To the Editor
Chronic myelomonocytic leukemia (CMML) is an aggressive hematologic malignancy with overlapping features of myelodysplastic syndromes (MDS) and myeloproliferative neoplasms (MPN).[1, 2] Patients with CMML, display between 10-15 mutations per kilobase of coding DNA regions; with majority of these (>90%) involving epigenetic regulator genes (TET2 60%, ASXL1 40%), splicing machinery (SRSF2 40%) and cell signalling (oncogenic RAS pathway 30%).[3-5] Nucleophosmin 1 (NPM1,chromosome 5q35) is a gene encoding for the NPM1 protein that primarily resides in the nucleus, but has the ability to shuttle between the nucleus and the cytoplasm.[6] NPM1 is required for critical cellular functions including; ribosome biogenesis, regulation of genomic stability, p53/MDM2 (mouse double minute 2 homolog) dependant stress response and the modulation of growth suppressive pathways.[6] NPM1 mutations are seen in ≈ 30% of patients with acute myeloid leukemia (AML) with normal cytogenetics, with common phenotypic associations being; high bone marrow (BM) blasts, high white blood cell counts, reduced CD34 expression and the presence of extramedullary manifestations.[6, 7] The presence of NPM1 mutations in the absence of FLT3-ITD (fms related tyrosine kinase 3- internal tandem duplications) in cytogenetically normal AML is associated with favorable outcomes. [8] NPM1 mutations are infrequent in CMML (<5%) and MDS (<5%) and have an unclear prognostic impact.[9-11] We carried out this study to assess the i) frequency and clinical correlates, ii) prognostic impact and iii) survival outcomes related to NPM1 mutations in CMML.
Patients were selected according to the 2016 World Health Organization (WHO) diagnostic criteria for CMML and identified from the institutional database.[1] All patients had BM biopsies and cytogenetic studies performed at diagnosis. A 29 gene panel next generation sequencing assay was carried out on BM DNA specimens obtained at diagnosis for the following genes; TET2, DNMT3A, IDH1, IDH2, ASXL1, EZH2, SUZ12, SRSF2, SF3B1, ZRSR2, U2AF1, PTPN11, Tp53, SH2B3, RUNX1, CBL, NRAS, KRAS, JAK2, CSF3R, FLT3, KIT, CALR, MPL, NPM1, CEBPA, IKZF,ETNK1 and SETBP1, by previously described methods.[5, 11] Statistical analyses considered parameters obtained at time of CMML diagnosis. Differences in the distribution of continuous variables between categories were analyzed by either Mann-Whitney or Kruskal-Wallis tests. Patient groups with nominal variables were compared by chi-square test. Over-all survival (OS) was calculated from the date of first referral to date of death or last contact. Leukemia-free survival (LFS) was calculated from the date of first referral to date of leukemic transformation or death/last contact. OS and LFS curves were prepared by the Kaplan-Meier method and compared by the log-rank test. Cox proportional hazard regression model was used for multivariable analysis.
A total of 373 WHO-defined CMML patients were included in the study; median age 71 years (range, 20-95), 66% males. NPM1 mutations were identified in 8 (2%) patients (table 1). All (100%) NPM1 mutations occurred in patients with normal cytogenetics, involved exon 12, with 4 of 8 (50%) being the classical type A NPM1 mutations resulting in a duplication of TCTG (nucleotides 956-959) creating a 4 base pair insertion at position 960. Morphological subtypes included; CMML-0 2 (25%), CMML-1 3 (38%) and CMML-2 3 (38%). Risk stratification by the Mayo Molecular Model included; intermediate-1 4 (50%) and high risk 4 (50%). [4] In comparison to their wild type counterparts, NPM1 mutant CMML patients were more likely to be anemic (p=0.02), have a “dysplastic CMML subtype” (87%, p=0.04), have an increased incidence of DNMT3A (50%, p<0.0001), FLT3 ITD (13%, p=0.04) and a lower frequency of TET2 (13%, p=0.03) and ASXL1 (13%, p=0.03) mutations.
Table 1. Comparison of clinical and laboratory characteristics of 373 chronic myelomonocytic leukemia (CMML) patients obtained at time of diagnosis, stratified by the presence or absence of NPM1 mutations.
| Variables | All patients with CMML (n=373) | CMML Patients with NPM1 mutations (n=8) | CMML patients without NPM1 mutation (n=365) | P-value |
|---|---|---|---|---|
|
| ||||
| Age in years; median (range) | 71 (20-95) | 76 (48-87) | 71 (20-95) | 0.8 |
|
| ||||
| Males; n (%) | 246 (66%) | 4 (50%) | 242 (66%) | 0.34 |
|
| ||||
| Hemoglobin, g/dL; median (range) | 10.6 (1.4-17) | 9.5 (7.4-10.6) | 10.6 (1.4-17) | 0.02 |
|
| ||||
| WBC × 109/L; median (range) | 12.2 (1.3-265) | 11 (3.7-186) | 12 (1.3-265) | 0.56 |
|
| ||||
| ANC × 109 /L; median (range | 5.8 (0-151) | 5.6 (2.9-143 | 5.8 (0-151) | 0.63 |
|
| ||||
| ALC × 109 /L; median (range) | 1.8 (0-22) | 3.2 (0.4-6) | 1.7 (0-22) | 0.3 |
|
| ||||
| AMC × 109 /L; median (range) | 2.5 (1.0-84) | 1.7 (1.2-30) | 2.5 (1.0-84) | 0.6 |
|
| ||||
| Platelets × 109 /L; median (range) | 96 (7-1184) | 80 (23-293) | 96 (7-1184) | 0.62 |
|
| ||||
| Presence of circulating immature myeloid cells; n (%) | 155 (42%) | 3 (38%) | 152 (42%) | 0.8 |
|
| ||||
| PB blast %; median (range) | 0 (0-18) | 1 (0-12) | 0 (0-18) | 0.19 |
|
| ||||
| BM blast % ; median (range) | 2 (0-21) | 5 (0-15) | 2 (0-21) | 0.47 |
|
| ||||
| Lactate dehydrogenase levels IU/ml; median (range) |
225 (84-1824) | 285 (153-696) | 225 (84-1824) | 0.41 |
|
| ||||
| Cytogenetics (n=344) Abnormal; n (%) |
105 (32%) | 0 (0%) | 105 (31%) | 0.06 |
|
| ||||
| FAB; n (%) | 0.047 | |||
| Proliferative | 176 (47%) | 1 (13%) | 175 (48%) | |
| Dysplastic | 197 (53%) | 7 (87%) | 190 (52%) | |
|
| ||||
| Next generation sequencing analysis; n (%) | ||||
| 1. Epigenetic regulators | ||||
| TET2 | 116 (51%) | 1 (13%) | 115 | 0.03 |
| DNMT3A | 15 (7%) | 4 (50%) | 11 (5%) | <0.0001 |
| IDH1 | 3 (1%) | 0 | 3 (1%) | 0.74 |
| IDH2 | 13 (6%) | 0 | 13 (6%) | 0.5 |
| 2. Chromatin regulators | ||||
| ASXL1 | 113 (50%) | 1 (13%) | 112 (51%) | 0.03 |
| EZH2 | 6 (3%) | 0 | 6 (3%) | 0.63 |
| 3. Transcription factors | ||||
| RUNX1 | 21 (9%) | 0 | 21 (10%) | 0.4 |
| 4. Spliceosome components | ||||
| SF3B1 | 11 (5%) | 1 (13%) | 10 (5%) | 0.3 |
| SRSF2 | 98 (43%) | 2 (25%) | 97 (44%) | 0.3 |
| U2AF1 | 18 (8%) | 1 (13%) | 17(8%) | 0.62 |
| ZRSR2 | 4 (2%) | 0 | 4 (2%) | 0.7 |
| 5. Cell signaling | ||||
| JAK2 V617F | 18 (7%) | 0 | 18 (7%) | 0.43 |
| MPL | 2 (1%) | 0 | 2 (1%) | 0.79 |
| CBL | 29 (13%) | 0 | 29 (13%) | 0.27 |
| NRAS | 31 (14%) | 2 (25%) | 29 (13%) | 0.33 |
| KRAS | 12 (5%) | 0 | 11 (5%) | 0.52 |
| PTPN11 | 7 (3%) | 0 | 6 (3%) | 0.64 |
| CSF3R | 3 (1%) | 0 | 3 (1%) | 0.74 |
| C-KIT | 10 (4%) | 0 | 10 (5%) | 0.54 |
| FLT3 | 5 (2%) | 1 (13%) | 4 (2%) | 0.04 |
| CALR | 1 (0.5%) | 0 | 1 (0.5%) | 0.85 |
| 6. Tumor suppressor genes | ||||
| Tp53 | 6 (3%) | 0 | 6 (3%) | 0.64 |
| 7. Others | ||||
| SETBP1 | 28 (12%) | 0 | 28 (13%) | 0.28 |
| ETNK1 | 1 (0.5%) | 0 | 1 (0.5%) | 0.85 |
|
| ||||
| Spanish Cytogenetic risk stratification; n (%) (n=343) | 0.18 | |||
| Low | 242 (71%) | 8 (100%) | 235 (70%) | |
| Intermediate | 69 (20%) | 0 | 69 (21%) | |
| High | 32 (9%) | 0 | 31 (9%) | |
|
| ||||
| Mayo-French cytogenetic risk stratification; n (%) (n=343) | 0.19 | |||
| Low | 242 (71%) | 8 (100%) | 235 (70%) | |
| Intermediate | 80 (23%) | 0 | 79 (24%) | |
| High | 21 (6%) | 0 | 21 (6%) | |
|
| ||||
| Mayo prognostic model (n=372) | 0.76 | |||
| Low | 91 (24%) | 2 (25%) | 89 (24%) | |
| Intermediate | 136 (36%) | 2 (25%) | 134 (37%) | |
| High | 145 (40%) | 4 (50%) | 141 (39%) | |
|
| ||||
| MDAPS (n=372) | 0.17 | |||
| Low | 159 (43%) | 1 (13%) | 159 (44%) | |
| Intermediate-1 | 113 (30%) | 4 (50%) | 109 (30%) | |
| Intermediate-2 | 88 (24%) | 2 (25%) | 85 (23%) | |
| High | 12 (3%) | 1 (13%) | 11 (3%) | |
|
| ||||
| GFM CMML prognostic model (n=371) | 0.24 | |||
| Low | 174 (47%) | 6 (74%) | 168 (46%) | |
| Intermediate | 143 (38%) | 1 (13%) | 142 (39%) | |
| High | 54 (15%) | 1 (13%) | 53 (15%) | |
|
| ||||
| MMM (n=371) | 0.16 | |||
| Low | 36 (10%) | 0 | 36 (10%) | |
| Intermediate-1 | 121 (33%) | 4 (50%) | 117 (32%) | |
| Intermediate-2 | 107 (29%) | 0 | 107 (29%) | |
| High | 107 (29%) | 4 (50%) | 103 (28%) | |
|
| ||||
| Leukemic transformations; n (%) | 69 (18%) | 5 (63%) | 64 (18%) | 0.0012 |
|
| ||||
| Deaths; n (%) | 249 (67%) | 6 (75%) | 243 (67%) | 0.62 |
|
| ||||
| Median follow up in months; median (range) | 13.6 (0-169) | 9.4 (0.3-41) | 13.8 (0-169) | 0.22 |
Key: CMML- chronic myelomonocytic leukemia, NPM1 nucleophosmin 1, WBC, white blood cell count; ANC- absolute neutrophil count, ALC- absolute lymphocyte count, AMC, absolute monocyte count; PB, peripheral blood; BM, bone marrow; FAB- French American British Classifcation, EM, extramedullary involvement; MDAPS, MD Anderson Prognostic Scoring System; GFM- Groupe Français des Myélodysplasies, MMM- Mayo Molecular Model.
At last follow up (median, 14 months), 249 (67%) deaths and 69 (18%) leukemic transformations were documented. Median survival in NPM1 mutated CMML patients was 12.5 months vs. 20.5 months in their wild type counterparts (p=0.12, figure 1A). Five of 8 (62%) NPM1 mutated CMML patients underwent blast transformation (BT), at a median of 5 months (range, 1-16). In 2 (of 5) evaluable patients, NPM1 mutations were detected at both, CMML diagnosis and at BT, without additional cytogenetic or molecular clonal evolution. Four (of 5) patients with BT were treated with AML-like induction chemotherapy, with 2 patients subsequently undergoing allogeneic stem cell transplantation (HCT). Median survival after BT was 3 months (range, 1-12), with only one patient that underwent HCT being alive at last follow up. In univariate analysis, LFS was adversely impacted by age (p<0.0001), low hemoglobin (p=0.0009), high absolute lymphocyte count (p=0.02), higher serum LDH (lactate dehydrogenase) levels (p=0.023), PB blast % (p<0.0001), BM blast % (p<0.0001), 2016 WHO morphological subtype CMML-2 (p<0.0001), abnormal cytogenetics (p=0.002), Mayo-French cytogenetic risk stratification (p=0.01),[12] NPM1 (p=0.0004) and Tp53 (p=0.002) mutations. ASXL1, DNMT3A, TET2 and FLT3 ITD mutations did not impact LFS; in multivariable analysis that included the aforementioned significant variables, only PB blasts % (p<0.0001, HR 1.2, 95% CI 1.13-1.3), NPM1 mutations (p=0.03, HR 4.7, 95% CI 1.7-12.8) and Tp53 mutations (p=0.001, HR 5.4, 95% CI 1.6-18.4) remained significant.
Figure 1A.
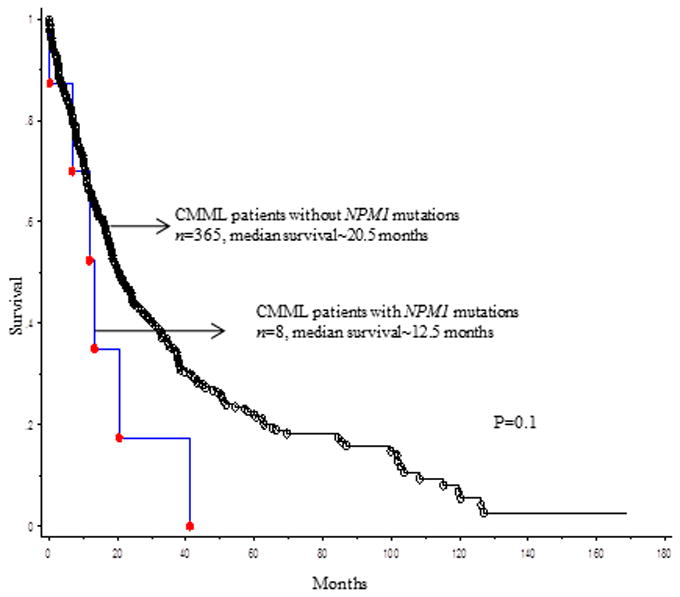
Overall survival of 373 patients with chronic myelomonocytic leukemia stratified by their NPM1 mutational status.
The current study confirms the infrequent occurrence of NPM1 mutations in CMML; also, the number of informative cases was large enough to allow demonstration of clustering of NPM1 mutations in CMML with normal cytogenetics, “dysplastic CMML phenotype” and DNTM3A mutations. More importantly, we show a significantly higher risk of blast transformation in patients with CMML harboring NPM1 mutations.
Figure 1B.
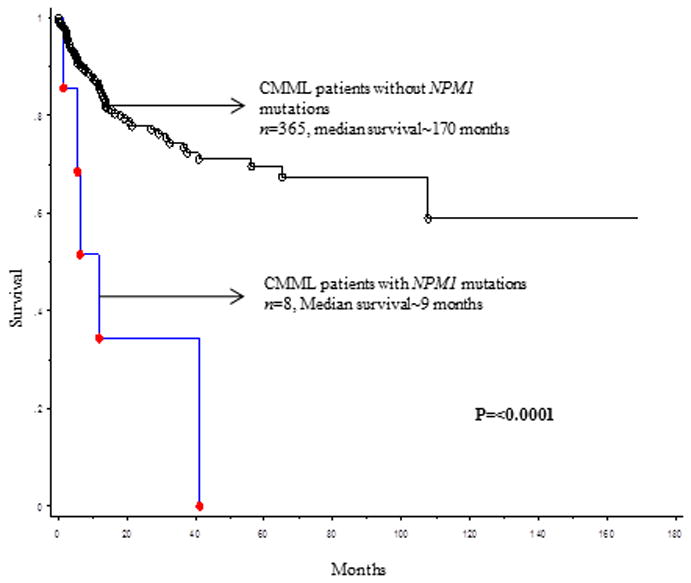
Leukemia-free survival of 373 patients with chronic myelomonocytic leukemia stratified by their NPM1 mutational status.
Acknowledgments
Current publication is supported in part by grants from the “The Henry J. Predolin Foundation for Research in Leukemia, Mayo Clinic, Rochester, MN, USA”.
This publication was supported by CTSA Grant Number KL2 TR000136 from the National Center for Advancing Translational Science (NCATS). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
References
- 1.Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–2405. doi: 10.1182/blood-2016-03-643544. [DOI] [PubMed] [Google Scholar]
- 2.Patnaik MM, Tefferi A. Chronic myelomonocytic leukemia: 2016 update on diagnosis, risk stratification, and management. Am J Hematol. 2016;91:631–642. doi: 10.1002/ajh.24396. [DOI] [PubMed] [Google Scholar]
- 3.Merlevede J, Droin N, Qin T, et al. Mutation allele burden remains unchanged in chronic myelomonocytic leukaemia responding to hypomethylating agents. Nat Commun. 2016;7:10767. doi: 10.1038/ncomms10767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patnaik MM, Itzykson R, Lasho TL, et al. ASXL1 and SETBP1 mutations and their prognostic contribution in chronic myelomonocytic leukemia: a two-center study of 466 patients. Leukemia. 2014;28:2206–2212. doi: 10.1038/leu.2014.125. [DOI] [PubMed] [Google Scholar]
- 5.Patnaik MM, Lasho TL, Vijayvargiya P, et al. Prognostic interaction between ASXL1 and TET2 mutations in chronic myelomonocytic leukemia. Blood Cancer J. 2016;6:e385. doi: 10.1038/bcj.2015.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heath EM, Chan SM, Minden MD, et al. Biological and clinical consequences of NPM1 mutations in AML. Leukemia. 2017;31:798–807. doi: 10.1038/leu.2017.30. [DOI] [PubMed] [Google Scholar]
- 7.Schlenk RF, Dohner K, Krauter J, et al. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358:1909–1918. doi: 10.1056/NEJMoa074306. [DOI] [PubMed] [Google Scholar]
- 8.Dohner K, Schlenk RF, Habdank M, et al. Mutant nucleophosmin (NPM1) predicts favorable prognosis in younger adults with acute myeloid leukemia and normal cytogenetics: interaction with other gene mutations. Blood. 2005;106:3740–3746. doi: 10.1182/blood-2005-05-2164. [DOI] [PubMed] [Google Scholar]
- 9.Peng J, Zuo Z, Fu B, et al. Chronic myelomonocytic leukemia with nucleophosmin (NPM1) mutation. Eur J Haematol. 2016;96:65–71. doi: 10.1111/ejh.12549. [DOI] [PubMed] [Google Scholar]
- 10.Bains A, Luthra R, Medeiros LJ, et al. FLT3 and NPM1 mutations in myelodysplastic syndromes: Frequency and potential value for predicting progression to acute myeloid leukemia. Am J Clin Pathol. 2011;135:62–69. doi: 10.1309/AJCPEI9XU8PYBCIO. [DOI] [PubMed] [Google Scholar]
- 11.Patnaik MM, Barraco D, Lasho TL, et al. DNMT3A mutations are associated with inferior overall and leukemia-free survival in chronic myelomonocytic leukemia. Am J Hematol. 2017;92:56–61. doi: 10.1002/ajh.24581. [DOI] [PubMed] [Google Scholar]
- 12.Wassie EA, Itzykson R, Lasho TL, et al. Molecular and prognostic correlates of cytogenetic abnormalities in chronic myelomonocytic leukemia: a Mayo Clinic-French Consortium Study. Am J Hematol. 2014;89:1111–1115. doi: 10.1002/ajh.23846. [DOI] [PubMed] [Google Scholar]


