ABSTRACT
A dynamic relationship between the gut microbiota and brain is pivotal in neonatal development. Dysbiosis of the microbiome may result in altered neurodevelopment; however, it is unclear which specific members of microbiota are most influential and what factors might mediate the relationship between the gut and the brain. Twenty-four vaginally-derived male piglets were subjected to magnetic resonance spectroscopy at 30 d of age. Ascending colon contents, feces, and blood were collected and analyzed for volatile fatty acids, microbiota relative abundance by 16s rRNA, and serum metabolites, respectively. A mediation analysis was performed to assess the mediatory effect of serum biomarkers on the relationship between microbiota and neurometabolites. Results indicated fecal Ruminococcus and Butyricimonas predicted brain N-acetylaspartate (NAA). Analysis of serum biomarkers indicated Ruminococcus independently predicted serum serotonin and cortisol. A 3-step mediation indicated: i) Ruminococcus negatively predicted NAA, ii) Ruminococcus negatively predicted cortisol, and iii) a significant indirect effect (i.e., the effect of fecal Ruminococcus through cortisol on NAA) was observed and the direct effect became insignificant. Thus, serum cortisol fully mediated the relationship between fecal Ruminococcus and brain NAA. Using magnetic resonance spectroscopy, this study used a statistical mediation analysis and provides a novel perspective into the potential underlying mechanisms through which the microbiota may shape brain development. This is the first study to link Ruminococcus, cortisol, and NAA in vivo, and these findings are substantiated by previous literature indicating these factors may be influential in the etiology of neurodevelopmental disorders.
KEYWORDS: autism, microbiome, N-acetylaspartate, neurodevelopment, pig, Ruminococcus
Introduction
The term microbiota-gut-brain axis is used to describe the bi-directional communication between the gut microbiota and the brain. Early in life the microbial community of the gut is rapidly evolving in parallel with brain development, providing a window during which highly dynamic changes in the brain could be susceptible to microbial alterations. Throughout this period, myelination, growth and expansion of neurons, and changes in neuro-metabolites are occurring, and the trajectory of these processes may be influenced by microbial dysbiosis.1,2 In fact, other findings from animal studies have suggested an influence of the microbiota on brain metabolites,3 behavior,4 and neurogenesis.5 Although the underlying mechanisms for these interactions are widely unknown, it has been suggested that bacterial metabolites could mediate the communication between the gut and the brain.6
Early-life changes in the structure and function of the brain commonly result in neurodevelopmental disorders, and there is increasing evidence linking the microbiome to psychiatric and neurodevelopmental disorders, including autism spectrum disorder (ASD). While the microbiome is suggested to play a role in the etiopathogenesis of neurodevelopmental disorders, it is unclear which gut bacteria might be most influential in the observed comorbid brain differences. Current literature is limited in the characterization of specific microbiota, which might modulate particular aspects of brain development. Accordingly, studies using a multimodal, interdisciplinary approach are necessary to understand the complex communication between the gut, the brain, and mediating factors. Use of magnetic resonance imaging (MRI) offers a novel, non-invasive approach to answering fundamental questions relating to the gut-brain-axis. To our knowledge, only 2 studies have used this technique, these studies showed that dietary probiotics alter brain function in adult women7 and brain neurometabolites in mice.3 By elucidating the effects of specific members of microbiota on measures of brain health using MRI, researchers can better understand the underlying mechanisms through which gut microbiota modulate brain development.
When studying the underlying mechanisms of host-microbe interactions in humans, researchers must account for genetic and environmental variation, including diet and supplement use (i.e., probiotics and prebiotics). The use of gnotobiotic and germ-free rodent models has significantly advanced mechanistic microbiome-gut-brain axis research; however, rodent models are limited by physiologic and metabolic differences when compared with humans. Pigs and humans exhibit strikingly similar physiology, immune system function, dietary patterns, and disease progression, thereby elevating the pig as optimal translational model for assessing the interactions between diet, micriobiota and host physiology.8 Moreover, the piglet is a well-established translational model for gastrointestinal9 and brain development,10 and its use allows for tightly controlled dietary, genetic, and environmental influences. The objective of this exploratory study was to apply an interdisciplinary approach using the piglet model to: 1) identify predictive relationships between microbiota and magnetic resonance spectroscopy-derived neurometabolites, 2) identify predictive relationships between microbiota and intermediary biomarkers, and 3) characterize how the identified biomarkers mediate the relationship between microbiota and brain metabolites. To our knowledge, this is the first time this multimodal approach has been applied to elucidate possible mechanisms between the microbiome-gut-brain axis.
Methods and materials
All animal care and experimental procedures were in accordance with the National Research Council Guide for the Care and Use of Laboratory Animals and approved by the University of Illinois at Urbana-Champaign Institutional Animal Care and Use Committee.
Animals and housing
Twenty-four vaginally-derived intact male Yorkshire piglets from the University of Illinois Imported Swine Research Laboratory were obtained 48 h after birth, to allow for adequate colostrum consumption, and were subsequently artificially reared over a 30 d trial period. Piglets were provided one of 2 experimental diets, over the course of the 30 d trial, a detailed description of dietary treatment was described previously,11,12 and diet was not significant in any of the outcomes assessed herein. A detailed description of animal husbandry was described previously,11,12 and details of the non-invasive neuroimaging procedures performed at 30 d of age are described below. At 31 d of age, piglets were anesthetized by intramuscular injection at 0.03 mL/kg bodyweight using a telzol:ketamine:xylazine solution [50.0 mg tiletamine plus 50.0 mg zolazepam reconstituted with 2.50 mL ketatmine (100 g/L) and 2.50 mL xylazine (100 g/L); Fort Dodge Animal Health]. Upon verifying anesthetic induction, piglets were euthanized via intracardiac administration of sodium pentobarbital (86.0 mg/kg bodyweight; Fatal Plus, Vortech Pharmaceuticals, Dearborn, MI, USA). Immediately following intracardiac injection, piglet heads were removed and trunk blood was collected. Whole blood was centrifuged and serum was collected and stored at −80°C for future analysis. Ascending colon (AC) (the proximal one third of the colon) contents and feces were snap-frozen in liquid nitrogen and stored at −80°C for volatile fatty acids and microbiota analyses.
Magnetic resonance imaging
Piglets were subjected to MRI procedures at 30 d-of-age, as described previously.11 All scans were acquired using a Siemens MAGNETOM Trio 3T Imager using a Siemens 32-channel head coil. Magnetic resonance spectroscopy (MRS) was used to non-invasively quantify metabolites both in the hippocampi and intervening tissue, as described previously.13,14 In short, an MRS spin-echo chemical shift sequence was used to assess a voxel which centered over the left and right dorsal hippocampi with a size of 12 mm × 25 mm × 12 mm. Sequence parameters used for MRS data acquisition are as follows: TR = 3000 ms; TE = 30 ms; signal averages = 128 (water-suppressed scan) and 8 (non-water supressed scan); vector size = 1024 point. Water-suppressed and non-water-suppressed data were acquired in institutional units, and all MRS data were analyzed using LC Model (version 6.3), using methods described previously.13,14 Only neurometabolites for which a single data point was obtained for all piglets on the study were analyzed. Thus, neurometabolites in this analysis include: creatine + phosphocreatine (Cr+Pcr), glycerophosphocholine + phosphocholine (GPC+PCh), myo-inositol (mI), n-acetylaspartate (NAA), n-acetylaspartate + n-acetylaspartylglutamate (NAA+NAAG).
Volatile fatty acid quantification
Concentrations of volatile fatty acids (VFA) in AC contents were determined as described previously.15 The VFA concentrations were quantified using a Hewlett-Packard 5890A Series gas chromatograph and a glass column (180 cm 3 4 mm i.d.) packed with 10% SP-1200/1% H3PO4 on 80/100 mesh Chromosorb WAW (Superlco). Volatile fatty acid production was calculated as VFA concentrations of substrate-containing tubes minus the VFA content of blank tubes divided by substrate weight expressed on a dry matter basis.
Pyrosequencing of bacterial 16S RRNA genes
DNA extraction, 16S rRNA gene sequencing and sequence analysis were performed as described previously.12 Briefly, DNA was isolated from AC contents and feces using the QIAamp DNA Stool Mini Kit (Qiagen, Valencia, CA) in combination with the FastPrep-24 System (MP Biomedicals, Carlsbad, CA). DNA concentration was determined with a NanoDrop 1000 spectrophotometer (NanoDrop Technologies). The V3 to V5 regions of the bacterial 16S rRNA genes were amplified for bacterial pyrosequencing with primers 341F and 907R (Klindworth et al 2013). Pyrosequencing was performed at Research Testing Laboratory (RTL; Lubbock, TX) using 454 Life Sciences Genome Sequencer FLX with Titanium series reagents and protocol (Roche Applied Science, Indianapolis, IN) (Dowd et al 2008, Handl et al 2011). Sequences processing and analysis were performed using a bioinformatic pipeline developed at RTL as described previously.12 Candidate bacteria used in subsequent regression analysis were limited to only bacterial genera for which data was present for each individual piglet, thus resulting in 18 candidate genera for analysis.
Metabolomics
Serum samples were prepared as described previously16 by a commercial laboratory (Metabolon, Inc., Research Triangle Park, NC). Briefly, sample extracts were divided into 5 fractions: 1) acidic positive ion conditions, chromatographically optimized for more hydrophilic compounds; 2) acidic positive ion conditions, chromatographically optimized for more hydrophobic compounds; 3) basic negative ion optimized conditions using a separate dedicated C18 column; 4) negative ionizations following elution from HILIC column; and 5) reserved for backup. Next, non-targeted mass spectrometry analyses were performed, where extracts were subjected to UPLC-MS/MS using methods optimized for each sample fraction; detailed methods described previously.16 Using a proprietary data analysis pipeline (Metabolon, Inc.), metabolites were characterized by ion features and compared with a reference library of chemical standards.17,18 Ion peaks were quantified using area-under-the-curve counts. Raw area counts were normalized to account for variation from instrument inter-day tuning differences by the median value for each run-day. As such the median value for each run day was set to 1.0, thus allowing for metabolites of different raw peak curves to be similarly compared.
Statistical analysis
Stepwise regression for genera and diet predicting brain neurometabolites
Statistical analysis was performed using SPSS Statistical Packages version 24 (SPSS, Inc., Chicago, IL, USA). First, a linear regression equation was created using the stepwise regression procedure in which candidate independent variables (i.e., relative abundances of AC bacterial genera and diet or fecal bacterial genera and diet) predicted the concentration of individual brain metabolites (i.e., derived from MRS); Table 1. Multicollinearity between independent variables was assessed using the variance inflation factor (VIF). Within each model, none of the independent variables exceeded VIF factors of 2, thus all candidate variables remained available for use in the final models. Upon construction of models, a Cook's D test was applied to each model to determine the influence of individual observations on the dependent variable; 3 models that met the criterion of a Cook's D value less than 0.5 and are reported herein. The Mallows' Cp value was also calculated and reported for each regression to assess goodness of fit. In the stepwise selection method, entry and stay α levels were set at 0.05 and 0.10, respectively, for independent variables to be included and remain in the final model. Only linear relationships were tested and unstandardized regression coefficients (β) ± SE are presented. This initial regression approach was used to characterize fecal microbiota which may serve as predictive fecal biomarkers for brain neurometabolites.
Table 1.
Candidate bacterial genera for inclusion in predictive models of MRS-derived metabolites.a
| Ascending Colon & Fecal Genera (Candidate Independent Variables) | MRS-derived Metabolite (Dependent Variables) | |
|---|---|---|
| Butyricimonas | Clostridium XIVa | Myo-Inositol (mI) |
| Parabacteroides | Clostridium XIVb | N-acetylaspartate (NAA) |
| Prevotella | Clostridium IV | Creatine+Phosphocreatine (CR+PCr) |
| Alistipes | Unclassified Lachnospiraceae | N-acetylaspartate+N-acetylaspartylglutamate (NAA+NAAG) |
| Bacteroides | Unclassified Ruminococcaceae | Glycerophosphocholine+Phosphocholine (GPC+PCh) |
| Eschericia Shigella | Ruminococcus | |
| Lactobacillus | Erysipelotrichaceae | |
| Clostridium Sensu Sctricto | Phascolartctobacterium | |
| Eubacterium | Blautia | |
Note.
The same genera were included in as candidate variables for both ascending colon and fecal microbiota (candidate independent variables) presented as a relative abundance, MRS-derived metabolites (candidate dependent variables) presented in parts per million.
Stepwise regression for individual genus and diet predicting serum serotonin, serum cortisol, and ascending colon volatile fatty acids
From the initial stepwise regressions, the following candidate fecal genera were identified as significant predictors of one or more neurometabolites and were used in subsequent regression analyses to identify possible mediating variables: Bacteroides, Clostridium XIVb, Ruminococcus, and Butyricimonas. A linear regression equation was created using the stepwise regression procedure in which candidate independent variables (i.e., individual ascending color or fecal bacterial genus and diet) predicted one of the dependent variables (i.e., serum serotonin, serum cortisol, or ascending colon volatile fatty acids). It is known that volatile fatty acids and microbiota are related, thus all volatile fatty acids (i.e., acetate, butyrate, proprionate, valterate, isobutyrate, and isovalerate) were assessed as possible mediators in this analysis. Previous research indicates serum serotonin and serum cortisol relate to microbiota, thus in an effort to limit the metabolites analyzed in as possible mediators, only serotonin and cortisol were analyzed herein.1,2 All selection criteria described above were applied to the regression described herein, and only models that met all specified criteria are presented.
Mediation analysis
A mediation analysis was performed using the Process macro designed for SPSS.19 The mediation model was used to characterize the relationship between relative abundances of fecal Ruminococcus, serum cortisol, and a MRS-derived brain metabolite (NAA) using a 3-step process. In the context of the following mediation, we tested whether the relationship between the fecal Ruminococcus and the MRS-derived brain NAA was mediated by serum cortisol. For a mediation to occur, there must have been a significant indirect effect,20 meaning the effect of the independent variable (fecal Ruminococcus) through the mediator (serum cortisol) on the dependent variable (spectroscopy-derived NAA) must be significant.
The following 3-step framework was used to determine the mediation:
-
1.
Path A: A regression model was used to describe the relationship between the independent variable (Ruminococcus) and the dependent variable (NAA).
-
2.
Path B: A regression model was used to describe the relationship between the independent variable (Ruminococcus) and the mediator (cortisol).
-
3.
In the third step, the Process macro was used and a bootstrapping method applied to estimate the mediation effects. The bootstrapping used 5000 samples with replacement from the data set to estimate the distribution for the direct and the indirect effects. The direct effect indicates the relationship between the indepdendent variable (Ruminococcus) and the dependent variable (NAA) (Path A’). The indirect effect describes the relationship from the independent variable (Ruminococcus) through the mediator (cortisol) to the dependent variable (NAA) (Path B × C).
A significant mediation occurred when the indirect effect did not include zero within the 95% bias-corrected confidence interval, and if the direct mediation effect did include zero within the 95% bias-corrected confidence interval.20 Unstandardized regression coefficients (β) ± SE and P-values for each regression relationship are presented herein.
Results
Piglet characteristics
One piglet was determined to be an outlier (i.e., >3 SD from the mean) in all analyses and was therefore excluded from the data set, thus 23 subjects were used to generate the results presented herein. Piglet growth assessment was described previously.12
Ascending colon eubacterium predicts brain NAA
Diet and 18 ascending colon genera (candidate independent variables) were entered into a stepwise regression procedure to generate individual linear models for 5 neurometabolites (dependent variables) (Table 1). Using a stepwise regression approach to identify candidate independent variables that predict brain NAA concentrations, Eubacterium (β1 = −0.412 ± 0.119; R2model = 0.364; Pmodel = 0.002) entered the model first; diet and all other candidate genera did not meet criteria to enter the model. Predictive models for brain mI, GPC+PCh, NAA+NAAG, and Cr+PCr did not meet criteria set forth above and were therefore excluded.
Specific fecal bacterial genera predict brain myo-Inositol (mI), N-acetylaspartate (NAA), and creatine+phosphocreatine (Cr+PCr)
Diet and 18 fecal genera (candidate independent variables) were entered into a stepwise regression procedure to generate individual linear models for 5 neurometabolites (dependent variables) (Table 1). Using a stepwise regression approach to identify candidate independent variables that predict brain mI concentration (Table 2), Bacteroides (β1 = 0.039 ± 0.011; Pβ1 = 0.001; Pmodel = 0.002) entered the model first and Clostridium XIVb (β2 = 0.127 ± 0.054; Pβ2 = 0.031; Pmodel = 0.001) entered the model second; diet and all other candidate genera did not meet criteria to enter the model. When identifying candidate variables that predict brain NAA (Table 2), Ruminococcus (β1 = −0.160 ± 0.045; Pβ1 = 0.002; Pmodel = 0.002) entered the model first and Butyricimonas (β2 = 0.155 ± 0.060; Pβ2 = 0.017; Pmodel = 0.001) entered the model second; diet and all other candidate genera did not meet criteria to enter the model. Lastly, a linear model for brain Cr+PCr (Table 2) indicated that Bacteroides (β1 = 0.010 ± 0.004; Pβ1 = 0.013; Pmodel = 0.013) entered the model, and diet and all other candidate genera did not meet criteria to enter the model. Predictive models for brain GPC+PCh and NAA+NAAG did not meet criteria set forth above and were therefore excluded.
Table 2.
Fecal genera predict MRS-derived brain metabolites.a
| Dependent Variable | Step | Variable Entered | No. of variables included | Partial R2 | Model R2 | C(p) | F-value | P-value |
|---|---|---|---|---|---|---|---|---|
| myo-Inositol (mI)b | 1 | Bacteroides | 1 | 0.373 | 0.373 | −9.145 | 12.51 | 0.002 |
| 2 | Clostridium XIVb | 2 | 0.134 | 0.507 | −9.245 | 10.28 | 0.001 | |
| N-acetylaspartate (NAA)c | 1 | Ruminococcus | 1 | 0.367 | 0.367 | 9.530 | 12.17 | 0.002 |
| 2 | Butyricimonas | 2 | 0.160 | 0.527 | 4.331 | 11.13 | 0.001 | |
| Creatine + Phosphocreatine (Cr+PCr)d | 1 | Bacteroides | 1 | 0.261 | 0.261 | 22.47 | 7.424 | 0.013 |
Note.
Fecal microbiota (independent variable) presented as a relative abundance, MRS-derived neurometabolite (dependent variable) presented in parts per million, diet and other candidate genera did not meet criterion for inclusion in the model, N = 23.
Final model equation predicting brain mI, presented with unstandardized coefficients: Y = 7.180 + 0.039x1 + 0.127x2.
Final model equation predicting brain NAA, presented with unstandardized coefficients: Y = 4.997 – 0.160x1 + 0.155x2.
Final model equation predicting brain Cr+PCr, presented with unstandardized coefficients: Y = 3.778 + 0.010x1.
Identification of candidate mediating variables for individual genus and MRS metabolite relationships
From the initial stepwise regressions, the following candidate genera were identified as predictors of one or more brain metabolites: ascending colon Eubacterium, fecal Bacteroides, fecal Clostridium XIVb, fecal Ruminococcus, and fecal Butyricimonas. Candidate mediating variables in this analysis included: serum serotonin, serum cortisol, and ascending colon volatile fatty acids. Individual stepwise regressions indicated fecal Bacteroides was predictive of: serum serotonin (β1 = 0.016 ± 0.007; Pmodel = 0.038; Table 3), ascending colon valerate (β1 = −0.174 ± 0.048; Pmodel = 0.002; Table 4), and total branched-chain fatty acids (β1 = −0.317 ± 0.103; Pmodel = 0.006; Table 4); diet did not meet criteria to enter any of the described models. Similarly, stepwise linear models indicated fecal Ruminococcus was predictive of serotonin (β1 = −0.169 ± 0.057; Pmodel = 0.008; Table 3) and cortisol (β1 = −0.109 ± 0.032; Pmodel = 0.002; Table 3); diet did not meet criteria to enter any of the described models. Individual regressions with ascending colon Eubacterium or fecal Clostridium XIVb and Butryricimonas did not meet selection criteria for inclusion into stepwise predictive models of serum serotonin, serum cortisol, or volatile fatty acids, and were therefore excluded. Previous research indicates serum cortisol and serum serotonin influence the expression of one another, however there was no observed correlation (P = 0.148) between cortisol and serotonin in this study.
Table 3.
Fecal Bacteroides and Ruminococcus predict serum serotonin and cortisol.a
| Dependent Variable | Step | Variable Entered | No. of variables included | Model R2 | C(p) | F-value | P-value |
|---|---|---|---|---|---|---|---|
| Serotoninb | 1 | Bacteroides | 1 | 0.189 | 2.631 | 4.885 | 0.038 |
| Serotoninc | 1 | Ruminococcus | 1 | 0.292 | 2.180 | 8.658 | 0.008 |
| Cortisold | 1 | Ruminococcus | 1 | 0.360 | 1.205 | 11.81 | 0.002 |
Note.
Fecal microbiota (independent variable) presented as a relative abundance, serum metabolite (dependent variable) presented as scaled import measure, diet did not meet criterion for inclusion in the model, N = 23.
Final model equation predicting serum serotonin, presented with unstandardized coefficients: Y = 0.642 + 0.016x1.
Final model equation predicting serum serotonin, presented with unstandardized coefficients: Y = 1.188 – 0.169x1.
Final model equation predicting serum cortisol, presented with unstandardized coefficients: Y = 1.138 – 0.109x1.
Table 4.
Fecal Bacteroides predicts ascending colon valerate and total branched-chain fatty acid concentrations.a
| Dependent Variable | Step | Variable Entered | No. of variables included | Model R2 | C(p) | F-value | P-value |
|---|---|---|---|---|---|---|---|
| Valerateb | 1 | Bacteroides | 1 | 0.393 | 1.354 | 12.97 | 0.002 |
| Total branched-chain fatty acidsc | 1 | Bacteroides | 1 | 0.321 | 1.004 | 9.456 | 0.006 |
Note.
Fecal microbiota (independent variable) presented as a relative abundance, ascending total branched chain fatty acids (dependent variable) presented in µmol/g (DMB), diet did not meet criterion for inclusion in the model, N = 23.
Final model equation predicting ascending colon valerate, presented with unstandardized coefficients: Y = 13.042 – 0.174x1.
Final model equation predicting ascending colon total branched-chain fatty acids, presented with unstandardized coefficients: Y = 29.822 – 0.317x1.
Serum cortisol fully mediates the relationship between fecal Ruminococcus and MRS-derived brain NAA
A formal mediation analysis was performed to assess the relationship between Ruminococcus and NAA through cortisol. The following regression framework was used to elucidate the characteristics of this relationship.
Higher Ruminococcus predicted decreased cortisol (Table 3), therefore cortisol was considered as a candidate mediating variable for the previously identified relationship between Ruminococcus and NAA (Table 2). Using the 3-step framework for a formal mediation analysis, higher Ruminococcus predicted decreased NAA concentrations (β = −0.160 ± 0.045; R2model = 0.367; Pmodel = 0.002, Fig. 1 path A). Next, higher Ruminococcus predicted decreased cortisol concentrations (β = −0.109 ± 0.032; R2model = 0.360; Pmodel = 0.002, Fig. 1 path B). Lastly, the formal mediation model, assessing the effects of serum cortisol concentrations on the relationship between Ruminococcus and NAA, was significant (R2model = 0.450, Pmodel = 0.003). The indirect pathway of the mediation was significant ([95%CI: −0.150 − −0.009; Fig. 1 path B × C] [β = 0.573 ± 0.330; Pβ = 0.098; Fig. 1 path C]), and the direct pathway of the mediation was not significant (95%CI: −0.238 – 0.012; β = −0.113 ± 0.060; Pβ = 0.075; Fig. 1 path A’). Thus, cortisol concentrations fully mediated the relationship between fecal Ruminococcus and brain NAA concentrations.
Figure 1.
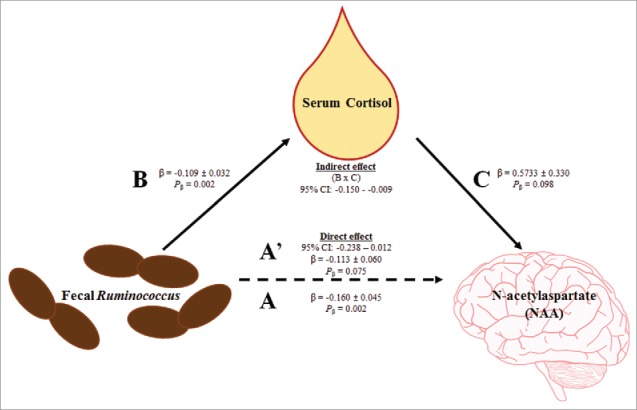
Serum cortisol fully mediates the relationship between fecal Ruminococcus and MRS-derived brain N-acetylaspartate. First, fecal Ruminococcus negatively predicted MRS-derived brain NAA concentrations (path A). Second, fecal Ruminococcus negatively predicted serum cortisol concentrations (path B). Third, mediation effects were assessed for serum cortisol concentrations. The indirect effect (i.e., the effect of fecal Ruminococcus through serum cortisol on MRS-derived brain NAA; path B × C) was significant, and the direct effect (path A’) was not significant. Thus, serum cortisol fully mediates the relationship between fecal Ruminococcus and MRS-derived brain NAA. Abbreviation: N-acetylaspartate (NAA).
Discussion
This study uses multimodal methodology to provide a novel insight into the complex interactions between the microbiome-gut-brain axis in early development. Due to their strikingly similar physiology, immune system function, dietary patterns, and disease progression, pigs have been identified as an optimal translational model for assessing the interactions between diet, micriobiota and host physiology.8 The piglet is considered the best model for human gastrointestinal9 and brain development10 with proven clinical translatability. Recent studies have used gnotobiotic piglet models,21,22 thereby establishing the piglet as a useful model for assessing the influence of specific microbiota throughout development. Using the biomedical piglet model, these findings provide a previously unidentified link between relative abundances of specific bacterial genera, neurometabolites and serum metabolites. In doing so, these results may identify a novel mechanism through which the microbiome may influence neurodevelopment. However, it should be noted that the proposed mechanism described herein should be further substantiated through empirical studies, thus this finding is reported as an exploratory analysis and should be considered as hypothesis generating evidence for future work. In the following paragraphs, we will discuss: i) predictive models between fecal microbiota and brain metabolites, ii) predictive models between fecal microbiota and serum/colonic biomarkers, and iii) a mediation model through which serum biomarkers mediate the relationship between fecal microbiota and brain metabolites.
Fecal microbiota predict neurometabolites
The first objective of this analysis was to identify novel relationships between fecal microbiota and neurometabolites. Our results indicated brain mI, a marker for glial cells and pre-myelinating events in the developing brain,23 was positively predicted by both Bacteroides and Clostridium XIVb. Additionally, brain Cr+PCr, an indicator of energy metabolism,24 was positively predicted by Bacteroides. Lastly, these analyses indicated brain NAA, a marker of neuronal health,24 was negatively predicted by Ruminococcus but positively predicted by Butyricimonas. Concentrations of these brain metabolites change over the course of development23 and our research suggests gut colonization of microbiota may play a pivotal role in this phenomenon. Dysbiosis in the gut microbiota is known to alter behavior in both humans and animal models,,4,25,26 and changes in neurometabolites may underlie these behavioral phenotypes. Interestingly, altered brain Cr+PCr,27-29 NAA,30 and mI27,31 have been reported in subjects diagnosed with ASD, yet no studies have identified specific links between gut microbes and these metabolites. Mice supplemented with a daily dose of Lactobacillus rhamnosus for 2 weeks showed an increase in brain NAA quantified by MRS, thus providing evidence that brain metabolite concentrations can be modulated by microbial shifts in the gut.3 Although Lactobacillus was not identified as a predictive bacteria for neurometabolites in this study, we assessed the interaction of Lactobacillus and brain NAA and the correlation was not significant, data not shown.
Fecal microbiota predict serum/colonic biomarkers
The next step of the analysis framework was to identify possible mediatory biomarkers that relate to the previously identified fecal bacterial genera. This analysis showed that Bacteroides positively predicted serum serotonin, whereas Ruminococcus negatively predicted both serum serotonin and cortisol. These findings have not previously been reported in the literature. However, correlations between operational taxonomic units that belong to Ruminococcus and daily cortisol levels in depressed patients have been reported.32 Previous research indicates both serotonin and cortisol release can be influenced by bacterial strains.25 In a probiotic study, supplementation of Lactobacillus helveticus R0052 and Bifidobacterium longum R0175 decreased urinary cortisol levels in healthy human volunteers, further indicating bacterial strains can influence cortisol levels.33 Although Lactobacillus was not identified as a predictive bacteria for neurometabolites in this study, we assessed the interaction of Lactobacillus and serum cortisol and the correlation was not significant, data not shown. Interestingly, alterations in serum serotonin and cortisol, as well as fecal Bacteroides and Ruminococcus levels, have been described in patients with ASD.6
Serum cortisol as a mediator between fecal ruminococcus and brain NAA
Given the relationships reported in our predictive models, we used a mediation model to characterize the mediatory effects of serum biomarkers on the relationship between fecal microbiota and neurometabolites. Our results provided clear evidence that serum cortisol mediated the relationship between fecal Ruminococcus and brain NAA. Interestingly, Ruminococcus, cortisol, and NAA have all independently been implicated in ASD; however, this is the first time a relationship between the 3 has been reported.
First, higher Ruminococcus predicted lower NAA. Previous research indicates elevated fecal Ruminococcus in subjects with ASD,34,35 and fecal Ruminococcus positively correlated with autistic-like behaviors in a mouse model of ASD.36 In addition, MRS indicates NAA was decreased in the cerebellum,37 hippocampus and amygdala,38 temporal lobe,39 gray matter and white matter,28 and whole brains27 of ASD patients. To our knowledge, no other study has reported a link between Ruminococcus and NAA.
Second, higher Ruminococcus predicted lower cortisol concentrations. This finding is supported by previous research in which decreased serum cortisol was observed in individuals with ASD.40 It should be noted that the implications of cortisol in ASD vary widely, with some research suggesting elevated cortisol in ASD patients.41 Although these inconsistent results could be attributed to differences in methodological approach (i.e., salivary vs. serum cortisol) as well as the heterogeneity of symptoms among individuals with ASD, it is important to note that diverse microbial populations may differentially influence serum cortisol concentrations. As stated previously, supplementation of specific bacterial strains resulted in reduced cortisol in healthy human volunteers.33 These findings further suggest that specific bacterial strains may differentially impact serum cortisol concentrations, rather than attributing the cortisol changes to a particular disease state.
Third, the indirect pathway of the mediation indicated cortisol has a mediatory effect on the relationship between Ruminococcus and NAA. Previous research shows salivary cortisol positively relates to hippocampal NAA, although it should be noted this observation was made in patients exhibiting posttraumatic stress disorder.42 Thus, it follows that lower levels of cortisol would relate to lower levels of NAA, thereby supporting our finding. Given the independent reports on Ruminococcus, cortisol, and NAA in individuals with ASD (Fig. 2), this mediation analysis could provide a possible mechanism underlying the complex relationship between the microbiota and neurodevelopment in the context of ASD. Interestingly, provision of fructo-oligosaccharides and galacto-oligosaccharides in mice resulted in a reduction in cecal Ruminoccocus among other bacterial genera, indicating a possible dietary approach to reducing Ruminococcus concentrations.43
Figure 2.
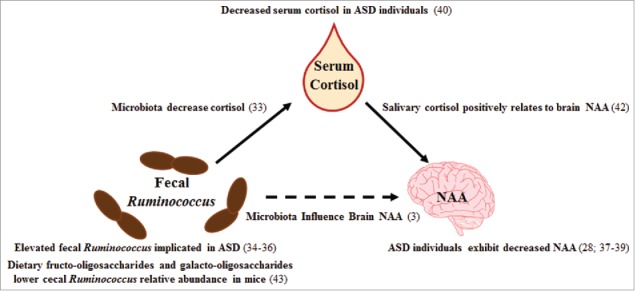
Cited literature for mediation results. Previous studies which support the findings of our mediation analysis are presented in the figure with appropriate literature citations. Abbreviations: autism spectrum disorder (ASD), N-acetylaspartate (NAA).
Conclusion
This study used an interdisciplinary approach through the use of fecal microbiota measures, serum biomarkers, and MRS. To our knowledge, this approach has not been undertaken in previously published studies. The relationships identified herein provide several novel perspectives on the possible mechanisms of microbiome-gut-brain-axis. First, due to the high diversity of the gut microbiota, unravelling specific relationships between bacterial genera and particular brain metabolites is difficult; however, these predictive models identify the most influential genera on individual brain metabolites. Second, identification of these relationships permits a focused approach for selection of mediatory biomarkers. Third, identification of predictive microbes on brain metabolites enables sensitive targeting of the gut microbiota to alter neurometabolites, thereby influencing neurodevelopment. This study reported a predictive relationship between levels of fecal Ruminococcus, serum cortisol, and brain NAA, 3 observations that have independently been reported in patients with ASD. While the initial aim of this study was not to focus on ASD etiology, these results provide preliminary data for future research aiming to investigate the microbiota-brain communication in ASD and to develop targeted treatment options for a subgroup of affected individuals.
Our study was limited to assessing fecal microbiota at the genera taxonomic level; thus, future work assessing the impact of specific microbial species may reveal a more detailed microbial signature as it relates to specific aspects of neurodevelopment. Provided the continued maturation of the brain and the gastrointestinal tract into childhood, longitudinal assessment in the pig model may provide key evidence of critical developmental windows during which microbiota are most influential. Lastly, this novel approach can be implemented in the assessment of other neurodevelopmental disorders that may be influenced by microbiota. While these findings do not specifically identify mechanisms of action between the observed relationships, this exploratory study used a statistical approach that aids in identifying potential mechanisms, which can set the framework for future interventional studies.
From our results we: i) provide a novel perspective into the interaction between fecal microbiota and brain metabolites, ii) provide novel links between fecal microbiota and serum biomarkers implicated in ASD, and iii) bring together 3 independently reported observations in ASD patients to provide a potential mechanism for the communication between one bacterial genera and a specific brain metabolite. The relationships identified in this study may serve as hypothesis-generating findings from which future empirical studies can aim to further investigate the underlying mechanisms proposed in this paper. Additionally, these novel findings may serve as preclinical evidence through which future dietary intervention can be implemented to alter microbial populations and influence neurodevelopment.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors would like to thank Jonathon Mosley from the University of Illinois Imported Swine Research Laboratory for his help in execution of the animal phase of this study. We would also like to acknowledge Nancy Dodge and Holly Tracy from the Beckman Institute Biomedical Imaging Center for their contributions during MRI data acquisition. Additionally, we would like to thank Dr. Dustin Bohler for his expertise in statistical methodology.
Funding
This project was funded by Mead Johnson Nutrition (Evansville, IN). Drs. Sharon Donovan and Ryan Dilger have received grant and consultant funding from Mead Johnson Nutrition. Dr. Mei Wang has received consultant funding from Mead Johnson Nutrition. Austin Mudd and Kirsten Berding have no financial disclosures to report.
Author contribution
SMD and RND were involved in initial project conceptualization. ATM was involved in daily project activities. ATM, KB, MW, SMD, and RND were involved in data analysis. All authors were involved in data interpretation and manuscript preparation.
References
- [1].Borre YE, O'Keeffe GW, Clarke G, Stanton C, Dinan TG, Cryan JF. Microbiota and neurodevelopmental windows: Implications for brain disorders. Trends Mol Med 2014; 20(9):509-18; PMID:24956966; doi: 10.1016/j.molmed.2014.05.002 [DOI] [PubMed] [Google Scholar]
- [2].Clarke G, O'Mahony SM, Dinan TG, Cryan JF. Priming for health: gut microbiota acquired in early life regulates physiology, brain and behaviour. Acta Paediatr 2014; 103(8):812-9; PMID:24798884; doi: 10.1111/apa.12674 [DOI] [PubMed] [Google Scholar]
- [3].Janik R, Thomason LAM, Stanisz AM, Forsythe P, Bienenstock J, Stanisz GJ. Magnetic resonance spectroscopy reveals oral Lactobacillus promotion of increases in brain GABA, N-acetyl aspartate and glutamate. Neuroimage 2016; 125:988-95; PMID:26577887; doi: 10.1016/j.neuroimage.2015.11.018 [DOI] [PubMed] [Google Scholar]
- [4].Sampson TR, Mazmanian SK. Control of brain development, function, and behavior by the microbiome. Cell Host Microbe 2015; 17(5):565-76; PMID:25974299; doi: 10.1016/j.chom.2015.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ogbonnaya ES, Clarke G, Shanahan F, Dinan TG, Cryan JF, O'Leary OF. Adult hippocampal neurogenesis is regulated by the microbiome. Biol Psychiatry 2015; 78(4):e7-e9; PMID:25700599; doi: 10.1016/j.biopsych.2014.12.023 [DOI] [PubMed] [Google Scholar]
- [6].Berding K, Donovan SM. Microbiome and nutrition in autism spectrum disorder: current knowledge and research needs. Nutr Rev 2016; 74(12):723-36; PMID:27864534; doi: 10.1093/nutrit/nuw048 [DOI] [PubMed] [Google Scholar]
- [7].Tillisch K, Labus J, Kilpatrick L, Jiang Z, Stains J, Ebrat B, Guyonnet D, Legrain-Raspaud S, Trotin B, Naliboff B, et al.. Consumption of fermented milk product with probiotic modulates brain activity. Gastroenterology 2013; 144(7):1394-401.e4; PMID:23474283; doi: 10.1053/j.gastro.2013.02.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Leulier F, MacNeil LT, Lee W, Rawls JF, Cani PD, Schwarzer M, Zhao L, Simpson SJ. Integrative physiology: at the crossroads of nutrition, microbiota, animal physiology, and human health. Cell Metab 2017; 25(3):522-534; PMID:28273475; doi: 10.1016/j.cmet.2017.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Jacobi SK, Odle J. Nutritional factors influencing intestinal health of the neonate. Adv Nutr 2012: 687-96; PMID:22983847; doi: 10.3945/an.112.002683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Mudd AT, Dilger RN. Early-Life nutrition and neurodevelopment: Use of the piglet as a translational model. Adv Nutr 2017; 8:92-104; PMID:28096130; doi: 10.3945/an.116.013243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Mudd A, Alexander L, Berding K, Waworuntu R, Berg B, Donovan S, Dilger R. Dietary prebiotics, milk fat globule membrane and lactoferrin affects structural neurodevelopment in the young piglet. Front Pediatr 2016; 4(4):1-10; PMID: 26835439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Berding K, Wang M, Monaco MH, Alexander LS, Mudd AT, Chichlowski M, Waworuntu RV, Berg BM, Miller MJ, Dilger RN, et al.. Prebiotics and bioactive milk fractions affect gut development, microbiota and neurotransmitter expression in piglets. J Pediatr Gastroenterol Nutr 2016; 63(6):688-97; PMID:27031373; doi: 10.1097/MPG.0000000000001200 [DOI] [PubMed] [Google Scholar]
- [13].Jacob RM, Mudd AT, Alexander LS, Lai C-S, Dilger RN. Comparison of brain development in sow-reared and artificially reared piglets. Front Pediatr 2016; 4:95; PMID:27672632; doi: 10.3389/fped.2016.00095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Radlowski EC, Conrad MS, Lezmi S, Dilger RN, Sutton B, Larsen R, Johnson RW. A neonatal piglet model for investigating brain and cognitive development in small for gestational age human infants. PLoS One 2014; 9(3):e91951; PMID:24637829; doi: 10.1371/journal.pone.0091951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Li M, Bauer LL, Chen X, Wang M, Kuhlenschmidt TB, Kuhlenschmidt MS, Fahey GC, Donovan SM. Microbial composition and in vitro fermentation patterns of human milk oligosaccharides and prebiotics differ between formula-fed and sow-reared piglets. J Nutr 2012; 142:681-9; PMID:22399522; doi: 10.3945/jn.111.154427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Evans A, Bridgewater B, Liu Q, Mitchell M, Robinson R, Dai H, Stewart S, DeHaven C, Miller L. High resolution mass spectrometry improves data quantity and quality as compared to unit mass resolution mass spectrometry in high-throughput profiling metabolomics. Metabolomics 2014; 4(132):S24-36. [Google Scholar]
- [17].Evans AM, DeHaven CD, Barrett T, Mitchell M, Milgram E. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal Chem 2009; 81(16):6656-67; PMID:19624122; doi: 10.1021/ac901536h [DOI] [PubMed] [Google Scholar]
- [18].Dehaven CD, Evans AM, Lawton KA. Organization of GC / MS and LC / MS metabolomics data into chemical libraries. J Cheminform 2010; 2(9):1-12; PMID:20298528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Preacher KJ, Hayes AF. Asymptotic and resampling strategies for assessing and comparing indirect effects in multiple mediator models. Behav Res Methods 2008; 40(3):879-91; PMID:18697684; doi: 10.3758/BRM.40.3.879 [DOI] [PubMed] [Google Scholar]
- [20].Zhao X, Lynch JG Jr, Chen Q. Reconsidering Baron and Kenny: myths and truths about mediation analysis. J Consum Res 2010; 37(2):197-206; doi: 10.1086/651257 [DOI] [Google Scholar]
- [21].Laycock G, Sait L, Inman C, Lewis M, Smidt H, van Diemen P, Jorgensen F, Stevens M, Bailey M. A defined intestinal colonization microbiota for gnotobiotic pigs. Vet Immunol Immunopathol 2012; 149(3–4):216-24; PMID:22868203; doi: 10.1016/j.vetimm.2012.07.004 [DOI] [PubMed] [Google Scholar]
- [22].Charbonneau MR, O'Donnell D, Blanton LV, Totten SM, Davis JCC, Barratt MJ, Cheng J, Guruge J, Talcott M, Bain JR, et al.. Sialylated milk oligosaccharides promote microbiota-dependent growth in models of infant undernutrition. Cell 2016; 164(5):859-71; PMID:26898329; doi: 10.1016/j.cell.2016.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Bluml S, Wisnowski JL, Nelson MD, Paquette L, Gilles FH, Kinney HC, Panigrahy A. Metabolic maturation of the human brain from birth through Aadolescence: insights from in vivo magnetic resonance spectroscopy. Cereb Cortex 2013; 23(12):2944-55; PMID:22952278; doi: 10.1093/cercor/bhs283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Rae CD. A guide to the metabolic pathways and function of metabolites observed in human brain 1H magnetic resonance spectra. Neurochem Res 2014; 39(1):1-36; PMID:24258018; doi: 10.1007/s11064-013-1199-5 [DOI] [PubMed] [Google Scholar]
- [25].Cryan JF, Dinan TG. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci 2012; 13(10):701-12; PMID:22968153; doi: 10.1038/nrn3346 [DOI] [PubMed] [Google Scholar]
- [26].Dinan TG, Stilling RM, Stanton C, Cryan JF. Collective unconscious: How gut microbes shape human behavior. J Psychiatr Res 2015; 63:1-9; PMID:25772005; doi: 10.1016/j.jpsychires.2015.02.021 [DOI] [PubMed] [Google Scholar]
- [27].Friedman SD, Shaw DW, Artru AA, Richards TL, Gardner J, Dawson G, Posse S, Dager SR. Regional brain chemical alterations in young children with autism spectrum disorder. Neurology 2003; 60:100-7; PMID:12525726; doi: 10.1212/WNL.60.1.100 [DOI] [PubMed] [Google Scholar]
- [28].Friedman SD, Shaw DW, Artru AA, Dawson G, Petropoulos H, Dager SR. Gray and white matter brain chemistry in young children with autism. Arch Gen Psychiatry 2006; 63(7):786-94; PMID:16818868; doi: 10.1001/archpsyc.63.7.786 [DOI] [PubMed] [Google Scholar]
- [29].Ipser JC, Syal S, Bentley J, Adnams CM, Steyn B, Stein DJ. 1H-MRS in autism spectrum disorders: a systematic meta-analysis. Metab Brain Dis 2012; 27(3):275-87; PMID:22426803; doi: 10.1007/s11011-012-9293-y [DOI] [PubMed] [Google Scholar]
- [30].Aoki Y, Kasai K, Yamasue H. Age-related change in brain metabolite abnormalities in autism: a meta-analysis of proton magnetic resonance spectroscopy studies. Transl Psychiatry 2012; 2(1):e69; PMID:22832731; doi: 10.1038/tp.2011.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Gabis L, Huang W, Azizian A, Devincent C, Tudorica A, Kesner-baruch Y, Roche P, Pomeroy J. H-magnetic resonance spectroscopy markers of cognitive and language ability in clinical subtypes of autism spectrum disorders. J Child Neurol 2008; 23(7):766-74; PMID:18487520; doi: 10.1177/0883073808315423 [DOI] [PubMed] [Google Scholar]
- [32].Naseribafrouei A. Correlation between gut microbiota and depression (Masters Thesis). Hedmark University College; 2013. Retrieved from BRAGE Hihm. [Google Scholar]
- [33].Messaoudi M, Lalonde R, Violle N, Javelot H, Desor D, Nejdi A, Bisson J-F, Rougeot C, Pichelin M, Cazaubiel M, et al.. Assessment of psychotropic-like properties of a probiotic formulation (Lactobacillus helveticus R0052 and Bifidobacterium longum R0175) in rats and human subjects. Br J Nutr 2011; 105(5):755-64; PMID:20974015; doi: 10.1017/S0007114510004319 [DOI] [PubMed] [Google Scholar]
- [34].Finegold SM, Molitoris D, Song Y, Liu C, Vaisanen M, Bolte E, McTeague M, Sandler R, Wexler H, Marlowe EM, et al.. Gastrointestinal microflora studies in late‐onset autism. Clin Infect Dis 2002; 35(s1):S6-16; PMID:12173102; doi: 10.1086/341914 [DOI] [PubMed] [Google Scholar]
- [35].Wang L, Christophersen CT, Sorich MJ, Gerber JP, Angley MT, Conlon MA. Increased abundance of Sutterella spp. and Ruminococcus torques in feces of children with autism spectrum disorder. Mol Autism 2013; 4(1):42; PMID:24188502; doi: 10.1186/2040-2392-4-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Davis DJ, Hecht PM, Jasarevic E, Beversdorf DQ, Will MJ, Fritsche K, Gillespie CH. Sex-specific effects of docosahexaenoic acid (DHA) on the microbiome and behavior of socially-isolated mice. Brain Behav Immun 2017; 59:38–48; PMID:27621225; doi: 10.1016/j.bbi.2016.09.003. [DOI] [PubMed] [Google Scholar]
- [37].Chugani DC, Sundram BS, Behen M, Lee M-L, Moore GJ. Evidence of altered energy metabolism in autistic children. Prog Neuro-Psychopharmacology Biol Psychiatry 1999; 23(4):635-41; doi: 10.1016/S0278-5846(99)00022-6 [DOI] [PubMed] [Google Scholar]
- [38].Otsuka H, Harada M, Mori K, Hisaoka S, Nishitani H. Brain metabolites in the hippocampus-amygdala region and cerebellum in autism: An 1H-MR spectroscopy study. Neuroradiology 1999; 41(7):517-9; PMID:10450847; doi: 10.1007/s002340050795 [DOI] [PubMed] [Google Scholar]
- [39].Hisaoka S, Harada M, Nishitani H, Mori K. Regional magnetic resonance spectroscopy of the brain in autistic individuals. Neuroradiology 2001; 43(6):496-8; PMID:11465765; doi: 10.1007/s002340000520 [DOI] [PubMed] [Google Scholar]
- [40].Ćurin JM, Terzić J, Petković ZB, Zekan L, Terzić IM, Šušnjara IM. Lower cortisol and higher ACTH levels in individuals with autism. J Autism Dev Disord 2003; 33(4):443-8; PMID:12959423; doi: 10.1023/A:1025019030121 [DOI] [PubMed] [Google Scholar]
- [41].Tordjman S, Anderson GM, Kermarrec S, Bonnot O, Geoffray MM, Brailly-Tabard S, Chaouch A, Colliot I, Trabado S, Bronsard G, et al.. Altered circadian patterns of salivary cortisol in low-functioning children and adolescents with autism. Psychoneuroendocrinology 2014; 50(August):227-45; PMID:25244637; doi: 10.1016/j.psyneuen.2014.08.010 [DOI] [PubMed] [Google Scholar]
- [42].Neylan TC, Schuff N, Lenoci M, Yehuda R, Weiner MW, Marmar CR. Cortisol levels are positively correlated with hippocampal N-acetylaspartate. Biol Psychiatry 2003; 54:1118-21; PMID:14625155; doi: 10.1016/S0006-3223(02)01974-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Burokas A, Arboleya S, Rachel D, Peterson VL, Murphy K, Clarke G, Stanton C, Timothy G, Cryan JF. Targeting the microbiota-gut-brain axis: prebiotics have anxiolytic and antidepressant-like effects and reverse the impact of chronic stress in mice. Biol Psychiatry [in press]. PMID:28242013; doi: 10.1016/j.biopsych.2016.12.031 [DOI] [PubMed] [Google Scholar]


