Abstract
To investigate the kinetics of Cas9-mediated double strand break generation and repair in vivo, we developed two new tools. The first, chemically inducible Cas9 (ciCas9), is a rapidly-activated, single-component Cas9 variant engineered using a novel domain replacement strategy. ciCas9 can be activated in a matter of minutes, and the level of ciCas9 specificity and activity can be tuned. The second tool, DSB-ddPCR, is a droplet digital PCR-based assay for double strand breaks. DSB-ddPCR is the first assay to demonstrate time-resolved, highly quantitative and targeted measurement of DSBs. Combining these tools facilitated an unprecedented exploration of the kinetics of Cas9-mediated DNA cleavage and repair. We find that sgRNAs targeting different sites generally produce cleavage within minutes and repair within an hour or two. However, we observe distinct kinetic profiles, even for proximal sites, suggesting that target sequence and chromatin state modulate cleavage and repair kinetics.
Introduction
Insights into the processes of DNA cleavage and repair have enabled the manipulation of Cas9, sgRNA, or experimental conditions to improve editing efficiency, specificity, and repair1–5. Nevertheless, large gaps remain in our understanding of the Cas9 editing process. In particular, little is known about the kinetics of Cas9-mediated DSB generation and repair in vivo. In vitro, the Cas9/sgRNA complex remains bound to cleaved DNA for ~6 hours, which may delay surveillance and repair1,6. In cells, progress has been made in characterizing the target search dynamics of nuclease-null Cas97. How these findings relate to cleavage and repair kinetics of nuclease-competent Cas9 in vivo remains unclear.
Investigating Cas9 cleavage and repair kinetics in cells requires rapid and precise temporal control of Cas9 activity, so that cleavage can be activated at a defined point in time. With reported activation times ranging from two to many hours, it is unclear whether existing inducible Cas9 variants become active rapidly enough (Supplementary Table 1)8–16. Additionally, a precise and temporally-resolved method for quantifying DSBs is required, so that DNA cleavage can be measured in parallel with DNA editing. Existing methods either qualitatively or semi-quantitatively evaluate DSBs on a locus-specific or genome-wide basis, but none are suitable for tracking Cas9 cleavage kinetics (Supplementary Table 2)17–24.
To overcome these limitations, we developed two new tools. The first, chemically inducible Cas9 (ciCas9), is a rapidly-inducible, single-component Cas9 variant engineered using a novel domain replacement strategy. ciCas9’s level of basal activity is low, and it can be activated within minutes in a dose-dependent fashion. The architecture of ciCas9 also allows us to generate variants with increased specificity for on-target sites or even lower basal activity. The second tool, DSB-ddPCR, is a droplet digital PCR-based assay for double strand breaks. DSB-ddPCR is the first assay to demonstrate time-resolved, highly quantitative, and targeted measurement of DSBs. The combined application of these tools facilitated an unprecedented exploration of the kinetics of Cas9-mediated DSB generation and repair. We find that DSBs are generated rapidly, within ten minutes for some sgRNAs, and that indels generally appear within an hour or two. sgRNAs targeting different sites produce distinct DNA cleavage and repair kinetics, even when the sites are proximal. These findings suggest that target sequence and chromatin state modulate cleavage and repair kinetics.
Results
Engineering a rapidly inducible Cas9 variant
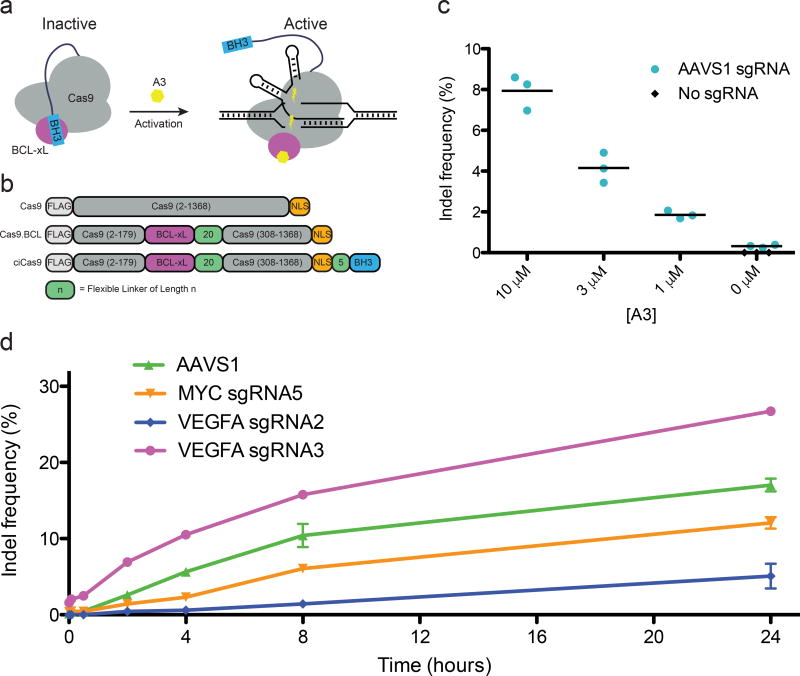
To generate a single-component, chemically inducible Cas9 (ciCas9) with rapid activation kinetics, we used the interaction between BCL-xL and a BH3 peptide as an intramolecular autoinhibitory switch25. Disruption of the BCL-xL/BH3 interaction by addition of a small molecule, A-385358 (“A3”), results in release of autoinhibition and activation of Cas9 (Fig. 1a). Modeling suggested the non-essential Cas9 REC2 domain could be replaced by BCL-xL, which is similar in size (Supplementary Fig. 1). Fusion of BH3 to either terminus would likely result in formation of a BCL-xL/BH3 complex that prevents binding to guide RNA or DNA. Thus, we replaced the REC2 domain with BCL-xL, creating Cas9.BCL, which retains activity (Fig. 1b, Supplementary Fig. 2)26. A BH3 peptide was appended to either terminus of Cas9.BCL via linkers of 5 to 30 residues to introduce autoinhibition (Supplementary Fig. 3, Supplementary Note 1). The activity of each construct was assessed at the neutral locus AAVS1 in the absence or presence of A3. High-throughput sequencing revealed that C-terminal BH3 fusions resulted in A3-activated editing (Supplementary Fig. 4). We selected the shortest, five-residue linker for further study, and subsequently refer to it as ciCas9.
Figure 1. Development of a chemically inducible Cas9 (ciCas9).
(a) A schematic depiction of the strategy to engineer a single-component, chemically inducible Streptococcus pyogenes Cas9 variant is shown. (b) The REC2 domain was replaced with BCL-xL and a BH3 peptide was appended to the C-terminus via flexible linkers of varying lengths. (c) Indel frequency at the AAVS1 locus 24 hours after activation of ciCas9 activity is shown for different concentrations of A3. Black bars depict means (n = 3 cell culture replicates). (d) Indel frequency at different times following activation of ciCas9 with A3 is shown for four sgRNAs at three different loci. Error bars depict s.e.m. (n = 3 cell culture replicates).
Expression of ciCas9 in the absence A3 resulted in minimal editing (0.32%, s.e.m. = 0.039%), compared to a no sgRNA control (0.0033%, s.e.m. = 0.0010%) (Fig. 1c). Addition of A3 generated a dose-dependent increase in editing, meaning activity can be tuned by varying drug concentration. The highest concentration of A3, 10 µM, yielded a 24.7-fold (s.e.m. = 3.34) increase in editing compared to the no drug control. This degree of activation is considerably higher than reported for other inducible Cas9 systems8,10. To demonstrate the generality of ciCas9, we assessed editing in HCT116 and U2OS cells. We observed A3-dependent editing, with minimal activity in the absence of drug (Supplementary Fig. 5).
ciCas9 activation rapidly produces indels
We next profiled ciCas9 indel kinetics with four sgRNAs at three distinct loci. (Fig. 1d). AAVS1 and VEGFA sgRNA3 showed significant increases in indels within 30 minutes of A3 addition, and a significant increase in indels was present at all sites by two hours (Supplementary Tables 3, 4). Thus, editing kinetics differ between sgRNA target sites at distinct loci. Kinetics also differ between sgRNA target sites at the same locus. The two VEGFA cleavage sites are only 1,091 nucleotides apart but have markedly different editing rates and total edits at 24 hours, which could be due to sequence and chromatin features that influence editing27–29. For example, VEGFA sgRNA3 has higher guanine content than VEGFA sgRNA2 (55% vs. 10%) and a higher predicted efficacy score based on the target and surrounding sequence (0.49 vs. 0.29, Azimuth 2.0 on-target scoring model)30.
We next compared ciCas9 kinetics to other inducible systems (Supplementary Table 1). Some of these systems are much slower. For example, indels were reported after nine hours following activation of photoactivatable split Cas9 (paCas9)9. After activation of ciCas9, indels were detectable by two hours using the same sgRNA and cell line (Supplementary Fig. 6a, b). To facilitate a comparison between ciCas9 and a recently described 4-hydroxytamoxifen-activated Cas9 (iCas9)8 that is reported to be faster than intein-Cas910 and split-Cas911, we measured ciCas9 editing kinetics using the same sgRNA. ciCas9 generated a detectable increase in indels within 30 minutes (one-sided t-test, n = 3, p = 0.016) of A3 addition. The first measured time point for iCas9 was two hours, but it is unclear if the observed increase in indels is significant because no statistical analyses were reported (Supplementary Fig. 6c, d).
DSB-ddPCR enables precise, site-specific quantitation of DSBs
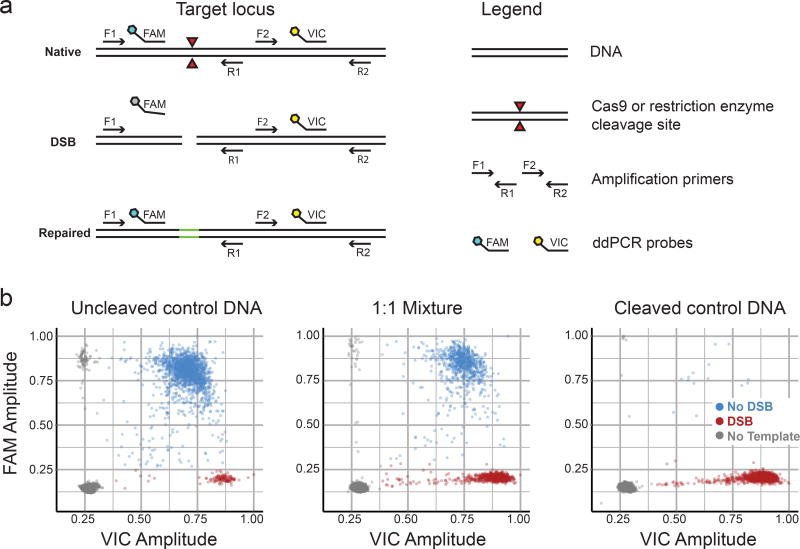
In vivo, Cas9-mediated cleavage of DNA target sites is generally measured indirectly, by quantifying indels. To directly and quantitatively measure DSBs at a target site, we developed a droplet digital PCR assay, DSB-ddPCR. DSB-ddPCR uses a primer pair that spans the sgRNA target site and another proximal pair that does not (Fig. 2a, https://www.nature.com/protocolexchange/protocols/6021/). Amplification is detected using primer pair-specific probes (FAM-labeled probe: uncleaved target present; VIC-labeled probe: template present). Droplets containing an uncleaved or repaired template yield products for both primer pairs. Droplets containing a template that has been cleaved by ciCas9 but not yet repaired are negative for the target amplicon.
Figure 2. DSB-ddPCR is an accurate and precise way to quantify DSBs.
(a) A schematic illustrates the DSB-ddPCR assay. The assay relies on two locus-specific PCR amplicons, the first of which spans the cleavage junction (primers F1, R1) and the second of which is adjacent (primers F2, R2). Amplicons are detected by the activation of amplicon-specific fluorescent hydrolysis probes. Template that has been cleaved by Cas9 or another nuclease does not generate the first amplicon or activate the first probe. Uncleaved control DNA was digested with a restriction enzyme to create cleaved control DNA. Uncleaved and cleaved control DNA was mixed in specific proportions. (b) Representative droplet FAM and VIC probe intensities are shown for uncleaved control MYC locus template DNA (left panel), a 1:1 mixture of uncleaved:cleaved control DNA (middle panel), and cleaved control DNA (right panel). Colors indicate droplets with no template (gray), intact template (blue), and cleaved template (red). Droplet populations in the dot plots are: uncleaved control DNA (DSB: 120, no DSB: 1,724, no template: 12,360), 1:1 Mixture (DSB: 799, no DSB: 914, no template: 12,305), cleaved control DNA (DSB: 1,757, no DSB: 14, no template: 13,247).
We evaluated the performance of DSB-ddPCR at three loci. We employed endonucleases that cleaved the template within the first primer pair, simulating a Cas9-mediated DSB. As expected, uncleaved control DNA yields probe signal for both amplicons, whereas fully digested control DNA yields signal only for the template amplicon (Fig. 2b). The relationship between observed and expected DSB frequency is linear (Pearson’s r2 > 0.99, Supplementary Fig. 7). DSB-ddPCR is also precise: technical replicates are essentially indistinguishable (Supplementary Fig. 7).
We next compared DSB-ddPCR to other methods for detecting DSBs (Supplementary Table 2). Immunodetection methods and the comet assay quantify overall DSB levels, but do not reveal where DSBs occur17,18. Sequencing-based methods like BLESS, DSB-capture, and GUIDE-Seq identify DSB locations genome-wide, but are complex, expensive, and semi-quantitative20,24,31. qPCR can reveal the prevalence of DSBs at a locus of interest relative to a distal control locus21. However, absolute quantitation of DSBs, as we show for DSB-ddPCR (Supplementary Fig. 7), has not been demonstrated. Furthermore, ddPCR offers several inherent advantages over qPCR, including absolute quantification without references, less sensitivity to PCR efficiency, greater precision and improved reproducibility32. Thus, DSB-ddPCR represents a new and more quantitative way to detect DSBs at specific loci.
Characterization of DNA cleavage and repair kinetics
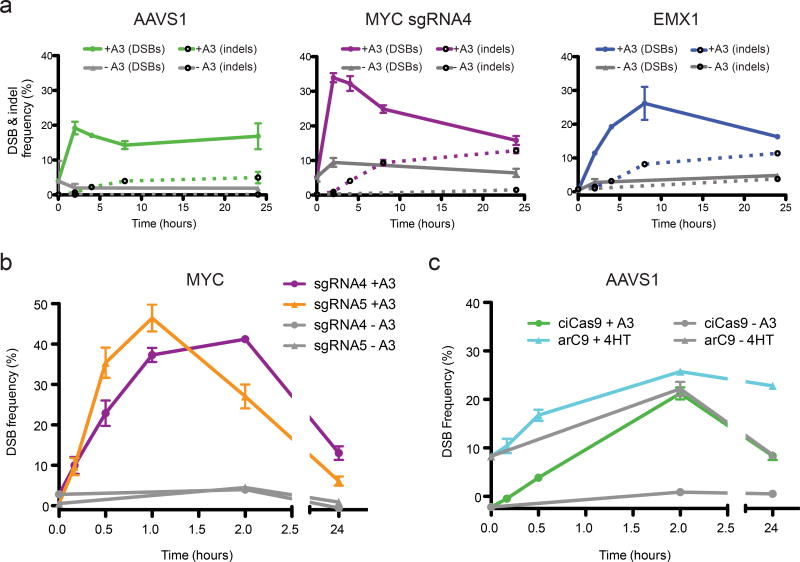
Cas9-mediated editing depends on DSB generation and subsequent repair, yet the temporal relationship between these two processes is unclear. To explore this relationship, we conducted time course experiments using ciCas9 to generate DSBs, DSB-ddPCR to quantify DSBs, and sequencing to quantify indels. To ensure that protein-DNA complexes, including ciCas9-bound DNA, did not interfere with DSB quantitation, we implemented an hour-long proteinase K digestion (Supplementary Fig. 8). We first examined DSB kinetics at the AAVS1, EMX1, and MYC loci (Fig. 3a, Supplementary Fig. 9). In this first-ever look at the kinetics of Cas9-mediated DSB generation in vivo, we observe some features that are common to all loci. For example, DSBs appear relatively rapidly, and reach a maximum well before 24 hours. In addition, at all three loci, a large number of unresolved DSBs are present. However, each locus has a distinct DSB kinetic profile. For example, at the AAVS1 locus, a maximum of 19.2% (s.e.m. = 1.8%) of templates have DSBs after two hours, whereas at the EMX1 locus, 26.2% (s.e.m. = 4.9%) of templates have DSBs after eight hours.
Figure 3. Investigation of DSB and indel kinetics with ciCas9 and DSB-ddPCR.
(a) Time courses of the frequency of DSBs and indels at three loci following activation of ciCas9 are shown. Solid lines indicate DSB frequency, dashed lines indicate indel frequency. DSBs were quantified using DSB-ddPCR and indels were quantified by high-throughput sequencing. Error bars depict s.e.m. (n = 3 cell culture replicates). (b) A DSB time course focused on the first two hours following ciCas9 activation is shown for two sgRNAs targeting the MYC locus. Error bars depict s.e.m. (n = 3 cell culture replicates). (c) A DSB time course comparing ciCas9 to a 4-hydroxytamoxifen-activated Cas9 variant, arC9, is shown. Error bars depict s.e.m. (n = 3 cell culture replicates).
We next examined the relationship between DSB and indel appearance at these three loci (Fig. 3a). Generally, indels are generated more slowly than DSBs, appearing in an approximately linear fashion over the first eight hours after ciCas9 activation. However, the rate of indel appearance is comparatively slow at the AAVS1 locus, which might be explained by the lower frequency of DSBs. The MYC and EMX1 loci share similar indel kinetics, but exhibit distinct DSB profiles. Most indels have appeared at both loci by eight hours, but, during this period, DSB frequency declines appreciably at the MYC locus whereas it increases at the EMX1 locus. The precise meaning of these different kinetic profiles remains unclear, but, at least for these three loci, Cas9 cleavage and DNA repair rates appear to differ.
To investigate events occurring shortly after activation of ciCas9, we performed high-resolution time courses using two adjacent sgRNAs targeting the MYC locus (Fig. 3b). DSBs are present for both sgRNAs after ten minutes (one-sided t-test, n = 3; MYC sgRNA4 p = 0.014; MYC sgRNA5 p = 0.007). Interestingly, although MYC sgRNA5 cleaves only ten nucleotides downstream of sgRNA4 on the same strand, we observe distinct kinetic profiles at each target site. Over the first 30 minutes, cleavage with MYC sgRNA5 is approximately twice as fast as with MYC sgRNA4 (sgRNA5 cleavage = 1.2 ± 0.1 percent per minute; sgRNA4 = 0.66 ± 0.09; Supplementary Fig. 10). DSB frequency peaks after one hour at the sgRNA5 site. DSB frequency peaks after two hours at the sgRNA4 site, by which time DSB frequency at the sgRNA5 site has already declined considerably. Despite the greater initial DSB generation rate, higher maximum, and earlier decline in DSB frequency, sgRNA5 yields fewer indels after 24 hours than does MYC sgRNA4 (Supplementary Fig. 11). One possible explanation for this difference between sgRNAs is that repair kinetics are distinct. Another possibility is that error-free repair is more prevalent for DSBs generated by sgRNA5. Differences in apparent kinetics and repair outcomes might inhere to differences in the guide sequences themselves, or the immediately adjacent sequence. Finally, loss of MYC impairs cell proliferation33, and both sgRNAs target the 5’ UTR of this gene. Thus, the differences may relate to proliferation, although any such effects are unlikely to appreciably impact early time points.
We next compared the DSB generation kinetics of ciCas9 to another inducible Cas9 variant, arC912. arC9 might also be rapidly activatable because, like ciCas9, it is a single-component system activated by a small molecule, 4-hydroxytamoxifen. We directly compared ciCas9 to arC9 at the AAVS1 locus (Fig. 3c). ciCas9, but not arC9, yielded a small but significant increase in DSBs after ten minutes (one-sided t-test, n = 3; ciCas9, p = 0.011; arC9, p = 0.13). Furthermore, arC9 suffered from higher basal activity and lower fold-activation.
Tuning ciCas9 specificity, basal activity and chemical activation
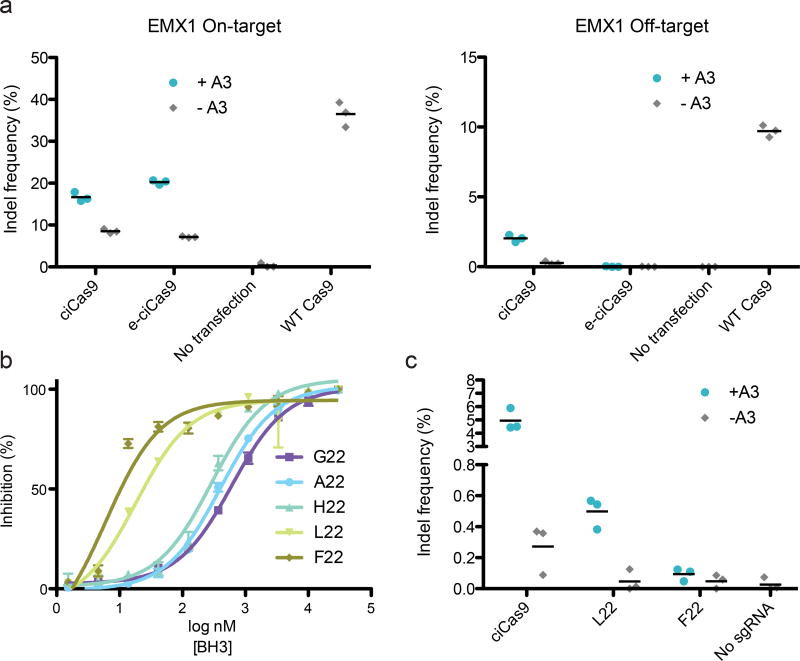
We explored the possibility of tuning ciCas9’s performance, focusing first on editing specificity. To determine the specificity of ciCas9 relative to wild type Cas9, we measured specificity ratios (i.e. on-target:off-target indel frequency ratios) for four sgRNAs at a total of five off-target sites. We found that ciCas9 had a higher specificity ratio than wild type Cas9 at four of five on-target:off-target pairs (Figure 4a, Supplementary Fig. 12). Despite the improved specificity of ciCas9 over wild type Cas9, off-target activity was still detectable at all off-target sites tested. To further improve on-target specificity, we introduced previously-identified, specificity-enhancing mutations3 to yield enhanced-specificity ciCas9 (e-ciCas9). e-ciCas9 had no detectable off-target activity for two of three on-target:off-target pairs, dramatically improving the specificity ratios (Fig. 4a, Supplementary Figure 13). For the third on-target:off-target pair, off-target editing was reduced but still detectable.
Figure 4. ciCas9 specificity and basal activity can be tuned.
(a) Editing is shown after 24 hours at the EMX1 on-target site (left panel) and an off-target site (right panel). ciCas9 and an enhanced-specificity variant (e-ciCas9) in the presence and absence of A3 are compared to wild type Cas9. Off-target editing with e-ciCas9 was not significantly increased relative to the no transfection control (one-sided t-test, n = 3, p = 0.21). Black bars depict means (n = 3 cell culture replicates). (b) Fluorescence polarization competitions between BH3 peptide variants and BODIPY-labeled BAK peptide for binding to BCL-xL. Data shown as inhibition of BODIPY-BAK binding. Error bars depict s.e.m. (n = 3 technical replicates). (c) Editing at the AAVS1 locus is shown for ciCas9 and two variants, L22 and F22, after 24 hours in the presence and absence of A3. Black bars depict means (n = 3 cell culture replicates).
To minimize the basal activity of ciCas9, we strengthened the BCL-xL/BH3 peptide interaction using a panel of BH3 variants with increased affinity for BCL-xL (Fig. 4b, Supplementary Fig. 14, Supplementary Table 5). The highest-affinity variants, ciCas9(L22) and ciCas9(F22), demonstrated significantly lower basal editing relative to ciCas9 (Fig. 4c; one-sided t-test; n = 3; L22, p = 0.044; F22, p = 0.039). However, the level of A3-activated editing for both the ciCas9(L22) and ciCas9(F22) was reduced relative to ciCas9. Thus, strengthening the BCL-xL/BH3 interaction reduced basal activity and activation by A3.
One concern regarding the use of small molecules that target BCL-xL, a member of the anti-apoptotic BCL-2 family, is the potential for toxicity through the release of pro-apoptotic factors. However, due to functional redundancy among BCL-2 family members, BCL-2/xL-selective disruptors are not generally cytotoxic34. For example, ABT-737 is a BCL-2/xL disruptor that is well-tolerated in mice for extended durations and is generally only toxic to cell lines that are primed for death35,36. We chose A3 for this study because it is even more selective for BCL-xL over BCL-2 and MCL-1 than ABT-737. BCL-xL-selective disruptors like A3 are well tolerated in mice, with reversible thrombocytopenia being the sole observed phenotypic effect37–40. Furthermore, we confirmed that A3 does not sensitize cells to the added stress of Cas9-mediated cleavage, and has no appreciable effect on the level of DSBs or indels generated by wild type Cas9 (Supplementary Fig. 15). Thus, ciCas9 and A3 could be used to investigate Cas9-mediated cleavage and repair in most cell lines or in mice. If the toxicity of BCL-2/xL disruptors has not been determined for a cell line of interest, users should perform cell proliferation and viability studies before proceeding.
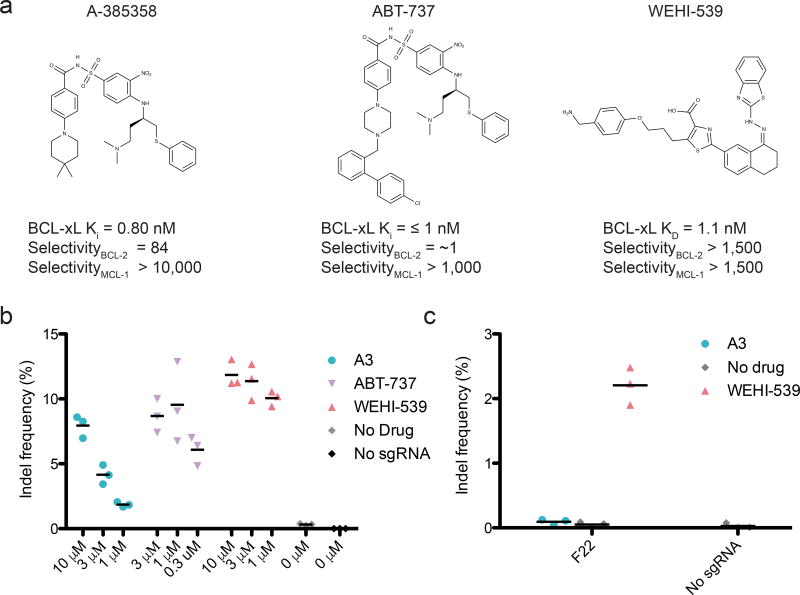
Because BCL-2/xL disruptors are being developed for clinical use, compounds with improved selectivity and drug-like properties continue to appear34. Thus, we investigated the effectiveness of different BCL-2 family disruptors to activate ciCas9. Two disruptors, ABT-73741 and WEHI-53942, resulted in robust editing at lower concentrations than A3, which is consistent with their increased cellular potency against BCL-xL (Fig. 5a, b). The enhanced cellular potency of WEHI-539 also facilitated editing with the most tightly autoinhibited ciCas9 variant, ciCas9(F22) (Fig. 5c). WEHI-539’s greater selectivity for BCL-xL and efficacy in activating ciCas9(F22) makes it an attractive candidate when low basal activity is required. While unfavorable pharmacokinetic properties preclude the use of WEHI-539 in animal models, derivatives like A-1155463 and A-1331852 have comparable BCL-xL affinity and specificity, and can be used in mice38,39. Furthermore, plasma concentrations of BCL-xL disruptors that are sufficient to potently activate ciCas9 can be achieved in mice39,43 and humans44 (Supplementary Table 6).
Figure 5. ciCas9 can be activated by a variety of BCL-xL disruptors.
(a) Structures of the three BCL-xL disruptors used in this study are shown with their reported Kd or Ki for BCL-xL, and their selectivity for BCL-xL over BCL-2 or MCL-137,40–42. (b) Editing at the AAVS1 locus 24 hours after ciCas9 activation with different concentrations of the three disruptors is shown. The A3 data are also shown in Fig. 1c. Black bars depict means (n = 3 cell culture replicates). (c) Editing is shown at the AAVS1 locus 24 hours after activation of the ciCas9(F22) variant with 10 µM of each disruptor. Black bars depict means (n = 3 cell culture replicates).
Discussion
We developed a rapidly chemically inducible Cas9 variant using a novel engineering strategy: intramolecular autoinhibition facilitated through REC2 domain replacement. We anticipate that REC2 domain replacement will complement other approaches like insertion scanning12, greatly expanding our ability to control Cas9 activity. ciCas9 demonstrated faster activation kinetics than other inducible Cas9s, enabling temporally resolved creation of DSBs. ciCas9 can also be rationally engineered to increase on-target specificity or reduce basal activity. These properties, along with other advantages like small size and high specificity, suggest that ciCas9 will be broadly useful.
We also developed DSB-ddPCR, an assay to precisely quantify DSBs at targeted sites with high temporal resolution. DSB-ddPCR is the first application of ddPCR to measure DSBs, and could be used to monitor other causes of cleavage. By combining ciCas9 and DSB-ddPCR, we were able to investigate the previously unexplored kinetic relationship between Cas9-mediated DNA cleavage and repair. We found that DSBs are generated in a matter of minutes, and indels appear in as little as half an hour. While further work is needed to precisely quantify DSB repair rates, our results suggest that repair occurs more rapidly than the long half-life of the Cas9-DSB complex measured in vitro might imply1,6. We also observe an excess of DSBs relative to indels, which might be due to error-free DSB repair45.
Our tools give a tantalizing first look at the kinetics of Cas9-mediated DSB generation and repair. However, many challenges remain. In addition to the action of ciCas9 and DNA repair machinery, other processes influence the frequency of DSBs and indels we observed. For example, cell proliferation during these experiments may be impacted by transfection or increasing density, which could in turn affect cleavage and repair kinetics. Disruption of some loci, such as MYC, might influence cell proliferation and thus DSB and indel frequency. These additional processes could be accounted for either experimentally or by modeling, enabling collection and interpretation of longer time courses. A ciCas9 variant that could be inactivated rapidly would enable pulsed DNA cleavage, which would facilitate precise estimation of DSB repair rates, and could reveal the prevalence of error-free repair. Furthermore, chemical or genetic perturbation of specific DNA-repair pathways could yield mechanistic insight into cleavage and repair kinetics. Ultimately, we envision using ciCas9 and DSB-ddPCR to dissect the influence of chromatin structure, target sequence, and cellular state on the kinetics of Cas9-mediated DSB generation and subsequent repair.
ONLINE METHODS
Modeling Cas9 REC2 domain replacement with BCL-xL
Volume estimates for the REC2 (residues 180–307) and BCL-xL (residues 4–198) domains were performed using the 3V volume calculator4,46. Computational modeling of REC2 replacement with BCL-xL was performed using RosettaRemodel47. Briefly, BCL-xL (PDB ID: 2BZW) was treated as a rigid-body inserted after Cas9 residues 179 and connected to 308 of Cas9 with a flexible linker (PDB ID: 4OO8). A blueprint file defined regions to rebuild in a modeling task. The functionality for fusing one structure into another (which is turned on with the -insert_segment_from_pdb flag) uses “0 × I” entries, which denote extensions of the peptide chain from the inserted PDB, in the range that corresponds to the placement of the BCl-xL domain. The flexible linker was composed of repeating glycine-(serine/threonine) residues, and backbone conformations were sampled by introducing random combinations of torsions from loop fragments. To facilitate the conformational search, a random break was made in the flexible linker to be reconnected via both random fragment moves and chain-closure algorithms guided by the Rosetta energy function; trajectories that properly reconnected the chain were considered successful. The lowest energy model from each successful trajectory was saved as a PDB file.
The flags to run the calculations are as follows:
-
-
database [rosetta database location]
-
-
s [template Cas9 PDB file] #4OO8, after deletion of REC2, nucleic acids
-
-
remodel:blueprint [blueprint files]
-
-
insert_segment_from_pdb [PDB formatted BCL-xL domain]
-
-
num_trajectory 10
-
-
save_top 1
-
-
remodel:quick_and_dirty
-
-
use_clusters false
-
-
vall debug1000vall
-
-
overwrite
300 independent trajectories were sampled in 30 parallel runs that used the flags above.
Cas9, ciCas9, arC9, and sgRNA expression plasmids
An N-terminal FLAG tag sequence was appended via Gibson Assembly Cloning (New England Biosciences) to a human codon optimized Cas9 (sub-cloned from hCas9, a gift from George Church, Harvard, Addgene plasmid #41815) with a single C-terminal NLS expressed from a pcDNA3.3-TOPO vector. BCL-xL (residues 4–198, Uniprot Q07817-1), BH3 peptide (APPNLWAAQRYGRELRRMADEGEGSFK), and linker sequences (Supplementary Fig. 3) were introduced using gBlocks (Integrated DNA Technologies) and Gibson Assembly Cloning. ciCas9 was subsequently sub-cloned into the pcDNA5/FRT/TO vector (ThermoFisher). Enhanced specificity-ciCas9, ciCas9(L22), and ciCas9(F22) were generated via restriction digest and NEBuilder HiFi Gibson Cloning (New England Biosciences). To create ciCas9(L22) and ciCas9(F22), we used higher-affinity BH3 sequences (Supplementary Table 5). The specificity enhancing mutations from eSpCas9(1.1) (gift from Feng Zhang, Broad Institute, Addgene plasmid # 71814) were subcloned into ciCas9 to create enhanced-specificity ciCas9 (e-ciCas9). sgRNA target sequences (Supplementary Data)—except for VEGFA sgRNAs—were cloned into the gRNA_Cloning Vector according to the hCRISPR gRNA Synthesis protocol (https://www.addgene.org/static/data/93/40/adf4a4fe-5e77-11e2-9c30-003048dd6500.pdf). gRNA_Cloning Vector was a gift from George Church, Harvard (Addgene plasmids # 41824). VEGFA site#2 (“VEGFA sgRNA2”) and VEGFA Site#3 (“VEGFA sgRNA3) were gifts from Keith Joung, Massachusetts General Hospital (Addgene plasmids # 47506 and # 47507). arC9 (containing NLS) expression plasmid was a gift from David Savage, UC Berkeley.
Cell culture
HEK-293T cells (293T/17, ATCC) were maintained in high glucose DMEM, 10% FBS, 4 mM L-glutamine (Life Technologies). U2OS (ATCC) and HCT116 (ATCC) cells were maintained in McCoy’s 5A (Modified) medium, 10% FBS (Life Technologies). Cells were tested and certified free of mycoplasma monthly.
Editing of genomic loci
Cells were plated in 12-well plates at the following densities: HEK-293T 3.25 × 105 cells/well; U2OS 7.5 × 104 cells/well; HCT116 3.00 × 105 cells/well. The following day, cells were transfected with Turbofectin 8.0 (Origene), except in Fig. 1d, Supplementary Fig. 2 and 6d, in which cells were transfected with X-tremeGENE HP. All cells were transfected with 3 µL transfection reagent and 1 µg plasmid DNA per well unless otherwise noted. The plasmid DNA mixture consisted of 450 ng sgRNA plasmid, 450 ng of Cas9 construct plasmid and 100 ng pMAX-GFP as a transfection control. 24 hours after transfection, ciCas9 was activated with the indicated concentrations of A-385358, WEHI-539 (ApexBio Technology), or ABT-737 (Selleck Chemicals). A-385358 was synthesized according to previously reported procedure1–5. The identity was confirmed by 1H-NMR and mass spectrometry (Bruker Esquire Ion Trap MS). >95% purity was confirmed by analytical HPLC. arC9 was activated with 1 µM 4-hydroxytamoxifen (Selleck Chemicals). Compounds were stored as 10 mM stocks in DMSO at −20°C. Final DMSO concentration in drug treated wells was 0.1%.
To harvest HEK-293T and HCT116 cells, wells were washed with 1 mL/well ice cold DPBS. Cells were subsequently harvested via trituration with 600 µL ice cold DPBS, spun down at 1500 × g for 10 min at 4°C, decanted, and cell pellets stored at −80°C. To enable temporal precision, plates for time course experiments were placed on ice prior to harvesting. To harvest U2OS cells, wells were washed with 1 mL pre-warmed DPBS, trypsinized (0.25% trypsin–EDTA, Life Technologies), and quenched with 10% FBS supplemented DMEM. Cells were spun down at 1500 × g for 10 min at 4°C, washed with ice cold DPBS, spun down at 1500 × g for 10 min at 4°C, decanted, and cell pellets stored at −80°C. For DSB-ddPCR experiments (Fig. 3, Supplementary Fig. 6c, 9–11) transfections were carried out as above except cells were transfected using 1.5µLTurbofectin 8.0 transfection reagent using 225 ng sgRNA, 225 ng ciCas9, and 50 ng pMAX-GFP plasmids per well in 12-well plates, which was found to result in low background signal in the absence of Cas9 activity (Supplementary Fig. 9). For cell viability experiments (Supplementary Fig. 15), cells were transfected using 1.5 µLTurbofectin 8.0 transfection reagent using 225 ng sgRNA, 225 ng ciCas9, and 50 ng pMAX-GFP plasmids per well in 24-well plates (plating density was scaled to 1.625 × 105 cells/well).
Indel quantification with high-throughput sequencing
Genomic DNA was isolated using the DNeasy Blood and Tissue kit (Qiagen) according to the manufacturer’s instructions with the following modification: proteinase K digestion was extended to 1h at 56 °C. 20 cycles of primary PCR to amplify the region of interest was performed using 2 µL of DNeasy eluate (~100–300 ng template) in a 5 µL Kapa HiFi HotStart polymerase reaction (Kapa Biosystems, for primers see Supplementary Data Set 1). The PCR reaction was diluted with 45 µL DNAse free water (Ambion). Illumina adapters and indexing sequences were added via 25 cycles of secondary PCR with 3 µL of diluted primary PCR product in a 10 µL Taq polymerase reaction (New England Biosciences, for primers see Supplementary Data Set 1). The final amplicons were run on a TBE-agarose gel (0.7%), and the product band was excised and extracted using the Freeze and Squeeze Kit according to the manufacturer’s instructions (Bio-Rad). Gel purified amplicons were quantified by qPCR using the Illumina Library Quantification Kit (Kapa Biosystems). Then, up to 300 indexed amplicons were pooled and sequenced on a MiSeq (MiSeq 150 V3 kit, Illumina, for primers see Supplementary Data Set 1).
After demultiplexing of reads (bcl2fastq, Illumina), indels were quantified with a custom Python script, which is freely available upon request. Briefly, 8-mer sequences were identified in the reference sequence located 20 base pairs up- and downstream of the target sequence. Sequence distal to these 8-mers was trimmed. Reads lacking these 8-mers were discarded. For the VEGFA sgRNA3 OT1 locus, the process was the same, except 20-mer sequences located 10 base pairs up- and downstream of the target sequence were used. The trimmed reads were then evaluated for indels using the Python difflib package. Indels were defined as trimmed reads which differed in length from the trimmed reference and for which an insertion or deletion operation spanning or within 1 bp of the predicted Cas9 cleavage site was present. Indel quantification for the initial Cas9.BCL screen (Supplementary Fig. 2) was performed using an analysis method similar to that previously described48. Briefly, reads were aligned to reference using Bowtie 249. SAMtools50 was used to generate pileup files, which were used to map and quantify indels.
DSB-ddPCR
A step-by-step protocol is available in Protocol Exchange (https://www.nature.com/protocolexchange/protocols/6021/).Two amplicons were designed for digital droplet PCR (ddPCR), one including the sgRNA binding/Cas9 cut site, the other a proximal control uncut site. For the MYC locus, a TaqMan® probe (Thermo Fisher Scientific) was designed for each amplicon using Primer 3 (http://bioinfo.ut.ee/primer3-0.4.0/). Dual-quenched probes were used for the EMX1 and AAVS1 loci (Integrated DNA Technologies). Droplets were created using Droplet generating oil for probes, DG8 cartridges, DG8 Gaskets and the QX200™ Droplet generator (Bio-Rad), amplification was performed using ddPCR Supermix for Probes (Bio-Rad). The ddPCR Supermix amplification reactions were set up according to the manufacturer’s specifications (Bio-Rad). 1µL of DNeasy eluate (~50–150 ng), obtained as described above, was added to a 20 µL amplification reaction with final primer and probe concentrations of 900 nM and 250 nM respectively. The AAVS1 locus required addition of SmaI and AvrII restriction enzymes (New England Biolabs) to the master mix for better separation of target and template amplicon probe signals (see Bio-Rad instructions). The reaction was divided into droplets for amplification following the manufacturer’s protocol (Bio-Rad). Droplets were transferred to a 96 well PCR plate and heat-sealed using PX1 PCR plate sealer (Bio-Rad). Droplets were amplified using the following cycling conditions: MYC 95 °C × 10 minutes, 40 cycles (94 °C × 30 seconds, 55 °C × 60 seconds), 98 °C × 10 minutes, all other amplicons 95 °C × 10 minutes, 40 cycles (94 °C × 30 seconds, 60 °C × 60 seconds), 98 °C × 10 minutes. Following thermal cycling, droplets were individually scanned using the QX200™ Droplet Digital™ PCR system (Bio-Rad). Positive and negative droplets in each fluorescent channel (VIC/FAM) were distinguished on the basis of fluorescence amplitude using a global threshold, set by the minimal, intrinsic fluorescence signal resulting from imperfect quenching of the fluorogenic probes (negative droplets) as compared to the strong fluorescence signal from cleaved probes in droplets with amplified template(s). Cleaved control DNA was created by digesting genomic DNA using the restriction enzyme Sfc-I for the MYC locus, DrdI for the EMX1 locus and BSU36I for the AAVS1 locus (New England Biolabs) according to the manufacturer’s instructions. The DSB frequency in the control and experimental samples was calculated as:
To generate standard curves, restriction enzyme digested control DNA was mixed with uncleaved control DNA in defined amounts, DSB frequency was quantified using the ddPCR protocol and a linear regression was performed. For the time course experiments, absolute DSB frequencies were calculated by fitting raw frequencies to standard curves.
Western blotting
Cells were harvested as above then lysed in a modified RIPA buffer (50 mM Tris-HCL, pH 7.8, 1% IGEPAL CA-630, 150 mM NaCl, 1mM EDTA, Pierce Protease Inhibitor Tablet). Lysates were cleared by centrifugation at 17,000 × g for 10 minutes at 4°C. Then, samples were incubated at 98 C for 7 minutes, subjected to SDS-PAGE (MOPS/SDS running buffer, Criterion and Mini-PROTEAN TGX gels, Bio-Rad) and transferred to nitrocellulose. Blocking and antibody incubation were done in TBS with 0.1% Tween-20 (v/v) and blocking buffer (Odyssey). Primary antibodies were diluted: FLAG (1:1,000, Cell Signaling Technology #8146; or 1:1,000 or Sigma M2 #F1804) and GAPDH (1:2,000, Cell Signaling Technology #2118). Blots were washed in TBS with 0.1% Tween-20. Antibody binding was detected by using near-infrared-dye-conjugated secondary antibodies and visualized on the LI-COR Odyssey scanner.
For evaluation of proteinase K digestion efficacy (Supplementary Fig. 8), cells from a 12-well plate were split into thirds. One third was processed as described above for DSB-ddPCR samples. One third was processed as described, but was not run on DNeasy column. One third of the cells underwent only the first step in processing—resuspension in PBS and Buffer AL (DNeasy kit)—but not treated with Proteinase K or run on DNeasy column. Protein was isolated from the three samples using methanol chloroform precipitation, resuspended in 30 µL LDS Sample buffer (ThermoFisher Scientific) supplemented with DTT and heated to 98°C for seven minutes. 10 µL of each sample was run on a Criterion gel (Bio-Rad) and blots were processed and visualized as described above.
Fluorescence anisotropy BH3 competition assay
Assays were performed as described previously51. The BH3 peptide variants (initial concentration = 10 µM, 3-fold serial dilutions, 10 data points) G22, V22, and A22 (GenScript) were assayed against 38 nM BCL-xL in the presence of 35 nM Bak-BODIPY. The BH3 peptide variants (initial concentration = 10 µM, 3-fold serial dilutions, 10 data points) L22 and F22 (GenScript) were assayed against 12 nM BCL-xL in the presence of 35 nM Bak-BODIPY. Competition assays were incubated at room temperature for 7 hours. The amount of polarization was measured in millipolarization (mP) units with an excitation of 485 nm and emission of 530 nm. IC50 values were determined using Graphpad Prism software (non-linear regression analysis). For each BH3 variant titration, three technical replicates were performed. The calculated IC50 values were used to determine KI values (classic model, http://botdb.abcc.ncifcrf.gov/toxin/kiConverter.jsp)52. KI values shown in Supplementary Table 5 are the average of assays performed in triplicate ± s.e.m. (n = 3 technical replicates).
Statistical Analysis
Statistical tests of indel and DSB frequency were performed in Excel using a one-sided two-sample Student’s t-test. Statistical tests of linear regression slopes were performed in Prism software.
Data Availability
Sequencing data are available at the NCBI Short Read Archive (Accession #XX). All other data that support the findings of this study are available from the corresponding authors upon request.
Supplementary Material
Acknowledgments
We thank M. Dickinson for assistance generating reagents. This work was supported by the NIH (R01GM086858 (D.J.M), R01GM109110 (D.M.F), F30CA189793 (J.C.R)), NSF (0954242 (D.J.M.)), and the Alzheimer’s Association (NIRG304150) (D.M.F).
Footnotes
Author Contributions
J.C.R., D.M.F, and D.J.M conceived the study and designed the experiments. B.M.T assisted in design of ciCas9. J.C.R. and J.J.S. performed the experiments. J.J.S. prepared samples for high-throughput sequencing and ddPCR. H.V.D. performed fluorescence BH3 competition experiments under the supervision of J.C.R. and D.J.M. W.J.V helped develop the DSB-ddPCR assay and analyze data under the supervision of J.H.B. J.C.R and D.M.F prepared custom Python scripts. J.C.R., D.M.F, and D.J.M wrote and edited the manuscript. All authors approved the final manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Richardson CD, Ray GJ, DeWitt MA, Curie GL, Corn JE. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat. Biotechnol. 2016;34:339–344. doi: 10.1038/nbt.3481. [DOI] [PubMed] [Google Scholar]
- 2.Lin S, Staahl BT, Alla RK, Doudna JA. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. Elife. 2014;3:e04766. doi: 10.7554/eLife.04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Slaymaker IM, et al. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kleinstiver BP, et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suzuki K, et al. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature. 2016;540:144–149. doi: 10.1038/nature20565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knight SC, et al. Dynamics of CRISPR-Cas9 genome interrogation in living cells. Science. 2015;350:823–826. doi: 10.1126/science.aac6572. [DOI] [PubMed] [Google Scholar]
- 8.Liu KI, et al. A chemical-inducible CRISPR-Cas9 system for rapid control of genome editing. Nat. Chem. Biol. 2016;12:980–987. doi: 10.1038/nchembio.2179. [DOI] [PubMed] [Google Scholar]
- 9.Nihongaki Y, Kawano F, Nakajima T, Sato M. Photoactivatable CRISPR-Cas9 for optogenetic genome editing. Nat. Biotechnol. 2015;33:755–760. doi: 10.1038/nbt.3245. [DOI] [PubMed] [Google Scholar]
- 10.Davis KM, Pattanayak V, Thompson DB, Zuris JA, Liu DR. Small molecule-triggered Cas9 protein with improved genome-editing specificity. Nat. Chem. Biol. 2015;11:316–318. doi: 10.1038/nchembio.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zetsche B, Volz SE, Zhang F. A split-Cas9 architecture for inducible genome editing and transcription modulation. Nat. Biotechnol. 2015;33:139–142. doi: 10.1038/nbt.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oakes BL, et al. Profiling of engineering hotspots identifies an allosteric CRISPR-Cas9 switch. Nat. Biotechnol. 2016;34:646–651. doi: 10.1038/nbt.3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nguyen DP, et al. Ligand-binding domains of nuclear receptors facilitate tight control of split CRISPR activity. Nat. Commun. 2016;7:12009. doi: 10.1038/ncomms12009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng J, et al. A general strategy to construct small molecule biosensors in eukaryotes. Elife. 2015;4:4004. doi: 10.7554/eLife.10606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Senturk S, et al. Rapid and tunable method to temporally control gene editing based on conditional Cas9 stabilization. Nat. Commun. 2017;8:14370. doi: 10.1038/ncomms14370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maji B, et al. Multidimensional chemical control of CRISPR-Cas9. Nat. Chem. Biol. 2017;13:9–11. doi: 10.1038/nchembio.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ivashkevich A, Redon CE, Nakamura AJ, Martin RF, Martin OA. Use of the γ-H2AX assay to monitor DNA damage and repair in translational cancer research. Cancer Lett. 2012;327:123–133. doi: 10.1016/j.canlet.2011.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olive PL, Banáth JP. The comet assay: a method to measure DNA damage in individual cells. Nat Protoc. 2006;1:23–29. doi: 10.1038/nprot.2006.5. [DOI] [PubMed] [Google Scholar]
- 19.Hanada K, et al. The structure-specific endonuclease Mus81 contributes to replication restart by generating double-strand DNA breaks. Nat Struct Mol Biol. 2007;14:1096–1104. doi: 10.1038/nsmb1313. [DOI] [PubMed] [Google Scholar]
- 20.Tsai SQ, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang Z, Sunder S, Nallasivam S, Wilson TE. Overhang polarity of chromosomal double-strand breaks impacts kinetics and fidelity of yeast non-homologous end joining. Nucleic Acids Res. 2016;44:2769–2781. doi: 10.1093/nar/gkw013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Furda AM, Bess AS, Meyer JN, Van Houten B. Analysis of DNA damage and repair in nuclear and mitochondrial DNA of animal cells using quantitative PCR. Methods Mol. Biol. 2012;920:111–132. doi: 10.1007/978-1-61779-998-3_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grégoire M-C, et al. Quantification and genome-wide mapping of DNA double-strand breaks. DNA Repair (Amst.) 2016;48:63–68. doi: 10.1016/j.dnarep.2016.10.005. [DOI] [PubMed] [Google Scholar]
- 24.Lensing SV, et al. DSBCapture: in situ capture and sequencing of DNA breaks. Nat. Meth. 2016;13:855–857. doi: 10.1038/nmeth.3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rose JC, et al. A computationally engineered RAS rheostat reveals RAS-ERK signaling dynamics. Nat. Chem. Biol. 2017;13:119–126. doi: 10.1038/nchembio.2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nishimasu H, et al. Crystal Structure of Cas9 in Complex with Guide RNA and Target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moreno-Mateos MA, et al. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat. Meth. 2015;12:982–988. doi: 10.1038/nmeth.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Isaac RS, et al. Nucleosome breathing and remodeling constrain CRISPR-Cas9 function. Elife. 2016;5:e13450. doi: 10.7554/eLife.13450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen X, et al. Probing the impact of chromatin conformation on genome editing tools. Nucleic Acids Res. 2016;44:6482–6492. doi: 10.1093/nar/gkw524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doench JG, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ran FA, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hindson CM, et al. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat. Meth. 2013;10:1003–1005. doi: 10.1038/nmeth.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hart T, et al. High-Resolution CRISPR Screens Reveal Fitness Genes and Genotype-Specific Cancer Liabilities. Cell. 2015;163:1515–1526. doi: 10.1016/j.cell.2015.11.015. [DOI] [PubMed] [Google Scholar]
- 34.Ashkenazi A, Fairbrother WJ, Leverson JD, Souers AJ. From basic apoptosis discoveries to advanced selective BCL-2 family inhibitors. Nat. Rev. Drug Discov. 2017;16:273–284. doi: 10.1038/nrd.2016.253. [DOI] [PubMed] [Google Scholar]
- 35.Konopleva M, et al. Mechanisms of apoptosis sensitivity and resistance to the BH3 mimetic ABT-737 in acute myeloid leukemia. Cancer Cell. 2006;10:375–388. doi: 10.1016/j.ccr.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 36.Wei G, et al. Chemical Genomics Identifies Small-Molecule MCL1 Repressors and BCL-xL as a Predictor of MCL1 Dependency. Cancer Cell. 2012;21:547–562. doi: 10.1016/j.ccr.2012.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shoemaker AR, et al. A small-molecule inhibitor of Bcl-XL potentiates the activity of cytotoxic drugs in vitro and in vivo. Cancer Res. 2006;66:8731–8739. doi: 10.1158/0008-5472.CAN-06-0367. [DOI] [PubMed] [Google Scholar]
- 38.Leverson JD, et al. Exploiting selective BCL-2 family inhibitors to dissect cell survival dependencies and define improved strategies for cancer therapy. Sci. Transl. Med. 2015;7:279ra40. doi: 10.1126/scitranslmed.aaa4642. [DOI] [PubMed] [Google Scholar]
- 39.Tao Z-F, et al. Discovery of a Potent and Selective BCL-XL Inhibitor with in Vivo Activity. ACS Med. Chem. Lett. 2014;5:1088–1093. doi: 10.1021/ml5001867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wendt MD, et al. Discovery and structure-activity relationship of antagonists of B-cell lymphoma 2 family proteins with chemopotentiation activity in vitro and in vivo. J. Med. Chem. 2006;49:1165–1181. doi: 10.1021/jm050754u. [DOI] [PubMed] [Google Scholar]
- 41.Oltersdorf T, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 42.Lessene G, et al. Structure-guided design of a selective BCL-XL inhibitor. Nat. Chem. Biol. 2013;9:390–397. doi: 10.1038/nchembio.1246. [DOI] [PubMed] [Google Scholar]
- 43.Tse C, et al. ABT-263: A Potent and Orally Bioavailable Bcl-2 Family Inhibitor. Cancer Res. 2008;68:3421–3428. doi: 10.1158/0008-5472.CAN-07-5836. [DOI] [PubMed] [Google Scholar]
- 44.Wilson WH, et al. Navitoclax, a targeted high-affinity inhibitor of BCL-2, in lymphoid malignancies: a phase 1 dose-escalation study of safety, pharmacokinetics, pharmacodynamics, and antitumour activity. Lancet Oncol. 2010;11:1149–1159. doi: 10.1016/S1470-2045(10)70261-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Geisinger JM, Turan S, Hernandez S, Spector LP, Calos MP. In vivo blunt-end cloning through CRISPR/Cas9-facilitated non-homologous end-joining. Nucleic Acids Res. 2016;44:e76–e76. doi: 10.1093/nar/gkv1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Voss NR, Gerstein M. 3V: cavity, channel and cleft volume calculator and extractor. Nucleic Acids Res. 2010;38:W555–W562. doi: 10.1093/nar/gkq395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang P-S, et al. Rosetta Remodel: a generalized framework for flexible backbone protein design. PLoS ONE. 2011;6:e24109. doi: 10.1371/journal.pone.0024109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kiani S, et al. Cas9 gRNA engineering for genome editing, activation and repression. Nat. Meth. 2015;12:1051–1054. doi: 10.1038/nmeth.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Meth. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li H, et al. The Sequence Alignment/Map format and SAMtools. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goreshnik I, Maly DJ. A Small Molecule-Regulated Guanine Nucleotide Exchange Factor. J. Am. Chem. Soc. 2010;132:938–940. doi: 10.1021/ja907886v. [DOI] [PubMed] [Google Scholar]
- 52.Cer RZ, Mudunuri U, Stephens R, Lebeda FJ. IC50-to-Ki: a web-based tool for converting IC50 to Ki values for inhibitors of enzyme activity and ligand binding. Nucleic Acids Res. 2009;37:W441–5. doi: 10.1093/nar/gkp253. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data are available at the NCBI Short Read Archive (Accession #XX). All other data that support the findings of this study are available from the corresponding authors upon request.