Abstract
Doubletime (DBT) has an essential circadian role in Drosophila melanogaster because it phosphorylates Period (PER). In order to determine if DBT antagonism can produce distinct effects in the cytosol and nucleus, forms of a dominant negative DBTK/R with these two alternative localizations were produced. DBT has a putative nuclear localization signal (NLS), and mutation of this signal confers cytosolic localization of DBT in the lateral neurons of Drosophila clock cells in the brain. By contrast, addition of a strong NLS domain (e.g. SV40 NLS) to DBT’s C terminus leads to more nuclear localization. Expression of DBTK/R with the mutated NLS (DBTK/R NLS−) using a timGAL4 driver does not alter the circadian period of locomotor activity, and the daily oscillations of PER detected by immunoblot and immunofluorescence persist, like those of wild type flies. By contrast, expression of DBTK/R with the strong NLS (DBTK/R stNLS) using the timGAL4 driver lengthens period more strongly than DBTK/R, with damped oscillations of PER phosphorylation and localization. Both DBTK/R and DBTWT without the NLS fail to interact with Bride of Doubletime protein (BDBT), which is related to FK506-binding proteins and shown to interact with DBT to enhance its circadian function. This result suggests that the DBTK/R NLS− has lost its dominant negative property because it does not form normal clock protein complexes. DBTWT proteins with the same changes (NLS- and stNLS) also produce equivalent changes in localization that do not produce opposite period phenotypes. Additionally, a DBTK/R protein with both the stNLS and NLS− mutation does not affect circadian period although it is nuclear, demonstrating that the lack of a dominant negative for the DBTK/R NLS− is not due to failure to localize to nuclei. Finally, bdbt RNAi increases the cytosolic localization of DBTK/R but not of DBTWT, suggesting a role for BDBT in DBT-kinase dependent nuclear localization of DBT.
Keywords: DBTK/R, PER phosphorylation, circadian rhythm, Drosophila clock, protein degradation
Introduction
Circadian rhythms are biological processes that are normally synchronized (or entrained) by environmental cues and therefore exhibit oscillations of 24 hours (h). However, in the absence of these cues, these oscillations persist with a period of only approximately 24 h, thereby establishing the role of an endogenous circadian clock (Pittendrigh, 1960). Genetic and biochemical studies in both Drosophila and mammals have revealed that the clock mechanism is highly conserved (Allada et al, 2001; Hardin, 2011). Integral to the clock mechanism are the oscillations of clock gene products and their control through both positive and negative feedback loops. In Drosophila, Period (PER) and Timeless (TIM) proteins accumulate in the cytoplasm through the course of the night, where they initially form a heterodimer, and then translocate to the nucleus, apparently independently. Once in the nucleus PER represses the transcription of per and tim genes by binding to a transcription factor complex composed of dCLOCK (dCLK) and dCYCLE (dCYC) proteins. The dCLK/dCYC complex is responsible for the transcription of per, tim and other genes that produce the diverse outputs of the circadian clock. In another transcriptional feedback loop, the dCLK/dCYC complex also negatively regulates the transcription of the dclk mRNA because it induces the expression of a VRI, a repressor of dClk transcription.
Post-translational regulation is important for both of these transcriptional feedback loops. At the onset of day, the light signal is transduced by the CRY photoreceptor protein, and this targets TIM for degradation (Ceriani et al, 1999; Emery et al, 1998; Stanewsky et al, 1998). Doubletime protein (DBT), the Drosophila ortholog of Casein kinase Iδ (CKIδ) and Casein kinase Iε (CKIε), phosphorylates PER and targets it for degradation in the absence of TIM (Kloss et al, 1998; Ko et al, 2002; Muskus et al, 2007; Price et al, 1998). Other kinases and phosphatases, including SGG, CKII, NMO, PP2A and PP1, also function cooperatively with DBT to produce circadian oscillations and determine circadian period (Weber et al, 2011). DBT has been proposed to inhibit the accumulation of PER in the nucleus by targeting it for degradation in the cytosol, and subsequently to relieve the repression of PER on its own transcription by targeting PER for degradation in the nucleus (Kloss et al, 1998; Price et al, 1998). In such a model, DBT activity would slow the pace of the clock in the cytoplasm by delaying its nuclear accumulation and increase the pace of the clock in the nucleus by targeting PER for elimination after it has accumulated there. However, strong inhibition of DBT kinase activity, produced in both flies and mammals by expression of catalytically inactive mutations of DBT (DBTK/R) (Lee et al, 2009; Muskus et al, 2007), or in mammals by tissue specific knockouts and pharmacological inhibition of CKIδ (Chen et al, 2012; Eide et al, 2005; Etchegaray et al, 2009; Hirota et al, 2010; Isojima et al, 2009; Meng et al, 2010; Miyazaki et al, 2004; Walton et al, 2009; Xu et al, 2009; Xu et al, 2007), only lengthens period, suggesting that the antagonized kinase activity normally increases the period of the clock.
Mutations which do not eliminate the kinase activity of DBT can either lengthen or shorten circadian period (Bao et al, 2001; Fan et al, 2009; Kloss et al, 1998; Muskus et al, 2007; Preuss et al, 2004; Price, 2004; Price et al, 1998; Suri et al, 2000). It has been suggested that these different effects on period are produced because the mutations may affect the activity of DBT towards different target sites in PER, and that short-period mutations of dbt (or mammalian CKIε) are gain-of-function on a subset of phosphorylation sites that shorten period or loss-of-function on sites that lengthen period (Bao et al, 2001; Fan et al, 2009; Gallego et al, 2006; Meng et al, 2008; Muskus et al, 2007; Preuss et al, 2004; Syed et al, 2011; Vanselow et al, 2006; Xu et al, 2007). Since, as noted above, reduced PER phosphorylation in the cytosol and nucleus might have opposite effects on circadian period, an alternative or additional difference between the short- and long-period dbt mutants might be that they reduce cytosolic or nuclear phosphorylation events, respectively.
To test the validity of this premise, we set out to determine if decreasing the activity of DBT in just the nuclear or cytosolic compartment has opposite effects on period. DBT has a putative Nuclear Localization Signal (NLS) which we mutated to reduce or eliminate nuclear localization of a catalytically inactive DBT (DBTK/R). Alternatively, a strong NLS sequence (SV40 NLS sequence; stNLS) was added to the DBT C-terminus to increase nuclear localization. While these mutations did alter the localization of DBTK/R as predicted, only the DBTK/R with the strong NLS strongly altered the wild type period (by lengthening it) and damped PER oscillations in electrophoretic mobility and subcellular localization. While this result alone appears to suggest that DBT plays a vital role in the nucleus rather than the cytoplasm to promote PER phosphorylation, we determined that the NLS mutation also reduced the interaction of DBT with Bride of Doubletime protein (BDBT), a DBT interactor and circadian modulator related to FK506-binding proteins (Fan et al, 2013; Price et al, 2015). Restoring some nuclear localization to this DBTK/R NLS mutant protein with the addition of a stNLS also produced a protein that did not lengthen circadian period, and eliminating the NLS from DBTWT produced a protein that exhibited loss of the BDBT/DBT interaction and shortened circadian period while adding a stNLS to DBTWT did not affect the outcome of DBTWT overexpression. Finally, RNAi knock-down of BDBT also produced alterations in DBTK/R localization. These results demonstrate that BDBT interacts with DBT in a manner requiring the DBT NLS for nuclear localization, and that the interaction is required to produce a fully functional circadian complex.
MATERIALS AND METHODS
Site directed Mutagenesis
The mutants carrying the dbtK/R NLS−, dbtWT NLS−, dbtK/R stNLS, dbtWT stNLS and dbtK/R stNLS NLS− constructs were generated in the S2 cell expression vector pMT carrying dbtK/R or dbtWT with a MYC epitope at the C terminus (Muskus et al, 2007). To generate the NLS− constructs, the dbt construct was further mutagenized with the QUICK CHANGE (QC) (Stratagene, CA) procedure using oligonucleotides primers encoding the NLS mutation: the QCNLS forward primer, 5′-GCTTAAAGGCAGCCAACAACAATCAAAACTACGAGAGGATCTCGG-3′, and the QCNLS reverse primer, 5′-CCGAGATCCTCTCGTAGTTTTGATTGTTGTTGGCTGCCTTTAAGC-3′.
For the insertion of a strong SV40 NLS sequence at the C terminus of dbtK/R, dbtWT, or dbtK/R NLS− the following primers were used: dbt stNLS Forward, 5′-CCGGTCCAAAGAAAAAGCGTAAAGTCTGAGTTT-3′, dbt stNLS Reverse, 5′-AAACTCAGACTTTACGCTTTTTCTTTGGA-3′. These primers were annealed to create an NLS flanked by restriction sites (PmeI and AgeI). The pMT-dbtK/R plasmid DNA was digested with the restriction enzymes PmeI and AgeI, and the strong NLS sequence was introduced at the C terminus following the MYC epitope. To make the dbtK/R stNLS NLS− construct, the dbtK/R stNLS and the dbtK/R NLS− constructs were each digested with EcoRI and AgeI, and the EcoRI/AgeI fragment (encoding DBT) from the NLS− construct was ligated in place of the one from the original dbtK/R stNLS construct. All constructs were confirmed by DNA sequencing (MU Genomic Sequencing Facility).
Generation of Transgenic Flies
The transgenic constructs were made by using the Φc31 integrase system (Bischof et al, 2007). In order to clone the dbt constructs (dbtK/R, dbtK/R NLS−, dbtK/R stNLS, dbtWT NLS−, dbtWT st NLS and dbtKR stNLS NLS−) into insertion vector pUAST-attB, the pUAST-attB polylinker was first modified by inserting a PmeI site as described elsewhere (Muskus et al, 2007). The pUAST-attB polylinker was then digested with EcoRI and PmeI for insertion of the various pMT-dbtK/R-myc constructs. The cloning placed the dbt genes under control of the upstream activation sequence (UAS) promoter. Transformants were produced by Model System Genomics of Duke University (Durham, NC) at the attP2 locus on chromosome III. The lines were generated, mapped by standard procedures to the third chromosome and balanced with the TM3SbSer balancer chromosome.
Fly Entrainment and Locomotor Assays
The transgenic dbt lines were crossed to a timGAL4 driver - yw; P (w [+mC] = GAL4-timE62 (from Bloomington stock center, stock number 7126) - limiting expression to clock specific neurons because the GAL4 expression was dependent on the activity of the timeless promoter. The crosses were entrained at 23.5°C under a 12h: 12h light-dark (LD) cycle with cool white fluorescent bulbs (ca 3000 lx). The F1 progeny containing the timGAL4 driver and the UAS-dbt responder were collected and were entrained for a further 72 hrs in separate vials at 23.5°C. Males were used for locomotor assays (because of their more robust locomotor activity) while females and males were used for immunoblot analysis of head extracts or for immunofluorescence analysis of brains. The male flies were loaded into individual cuvettes and were placed in a monitoring device connected to a computer (Trikinetics Inc, Waltham, Mass) and data was recorded and analyzed using the method described previously (Muskus et al, 2007). Responder-gene-only controls were generated by crossing each UAS-responder line to wild type Canton S wild type flies, while timGAL4>+ flies were generated by crossing timGAL4 to Canton S wild type flies. For demonstration of non-developmental effects of DBTK/R expression in Table S1, lines 7018 from the Bloomington Drosophila Stock Center (with a third chromosome tubGAL80ts repressor) and line 7108 (with a second chromosome tubGAL80ts repressor) were crossed to a timGAL4>UAS-dbtK/R line at 23°C (functional repressor). The progeny flies were tested first at 23°C and showed no effect on locomotor activity, and then they were tested for a week at 31°C and a week at 33°C to reduce tubGAL80 repressor activity, thereby revealing long periods and reduced rhythmicity.
Immunoprecipitation and Immunoblot Analysis
For immunoprecipitations, fly heads from appropriate genotypes were chopped off with a razor blade, homogenized, treated with anti-MYC beads, centrifuged and washed as previously described (Fan et al, 2013; Price et al, 2015). For analysis of proteins in extracts without immunoprecipitation, heads were homogenized with 7μl of 1.1X sodium dodecyl sulfate (SDS) Laemmli gel loading buffer per fly with a Kontes pellet pestle homogenizer, and incubated at 95°C for 5 min. 3–5 μl of the extracts were analyzed by SDS-PAGE (10% acrylamide for DBT/BDBT/Tubulin or 5.7% for PER) followed by immunoblot analysis. For analysis of DBT, the membranes were incubated with a 1:1000 dilution of mouse anti-MYC antibody (Invitrogen, Carlsbad, CA) or 1:2000 dilution of rabbit anti-DBT antibody (Muskus et al, 2007). The DBT blots were also probed with a 1:10000 dilution of mouse anti-tubulin antibody (Covance) used as a loading control. For the analysis of PER, the blots were probed with a 1:25000 dilution of rabbit anti-PER (Muskus et al, 2007), and for analysis of BDBT the blots were probed with a 1:5000 dilution of anti-BDBT (Fan et al, 2013). The blots were then incubated with a 1:5000 dilution of appropriate secondary antibodies labeled with horse-radish peroxidase (American Qualex). The blots were visualized with ECL2 Western blot detection solution (Pierce) and the signals were quantified with TotalLab Quant (Total Lab, Ltd, Newcastle upon Tyne, UK) software using the minimum background method. The signals for each DBT protein was normalized to the amount of tubulin in the lane, and this signal was then normalized to the amount of DBTK/R without NLS mutations/tubulin for all DBTK/R proteins or to DBTWT/tubulin for the DBTWT NLS− protein.
Immunofluorescence
The brains of timGAL4>UAS-dbt-myc flies were dissected at the indicated times. For demonstration of the effects of bdbt RNAi on DBT-MYC localization, timGAL4>UAS-dbtWT or timGAL4>UAS-dbtK/R flies were crossed to line 100028 (UAS-bdbt RNAi) from the Vienna Drosophila RNAi center, and progeny carrying all three transgenes were collected at the indicated times. The fly brains were analyzed as previously described (Muskus et al, 2007). For DBT-MYC detection and localization, the brains were then probed with a 1:5000 dilution of mouse anti-PDF (Developmental Studies Hybridoma Bank, Iowa City, IA) and a 1:1000 dilution of goat anti-MYC (Santa Cruz Biotech, Santa Cruz CA) overnight. The secondary antibodies used were Alexa-Fluor 488 anti-goat immunoglobulin G (Jackson ImmunoResearch Laboratories, West Grove, PA) and Alexa-Fluor 568 anti-mouse immunoglobulin G in 1:1000 dilutions (Invitrogen, Carlsbad, CA). For PER detection and localization, the brains were probed with a 1:5000 dilution of mouse anti- PDF (Developmental Studies Hybridoma Bank, Iowa City, IA) and a 1:10000 dilution of rabbit anti-PER overnight. The secondary antibodies used were Alexa-Fluor 488 anti-rabbit immunoglobulin G and Alexa-Fluor 568 anti-mouse immunoglobulin G in appropriate dilutions (Invitrogen, Carlsbad, CA). Z-stacks were acquired on an Olympus BX61W1 confocal microscope with a 60X water immersion lens.
RESULTS
Restriction of DBTK/R to Either the Cytoplasm or Nucleus
Whether and how DBT affects the core clock in the cytosol and the nucleus is still unclear. We have previously shown that the dominant negative form of DBT (DBTK/R) lengthens period and produces hypophosphorylated PER at all time points when expressed in all clock cells (Muskus et al, 2007). The expression of DBTK/R antagonizes the activity of the endogenous DBT kinase in a dose-dependent manner, leading to increased lengthening of period with higher levels of expression. The longer periods are due to a direct effect on the circadian clock rather than a developmental effect, because they are prevented if the flies are raised at 23°C with a temperature sensitive GAL80 repressor expressed from the tubulin promoter, and are then produced progressively if the flies are raised to 31°C and 33°C, which inactivates the repressor and allows the DBTK/R to be expressed (Supplementary Table I).
We decided to limit expression of DBTK/R to either the cytosol or nucleus to test the capacity of DBTK/R to antagonize endogenous DBT in each of these compartments. Since DBT has a putative Nuclear Localization Signal (NLS) (Fig. 1), three positively charged residues in the NLS sequence were mutated to asparagine. Alternatively, a strong NLS sequence (SV40 NLS sequence) was added to the DBT C-terminus. The mutations were produced both in the context of DBTK/R-MYC and DBTWT−MYC.
Figure 1.
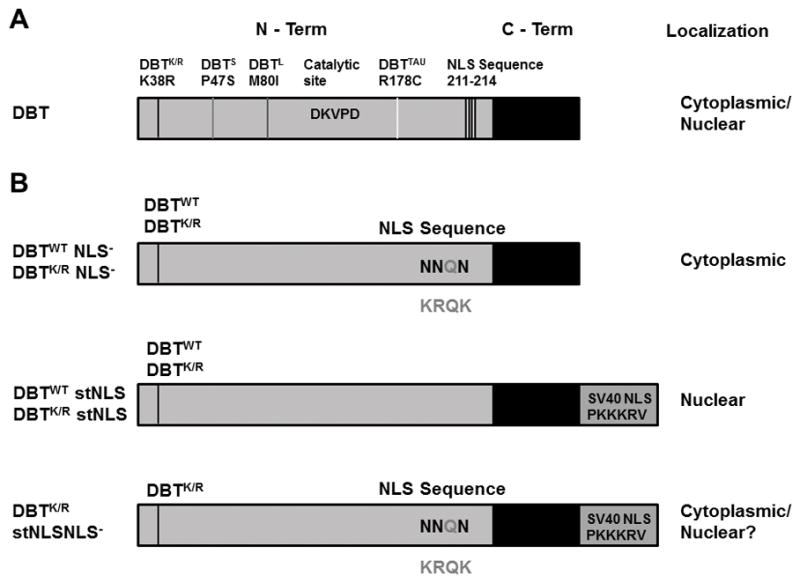
Domain structure of DBT and sites of mutations.
A) Location of previously characterized period-altering mutations of dbt, the catalytic site and the NLS sequence. The NLS mutation is away from the catalytic region of DBT. B) DBT mutant proteins analyzed in this study. The residues outside the proteins indicate the amino acids that are part of the putative NLS sequence. The residues in the NLS mutated to Asparagine are the darker ones, as shown inside the protein. The addition of strong SV40 NLS sequence is shown below the NLS− mutant. Predicted subcellular localizations are shown to the right; the DBTK/R stNLS NLS− mutant was predicted to be nuclear because of the addition of the stNLS but in fact is less nuclear than the DBTK/R stNLS protein (hence the “?”),.
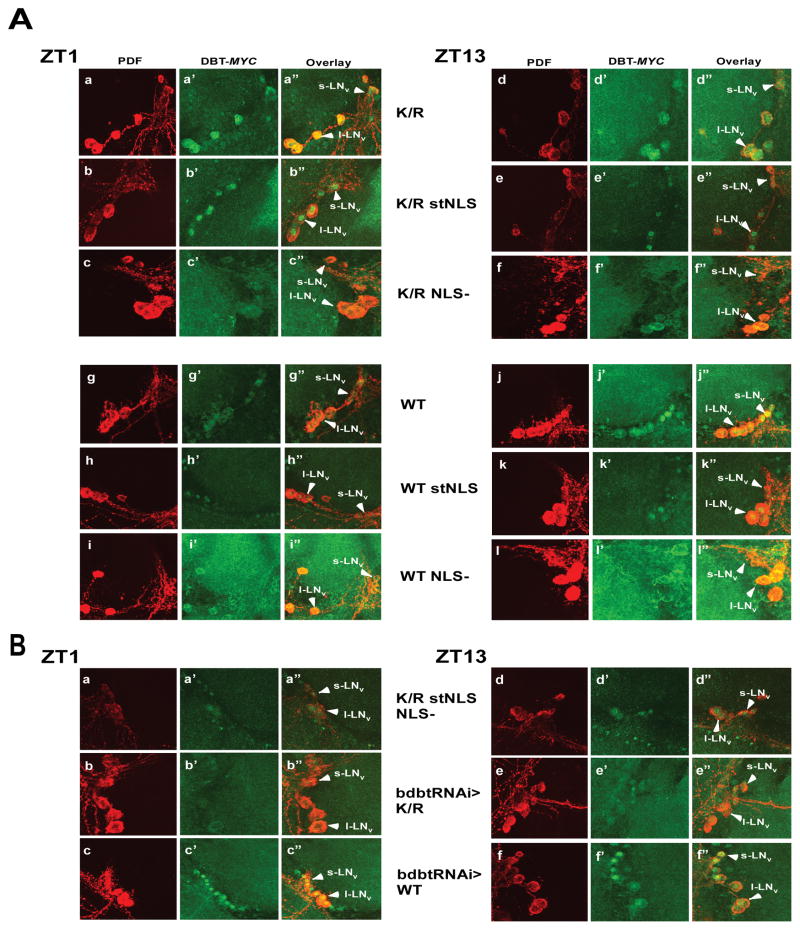
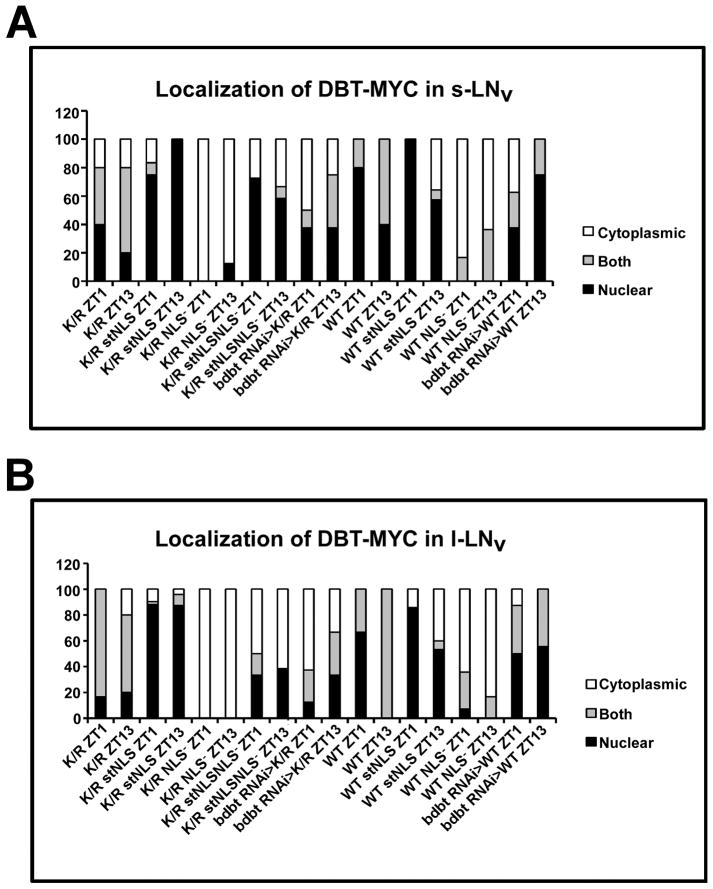
To use the mutant DBT in flies, we generated site-specific recombinant fly lines containing these mutants of DBT under UAS control and crossed them to a timGAL4 driver that drives the expression of the corresponding UAS-dbt in the clock neurons. The neuropeptide PDF localizes to the cytoplasm of the small and large lateral neurons (sLNv and lLNv, respectively; the adult brain neurons which drive circadian behavior)(Helfrich-Forster, 1997), and its detection with an anti-PDF antibody was employed to differentiate cytosolic versus nuclear localization. The localization of mutant DBT in adult brains at ZT1 and ZT13 was assessed to determine if the localization was restricted to just one compartment. The original DBTWT-MYC and DBTK/R–MYC proteins localize to both compartments at both time points (Fig. 2A ZT1: a-a″ and g-g″; ZT13: d- d″ and j-j″; Fig. 3A&B). By contrast, DBTWT stNLS and DBTK/R stNLS localize mostly to the nucleus at both time points (Fig. 2A ZT1: b-b″ and h-h″; ZT13: e-e″ and k-k″; Fig. 3A&B). Alternatively, DBTWT NLS− and DBTK/R NLS− localize mostly to the cytosol, where they sometimes exhibit faint cytosolic rings but more often are difficult to detect, in small and large lateral neurons of the adult brains (Fig. 2A ZT1: c-c″ and i-i″; ZT13: f-f″ and l-l″; Fig. 3A&B).
Figure 2.
Confocal images of adult brain hemispheres show different subcellular localizations for various DBT-MYC mutants and DBT-MYC with bdbt RNAi. (A) timGAL4>UAS-dbt-myc flies with the indicated dbt-myc genotypes were collected at ZT1 or ZT13, and their brains were incubated with anti-PDF and anti-MYC antibodies. Representative images are shown. Solid arrowheads labeled “I-LNvs” indicate the positions of large ventral lateral neurons, and solid arrowheads labeled “s-LNvs” show small ventral lateral neurons (both express PDF). The nuclear localization signal mutants were assessed in the context of both DBTK/R-MYC and DBTWT-MYC. Multiple confocal images of each genotype were scored by an observer blinded to the genotype and time, and the results are tabulated in figure 3. (B) Representative images of timGAL4> flies expressing DBTK/R stNLS NLS− (a double mutant), or DBT of the indicated genotype in the presence of bdbt RNAi. Collections, treatments and labeling were the same as for panel A.
Figure 3.
Quantification of DBT-MYC localization in s-LNv cells (A) and l-LNvs (B). Images were scored blind to the identity of the samples and the time of collection for localization of DBT. PDF was used as a marker for cytoplasmic localization. Brain hemispheres showing DBT signal that was all through most of the PDF+ s-LNv or l-LNv were scored as “both.” Brain hemispheres with DBT signal that colocalized only with PDF in most s-LNv or l-LNv or was not visible were scored as cytoplasmic, while those in which the signal did not colocalize with PDF and was concentrated centrally were scored as nuclear. The width of the differently shaded bars is proportional to the percentage of brains with the indicated score.
NLS Mutant Forms of DBTK/R are Expressed at Comparable Levels to Those of DBTK/R
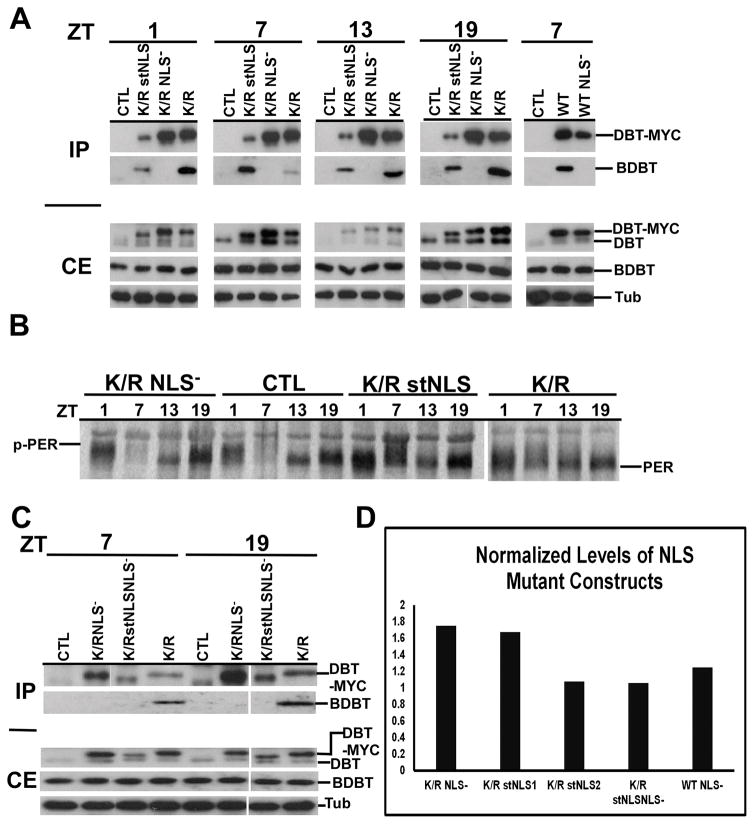
Head extracts were collected at ZT1 (1 hr after lights on), ZT7, ZT13 and ZT19 from multiple lines of each dbtK/R genotype and were subjected to immunoblot analysis to determine the levels of DBT expression. The relative levels of expression of DBTK/R NLS− and DBTK/R stNLS were comparable to those of DBTK/R (Fig. 4A, CE) when detected with an antibody that was raised against the entire C terminal domain of DBT (and not the MYC epitope), and repeated quantification of the levels by western blot analysis confirmed this (Fig; 4D). When an anti-MYC antibody was used to detect the transgenic DBT, the level of detection for DBTK/R stNLS-MYC was much lower than for DBTK/R NLS− MYC and DBTK/R MYC (Figure S1). Since the antibody directed against the C terminus does not detect strongly reduced levels and the levels of DBTK/R stNLS-MYC detected in brain nuclei with anti-MYC are quite high (Fig. 2), it is likely that the strongly reduced detection on immunoblots with anti-MYC results from the effect of the NLS sequences adjacent to the MYC epitope; the proximity of the NLS is likely to reduce the affinity of the anti-MYC antibody for the MYC epitope. While it is possible the strong NLS at the C terminus of DBT destabilizes DBT to some extent, this effect would only make the strong effects of DBTK/R stNLS described below even more striking. Levels of transgenic DBT-MYC were equivalent at ZT 1, ZT7, ZT13 and ZT19 (Fig. 4A), ruling out a circadian effect on the different levels detected with anti-MYC. Moreover, the differences in detection levels for DBT were not statistically significant for different lines within a particular genotype (Fig. 4D), as was expected given the site-specific recombination that inserts all of the transgenes at the same locus.
Figure 4.
Immunoblot analysis of head extracts for DBT-MYC demonstrates comparable overall levels of expression of different transgenic DBTs, a lack of interaction between DBTs carrying the NLS mutation and BDBT, and different effects on PER protein oscillations.
(A) Immunoblot analysis of fly head extracts for DBT. The F1 progeny with the indicated genotype of timGAL4/+>UAS-dbt-myc/+ were collected for different mutant genotypes, head extracts were collected and immunoprecipitated with anti-MYC antibody, and blots of SDS PAGE were probed to detect DBT and BDBT levels in the crude extracts (CE) and the immunoprecipitates (IP). DBT was detected using a 1:2000 dilution of anti-DBT-C, which detects both endogenous DBT (lower band) and transgenic DBT-MYC (upper band). The CE blot was then stripped and reprobed with anti-tubulin (lower panel). No clear genotype-specific or circadian changes in DBT-MYC level were seen. Both DBTWT and DBTK/R proteins carrying the NLS mutation failed to interact with BDBT. (B) Immunoblot analysis of fly head extracts for PER. Fly heads were collected for different mutant genotypes at the indicated time points (ZT1: one hour after lights on; ZT7: 7 hour after lights on; ZT13: one hour after lights off; ZT19: 7 hour after lights off) and the blot was probed for PER using a 1:25000 dilution of anti-PER. dbtK/R and dbtK/R stNLS flies produce high levels of hypophosphorylated PER at all four time points, while dbtK/R NLS− flies produce a wild type PER rhythm, compared with wild type flies that inherited a TM3 balancer chromosome instead of a chromosome with the UAS responder gene (CTRL). C) Head extracts of the indicated genotypes were analyzed for DBT expression with an anti-DBT antibody at the indicated circadian times, and they were immunoprecipitated with anti-MYC for detection of DBT and co-immunoprecipitating BDBT. DBTK/R stNLS NLS− protein carries both the NLS− and stNLS mutations, is expressed at comparable levels to the other transgenic DBTs, and like DBTK/R -NLS− and DBTWT NLS− it does not co-immunoprecipitate BDBT. (D) The levels of DBT-MYC expression were assessed for representative lines of each genotype at ZT7 and were found to be equivalent within the range of the measurements. The DBT-MYC signal was quantified for each lane on a gel and normalized to the level of tubulin signal in that lane. Then, this signal was further normalized to the level of DBTK/R/tubulin on the blot for DBTK/R proteins or to DBTWT/tubulin for the DBTWT NLS- construct. Relative signals varied between 1 and 1.8-fold of the DBTK/R or DBTWT baseline signal, and the differences were not statistically significant by one-way ANOVA [F(4,6)=2.8, p>0.1]. Two dbtK/R stNLS lines were tested – lines 2MB (line 1) and line 1MB (line 2). The difference in measured DBT level runs counter to the difference in their average periods (Table 1), but neither of these differences is statistically significant.
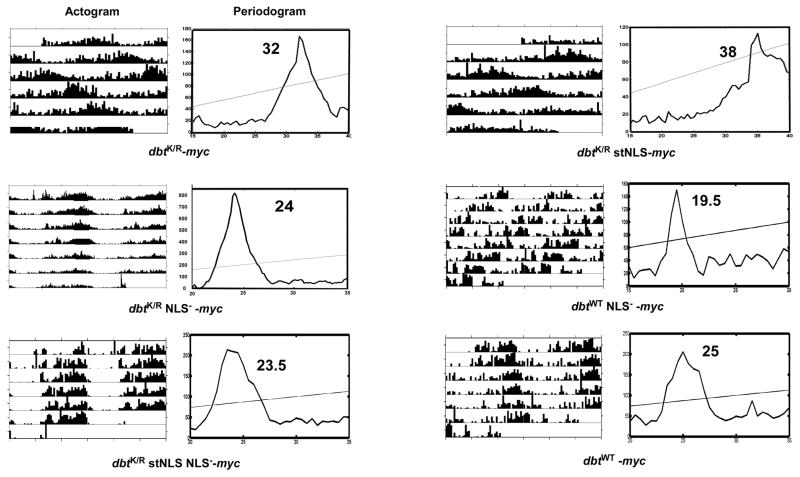
Circadian Period is Lengthened with the Overexpression of DBTK/R stNLS but not by DBTK/R NLS−
As previously shown (Muskus et al, 2007), expression of DBTK/R resulted in a lengthening of period of approximately 8 h relative to wild type flies (dbtK/R: 31.6 h, without driver: 23.5 h; Table 1 and Figure 5) and reduced rhythmicity. When the NLS region of dbtK/R was mutated (dbtK/R NLS−) and the mutant protein expressed in flies, it did not alter the period relative to wild type flies (dbtK/R NLS−: 23.8 h, without driver: 23.5 hrs; Table 1 and Figure 5). Alternatively, expression of the DBTK/R with a strong SV40 NLS (DBTK/R stNLS) resulted in the lengthening of period by about 15 hrs compared to wild type flies and about 7 h compared to dbtK/R flies (dbtK/R stNLS: 39 h, dbtK/R: 31.6 h; Figure 5 and Table 1), with reduced rhythmicity. The differences in circadian period were statistically significant for both the dbtK/R and the dbtK/R stNLS mutants compared to the wild type flies, and all three dbtK/R genotypes differed significantly from each other. Individual flies from each of the lines showed substantial differences from one another in their circadian periods (note the large standard deviations for each line) and were often arrhythmic, and so within a genotype the reported differences in average period are generally not statistically significant (Table 1). All of the individual dbtK/R and dbtK/R stNLS lines analyzed herein had longer periods than the shorter period line (28 h) previously used to demonstrate the effect on period of DBTK/R expression levels (Muskus et al, 2007), so the expression levels of DBTK/R are essentially in the saturation range in this current study.
Table 1.
Effects of NLS mutant DBTs on locomotor activity in DD.
| DBT | Line | Genotype | Avg Period (h)±SEM (SD) | %Rhythmicity (N) |
|---|---|---|---|---|
| DBTK/R | 31.6 ±0.4 (0.6)*** | 47 (3) | ||
| 1MA | timGAL4>UAS- dbtK/R | 31.9 ±0.5 (2.9) | 48 (58) | |
| 10FB | timGAL4>UAS- dbtK/R | 32 ±0.9 (3.0) | 43 (28) | |
| 4MA | timGAL4>UAS- dbtK/R | 30.9 ±0.8 (2.4) | 50 (16) | |
| >UAS- dbtK/R | 23.5 ±0.1(0.3)* | 100 (21) | ||
| DBTK/R NLS- | 23.8 ±0.1 (0.2)* | 82 (5) | ||
| 1MB | timGAL4>UAS- dbtK/R NLS- | 23.9 ±0.2 (0.7) | 82 (22) | |
| 11MB | timGAL4>UAS- dbtK/R NLS- | 23.6 ±0.1 (0.3) | 75 (16) | |
| 2MB | timGAL4>UAS- dbtK/R NLS- | 23.7 ±0.1 (0.2) | 81 (16) | |
| 3MB | timGAL4>UAS- dbtK/R NLS- | 23.6 ±0.1 (0.2) | 75 (16) | |
| B4 | timGAL4>UAS- dbtK/R NLS- | 24.0 ±0.1 (0.5) | 97 (33) | |
| >UAS- dbtK/R NLS- | 23.5 ±0.1 (0.4)* | 95 (42) | ||
| DBTK/R stNLS | 39.0 ±1.0 (2.0)*** | 34 (4) | ||
| 1MB | timGAL4>UAS- dbtK/R stNLS | 40.3 ±1.5 (4.3) | 32 (28) | |
| 2MB | timGAL4>UAS- dbtK/R stNLS | 38.7 ±1.6 (4.0) | 29 (21) | |
| 10MB | timGAL4>UAS- dbtK/R stNLS | 36.3 ±1.4 (5.2) | 41 (34) | |
| 6FB | timGAL4>UAS- dbtK/R stNLS | 40.6 ±0.7 (1.5) | 33 (15) | |
| >UAS- dbtK/R stNLS | 23.8 ±0.1 (0.5)* | 89 (47) | ||
| DBTWT | 25.0 ±0.1 (0.1)* | 72 (2) | ||
| 6M3B | timGAL4>UAS- dbtWT | 25.1 ±0.5 (1.1) | 44 (9) | |
| 21M1C | timGAL4>UAS- dbtWT | 24.9 ±0.2 (0.9) | 100 (30) | |
| >UAS- dbtWT | 23.5 ±0.1 (0.2)* | 100 (12) | ||
| DBTWT NLS | 20.7 ±0.6 (1.0)** | 83 (3) | ||
| A17 | timGAL4>UAS- dbtWT NLS | 21.8 ±0.2 (0.6) | 85 (13) | |
| A2 | timGAL4>UAS- dbtWT NLS | 20.5 ±0.3 (0.6) | 100 (5) | |
| A7 | timGAL4>UAS- dbtWT NLS | 19.8 ±0.1 (0.5) | 65 (23) | |
| >UAS- dbtWT NLS | 23.7 ±0.1 (0.5)* | 92 (12) | ||
| DBTWT stNLS | 24.1 ±0.03 (0.1)* | 75 (3) | ||
| D2 | timGAL4>UAS- dbtWT stNLS | 24.0 ±0.2 (0.8) | 94 (16) | |
| D3 | timGAL4>UAS- dbtWT stNLS | 24.1 ±0.1(0.5) | 82 (57) | |
| D4 | timGAL4>UAS- dbtWT stNLS | 24.1 ±0.3 (1.0) | 48 (21) | |
| >UAS- dbtWT stNLS | 23.5 ±0.1 (0.2)* | 94 (16) | ||
| DBTK/R stNLS NLS- | 23.9 ±0.03 (0.1) * | 73 (3) | ||
| C7MA | timGAL4>UAS- dbtK/R stNLSNLS- | 24.0 ±0.3(1.0) | 75 (12) | |
| C6M | timGAL4>UAS- dbtK/R stNLSNLS- | 23.9 ±0.2 (0.7) | 80 (10) | |
| B1 | timGAL4>UAS- dbtK/R stNLSNLS- | 23.9 ±0.3 (0.7) | 63 (8) | |
| >UAS- dbtK/R stNLSNLS- | 23.8 ±0.1 (0.2)* | 100 (27) | ||
| DBT+ | ||||
| timGAL4> | 24.0 ±0.1 (0.5)* | 93 (28) | ||
Lines containing the indicated type of insertion were crossed to flies containing the timGAL4 driver, and progeny hemizygous for both the driver and responder were assayed in DD for locomotor activity. Each line contained an independent insertion of the responder UAS-dbt gene, generated at the AttP2 locus (at 68A4) by phiC31-mediated integration, except for the wild type controls. The circadian period was determined by chi-square periodogram analysis. Rhythmic flies produced single strong peaks in the periodogram analysis and rhythmicity that was obvious by inspection of actograms. The mean period ± SEM (SD) and the mean percentage of rhythmicity (N, number of lines for genotype averages and number of flies for line averages; only flies that lived for the entire assay were included in N and the denominator for determination of % rhythmicity). One way ANOVA of all the individual line averages showed a significant effect of genotype on period [F (14,16) = 74.0, p<0.001].
The average periods of lines with these genotypes were significantly different from the average period of all other genotypes with p<0.001 by Tukey HSD.
The average period of lines with this genotype were significantly different from the average periods of all other lines expressing a UAS-dbt transgene with the timGAL4 driver by Tukey HSD (p<0.05).
The average periods of genotypes without the timGAL4 driver, with the timGAL4 driver only, expressing the dbtWT transgene, expressing the dbtWT transgene with a stNLS or expressing the dbtK/R transgene with an NLS mutation did not exhibit statistically significant differences from each other. One way ANOVA of individual fly periods for all the flies also demonstrated a significant effect of line identity on period [F(30, 487) = 142, p<0.0001), but no lines with the same UAS-dbt genotype showed statistically significant differences from each other except UAS-dbtK/R stNLS line 10M1B, which did differ from UAS-dbtK/R stNLS lines 1MB and 6FB by post-hoc Tukey (p< 0.0001).
Figure 5.
Behavioral analysis of dbt NLS mutants demonstrates strong effects of dbtK/R stNLS on behavior, no effects for dbtK/R NLS− and dbtK/R stNLSNLS− flies and shorter periods for dbtWT NLS− flies. Flies with the indicated genotypes (timGAL4>/+; UAS-dbt-myc/+) were entrained for at least three days in LD 12hr: 12hr and then released into DD. Representative actograms and periodograms for the indicated genotypes are shown. All of the dbtK/R NLS− and dbtK/R stNLS NLS− mutants exhibited normal circadian periods, while the dbtK/R stNLS mutants exhibited lengthened periods even compared to dbtK/R, and the dbtWT NLS− flies exhibited short circadian periods. See Table 1 for tabulation of these results.
Molecular Oscillations of PER are Blunted by Expression of the DBTK/R stNLS but not by Expression of the DBTK/R NLS−
The oscillations of PER level and phosphorylation state (as manifested in electrophoretic mobility shifts on SDS-PAGE) are indicative of the state of the cellular clock that drives circadian behavior and physiology (Edery et al, 1994). To assess these oscillations, head extracts were collected at ZT1 (1 hr after lights on), ZT7 (7 hrs after light on), ZT13 (1 hr after lights off) and ZT19 (7 hrs after lights off), and subjected to immunoblot analysis. The dbtK/R stNLS mutants produced hypophosphorylated PER (i.e., fast mobility) at all time points (Fig. 4B), similar to that of the dbtK/R lines. By contrast, the dbtK/R NLS− mutants produced robust oscillations of PER similar to that of the wild type control (flies which inherited the TM3 chromosome instead of the UAS-dbt responder gene; Fig. 4B). The electrophoretic mobility differences were consistent (See Fig. S1B). The hypophosphorylated PER in the dbtK/R and dbtK/R stNLS mutants is consistent with the strong lengthening of circadian period and arrhythmicity for the behavioral rhythms (Fig. 5 and Table 1).
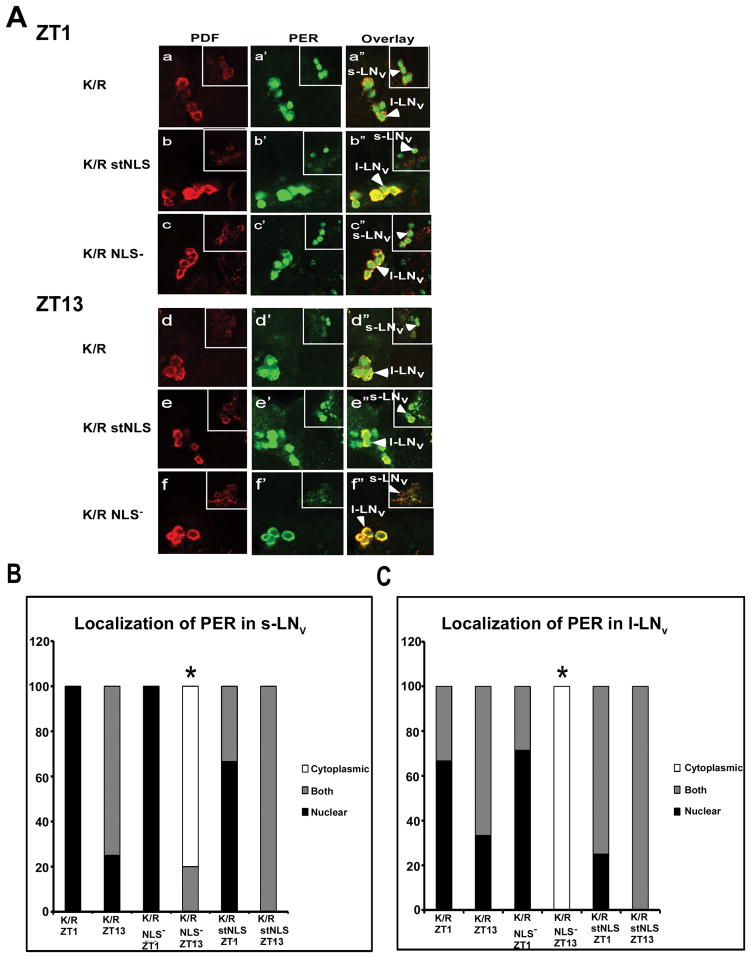
Next we analyzed the subcellular localization of PER in the adult brains to determine if the normal circadian oscillations (Zerr et al, 1990) are affected due to the expression of DBTK/R in the cytosol or the nucleus. Expression of DBTK/R with no nuclear localization mutations in the clock neurons resulted in PER which localized to nuclei of PDF+ LNv neurons, or to both the nucleus and the cytoplasm ((Muskus et al, 2007) and Fig. 6A panels a-a″ and d-d″; Fig. 6B&C). Expression of DBTK/R stNLS in the nucleus of clock neurons resulted in a phenotype similar to that of dbtK/R, wherein PER existed in the nuclei or both the nuclei and cytosol of the clock cells at ZT1 and ZT13 (Fig. 6A panels b-b″, e-e″; Fig. 6B&C). By contrast, in dbtK/R NLS− flies, PER exhibited strong nuclear/cytoplasmic oscillation, with mostly nuclear localization at ZT1 and cytoplasmic localization (or undetectable PER) at ZT13 in the LNvs (Fig. 6A panels c-c‴′, f-f″; Fig. 6B&C), as do wild type flies. Taken together, the molecular and behavioral analysis presented herein suggests that the suppression of PER phosphorylation by the endogenous DBT with expression of dbtK/R stNLS recapitulates the original dbtK/R phenotype (long periods and arrhythmicity), while expression of dbtK/R NLS− does not.
Figure 6.
Confocal images of adult brain hemisphere show damped oscillations for PER subcellular localization in dbtK/R and dbtK/R stNLS flies, but robust oscillations in dbtK/R NLS− flies. (A) Brains were collected from the indicated genotypes at ZT1 and ZT13, and both PER (green) and PDF (red) were detected with antibodies as indicated in Materials and Methods. Small lateral neurons (s-LNV) and large lateral neurons (L-LNv) were imaged in separate optical sections in which the most neurons of each type could be visualized and are presented separately for each image, with s-LNvs in the smaller window. dbtK/R flies exhibit high levels of PER localized to nuclei or to both nuclei and cytosol at both ZT1 (a-a″) and ZT13 (d-d″). dbtK/R stNLS flies also exhibit high levels of PER localized to nuclei or to both nuclei and cytosol at both ZT1 (b-b″) and ZT13 (e-e″). dbtK/R NLS− flies exhibit robust rhythms of PER subcellular localization, with mostly nuclear PER at ZT1 (c-c″) and cytosolic or no PER detection at ZT13 (f-f″). Quantification of PER localization in PDF+ s-LNvs (B) and l-LNvs (C). Images were scored blinded to sample identity and time for localization of PER. PDF was used as a marker for cytoplasmic localization. Brain hemispheres with PDF+ s-LNV or l-LNv cells that showed PER signal throughout the cell were scored as both. Brain hemispheres with PDF+ s-LNv or l-LNv cells that showed PER signal colocalized only with PDF or no PER detection were scored as cytoplasmic, and brain hemispheres with PDF+ s-LNv or l-LNv cells showing PER signal that did not colocalize with PDF and was centrally located were scored as nuclear. * PER localization scores in dbtK/R NLS− mutant flies at ZT13 in the s-LNv or l- LNv differed significantly from PER localization in dbtK/RNLS− flies at ZT1 (P<0.01), by Kruskal-Wallis nonparametric H ANOVA (H (11, N= 67) =51.7), with a multiple comparisons of mean ranks for all groups.
Mutants in DBT Lacking the NLS Do Not Interact with Drosophila BDBT
The wild type result with the dbt NLS− mutant was surprising, as we had predicted that this mutant would shorten circadian period by facilitating earlier accumulation of PER in the cytosol. However, it was also possible that it disrupted assembly of DBT into circadian complexes and thereby eliminated the dominant negative property of the DBTK/R construct. Recently, our lab had identified the BDBT protein as a noncanonical FK506-binding protein that interacts with DBT to facilitate its targeting of PER for degradation (Fan et al, 2013; Price et al, 2015), and so the effect of the NLS mutation on the DBT interaction with BDBT was assessed in circadian clock cells by BDBT coimmunoprecipitation with MYC-tagged DBT. The NLS− mutation completely eliminated the co-immunoprecipitation of BDBT with DBTK/R while the stNLS mutation did not (Fig. 4A, IP). Note that DBTK/R stNLS-MYC immunoprecipitates less readily than the other DBTK/R-MYC proteins (presumably because the proximity of the MYC epitope to the stNLS reduces the interaction of the MYC antibody, as noted above) but nevertheless co-immunoprecipitates more BDBT that DBTK/R NLS− MYC does.
We also observed a similar effect of the NLS− mutation on the BDBT/DBTWT interaction (DBTWT interacts with BDBT while DBTWT NLS− does not) with no effect on the expression level of DBT (Fig. 4A, rightmost lanes, and Fig. 4D). Moreover, the elimination of the NLS from DBTWT produced a short-period rhythm (Table I and Fig. 5). Finally, the DBTWT stNLS protein mildly lengthened period and reduced rhythmicity (Table 1), as does expression of DBTWT; our argument for the lack of a difference from DBTWT expression is that overexpression of a wild type protein simply replaces endogenous wild type DBT from circadian complexes and therefore has little effect, whether it occurs in the cytoplasm, the nucleus or both.
A DBT Protein with Both stNLS and NLS− Mutations Exhibits Partial Nuclear Localization but No Dominant Negative Property
If the NLS mutation were compromising the dominant negative capacity of DBTK/R by eliminating its nuclear localization, localization of this construct to the nucleus would restore its dominant negative capacity. In order to test this hypothesis, a stNLS was added to the C terminal domain of DBTK/R NLS−. This protein was expressed at comparable levels to the other DBTK/R proteins (Fig. 4C&D), and the addition of the stNLS restored some degree of nuclear localization to PDF+ cells, although the nuclear localization was not as bright and as consistent as the ones detected with the DBTK/R stNLS (Fig. 2B, a-a″ and d-d″; Fig. 3A&B). Nevertheless, the addition of the stNLS did rescue some nuclear localization of the DBTK/R stNLSNLS− protein, but like expression of DBTK/R NLS−, expression of this protein in circadian cells did not alter the circadian period (Fig. 5 and Table 1). Moreover, DBTK/R stNLS NLS− protein failed to produce interactions with BDBT (Fig. 4C) despite its nuclear entry, demonstrating that nuclear entry is not sufficient for DBT/BDBT interactions and supporting the conclusion that the NLS mutation disrupts the DBT/BDBT interaction. Hence, the absence of a dominant negative function for the DBTK/R NLS− and DBTK/R stNLSNLS− proteins is more likely to be due to their failure to form proper protein/protein interactions than failure to localize to nuclei.
Loss of BDBT leads to a kinase-dependent reduction in DBT localization
Since BDBT requires the DBT NLS site for its interaction with DBT and addition of a C-terminal stNLS did not fully restore nuclear localization to DBT, it is possible that BDBT participates in the nuclear localization of DBT. In order to test this possibility, a bdbt RNAi knock-down transgene was introduced into the timGAL4>UAS-dbtK/R and >UAS-dbtWT genotypes and the localization of DBT-MYC was assessed. For DBTK/R-MYC, bdbt RNAi enhanced cytosolic/no detection of DBT-MYC (particularly in the lLNv; Fig 2B, b-b″ and e-e″; Fig. 3A&B), while for DBTWT –MYC the effect was more marginal (if anything, somewhat more nuclear localization at ZT13; fig 2Bc-c‴′ and f-f″; Fig 3A&B). The bdbt RNAi effect is rather weak (Fan et al., 2013), and these effects may not indicate the full extent of BDBT involvement in nuclear localization, but they suggest that BDBT may be involved in DBT kinase activity-dependent nuclear localization of clock protein complexes.
DISCUSSION
The original view of DBT-dependent post-translational modifications holds that they modulate the length of phase delays essential for circadian oscillations (Kloss et al, 1998; Price et al, 1998). The first phase delay occurs during day and early evening, when per mRNA is accumulating but PER protein is not accumulating. Because PER accumulates to high levels late at night, many hours after per mRNA levels peak, and this phase delay in accumulation is DBT-dependent, it seems likely that DBT phosphorylates PER and targets it for degradation during the day and early evening (from ZT7 - ZT13) before it accumulates in the nucleus. The second phase delay occurs after PER becomes stabilized and moves to the nucleus. During this period, PER represses CLK/CYC-dependent transcription (Abruzzi et al, 2011; Allada et al, 1998; Zeng et al, 1994) as it becomes progressively phosphorylated by DBT. After PER becomes hyperphosphorylated, it is thought to be targeted by this phosphorylation for degradation, and this degradation ends the period of PER-dependent transcriptional repression, thereby allowing another circadian cycle of CLK/CYC-dependent transcription. Without the two phase delays that are postulated to be produced or terminated by DBT, negative feedback by PER would be immediate and continuous, producing a constitutively repressed state for the per and tim genes (as seen in the dbt loss-of-function mutants during DD (Price et al, 1998)) rather than oscillations of their mRNAs. This original view therefore posits roles for DBT in both the cytosol and the nucleus and is consistent with the observed movement of DBT to the nucleus during the late night to early day (Kloss et al, 2001) when PER also accumulates in the nucleus.
In light of this original view, our finding of altered circadian periods by nuclear-localized DBTK/R (dbtK/R stNLS) but not by cytosolic DBTK/R (dbtK/R NLS−) was surprising. It is likely explained by the reduced interaction capacity of DBTK/R NLS− protein with BDBT and possibly other circadian proteins. The long periods that are produced by DBTK/R stNLS are predicted by the reduced phosphorylation of PER. It is possible that the extremely long periods produced by dbtK/R stNLS are the consequence of eliminating a period-shortening activity that is produced by DBTK/R in the cytosol, and that the cytosolic DBTK/R NLS− protein is unable to produce this period-shortening because it has eliminated protein/protein interactions needed for its dominant negative properties. The possibility that it does not produce dominant negative properties because it is not in nuclei where dominant negative interactions must occur is negated by the finding that dbtK/R stNLS NLS− mutant also lacks dominant negative properties, despite exhibiting significantly more nuclear localization. The dbtK/R stNLS NLS− double mutant also does not interact with BDBT, thereby suggesting that it is the loss of this interaction and possibly others that compromise its dominant negative properties. Finally, our previous work has detected BDBT in the cytosol and not the nuclei of photoreceptors (Fan et al., 2013), which is not consistent with a nuclear DBT/BDBT interaction. The most parsimonious interpretation of our results is that dbtK/R NLS− and dbtK/R stNLSNLS− genotypes do not affect the circadian oscillations of PER because they have eliminated the dominant negative interactions of the protein.
It is not clear at the moment if BDBT directly binds the NLS site or interacts with something else (a nuclear importin, for example; see (Jang et al, 2015)) that bridges the interaction. Since a catalytically active (wild type) form of DBT with the NLS mutation does shorten period, the NLS mutation does not produce a complete null but seriously compromises the capacity of DBTK/R to exert its dominant negative phenotype. The requirement of the NLS for a normal interaction between DBT and BDBT and the reduced nuclear localization of DBT proteins carrying this mutation suggest that BDBT might normally contribute to the nuclear localization of DBT-containing complexes, which is therefore compromised in the NLS mutants. This interpretation is supported by our observation that bdbt RNAi increases cytosolic localization of DBTK/R but not of DBTWT; the difference between DBTK/R and DBTWT suggests that the localization of DBT may require some DBT activity, which BDBT loss of function should reduce further in the dbtK/R mutant (Fan et al, 2013). This suggestion is also intriguing because FK506-binding proteins are involved in HSP-90 dependent nuclear localization of the glucocorticoid receptor in response to glucocorticoids (Davies et al, 2002).
Because DBTWT NLS− does shorten circadian period, it seems unlikely that the DBTK/R NLS− fails to affect circadian period because there is no effect of DBT in the cytosol. It is more likely that a dominant negative effect is more sensitive to lack of interactions between DBT and other factors like BDBT, precluding an effect of DBTK/R NLS− in the cytosol because it does not undergo all of these interactions. Moreover, the short-period circadian rhythms produced by DBTWT NLS− are likely to come from other effects than simply the elimination of its interaction with BDBT, as reduced BDBT leads to long-periods and arrhythmicity (Fan et al, 2013). Because overexpression of DBTWT in nuclei (with DBTWT stNLS) does not lengthen period, it is also not possible that short periods and long periods are alternative outcomes for DBTWT overexpression in the cytosol and nuclei respectively. DBTWT expressed in both compartments or the nucleus produces the minimally altered periods, so it is more likely the NLS mutation is affecting other DBT interactions rather than affecting nuclear localization to shorten period in the context of DBTWT.
Prior work has shown that both DBTWT and DBTK/R mediate targeting of a kinase which targets CLK for phosphorylation (Yu et al, 2009), and potentially other clock components besides CLK and BDBT might require DBT for recruitment as well. In the absence of a critical component, DBTK/R NLS− may no longer compete well with endogenous DBT for assembly into clock protein complexes and may therefore not function as a dominant negative, or the complexes containing DBTK/R may be altered in some way to not lengthen the pace of the clock (e.g., by loss of a regulator which slows the pace of the clock).
It is intriguing that PER is neither strongly nuclear nor cytosolic in flies expressing DBTK/R or DBTK/R stNLS. Heterogeneity in PER subcellular localization was observed in our initial analysis of the dbtK/R mutant (Muskus et al, 2007). As well, previous analysis of DBT-dependent regulation of PER nuclear localization has produced rather conflicting results, with some studies suggesting a role in the inhibition of nuclear localization and others a role in induction of nuclear localization (Bao et al, 2001; Cyran et al, 2005; Kim et al, 2007; Muskus et al, 2007; Nawathean & Rosbash, 2004; Nawathean et al, 2007). It is possible that some phosphorylation sites induce nuclear localization and others inhibit it, and that a decreases in general phosphorylation state for PER leads to a more dispersed subcellular localization.
Because our dbtK/R NLS− mutant does not exhibit dominant negative properties, it is possible that other short-period dbt mutants could be produced that specifically inhibit cytosolic phosphorylation and PER degradation events in a dominant negative manner. On the other hand, our dbtWT NLS− mutant exhibits shorter (rather than longer) circadian periods with potentially elevated levels of cytosolic DBT. It is more likely that the existing dbtS mutant specifically affects a subset of sites to shorten period, or alters something besides DBT kinase activity or PER degradation (e.g., its interaction with targets or regulators) to shorten period (Fan et al, 2009; Gallego et al, 2006; Muskus et al, 2007; Vanselow et al, 2006; Xu et al, 2007). In support of this point are the studies by Syed and collaborators showing that DBTs proteins did not alter PER stability, and who therefore proposed that the dbtS mutation shortens circadian period in a manner that does not involve a direct alteration in capacity to target PER for degradation (Syed et al, 2011). It will be important to identify regulatory mechanisms that modulate DBT activity in a site-specific, time-of-day-specific or subcellular compartment-specific manner to understand the post-translational regulation that underlies the circadian clock mechanism.
Supplementary Material
Acknowledgments
We acknowledge the Bloomington stock center for stocks, the Model Systems Genomics Center at Duke University for the generation of transgenic flies, and the Developmental Studies Hybridoma Bank for several of the antibodies used herein. This work was supported by R01GM090277 (to J.L.P) from the NIH.
Footnotes
CONFLICT OF INTEREST STATEMENT
The authors have no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- Abruzzi KC, Rodriguez J, Menet JS, Desrochers J, Zadina A, Luo W, Tkachev S, Rosbash M. Drosophila CLOCK target gene characterization: implications for circadian tissue-specific gene expression. Genes Devel. 2011;25:2374–2386. doi: 10.1101/gad.178079.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allada R, Emery P, Takahashi JS, Rosbash M. Stopping time: the genetics of fly and mouse circadian clocks. Ann Rev Neurosci. 2001;24:1091–1119. doi: 10.1146/annurev.neuro.24.1.1091. [DOI] [PubMed] [Google Scholar]
- Allada R, White NE, So WV, Hall JC, Rosbash M. A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell. 1998;93:791–804. doi: 10.1016/s0092-8674(00)81440-3. [DOI] [PubMed] [Google Scholar]
- Bao S, Rihel J, Bjes E, Fan JY, Price JL. The Drosophila double-timeS mutation delays the nuclear accumulation of period protein and affects the feedback regulation of period mRNA. Journ Neurosci. 2001;21:7117–7126. doi: 10.1523/JNEUROSCI.21-18-07117.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci U S A. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceriani MF, Darlington TK, Staknis D, Mas P, Petti AA, Weitz CJ, Kay SA. Light-dependent sequestration of TIMELESS by CRYPTOCHROME. Science. 1999;285:553–556. doi: 10.1126/science.285.5427.553. [DOI] [PubMed] [Google Scholar]
- Chen Z, Yoo SH, Park YS, Kim KH, Wei S, Buhr E, Ye ZY, Pan HL, Takahashi JS. Identification of diverse modulators of central and peripheral circadian clocks by high-throughput chemical screening. Proc Natl Acad Sci U S A. 2012;109:101–106. doi: 10.1073/pnas.1118034108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyran SA, Yiannoulos G, Buchsbaum AM, Saez L, Young MW, Blau J. The double-time protein kinase regulates the subcellular localization of the Drosophila clock protein period. Journ Neurosci. 2005;25:5430–5437. doi: 10.1523/JNEUROSCI.0263-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies TH, Ning YM, Sanchez ER. A new first step in activation of steroid receptors: hormone-induced switching of FKBP51 and FKBP52 immunophilins. Journ Biol Chem. 2002;277:4597–4600. doi: 10.1074/jbc.C100531200. [DOI] [PubMed] [Google Scholar]
- Edery I, Zwiebel LJ, Dembinska ME, Rosbash M. Temporal phosphorylation of the Drosophila period protein. Proc Natl Acad Sci U S A. 1994;91:2260–2264. doi: 10.1073/pnas.91.6.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide EJ, Woolf MF, Kang H, Woolf P, Hurst W, Camacho F, Vielhaber EL, Giovanni A, Virshup DM. Control of mammalian circadian rhythm by CKIepsilon-regulated proteasome-mediated PER2 degradation. Mol Cell Biol. 2005;25:2795–2807. doi: 10.1128/MCB.25.7.2795-2807.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery P, So WV, Kaneko M, Hall JC, Rosbash M. CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell. 1998;95:669–679. doi: 10.1016/s0092-8674(00)81637-2. [DOI] [PubMed] [Google Scholar]
- Etchegaray JP, Machida KK, Noton E, Constance CM, Dallmann R, Di Napoli MN, DeBruyne JP, Lambert CM, Yu EA, Reppert SM, Weaver DR. Casein kinase 1 delta regulates the pace of the mammalian circadian clock. Mol Cell Biol. 2009;29:3853–3866. doi: 10.1128/MCB.00338-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan JY, Agyekum B, Venkatesan A, Hall DR, Keightley A, Bjes ES, Bouyain S, Price JL. Noncanonical FK506-Binding Protein BDBT Binds DBT to Enhance Its Circadian Function and Forms Foci at Night. Neuron. 2013;80:984–996. doi: 10.1016/j.neuron.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan JY, Preuss F, Muskus MJ, Bjes ES, Price JL. Drosophila and vertebrate casein kinase Idelta exhibits evolutionary conservation of circadian function. Genetics. 2009;181:139–152. doi: 10.1534/genetics.108.094805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego M, Eide EJ, Woolf MF, Virshup DM, Forger DB. An opposite role for tau in circadian rhythms revealed by mathematical modeling. Proc Natl Acad Sci U S A. 2006;103:10618–10623. doi: 10.1073/pnas.0604511103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin PE. Molecular genetic analysis of circadian timekeeping in Drosophila. Advanc Genetics. 2011;74:141–173. doi: 10.1016/B978-0-12-387690-4.00005-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich-Forster C. Development of pigment-dispersing hormone-immunoreactive neurons in the nervous system of Drosophila melanogaster. Journ Comp Neurol. 1997;380:335–354. doi: 10.1002/(sici)1096-9861(19970414)380:3<335::aid-cne4>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Hirota T, Lee JW, Lewis WG, Zhang EE, Breton G, Liu X, Garcia M, Peters EC, Etchegaray JP, Traver D, Schultz PG, Kay SA. High-throughput chemical screen identifies a novel potent modulator of cellular circadian rhythms and reveals CKIalpha as a clock regulatory kinase. PLoS Biol. 2010;8:e1000559. doi: 10.1371/journal.pbio.1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isojima Y, Nakajima M, Ukai H, Fujishima H, Yamada RG, Masumoto KH, Kiuchi R, Ishida M, Ukai-Tadenuma M, Minami Y, Kito R, Nakao K, Kishimoto W, Yoo SH, Shimomura K, Takao T, Takano A, Kojima T, Nagai K, Sakaki Y, Takahashi JS, Ueda HR. CKIepsilon/delta-dependent phosphorylation is a temperature-insensitive, period-determining process in the mammalian circadian clock. Proc Natl Acad Sci U S A. 2009;106:15744–15749. doi: 10.1073/pnas.0908733106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang AR, Moravcevic K, Saez L, Young MW, Sehgal A. Drosophila TIM binds importin alpha1, and acts as an adapter to transport PER to the nucleus. PLoS Genetic. 2015;11:e1004974. doi: 10.1371/journal.pgen.1004974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EY, Ko HW, Yu W, Hardin PE, Edery I. A DOUBLETIME kinase binding domain on the Drosophila PERIOD protein is essential for its hyperphosphorylation, transcriptional repression, and circadian clock function. Mol Cell Biol. 2007;27:5014–5028. doi: 10.1128/MCB.02339-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloss B, Price JL, Saez L, Blau J, Rothenfluh A, Wesley CS, Young MW. The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Ie. Cell. 1998;94:97–107. doi: 10.1016/s0092-8674(00)81225-8. [DOI] [PubMed] [Google Scholar]
- Kloss B, Rothenfluh A, Young MW, Saez L. Phosphorylation of period is influenced by cycling physical associations of double-time, period, and timeless in the Drosophila clock. Neuron. 2001;30:699–706. doi: 10.1016/s0896-6273(01)00320-8. [DOI] [PubMed] [Google Scholar]
- Ko HW, Jiang J, Edery I. Role for Slimb in the degradation of Drosophila Period protein phosphorylated by Doubletime. Nature. 2002;420:673–678. doi: 10.1038/nature01272. [DOI] [PubMed] [Google Scholar]
- Lee H, Chen R, Lee Y, Yoo S, Lee C. Essential roles of CKIdelta and CKIepsilon in the mammalian circadian clock. Proc Natl Acad Sci U S A. 2009;106:21359–21364. doi: 10.1073/pnas.0906651106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng QJ, Logunova L, Maywood ES, Gallego M, Lebiecki J, Brown TM, Sladek M, Semikhodskii AS, Glossop NR, Piggins HD, Chesham JE, Bechtold DA, Yoo SH, Takahashi JS, Virshup DM, Boot-Handford RP, Hastings MH, Loudon AS. Setting clock speed in mammals: the CK1 epsilon tau mutation in mice accelerates circadian pacemakers by selectively destabilizing PERIOD proteins. Neuron. 2008;58:78–88. doi: 10.1016/j.neuron.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng QJ, Maywood ES, Bechtold DA, Lu WQ, Li J, Gibbs JE, Dupre SM, Chesham JE, Rajamohan F, Knafels J, Sneed B, Zawadzke LE, Ohren JF, Walton KM, Wager TT, Hastings MH, Loudon AS. Entrainment of disrupted circadian behavior through inhibition of casein kinase 1 (CK1) enzymes. Proc Natl Acad Sci U S A. 2010;107:15240–15245. doi: 10.1073/pnas.1005101107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki K, Nagase T, Mesaki M, Narukawa J, Ohara O, Ishida N. Phosphorylation of clock protein PER1 regulates its circadian degradation in normal human fibroblasts. Biochem J. 2004;380:95–103. doi: 10.1042/BJ20031308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muskus MJ, Preuss F, Fan JY, Bjes ES, Price JL. Drosophila DBT lacking protein kinase activity produces long-period and arrhythmic circadian behavioral and molecular rhythms. Mol Cell Biol. 2007;27:8049–8064. doi: 10.1128/MCB.00680-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawathean P, Rosbash M. The doubletime and CKII kinases collaborate to potentiate Drosophila PER transcriptional repressor activity. Mol Cell. 2004;13:213–223. doi: 10.1016/s1097-2765(03)00503-3. [DOI] [PubMed] [Google Scholar]
- Nawathean P, Stoleru D, Rosbash M. A small conserved domain of Drosophila PERIOD is important for circadian phosphorylation, nuclear localization, and transcriptional repressor activity. Mol Cell Biol. 2007;27:5002–5013. doi: 10.1128/MCB.02338-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittendrigh CS. Circadian rhythms and the circadian organization of living systems. Cold Spring Harbor Symposia Quant Biol. 1960;25:159–182. doi: 10.1101/sqb.1960.025.01.015. [DOI] [PubMed] [Google Scholar]
- Preuss F, Fan JY, Kalive M, Bao S, Schuenemann E, Bjes ES, Price JL. Drosophila doubletime mutations which either shorten or lengthen the period of circadian rhythms decrease the protein kinase activity of casein kinase I. Mol Cell Biol. 2004;24:886–898. doi: 10.1128/MCB.24.2.886-898.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price JL. Drosophila melanogaster: a model system for molecular chronobiology. In: Sehgal A, editor. Molecular Biology of Circadian Rhythms. Hoboken, NJ: Wiley; 2004. pp. 33–74. [Google Scholar]
- Price JL, Blau J, Rothenfluh A, Abodeeley M, Kloss B, Young MW. Double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell. 1998;94:83–95. doi: 10.1016/s0092-8674(00)81224-6. [DOI] [PubMed] [Google Scholar]
- Price JL, Fan JY, Keightley A, Means JC. The role of casein kinase I in the Drosophila circadian clock. Method Enzymol. 2015;551:175–195. doi: 10.1016/bs.mie.2014.10.012. [DOI] [PubMed] [Google Scholar]
- Stanewsky R, Kaneko M, Emery P, Beretta B, Wagner-Smith K, Kay SA, Rosbash M, Hall JC. The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell. 1998;95:681–692. doi: 10.1016/s0092-8674(00)81638-4. [DOI] [PubMed] [Google Scholar]
- Suri V, Hall JC, Rosbash M. Two novel doubletime mutants alter circadian properties and eliminate the delay between RNA and protein in Drosophila. Journ Neurosci. 2000;20:7547–7555. doi: 10.1523/JNEUROSCI.20-20-07547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed S, Saez L, Young MW. Kinetics of doubletime kinase-dependent degradation of the Drosophila period protein. Journ Biol Chem. 2011;286:27654–27662. doi: 10.1074/jbc.M111.243618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanselow K, Vanselow JT, Westermark PO, Reischl S, Maier B, Korte T, Herrmann A, Herzel H, Schlosser A, Kramer A. Differential effects of PER2 phosphorylation: molecular basis for the human familial advanced sleep phase syndrome (FASPS) Gene Develop. 2006;20:2660–2672. doi: 10.1101/gad.397006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton KM, Fisher K, Rubitski D, Marconi M, Meng QJ, Sladek M, Adams J, Bass M, Chandrasekaran R, Butler T, Griffor M, Rajamohan F, Serpa M, Chen Y, Claffey M, Hastings M, Loudon A, Maywood E, Ohren J, Doran A, Wager TT. Selective inhibition of casein kinase 1 epsilon minimally alters circadian clock period. Journal Pharmacol Experimen Therapeut. 2009;330:430–439. doi: 10.1124/jpet.109.151415. [DOI] [PubMed] [Google Scholar]
- Weber F, Zorn D, Rademacher C, Hung HC. Post-translational timing mechanisms of the Drosophila circadian clock. FEBS letters. 2011;585:1443–1449. doi: 10.1016/j.febslet.2011.04.008. [DOI] [PubMed] [Google Scholar]
- Xu Y, Toh KL, Jones CR, Shin JY, Fu YH, Ptacek LJ. Modeling of a human circadian mutation yields insights into clock regulation by PER2. Cell. 2007;128:59–70. doi: 10.1016/j.cell.2006.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Zheng H, Price JL, Hardin PE. DOUBLETIME plays a noncatalytic role to mediate CLOCK phosphorylation and repress CLOCK-dependent transcription within the Drosophila circadian clock. Mol Cell Biol. 2009;29:1452–1458. doi: 10.1128/MCB.01777-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H, Hardin PE, Rosbash M. Constitutive overexpression of the Drosophila period protein inhibits period mRNA cycling. EMBO Journ. 1994;13:3590–3598. doi: 10.1002/j.1460-2075.1994.tb06666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerr DM, Hall JC, Rosbash M, Siwicki KK. Circadian fluctuations of period protein immunoreactivity in the CNS and the visual system of Drosophila. Journ Neurosci. 1990;10:2749–2762. doi: 10.1523/JNEUROSCI.10-08-02749.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.