Abstract
Aims
PLAG1 gene rearrangement is the most common genetic abnormality in pleomorphic adenoma (PA), resulting in overexpression of PLAG1 protein. PA and carcinoma ex pleomorphic adenoma (CA ex-PA) can mimic various benign and malignant salivary gland tumors. The aims of this study are to evaluate the sensitivity and specificity of PLAG1 immunohistochemistry (IHC) in the differential diagnosis of PA and CA ex-PA and to compare the PLAG1 immunohistochemical results to PLAG1 gene abnormalities as detected by fluorescence in situ hybridization (FISH).
Methods and Results
PLAG1 immunostaining was performed on 83 salivary gland tumors including 23 PA, 15 CA ex-PA, and 45 other salivary gland tumors. In addition, PLAG1 FISH was performed in 44 cases for the presence of gene rearrangements/amplifications. The results showed high sensitivity of PLAG1 IHC in 96% of PA; however, discordant results between PLAG1 FISH abnormalities and IHC were noted in 15/44 cases (34%). Seven PA, four de novo myoepithelial carcinomas and one basal cell adenocarcinoma had negative FISH results, but were positive for IHC; while 3 salivary duct carcinomas (SDC) ex-PA were positive for FISH but negative for IHC. PLAG1 IHC can differentiate CA ex-PA from de novo SDC (p=0.02), but not from de novo myoepithelial carcinoma. PLAG1 IHC is a sensitive marker for PA. This could be due to PLAG1 gene abnormalities beyond FISH resolution.
Conclusions
A negative PLAG1 IHC might be helpful in excluding a PA diagnosis. Interestingly in the context of CA ex-PA, FISH is more sensitive than IHC in detecting PLAG1 abnormalities.
Keywords: PLAG1, immunohistochemistry, fluorescence in situ hybridization (FISH), pleomorphic adenoma, salivary gland
INTRODUCTION
Pleomorphic adenoma (PA) is the most common salivary gland neoplasm, accounting for approximately 60% of all epithelial salivary gland tumors 1. Histologically, PA is typically composed of three components of various proportions: epithelial/ductal cells, myoepithelial cells, and myxoid/chondroid stroma 2. However, as PA is known to be associated with marked morphological diversity, distinguishing PA from other salivary gland neoplasms, benign or malignant, may not always be straightforward and may require additional ancillary studies (e.g. immunohistochemistry, cytogenetics, and molecular analysis).
Pleomorphic adenoma gene 1 (PLAG1) is a zinc finger transcription factor and a proto-oncogene located on chromosome 8q12. Fusions involving PLAG1 locus were first discovered in PA in 1997 3, and has since been reported in 24 to 88% of PA and carcinoma ex-pleomorphic adenoma (CA ex-PA) 2, 4–7. PLAG1 fusion appears to be highly specific for PA and CA ex-PA as it has not been detected in other benign or malignant salivary gland neoplasms 2, 4–8. The high prevalence and near 100% specificity of PLAG1 fusion make it an attractive potential target of ancillary diagnostic tests. As PLAG1 primers for polymerase chain reaction (PCR) and PLAG1 probes for fluorescence in situ hybridization (FISH) are not readily available commercially and as molecular and cytogenetics diagnostic laboratory services are usually provided only in large academic centers, there is a demand to seek for an affordable and feasible alternative testing method for PLAG1 translocation.
Fusions between PLAG1 gene and various partners, including CTNNB1 (β-catenin), LIFR (leukemia inhibitory factor receptor), FGFR1 (fibroblast growth factor receptor 1), and CHCHD7 (Coiled-Coil-Helix-Coiled-Coil-Helix Domain Containing 7), leads to overexpression of PLAG1 oncoprotein 2, 4–7. PLAG1 overexpression mediate multiple downstream factors, including the insulin-like growth factor 2 (IGF-2) mitogenic signaling pathway, growth factors and their receptors, tumor suppressors, cell cycle-related proteins, and apoptosis-related proteins, which subsequently influence cell proliferation and tumorigenesis 9–13. Additionally, PLAG1 alteration has been reported in skin and soft tissue myoepithelioma 14, 15, lipoblastoma 16, hepatoblastoma 17, uterine leiomyoma 18, uterine endometrial stromal sarcoma 19, and certain types of acute myeloid leukemia 10.
Several recent studies have shown that PLAG1 overexpression can be detected by immunohistochemistry (IHC), and that PLAG1 IHC is a relatively sensitive marker for PA in surgical and cytologic samples 4, 20–26. However, most of these studies did not include molecular or cytogenetics assays as the gold standard reference test to confirm the presence of PLAG1 fusion and to evaluate the sensitivity and specificity of PLAG1 IHC in detecting PLAG1 fusion.
In the present study, we aimed to characterize PLAG1 IHC in a large cohort of 83 salivary gland tumors, including PA, CA ex-PA, and other types of salivary gland tumors. Moreover, a significant subset of these tumors was examined for PLAG1 fusion in order to determine the sensitivity and specificity of PLAG1 IHC in detecting PLAG1 fusion and in differentiating PA and CA ex-PA from other potential mimickers.
MATERIAL AND METHODS
Case selection and study cohort
Eighty-three patients with epithelial salivary gland neoplasms who had surgery at Memorial Sloan Kettering Cancer Center (New York, NY) between 1995 and 2014 with appropriate material for subsequent IHC and FISH studies were included. The histologic slides were reviewed by a head and neck pathologist (NK) to confirm the diagnosis. The study was approved by the Institutional Review Board of MSKCC. Informed consent was not required for this retrospective study.
Tumor histology
The study cohort was composed of 23 PAs, 15 CAs ex-PA, and a control group of 45 other types of salivary gland carcinomas without a PA component (including ten de novo myoepithelial carcinomas (MECA), six de novo salivary duct carcinomas (SDC), four basal cell adenocarcinomas, eight polymorphous adenocarcinoma (PAC), two epithelial-myoepithelial carcinomas (EMC), four mucoepidermoid carcinomas (MEC), five adenoid cystic carcinomas (ACC), five acinic cell carcinomas (AciCC), and one secretory carcinoma (previously known as mammary analogue secretory carcinoma) (Table 2). The histologic subtypes of the CAs ex-PA were as follow: SDC (n = 9), MECA (n = 3), carcinoma with squamous and glandular features (n = 1), EMC (n = 1), and one adenocarcinoma not otherwise specified (NOS) (n =1).
Table 2.
Sensitivity and specificity of PLAG1 IHC in predicting PLAG1 fusion in PA, CA ex-PA and other salivary gland neoplasms.
| PLAG1 IHC (+) |
PLAG1 IHC (−) | Sensitivity | Specificity | ||
|---|---|---|---|---|---|
| Overall | PLAG1 fusion (+) | 12 | 3 | 80% | 59% |
| PLAG1 fusion (−) | 12 | 17 | |||
| PA | PLAG1 fusion (+) | 5 | 0 | 100% | 30% |
| PLAG1 fusion (−) | 7 | 3 | |||
| CA ex-PA | PLAG1 fusion (+) | 7 | 3 | 70% | 100% |
| PLAG1 fusion (−) | 0 | 2 | |||
| Others | PLAG1 fusion (+) | 0 | 0 | NA | 71% |
| PLAG1 fusion (−) | 5 | 12 | |||
PLAG1 immunohistochemistry
Immunohistochemical stains for PLAG1 were performed using monoclonal antibody clone 3B7 (4ug/ml; Novus Biologicals, Littleton, CO). All immunostains were done on a Leica Bond-3 (Leica, Buffalo Grove, IL) automated stainer platform. Prior to immunohistochemical staining, heat-based antigen retrieval employing a high pH buffer (Leica, ER2) was performed on all slides. As a secondary system, a polymeric detection kit (Refine, Leica) was used. PLAG1 immunopositivity was detected in the nuclei of tumor cells. Both ductal and myoepithelial cells showed positive PLAG1 staining but the immunopositivity was more prevalent in the myoepithelial cells. A tumor was considered as positive for PLAG1 IHC when nuclear immunostain was noted in > 5% of tumor cells.
PLAG1 fluorescence in situ hybridization (FISH)
FISH on interphase nuclei from paraffin-embedded 4-µm sections was performed using custom probes of bacterial artificial chromosomes (BACs) flanking PLAG1 on chromosome 8q12 (Supplementary Table 1) was performed in 44 cases as previously described 2. These 44 cases included 15 PA, 12 CAs ex-PA, 3 de novo SDCs, six de novo MECAs, two basal cell adenocarcinoma, and six PACs.
Two hundred successive nuclei were examined for the presence of PLAG1 gene rearrangements/amplifications using a Zeiss fluorescence microscope (Zeiss Axioplan, Oberkochen, Germany), controlled by Isis 5 software (Metasystems, Waltham, MA). A positive FISH score was interpreted when at least 20% of the nuclei showed a break-apart signal. Nuclei with incomplete set of signals were omitted from the score.
Statistical analysis
All statistical analyses were performed using the SPSS software 22.0 (IBM Corporation, New York, NY, U.S.). The frequency of tumors with PLAG1 immunopositivity as well as the percentage of positive tumor cells in different tumor types was compared using Fisher’s exact test and two-tailed student t test respectively. P values less than 0.05 were considered to be statistically significant.
RESULTS
The results of PLAG1 IHC and PLAG1 FISH are shown in Table 1. PLAG1 IHC was positive in 22 out of 23 (96%) PAs (Figure 1 A-C) and nine out of 15 (60%) CAs ex-PA (Figure 2 A-D). The sensitivity of PLAG1 immunostaining in predicting PA and CA ex-PA were 96% and 60%, respectively. Five out of nine (56%) SDCs ex-PA were positive for PLAG1 IHC, while all six tested de novo SDCs were negative. PLAG1 IHC could differentiate CA ex-PA from de novo SDC (Fisher’s exact test, p = 0.020). Three out of three (100%) MECAs ex-PA and seven out of ten (70%) de novo MECAs were positive for PLAG1 IHC (Figure 2 E-F). PLAG1 was not a useful marker to distinguish CA ex-PA from de novo MECA (Fisher’s exact test, p = 0.601).
Table 1.
Results of PLAG1 Immunohistochemistry and FISH in salivary gland neoplasms.
|
PLAG1 rearrangements by FISHa |
PLAG1 IHCa |
Percentage of tumor cells positive for PLAG1 (mean ± SEM) |
PLAG1 intensity in positive cases (N) |
|||
|---|---|---|---|---|---|---|
| Weak | Moderate | Strong | ||||
| Pleomorphic adenoma (PA) | 5/15 (33%) | 22/23 (96%) | 50±4% | 5 | 4 | 11 |
| Carcinoma ex pleomorphic adenoma (CA ex-PA) | 10/12 (83%) | 9/15 (60%) | 36±7% | 1 | 5 | 3 |
| De novo myoepithelial carcinoma | 0/6 (0%) | 7/10 (70%) | 65±10% | 1 | 1 | 5 |
| De novo salivary duct carcinoma | 0/3 (0%) | 0/6 (0%) | NA | NA | NA | NA |
| Basal cell adenocarcinoma | 0/2 (0%) | 2/4 (50%) | 16±9% | 1 | 1 | NA |
| Polymorphous adenocarcinoma | 0/6 (0%) | 0/8 | NA | NA | NA | NA |
| Epithelial-myoepithelial carcinoma | ND | 1/2 (50%) | 50% | NA | NA | 1 |
| Mucoepidermoid carcinoma | ND | 1/ 4 (25%) | 15% | NA | 1 | NA |
| Adenoid cystic carcinoma | ND | 0/5 | NA | NA | NA | NA |
| Acinic cell carcinoma | ND | 0/5 | NA | NA | NA | NA |
| Secretory carcinoma | ND | 0/1 | NA | NA | NA | NA |
Values are expressed as number of positive cases/total number of cases tested (% of positivity).
IHC: immunohistochemistry; FISH: fluorescence in situ hybridization, NA: not applicable, ND: not performed, SEM: standard error of mean N, number of cases tested.
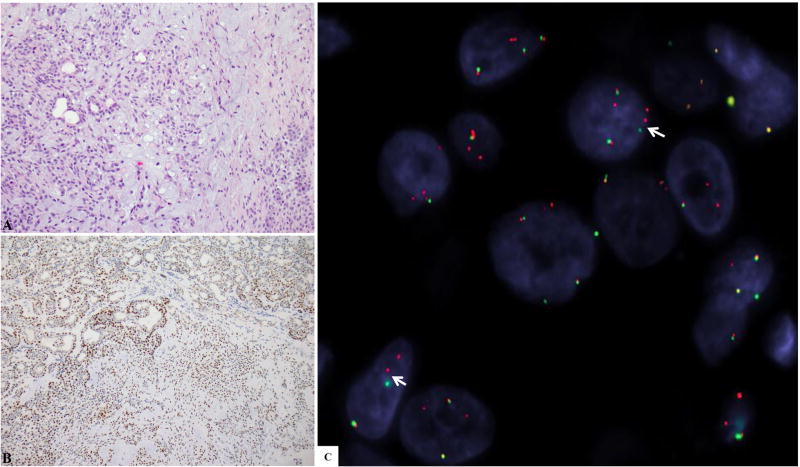
Figure 1.
(A-C): Pleomorphic adenoma (PA). A: H&E stain showing typical histology of PA with ductal structures (epithelial component), myoepithelial cells arranged as cords and nests, and myxoid stroma. B: PLAG1 IHC showing diffuse nuclear labeling. C: FISH for PLAG1 showing break-apart signal (arrow) (red, centromeric; green, telomeric).
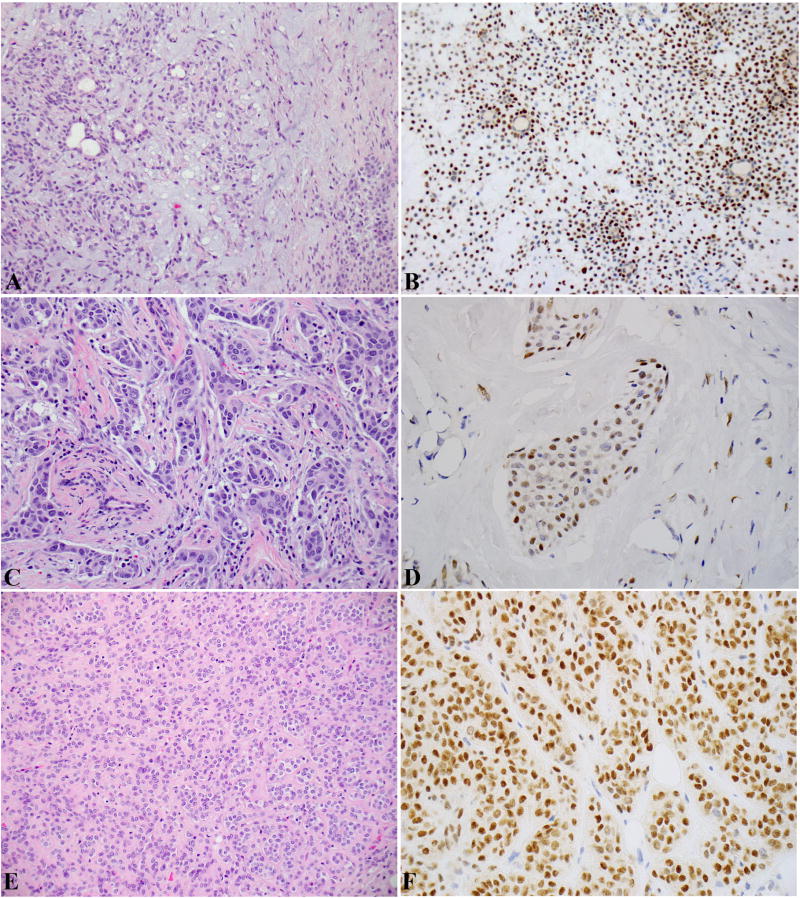
Figure 2.
(A-D): Carcinoma ex-PA (CA ex-PA). A-B: the PA component: histology in (A) and PLAG1 IHC in (B). C-D: the carcinoma component: histology in (C) and PLAG1 IHC in (D). The carcinoma component is a salivary duct carcinoma with typical apocrine cytomorphology and marked nuclear atypia. (E-F): De novo myoepithelial carcinoma: histology in (A) and PLAG1 IHC in (B).
Among the tumors in the control group, 11 out of 45 tumors demonstrated positive PLAG1 immunostaining, showing a specificity of PLAG1 IHC in predicting the PA component of 74%. The histologic types of these 11 tumors were as follow: seven out of ten (70%) de novo MECAs, two out of four (50%) basal cell adenocarcinomas, one out of two (50%) EMCs, and one out of four (25%) MECs. None of the de novo SDCs (0/6), PACs (0/8), ACCs (0/5), AciCCs (0/5), or secretory carcinomas (0/1) showed positive staining for PLAG1.
The majority of the PAs (15/23), CAs ex-PA (8/15), and de novo MECAs (6/10) exhibited moderate or strong PLAG1 immunolabeling. The percentage of positive tumor cells for PLAG1 IHC expressed as mean ± standard error of mean were 50±4%, 36±7%, and 65±10% in PAs, CAs ex-PA, and de novo MECAs, respectively. The percentage of positive staining was significantly lower in CA ex-PA compared to PA (two tailed Student t test, p = 0.008), while there was no significant difference between de novo MECA and PA (p = 0.90), or between MECA and CA ex-PA (p = 0.07).
The FISH test was previously performed on 44 cases, and the results were previously reported by our group 2. The correlation between PLAG1 immunoreactivity and PLAG1 fusion status is shown in Table 2. Overall, PLAG1 IHC had a sensitivity of 80% and specificity of 59% in predicting PLAG1 fusion. Three of 15 tumors with proven PLAG1 fusion on FISH analysis were falsely negative on PLAG1 IHC study. All three cases were CAs ex-PA with the carcinoma component being SDC (n = 2) and carcinoma with squamous and glandular features (or unclassified) (n = 1). Twelve of 30 fusion-negative neoplasms exhibited PLAG1 immunoreactivity, including seven PAs, four de novo MECAS, and one basal cell adenocarcinoma. The specificity of PLAG1 IHC in predicting PLAG1 fusion status was 30% in PA and 100% in CA ex-PA.
DISCUSSION
In 1997, Kas et al. were the first to report PLAG1-CTNNB1 fusion as the key event in the tumorigenesis of pleomorphic adenoma 3, 27. Since then, multiple groups have confirmed PLAG1 rearrangements as the most prevalent molecular event in pleomorphic adenoma and carcinoma ex-pleomorphic adenoma, affecting 24–88% of the tumors 2, 4–8, 27. In these studies, the PLAG1 translocation was detected using techniques that might not be readily available in daily pathology practices, e.g. Northern blot analysis 5, 6, RT-PCR 4, or FISH 2, 7, 8, 27. PLAG1 immunohistochemistry might serve as an accessible and feasible alternative. However, the sensitivity and specificity of PLAG1 IHC in detecting PA and PLAG1 fusion has not yet been well established.
Fusion involving PLAG1 locus results in overexpression of PLAG1 oncoprotein, which can be detected by immunohistochemistry 4, 27. Several recent reports have investigated the utility of PLAG1 IHC as an ancillary tool in diagnosing PA and CA ex-PA in surgical and cytologic specimens. The results of these reports are summarized in Table 3 4, 20–27. All but one study have demonstrated that PLAG1 IHC was a highly sensitive marker for PA in surgical specimens, showing positivity in 93% to 100% of PAs. Only one group has reported a low PLAG1 IHC sensitivity of 62% in 22 lacrimal gland PAs, using the commercially available 3B7 PLAG1 monoclonal antibody (Novus Biologicals, Littleton, CO, US) 26. However, the same group has previously reported 100% PLAG1 immunopositivity in the same cohort using a customized PLAG1 antibody 25. In cytologic samples, the reported percentage of PLAG1 immunoreactivity in PA was relatively low, being 55 – 73%, which might be in part attributed to different preparation methods and/or storage media that were utilized in cytology.
Table 3.
A comprehensive literature review of PLAG1 immunohistochemistry in PA, Ca ex-PA and other salivary gland tumors.
| Ref. | Tumors tested (n) |
Antibody clone |
Criteria for PLAG1 IHC positivity |
PLAG1 immunopositivity in PA/CA ex-PA |
PLAG1 immunopositivity in non-PA |
PLAG1 Fusion, detecting technique |
Sensitivity and specificity of PLAG1 IHC in detecting PLAG1 fusion in PA |
|---|---|---|---|---|---|---|---|
| PLAG1 IHC in pleomorphic adenoma (PA) (surgical specimens) | |||||||
| a | PA (23) Non-PA (45) | Novus 3B7 | > 5% positive cells | 22/23 (96%) | 13/45 (29%) | 5/15 (33%), FISH | Sensitivity: 100% Specificity: 30% |
| 4 | PA (45) Non-PA (42) | Sigma-Aldrich Co., 1:500 | NA | 45/45 (100%) | 3/42 (7%) | 11/45 (24%), RT-PCR | Sensitivity 100% Specificity 0% |
| 20 | PA (36) Non-PA (37) | Novus 3B7 1:20 | Nuclear staining > 5 (intensity X %) | 33/36 (92%) | 17/37 (46%) | NA | NA |
| 21 | PA (40) | Novus 3B7 1:100 | > 10% positive cells | 37/40 (93%) | NA | NA | NA |
| 22b | PA (76) | Novus 3B7 1:100 | > 10% positive cells | 71/76 (94%) | NA | NA | NA |
| 23 | PA (36) Non-PA (64) | Novus 3B7 1:25 | Any nuclear staining | 34/36 (94%) | 1110/64 (16%) | NA | NA |
| 25, 26 | PA (26) | Astrom et al. 2000 16 | Any nuclear staining | 26/26 (100%) | NA | NA | NA |
| PA (28) | Novus 3B7 | NA | 16/26 (62%) | NA | NA | NA | |
| PLAG1 IHC in pleomorphic adenoma (PA) (cytologic specimens) | |||||||
| 24 | PA (30) Non-PA (39) | Novus 3B7 1:100 | Moderate to strong nuclear staining | 22/30 (73%) | 4/39 (10%) | NA | NA |
| 20 | PA (40) Non-PA (12) | Novus 3B7 1:20 | Nuclear staining > 5 (intensity X %) | 22/40 (55%) | 3/12 (25%) | NA | NA |
| PLAG1 IHC in carcinoma ex pleomorphic adenoma | |||||||
| a | CA ex-PA | Novus 3B7 1:20 | > 5% positive cells | 9/15 (60%) | NA | 12/14 (86%) FISH | Sensitivity: 70% Specificity: 100% |
| 27 | CA ex-PA (22) Non-PA (39) | Novus 3B7 | >50% positive cells | 17/22 (77%) | NA | 12/18 (67%), FISH | Sensitivity: 92% Specificity: 17% |
| 4 | CA ex-PA (4) | Sigma-Aldrich Co., 1:500 | NA | 4/4 (100%) | NA | 0/4 (0%) RT-PCR | Sensitivity: NA Specificity: 0% |
| 23 | CA ex-PA (1) | Novus 3B7 1:25 | Any nuclear staining | 1/1 (100%) | NA | NA | NA |
| 21 | CA ex-PA (40) | Novus 3B7 1:100 | > 10% positive cells | 14/40 (35%) | NA | NA | NA |
| 25, 26 | CA ex-PA (5) | Novus 3B7 | NA | 1/5 (20%) | NA | NA | NA |
| CA ex-PA (5) | Astrom et al 2000 16 | Any nuclear staining | 3/5 (60%) | NA | NA | NA | |
Data from the present study.
These 76 patients included the 40 patients studied in the above row.
NA: Not available, RT-PCR: reverse-transcription polymerase chain reaction.
Compared to PA, the reported PLAG1 IHC immunopositivity appeared to be more variable in CA ex-PA, ranging from 20% to 100% 4, 21, 23, 25–27. In this study, PLAG1 IHC was positive in 60% of the tested CA ex PA cases. Several mechanisms may explain the wide range of PLAG1 immunopositivity in CA ex-PA. First, many types of salivary gland tumors can show hyalinizing stroma mimicking the PA component of CA ex PA; therefore, some of the reported negative PLAG1 IHC cases of CA ex-PA could have been misclassified. Second, PLAG1 oncoprotein overexpression may be lost during the process of tumor progression or malignant transformation, leading to low PLAG1 expression and subsequently negative PLAG1 IHC. Taken together, PLAG1 IHC emerges as a highly sensitive marker for PA and but less reliable in detecting CA ex-PA.
The reported specificity of PLAG1 IHC as a marker for PA and CA ex PA seemed to be relatively low. Several previous studies have reported positive PLAG1 IHC in non-PA and non-CA ex-PA tumors (7% and 46%, respectively) 4, 20, 23, 24. In this study, PLAG1 IHC was positive in 29% of other types of tumors. PLAG1 positive staining has been reported in various salivary gland neoplasms, malignant or benign, including myoepithelioma, basal cell adenoma, MECA, ACC, PAC AciCC, and poorly differentiated basaloid carcinoma 4, 20, 23, 24. In the present study, PLAG1 IHC was found positive in a majority (7/10, 70%) of de novo MECA carcinoma, despite that none of these tumors showed PLAG1 gene rearrangements by FISH. This result limits the utility of PLAG IHC in distinguishing de novo MECA from PA which is not always easily done by morphology. Additionally, we found PLAG1 to be positive in a proportion of basal cell adenocarcinoma, EMC, and mucoepidermoid carcinoma, but not in de novo SDC, polymorphous adenocarcinoma, ACC, AciCC, and secretory carcinoma. In our hands, PLAG1 IHC seems to a helpful immunomarker in distinguishing between de novo SDC and CA ex-PA, but not between de novo MECA and CA ex-PA.
Two prior studies have correlated PLAG1 immunoreactivity with PLAG1 fusion status 4, 27. In one study, using RT-PCR with customized primers for PLAG1 and PLAG1 IHC, Matsuyama et al. have reported that the sensitivity and specificity of PLAG1 IHC in detecting PLAG1 fusion were 100% and 0%, respectively. In the second study, Bahrami et al. 27 have reported 67% (12/18) of CA ex-PA with PLAG1 fusion and 77% (17/22) with positive PLAG1 by IHC. The sensitivity and specificity of PLAG1 IHC in detecting PLAG1 fusion in CA ex-PA were 92% and 17%, respectively. In the present study, the sensitivity and specificity of PLAG1 IHC compared to PLAG1 fusion were 100% and 30% in PA and 70% and 100% in CA ex-PA. Taken together, it appears that PLAG1 IHC is a sensitive but not specific test in predicting PLAG1 fusion. A negative PLAG1 IHC might be helpful in excluding PLAG1 fusion and a PA diagnosis, but a positive PLAG1 IHC may not always predict the existence of PLAG1 fusion.
CONCLUSIONS
PLAG1 IHC is a sensitive marker and a valuable ancillary test for PA and CA ex-PA, and may point to PLAG1 gene abnormalities that are characteristics in these tumors. Thus, a negative PLAG1 IHC might be more reliable in excluding a PA diagnosis. However, the reverse remains to be determined if PLAG1 IHC expression in the absence of a positive FISH result is non-specific or implies alternative genetic or epigenetic mechanisms beyond the FISH resolution.
Supplementary Material
Acknowledgments
Source of Funding:
Research reported in this publication was supported in part by the Cancer Center Support Grant of the National Institutes of Health/National Cancer Institute under award number P30CA008748 and P50 CA140146-01 (CRA). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflicts of Interest:
The authors have disclosed that they have no significant relationships with, or financial interest in any commercial companies pertaining to this article.
Authors’ contributions:
Nora Katabi: Study design, pathology review, PLAG1 IHC evaluation, review and revise the manuscript
Bin Xu: Database management, statistics, draft the manuscript
Achim A. Jungbluth: Pathology review, perform and evaluate PLAG1 IHC
Lei Zhang: Perform and evaluate PLAG1 FISH
Sung Yun Shao: Perform and evaluate PLAG1 FISH
Jason Lane: Pathology review, perform and evaluate PLAG1 IHC
Ronald Ghossein: Pathology review and review and revise the manuscript
Cristina R Antonescu: Study design, perform and evaluate PLAG1 FISH, review and revise the manuscript
References
- 1.Barnes EL, Eveson JW, Reichart P, Sidransky D. World health organization classification of tumours: Pathology and genetics of head and neck tumours. Lyon: International Agency for Research on Cancer (IARC); 2005. p. 430. [Google Scholar]
- 2.Katabi N, Ghossein R, Ho A, et al. Consistent plag1 and hmga2 abnormalities distinguish carcinoma ex-pleomorphic adenoma from its de novo counterparts. Hum Pathol. 2015;46:26–33. doi: 10.1016/j.humpath.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kas K, Voz ML, Roijer E, et al. Promoter swapping between the genes for a novel zinc finger protein and beta-catenin in pleiomorphic adenomas with t(3;8)(p21;q12) translocations. Nature genetics. 1997;15:170–174. doi: 10.1038/ng0297-170. [DOI] [PubMed] [Google Scholar]
- 4.Matsuyama A, Hisaoka M, Nagao Y, Hashimoto H. Aberrant plag1 expression in pleomorphic adenomas of the salivary gland: A molecular genetic and immunohistochemical study. Virchows Arch. 2011;458:583–592. doi: 10.1007/s00428-011-1063-4. [DOI] [PubMed] [Google Scholar]
- 5.Astrom AK, Voz ML, Kas K, et al. Conserved mechanism of plag1 activation in salivary gland tumors with and without chromosome 8q12 abnormalities: Identification of sii as a new fusion partner gene. Cancer Res. 1999;59:918–923. [PubMed] [Google Scholar]
- 6.Voz ML, Astrom AK, Kas K, Mark J, Stenman G, Van de Ven WJ. The recurrent translocation t(5;8)(p13;q12) in pleomorphic adenomas results in upregulation of plag1 gene expression under control of the lifr promoter. Oncogene. 1998;16:1409–1416. doi: 10.1038/sj.onc.1201660. [DOI] [PubMed] [Google Scholar]
- 7.Martins C, Fonseca I, Roque L, et al. Plag1 gene alterations in salivary gland pleomorphic adenoma and carcinoma ex-pleomorphic adenoma: A combined study using chromosome banding, in situ hybridization and immunocytochemistry. Mod Pathol. 2005;18:1048–1055. doi: 10.1038/modpathol.3800386. [DOI] [PubMed] [Google Scholar]
- 8.Chiosea SI, Thompson LD, Weinreb I, et al. Subsets of salivary duct carcinoma defined by morphologic evidence of pleomorphic adenoma, plag1 or hmga2 rearrangements, and common genetic alterations. Cancer. 2016 doi: 10.1002/cncr.30179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, Shang W, Lei X, et al. Opposing functions of plag1 in pleomorphic adenoma: A microarray analysis of plag1 transgenic mice. Biotechnol Lett. 2013;35:1377–1385. doi: 10.1007/s10529-013-1213-7. [DOI] [PubMed] [Google Scholar]
- 10.Van Dyck F, Declercq J, Braem CV, Van de Ven WJ. Plag1, the prototype of the plag gene family: Versatility in tumour development (review) Int J Oncol. 2007;30:765–774. [PubMed] [Google Scholar]
- 11.Declercq J, Van Dyck F, Braem CV, et al. Salivary gland tumors in transgenic mice with targeted plag1 proto-oncogene overexpression. Cancer Res. 2005;65:4544–4553. doi: 10.1158/0008-5472.CAN-04-4041. [DOI] [PubMed] [Google Scholar]
- 12.Juma AR, Damdimopoulou PE, Grommen SV, Van de Ven WJ, De Groef B. Emerging role of plag1 as a regulator of growth and reproduction. J Endocrinol. 2016;228:R45–56. doi: 10.1530/JOE-15-0449. [DOI] [PubMed] [Google Scholar]
- 13.Voz ML, Agten NS, Van de Ven WJ, Kas K. Plag1, the main translocation target in pleomorphic adenoma of the salivary glands, is a positive regulator of igf-ii. Cancer Res. 2000;60:106–113. [PubMed] [Google Scholar]
- 14.Antonescu CR, Zhang L, Shao SY, et al. Frequent plag1 gene rearrangements in skin and soft tissue myoepithelioma with ductal differentiation. Genes Chromosomes Cancer. 2013;52:675–682. doi: 10.1002/gcc.22063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bahrami A, Dalton JD, Krane JF, Fletcher CD. A subset of cutaneous and soft tissue mixed tumors are genetically linked to their salivary gland counterpart. Genes Chromosomes Cancer. 2012;51:140–148. doi: 10.1002/gcc.20938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Astrom A, D'Amore ES, Sainati L, et al. Evidence of involvement of the plag1 gene in lipoblastomas. Int J Oncol. 2000;16:1107–1110. doi: 10.3892/ijo.16.6.1107. [DOI] [PubMed] [Google Scholar]
- 17.Zatkova A, Rouillard JM, Hartmann W, et al. Amplification and overexpression of the igf2 regulator plag1 in hepatoblastoma. Genes Chromosomes Cancer. 2004;39:126–137. doi: 10.1002/gcc.10307. [DOI] [PubMed] [Google Scholar]
- 18.Mehine M, Kaasinen E, Heinonen HR, et al. Integrated data analysis reveals uterine leiomyoma subtypes with distinct driver pathways and biomarkers. Proc Natl Acad Sci U S A. 2016;113:1315–1320. doi: 10.1073/pnas.1518752113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davidson B, Abeler VM, Hellesylt E, et al. Gene expression signatures differentiate uterine endometrial stromal sarcoma from leiomyosarcoma. Gynecol Oncol. 2013;128:349–355. doi: 10.1016/j.ygyno.2012.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Avadhani V, Cohen C, Siddiqui MT. Plag1: An immunohistochemical marker with limited utility in separating pleomorphic adenoma from other basaloid salivary gland tumors. Acta Cytol. 2016;60:240–245. doi: 10.1159/000447622. [DOI] [PubMed] [Google Scholar]
- 21.de Brito BS, Giovanelli N, Egal ES, et al. Loss expression of plag1 in malignant transformation from pleomorphic adenoma to carcinoma ex-pleomorphic adenoma. Hum Pathol. 2016 doi: 10.1016/j.humpath.2016.07.011. [DOI] [PubMed] [Google Scholar]
- 22.de Brito BS, Gaspar NG, Egal ES, et al. Plag1 expression is maintained in recurrent pleomorphic adenoma. Virchows Arch. 2016;469:477–481. doi: 10.1007/s00428-016-1980-3. [DOI] [PubMed] [Google Scholar]
- 23.Rotellini M, Palomba A, Baroni G, Franchi A. Diagnostic utility of plag1 immunohistochemical determination in salivary gland tumors. Appl Immunohistochem Mol Morphol. 2014;22:390–394. doi: 10.1097/PAI.0b013e3182936ea7. [DOI] [PubMed] [Google Scholar]
- 24.Foo WC, Jo VY, Krane JF. Usefulness of translocation-associated immunohistochemical stains in the fine-needle aspiration diagnosis of salivary gland neoplasms. Cancer Cytopathol. 2016;124:397–405. doi: 10.1002/cncy.21693. [DOI] [PubMed] [Google Scholar]
- 25.von Holstein SL. Tumours of the lacrimal gland. Epidemiological, clinical and genetic characteristics. Acta Ophthalmol. 2013;91 Thesis 6:1–28. doi: 10.1111/aos.12271. [DOI] [PubMed] [Google Scholar]
- 26.von Holstein SL, Fehr A, Persson M, et al. Lacrimal gland pleomorphic adenoma and carcinoma ex pleomorphic adenoma: Genomic profiles, gene fusions, and clinical characteristics. Ophthalmology. 2014;121:1125–1133. doi: 10.1016/j.ophtha.2013.11.037. [DOI] [PubMed] [Google Scholar]
- 27.Bahrami A, Dalton JD, Shivakumar B, Krane JF. Plag1 alteration in carcinoma ex pleomorphic adenoma: Immunohistochemical and fluorescence in situ hybridization studies of 22 cases. Head Neck Pathol. 2012;6:328–335. doi: 10.1007/s12105-012-0353-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.