Using ND and CND
The ND and CND trained on the BraTS dataset were used on the ISLES dataset to detect ischemic lesions. Segmentation of the lesion was achieved by binarizing the error maps generated by ND and CND. ND error maps were binarized using Otsu’s thresholding, whereas CND error maps were binarized by setting a threshold one standard deviation away from the mean.
4. Results and Discussion
For segmentation-related tasks, Dice score or Dice similarity coefficient (DSC) is the commonly used metric to measure the performance of the algorithm. DSC is given by Eq. (5), where TP, FP, and FN are the number of true positives, false positives, and false negatives calculated upon comparing the segmentations generated by the algorithm and the associated ground truth. The value of DSC ranges between 0 and1, and a DSC equivalent to 1 depicts the ability of the algorithm to perform segmentations as good as an expert:
| (5) |
4.1. Hyperparameter optimization
Hyperparameters, such as patch size, optimizer, number of neurons in each layer, amount of data augmentation, etc., were set by random search. A base model with four hidden layers (2500, 1000, 500, and 500) was trained with various combinations of the hyperparameters and was tested on the stand out test set. The hyperparameter corresponding to the best Dice score following postprocessing by ND was identified. In all experiments, the hyperparameter of choice was found by retaining all other hyperparameters, which were found to have positive impact on performance (Table 5).
Table 5.
Hyperparameter selection.
| Architecture | Hyperparameter | WT | TC | AT |
|---|---|---|---|---|
| 1764-2500-1000-500-500-5* | Masking noise 10%, data augmentation | |||
| 1764-2500-1000-500-500-5* | Masking noise 25%, data augmentation | |||
| 1764-2500-1000-500-500-5* | Masking noise 25%, no data augmentation | |||
| 1764-3500-2000-1000-500-5* | Masking noise 25%, data augmentation | |||
| 1764-3500-2000-1000-1000-5* | Masking noise 25%, data augmentation | |||
| 1764-3500-2000-1000-1000-5** | Rotation of patches using larger patches () | — | ||
| 1764-3500-2000-1000-1000-5** | Rotation of patches using patches | — |
Note: WT, mean whole tumor Dice score; TC, mean tumor core Dice; AT, mean active tumor Dice score. Results in bold indicate improvement in Dice score, * indicates test carried out on local HGG test data (), ** indicate test carried out on local LGG test data (), and hence active tumor Dice score not was calculated.
The inputs to the base model were corrupted by 10% and 25% masking noise, respectively; it was observed that the latter provided better whole tumor and tumor core segmentation when compared to corrupting the input by a lower amount of noise. Data augmentation plays a pivotal role in the network’s performance and a dip in all compartments was observed upon eliminating the data augmentation scheme from the training regime.
Increasing the number of neurons per layer supplements the discriminative power of the network. On HGG data, increasing the number of the neurons in the hidden layers to (3500, 2000, 1000, and 500) led to an increase of 0.03, 0.01, and 0.02 in mean whole tumor, tumor core, and active tumor Dice score, respectively. Increasing the number of neurons in the penultimate layer to 1000 had negligible improvement and was thus discarded.
For the LGG network, due to limited training data, the patches were rotated by 90 deg, , 180 deg, 45 deg, and . Rotation of patches by 45 deg and could be carried out by either:
-
•
extracting larger patches of size (42,42), rotating by required angles, and cropping the rotated patches to or
-
•
rotating the extracted patches by 45 deg and .
Of the two choices, the latter introduces zero padding into the patches, whereaas the former prevents zero padding at the cost of being memory intensive. From the experiments carried out, it was observed that both networks performed equally well and, in the event, 45-deg rotations were carried out on the extracted patches.
4.2. Performance on the Training Data
On ST point patients in the training data (), the proposed technique achieves whole tumor, tumor core, and active tumor Dice scores of , , and , respectively. The performance of the algorithm on the ST point patients is shown in Figs. 3(a)–3(d).
Fig. 3.
Performance of proposed networks: (a) ground truth, (b) prediction, (c) ground truth, (d) prediction, (e) ground truth, (f) prediction, (g) ground truth, (h) prediction, (i) raw prediction, (j) Otsu’s mask, (k) prediction after postprocessing, (l) ground truth, (m) FLAIR, (n) ND error map, (o) binarized error map, and (p) ground truth. In all images, orange indicates edema; yellow indicates nonenhancing tumor; red indicates necrotic region; white indicates enhancing tumor; and green indicates Otsu’s mask. In image (o), white indicates binarized error map.
Figures 3(e)–3(h) show the performance of the network on a longitudinal patient data at two different time points. By including LT point patients as training data, the network attained the capability to capture the tumor region across time points. The whole tumor, tumor core, and active tumor Dice score corresponding to the longitudinal patients () in the dataset were , , and , respectively.
Table 6 compares the performance of the proposed technique against various top performing automatic techniques18,30 and semiautomatic technique48 on the entire BraTS 2015 training data (). It was observed that the proposed technique produced comparable results despite being trained on limited labeled data.
Table 6.
Comparison of proposed technique against top performing contribution in BraTS challenge in 2015, 2016 on the BraTS training data ().
| Technique | Whole tumor | Tumor core | Active tumor | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Spec | Sens | Spec | Sens | Spec | Sens | ||||
| DSSN (no postprocessing) | 0.78 | 0.85 | 0.77 | 0.71 | 0.70 | 0.74 | |||
| DSSN+FLAIR thresholding | 0.79 | 0.85 | 0.77 | 0.71 | 0.70 | 0.74 | |||
| DSSN+ ND (proposed) | 0.85 | 0.85 | 0.81 | 0.71 | 0.76 | 0.74 | |||
| DSSN+CND (proposed) | 0.85 | 0.85 | 0.81 | 0.71 | 0.76 | 0.74 | |||
| DeepMedic [ensemble CNN + conditional random fields (CRF)] | * | * | 0.89 | * | * | 0.72 | * | * | 0.74 |
| Sergio (CNN) | * | * | 0.86 | * | * | 0.77 | * | * | 0.70 |
| Bakas (GLISTRBoost) | * | * | 0.89 | * | * | 0.76 | * | * | 0.75 |
Note: Std, standard deviation; Spec, specificity; Sens, sensitivity; *, information not available.
4.3. Effect of ND and CND on BraTS Dataset
Figures 3(i)–3(l) demonstrate the reduction in FP voxels using ND. Postprocessing using the ND mask led to good improvements in glioma segmentation. On the entire training data (), the improvement in performance was in the order of 0.04 for whole tumor Dice score, 0.03 for tumor core, and 0.04 for active tumor (Table 6).
Figure 3(n) shows the reconstruction error heat map for a sample slice [Fig. 3(m)]. A bimodal distribution could be inferred on visual inspection of the error map. The mean square reconstruction error corresponds to maximizing the log-likelihood of the training data assuming a Gaussian distribution. Given this interpretation, any data input that does not correspond to the training data distribution can be expected to give rise to a large mean square error enabling lesion patch detection. Cross-validation can be used to determine the ideal threshold but would have to be changed depending on the lesion. Binarizing individual error volume using Otsu’s technique guarantees a robust threshold for each patient irrespective of the lesion, without the need for cross-validation.
A sample ND error map binarized using Otsu’s thresholding is shown in Fig. 3(o). The mean square error of a patch is assigned to the center pixel of the patch; consequently, the voxels toward the boundaries of the lesions will get assigned a much lower error than the voxels near the center of the lesion. Thus, the ND error map underestimates the size of the lesion. For every voxel in the volume, CNDs take into account the reconstruction error from patches centered on its neighbors; therefore, the degree of undersegmentation of the lesion is lower in CNDs when compared with NDs [Figs. 4(a)–4(c) and 4(k)–4(o)].
Fig. 4.
Performance of ND and CND on BraTS dataset. (a) ND error map, (b) CND error map, (c) ground truth, (d) CND error map, (e) ground truth, (f) FLAIR, (g) T2, (h) CND error map, (i) ground truth, (j) binarized CND error map, (k) FLAIR, (l) T2, (m) binarized ND error map, (n). binarized CND error map, (o) ground truth in images (c), (e), (i) and (o), orange indicates edema; yellow indicates nonenhancing tumor; red indicates necrotic region; and white indicates enhancing tumor. In images (m) and (n), green indicates binarized error map.
Qualitatively, it was observed that CNDs produce a unique error distribution for various constituents of the lesion [Figs. 4(d), 4(e), 4(h), and 4(i)]. From the CND error map, the necrotic region would be easily delineated from edema and enhancing tumor. It is noteworthy that even though CND was trained on FLAIR and T2 and not T1c, it was able to delineate enhancing tumor regions. However, this was only possible if there existed a corresponding hyperintensity profile in either of its input sequences. No changes in Dice scores were observed upon the inclusion of T1c sequence as an additional input to ND and CND. Furthermore, addition of the T1c sequence increases the number of parameters of the network; thus, in the interest of minimizing the number of parameters without affecting the performance, ND and its variant were trained on FLAIR and T2 sequences.
The CND-generated error map could be used as an initialization point for various generative techniques. The whole lesion could be segmented from the CND error map by setting the threshold to be one standard deviation away from the mean [Figs. 4(f)–4(j)]. ND and its variant help in localizing the lesion in the volume and apply a spatial constraint of the prediction made by the SDAE network, thus reducing the FPs.
Due to the hyperintense appearance of lesion in the FLAIR sequence, thresholding of the sequence is an often used technique to localize possible lesion candidates. Table 6 shows the mean Dice score of the raw predictions (no postprocessing) and the performance after reduction of FPs using thresholding of FLAIR sequence, ND, and CND. Postprocessing using binarized FLAIR sequence yields no improvement in scores. Figures 5(a)–5(d) showcase a scenario wherein thresholding of FLAIR retains the FPs. The ND considers all voxels in the slice as nonlesion and thereby assigns a low reconstruction error [Fig. 5(e)], in the process of helping in reduction of FPs [Fig. 5(f)] and improving the overall performance (specificity and Dice score) of the classifier.
Fig. 5.
Performance of various FP reduction technique. (a) FLAIR, (b) raw prediction, (c) binarized FLAIR, (d) postprocessed output using binarized FLAIR, (e) reconstruction error map using ND, (f) postprocessed output using ND error map. Images (b)–(d) and (f) are overlaid on top of FLAIR.
The postprocessing technique explained in Secs. 3.5.1.3 and 3.5.2.1 aids in the reduction of FPs by retaining the connected components in the label map that had a nonempty intersection with the binarized error map generated by ND or CND. Compared with CND, though the binarized error map generated by ND undersegments the lesion boundaries, it still maintains a nonempty intersection with the components in the label map. The reason stated above explains how postprocessing using ND achieves performance on par with CND despite undersegmenting the lesion (Table 6).
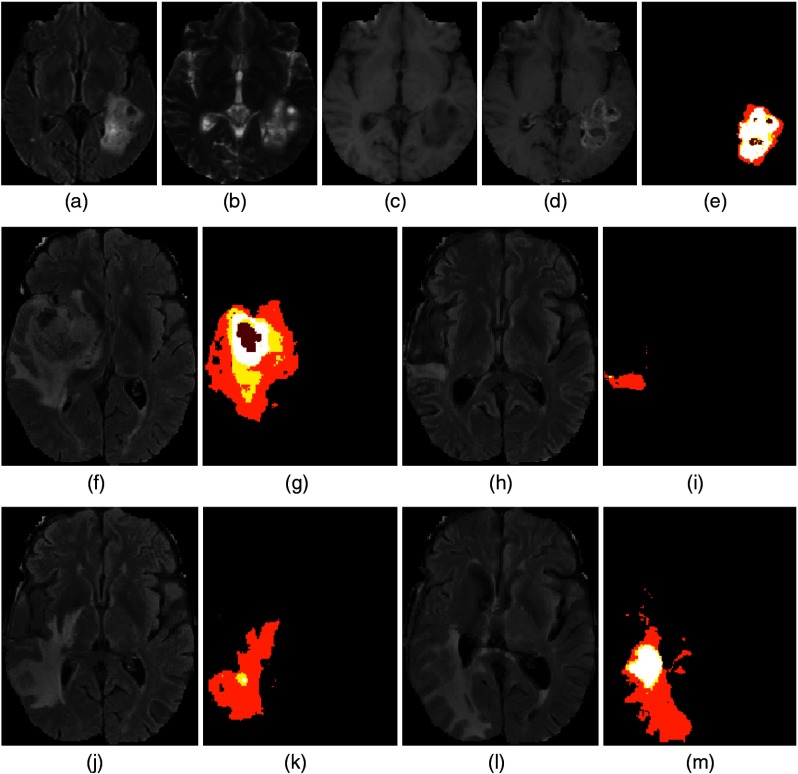
4.4. Performance of ND and CND on the ISLES Dataset
As ND and its variant were trained exclusively on nonlesion patches, apart from reduction of FPs, they could also be used to localize possible lesion candidates from MR scans. The ND and CND were tested on the ISLES dataset () to measure its ability to capture ischemic lesions. On patients with lesions that constitute more than 1% of the total number of voxels (), ND achieved a Dice score of [Figs. 6(a)–6(d), 6(h)–6(k), and 6(o)–6(r)]. By contrast, CND achieved a much higher Dice score of [Figs. 6(e), 6(f), 6(l), 6(m), 6(s), and 6(t)]. The improved performance of CND was due to the difference between ND and CND error map estimation. For the patches centered on the boundary of a lesion, straddling normal tissue, and tumor tissues, the ND error estimates are lower as the reconstruction error from the normal tissue component tends to be low. However, for the CND, there is a contribution to the reconstruction error of the boundary pixels from patches centered on the neighboring pixels inside the lesion tissue class. This leads to an increased reconstruction error of the boundary pixels thus preventing undersegmentation.
Fig. 6.
Segmentation of ischemic lesion using ND and CND. (a) FLAIR, (b) ND errormap, (c) binarized ND error map, (d) CND error map, (e) binarized CND error map, (f) ground truth, (g) FLAIR, (h) T2, (i) ND error map, (j) binarized ND error map, (k) CND error map, (l) binarized CND error map, (m) ground truth, (n) FLAIR, (o) T2, (p) ND error map, (q) binarized ND error map, (r) CND error map, (s) binarized CND error map, and (t) ground truth. In images (c), (e), (j), (l), (q), and (s), green indicates binarized error map. In images (f), (m), and (t), red indicates ischemic lesion.
Similar to glioma segmentation, the error map picks up the location of the ischemic lesions accurately; however, ND and CND miss lesions that constitute less than 1% of total number of voxels in the volume due to resolution mismatch between the ISLES and BraTS database. The T2-weighted sequences in the ISLES dataset were acquired in the sagittal plane as opposed to the axial plane acquisition in the BraTS dataset. Since the ND and CND were trained using the BraTS dataset, the resolution mismatch would lead to poor performance in detecting small lesions. These results imply that the ND and its variant can be trained using data from healthy volunteers or other imaging studies comprising relevant MR sequences.
The results provided above are on a limited number of patients, i.e., with lesion burden , and hence cannot be compared with state-of-the-art techniques presented as part of the ISLES challenge.
4.5. Prediction with Missing Sequences
The performance of the network upon blocking individual sequences from the input is shown in Table 7. Prediction with a missing sequence was expected to lower the Dice scores; however, the magnitude of the decline was dependent on the sequence dropped. The results were also informative, indicating the relative importance of the sequences. Removing T1 had negligible impact on the whole tumor score while removing T2 and FLAIR lead to the maximum change, i.e., decline. The change in Dice scores of enhancing tumor or active tumor was the largest when T1c was removed, which can be expected. Based on the decline in performance, one can conclude that FLAIR plays an important role in delineating lesion from normal tissues.
Table 7.
Performance of DSSN performance with missing sequences (MS).
| MS | Whole tumor | Tumor core | Active tumor | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Mean | Std | Median | Mean | Std | Median | Mean | Std | Median | |
| FLAIR | 0.35 | 0.23 | 0.35 | 0.55 | 0.29 | 0.63 | 0.58 | 0.31 | 0.68 |
| T2 | 0.79 | 0.13 | 0.82 | 0.61 | 0.29 | 0.69 | 0.70 | 0.26 | 0.80 |
| T1 | 0.80 | 0.16 | 0.85 | 0.61 | 0.25 | 0.66 | 0.58 | 0.29 | 0.60 |
| T1c | 0.81 | 0.13 | 0.86 | 0.40 | 0.22 | 0.38 | 0.00 | 0.00 | 0.00 |
| None | 0.86 | 0.12 | 0.90 | 0.76 | 0.24 | 0.83 | 0.79 | 0.23 | 0.86 |
Note: Std, standard deviation.
4.6. Weakly Supervised Learning
The minimum amount of data required for the network to maintain its level of performance was tested by fine-tuning the LGG and HGG networks with a lower number of patient data (leading to a decreasing number of patches). The results in Table 8 show that, training with patches drawn from only 20 patients, the HGG network had marginal decline in Dice scores, which are comparable to results obtained when the networks were trained on patches drawn from a larger number of patients. It is also notable that if the number of extracted patches is increased from a limited number of patients then the network performance rebounds, as shown in Table 8. The structures in the brain appear similar across different brain MR images. Drawing patches from a limited number of patient volumes coupled with data augmentation would still provide enough samples for the network to learn and maintain prediction performance.
Table 8.
Performance of network on local test set () based on number of training patients used (), (20 M) is 20 patients with more patches.
| N | Whole tumor | Tumor core | Active tumor | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Mean | Std | Median | Mean | Std | Median | Mean | Std | Median | |
| 20 | 0.84 | 0.13 | 0.89 | 0.72 | 0.24 | 0.81 | 0.74 | 0.25 | 0.84 |
| 40 | 0.85 | 0.13 | 0.90 | 0.75 | 0.23 | 0.83 | 0.78 | 0.23 | 0.87 |
| 65 | 0.84 | 0.15 | 0.89 | 0.75 | 0.23 | 0.83 | 0.78 | 0.24 | 0.86 |
| 20M | 0.86 | 0.12 | 0.90 | 0.76 | 0.24 | 0.83 | 0.79 | 0.23 | 0.86 |
Note: Std, standard deviation.
4.7. Transfer Learning for LGG
The LGG network was trained by fine-tuning the pretrained HGG network. Compared with the authors’ submission49 to the BraTS 2015 challenge, an 8% improvement was observed in the tumor core Dice for LGG volumes (Table 9).
Table 9.
Performance of DSSN on challenge datasets compared against the authors’ original submission to BraTS 2015 challenge and other top performing algorithms.
| Year | Nw | G | Whole tumor | Tumor core | Active tumor |
|---|---|---|---|---|---|
| 2013 | DSSN | All | |||
| 2015 | PS | HGG | |||
| LGG | * | ||||
| All | |||||
| DSSN | HGG | ||||
| LGG | * | ||||
| All | |||||
| 2015 | Sergio (CNN) | All | ** | ** | ** |
| 2015 | DeepMedic (ensemble of CNN and CRF) | All | ** | ** | ** |
Note: Nw, network; G, grade of tumor; PS, previous submission; DSSN, current submission. Based on the Wilcoxon rank-sum test (), results in bold show improvement in Dice scores in the proposed technique when compared with the author’s previous submission. * indicates Dice scores were not calculated for a particular class and ** indicate information not available.
In the author’s previous submission, preprocessing of data comprises histogram matching, -score normalization, and clipping of extreme intensity values. Clipping of intensity values led to saturation of intensities within the lesion, enhancing the lesion pixels compared with the normal tissue. This resulted in LGG whole tumor Dice scores comparable to the HGG Dice scores. However, the saturation led to loss of contrast between the different tissue classes in the tumor and, consequently, a lower tumor core Dice score.
It was observed that removing clipping of extreme intensities from the preprocessing pipeline and training the LGG network using a transfer learning approach improved the capability of the network to classify structures within the tumor. On the test data, compared with LGG, the proposed algorithm performs better on HGG patients, which indicates that pretraining the LGG network with a collection of unlabeled LGG volumes would improve the LGG predictions.
4.8. Performance on Challenge Dataset
The networks were tested on two different challenge test data, namely, BraTS 2013 challenge test data and BraTS 2015 test data. The performance of networks on BraTS 2013 challenge data and BraTS 2015 test data is given in Table 9.
On the entire 2015 BraTS test set, the proposed network improves the whole tumor, tumor core, and active tumor Dice score by order of 2, 5, and 10%, respectively. Based on the Wilcoxon rank-sum test (), our current approach has a significant positive impact on the active tumor Dice scores.
Table 9 compares the performance of the proposed technique against top performing techniques, such as DeepMedic30 and Serigo.18 It was observed that, though DSSN had fewer layers when compared with the aforementioned CNN-based techniques, for the task of segmenting enhancing tumor from the lesion, DSSN was comparable with DeepMedic. Similarly, for whole tumor segmentation, the performance of DSSN was found to be comparable with Sergio. Typically, the improved performance of CNNs is due to the design of their kernels/filters to enable them to learn spatial information in an image/patch; however, autoencoders fail to learn spatial context in the data as rasterization of inputs could lead to loss of spatial information. The winning algorithm at the BraTS 2015 challenge (GLISTRBoost),48 a semiautomatic technique based on gradient boosting, was excluded from Table 9 due to unavailability of the performance metrics on the challenge data.
5. Conclusion
In this paper, we propose a completely automated brain tumor segmentation technique with a FP/candidate detection method based on DAEs.
-
•
Despite differences in acquisition resolution, ND trained using nonlesion patches (BraTS data) was able to learn the normal brain structure and detect ischemic lesions (ISLES data). A variant of ND (CND), wherein a cumulative error map was calculated for every voxel, was able to significantly improve lesion detection performance on ISLES data. In addition, CND error maps assigned different error distributions to various constituents of glioma, making it an ideal tool to construct tumor atlases. This can also serve as a good initialization for various segmentation techniques.
-
•
The performance of the proposed technique does not surpass state-of-the-art CNN-based techniques; however, the paper clearly demonstrates the ability of SDAEs to produce good segmentation using a minimal number of patient data. An approach using SDAEs would be an ideal choice on applications with a limited amount of labeled data.
-
•
The results presented are the prediction of a single network with minimal data preprocessing and postprocessing. The N4 bias correction technique, which is an often used preprocessing step, was eliminated. Histogram matching to reference data was still done, and future work would be to eliminate the same by appropriate data normalization. Skull stripping (BraTS data and ISLES challenge data were skull stripped) could potentially be eliminated as a separate step using ND. The idea is to enable prediction on MR images without expensive preprocessing.
In summary, the work presented applies SDAEs for the brain lesion detection and segmentation task using a limited number of training data. The ND concept allows for efficient elimination of FPs and candidate detection, making it a valuable CAD tool.
Biographies
Ganapathy Krishnamurthi is an assistant professor at the Indian Institute of Technology Madras. He received his BS and MS degrees in physics from the University of Madras (India) and Indian Institute of Technology Madras (India), respectively, and his PhD from Purdue University (USA). His current research interests include medical image analysis and small animal imaging.
Biographies for the other authors are not available.
Appendix A: Data
In this paper, publicly available BraTS 2015 and ISLES 2015 datasets were used. SDAEs were trained and tested on the BraTS dataset. ND was trained on the BraTS dataset and tested on the BraTS and ISLES database.
A.1. BraTS Dataset
The BraTS 2015 training dataset comprises 274 cases of which 220 cases and 54 cases correspond to HGG and LGG, respectively. Each training case comprises four MRI sequences (T2 weighted FLAIR, T2, T1, T1c) and the associated ground truth [Figs. 7(a)–7(e)]. Each sequence gives complimentary information about the lesion. For example, FLAIR and T2 help in localizing the lesion while T1 postsequence helps in detecting rupture in the blood brain barrier (BBB) due to the lesion and the necrotic region. In cases of gliomas, rapture of the BBB is called enhancing part of the tumor.
Fig. 7.
BraTs dataset. (a) FLAIR, (b) T2, (c) T1, (d) T1c images, (e) associated ground truth, (f) FLAIR at , (g) associated ground truth, (h) FLAIR at , (i) associated ground truth, (j) FLAIR at . (k) associated ground truth, (l) FLAIR at , (m) associated ground truth. In all images, orange indicates edema; yellow indicates nonenhancing tumor; white indicates enhancing tumor; red indicates necrotic region.
The HGG dataset is composed of patients imaged only once (ST point) and patients who are scanned over a period of time to study the growth of tumor (longitudinal data) or to study treatment response. The HGG dataset comprises 123 ST point patients and 97 longitudinal patients, while no longitudinal studies were found in the LGG dataset. Figures 7(f)–7(m) show the lesion at four different time points.
The majority of sequences in the dataset are acquired axially and thus have highest resolution along axial plane. Therefore, the SDAE (SDAEs) and ND (ND) were trained on patches extracted from the axial plane of the data.
A.2. ISLES Dataset
The ISLES training dataset comprises 28 patient volumes, each consisting of four MR sequences (FLAIR, T2, T1, and diffusion weighted image) and the associated ground truth.
Unlike the BraTS dataset, the FLAIR sequence had the highest resolution along the axial plane while T2 had the highest resolution along the sagittal plane. The axial and sagittal views of the various sequences are given in Figs. 8(a)–8(e), 8(f)–8(j), respectively. Poor resolution of the T2 weighted image along the axial plane hampers the performance of the ND since the ND was trained on axially acquired FLAIR and T2 volumes.
Fig. 8.
Axial and sagittal view of ISLES dataset. (a) DWI, (b) FLAIR, (c) T1, (d) T2, (e) ground truth, (f) DWI, (g) FLAIR, (h) T1, (i) T2, and (j) ground truth.
Appendix B: Autoencoders
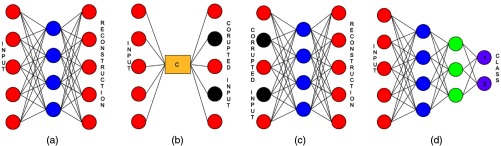
Autoencoders are fully connected neural networks that are trained to reconstruct the input. A traditional autoencoder comprises three layers, namely, the input layer, hidden layer, and output layer [Fig. 9(a)]. The input layer and the hidden layer form the encoder part of an autoencoder, whereas the hidden layer and the output layer form the decoder part of an autoencoder.
Fig. 9.
(a) Autoencoders—traditional autoencoder; (b) corruption of input by masking noise; (c) denoising autencoder; and (d) deep neural network.
The weights () and biases () in the encoder map the input to a different dimension, which is called the hidden representation of the data [Eq. (6)]. The decoder maps the hidden representation to the input space [Eq. (7)], where and are weights and biases in the decoder, respectively. In other words, the decoder tries to reconstruct the input from hidden representations. The parameters, i.e., the weights and biases connecting the input, hidden, and output layers are optimized by minimizing the error between the input and reconstruction [Eq. (8)], where is the input and is the reconstruction. Since the training procedure involves minimization of reconstructional error, autoencoders tend to learn the distribution of the data:
| (6) |
| (7) |
| (8) |
The hidden layer learns strong features in the data, such as edges, patterns, texture, etc., which hence are considered feature extractors. The features learned by a network depend on the constraints added to a network, such as number of neurons, activation functions, L2 penalty, etc. Table 10 shows the transforms learned by an autoencoder. Autoencoders can approximate principal component analysis (PCA) by using a linear activation function and setting the number of neurons in the hidden layer to be less than the number of neurons in the input layer. Sigmoid and other nonlinear activation functions perform a nonlinear PCA on the input data. When the number of neurons in the hidden layer is greater than the number of input neurons, the network learns sparse representation of the data. Networks with fewer neurons in the hidden layer when compared with input neurons are called undercomplete autoencoders, and those with more neurons in hidden layer when compared to the input layer are called overcomplete autoencoders.
Table 10.
Various transforms learned by autoencoders.
| Size | Activation | Transform |
|---|---|---|
| Linear | Identity | |
| Linear | PCA | |
| Sigmoid | Sparse |
Hidden layers are stacked to form deep autoencoders/stacked autoencoders to capture higher order features from the data. Each layer in a deep autoencoder is pretrained in a greedy unsupervised manner and the input is from the layer below it. As the depth of the network increases, the representations learned at each layer are more abstract than in the preceding layers.
B.1. Denoising Autoencoder
In DAEs, the network is forced to reconstruct the uncorrupted/original input from partially corrupted input. The input is artificially corrupted by injecting noise such as gaussian noise, salt and pepper noise, or masking noise. Corruption of input is given by Eq. (9), where is the input, is the type of noise used, and is corrupted input [Fig. 9(b)]:
| (9) |
The encoder and decoder of a DAE are similar to those of a conventional autoencoder. The encoding and decoding functions are given by Eqs. (10) and (11), respectively:
| (10) |
| (11) |
The weights and biases of the network are learned by minimizing the least square error between the reconstruction () and the uncorrupted input [; Eq. (12)]:
| (12) |
Due to the denoising of data during the training phase, DAEs learn the manifold of the data, and the features learned by a DAE are robust to noisy input. For example, on natural images, with appropriate noising, DAEs learn Gabor like edge detectors, while a L2 regularized conventional undercomplete autoencoder learns local blob detectors and features learned by conventional overcomplete network had no recognizable structure.21
DAE pretraining is done using unlabeled data [Fig. 9(c)]. Apart from learning higher level representations of input at each layer, pretraining also helps in attaining good initialization for the deep classifier.50 For classification related tasks, SDAEs are fine-tuned with class label outputs. Encoders of all layers are stacked to form a deep multilayer perceptron (MLP). An additional layer is concatenated to the MLP, and the number of neurons in this layer equals the number of classes in the dataset [Fig. 9(d)]. The parameters in the network are learned by minimizing the negative log likelihood loss function [Eq. (13)], where are parameters of the network, is size of the dataset, is the th datapoint in the dataset, and is the label associated to it:
| (13) |
SDAEs can be pretrained on a large amount of unlabeled data to a good initialization for a deep network. Fine tuning is then carried out using a small set of labeled data.
Appendix C: Effect of Patch Size on Novelty Detector Performance
The patch size decides the amount of the data that is fed into the network and is one of the most important hyperparameters of the network. ND underestimates the size of the lesion on both glioma and ischemic lesion. Since the mean square error of a patch is assigned to the center voxel of the patch, the voxels towards the boundaries of the lesions are assigned with a much lower error value than the voxels near the center of the lesion. ND trained with smaller patch sizes fails to learn the various structures in the brain such as the ventricles and thus creates a erroneous results. The performance of ND with various patch sizes on the ISLES and BraTS datasets is given in Figs. 10(a)–10(d), 10(f)–10(i), respectively. No significant improvement in performance was noted upon using a patch size of when compared with a patch size of . A 3-D version of ND () yielded similar results compared with the 2-D variant (). The ratio of the number of neurons in the hidden and input layers was maintained for different patch sizes.
Fig. 10.
Effect of ND based on input patch size (a–g) on ISLES dataset and (i–n) on BraTS dataset. (a) Patch size 11, (b) patch size 15, (c) patch size 21, (d) patch size 31, (e) two layer ND, (f) three layer ND, (g) ground truth, (h) patch size 11, (i) patch size 15, (j) patch size 21, (k) patch size 31, (l) two layer ND, (m) three layer ND, and (n) ground truth. In image (g), red indicates ischemic lesion. In image (n), orange indicates edema, yellow indicates nonenhancing tumor; red indicates necrotic region; and white indicates enhancing tumor.
Deeper variants of the ND with two hidden layers (3500 and 2000 hidden units) and three hidden layers (3500, 2000, and 1000 hidden units) were found to produce similar results as compared with a one layer DAE. Thus, in the interest of minimizing the amount of network parameters without affecting the performance, a one layer ND trained on patches of size was retained.
Disclosures
Authors have no competing interests to declare.
References
- 1.Menze B. H., et al. , “The multimodal brain tumor image segmentation benchmark (BRATS),” IEEE Trans. Med. Imaging 34(10), 1993–2024 (2015). 10.1109/TMI.2014.2377694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Corso J. J., et al. , “Efficient multilevel brain tumor segmentation with integrated Bayesian model classification,” IEEE Trans. Med. Imaging 27(5), 629–640 (2008). 10.1109/TMI.2007.912817 [DOI] [PubMed] [Google Scholar]
- 3.Görlitz L., et al. , “Semi-supervised tumor detection in magnetic resonance spectroscopic images using discriminative random fields,” in Joint Pattern Recognition Symp., pp. 224–233 (2007). [Google Scholar]
- 4.Schmidt M., et al. , “Segmenting brain tumors using alignment-based features,” in Proc. Fourth Int. Conf. on Machine Learning and Applications, p. 6, IEEE; (2005). 10.1109/ICMLA.2005.56 [DOI] [Google Scholar]
- 5.Prastawa M., et al. , “A brain tumor segmentation framework based on outlier detection,” Med. Image Anal. 8(3), 275–283 (2004). 10.1016/j.media.2004.06.007 [DOI] [PubMed] [Google Scholar]
- 6.Menze B. H., et al. , “A generative probabilistic model and discriminative extensions for brain lesion segmentation with application to tumor and stroke,” IEEE Trans. Med. Imaging 35(4), 933–946 (2016). 10.1109/TMI.2015.2502596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Leemput K., et al. , “Automated model-based bias field correction of MR images of the brain,” IEEE Trans. Med. Imaging 18(10), 885–896 (1999). 10.1109/42.811268 [DOI] [PubMed] [Google Scholar]
- 8.Meier R., et al. , “A hybrid model for multimodal brain tumor segmentation,” in NCI-MICCAI Challenge on Multimodal Brain Tumor Segmentation. Proc. of NCI-MICCAI BRATS, pp. 31–37 (2013). [Google Scholar]
- 9.Meier R., et al. , “Appearance-and context-sensitive features for brain tumor segmentation,” in Proc. of MICCAI BRATS Challenge, pp. 020–026 (2014). [Google Scholar]
- 10.Zikic D., et al. , “Decision forests for tissue-specific segmentation of high-grade gliomas in multi-channel MR,” in Medical Image Computing and Computer-Assisted Intervention–MICCAI, pp. 369–376 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Bauer S., et al. , “Segmentation of brain tumor images based on integrated hierarchical classification and regularization,” in MICCAI BraTS Workshop, Miccai Society, Nice: (2012). [Google Scholar]
- 12.Reza S., Iftekharuddin K., “Multi-fractal texture features for brain tumor and edema segmentation,” Proc. SPIE 9035, 903503 (2014). 10.1117/12.2044264 [DOI] [Google Scholar]
- 13.Tustison N. J., et al. , “Optimal symmetric multimodal templates and concatenated random forests for supervised brain tumor segmentation (simplified) with ANTsR,” Neuroinformatics 13(2), 209–225 (2015). 10.1007/s12021-014-9245-2 [DOI] [PubMed] [Google Scholar]
- 14.Geremia E., Menze B. H., Ayache N., “Spatially adaptive random forests,” in IEEE 10th Int. Symp. on Biomedical Imaging (ISBI ’13), pp. 1344–1347, IEEE; (2013). 10.1109/ISBI.2013.6556781 [DOI] [Google Scholar]
- 15.Pinto A., et al. , “Brain tumour segmentation based on extremely randomized forest with high-level features,” in 37th Annual Int. Conf. of the IEEE Engineering in Medicine and Biology Society (EMBC ’15), pp. 3037–3040, IEEE; (2015). 10.1109/EMBC.2015.7319032 [DOI] [PubMed] [Google Scholar]
- 16.Bauer S., Nolte L.-P., Reyes M., “Fully automatic segmentation of brain tumor images using support vector machine classification in combination with hierarchical conditional random field regularization,” in Int. Conf. on Medical Image Computing and Computer-Assisted Intervention, pp. 354–361, Springer; (2011). [DOI] [PubMed] [Google Scholar]
- 17.Lee C.-H., et al. , “Segmenting brain tumors using pseudo–conditional random fields,” in Medical Image Computing and Computer-Assisted Intervention (MICCAI ’08), pp. 359–366 (2008). [PubMed] [Google Scholar]
- 18.Pereira S., et al. , “Brain tumor segmentation using convolutional neural networks in MRI images,” IEEE Trans. Med. Imaging 35(5), 1240–1251 (2016). 10.1109/TMI.2016.2538465 [DOI] [PubMed] [Google Scholar]
- 19.Krizhevsky A., Sutskever I., Hinton G. E., “ImageNet classification with deep convolutional neural networks,” in Advances in Neural Information Processing Systems, pp. 1097–1105 (2012). [Google Scholar]
- 20.Hinton G. E., Salakhutdinov R. R., “Reducing the dimensionality of data with neural networks,” Science 313(5786), 504–507 (2006). 10.1126/science.1127647 [DOI] [PubMed] [Google Scholar]
- 21.Vincent P., et al. , “Stacked denoising autoencoders: learning useful representations in a deep network with a local denoising criterion,” J. Mach. Learn. Res. 11, 3371–3408 (2010). [Google Scholar]
- 22.Havaei M., et al. , “Brain tumor segmentation with deep neural networks,” Med. Image Anal. 35, 18–31 (2017). 10.1016/j.media.2016.05.004 [DOI] [PubMed] [Google Scholar]
- 23.Zikic D., et al. , “Segmentation of brain tumor tissues with convolutional neural networks,” in Proc of BRATS-MICCAI (2014). [Google Scholar]
- 24.Urban G., et al. , “Multi-modal brain tumor segmentation using deep convolutional neural networks,” in Proc., Winning Contribution, MICCAI BraTS (Brain Tumor Segmentation) Challenge, pp. 31–35 (2014). [Google Scholar]
- 25.Lyksborg M., et al. , “An ensemble of 2D convolutional neural networks for tumor segmentation,” in Scandinavian Conf. on Image Analysis, pp. 201–211, Springer; (2015). [Google Scholar]
- 26.Rao V., Shari Sarabi M., Jaiswal A., “Brain tumor segmentation with deep learning,” in MICCAI Multimodal Brain Tumor Segmentation Challenge (BraTS), pp. 56–59 (2015). [Google Scholar]
- 27.Dvorak P., Menze B., “Structured prediction with convolutional neural networks for multimodal brain tumor segmentation,” in Proc. of the Multimodal Brain Tumor Image Segmentation Challenge, pp. 13–24 (2015). [Google Scholar]
- 28.Akkus Z., et al. , “Deep learning for brain MRI segmentation: state of the art and future directions,” J. Digital Imaging 30(4), 449–459 (2017). 10.1007/s10278-017-9983-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao X., et al. , “Brain tumor segmentation using a fully convolutional neural network with conditional random fields,” Lect. Notes Comput. Sci. 10154, 75–87 (2016). 10.1007/978-3-319-55524-9 [DOI] [Google Scholar]
- 30.Kamnitsas K., et al. , “DeepMedic for brain tumor segmentation,” In Int. Workshop on Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries, pp. 138–149, Springer, Cham: (2016). [Google Scholar]
- 31.Havaei M., et al. , “Deep learning trends for focal brain pathology segmentation in MRI,” in Machine Learning for Health Informatics, pp. 125–148, Springer International Publishing; (2016). [Google Scholar]
- 32.Salakhutdinov R., Hinton G., “Deep Boltzmann machines,” in Artificial Intelligence and Statistics, pp. 448–455 (2009). [Google Scholar]
- 33.Agn M., et al. , “Brain tumor segmentation using a generative model with an RBM prior on tumor shape,” in Int. Workshop on Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries, pp. 168–180, Springer; (2015). [Google Scholar]
- 34.Sheet D., et al. , “Deep learning of tissue specific speckle representations in optical coherence tomography and deeper exploration for in situ histology,” in IEEE 12th Int. Symp. on Biomedical Imaging (ISBI ’15), pp. 777–780, IEEE; (2015). [Google Scholar]
- 35.Shin H.-C., et al. , “Stacked autoencoders for unsupervised feature learning and multiple organ detection in a pilot study using 4d patient data,” IEEE Trans. Pattern Anal. Mach. Intell. 35(8), 1930–1943 (2013). 10.1109/TPAMI.2012.277 [DOI] [PubMed] [Google Scholar]
- 36.Xu J., et al. , “Stacked sparse autoencoder (SSAE) for nuclei detection on breast cancer histopathology images,” IEEE Trans. Med. Imaging 35(1), 119–130 (2016). 10.1109/TMI.2015.2458702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carreira P., Hinton G., “On contrastive divergence learning,” Aistats 10, 33–40 (2005). [Google Scholar]
- 38.Hinton G., “A practical guide to training restricted Boltzmann machines,” Momentum 9(1), 926 (2010). [Google Scholar]
- 39.Japkowicz N., et al. , “A novelty detection approach to classification,” in Int. Joint Conf. on Artificial Intelligence, Vol. 1, pp. 518–523 (1995). [Google Scholar]
- 40.Eskin E., “Anomaly detection over noisy data using learned probability distributions,” in Proc. of the Int. Conf. on Machine Learning, Citeseer; (2000). [Google Scholar]
- 41.Kistler M., et al. , “The virtual skeleton database: an open access repository for biomedical research and collaboration,” J. Med. Internet Res. 15(11), e245 (2013). 10.2196/jmir.2930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maier O., et al. , “Extra tree forests for sub-acute ischemic stroke lesion segmentation in MR sequences,” J. Neurosci. Methods 240, 89–100 (2015). 10.1016/j.jneumeth.2014.11.011 [DOI] [PubMed] [Google Scholar]
- 43.Nyúl L. G., Udupa J. K., Zhang X., “New variants of a method of MRI scale standardization,” IEEE Trans. Med. Imaging 19(2), 143–150 (2000). 10.1109/42.836373 [DOI] [PubMed] [Google Scholar]
- 44.Tieleman T., Hinton G., “Lecture 6.5-rmsprop: divide the gradient by a running average of its recent magnitude,” COURSERA: Neural Networks Mach. Learn. 4(2), 26–31 (2012). [Google Scholar]
- 45.Srivastava N., et al. , “Dropout: a simple way to prevent neural networks from overfitting,” J. Mach. Learn. Res. 15(1), 1929–1958 (2014). [Google Scholar]
- 46.Wen P. Y., et al. , “Updated response assessment criteria for high-grade gliomas: response assessment in neuro-oncology working group,” J. Clin. Oncol. 28(11), 1963–1972 (2010). 10.1200/JCO.2009.26.3541 [DOI] [PubMed] [Google Scholar]
- 47.Otsu N., “A threshold selection method from gray-level histograms,” IEEE Trans. Syst. Man Cybern. 9(1), 62–66 (1979). 10.1109/TSMC.1979.4310076 [DOI] [Google Scholar]
- 48.Bakas S., et al. , “Glistrboost: combining multimodal MRI segmentation, registration, and biophysical tumor growth modeling with gradient boosting machines for glioma segmentation,” in Int. Workshop on Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries, pp. 144–155 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vaidhya K., et al. , “Multi-modal brain tumor segmentation using stacked denoising autoencoders,” in Int. Workshop on Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries, pp. 181–194, Springer; (2015). [Google Scholar]
- 50.Bengio Y., et al. , “Greedy layer-wise training of deep networks,” in Advances in Neural Information Processing Systems, Vol. 19, p. 153 (2007). [Google Scholar]