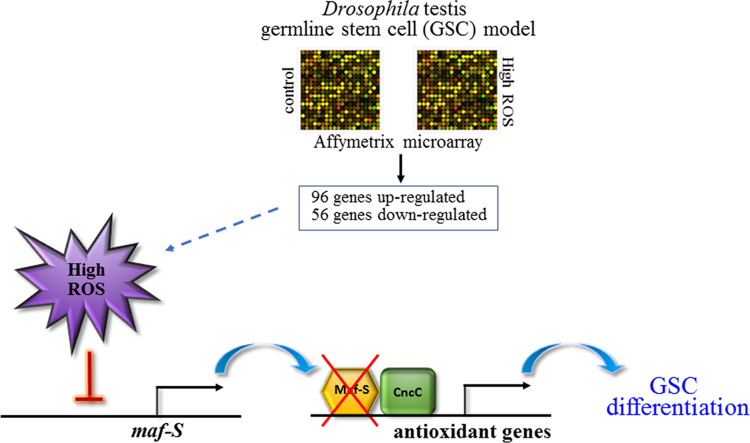
Abstract
Reactive oxygen species (ROS) are byproducts generated during normal cellular metabolism, and redox states have been shown to influence stem cell self-renewal and lineage commitment across phyla. However, the downstream effectors of ROS signaling that control stem cell behavior remain largely unexplored. Here, we used the Drosophila testis as an in vivo model to identify ROS-induced effectors that are involved in the differentiation process of germline stem cells (GSCs). In the Affymetrix microarray analysis, 152 genes were either upregulated or downregulated during GSC differentiation induced by elevated levels of ROS, and a follow-up validation of the gene expression by qRT-PCR showed a Spearman's rho of 0.9173 (P<0.0001). Notably, 47 (31%) of the identified genes had no predicted molecular function or recognizable protein domain. These suggest the robustness of this microarray analysis, which identified many uncharacterized genes, possibly with an essential role in ROS-induced GSC differentiation. We also showed that maf-S is transcriptionally downregulated by oxidative stress, and that maf-S knockdown promotes GSC differentiation but Maf-S overexpression conversely results in an over-growth of GSC-like cells by promoting the mitotic activity of germ cell lineage. Together with the facts that Maf-S regulates ROS levels and genetically interacts with Keap1/Nrf2 in GSC maintenance, our study suggests that Maf-S plays an important role in the Drosophila testis GSC maintenance by participating in the regulation of redox homeostasis.
Keywords: Reactive oxygen species, Drosophila, Germline stem cell, Small Maf, Keap1/Nrf2, Microarray
Graphical abstract
Highlights
-
•
High ROS promote germline stem cell (GSC) differentiation in Drosophila testis.
-
•
152 genes are transcriptionally regulated during GSC differentiation.
-
•
maf-S is transcriptionally downregulated by oxidative stress.
-
•
Maf-S genetically interacts with Keap1/Nrf2 and is required for GSC maintenance.
1. Introduction
Reactive oxygen species (ROS), including superoxide anion (O2·-), hydrogen peroxide (H2O2) and hydroxyl free radical (HO·), are the by-products of cellular metabolism and homeostasis. The reactive oxygen anion O2·- is converted throughout a series of enzymatic reactions to H2O2, which is then further catalyzed to release highly reactive HO·. On the other hand, the scavenger antioxidative molecules such as superoxide dismutase, catalase, glutathione peroxidase and peroxiredoxin convert O2·- to H2O2, and subsequently convert H2O2 to H2O and dioxide (O2) [1]. ROS were initially considered as a destructive byproduct of metabolism that are highly associated with a wide variety of human diseases such as cancer and neurodegenerative disorders [2], [3], [4]. However, increasing evidence has shown that ROS can be generated on purpose to benefit various physiological and biological processes such as cell growth, survival, signal transduction and protein-folding [5], [6], [7].
Stem cells are essential for the development of an organism, as well as to replenish damaged tissue lost throughout life. The characteristics of stem cells include the ability to self-renew and differentiate into specialised cell types. Importantly, stem cell homeostasis must be tightly regulated to prevent unfavourable balance between stem cells and differentiated cells, as dysregulated stem cell behavior is closely associated with various human diseases such as cancers, neurological diseases and irritable bowel syndrome (IBS) [8], [9], [10]. Intriguingly, intracellular ROS levels have been reported to play important roles in balancing self-renewal and differentiation of various stem cell populations across phyla. Normal stem cells are known to reside in the stem cell niches characterized by a low ROS environment so that stem cells remain in a quiescent state, a property that is essential for their self-renewal capacity [11]. Consistently, it was shown that low levels of ROS in stem cell niches are of importance to maintain the stem identity of hematopoietic stem cells [12], which were reported to lose their stem identity when ROS levels become excessive [13]. Embryonic stem cells (ESCs) are also known to maintain their stemness and pluripotency under low levels of ROS, but they undergo apoptosis or senescence when exposed to prolonged ROS [14], [15]. On the other hand, elevated ROS levels were detected during the differentiation process of human adipose tissue-derived multipotent adult stem cells into a neural phenotype [16]. However, despite the essential role of redox states in stem cell homeostasis, ROS-induced downstream effectors that regulate stem cell behavior are not fully characterized.
The Drosophila testis germline stem cell (GSC) system is one of the best understood adult stem cell models for studying and understanding the fundamental cellular mechanisms of stem cell behavior, as stem cells and their progenies can be easily identified, traced, imaged and genetically manipulated in vivo [17]. Importantly, we previously showed that high levels of ROS facilitate GSC differentiation through the activation of EGFR signaling, whereas decreased ROS levels conversely promote the proliferation of GSC-like cells in the Drosophila testes [18]. In this study, we performed Affymetrix microarray analysis using the Drosophila testes to identify the downstream effectors of ROS-mediated GSC differentiation. 152 genes were found to be differentially expressed during GSC differentiation. Several genes such as maf-S (small maf) and lox2 (lipoxygenases 2), whose products are implicated in redox signaling, were identified, suggesting the validity of this genome-wide approach. Notably, many of the identified genes have not yet been characterized. Lastly, our genetic analyses revealed that maf-S knockdown promotes GSC differentiation, but Maf-S overexpression conversely facilitates the proliferation of GSC-like early-stage germ cells. Since Maf-S genetically interacted with Keap1/Nrf2 in GSC homeostasis, our study suggests that Maf-S functions in the regulation of ROS-associated stem cell behavior in the Drosophila testis.
2. Material and methods
2.1. Fly strains and fly husbandry
UAS-Maf-S (II), UAS-Keap1RNAi, UAS-CncC, UAS-CncCRNAi, keap1036, keap1EY5 and GstD1-GFP fly lines were obtained from D. Bohmann [19]. UAS-ND75RNAi and UAS-ND42RNAi were obtained from the NIG-FLY Stock Center. Nanos (nos)-Gal4, esgM5-4-lacZ was obtained from S. Dinardo [20], and bam-GFP transgenic line was obtained from D.M. Mckearin [21]. UAS-maf-SRNAi (BL#40853) was obtained from the Bloomington Drosophila Stock Center. For UAS/Gal4 experiments, eclosed F1 adult male flies were incubated at 30 °C to maximize the Gal4 activity. All fly stocks were maintained on a standard diet at room temperature.
2.2. Immunohistochemistry
The Drosophila testes were dissected on a glass slide with dissection buffer at pH 7.2 (130 mM NaCl; 1.9 mM CaCl2; 4.7 mM KCl; 10 mM HEPES) and fixed with 4% paraformaldehyde for 20 min, followed by washing with PBST (1XPBS with 0.3% Triton-X) for three times, 20 min each. The testes were then incubated with primary antibodies at 4 °C for overnight. Testes were washed and subsequently incubated with secondary antibodies at room temperature for 2 h. Images were taken using the Olympus FluoView ™ FV1000 Confocal Laser Microscope. ImajeJ was used to measure the distance of cells. Primary antibodies used were: rat anti-Vasa (Developmental Studies Hybridoma Bank [DSHB], 1:100), mouse anti-Fasciclin III (DSHB, 1:100), rabbit anti-GFP Alexa Fluor® 488 conjugate (Molecular Probes®, 1:500), mouse anti-1B1 (DSHB, 1:150), mouse anti-β-gal (β-galactosidase) (Sigma Aldrich [SA] #G4644, 1:200) and rabbit anti-pH3 (Cell Signaling Technology #9701, 1:200). Secondary antibodies used were: Alexa Fluor® 488-AffiniPure Donkey Anti-Mouse IgG (Jackson ImmunoResearch Laboratories Inc. [JIR] #715-545-150, 1:300), Alexa Fluor® 594-AffiniPure Goat Anti-Rat IgG (JIR #112-585-003, 1:300) and Goat anti-rabbit Alexa Fluor® 488 (ThermoFisher Scientific [TFS] #R37116, 1:200).
2.3. DHE assay
Testes were dissected into 1 ml of Schneider media with 10% FBS. 1ul of reconstituted DHE dye (TFS) was added and allowed to rock for 5 min in the dark. Testes were then washed three times with Schneider media for 5 min each, followed by fixation with 4% paraformaldehyde for 10 min. Testes were mounted and immediately viewed under the Olympus FluoView™ FV1000 Confocal Laser Scanning Biological Microscope. The intensity of DHE staining was quantified using ImageJ.
2.4. Sample preparation for microarray
Testes expressing ND75RNAi under the control of nanos-Gal4 driver were used as an experimental group (high ROS levels), whereas testes carrying the driver alone served as a control group (physiological/moderate ROS levels). Three biological replicates for each group were prepared. For each replicate, approximately 200 adult male flies were dissected in ice cold Schneider media, and the resulting testes were washed with ice cold 1XPBS for three times. 800μl of a solution containing 0.25% collagenase (TFS) and 0.5% trypsin (TFS) prepared in 1XPBS were used to dissociate the testes by rocking for 15 min. The solution was then filtered through a 40 µm mesh, and the reaction of cell dissociation was stopped by the addition of 500μl Schneider media. Cell pellet was then obtained by centrifugation at 600g for 10 min at 4 °C, followed by RNA extraction by RNeasy microarray tissue Kit (Qiagen). The quality of the RNA was determined using an Agilent Bioanalyzer.
2.5. Microarray
100 ng of RNA were converted into double-stranded cDNA, which was then amplified to cRNA by in vitro transcription. The cRNA was purified and subjected to 2nd-cycle single-stranded sense cDNA synthesis, followed by fragmentation and terminal labelling (Affymetrix GeneChip WT PLUS Reagent Kit) before hybridization to Affymetrix Drosophila Gene 1.0 ST array. The arrays were washed and stained using GeneChip Hybridization, Wash and Stain Kit (Affymetrix), and subsequently scanned by Affymetrix 3000 7 G scanner. The differentially expressed genes between control and experimental group were determined using the Transcriptome Analysis Console 3.0 software (Affymetrix), with the criteria of at least a 1.5- or −1.5-fold change, and FDR p-value <0.05.
2.6. Quantitative RT-PCR
RNA was extracted from testes with different genotypes, using TRIzol Reagent (TFS). 1 μg of RNA was converted to cDNA using the RevertAid First Strand cDNA Synthesis Kit (TFS). RT-qPCR analysis was then performed using FAST SYBR Green Master Mix (TFS) and the AB7900HT Fast Real Time PCR system (Applied Biosystems). The list of primer sets used is described in the Supplementary Table 1.
2.7. Statistical analysis
Data was presented as mean ± standard error of the mean (SEM). Statistical analysis was performed using the GraphPad Prism 6.0 software (La Jolla, CA, USA). The Student's t-test was used to compare between two groups. For comparisons among three or more groups, one-way ANOVA was carried out followed by post-hoc Bonferroni test. Association between two variables was evaluated using Spearman rank correlation. P-values below 0.05 were deemed to be statistically significant.
3. Results and discussion
3.1. Inhibition of ND75 promotes GSC differentiation by increasing ROS levels in the Drosophila testis
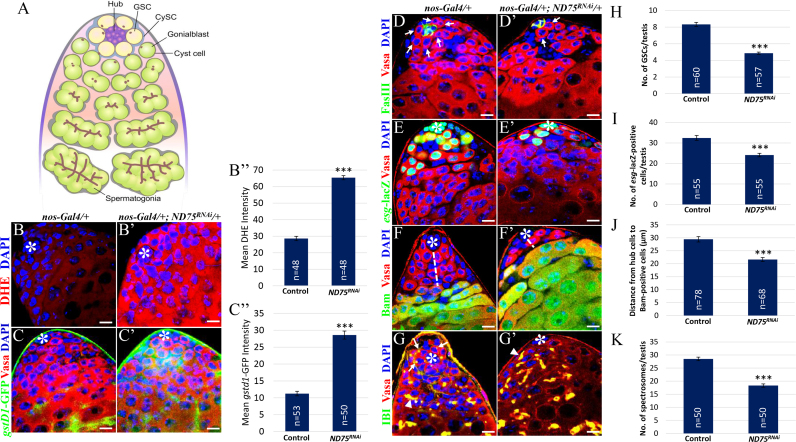
Oxidative stress has been shown to influence stem cell behavior by facilitating the proliferation, differentiation or apoptosis of stem cell populations [1]. In the Drosophila testis, redox states regulated by Keap1/Nrf2 activity play important roles in GSC maintenance [18]. In an attempt to identify molecules that are associated with GSC differentiation induced by high levels of ROS, we first confirmed the effects of increased intracellular ROS levels by knocking down ND75 on GSC homeostasis. The mitochondrial electron transport chain (ETC) is the major consumer of O2 and thus serves as a key contributor to ROS generation in mammalian cells [22]. ND75 is the Drosophila homolog of human mitochondrial complex I subunit NDUFS1 (NADH: Ubiquinone Oxidoreductase Core Subunit S1), which has NADH (nicotinamide adenine dinucleotide, reduced) dehydrogenase and oxidoreductase activities, and whose inhibition was shown to increase ROS levels [23], [24], [25]. To assess if altered ND75 activity in the Drosophila testis can affect ROS levels and thus disrupt GSC behavior, testes expressing ND75RNAi under the control of nos-Gal4, which is expressed in early-stage germ cells, were stained with DHE dye to monitor O2·- levels. In the Drosophila testis, 6–12 GSCs are present, and each is enclosed by a pair of somatic cyst stem cells (CySCs); both GSCs and CySCs attach and surround 10–15 post-mitotic hub cells, forming the stem cell niche (Fig. 1A). GSCs undergo asymmetric division to produce one daughter cell that remains as a stem cell, and another differentiating daughter cell called gonialblast that is displaced away from hub cells. The gonialblast then proceeds with the four rounds of synchronous transit-amplifying (TA) divisions, giving rise to a cyst of 16 spermatogonia which go on to eventually differentiate into mature sperms [17]. Notably, testes expressing ND75RNAi showed a higher intensity of DHE staining as compared to control testes (Fig. 1B, B’ and B”), suggesting that ND75 knockdown can cause an increase in ROS levels. This finding was further validated by using transgenic flies carrying an independent oxidative stress reporter gene gstD1-GFP [19]. While GFP expression was barely detected at the apical tip of control testes in which GSCs and early-stage germ cells reside, an intense GFP expression was observed at the apical tip of ND75 knockdown testes (Fig. 1C, C’ and C”).
Fig. 1.
ND75 knockdown increases ROS levels and promotes GSC differentiation in the Drosophila testis. (A) Schematic of the Drosophila testis. (B, B’ C and C’) ROS levels were monitored by using DHE probe and the in vivo ROS reporter GstD1-GFP in (B and C) control testes and (B’ and C’) testes expressing ND75RNAi. (B” and C”) The intensity of DHE staining and GFP was quantified using the ImageJ software. (D, E, F and G) Control and (D’, E’, F’ and G’) ND75 knockdown testes. (D and D’) Testes stained for FasIII and Vasa, which mark hub cells and germ cell lineage, respectively (arrows indicate GSCs attached to hub cells). (E and E’) Testes stained for esg-lacZ, which marks early-stage germ cells. (F and F’) Testes stained for the differentiation marker Bam. Dashed lines indicate the distance between hub cells and differentiating germ cells. (G and G’) Testes stained with 1B1, which stains spectrosomes and branching fusomes. Arrows indicate spectrosomes and arrowheads indicate branched fusomes. Quantification of (H) GSCs, (I) esg-lacZ-positive cells, (J) the distance between hub cells and Bam-positive germ cells, and (K) spectrosomes. Error bar is SEM from three independent experiments. Significance was assessed using unpaired t-test (***p<0.001). GSC: germline stem cell. CySC: (somatic) cyst stem cell. Scale bar, 10 µM. *hub cells.
We next examined whether high levels of ROS induced by ND75RNAi can affect GSC maintenance. Testes were stained with Vasa antibody, which marks germ cell lineage. As compared to control testes, ND75 knockdown testes showed a reduced number of GSCs, which are Vasa-positive cells directly attached to hub cells and are arranged in a rosette pattern (Fig. 1D and D’, arrows). While testes expressing ND75RNAi had an average of 4.9 GSCs per testis, control testes had an average of 8.3 GSCs (Fig. 1H). Consistently, in testes expressing ND75RNAi much less Vasa-positive cells were found to be positive for esg (escargot)-lacZ, which marks only early-stage germ cells such as GSCs and gonialblasts, as compared to control (Fig. 1E, E’ and I). To further confirm the finding that ND75 inhibition decreases GSC number, we next examined the expression pattern of the differentiation marker Bam (Bag of marble), which is normally expressed in 4- to early 16-germ cells located several cell diameters away from the hub cells. In testes expressing ND75RNAi, Bam was detected much closer to the hub cells as compared to control (Fig. 1F, F’ and J), indicative of premature GSC differentiation. The inhibitory effects of ND75 on GSC differentiation was further confirmed by the decreased number of spectrosomes, which appear a small dot in GSCs and GSC/gonialblast pairs, in testes expressing ND75RNAi as compared to control testes (Fig. 1G, G’ and K, arrows). All these findings are in accordance with our previous observation that high levels of ROS induced by the oxidant paraquat promote GSC differentiation [18], and suggest that testes with elevated ROS by ND75 knockdown can be utilized for genome-wide studies to identify the downstream effectors of high ROS-induced GSC differentiation.
3.2. Microarray and data analysis
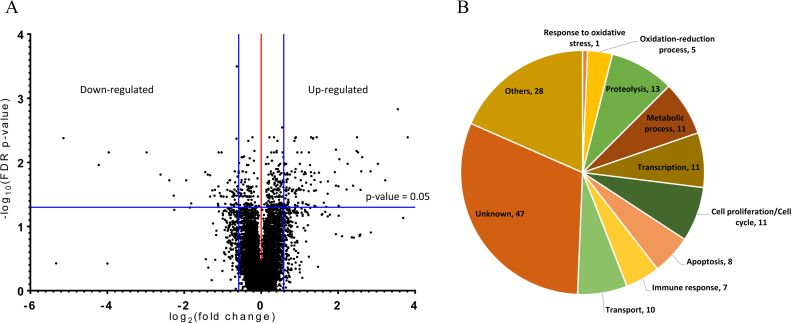
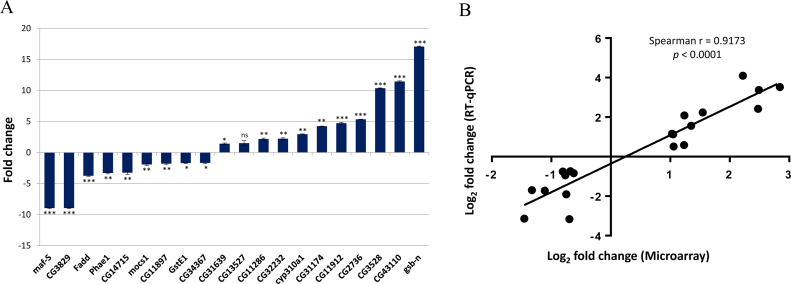
The regulators of redox homeostasis within stem cells remain largely unexplored. In particular, ROS-induced effectors that control stem cell behavior are not fully understood. To identify those that are involved in GSC differentiation facilitated by high levels of ROS, we performed Affymetrix genechip microarray analyses to establish gene expression profiling of testes with moderate (control) and high (knockdown of ND75) ROS levels. 152 genes were found to be differentially expressed in testes with ND75 knockdown as compared to control testes; 96 genes were upregulated and 56 genes were downregulated by more than 1.5 folds (Fig. 2A, Table 1). We identified several genes, including maf-S (small maf), GstE1 (Glutathione S transferase E1) and lox2 (lipoxygenases 2), whose products have yet been implicated in redox homeostasis as a candidate. In an attempt to predict functions and generate testable hypotheses for the characterization of the 152 genes, we have assigned these genes to categories based on their predicted molecular functions, protein domains, and reports from the ontology database in the Flybase (http://flybase.org/). These categories include (1) oxidation-reduction process, (2) metabolic process, (3) transcription, and (4) cell proliferation (Fig. 2B). Notably, 31% of the identified genes showed no predicted molecular function and/or recognizable protein domain, suggesting that this genome-wide microarray analysis identified many uncharacterized genes, possibly with an essential role in ROS-induced GSC differentiation. Importantly, we randomly selected 20 genes among the candidates for further validation by quantitative RT-PCR analyses, and found that expression pattern of the 20 genes tested was highly correlated to that obtained from microarray analysis, with the Spearman's rho of 0.9173 (P<0.0001) (Fig. 3A and B).
Fig. 2.
Affymetrix microarray data analysis. (A) A volcano plot. 152 genes were found to be differentially expressed by more than 1.5 folds in testes with high levels of ROS as compared to control testes. (B) The identified genes were assigned to categories based on their predicted molecular functions, protein domains, and reports from the ontology database in the Flybase.
Table 1.
A list of genes transcriptionally modulated during GSC differentiation.
| Up-regulated genes |
Down-regulated genes |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Transcript Cluster ID | Fly Gene Symbol | Human Ortholog | Fold Change | FDR p-value | Transcript Cluster ID | Fly Gene Symbol | Human Ortholog | Fold Change | FDR p-value |
| 18197227 | CG14369 | N/A | 14.03 | 0.004053 | 18143346 | Ref2 | ALYREF | −1.5 | 0.013804 |
| 18200889 | Gr93d | N/A | 9.34 | 0.019055 | 18198373 | CG17186 | N/A | −1.5 | 0.014708 |
| 18164680 | CG43110 | GZMH | 7.17 | 0.013804 | 18133084 | CG5440 | UBE2E3 | −1.51 | 0.030002 |
| 18199988 | CG10000 | GALNT14 | 6.17 | 0.008274 | 18194131 | wts | LATS2 | −1.51 | 0.02603 |
| 18145258 | CG33307 | N/A | 5.99 | 0.0268 | 18188101 | sosie; CR45216 | N/A | −1.53 | 0.016607 |
| 18141312 | CG3528 | C4orf22 | 5.62 | 0.004053 | 18171734 | CG33217 | PELP1 | −1.54 | 0.023726 |
| 18162277 | CG2736 | SCARB2 | 5.57 | 0.006617 | 18188937 | CG11897 | ABCC4 | −1.54 | 0.045327 |
| 18138530 | CG43402 | N/A | 4.75 | 0.010525 | 18162274 | Phk-3 | N/A | −1.55 | 0.033 |
| 18156218 | gsb-n | PAX3 | 4.66 | 0.02644 | 18194371 | mia | TAF6L | −1.56 | 0.028082 |
| 18134734 | dpr19 | USH2A | 4.09 | 0.008924 | 18169211 | Tgi | VGLL4 | −1.56 | 0.004262 |
| 18144843 | CG31777 | N/A | 4.04 | 0.047532 | 18145722 | del | N/A | −1.57 | 0.042923 |
| 18185346 | CG18473 | PTER | 4.02 | 0.023605 | 18189333 | CG11333 | ISOC1 | −1.6 | 0.020327 |
| 18136286 | Gr32a | GZMH | 3.85 | 0.006331 | 18145631 | CG34367 | SHOX2 | −1.6 | 0.020466 |
| 18151208 | CG10764 | N/A | 3.85 | 0.023075 | 18142936 | CG5676 | FUNDC1 | −1.61 | 0.038272 |
| 18180564 | CG34025 | N/A | 3.31 | 0.030731 | 18151843 | maf-S | MAFK | −1.62 | 0.032486 |
| 18132093 | Acyp | ACYP2 | 3.23 | 0.049199 | 18146894 | a | PDZD2 | −1.62 | 0.045041 |
| 18197785 | CG14905 | CCDC63 | 3.09 | 0.042576 | 18155416 | mi | N/A | −1.62 | 0.02644 |
| 18141034 | CG11912 | PRTN3 | 2.92 | 0.020284 | 18133793 | CG18269 | MCMBP | −1.63 | 0.049199 |
| 18136753 | CG31776 | GALNTL6 | 2.83 | 0.01447 | 18134015 | CG3430 | N/A | −1.63 | 0.047739 |
| 18157091 | imd | PIDD1 | 2.72 | 0.004053 | 18135993 | DCTN5-p25 | DCTN5 | −1.63 | 0.030878 |
| 18139932 | cutlet | CHTF18 | 2.68 | 0.027276 | 18161200 | Or56a | N/A | −1.64 | 0.046053 |
| 18163569 | sprt | PARD3B | 2.66 | 0.048525 | 18151370 | CG14500 | N/A | −1.64 | 0.031712 |
| 18177597 | CG10361 | GCAT | 2.61 | 0.018769 | 18165004 | CG43668 | N/A | −1.65 | 0.029659 |
| 18135200 | CG16820 | N/A | 2.61 | 0.047532 | 18202690 | CG42487; CG4884 | C6orf203 | −1.65 | 0.034006 |
| 18154275 | Idgf5 | CHIT1 | 2.6 | 0.027711 | 18187489 | Fadd | PIDD1 | −1.68 | 0.034006 |
| 18143767 | Cyp310a1 | TBXAS1 | 2.55 | 0.004184 | 18168671 | CG3335 | RBM19 | −1.69 | 0.028633 |
| 18135886 | CG1421 | N/A | 2.54 | 0.004262 | 18173465 | Mocs1 | MOCS1 | −1.7 | 0.046053 |
| 18156154 | Cyp6a2 | TBXAS1 | 2.47 | 0.016296 | 18202203 | Nlg3 | NLGN4Y | −1.73 | 0.045041 |
| 18141305 | CG18641 | LIPI | 2.45 | 0.022122 | 18183067 | GstD5 | VARS | −1.75 | 0.028009 |
| 18168480 | CG13309 | N/A | 2.45 | 0.01418 | 18151405 | GstE1 | GSTT2B | −1.75 | 0.018895 |
| 18201360 | CG31174 | DCTN3 | 2.36 | 0.006981 | 18178062 | CG18649 | N/A | −1.77 | 0.019055 |
| 18152278 | CG13527 | N/A | 2.35 | 0.028082 | 18137896 | gkt | TDP1 | −1.83 | 0.020721 |
| 18165049 | CG43742 | N/A | 2.35 | 0.046053 | 18191472 | tal-AA; tal-1A; tal-2A; tal-3A | N/A | −1.84 | 0.0268 |
| 18188796 | CG1894 | KAT8 | 2.32 | 0.04604 | 18161799 | RpL37b | RPL37 | −1.91 | 0.044144 |
| 18197860 | CG17477 | KLK14 | 2.24 | 0.044589 | 18179059 | CG7298 | N/A | −1.97 | 0.034066 |
| 18136253 | Tep3 | CD109 | 2.23 | 0.015042 | 18195064 | Aats-met-m | MARS2 | −2.04 | 0.006981 |
| 18202255 | CG34283 | GTSF1L | 2.2 | 0.01418 | 18132382 | CG17834 | N/A | −2.05 | 0.034006 |
| 18191104 | CG34436 | N/A | 2.16 | 0.015331 | 18199863 | CG5402 | N/A | −2.07 | 0.033 |
| 18131563 | Ance | ACE | 2.15 | 0.02719 | 18139318 | prd | PAX3 | −2.11 | 0.02995 |
| 18130728 | salr | SALL4 | 2.11 | 0.043396 | 18141809 | Vps52 | VPS52 | −2.15 | 0.006981 |
| 18179578 | CG13053 | N/A | 2.11 | 0.041534 | 18138505 | Phae1 | PRSS53 | −2.16 | 0.023724 |
| 18141944 | CG9107 | RRP7A | 2.09 | 0.023266 | 18193273 | Blm | BLM | −2.17 | 0.047191 |
| 18144661 | CG31639 | UCHL5 | 2.08 | 0.013804 | 18187075 | CG16727 | SLC22A24 | −2.18 | 0.044165 |
| 18195851 | CG11286 | LAPTM5 | 2.07 | 0.006981 | 18133474 | CG2772 | LIPN | −2.3 | 0.0268 |
| 18180053 | CG32232 | TUBGCP5 | 2.04 | 0.018899 | 18190914 | CG34278 | N/A | −2.32 | 0.033063 |
| 18201456 | CG31244 | TACO1 | 2.04 | 0.026546 | 18160388 | CG8195 | SLC35F5 | −2.41 | 0.027265 |
| 18186923 | CG7694 | RNF181 | 2.03 | 0.0268 | 18185768 | CG14715 | FKBP2 | −2.51 | 0.034006 |
| 18201815 | CG32945 | N/A | 2.01 | 0.010525 | 18199680 | TwdlP | N/A | −2.51 | 0.015833 |
| 18200489 | CG15563 | N/A | 1.97 | 0.025277 | 18206397 | Cp7Fb | N/A | −2.62 | 0.015445 |
| 18146064 | CG42399 | FAM179B | 1.97 | 0.02644 | 18152912 | CG3829 | SCARB2 | −2.74 | 0.01418 |
| 18185352 | CG8129 | SRR | 1.95 | 0.042771 | 18178266 | CG13047 | N/A | −3.53 | 0.043603 |
| 18136832 | Qtzl | SCLY | 1.92 | 0.047532 | 18186763 | CG17283 | REN | −4.83 | 0.033 |
| 18167562 | CG12017 | N/A | 1.92 | 0.006981 | 18170763 | CG11458 | N/A | −5.23 | 0.019055 |
| 18142751 | CG4438 | PDAP1 | 1.89 | 0.0268 | 18169512 | CG13445 | N/A | −6.13 | 0.015445 |
| 18175784 | CG2211 | JMJD6 | 1.88 | 0.027711 | 18169271 | CG8100 | N/A | −15.57 | 0.006981 |
| 18190415 | CG31465 | N/A | 1.87 | 0.004053 | 18171820 | CG33500 | N/A | −18.66 | 0.010997 |
| 18186284 | CG3259 | TRAF3IP1 | 1.86 | 0.01269 | |||||
| 18138632 | CG43707 | RHPN2 | 1.86 | 0.030878 | |||||
| 18136825 | ZnT33D | SLC30A3 | 1.85 | 0.017284 | |||||
| 18133131 | Der-1 | DERL1 | 1.85 | 0.004184 | |||||
| 18164674 | CG43108 | N/A | 1.84 | 0.046581 | |||||
| 18131918 | SA | STAG1 | 1.84 | 0.046053 | |||||
| 18131649 | Try29F | PRSS3 | 1.84 | 0.01418 | |||||
| 18204185 | CG32006 | FOXJ1 | 1.83 | 0.029567 | |||||
| 18143367 | CG17036 | SLC19A2 | 1.82 | 0.004625 | |||||
| 18183737 | Gcn2 | EIF2AK4 | 1.78 | 0.02644 | |||||
| 18158424 | CG14591 | TMEM164 | 1.75 | 0.02669 | |||||
| 18153402 | CG30056 | N/A | 1.67 | 0.025134 | |||||
| 18181298 | nbs | NBN | 1.65 | 0.049927 | |||||
| 18199613 | CG4960 | REEP5 | 1.64 | 0.041962 | |||||
| 18198659 | CG6475 | UGT2B10 | 1.63 | 0.038221 | |||||
| 18145247 | Muc30E | MPV17L2 | 1.62 | 0.041534 | |||||
| 18180076 | CG32263 | N/A | 1.62 | 0.045041 | |||||
| 18142588 | Ostgamma | TUSC3 | 1.61 | 0.015445 | |||||
| 18177436 | Ufd1-like | UFD1L | 1.61 | 0.018179 | |||||
| 18203120 | CG42778 | N/A | 1.61 | 0.043368 | |||||
| 18192979 | Dip-C | PEPD | 1.6 | 0.046053 | |||||
| 18160269 | Adgf-E | CECR1 | 1.58 | 0.020284 | |||||
| 18138273 | CG43050 | MAIP1 | 1.58 | 0.018766 | |||||
| 18152420 | CG9863 | N/A | 1.58 | 0.046581 | |||||
| 18134839 | CG6094 | MRPL58 | 1.57 | 0.028082 | |||||
| 18164795 | CG43326; CG43325 | N/A | 1.57 | 0.03253 | |||||
| 18149666 | Cyp4p3 | CYP4Z1 | 1.57 | 0.010525 | |||||
| 18177352 | CG6674 | TSSC4 | 1.56 | 0.046581 | |||||
| 18161537 | lox2 | LOXL3 | 1.56 | 0.026405 | |||||
| 18185958 | Gnmt | GNMT | 1.55 | 0.0365 | |||||
| 18134385 | CG13090 | MOCS3 | 1.55 | 0.027298 | |||||
| 18175967 | CG5687 | SLC5A6 | 1.55 | 0.015445 | |||||
| 18199611 | CG4956 | ZDHHC24 | 1.54 | 0.04802 | |||||
| 18153798 | Mal-A6 | SLC3A1 | 1.53 | 0.010525 | |||||
| 18196850 | CG3313 | DCAF12 | 1.52 | 0.010997 | |||||
| 18153174 | Obp56i | N/A | 1.51 | 0.044165 | |||||
| 18163701 | CG34216 | N/A | 1.51 | 0.014708 | |||||
| 18142869 | CG5846 | SMOX | 1.5 | 0.046053 | |||||
| 18177135 | CG5653 | RFXANK | 1.5 | 0.020284 | |||||
| 18162351 | CG16926 | N/A | 1.5 | 0.048525 | |||||
Fig. 3.
Validation of microarray data by qRT-PCR analysis. (A) qRT-PCR analysis. Randomly selected 20 genes were validated by qRT-PCR analyses using the same RNA extracted for microarray analysis. Error bar is SEM from three independent experiments. Significance was assessed using unpaired t-test (*p<0.05, ***p<0.01, ***p<0.001). (B) Spearman's rho. Expression pattern of the 20 genes tested by qRT-PCR is highly correlated to that observed in microarray analysis, with a Spearman's correlation coefficient value of 0.9173 (P<0.0001).
Interestingly, mal-A6 (maltase A6), the Drosophila homolog of the cysteine transporter gene Slc3a1 (solute carrier family 3 member A1), was identified whose product has been reported to function in carbohydrate process (Table 1). Notably, high SLC3A1 expression was shown to promote the cysteine uptake and the accumulation of reductive glutathione (GSH), leading to a decrease in ROS levels and an activation of Akt signaling [26]. By contrast, knockdown of Slc3a1 was reported to decrease intracellular GSH and thus increase intracellular ROS activity [27]. These may suggest the potential role of Akt signaling in ROS-associated GSC behavior, and the presence of a feedback loop between ROS signaling and Mal-A6, in the Drosophila testis. We also found wts, the Drosophila homolog of Lats (Large tumor suppressor kinase), to be transcriptionally regulated upon high levels of ROS (Table 1). LATS is a central component of the Hippo pathway that negatively regulates the activity of the transcriptional co-factor YAP (Yes-associated protein) [28]. Notably, YAP has been reported to form a heterodimer with FoxO1, binding to the promoters of the catalase and manganese superoxide dismutase (MnSod) antioxidant genes and activating their transcription [29], suggesting the potential role of the Hippo signaling pathway in redox homeostasis and ROS-mediated biological processes such as stem cell maintenance. All these observations suggest the validity and reliability of our microarray analysis to identify ROS-associated effectors that may regulate GSC behavior in the Drosophila testis.
3.3. The gene maf-S is transcriptionally downregulated by high levels of ROS
Keap1 (Kelch-like ECH-associated protein 1)/Nrf2 (NF-E2-related factor 2) signaling acts as a key regulator of redox homeostasis. Upon electrophilic and oxidative stress, Keap1/Cul3 E3 ubiquitin ligase-mediated proteosomal degradation of Nrf2 becomes disrupted, allowing Nrf2 to be stable and translocate into the nucleus to form a hetero-dimerization with the basic region leucine zipper (bZIP)-type transcription factor Maf for the transcriptional activation of many phase II detoxification enzyme genes such as thioredoxin reductase and glutathione reductase [30], [31], [32], [33], [34], [35]. The small Mafs (sMafs) comprise of MafF, MafG and MafK. They form heterodimers with cap ‘n’ collar (CNC) proteins such as Nrf1 and Nrf2, and bind to the antioxidant/electrophile response element (ARE/EpRE), indicating the role of sMafs in redox balance. However, despite their putative role in the regulation of antioxidant gene expression, their function in stem cell homeostasis is not fully characterized.
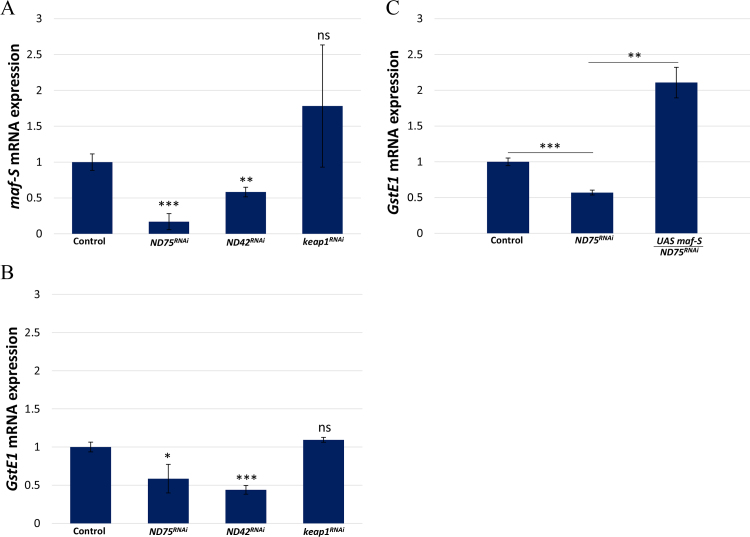
Keap1/Nrf2 signaling is evolutionarily well conserved in Drosophila [19]. In this study, Maf-S, a sole small Maf family member in Drosophila, was found to be downregulated in testes with high levels of ROS. To determine whether Maf-S functions in GSC maintenance, we further confirmed the transcription of maf-S by qRT-PCR using RNA extracted from testes with increased ROS levels induced by the suppression of ND75 or ND42 (the Drosophila homolog of NDUFA10, a mitochondria complex 1 subunit), and testes with decreased ROS levels induced by keap1 knockdown (keapRNAi). Consistent with the microarray data, we found that maf-S mRNA levels are significantly reduced in testes with high levels of ROS (Fig. 4A). On the other hand, we found that decreased levels of ROS induced by keap1 knockdown do not cause any significant alterations in maf-S transcription (Fig. 4A), suggesting that Maf-S expression is responsive to oxidative stress in the Drosophila testis. Interestingly, we found that the expression of the oxidative stress response gene GstE1 is also downregulated upon high ROS (Table 1, Fig. 4B), whereas the expression of other oxidative stress response genes such as GstE9, Gclc (Glutamate-cysteine ligase catalytic subunit) and Gclm (Glutamate-cysteine ligase modifier subunit) showed a trend of increase upon high ROS in our microarray experiment (data not shown). In an attempt to show that GstE1 expression is related to Maf-S, we assessed whether overexpression of Maf-S can restore the decreased expression of GstE1 caused by ND75 knockdown. We found that ectopic expression of Maf-S efficiently restores GstE1 expression, suggesting that GstE1 expression is associated with Maf-S (Fig. 4C). It has been reported that many stimuli such as H2O2, cadmium and zinc, which can induce Nrf2 activity, enhance the expression of MafG [36], [37], [38]. On the other hand, exogenous H2O2 decreased c-Maf expression in human adipose tissue-derived mesenchymal stem cells (hAMSCs) [39]. These observations may suggest that maf genes are transcriptionally modulated upon redox states in a cell, tissue and/or context-specific manner.
Fig. 4.
The gene maf-S is transcriptionally downregulated upon oxidative stress. The mRNA levels of (A) maf-S and (B) GstE1 are downregulated upon increased levels of ROS induced by knockdown of ND75 or ND42, but are not affected upon decreased levels of ROS induced by knockdown of keap1. (C) Overexpression of Maf-S restores the decreased expression of GstE1 caused by ND75 knockdown. Error bar is SEM from three independent experiments. Significance was assessed using unpaired t-test (*p<0.05, ***p<0.01, ***p<0.001).
3.4. Knockdown of maf-S promotes GSC differentiation
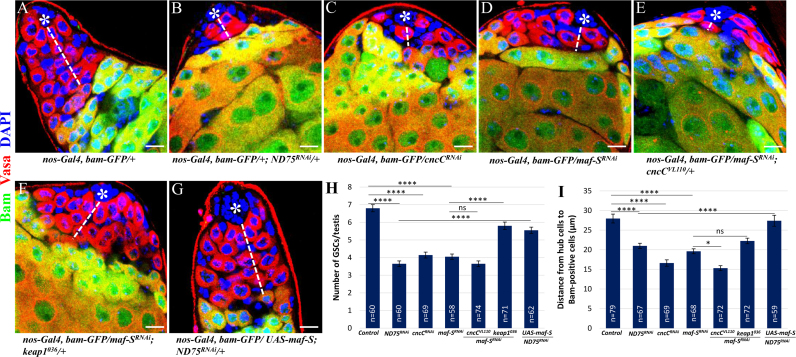
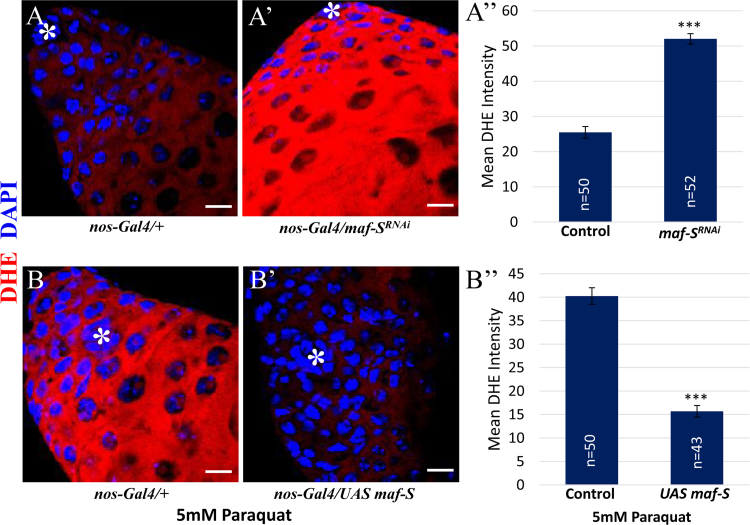
To examine the potential role of Maf-S in GSC maintenance, we first assessed if Maf-S can function in redox homeostasis by quantifying DHE staining in testes ectopically expressing maf-SRNAi or Maf-S under the control of nos-Gal4. While maf-S knockdown significantly increased ROS levels, ectopic expression of Maf-S decreased ROS levels, suggesting that Maf-S plays a role in the maintenance of redox homeostasis (Supplementary Fig. 1). Since high levels of ROS downregulate maf-S transcription and decrease GSC number by promoting GSC differentiation, we asked whether knockdown of maf-S is indeed associated with the decrease in GSCs. As a positive control, we inhibited ND75 under the control of nos-Gal4 in testes to show that high levels of ROS decrease GSC number as compared to control (Fig. 5A, B and H). To confirm the effects of high ROS on GSC number, we disrupted the activity of CncC, the Drosophila homolog of Nrf2, by expressing cncCRNAi, and found that CncC inhibition also results in a decrease in GSC number as compared to control (Fig. 5A, C and H). Interestingly, maf-S knockdown also led to a significant decrease in GSC number, indicating that Maf-S is a downstream effector of ROS-induced GSC differentiation and is required for GSC maintenance (Fig. 5A, D and H). To examine if Maf-S genetically interacts with Keap1/Nrf2 (CncC) in GSC homeostasis, Maf-S was inhibited in testes heterozygous for cncC (cncCVL110) or keap1 (keap1036). The inhibitory effect of Maf-S on GSC number was not enhanced in cncC+/- testes, but significantly suppressed in keap1+/- testes (Fig. D, E, F and H). Since Maf-S is a downstream effector of ROS signaling, we hypothesized that ectopic expression of Maf-S would suppress the phenotype of GSC loss induced by oxidative stress. As expected, the effect of ND75 inhibition on GSC number was greatly bypassed by Maf-S overexpression (Fig. 5B, G and H).
Fig. 5.
Maf-S inhibition causes a decrease in GSC number by promoting GSC differentiation. (A, B, C and H) GSC number in testes with ND75 or CncC knockdown decreases as compared to that in control testes. (D and H) maf-S knockdown testes also show decreased number of GSCs, suggesting that Maf-S is required for GSC homeostasis. (E, F and H) Inhibitory effects of Maf-S on GSC number in cncC+/- and in keap1+/- testes. (G and H) Ectopic expression of Maf-S rescues the inhibitory effects of ND75 on GSC number, suggesting that Maf-S acts as a downstream effector of high ROS-associated GSC differentiation. (A, B, C, D, I) Bam-positive germ cells in testes expressing ND75RNAi, cncCRNAi or maf-SRNAi are detected closer to hub cells as compared to those in control testes. (E, F and I) Inhibitory effects of Maf-S on GSC differentiation in cncC+/- and in keap1+/- testes. (G and I) Ectopic expression of Maf-S rescues the ND75RNAi phenotype of GSC differentiation. Error bar is SEM from more than three independent experiments. Significance was assessed using one-way ANOVA with post-hoc Bonferroni test (*p<0.05, ****p<0.0001). Scale bar, 10 µM. *hub cells.
We next examined the expression of Bam to confirm the above mentioned findings. Bam expression was detected much closer to hub cells in testes expressing either ND75RNAi, cncCRNAi or maf-SRNAi, as compared to control, suggesting that elevated ROS levels decreased GSC number by promoting their differentiation (Fig. 5A, B, C, D and I). Furthermore, we found that Maf-S inhibition-promoted GSC differentiation was further enhanced by removing one copy of cncC alleles, but was not significantly suppressed by removing one copy of keap1 alleles (Fig. 5D, E, F and I). Importantly, Bam-positive cells were detected further away from hub cells in testes co-expressing ND75RNAi and Maf-S, as compared to those in testes expressing ND75RNAi alone (Fig. 5B, G and I). It is worth to note that knockout of all three sMafs in mouse embryonic fibroblasts was shown to interfere with the induction of antioxidant genes such as thioredoxin reductase 1, suggesting the essential role of sMafs in scavenging excessive ROS [38]. The oxidative stress-sensitive c-Maf has also been shown to be downregulated by exogenous H2O2 which is associated with H2O2-mediated reduced pluripotency and adipogenic differentiation of hAMSCs [39]. Together with the facts that Maf-S functions in the maintenance of redox homeostasis and genetically interacts with Keap1/CncC in GSC behavior, our data suggests that Maf-S plays an important role in GSC maintenance by engaging in the regulation of redox balance in the Drosophila testis.
3.5. Ectopic expression of Maf-S causes an over-growth of GSC-like cells
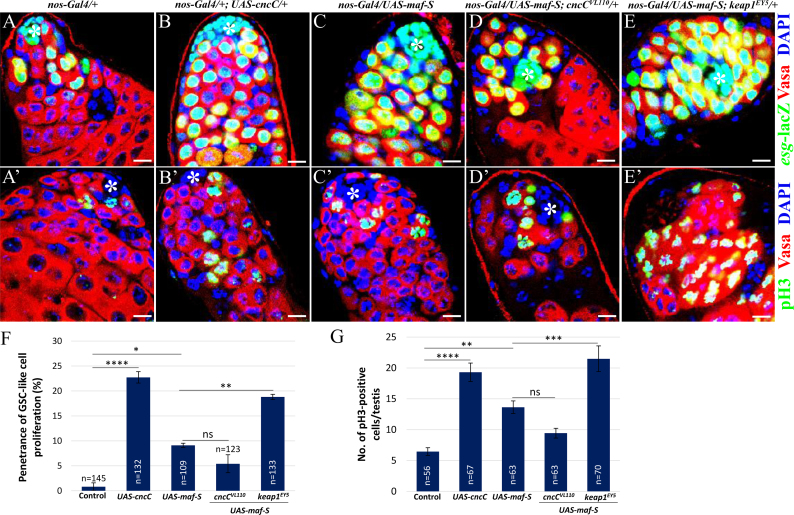
Since loss of function of Maf-S promoted GSC differentiation, we hypothesized that ectopic expression of Maf-S may conversely promote GSC growth by decreasing intracellular ROS levels. We previously showed that germ cells mutant for cncC exhibit higher ROS levels than neighbouring control cells, suggesting that ectopic expression of CncC decreases ROS levels [18]. Interestingly, testes overexpressing CncC contained a dramatically increased number of Vasa- and esg-LacZ-positive cells, which are distributed widely throughout the testes and appear as single cells, whereas esg-LacZ-positive cells were found to be restricted to the apical tip of control testes (Fig. 6A and B). Indeed, 22.7% of testes (n=132) overexpressing CncC showed the phenotype of GSC-like cell proliferation (Fig. 6F). We also observed the similar phenotype in 9.1% of testes (n=109) overexpressing Maf-S, suggesting that Maf-S overexpression can promote the proliferation of GSC-like cells, possibly through lowering ROS levels via activating the transcription of antioxidant genes (Fig. 6C and F). We next overexpressed Maf-S in cncC+/- testes to examine their genetic interaction in the growth of GSC-like cells, and found a trend of almost two-fold decrease in the penetrance by removing one copy of cncC alleles, although the result did not achieve statistical significance at the 0.05 level (Fig. 6C, D and F). In a keap1-null background, many of the ARE-dependent genes have been shown to be persistently activated in an Nrf2-dependent manner [38]. Furthermore, we previously showed that decreased levels of ROS by Keap1 inhibition causes an over-growth of GSC-like cells [18]. Thus, we next overexpressed Maf-S in testes heterozygous for keap1 (keap1EY5), expecting that more ARE-dependent gene products become available. As expected, the penetrance was drastically elevated upon removing one copy of keap1 alleles as compared to that observed in testes expressing Maf-S alone (9.1% vs. 18.8%) (Fig. 6C, E and F), suggesting that the over-growth of GSC-like cells observed in testes expressing Maf-S is associated with the downregulation of ROS levels.
Fig. 6.
Ectopic expression of Maf-S causes an over-growth of GSC-like cells. (A, B, C, D and E) Testes stained for esg-lacZ. As compared to (A) control, ectopic expression of (B) CncC or (C) Maf-S causes a drastic increase in the number of esg-lacZ-positive early-stage germ cells. (D and E) Effects of Maf-S overexpression on GSC-like cells in cncC+/- and in keap1+/- testes. (F) Quantification of the penetrance of GSC-like cell proliferation. (A’, B’, C’, D’ and E’) Testes stained for pH3, which acts as a mitotic marker. As compared to (A’) control, ectopic expression of (B’) CncC or (C’) Maf-S significantly increases the number of pH3-positive germ cells. (D’ and E’) Effects of Maf-S overexpression on pH3-positive cell number in cncC+/- and in keap1+/- testes. (G) Quantification of pH3-postive cell number. Error bar is SEM from more than three independent experiments. Significance was assessed using one-way ANOVA with post-hoc Bonferroni test (*p<0.05, **p<0.01, ***p<0.001 and ****p<0.0001). Scale bar, 10 µM. *hub cells.
In an attempt to demonstrate that the over-growth phenotype of GSC-like cells is a cause of increased mitotic activity of germ cells, we next examined the expression of phospho-Histone H3 (pH3), a mitotic marker detected only in actively dividing cells [40]. In control testes, pH3 staining is detected only in self-renewing GSCs directly attached to hub cells, as well as in transit-amplifying spermatogonia (Fig. 6A’). However, testes overexpressing CncC or Maf-S showed a significantly increased number of both Vasa- and pH3-positive cells throughout the testes (Fig. 6B’ and C’), suggesting that germ cell lineage underwent abnormal cell division. Notably, the over-growth of germ cells caused by Maf-S overexpression was not significantly affected in cncC+/- testes, but greatly enhanced in keap1+/- testes (Fig. 6C’, D’, E’ and G).
In mammals, sMafs form heterodimers with other specific bZIP transcription factors, such as CNC and Bach family members. Notably, sMafs were reported to be an indispensable partner of Nrf2 in various cellular processes such as the transcriptional activation of antioxidant enzymes and keratinocyte differentiation [38], [41]. In Drosophila, Maf-S was shown to interact both physically and genetically with CncC [42], [43]. Overexpression of Maf-S or its dimerization partner, CncC, could restore the locomotor activity in the Drosophila model of Parkinson's disease, which is the most common neurodegenerative movement disorder highly associated with oxidative stress [44]. Furthermore, it was reported that ectopic expression of Maf-S can restore the age-associated decline in the oxidative stress resistance and can upregulate the expression of the CncC-target antioxidant genes such as gstD1 (Glutathione S transferase D1), gclc and gclm [43]. Importantly, these observations are in accordance with our finding that Maf-S genetically interacts with Keap1/CncC in GSC homeostasis, and ectopic expression of Maf-S or CncC results in the similar phenotype of GCS-like cell proliferation in the Drosophila testis.
4. Conclusion
The disruption of redox balance interferes with the maintenance and self-renewal of various stem cell populations by affecting their proliferation, differentiation and senescence. Furthermore, oxidative stress has been implicated in a wide variety of human diseases, including neurodegenerative diseases, cardiovascular diseases, diabetes mellitus type 2 and cancers. Hence, it is essential to elucidate molecular mechanisms by which altered intracellular ROS levels induce these biological processes. Here, we performed Affymetrix microarray analysis to identify the downstream effectors of ROS signaling that may control GSC differentiation in the Drosophila testis. We demonstrated that 1) 152 genes are transcriptionally modulated during GSC differentiation, and 31% of the identified genes have no predicted molecular function and protein domains, suggesting that our analysis identified many uncharacterized genes, possibly with essential roles in ROS-associated stem cell homeostasis; 2) maf-S transcription is downregulated upon oxidative stress; 3) Maf-S functions in the maintenance of redox homeostasis; 4) maf-S knockdown and Maf-S overexpression promotes premature GSC differentiation and the over-growth of GSC-like cells, respectively; 5) premature GSC differentiation induced by oxidative stress is suppressed by the ectopic expression of Maf-S, indicating that Maf-S acts as a downstream effector of ROS signaling; 6) Maf-S genetically interacts with Keap1/CncC in GSC homeostasis. Taken altogether, our study reveals that Maf-S is one of the key downstream effectors of redox signaling that controls GSC behavior in the Drosophila testis.
Acknowledgements
We thanks D. Bohmann, L. Liu, S. Dinardo, D.M. Mckearin, H. Jasper, NIG-FLY Stock Center and BDSC for fly stocks; DSHB for antibodies. We also thanks S.L. Bay for her assistance in preparing the illustration. This work was supported by MOE TIER1 (R-181-000-172-112).
Acknowledgments
Author contribution
W.S.T. performed the experiments and wrote the manuscript. G.W. Y. analysed the microarray data. T.S. proofread the manuscript. G.H.B designed the experiments and wrote the manuscript.
Conflict of interest statement
The authors have no conflict of interest to declare.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2017.12.002.
Appendix A. Supplementary material
Supplementary material. A list of primers used for qRT-PCR analysis.
Fig. S1.
(A and A’) Maf-S knockdown significantly increases ROS levels compared to control as determined by DHE staining. (B and B’) Ectopic expression of Maf-S reverts the increased ROS levels caused by paraquat treatment. (A” and B”) Quantification of DHE staining intensity. Scale bar, 10 µM. *hub cells.
References
- 1.Bigarella C.L., Liang R., Ghaffari S. Stem cells and the impact of ROS signaling. Development. 2014;141(22):4206–4218. doi: 10.1242/dev.107086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liou G.Y., Storz P. Reactive oxygen species in cancer. Free Radic. Res. 2010;44(5):479–496. doi: 10.3109/10715761003667554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trachootham D., Alexandre J., Huang P. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nat. Rev. Drug Discov. 2009;8(7):579–591. doi: 10.1038/nrd2803. [DOI] [PubMed] [Google Scholar]
- 4.Shukla V., Mishra S.K., Pant H.C. Oxidative stress in neurodegeneration. Adv. Pharmacol. Sci. 2011;2011:572634. doi: 10.1155/2011/572634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trachootham D., Lu W., Ogasawara M.A., Nilsa R.D., Huang P. Redox regulation of cell survival. Antioxid. Redox Signal. 2008;10(8):1343–1374. doi: 10.1089/ars.2007.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Finkel T. Signal transduction by reactive oxygen species. J. Cell Biol. 2011;194(1):7–15. doi: 10.1083/jcb.201102095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Janssen-Heininger Y.M., Mossman B.T., Heintz N.H., Forman H.J., Kalyanaraman B., Finkel T. Redox-based regulation of signal transduction: principles, pitfalls, and promises. Free Radic. Biol. Med. 2008;45(1):1–17. doi: 10.1016/j.freeradbiomed.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reya T., Morrison S.J., Clarke M.F., Weissman I.L. Stem cells, cancer, and cancer stem cells. Nature. 2001;414(6859):105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 9.Ager R.R., Davis J.L., Agazaryan A., Benavente F., Poon W.W., LaFerla F.M. Human neural stem cells improve cognition and promote synaptic growth in two complementary transgenic models of Alzheimer's disease and neuronal loss. Hippocampus. 2015;25(7):813–826. doi: 10.1002/hipo.22405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ratanasirintrawoot S., Israsena N. Stem Cells in the Intestine: Possible Roles in Pathogenesis of Irritable Bowel Syndrome. J. Neurogastroenterol. Motil. 2016;22(3):367–382. doi: 10.5056/jnm16023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liang R., Ghaffari S. Stem cells, redox signaling, and stem cell aging. Antioxid. Redox Signal. 2014;20(12):1902–1916. doi: 10.1089/ars.2013.5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jang Y.Y., Sharkis S.J. A low level of reactive oxygen species selects for primitive hematopoietic stem cells that may reside in the low-oxygenic niche. Blood. 2007;110(8):3056–3063. doi: 10.1182/blood-2007-05-087759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naka K., Muraguchi T., Hoshii T., Hirao A. Regulation of reactive oxygen species and genomic stability in hematopoietic stem cells. Antioxid. Redox Signal. 2008;10(11):1883–1894. doi: 10.1089/ars.2008.2114. [DOI] [PubMed] [Google Scholar]
- 14.Forsyth N.R., Musio A., Vezzoni P., Simpson A.H., Noble B.S., McWhir J. Physiologic oxygen enhances human embryonic stem cell clonal recovery and reduces chromosomal abnormalities. Cloning Stem Cells. 2006;8(1):16–23. doi: 10.1089/clo.2006.8.16. [DOI] [PubMed] [Google Scholar]
- 15.Guo Y.L., Chakraborty S., Rajan S.S., Wang R., Huang F. Effects of oxidative stress on mouse embryonic stem cell proliferation, apoptosis, senescence, and self-renewal. Stem Cells Dev. 2010;19(9):1321–1331. doi: 10.1089/scd.2009.0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Domenis R., Bergamin N., Gianfranceschi G., Vascotto C., Romanello M., Rigo S. The redox function of APE1 is involved in the differentiation process of stem cells toward a neuronal cell fate. PLoS One. 2014;9(2):e89232. doi: 10.1371/journal.pone.0089232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Cuevas M., Matunis E.L. The stem cell niche: lessons from the Drosophila testis. Development. 2011;138(14):2861–2869. doi: 10.1242/dev.056242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tan S.W.S., Lee Q.Y., Wong B.S.E., Cai Y., Baeg G.H. Redox homeostasis plays important roles in the maintenance of the Drosophila testis germline stem cells. Stem Cell Rep. 2017;9(1):342–354. doi: 10.1016/j.stemcr.2017.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sykiotis G.P., Bohmann D. Keap1/Nrf2 signaling regulates oxidative stress tolerance and lifespan in Drosophila. Dev. Cell. 2008;14(1):76–85. doi: 10.1016/j.devcel.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Terry N.A., Tulina N., Matunis E., DiNardo S. Novel regulators revealed by profiling Drosophila testis stem cells within their niche. Dev. Biol. 2006;294(1):246–257. doi: 10.1016/j.ydbio.2006.02.048. [DOI] [PubMed] [Google Scholar]
- 21.Chen D., McKearin D. Dpp signaling silences bam transcription directly to establish asymmetric divisions of germline stem cells. Curr. Biol. 2003;13(20):1786–1791. doi: 10.1016/j.cub.2003.09.033. [DOI] [PubMed] [Google Scholar]
- 22.Tahara E.B., Navarete F.D., Kowaltowski A.J. Tissue-, substrate-, and site-specific characteristics of mitochondrial reactive oxygen species generation. Free Radic. Biol. Med. 2009;46(9):1283–1297. doi: 10.1016/j.freeradbiomed.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 23.Iuso A., Scacco S., Piccoli C., Bellomo F., Petruzzella V., Trentadue R. Dysfunctions of cellular oxidative metabolism in patients with mutations in the NDUFS1 and NDUFS4 genes of complex I. J. Biol. Chem. 2006;281(15):10374–10380. doi: 10.1074/jbc.M513387200. [DOI] [PubMed] [Google Scholar]
- 24.Sinenko S.A., Shim J., Banerjee U. Oxidative stress in the haematopoietic niche regulates the cellular immune response in Drosophila. EMBO Rep. 2011;13(1):83–89. doi: 10.1038/embor.2011.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lopez-Fabuel I., Le Douce J., Logan A., James A.M., Bonvento G., Murphy M.P. Complex I assembly into supercomplexes determines differential mitochondrial ROS production in neurons and astrocytes. Proc. Natl. Acad. Sci. USA. 2016;113(46):13063–13068. doi: 10.1073/pnas.1613701113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang Y., Cao Y., Wang Y., Li W., Liu X., Lv Y. Cysteine transporter SLC3A1 promotes breast cancer tumorigenesis. Theranostics. 2017;7(4):1036–1046. doi: 10.7150/thno.18005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amano Y., Mandai M., Yamaguchi K., Matsumura N., Kharma B., Baba T. Metabolic alterations caused by HNF1beta expression in ovarian clear cell carcinoma contribute to cell survival. Oncotarget. 2015;6(28):26002–26017. doi: 10.18632/oncotarget.4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harvey K.F., Zhang X., Thomas D.M. The Hippo pathway and human cancer. Nat. Rev. Cancer. 2013;13(4):246–257. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- 29.Shao D., Zhai P., Del Re D.P., Sciarretta S., Yabuta N., Nojima H. A functional interaction between Hippo-YAP signalling and FoxO1 mediates the oxidative stress response. Nat. Commun. 2014;5:3315. doi: 10.1038/ncomms4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Friling R.S., Bensimon A., Tichauer Y., Daniel V. Xenobiotic-inducible expression of murine glutathione S-transferase Ya subunit gene is controlled by an electrophile-responsive element. Proc. Natl. Acad. Sci. USA. 1990;87(16):6258–6262. doi: 10.1073/pnas.87.16.6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rushmore T.H., Morton M.R., Pickett C.B. The antioxidant responsive element. Activation by oxidative stress and identification of the DNA consensus sequence required for functional activity. J. Biol. Chem. 1991;266(18):11632–11639. [PubMed] [Google Scholar]
- 32.Motohashi H., O'Connor T., Katsuoka F., Engel J.D., Yamamoto M. Integration and diversity of the regulatory network composed of Maf and CNC families of transcription factors. Gene. 2002;294(1–2):1–12. doi: 10.1016/s0378-1119(02)00788-6. [DOI] [PubMed] [Google Scholar]
- 33.Kobayashi A., Kang M.I., Okawa H., Ohtsuji M., Zenke Y., Chiba T. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell Biol. 2004;24(16):7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y., Kern J.T., Walker J.R., Johnson J.A., Schultz P.G., Luesch H. A genomic screen for activators of the antioxidant response element. Proc. Natl. Acad. Sci. USA. 2007;104(12):5205–5210. doi: 10.1073/pnas.0700898104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kensler T.W., Wakabayashi N., Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev. Pharmacol. Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 36.Crawford D.R., Leahy K.P., Wang Y., Schools G.P., Kochheiser J.C., Davies K.J. Oxidative stress induces the levels of a MafG homolog in hamster HA-1 cells. Free Radic. Biol. Med. 1996;21(4):521–525. doi: 10.1016/0891-5849(96)00160-8. [DOI] [PubMed] [Google Scholar]
- 37.Suzuki T., Blank V., Sesay J.S., Crawford D.R. Maf genes are involved in multiple stress response in human. Biochem. Biophys. Res. Commun. 2001;280(1):4–8. doi: 10.1006/bbrc.2000.4064. [DOI] [PubMed] [Google Scholar]
- 38.Katsuoka F., Motohashi H., Ishii T., Aburatani H., Engel J.D., Yamamoto M. Genetic evidence that small maf proteins are essential for the activation of antioxidant response element-dependent genes. Mol. Cell Biol. 2005;25(18):8044–8051. doi: 10.1128/MCB.25.18.8044-8051.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen P.M., Lin C.H., Li N.T., Wu Y.M., Lin M.T., Hung S.C. c-Maf regulates pluripotency genes, proliferation/self-renewal, and lineage commitment in ROS-mediated senescence of human mesenchymal stem cells. Oncotarget. 2015;6(34):35404–35418. doi: 10.18632/oncotarget.6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tapia C., Kutzner H., Mentzel T., Savic S., Baumhoer D., Glatz K. Two mitosis-specific antibodies, MPM-2 and phospho-histone H3 (Ser28), allow rapid and precise determination of mitotic activity. Am. J. Surg. Pathol. 2006;30(1):83–89. doi: 10.1097/01.pas.0000183572.94140.43. [DOI] [PubMed] [Google Scholar]
- 41.Motohashi H., Yamamoto M. Nrf2–Keap1 defines a physiologically important stress response mechanism. Trends Mol. Med. 2004;10(11):549–557. doi: 10.1016/j.molmed.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 42.Veraksa A., McGinnis N., Li X., Mohler J., McGinnis W. Cap 'n' collar B cooperates with a small Maf subunit to specify pharyngeal development and suppress deformed homeotic function in the Drosophila head. Development. 2000;127(18):4023–4037. doi: 10.1242/dev.127.18.4023. [DOI] [PubMed] [Google Scholar]
- 43.Rahman M.M., Sykiotis G.P., Nishimura M., Bodmer R., Bohmann D. Declining signal dependence of Nrf2-MafS-regulated gene expression correlates with aging phenotypes. Aging Cell. 2013;12(4):554–562. doi: 10.1111/acel.12078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barone M.C., Sykiotis G.P., Bohmann D. Genetic activation of Nrf2 signaling is sufficient to ameliorate neurodegenerative phenotypes in a Drosophila model of Parkinson's disease. Dis. Model Mech. 2011;4(5):701–707. doi: 10.1242/dmm.007575. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material. A list of primers used for qRT-PCR analysis.