Abstract
The chaperone protein and guanine nucleotide exchange factor SmgGDS (RAP1GDS1) is a key promoter of cancer cell proliferation and tumorigenesis. SmgGDS undergoes nucleocytoplasmic shuttling, suggesting that it has both cytoplasmic and nuclear functions that promote cancer. Previous studies indicate that SmgGDS binds cytoplasmic small GTPases and promotes their trafficking to the plasma membrane. In contrast, little is known about the functions of SmgGDS in the nucleus, or how these nuclear functions might benefit cancer cells. Here we show unique nuclear localization and regulation of gene transcription pathways by SmgGDS. Strikingly, SmgGDS depletion significantly reduces expression of over 600 gene products that are targets of the DREAM complex, which is a transcription factor complex that regulates expression of proteins controlling the cell cycle. The cell cycle regulators E2F1, MYC, MYBL2 (B-Myb) and FOXM1 are among the DREAM targets that are diminished by SmgGDS depletion. E2F1 is well known to promote G1 cell cycle progression, and the loss of E2F1 in SmgGDS-depleted cells provides an explanation for previous reports that SmgGDS depletion characteristically causes a G1 cell cycle arrest. We show that SmgGDS localizes in nucleoli, and that RNAi-mediated depletion of SmgGDS in cancer cells disrupts nucleolar morphology, signifying nucleolar stress. We show that nucleolar SmgGDS interacts with the RNA polymerase I transcription factor upstream binding factor (UBF). The RNAi-mediated depletion of UBF diminishes nucleolar localization of SmgGDS and promotes proteasome-mediated degradation of SmgGDS, indicating that nucleolar sequestration of SmgGDS by UBF stabilizes SmgGDS protein. The ability of SmgGDS to interact with UBF and localize in the nucleolus is diminished by expressing DiRas1 or DiRas2, which are small GTPases that bind SmgGDS and act as tumor suppressors. Taken together, our results support a novel nuclear role for SmgGDS in protecting malignant cells from nucleolar stress, thus promoting cell cycle progression and tumorigenesis.
Introduction
The chaperone protein SmgGDS (RAP1GDS1) interacts with multiple small GTPases, including Rac1, K-Ras, RhoA, Rap1 and DiRas,1, 2, 3, 4, 5, 6, 7 and is overexpressed in lung,8 breast3 and prostate9 cancer. SmgGDS promotes malignancy by stimulating cell proliferation, colony formation, NF-κB activity and cell migration.1, 2, 3, 8, 9, 10 Two known isoforms of SmgGDS are expressed in cells; the longer isoform is named SmgGDS-607 (NCBI accession #NP_001093897, isoform 3) and a shorter splice variant is called SmgGDS-558 (NCBI accession #NP_001093899, isoform 5).1 The RNAi-mediated depletion of SmgGDS-558 significantly diminishes the malignant phenotype of lung, breast and pancreatic cancer cell lines,1, 3, 10 and slows tumorigenesis of human lung cancer and breast cancer xenografts in immunodeficient mice.3, 10 In contrast, the RNAi-mediated depletion of SmgGDS-607 has negligible effects on the malignant phenotype or on tumorigenesis in mouse models.1, 3, 10 This result might occur because SmgGDS-607 does not promote malignancy, or alternatively, the RNAi treatments used in previous studies did not reduce SmgGDS-607 levels low enough to detect biological effects. Despite the uncertain role of SmgGDS-607 in cancer, it is clear that SmgGDS-558 induces multiple events that enhance malignancy. Most notably, SmgGDS-558 has emerged as an important participant in the cell cycle of malignant cells.
SmgGDS-558 is a promoter of G1 cell cycle progression in lung, breast and pancreatic cancer.10 SmgGDS-558 promotes G1 progression in part by increasing expression of the pro-proliferative protein Cyclin D and decreasing expression of the anti-proliferative proteins p21 and p27,10 as well as through promotion of NF-κB transcriptional activity.2, 3 The molecular mechanisms utilized by SmgGDS-558 to enhance these proliferative events are not well defined. We previously proposed a model in which cytoplasmic SmgGDS-558 cooperates with SmgGDS-607 to promote the prenylation and subsequent membrane trafficking of small GTPases, potentially promoting malignancy through increased signaling by small GTPases at the plasma membrane.1 While the role of SmgGDS-558 in promoting proliferation is likely mediated through its cytoplasmic interactions with small GTPases, it may also be mediated by additional as-yet-unidentified mechanisms. In this study, we focused on the nuclear functions of SmgGDS-558, as SmgGDS-558 has a nuclear export sequence (NES) and undergoes nucleocytoplasmic shuttling.11
We report here that SmgGDS-558 protects cancer cells from nucleolar stress, providing a novel mechanism to explain why SmgGDS-558 is needed for cell cycle progression in malignant cells. Protection from nucleolar stress is crucial for the development and progression of malignancy, because the nucleolus provides specific functions that promote the proliferation and survival of malignant cells.12, 13, 14, 15, 16 In addition to being the site of ribosomal RNA (rRNA) generation needed for increased ribosome biogenesis during accelerated proliferation, the nucleolus acts as a major hub of oncogenic signaling by sequestering and releasing proteins involved in the p53, retinoblastoma (Rb) and NF-κB signaling pathways.17, 18, 19, 20, 21, 22 Both p53 and Rb regulate the cell cycle by controlling transcriptional targets that are tightly integrated with and regulated by the DREAM complex, which is a transcription factor complex that is a master coordinator of cell cycle-dependent gene expression.23, 24, 25, 26, 27 Remarkably, we observed that the RNAi-mediated depletion of SmgGDS-558 significantly reduces expression of over 600 gene products associated with the DREAM complex, including the major cell cycle regulators E2F1, Myc, B-Myb and FoxM1. This profound loss of gene expression upon depletion of SmgGDS-558 is most likely due to nucleolar stress, because we demonstrate for the first time that SmgGDS-558 is sequestered in the nucleolus by associating with the resident nucleolar protein upstream binding factor (UBF), and SmgGDS-558 depletion alters nucleolar structure indicative of nucleolar stress. Furthermore, we demonstrate that both the interaction of SmgGDS-558 with UBF, and the nucleolar sequestration of SmgGDS-558, are diminished by expression of DiRas1 and DiRas2, which are tumor suppressor GTPases that bind SmgGDS,2, 4 indicating that DiRas proteins might act as tumor suppressors by inhibiting nucleolar functions of SmgGDS-558. Taken together, our demonstration that targeting SmgGDS in cancer promotes nucleolar stress and impacts expression of DREAM complex target genes provides a novel and previously unsuspected mechanism by which SmgGDS promotes malignancy.
Results
SmgGDS depletion diminishes expression of DREAM target genes required for cell cycle progression
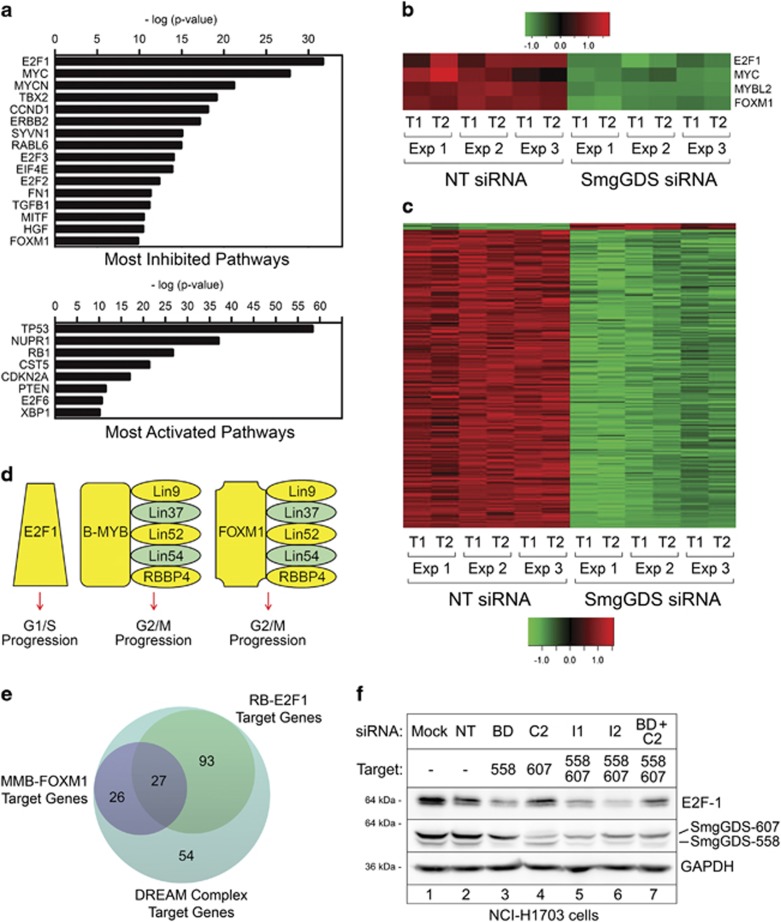
To begin defining the pathways that are regulated by SmgGDS, we used RNA-sequencing and bioinformatic analysis to identify gene networks that are disrupted by depleting SmgGDS-558 and SmgGDS-607 in NCI-H1703 cells using siRNA I1 (Supplementary Table 1). This siRNA has been well characterized in previous studies to diminish expression of both isoforms of SmgGDS.1, 3, 8, 9, 10 Ingenuity Pathway Analysis revealed that the highly oncogenic E2F1 and MYC pathways are the most significantly inhibited pathways in NCI-H1703 cells depleted of SmgGDS (E2F1, Z-score=−4.3, P=2 × 10−32; MYC, Z-score=−5.9, P=1.73 × 10−28; Figure 1a). The two major tumor suppressive pathways, TP53 and RB1, are among the top three most significantly activated pathways in these cells (TP53, Z-score=3.0, P=5.11 × 10−59; RB1, Z-score=4.3, P=2.12 × 10−27; Figure 1a). Among the most significantly downregulated genes in SmgGDS-depleted cells are E2F1, MYBL2 (B-Myb), FoxM1 and MYC, which are transcription factors that promote the cell cycle23, 25, 28, 29 (Figure 1b).
Figure 1.
Depletion of SmgGDS diminishes expression of DREAM complex target genes required for cell cycle progression. (a) Ingenuity Pathway Analysis of microarray data from NCI-H1703 cells depleted of both SmgGDS-558 and SmgGDS-607 using siRNA I1 shows significant effects on gene expression within the indicated pathways. (b, c) Heatmaps display changes in expression of the 200 DREAM target genes that were most significantly altered in NCI-H1703 cells treated with siRNA I1, generated in two technical replicates (T1 and T2) from three independent experiments (Exp 1, 2 and 3). (d) Protein complexes containing E2F1, B-MYB, FOXM1 and Lin family members promote cell cycle progression, and yellow highlights identify specific gene products that exhibit reduced expression in NCI-H1703 cells following depletion of SmgGDS with siRNA I1. (e) The 200 DREAM target genes that were most significantly altered in SmgGDS-depleted cells (shown in c) were analyzed using the targetgenereg.org24 database to predict which of these target genes are regulated by Rb-E2F1 or MMB-FOXM1 complexes. (f) Immunoblotting was used to examine E2F1 protein expression in NCI-H1703 cells following depletion of different SmgGDS isoforms using specific siRNAs (n=3). Mean normalized densitometry values are shown in Supplementary Figure S1b.
E2F1, B-Myb and FoxM1 are well-established targets of the DREAM complex, which regulates expression of genes associated with the cell cycle.23, 24, 25, 26, 27 Fischer et al.24 expanded the number of transcriptional targets regulated by the DREAM complex to 971 genes, and we found that 728 of these genes were differentially expressed following SmgGDS depletion (75%, P<2.2 × 10−16; Supplementary Figure S1a; Supplementary Table 2). Of these 728 genes, 629 were downregulated following SmgGDS depletion (Supplementary Table 2). Figure 1c shows a heatmap of the 200 DREAM targets that were most significantly changed upon SmgGDS depletion; remarkably, 98% of these 200 genes were downregulated in cells depleted of SmgGDS (Figure 1c).
E2F1, B-Myb and FoxM1 form unique protein complexes that act as transcriptional regulators with different roles in the cell cycle. E2F associates with Rb and regulates expression of proteins needed for G1 progression. In contrast, proteins needed for G2/M progression are transcriptionally regulated by B-Myb and/or FoxM1 in association with the MuvB complex, which consists of RBBP4 and multiple Lin family proteins.23, 24, 25, 26 SmgGDS depletion diminishes expression of the majority of the genes involved in these regulatory complexes (Figure 1d; Supplementary Table 2). To determine whether SmgGDS depletion preferentially diminishes expression of genes regulated by E2F-Rb or by MuvB, B-Myb and FOXM1 complexes (collectively designated as MMB-FOXM1), we utilized a database (targetgenereg.org) that predicts which genes are regulated by these different complexes.24 Of the 200 DREAM targets that are most significantly altered in SmgGDS-depleted cells (Figure 1c), we found that 120 genes (60%) are predicted to be regulated by E2F-Rb, 53 genes (26.5%) are predicted to be regulated by MMB-FOXM1 and 27 genes (13.5%) are predicted to be regulated by both MMB-FOXM1 and E2F-Rb (Figure 1e). Our observation that SmgGDS depletion preferentially diminishes expression of genes predicted to be regulated by E2F-Rb, which is a promoter of G1 progression (Figures 1d and e), is consistent with reports that SmgGDS depletion causes malignant cells to arrest in G1.8, 10
Previous studies indicate that G1 arrest is induced by depleting SmgGDS-558 alone,10 or both SmgGDS isoforms simultaneously,8, 10 but not SmgGDS-607 alone.10 To investigate the role of E2F1 in these differential effects of SmgGDS-558 and SmgGDS-607, we examined E2F1 protein levels in lung cancer cells after depleting one or both SmgGDS isoforms using siRNAs that deplete either SmgGDS-607 (siRNA C2) or SmgGDS-558 (siRNA BD) individually, or both isoforms simultaneously (siRNA I1 and I2).1, 3, 8, 9, 10 We found that E2F1 protein levels are significantly reduced by depleting SmgGDS-558 alone (Figure 1f, lane 3) or both SmgGDS isoforms simultaneously (Figure 1f, lanes 5–7), but not SmgGDS-607 alone (Figure 1f, lane 4; Supplementary Figure S1b). Taken together, these findings identify SmgGDS as an important regulator of DREAM target gene expression, providing an explanation for the loss of cell cycle progression when SmgGDS is depleted.1, 3, 8, 9, 10
Interestingly, gene expression regulated by the DREAM complex is tightly integrated with p53-dependent pathways,23, 24, 25, 26, 30 and p53-dependent pathways are commonly induced by nucleolar stress.16, 18, 19, 20, 21, 31, 32 Nucleolar stress responses can also occur independently of p53, and are characterized by reduced expression of specific genes, including E2F1 and c-Myc.21, 31, 32, 33 Our findings that SmgGDS depletion promotes expression of p53-associated pathways (Figure 1a), decreases DREAM target gene expression (Figure 1c) and causes loss of E2F1 and MYC (Figure 1b), coupled with the ability of SmgGDS to enter the nucleus,11 prompted us to investigate whether targeting SmgGDS in cancer cells induces nucleolar stress.
SmgGDS accumulates in the nucleolus and regulates nucleolar morphology
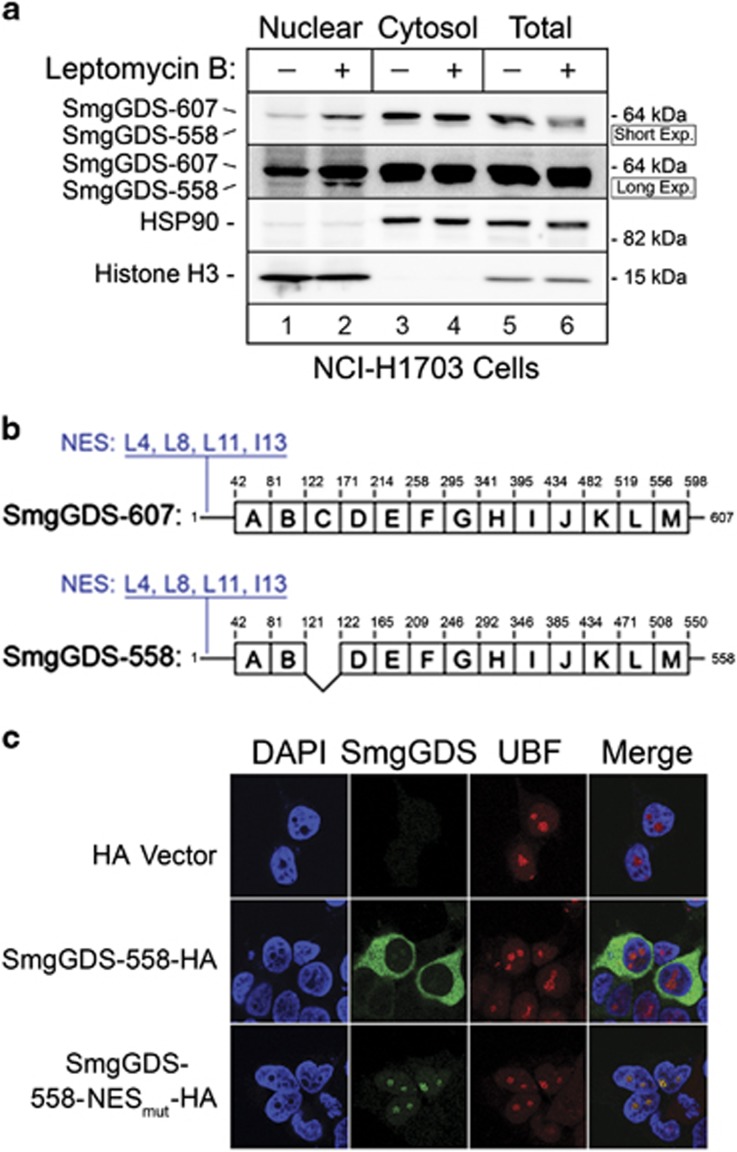
We reported11 that SmgGDS has a functional NES at its N terminus and hemagglutinin (HA)-tagged wild-type SmgGDS accumulates in the nucleus of cells treated with leptomycin B, which slows nuclear export by inhibiting exportin 1. Consistent with this report, endogenous SmgGDS accumulates in the nucleus approximately twofold in NCI-H1703 cells treated with leptomycin B (Figure 2a; Supplementary Figure S2). We mutated the NES in SmgGDS-558 to generate SmgGDS-558-NESmut (Figure 2b) and observed significant nucleolar accumulation of SmgGDS-558-NESmut, as evidenced by colocalization of SmgGDS-558-NESmut with the nucleolar marker UBF (Figure 2c). The nucleolar localization of SmgGDS-558 prompted us to examine how the RNAi-mediated depletion of SmgGDS impacts nucleolar events.
Figure 2.
SmgGDS undergoes nucleocytoplasmic shuttling and can localize in the nucleolus. (a) Immunoblotting was used to detect endogenous SmgGDS in the nuclear fraction (50 μg protein/lane), cytosolic fraction (10 μg protein/lane) and total cell lysates (10 μg protein/lane) prepared from NCI-H1703 cells treated with or without leptomycin B (25 nm, 8 h; n=3). Immunoblotting using antibodies to the cytosolic protein HSP90 and the nuclear protein Histone H3 indicated purity of the subcellular fractions. Two exposures of the SmgGDS immunoblot are shown to visualize both isoforms of SmgGDS. Mean normalized densitometry values are shown in Supplementary Figure S2. (b) Both isoforms of SmgGDS contain multiple armadillo repeats (labeled A–M)1 and an N-terminal NES. Four residues within the NES (L4, L8, L11 and I13) of SmgGDS-558 were mutated to alanine to generate SmgGDS-558-NESmut that has reduced nuclear export.11 (c) HEK293T cells expressing SmgGDS-558-HA, SmgGDS-558-NESmut-HA, or the HA vector (control cells) were immunofluorescently stained with HA antibody, UBF antibody and 4,6-diamidino-2-phenylindole (DAPI), and examined by confocal fluorescence microscopy (n=3).
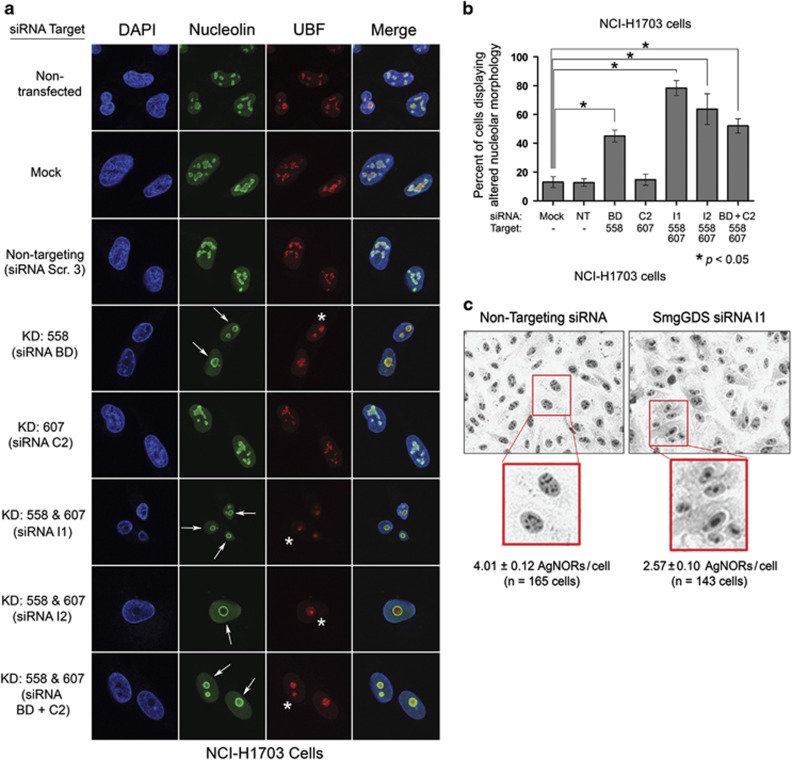
Nucleolar morphology in NCI-H1703 cells is disrupted by the RNAi-mediated depletion of SmgGDS-558 alone, or both SmgGDS-558 and SmgGDS-607 simultaneously (Figures 3a and b). Nucleolar disruption is indicated by circular redistribution of nucleolin to the outer edge of nucleoli (Figure 3a; Supplementary Figure S3). Depleting both SmgGDS isoforms simultaneously also causes UBF condensation and subsequent formation of UBF caps at the nucleolar periphery (Figure 3a). Interestingly, depleting SmgGDS-607 alone did not detectably disrupt nuclear morphology (Figures 3a and b), consistent with our finding that depleting SmgGDS-607 alone does not alter cell cycle progression3, 10 or E2F1 expression (Figure 1f; Supplementary Figure S1b). These changes in the distribution of nucleolin and UBF induced by SmgGDS-558 depletion signify nucleolar segregation, which is a well-known indicator of nucleolar stress.18, 19, 31
Figure 3.
The RNAi-mediated depletion of SmgGDS-558 disrupts nucleolar morphology. (a) NCI-H1703 cells were transfected with the indicated siRNAs to deplete SmgGDS isoforms, and 72 h later the cells were immunofluorescently stained with nucleolin antibody, UBF antibody and 4,6-diamidino-2-phenylindole (DAPI), and examined by confocal fluorescence microscopy. Arrows indicate circular redistribution of nucleolin, and asterisks indicate UBF cap formation (n=3). (b) NCI-H1703 cells treated the same as in a were scored for disruption of nucleolar morphology according to the key described in Supplementary Figure S3. Scoring was conducted without knowledge of the siRNAs used to treat the cells, and values represent the mean±s.e.m. from 100 cells scored in three independent experiments. Statistical significance was determined by one-way repeated measures analysis of variance and Dunnett’s multiple comparison test (*P<0.05), comparing each condition to non-targeting (NT) siRNA. (c) NCI-H1703 cells were transfected with siRNA I1 to deplete SmgGDS, or with NT siRNA, and 72 h later subjected to AgNOR staining. Phase contrast images were collected, and AgNORs/cell was counted without knowledge of the siRNAs used to treat the cells (n=3).
UBF1 is argyrophilic and participates in the formation of nucleolar organizing regions (NORs). The UBF1-containing NORs detected in silver-stained cells are called AgNORs, and reduced AgNOR levels are associated with altered nucleolar function and loss of cell proliferation.15, 34, 35, 36 Depleting both SmgGDS-558 and SmgGDS-607 simultaneously in NCI-H1703 cells reduces the mean number of AgNORs per cell, consistent with increased nucleolar stress and diminished cell proliferation (Figure 3c). Overall, these results identify SmgGDS as an important regulator of nucleolar morphology, and suggest that SmgGDS-558 has a greater role than SmgGDS-607 in protecting against nucleolar stress.
Nucleolar accumulation of SmgGDS-558 is increased by interactions with UBF and decreased by interactions with DiRas
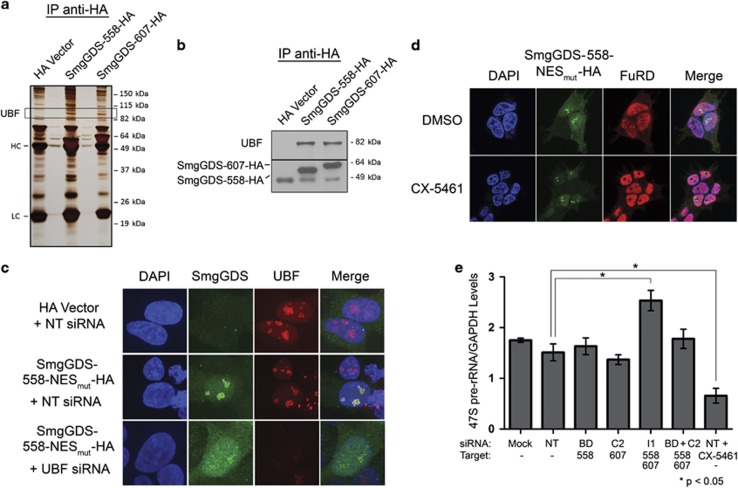
Utilizing mass spectrometry, we determined that UBF co-precipitates with both SmgGDS-558 and SmgGDS-607 in HEK293T cells (Figure 4a). These results are corroborated by immunoprecipitation experiments demonstrating association of endogenous UBF with both SmgGDS-558 and SmgGDS-607 (Figure 4b). Depletion of UBF decreases nucleolar localization of SmgGDS-558-NESmut while concomitantly increasing its nucleoplasmic localization (Figure 4c), suggesting that UBF sequesters SmgGDS-558 in the nucleolus.
Figure 4.
SmgGDS-558 physically interacts with UBF, and this interaction promotes the nucleolar accumulation of SmgGDS-558. (a) HEK293T cells were transfected with cDNAs encoding the HA vector or HA-tagged SmgGDS, followed by immunoprecipitation using HA antibody and silver staining to detect co-precipitating proteins. Mass spectrometry identified UBF as one of the co-precipitating proteins (HC and LC; heavy and light chains, respectively, of antibodies used in the immunoprecipitation). (b) Lysates from HEK293T cells transfected with cDNAs encoding the HA vector or HA-tagged SmgGDS were immunoprecipitated using HA antibody, followed by immunoblotting using antibodies to UBF and HA (n=3). (c) HEK293T cells were transfected with cDNAs encoding the HA vector or SmgGDS-558-NESmut-HA along with non-targeting (NT) siRNA or UBF siRNA. After 72 h, the cells were immunofluorescently stained with HA antibody, UBF antibody and 4,6-diamidino-2-phenylindole (DAPI), and examined by confocal fluorescence microscopy (n=3). (d) HEK293T cells expressing SmgGDS-558-NESmut-HA were treated with or without the RNA Pol I inhibitor CX-5461 (1 μm, 2 h), followed by FuRD (2 mm, 15 min), and immunofluorescently stained using antibodies to HA and BrdU (n=3). Images were obtained by confocal microscopy. (e) NCI-H1703 cells were transfected with the indicated siRNAs to deplete SmgGDS, and 72 h later quantitative PCR was conducted to examine 47S pre-rRNA levels (normalized to cellular GAPDH). Control cells were treated with CX-5461 (1 μm, 2 h) before collecting RNA. Error bars represent ±s.e.m. of three biological replicates, and statistical significance was determined by one-way analysis of variance and Holm-Sidak multiple comparisons test (*P<0.05).
Because UBF promotes rRNA synthesis,37, 38, 39, 40 we investigated how rRNA synthesis affects nucleolar accumulation of SmgGDS-558. Nucleolar SmgGDS-558-NESmut colocalizes with 5-fluorouridine (FuRD), which incorporates into newly synthesized RNA41 (Figure 4d), suggesting that SmgGDS-558-NESmut localizes at active sites of rRNA synthesis. Treatment with the RNA Pol I inhibitor CX-5461, which selectively inhibits Pol I but not Pol II,42, 43 diminishes FuRD incorporation in the nucleolus but not the nucleoplasm (Figure 4d), confirming that nucleolar FuRD detects sites of Pol I-induced rRNA synthesis. Interestingly, SmgGDS-558-NESmut continues to accumulate in nucleoli after CX-5461 inhibits rRNA synthesis (Figure 4d). SmgGDS-558-NESmut redistributes to just inside the edge of nucleoli in CX-5461-treated cells (Figure 4d), similar to the redistribution of UBF in CX-5461-treated cells (Supplementary Figure S4). These findings indicate that interaction of SmgGDS with UBF, but not active rRNA transcription, promotes nucleolar accumulation of SmgGDS.
The association of SmgGDS with UBF suggests that SmgGDS facilitates UBF-dependent rRNA synthesis. However, SmgGDS depletion does not inhibit pre-rRNA synthesis (Figure 4e), and [3H]-methyl methionine incorporation assays indicated no detectable effects of SmgGDS depletion on pre-rRNA processing (data not shown). Treatment with CX-5461 significantly reduced abundance of pre-rRNA, as anticipated (Figure 4e). On the basis of these results, nucleolar SmgGDS does not promote pre-rRNA generation.
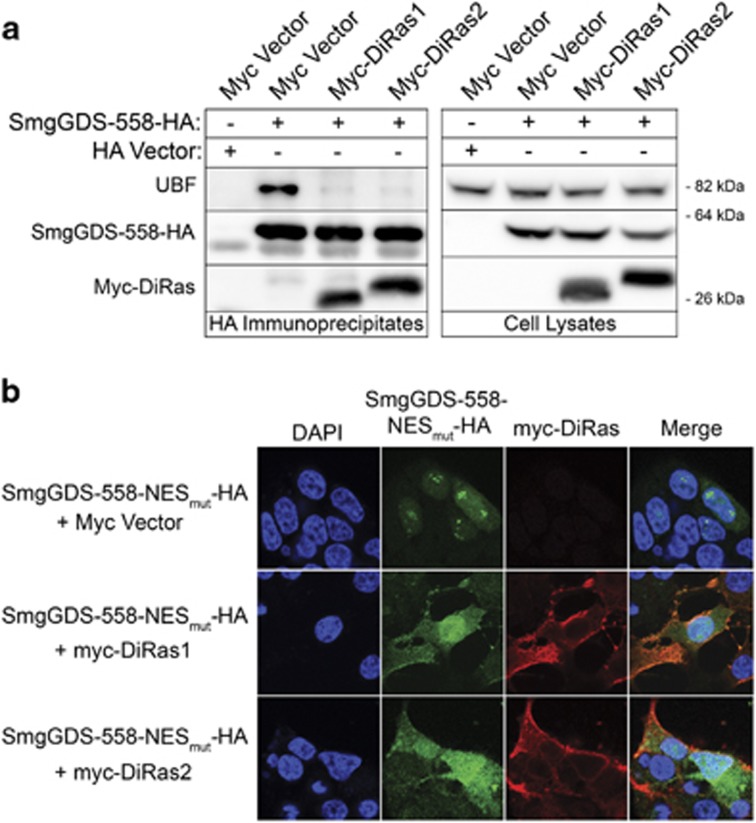
Co-expression of the tumor suppressor small GTPases DiRas1 or DiRas2 decreases binding of SmgGDS-558 to UBF (Figure 5a) and causes SmgGDS-558-NESmut to redistribute from nucleoli to the nucleoplasm (Figure 5b). This DiRas-induced redistribution of SmgGDS-558-NESmut is not due to UBF redistribution, because expression of DiRas1 or DiRas2 does not detectably alter UBF localization (Supplementary Figure S5). These results suggest that DiRas family members inhibit SmgGDS nucleolar localization by inhibiting SmgGDS interactions with UBF.
Figure 5.
DiRas1 and DiRas2 inhibit the interaction of SmgGDS-558 with UBF, and diminish the nucleolar accumulation of SmgGDS-558. (a) HEK293T cells transfected with the HA vector or SmgGDS-558-HA were co-transfected with the myc vector, myc-DiRas1 or myc-DiRas2. After 24 h, the cell lysates were immunoprecipitated using HA antibody, followed by immunoblotting using antibodies to UBF, HA and myc (n=4). (b) HEK293T cells transfected with SmgGDS-558-NESmut-HA were co-transfected with the myc vector, myc-DiRas1 or myc-DiRas2. After 24 h, the cells were immunofluorescently stained with HA antibody, myc antibody and 4,6-diamidino-2-phenylindole (DAPI), and examined by confocal fluorescence microscopy (n=3).
UBF-dependent nucleolar sequestration of SmgGDS-558 diminishes SmgGDS-558 degradation in the nucleoplasm
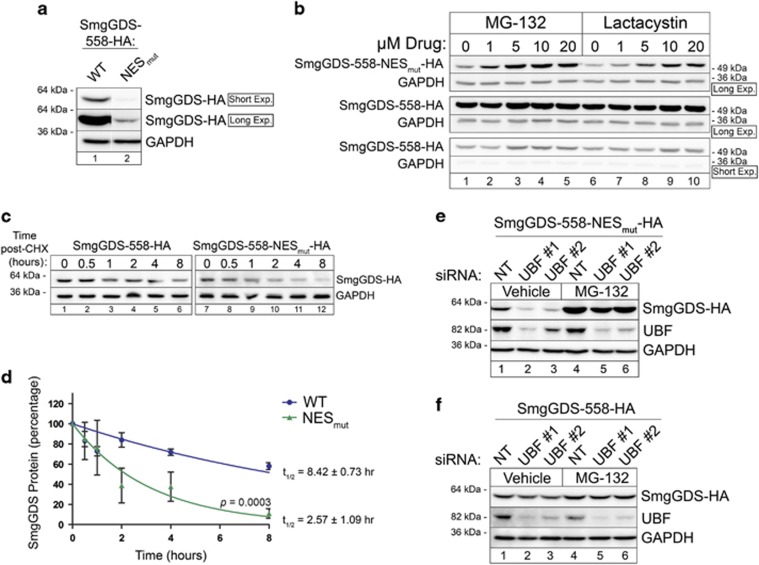
We observed that there is lower steady-state expression of transfected SmgGDS-558-NESmut than wild-type SmgGDS-558 (Figure 6a). Treatment with the proteasome inhibitors MG-132 or lactacystin significantly increases SmgGDS-558-NESmut expression, but only moderately increases expression of wild-type SmgGDS-558 (Figure 6b; Supplementary Figures S6a–d). Upon treatment with cycloheximide to halt protein translation, there is a faster decrease in abundance of SmgGDS-558-NESmut than wild-type SmgGDS (Figure 6c). Protein decay curves generated from cycloheximide-treated cells indicate that the half-life (t1/2) of wild-type SmgGDS-558 is 8.42±0.73 h, whereas t1/2 of SmgGDS-558-NESmut is 2.57±1.09 h (Figure 6d). Depletion of UBF further decreases levels of SmgGDS-558-NESmut (Figure 6e, lanes 1–3; Supplementary Figure S6e) but has little effect on levels of wild-type SmgGDS-558 (Figure 6f, lanes 1–3; Supplementary Fig. S6f). Proteasome inhibition in UBF-depleted cells restores expression of SmgGDS-558-NESmut to a greater extent than wild-type SmgGDS-558 (Figures 6e and f, lanes 4–6; Supplementary Figures S6e and f). Together, these results support the model that SmgGDS becomes destabilized when it is in the nucleoplasm, but it is protected from degradation by interacting with UBF in the nucleolus.
Figure 6.
The UBF-dependent nucleolar sequestration of SmgGDS-558 protects SmgGDS-558 from proteasome-mediated degradation in the nucleoplasm. (a) Lysates from HEK293T cells expressing SmgGDS-558-HA and SmgGDS-558-NESmut-HA were immunoblotted using HA antibody. Two exposures of the same immunoblot are shown; SmgGDS-558-NESmut-HA was detected only in the long exposure. Immunoblotting with GAPDH antibody was used as a loading control (n=3). (b) HEK293T cells were transfected with SmgGDS-558-HA or SmgGDS-558-NESmut-HA, and 56 h later the cells were incubated with the indicated concentrations of MG-132 or lactacystin. After 16 h, cell lysates were immunoblotted using HA and GAPDH antibodies (n=3). Long and short exposures of the immunoblots are shown. Mean normalized densitometry values are shown in Supplementary Figure S6. (c) HEK293T cells expressing SmgGDS-558-HA or SmgGDS-558-NESmut-HA were incubated with cycloheximide (CHX; 1 μg/ml) for the indicated times, and cell lysates were immunoblotted using HA and GAPDH antibodies (n=3). (d) Mean densitometry values obtained from three independent experiments shown in c were used to fit exponential regression curves and determine the half-life of SmgGDS-558-HA and SmgGDS-558-NESmut-HA. (e, f) HEK293T cells transfected with SmgGDS-558-NESmut-HA (e) or SmgGDS-558-HA (f) were co-transfected with non-targeting (NT) siRNA or UBF siRNAs, and 56 h later the cells were treated with or without MG-132 (5 μm, 16 h; n=3). Cell lysates were immunoblotted using HA, UBF and GAPDH antibodies. Mean normalized densitometry values are shown in Supplementary Figure S6.
Nuclear SmgGDS is detectable in patients’ tumors
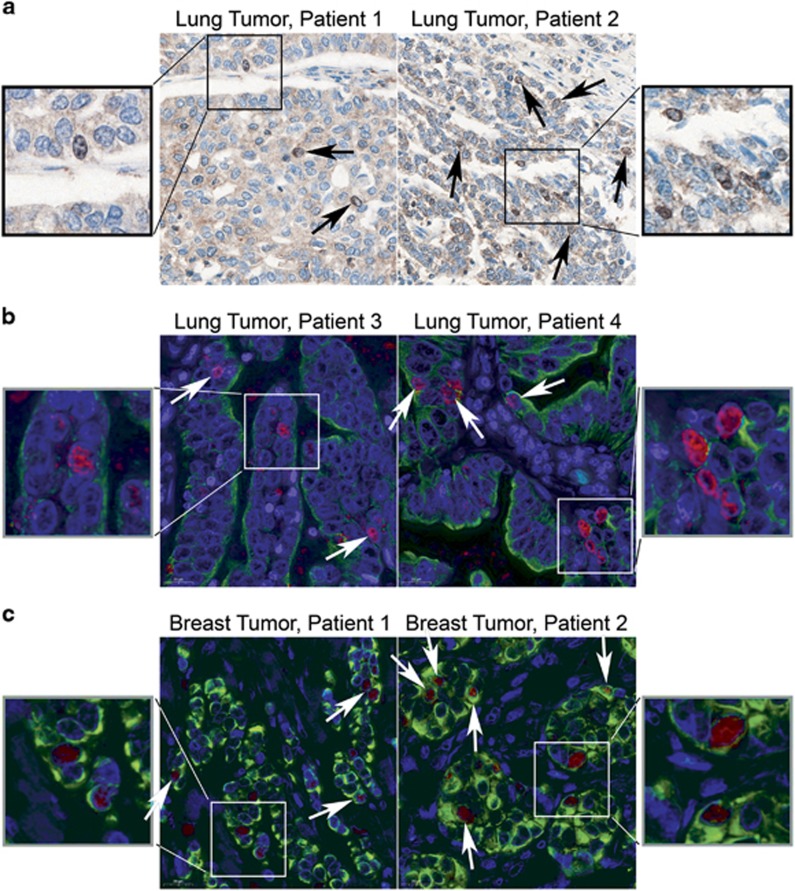
We previously reported that SmgGDS is overexpressed in lung and breast tumors,3, 8 but we did not analyze nuclear localization of SmgGDS in lung and breast tumors. We detect endogenous SmgGDS in the cytoplasm, nucleus and the nucleolus of human lung cancer (Figures 7a and b) and breast cancer (Figure 7c) tissue, consistent with SmgGDS having both cytoplasmic and nuclear functions in tumors. As anticipated, SmgGDS-558-NESmut accumulates in nucleoli and colocalizes with UBF in NCI-H1703 and NCI-H23 lung cancer cell lines, and MDA-MB-231 and MCF7 breast cancer cells, whereas wild-type SmgGDS-558 is predominantly cytosolic (Supplementary Figure S7).
Figure 7.
Nuclear SmgGDS is detectable in patients’ lung and breast tumors. (a) Representative immunohistochemical staining of SmgGDS in patients’ lung tumors is shown. Black arrows indicate cells with nuclear and nucleolar SmgGDS. (b, c) Representative immunofluorescent staining of SmgGDS (red) and cytokeratin (green) in patients’ lung tumors (b) and breast tumors (c) is shown. White arrows indicate cells with nuclear and nucleolar SmgGDS.
Discussion
This study defines SmgGDS as a previously unsuspected interacting partner of UBF in the nucleolus, identifying new regulatory networks by which SmgGDS might promote cancer. Our finding that SmgGDS is sequestered by UBF in the nucleolus and protects cells from nucleolar stress provides a new pathway to control nucleolar stress in malignant cells. The functional importance of the nucleolus in cell cycle progression and cancer biology is a rapidly expanding field of research, and our findings provide an additional link between nucleolar stress and loss of cell cycle progression.
Our observations that SmgGDS colocalizes with nucleolar UBF and that depletion of UBF releases SmgGDS from the nucleolus suggest that UBF acts as an anchor to sequester SmgGDS in the nucleolus. The sequestration of SmgGDS in the nucleolus may regulate nucleolar structure or function, suppress SmgGDS functions in the cytosol and regulate SmgGDS protein stability. In support of this latter idea, our findings indicate that SmgGDS protein is stable when localized to the cytoplasm, degraded when localized to the nucleoplasm and protected from degradation when bound to UBF in the nucleolus. Nucleolar sequestration is a common mechanism in which general protein stability is regulated; for example, in response to nucleolar stress, MDM2 is sequestered in the nucleolus by CDKN2A (p14ARF), resulting in stabilization of the tumor suppressor p53.14, 16, 31 Conversely, the oncogene c-Myc is targeted to the nucleolus for its ubiquitin-dependent degradation,44, 45, 46 although mechanisms that stabilize nucleolar-localized c-Myc also exist.47 Localization of RelA, a subunit of NF-κB, is regulated by ubiquitin-dependent mechanisms that promote either its degradation in the nucleus48 or its translocation to the nucleolus.22 The ability of UBF to sequester SmgGDS and protect it from degradation most likely contributes to unique nucleolar functions of SmgGDS.
Increased rRNA synthesis in the nucleolus occurs commonly in cancer,12, 13, 14, 16 and evidence suggests that this process actively promotes the malignant process.42 It was recently reported that the guanine nucleotide exchange factor Ect2 localizes in the nucleolus and promotes rRNA synthesis in lung cancer through interactions with UBF1, Rac1 and nucleophosmin.49 As SmgGDS also interacts with Rac111 and UBF, it is reasonable to postulate that SmgGDS promotes rRNA synthesis. However, we found that depleting SmgGDS in NCI-H1703 cells did not detectably alter rRNA synthesis, or surprisingly slightly increased rRNA synthesis (Figure 4e). On the basis of these results, it is less likely that SmgGDS promotes rRNA synthesis, but perhaps interactions of SmgGDS with UBF help maintain proper nucleolar structure. UBF1 binds and promotes chromatin decondensation of rDNA gene repeats,50, 51, 52, 53, 54, 55 and UBF1 is critical for the maintenance and morphological appearance of active NORs.51, 52, 56 Our observation that targeting SmgGDS in non-small-cell lung carcinoma cells alters nucleolar morphology and decreases AgNORs suggests that SmgGDS is important for maintenance of nucleolar architecture and integrity.
The ability of DiRas1 and DiRas2 to inhibit SmgGDS interactions with UBF and reduce SmgGDS nucleolar localization suggests that the tumor suppressive effects of these small GTPases57, 58, 59 involve loss of nuclear functions of SmgGDS. It is currently unclear how DiRas proteins regulate interactions of SmgGDS with UBF. We reported that DiRas1 outcompetes K-Ras4B, RhoA and Rap1A for binding to SmgGDS;2 thus, physical binding of DiRas to SmgGDS might outcompete UBF for binding to SmgGDS. It is also possible that DiRas-mediated signaling events inhibit SmgGDS interactions with UBF indirectly; for example, DiRas signaling might prevent nucleolar import and/or alter binding affinity of SmgGDS through post-translational modifications of SmgGDS. We did not detect alterations in nucleolar morphology indicative of nucleolar stress upon expression of DiRas1 or DiRas2. Expression of DiRas might induce low levels of nucleolar stress that are beneath our ability to detect. The participation of DiRas in the regulation of SmgGDS in the nucleolus warrants further investigation.
Numerous insults induce nucleolar stress, including genotoxic stress (DNA damage and ultraviolet irradiation), inhibition of RNA polymerase I/II, osmotic stress, viral infection and others,18, 19 and it is well known that p53 is a critical mediator of the nucleolar stress response, resulting in cell cycle arrest.20, 21, 31, 32 Induction of nucleolar stress is consistent with our RNA-sequencing data indicating that depletion of SmgGDS activates p53 signaling networks and reduces expression of DREAM complex target genes, including the key cell cycle-promoting transcription factors E2F1, MYBL2 (B-Myb), FOXM1 and MYC. In addition to these genes, SmgGDS depletion also diminishes expression of other DREAM targets that are reported to be important in cancer, including EZH260 (P=1.09 × 10−33), AURKA and AURKB61, 62, 63 (P=3.91 × 10−43 and 2.73 × 10−63, respectively) and Ect249, 64 (P=2.64 × 10−34) (Supplementary Table 2).
Important questions remain regarding these studies. For example, it will be important to understand the role of SmgGDS in regulating the DREAM complex and nucleolar stress in non-transformed cells. The observation that SmgGDS is overexpressed in multiple cancers3, 8, 9 suggests that SmgGDS is needed to protect malignant cells from nucleolar stress, inducing the phenomenon known as oncogene addiction65 to SmgGDS in malignant cells. If that is the case, knockdown of SmgGDS should have little impact on the proliferation of non-transformed cells. Consistent with this, we reported that knockdown of SmgGDS has a much more deleterious effect on the proliferation of lung cancer cells than the proliferation of normal human bronchial epithelial cells.8 These results support a potentially greater role for SmgGDS in diminishing nucleolar stress and regulating DREAM complex activity in cancer than in normal cells, but this relationship should be investigated further.
Another important question is the contribution of the cytoplasmic and nuclear functions of SmgGDS in protecting cells from nucleolar stress. We have not formally shown that nucleolar stress induced by loss of SmgGDS is mediated by the nucleolar pool of SmgGDS. For example, nucleolar stress may arise when SmgGDS is knocked down because of loss of cytoplasmic signaling by small GTPases that interact with SmgGDS. SmgGDS obviously has functions in the cytoplasm and the nucleus, and nucleocytoplasmic shuttling is important for SmgGDS to achieve these functions. Undoubtedly, there is a functional connection between nuclear and cytoplasmic roles of SmgGDS in cancer. For example, interactions of SmgGDS with small GTPases promote oncogenic signaling by those small GTPases,1, 3, 7 but additionally, interactions of SmgGDS with small GTPases, such as Rac1, also regulate nuclear localization of SmgGDS.11 We have been unable to separate the nuclear and cytoplasmic functions of SmgGDS, and because these functions are likely to be inter-related, it may be technically and biologically not feasible to do so, or difficult to interpret the results of these studies. Future studies are aimed at identifying the signals and the structural aspects of SmgGDS that have key roles in the subcellular localization of SmgGDS and its functions in different subcellular compartments.
Collectively, our discoveries support the model that targeting SmgGDS induces nucleolar stress, resulting in profound loss of DREAM target gene expression required for G1, S and G2 progression, and ultimately resulting in cell cycle arrest. These results greatly enhance our mechanistic understanding of the pathways that diminish cell cycle progression when cancer cells are depleted of SmgGDS, and further validate SmgGDS as a novel therapeutic target. The ability to induce nucleolar stress in cancer cells, perhaps by targeting SmgGDS, is a potentially promising therapeutic strategy for the treatment of cancer.
Materials and methods
Cell culture and transfection of cDNAs and siRNAs
NCI-H1703 and NCI-H23 human non-small-cell lung carcinoma cells, MCF7 and MDA-MB-231 human breast cancer cells, and HEK293T human embryonic kidney cells were obtained from the American Type Culture Collection (Manassas, VA, USA), which documents and guarantees their authentication. Cell lines were checked to confirm that they are free of mycoplasma contamination. The cells were cultured as previously described,10 and treated in some experiments with the following compounds: leptomycin B (L2913, Sigma, St Louis, MO, USA); MG-132 (Z-Leu-Leu-Leu-al, Sigma, C2211); lactacystin (Sigma, L6785); cycloheximide (239764, Calbiochem, San Diego, CA, USA); CX-5461 (Calbiochem, 509265); and FuRD (Sigma, F5130).
Previously described methods were used to generate pcDNA3.1 expression vectors encoding N-terminal myc-tagged DiRas1 (#NP_660156) or DiRas2 (#NP_060064),2 C-terminal HA-tagged SmgGDS-558 (#NP_001093899) or SmgGDS-607 (#NP_001093897),1 and the SmgGDS-558-NESmut-HA mutant that has alanine substitutions at amino acids L4, L8, L11 and I13.11 The cDNAs were transfected using Lipofectamine 2000 (ThermoFisher, Waltham, MA, USA; 11668019) as previously described.1, 2, 3
DharmaFECT 3 (T-2003, Dharmacon, Lafayette, CO, USA) was used to transfect cells with previously characterized siRNAs (25 nm) targeting SmgGDS.1, 3, 8, 9, 10 siRNAs I1 and I2 simultaneously deplete both splice variants of SmgGDS, whereas siRNA BD targets only SmgGDS-558 and siRNA C2 targets only SmgGDS-607.1, 3, 8, 9, 10 siRNA NT is a non-targeting siRNA. Two independent siRNAs to deplete UBF (UBF#1 and UBF#2) were also used as indicated. siRNA sequences are as follows: siRNA BD, 5′-ACGATAGCCATTCGCTTCA-3′; siRNA C2, 5′-GAACTATAGCAATGAGAAT-3′; siRNA I1, 5′-GCAAAGATGTTATCAGCTG-3′; siRNA I2, 5′-GTTAATAGATGCACAAGAA-3′; siRNA NT, 5′-UGGUUUACAUGUUUUCUGA-3′ siRNA UBF#1, 5′-TAACCAAGATTCTGTCCAA-3′ and siRNA UBF#2, 5′-GGACCGTGCAGCATATAAA-3′.
RNA sequencing
NCI-H1703 cells were transfected with siRNA I1 targeting both SmgGDS-558 and SmgGDS-607, and RNA was collected 72 h later using TRIzol Reagent (ThermoFisher, 15596-026). Total RNA (4 μg) was poly-A-purified, transcribed and chemically fragmented using Illumina’s TruSeq RNA library kit using the manufacturer’s protocol. Individual libraries were prepared for each sample, indexed for multiplexing and then sequenced on an Illumina HiSeq 2500 (Illumina, Inc., San Diego, CA, USA). Reads of each sample were aligned to NCBI Build GRCh38.p2 of the human transcriptome references using Bowtie2 version 2.2.3.66 Default parameters were used with the exception of a Bowtie2 offset of 1, trading index size for increased alignment speed. Sequences for all RNA transcripts were annotated using NCBI Homo sapiens Annotation Release 107. Expression abundances were quantified at the whole transcript-level as effect counts using eXpress version 1.5.1.67 The transcript-level count data were aggregated per gene and rounded to integers to produce the gene-level count matrix. Differential expression analysis was performed using the Bioconductor package DESeq2 version 1.12.468 to compute log2 fold changes and false discovery rate-adjusted P-values. Statistical significance was determined at a false discovery rate threshold of 0.05. Data were analyzed for molecular and functional pathway enrichment using the Ingenuity Pathway Analysis tool (Qiagen, Redwood City, CA, USA).
Subcellular fractionation, immunoprecipitation and immunoblotting
Whole-cell lysates were generated by lysing cells in 1% TX-100 (10 mm Tris (pH 7.4), 150 mm NaCl) containing protease inhibitors, phosphatase inhibitors and benzonase (50 units/ml; 37 ºC, 5 min). The lysates were diluted 1:1 with SDS lysis buffer (2% SDS, 50 mm Tris, pH 7.4), rocked, (4 ºC, 15 min) and centrifuged (16000 g, 10 min, 4 ºC) to generate a cleared lysate.
Nuclear and cytosolic fractions were generated by lysing cells in hypotonic lysis buffer (10 mm HEPES, 10 mm KCl, 1.5 mm MgCl2, 0.5% Nonidet-P40 and 0.5 mm dithiothreitol) with protease and phosphatase inhibitors. The lysates were incubated (4 ºC, 5 min) and centrifuged (500 g, 3 min, 4 ºC), and the supernatant was saved as the ‘cytosolic’ fraction. The pellets were resuspended in hypotonic lysis buffer, incubated (4 ºC, 5 min) and centrifuged again (500 g, 3 min, 4 ºC) to generate a pellet that was saved as the ‘nuclear’ fraction. The nuclear fraction was lysed in TX-100/SDS with benzonase, as described above for whole-cell lysates. Protein concentrations of whole-cell lysates and subcellular fractions were determined using the Pierce BCA protein assay kit (ThermoFisher, 23225).
For immunoprecipitation assays, cell lysates were prepared in 0.5% Nonidet-P40 with protease and phosphatase inhibitors.1 After centrifugation, the cleared lysates were immunoprecipitated using HA-conjugated beads (Sigma, A2095) as previously described.1
Immunoprecipitates, whole-cell lysates and subcellular fractions were analyzed by enhanced chemiluminescence immunoblotting as previously described69 using the following antibodies: RAP1GDS1 (sc-390003, Santa Cruz, Dallas, TX, USA); HSP90 (4877, Cell Signaling, Danvers, MA, USA); Histone H3 (Cell Signaling, 4499); E2F1 (Cell Signaling, 3742); GAPDH (Santa Cruz, sc-32233); UBF (Santa Cruz, sc-13125); HA (901503, Covance, San Diego, CA, USA); and myc (Santa Cruz, sc-40). Images of the immunoblots were acquired with an ImageQuant LAS4000 biomolecular imager and analyzed with ImageQuant LAS4000 software (GE Life Sciences, Pittsburgh, PA, USA).
Immunofluorescent colocalization of proteins and FuRD labeling
Cells plated on glass coverslips were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS; 20 min, 25 ºC), permeabilized with 0.1% TX-100 in PBS (10 min, 25 ºC) and incubated (1 h, 25 ºC) with the following primary antibodies diluted in PBS with 4% fetal bovine serum: mouse or rabbit anti-HA (Covance 901503 or 902302, respectively); mouse or rabbit anti-UBF (Santa Cruz sc-13125 or sc-9131, respectively); mouse anti-myc (Santa Cruz sc-40); or rabbit anti-Nucleolin (Santa Cruz sc-13057). For FuRD labeling, cells were incubated with FuRD (2 mm, 15 min, 37ºC)41 before fixing and staining with mouse anti-BrdU antibody (Sigma, B8434), as described above. After incubating with primary antibodies, the cells were incubated with fluoroscein isothiocyanate- or tetramethylrhodamine-conjugated anti-mouse or anti-rabbit secondary antibody (1 h, 25 ºC), mounted in mounting media (1 mg/ml p-phenylenediamene in 1:9 PBS:glycerol), and imaged using a Nikon A1 confocal microscope and Nikon NIS elements software (Nikon, Melville, NY, USA).
Staining of NORs (AgNORs)
Cells on glass coverslips in a 24-well plate were fixed in ethanol:glacial acetic acid (3:1; 30 min, 25 ºC) and incubated (30 min, 25 ºC) with staining solution (one part 2% gelatin/1% formic acid to two parts 50% silver nitrate). The washed coverslips were incubated with 5% sodium thiosulfate (10 min, 25 ºC), washed in dH2O, washed in 95% ethanol and examined by phase contrast microscopy.
Quantitative PCR
Cellular RNA was quantified using quantitative reverse-transcriptase PCR. NCI-H1703 cells were transfected with siRNAs targeting SmgGDS for 72 h. RNA was isolated with the RNeasy mini kit (Qiagen, 74104) and cDNA was generated with iScript cDNA synthesis kit (170-8890, BioRad, Hercules, CA, USA) using 1 μg of DNase-treated RNA. Quantitative PCR of cDNA was performed using iTaq universal SYBR green (BioRad, 172-5121), the 7900HT Fast Real-Time PCR system (ThermoFisher) and the following primers: 5′-ETS pre-rRNA (5′-GAACGGTGGTGTGTCGTTC-3′ and 5′-GCGTCTCGTCTCGTCTCACT-3′) and GAPDH (5′-CCCCTTCATTGACCTCAACTACAT-3′ and 5′-CGCTCCTGGAAGATGGTGA-3′).70 Relative abundance was determined using a standard curve consisting of 10-fold serial dilutions of one sample.
Immunohistochemical and immunofluorescent analysis of tumor tissues
Commercial, de-identified human lung cancer tissue microarrays (US Biomax, Derwood, MD, USA) were immunohistochemically stained for SmgGDS using antigen retrieval Low pH (citrate buffer (pH 6.1), Dako, Carpinteria, CA, USA), SmgGDS antibody (Santa Cruz, sc-390003, 1:50, 45 min) and Dako EnVision FLEX mini Kit, utilizing a Dako Autostainer Omnis (Dako). High-resolution digital images were captured at × 20 using a Pannoramic 250 Flash III slide scanner (3DHISTECH Ltd., Budapest, Hungary).
Immunofluorescent detection of SmgGDS (1:50) and cytokeratin (1:100; Dako, Z0622) in breast cancer tissue microarrays (US Biomax) and the lung cancer tissues was conducted as described above for immunohistochemistry, using the TSA Plus Fluorescence Kit (Perkin Elmer, Santa Clara, CA, USA) and secondary antibodies labeled with Cy5 or Alexa Fluor 555. Coverslips were mounted onto slides using Prolong Gold antifade reagent with 4,6-diamidino-2-phenylindole (Invitrogen, Carlsbad, CA, USA). Fluorescent images were captured in three channels (Alexa Fluor 555, Cy5 or 4,6-diamidino-2-phenylindole) at × 20 using the Pannoramic 250 Flash III slide scanner (3DHistech, Budapest, Hungary).
Statistical analysis
Statistical analyses were designed and performed in consultation with Dr Aniko Szabo (Division of Biostatistics, Medical College of Wisconsin), and using Graphpad Prism 5 (San Diego, CA, USA) software. For each experiment, three or more biological replicates were conducted, as indicated in each figure legend. Data are presented as mean±s.e.m. and analyzed by unpaired Student’s t-test or one-way analysis of variance followed by Dunnett’s or Holm-Sidak multiple comparisons post hoc tests, as indicated in the figure legends. Statistical significance was determined at P<0.05.
Acknowledgments
We thank Dr Aniko Szabo (Medical College of Wisconsin) for assistance with statistical analysis. This work was supported by NCI R01 CA188871 (CLW), NCI R01 CA193343 (MJF), NIAID R01 AI083281 (SST), NCI R01 CA188575 (HR) and Institutional Research Grant 86-004-26 from the American Cancer Society (CB). Additional support was provided by the Kathleen M Duffey Fogarty Eminent Scholar in Breast Cancer Research Award (CLW), the Mary Kay Foundation Award Grant No. 024.16 (MJF), the Nancy Laning Sobczak, PhD, Breast Cancer Research Award (CB and CLW), and by the National Center for Research Resources, the National Center for Advancing Translational Sciences and the Office of the Director of the National Institutes of Health through Grant 8KL2TR000056 (CB).
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
The authors declare no conflict of interest.
Supplementary Material
References
- Berg TJ, Gastonguay AJ, Lorimer EL, Kuhnmuench JR, Li R, Fields AP et al. Splice variants of SmgGDS control small GTPase prenylation and membrane localization. J Biol Chem 2010; 285: 35255–35266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergom C, Hauser AD, Rymaszewski A, Gonyo P, Prokop JW, Jennings BC et al. The tumor-suppressive small GTPase DiRas1 binds the noncanonical guanine nucleotide exchange factor SmgGDS and antagonizes SmgGDS interactions with oncogenic small GTPases. J Biol Chem 2016; 291: 10948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser AD, Bergom C, Schuld NJ, Chen X, Lorimer EL, Huang J et al. The SmgGDS splice variant SmgGDS-558 is a key promoter of tumor growth and RhoA signaling in breast cancer. Mol Cancer Res 2014; 12: 130–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogita Y, Egami S, Ebihara A, Ueda N, Katada T, Kontani K. Di-Ras2 protein forms a complex with SmgGDS protein in brain cytosol in order to be in a low affinity state for guanine nucleotides. J Biol Chem 2015; 290: 20245–20256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T, Kaibuchi K, Yamamoto T, Kawamura M, Sakoda T, Fujioka H et al. A stimulatory GDP/GTP exchange protein for smg p21 is active on the post-translationally processed form of c-Ki-ras p21 and rhoA p21. Proc Natl Acad Sci USA 1991; 88: 6442–6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T, Kaibuchi K, Mizuno T, Hiroyoshi M, Shirataki H, Takai Y. Purification and characterization from bovine brain cytosol of proteins that regulate the GDP/GTP exchange reaction of smg p21s, ras p21-like GTP-binding proteins. J Biol Chem 1990; 265: 16626–16634. [PubMed] [Google Scholar]
- Ntantie E, Gonyo P, Lorimer EL, Hauser AD, Schuld N, McAllister D et al. An adenosine-mediated signaling pathway suppresses prenylation of the GTPase Rap1B and promotes cell scattering. Sci Signal 2013; 6: ra39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tew GW, Lorimer EL, Berg TJ, Zhi H, Li R, Williams CL. SmgGDS regulates cell proliferation, migration, and NF-κB transcriptional activity in non-small cell lung carcinoma. J Biol Chem 2008; 283: 963–976. [DOI] [PubMed] [Google Scholar]
- Zhi H, Yang X, Kuhnmuench J, Berg T, Thill R, Yang H et al. SmgGDS is up‐regulated in prostate carcinoma and promotes tumour phenotypes in prostate cancer cells. J Pathol 2009; 217: 389–397. [DOI] [PubMed] [Google Scholar]
- Schuld NJ, Hauser AD, Gastonguay AJ, Wilson JM, Lorimer EL, Williams CL. SmgGDS-558 regulates the cell cycle in pancreatic, non-small cell lung, and breast cancers. Cell Cycle 2014; 13: 941–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanning CC, Ruiz-Velasco R, Williams CL. Novel mechanism of the co-regulation of nuclear transport of SmgGDS and Rac1. J Biol Chem 2003; 278: 12495–12506. [DOI] [PubMed] [Google Scholar]
- Hein N, Hannan KM, George AJ, Sanij E, Hannan RD. The nucleolus: an emerging target for cancer therapy. Trends Mol Med 2013; 19: 643–654. [DOI] [PubMed] [Google Scholar]
- Drygin D, Rice WG, Grummt I. The RNA polymerase I transcription machinery: an emerging target for the treatment of cancer. Annu Rev Pharmacol Toxicol 2010; 50: 131–156. [DOI] [PubMed] [Google Scholar]
- Quin JE, Devlin JR, Cameron D, Hannan KM, Pearson RB, Hannan RD. Targeting the nucleolus for cancer intervention. Biochim Biophys Acta 2014; 1842: 802–816. [DOI] [PubMed] [Google Scholar]
- Montanaro L, Treré D, Derenzini M. Nucleolus, ribosomes, and cancer. Am J Pathol 2008; 173: 301–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods SJ, Hannan KM, Pearson RB, Hannan RD. The nucleolus as a fundamental regulator of the p53 response and a new target for cancer therapy. Biochim Biophys Acta 2015; 1849: 821–829. [DOI] [PubMed] [Google Scholar]
- Boisvert F, van Koningsbruggen S, Navascués J, Lamond AI. The multifunctional nucleolus. Nat Rev Mol Cell Biol 2007; 8: 574–585. [DOI] [PubMed] [Google Scholar]
- Grummt I. The nucleolus—guardian of cellular homeostasis and genome integrity. Chromosoma 2013; 122: 487–497. [DOI] [PubMed] [Google Scholar]
- Boulon S, Westman BJ, Hutten S, Boisvert F, Lamond AI. The nucleolus under stress. Mol Cell 2010; 40: 216–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisenroth C, Zhang Y. Ribosome biogenesis surveillance: probing the ribosomal protein-Mdm2-p53 pathway. Oncogene 2010; 29: 4253–4260. [DOI] [PubMed] [Google Scholar]
- Donati G, Montanaro L, Derenzini M. Ribosome biogenesis and control of cell proliferation: p53 is not alone. Cancer Res 2012; 72: 1602–1607. [DOI] [PubMed] [Google Scholar]
- Thoms HC, Loveridge CJ, Simpson J, Clipson A, Reinhardt K, Dunlop MG et al. Nucleolar targeting of RelA(p65) is regulated by COMMD1-dependent ubiquitination. Cancer Res 2010; 70: 139–149. [DOI] [PubMed] [Google Scholar]
- Sadasivam S, DeCaprio JA. The DREAM complex: master coordinator of cell cycle-dependent gene expression. Nat Rev Cancer 2013; 13: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M, Grossmann P, Padi M, DeCaprio JA. Integration of TP53, DREAM, MMB-FOXM1 and RB-E2F target gene analyses identifies cell cycle gene regulatory networks. Nucleic Acids Res 2016; 44: 6070–6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller GA, Stangner K, Schmitt T, Wintsche A, Engeland K. Timing of transcription during the cell cycle: protein complexes binding to E2F, E2F/CLE, CDE/CHR, or CHR promoter elements define early and late cell cycle gene expression. Oncotarget epub ahead of print 28 July 2016; doi:10.18632/oncotarget.10888. [DOI] [PMC free article] [PubMed]
- Fischer M, Quaas M, Steiner L, Engeland K. The p53-p21-DREAM-CDE/CHR pathway regulates G2/M cell cycle genes. Nucleic Acids Res 2016; 44: 164–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litovchick L, Sadasivam S, Florens L, Zhu X, Swanson SK, Velmurugan S et al. Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Mol Cell 2007; 26: 539–551. [DOI] [PubMed] [Google Scholar]
- Sadasivam S, Duan S, DeCaprio JA. The MuvB complex sequentially recruits B-Myb and FoxM1 to promote mitotic gene expression. Genes Dev 2012; 26: 474–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertoli C, Skotheim JM, de Bruin RA. Control of cell cycle transcription during G1 and S phases. Nat Rev Mol Cell Biol 2013; 14: 518–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaas M, Muller GA, Engeland K. p53 can repress transcription of cell cycle genes through a p21(WAF1/CIP1)-dependent switch from MMB to DREAM protein complex binding at CHR promoter elements. Cell Cycle 2012; 11: 4661–4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg Olausson K, Nistér M, Lindström MS. p53-dependent and-independent nucleolar stress responses. Cells 2012; 1: 774–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James A, Wang Y, Raje H, Rosby R, DiMario P. Nucleolar stress with and without p53. Nucleus 2014; 5: 402–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donati G, Brighenti E, Vici M, Mazzini G, Trere D, Montanaro L et al. Selective inhibition of rRNA transcription downregulates E2F-1: a new p53-independent mechanism linking cell growth to cell proliferation. J Cell Sci 2011; 124(Pt 17): 3017–3028. [DOI] [PubMed] [Google Scholar]
- Derenzini M, Montanaro L, Treré D. What the nucleolus says to a tumour pathologist. Histopathology 2009; 54: 753–762. [DOI] [PubMed] [Google Scholar]
- Derenzini M, Ceccarelli C, Santini D, Taffurelli M, Trere D. The prognostic value of the AgNOR parameter in human breast cancer depends on the pRb and p53 status. J Clin Pathol 2004; 57: 755–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pich A, Chiusa L, Margaria E. Prognostic relevance of AgNORs in tumor pathology. Micron 2000; 31: 133–141. [DOI] [PubMed] [Google Scholar]
- Hamdane N, Stefanovsky VY, Tremblay MG, Nemeth A, Paquet E, Lessard F et al. Conditional inactivation of upstream binding factor reveals its epigenetic functions and the existence of a somatic nucleolar precursor body. PLoS Genet 2014; 10: e1004505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panov KI, Friedrich JK, Russell J, Zomerdijk JC. UBF activates RNA polymerase I transcription by stimulating promoter escape. EMBO J 2006; 25: 3310–3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovsky V, Langlois F, Gagnon-Kugler T, Rothblum LI, Moss T. Growth factor signaling regulates elongation of RNA polymerase I transcription in mammals via UBF phosphorylation and r-chromatin remodeling. Mol Cell 2006; 21: 629–639. [DOI] [PubMed] [Google Scholar]
- Stefanovsky VY, Pelletier G, Hannan R, Gagnon-Kugler T, Rothblum LI, Moss T. An immediate response of ribosomal transcription to growth factor stimulation in mammals is mediated by ERK phosphorylation of UBF. Mol Cell 2001; 8: 1063–1073. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Forys JT, Miceli AP, Gwinn AS, Weber JD. Identification of DHX33 as a mediator of rRNA synthesis and cell growth. Mol Cell Biol 2011; 31: 4676–4691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bywater MJ, Poortinga G, Sanij E, Hein N, Peck A, Cullinane C et al. Inhibition of RNA polymerase I as a therapeutic strategy to promote cancer-specific activation of p53. Cancer Cell 2012; 22: 51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drygin D, Lin A, Bliesath J, Ho CB, O'Brien SE, Proffitt C et al. Targeting RNA polymerase I with an oral small molecule CX-5461 inhibits ribosomal RNA synthesis and solid tumor growth. Cancer Res 2011; 71: 1418–1430. [DOI] [PubMed] [Google Scholar]
- Arabi A, Rustum C, Hallberg E, Wright AP. Accumulation of c-Myc and proteasomes at the nucleoli of cells containing elevated c-Myc protein levels. J Cell Sci 2003; 116(Pt 9): 1707–1717. [DOI] [PubMed] [Google Scholar]
- Arabi A, Wu S, Ridderstrale K, Bierhoff H, Shiue C, Fatyol K et al. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat Cell Biol 2005; 7: 303–310. [DOI] [PubMed] [Google Scholar]
- Welcker M, Orian A, Grim JE, Eisenman RN, Clurman BE. A nucleolar isoform of the Fbw7 ubiquitin ligase regulates c-Myc and cell size. Curr Biol 2004; 14: 1852–1857. [DOI] [PubMed] [Google Scholar]
- Sun XX, He X, Yin L, Komada M, Sears RC, Dai MS. The nucleolar ubiquitin-specific protease USP36 deubiquitinates and stabilizes c-Myc. Proc Natl Acad Sci USA 2015; 112: 3734–3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natoli G, Chiocca S. Nuclear ubiquitin ligases, NF-kappaB degradation, and the control of inflammation. Sci Signal 2008; 1: pe1. [DOI] [PubMed] [Google Scholar]
- Justilien V, Ali SA, Jamieson L, Yin N, Cox AD, Der CJ et al. Ect2-dependent rRNA synthesis is required for KRAS-TRP53-driven lung adenocarcinoma. Cancer Cell 2017; 31: 256–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Belmont AS, Huang S. Upstream binding factor association induces large-scale chromatin decondensation. Proc Natl Acad Sci USA 2004; 101: 15106–15111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McStay B, Grummt I. The epigenetics of rRNA genes: from molecular to chromosome biology. Annu Rev Cell Dev Biol 2008; 24: 131–157. [DOI] [PubMed] [Google Scholar]
- McStay B. Nucleolar organizer regions: genomic 'dark matter' requiring illumination. Genes Dev 2016; 30: 1598–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan AC, Sullivan GJ, McStay B. UBF binding in vivo is not restricted to regulatory sequences within the vertebrate ribosomal DNA repeat. Mol Cell Biol 2002; 22: 657–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanij E, Poortinga G, Sharkey K, Hung S, Holloway TP, Quin J et al. UBF levels determine the number of active ribosomal RNA genes in mammals. J Cell Biol 2008; 183: 1259–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanij E, Hannan RD. The role of UBF in regulating the structure and dynamics of transcriptionally active rDNA chromatin. Epigenetics 2009; 4: 374–382. [DOI] [PubMed] [Google Scholar]
- Mais C, Wright JE, Prieto JL, Raggett SL, McStay B. UBF-binding site arrays form pseudo-NORs and sequester the RNA polymerase I transcription machinery. Genes Dev 2005; 19: 50–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis CA, Vos MD, Howell H, Vallecorsa T, Fults DW, Clark GJ. Rig is a novel Ras-related protein and potential neural tumor suppressor. Proc Natl Acad Sci USA 2002; 99: 9876–9881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontani K, Tada M, Ogawa T, Okai T, Saito K, Araki Y et al. Di-Ras, a distinct subgroup of ras family GTPases with unique biochemical properties. J Biol Chem 2002; 277: 41070–41078. [DOI] [PubMed] [Google Scholar]
- Zhu YH, Fu L, Chen L, Qin YR, Liu H, Xie F et al. Downregulation of the novel tumor suppressor DIRAS1 predicts poor prognosis in esophageal squamous cell carcinoma. Cancer Res 2013; 73: 2298–2309. [DOI] [PubMed] [Google Scholar]
- Fillmore CM, Xu C, Desai PT, Berry JM, Rowbotham SP, Lin YJ et al. EZH2 inhibition sensitizes BRG1 and EGFR mutant lung tumours to TopoII inhibitors. Nature 2015; 520: 239–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldenson B, Crispino JD. The aurora kinases in cell cycle and leukemia. Oncogene 2015; 34: 537–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marumoto T, Zhang D, Saya H. Aurora-A—a guardian of poles. Nat Rev Cancer 2005; 5: 42–50. [DOI] [PubMed] [Google Scholar]
- Portella G, Passaro C, Chieffi P, Aurora B. a new prognostic marker and therapeutic target in cancer. Curr Med Chem 2011; 18: 482–496. [DOI] [PubMed] [Google Scholar]
- Fields AP, Justilien V. The guanine nucleotide exchange factor (GEF) Ect2 is an oncogene in human cancer. Adv Enzyme Regul 2010; 50: 190–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SV, Settleman J. Oncogene addiction: setting the stage for molecularly targeted cancer therapy. Genes Dev 2007; 21: 3214–3231. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012; 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A, Pachter L. Streaming fragment assignment for real-time analysis of sequencing experiments. Nat Methods 2013; 10: 71–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014; 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanning CC, Daddona JL, Ruiz-Velasco R, Shafer SH, Williams CL. The Rac1 C-terminal polybasic region regulates the nuclear localization and protein degradation of Rac1. J Biol Chem 2004; 279: 44197–44210. [DOI] [PubMed] [Google Scholar]
- Nguyen, le XT, Mitchell BS. Akt activation enhances ribosomal RNA synthesis through casein kinase II and TIF-IA. Proc Natl Acad Sci USA 2013; 110: 20681–20686. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.