Abstract
DEK is an oncoprotein that is overexpressed in many forms of cancer and participates in numerous cellular pathways. Of these different pathways, relevant interacting partners and functions of DEK are well described in regard to the regulation of chromatin structure, epigenetic marks, and transcription. Most of this understanding was derived by investigating DNA-binding and chromatin processing capabilities of the oncoprotein. To facilitate the generation of mechanism-driven hypotheses regarding DEK activities in underexplored areas, we have developed the first DEK interactome model using tandem-affinity purification and mass spectrometry. With this approach we confirmed IMPDH2 and DDX21 as novel DEK binding partners, hinting at new roles for the oncogene in de novo nucleotide biosynthesis and rRNA processing, respectively. Additionally, a hydroxyurea-specific interaction with RPA was observed, suggesting that a DEK-RPA complex may form in response to DNA replication fork stalling. Taken together, these findings highlight diverse activities for DEK across cellular pathways and support a model wherein this molecule performs a plethora of functions.
Keywords: DDX21, DEK, IMPDH2, interactome, mass spectrometry metabolism, RPA
INTRODUCTION
DEK is a DNA-binding and predominantly nuclear protein that was first identified as a DEK-NUP214 fusion protein in AML1. Since this discovery, DEK has been classified as an oncoprotein and shown to be overexpressed in many diverse tumor types2, wherein the degree of overexpression was linked to worse prognosis, advanced stage tumors, and chemotherapy resistance3–9. Cellular functions of the oncoprotein include activities in modifying chromatin structure2,10–13, histone chaperoning14,15, epigenetic modification and transcription regulation16–19, mRNA splicing20,21, DNA repair22,23, DNA replication fork restart24, mitotic non-disjunction events25, evasion of senescence and apoptosis19,26,27, proliferation28–30 cancer stem cell fitness and invasion28,31, inflammation32,33, and metabolic reprogramming34. Precise molecular mechanisms whereby DEK regulates these cellular processes remain unclear in many cases. For instance, while DEK is necessary for optimal non-homologous end joining DNA repair, the mechanism of action or interacting partners remain elusive23.
For an oncogene that is widely implicated in human carcinogenesis and outcome, determining how DEK operates is imperative. However, this has proven challenging as the oncoprotein DEK has no known enzymatic activity or paralogs2. While significant progress has been made to understand molecular DEK activities in transcription via chromatin remodeling2,10–13,16–19, there is a dearth of information regarding other functions. In part this is due to the limited number of published proteomic data sets which identify potential DEK interactors. Most proteome studies have not examined DEK interactions but instead explored differences in protein expression between DEK deficient versus proficient cells35, found DEK to be a protein secreted by cells36 and a component of the insoluble nuclear fraction37, and determined that the oncogene was hypophosphorylated during apoptosis38. The only proteomic report that identified interacting partners, to our knowledge, was conducted with drosophila-DEK and identified CKII and histones as interacting partners15. Thus, there is a need to develop a global DEK interactome.
Once confirmed, these binding partners can be used to localize where DEK may function in a given pathway and provide testable hypotheses for a mechanism of action. Further, since the literature has demonstrated DEK to be exceptionally multifunctional, it is possible that a proteomics approach may also identify novel pathways in which the protein may function. To this end we have used an established tandem affinity purification and mass spectrometry (TAP-MS) approach on a tagged DEK construct. Functional enrichment analysis revealed interactors in biological processes and pathways where DEK involvement is known and identified novel pathways where the oncogene is now implicated. These include nucleotide synthesis and rRNA processing, with IMPDH2 and DDX21 confirmed to interact with DEK, respectively. This approach also identified binding partners that may interact with DEK following DNA damage, and RPA was a validated binding partner. Additionally, four novel DEK phosphorylation sites were discovered by dissection of captured DEK peptides, and the most likely kinase candidates were predicted.
In summary, our proteomics approach has identified novel interacting factors, cellular roles, and phosphorylation sites regarding the DEK oncoprotein. The results of this analysis will hopefully provide a foundation for future mechanistic studies into known and novel functions of the human DEK oncoprotein.
MATERIALS AND METHODS
Cell culture conditions and retroviral constructs
HeLa cells were grown in Dubecco’s Modified Essential Medium (DMEM) completed with 10% fetal bovine serum (FBS) and antibiotics. The MIEG-His-FLAG DEK retroviral vector was described recently39, and cells were transduced with virus for >24hrs prior to GFP sorting on a BD-FACSAria II flow cytometer. To induce DNA damage, cells were treated with 1mM hydroxyurea (HU) for 15 hours prior to collection.
Tandem affinity purification and mass spectrometry (TAP-MS)
TAP was performed as described previously40. Briefly, pMIEG3 and pMIEG3-His-Flag-DEK transduced HeLa cells were homogenized in lysis buffer (10mM HEPES pH=7.9, 1.5mM MgCl2, 10mM KCl, 0.5mM PMSF, 0.5mM DTT, 1mM NaVO3, 10mM NaF, and 1x protease inhibitor cocktail), and the nuclear pellet was lysed in a nuclear lysis buffer (20mM HEPES pH=7.9, 400mM NaCl, 1% Triton, 0.1% NP-40, 10% Glycerol, 0.5mM PMSF, 0.5mM DTT, 1mM NaVO3, 10mM NaF, and 1x protease inhibitor cocktail) on ice. After ultracentifugation, the DEK-containing supernatant was loaded onto anti-FLAG M2 affinity gel (A2200, Sigma, St. Louis, MO, USA) and incubated overnight at 4°C. Following addition of Talon Resin Buffer (20mM HEPES pH=7.9, 300mM NaCl, 0.1% NP-40, 10% Glycerol, 10mM Imidazole) with 300ng/ml of 3x-FLAG, the eluate was added to TALON Superflow beads (635506, Clonetech, Mountain View, CA, USA) for 3hrs at 4°C. TALON beads were washed and then boiled in 2x Laemmli Buffer (12.5mM Tris-HCl, 20% Glycerol, 4% SDS, 0.004% Bromophenol Blue, 10% 2-β-mercaptoethanol.). The samples were ran by SDS-PAGE and silver stained to identify protein enrichment. A second SDS-PAGE was stained with Coomassie brilliant blue, the lanes excised, and sent to the Taplin Biological Mass Spectrometry Core Facility at Harvard Medical School for analysis. GeneVenn was utilized to sort hits in the MIEG and HF-DEK samples, and the MIEG hits were subtracted from the untreated and HU datasets prior to sorting with GeneVenn. Functional enrichment analysis (p-value <0.05) was performed using the ToppFun application of the ToppGene Suite to categorize the DEK interacting partners by enriched pathways and biological processes in gene ontology41. Network representation of selected enriched biological processes and pathways was done using Cytoscape (v3.4)42.
Phosphorylation site analysis
The mass spectrometry data was evaluated for phosphorylated peptides in DEK using the Mascot Search algorithm (Matrix Sciences, London, UK) against the SwissProt protein database (12/15/2011) of 533,657 sequences of which 20,323 corresponded to the Homo sapiens taxonomy. Searches included a fixed modification of carbamidomethyl-cysteine and variable modifications including oxidation of methionine and phosphorylation of serine, threonine or tyrosine. Phosphorylated peptide with a MASCOT significance threshold p<0.005 and a peptide false discovery rate of <5% are reported. Predicted kinases were calculated by Group-based Prediction System v3.0 (“GPS 3.0,” The CUCKOO Workgroup, Wuhan, Hubei, China)43.
Western blot analysis
Immunoprecipitated proteins were separated by 10% SDS-PAGE electrophoresis, and transferred onto a PVDF membrane for 1–2hr at 500mA. Antibodies used are listed as follows: DEK (610948, BD Bioscience, San Jose, CA, USA), Casein Kinase II (04-1129, EMD Millipore, Darmstadt, Germany), IMPDH2 (AP7390a, Abgent, San Diego, CA), DDX21 (AP174238a, Abgent), and RPA1 (AP14415b, Abgent).
RESULTS AND DISCUSSION
To identify DEK interacting partners, TAP-MS was utilized on HeLa cells transduced with an empty MIEG or a MIEG-His-FLAG-DEK vector. Nuclear extracts were collected, and His-FLAG DEK (HF-DEK) was enriched using an established FLAG and Talon TAP protocol40. As seen in Fig. 1A, HF-DEK forms a distinct band by Western blot, and is enriched after each serial pulldown. The presence of interacting partners was confirmed by silver stain (Fig. 1B), and gel slices from a paired Coomassie-stained gel were sent to the Taplin Mass Spectrometry Facility for analysis. Using a protein score cutoff score of 2, which reflects a high probability that the observed ions match the indicated protein as represented in values of −10log(p), 163 hits were identified in the HF-DEK sample compared to 47 for MIEG (Tables S-I and S-II). Visualization of the data with a Venn diagram revealed that 123 of the hits were unique to the HF-DEK sample and were investigated further (Fig. 1C and Table S-II with individual scores, rank, and peptide sequences available in Table S-I). Some of the identified proteins are well known DEK interactors thus validating the approach, including two that are represented in the top 20 proteins as ranked by protein score (Table I): casein kinase II, which phosphorylates DEK15,44, and histone 2A, which is chaperoned by the oncoprotein15,17.
Fig. 1. TAP-MS of His-FLAG DEK identified 167 potential DEK interacting partners.
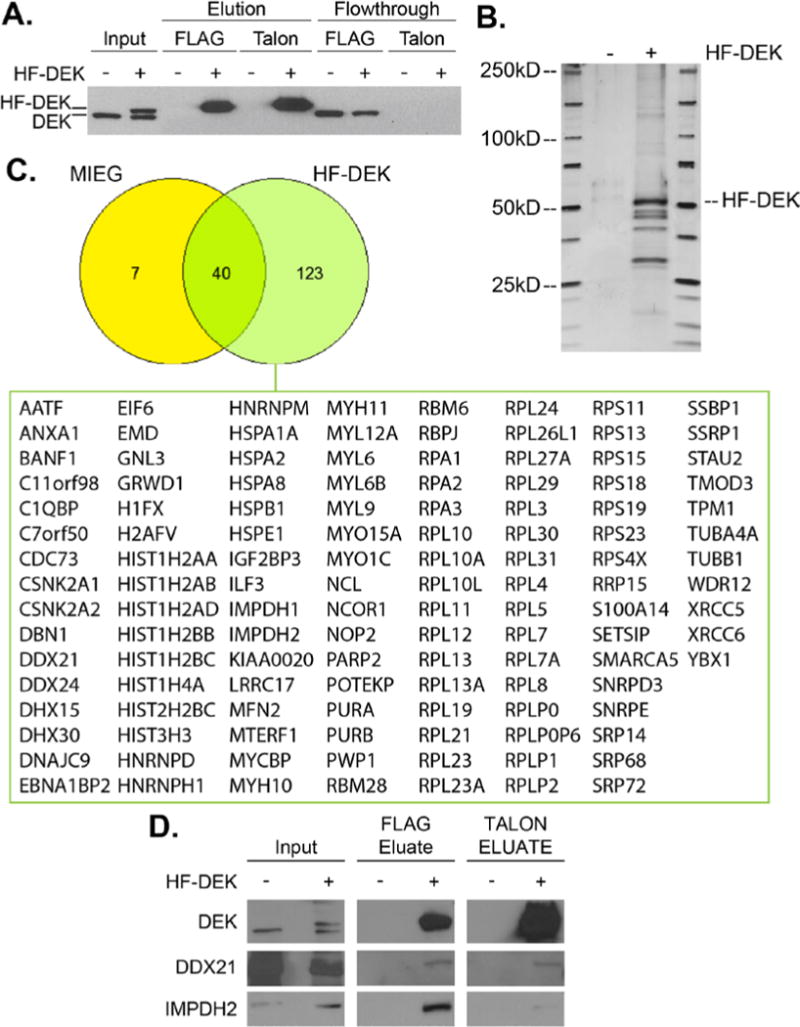
(A) Western blot analysis identified enrichment of HF-tagged DEK by tandem FLAG and Talon affinity purification. MIEG lysates are indicated by (−), HF-DEK by (+). (B) Silver stain of (A) indicated enrichment of HF-DEK and other proteins compared to MIEG control. (C) Lanes from MIEG and HF-DEK samples were sent to the Taplin Mass Spectrometry Facility for analysis. A total of 163 potential DEK interacting factors were identified by TAP-MS. Of these hits, 123 were unique to the HF-DEK sample and are listed below the Venn diagram. The number of peptides and scores for each hit are available in Table S-I. (D) DDX21 and IMPDH2 were confirmed to interact with DEK by Western blot analysis.
Table I.
Top 20 HF-DEK interacting factors identified by TAP-MS
| Rank | Gene Symbol | Unique Peptides | Total Peptides | Protein Score |
|---|---|---|---|---|
| 1 | YBX1 | 2 | 3 | 5.09 |
| 2 | C7orf50 | 1 | 1 | 5.03 |
| 3 | DDX24 | 1 | 1 | 5.00 |
| 4 | RPLP0 | 4 | 9 | 4.39 |
| 5 | C1QBP | 7 | 16 | 4.35 |
| 6 | WDR12 | 1 | 1 | 4.18 |
| 7 | RPL10 | 2 | 2 | 4.15 |
| 8 | CSNK2A1 | 22 | 143 | 4.02 |
| 9 | TMOD3 | 1 | 2 | 4.02 |
| 10 | SETSIP | 2 | 3 | 3.98 |
| 11 | HIST1H2AB | 2 | 6 | 3.94 |
| 12 | CSNK2A2 | 20 | 44 | 3.90 |
| 13 | AATF | 3 | 3 | 3.84 |
| 14 | IMPDH2 | 12 | 18 | 3.83 |
| 15 | GRWD1 | 5 | 5 | 3.83 |
| 16 | DBN1 | 1 | 1 | 3.80 |
| 17 | PURB | 2 | 2 | 3.77 |
| 18 | MYH11 | 5 | 10 | 3.72 |
| 19 | BANF1 | 3 | 3 | 3.69 |
| 20 | RPS23 | 1 | 1 | 3.69 |
Beyond known interactors, the highest scoring DEK binding proteins are predominantly involved in ribosome biosynthesis, function, and localization. Correspondingly, related pathways were most significantly represented in the gene ontology analysis (Tables II and S-III), and a distinct node for ribosome synthesis and rRNA processing was observed in the pathway analysis map (Fig. 2). The observed dramatic enrichment of these ribosome-associated factors is intriguing as no rRNA or ribosomal activity has been attributed to DEK to date. To provide further evidence for potential DEK activities in ribosome associated pathways, we validated the interaction with DDX21, a RNA helicase that promotes rRNA transcription and modification45 (Fig. 1D).
Table II.
Representative hits from gene ontology pathway analysis of HF-DEK interacting partners
| Rank | Name | q-value FDR B&H |
|---|---|---|
| 1 | SRP-dependent co-translational protein targeting to membrane | 2.01E-46 |
| 2 | Peptide chain elongation | 5.85E-45 |
| 3 | Viral mRNA Translation | 5.85E-45 |
| 4 | Selenocysteine synthesis | 1.55E-44 |
| 5 | Eukaryotic Translation Termination | 1.87E-44 |
| 6 | Eukaryotic Translation Elongation | 2.34E-44 |
| 7 | Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | 4.44E-44 |
| 8 | Formation of a pool of free 40S subunits | 5.42E-43 |
| 9 | L13a-mediated translational silencing of Ceruloplasmin expression | 2.53E-41 |
| 10 | GTP hydrolysis and joining of the 60S ribosomal subunit | 2.53E-41 |
| 17 | Influenza Life Cycle | 7.02E-40 |
| 21 | rRNA processing in the nucleus and cytosol | 1.19E-38 |
| 24 | Gene Expression | 2.06E-30 |
| 26 | Metabolism of amino acids and derivatives | 2.80E-23 |
| 29 | Telomere Maintenance | 6.15E-07 |
| 30 | Chromosome Maintenance | 6.63E-07 |
| 31 | Meiotic recombination | 1.23E-06 |
| 36 | DNA Double-Strand Break Repair | 6.06E-05 |
| 42 | Formation of the beta-catenin:TCF transactivating complex | 3.96E-04 |
| 47 | Nucleosome assembly | 1.04E-03 |
Fig. 2. Gene ontology pathway map of the HF-DEK interactome.
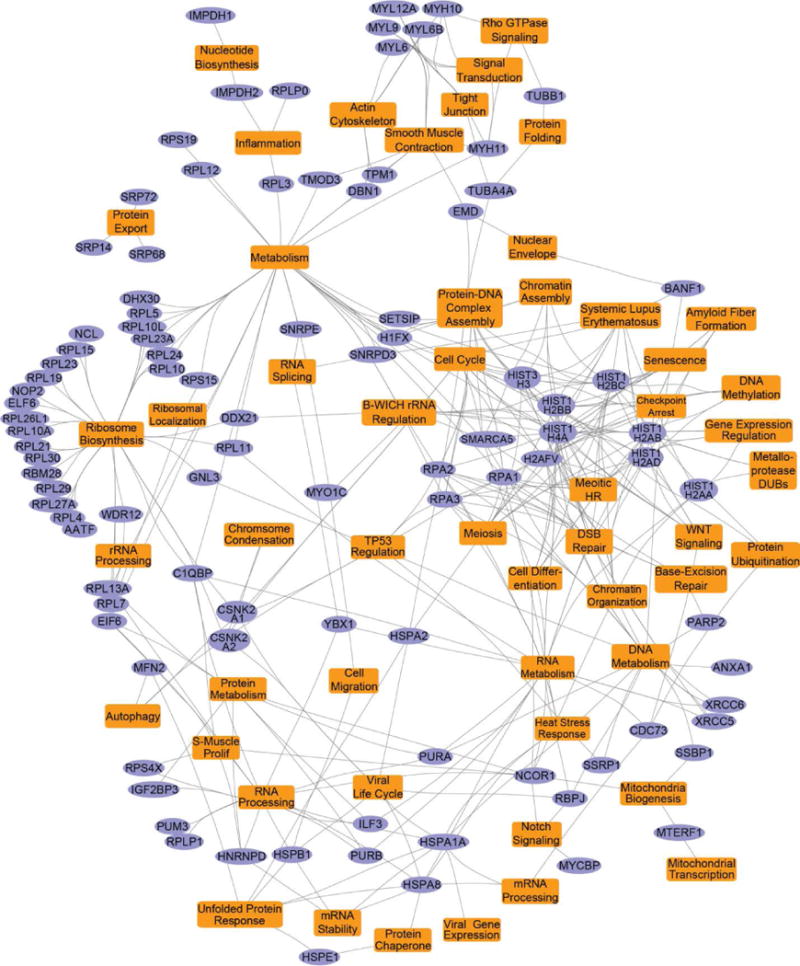
Gene ontology and pathway analysis was performed on the 123 interacting HF-DEK factors identified in Fig. 1C using the ToppFun application of ToppGene Suite. Select enriched pathways, biological processes, and their associated genes (as highlighted in Table S3) are represented as a network using Cytoscape (v3.4). In the map, DEK interacting factors discovered by TAP-MS are shown as purple elliptical nodes, and the enriched pathways and biological processes are indicated as orange colored rectangles.
Similarly, many interactors formed nodes that reflected DEK activities in metabolism, nucleotide synthesis, and inflammation (Fig. 2). Of these potential binding partners, we confirmed an interaction between IMPDH2 and DEK (Fig. 1D). Both IMPDH2 and the associated IMPDH1 isoform, that was also identified in the screen, catalyze the rate-limiting step of de novo guanine nucleotide synthesis46,47. Similar to DEK, elevated IMPDH2 expression has been linked to tumor progression and chemotherapy resistance in patients48,49. However, a mechanism for how IMPDH2 drives these cancer phenotypes is unknown. Should this DEK-IMPDH2 complex alter IMPDH2 activity, it may in part explain the proliferation advantages observed in some DEK overexpression models and the stagnation of growth in DEK knockdown systems26,29,30.
Other potential binding partners belonged to several pathways already known to be regulated by DEK (Fig. 1C and Fig. 2). Regarding chromatin structure, we newly identify histone 1 family member, H1FX, as a DEK interactor. Novel epigenetic modulators were also uncovered, including SMARCA5, a SWI/SNF-related and nucleosome-remodeling helicase factor that is part of the B-WICH complex50, and NCOR1, a transcriptional regulator that, like DEK, mediates histone deacetylase activity51. Additionally, mRNA splicing factors were detected by mass spectrometry, although U2AF, an established in vitro and cell-free DEK interactor21, was not observed.
In addition to the above pathways, the function of DEK in DNA double strand break (DSB) repair has become a topic of interest due to clinical associations between the oncoprotein and chemotherapy resistance in cancer3,8,9. Molecularly, DEK may mediate chemoresistance through several mechanisms. It allows the cell to overcome chemotherapy-induced DNA replication stress by facilitating the restart of stalled replication forks, the mechanism of which is uncertain24, and it is required for optimal repair of DNA double strand breaks (DSBs), a highly toxic lesion that is directly and indirectly induced by chemotherapeutics22,23. With regard to the DSB repair pathways, DEK was discovered to be essential for homologous recombination (HR), and may function through an interaction with RAD51. However, it remains unclear which RAD51 complex(es) contain DEK22. For a complementary DSB repair pathway, non-homologous end joining (NHEJ), DEK is necessary for optimal repair and may operate by promoting the timely recruitment of the Ku70/80 heterodimer, the pathway’s initiating factors23.
The TAP-MS analysis identified that Ku70/80 (XRCC5 and XRCC6) may physically interact with DEK (Fig. 1C and 2). A second HF-DEK TAP-MS conducted in the presence of 1mM hydroxyurea (HU) to induce DNA damage recognized Ku70/80 more readily following DNA damage induction compared to the untreated sample (Fig. 2A–B and Tables S-IV and S-V). Beyond DNA repair, the most common pathways identified by gene ontology analysis of the hits unique to HU treatment were RNA processing and HIV life cycle events (Fig. 3B, Tables III and S-VI). The observation of HIV life cycle events is not surprising as NHEJ, including Ku70/80, is required to circularize retro/lentiviral DNA or insert the provirus into the genome52.
Fig. 3. A small number of proteins were identified to specifically interact with DEK following DNA damage.
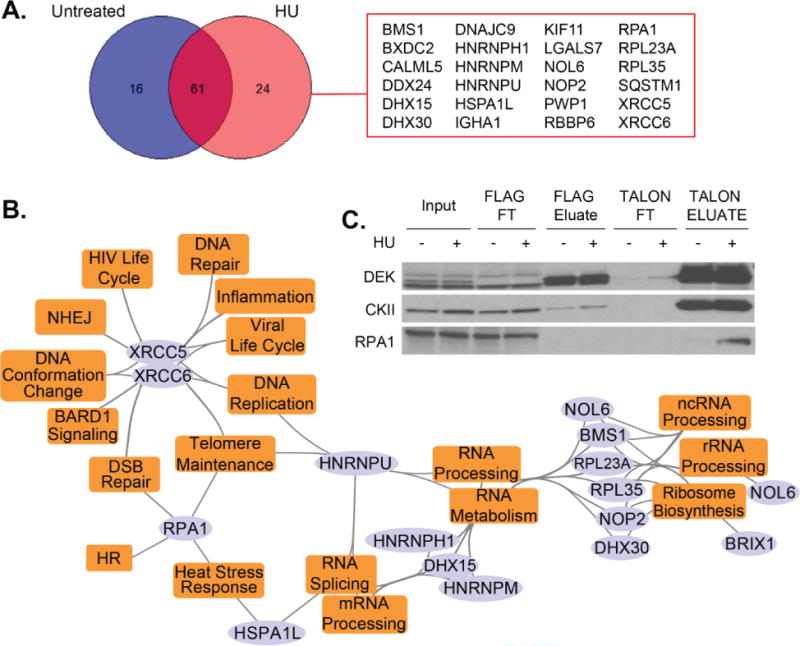
(A) TAP-MS was performed on HF-DEK overexpressing cells that were untreated or incubated with 1mM HU for 15 hours. Analyses revealed a small number of genes that specifically precipitated in HU-treated cells. (B) A pathway and biological process map, based on functional enrichment analysis (Table S3), was generated using the identified hits from the HU-treated sample. Purple colored elliptical nodes represent genes while orange colored rectangles represent enriched pathways and biological processes. (C) RPA1 was confirmed as an interacting partner whose affinity increased following HU treatment.
Table III.
Representative top hits from gene ontology and pathway analysis of HF-DEK interacting partners following HU treatment
| Rank | Pathway Name | q-value FDR B&H |
|---|---|---|
| 1 | Spliceosome | 1.07E-03 |
| 2 | mRNA processing | 1.07E-03 |
| 3 | Cytoskeletal regulation by Rho GTPase | 1.07E-03 |
| 4 | Double-Strand Break Repair | 1.07E-03 |
| 5 | Nicotinic acetylcholine receptor signaling pathway | 2.04E-03 |
| 7 | Non-homologous end joining (NHEJ) | 2.56E-03 |
| 11 | 2-LTR circle formation | 3.10E-03 |
| 13 | Integration of provirus | 6.74E-03 |
| 15 | IRF3-mediated induction of type I IFN | 7.33E-03 |
| 16 | STING mediated induction of host immune responses | 1.02E-02 |
| 17 | Telomeres, Telomerase, Cellular Aging, and Immortality | 1.02E-02 |
| 20 | Salmonella infection | 1.12E-02 |
Additionally members of the RPA heterotrimer family, proteins involved in single-strand DNA binding during HR and DNA replication53, were identified (Fig. 1C, and 3A–B, Table S-IV and S-V) and validated by Western blot analysis (Fig. 2B). Interestingly, the DEK-RPA complex was strongest following HU treatment with minimal interaction observed following treatment with 10Gy of γ-irradiation (Fig. S1). Considering that HU treatment induces stalled replication forks that collapse into DSBs54 and that γ-irradiation immediately generates DSBs55, DEK is most likely to interact with RPA in the context of replication fork restart instead of DSB-induced homologous recombination.
In addition to identifying potential protein interactors, each identified DEK peptide sequence was screened to identify novel phosphorylation sites on the protein and described in the Methods section. In total 77% of the protein sequence was covered, and four new phosphorylation sites were found on S2, S4, T13, and T342 (Fig. 4). For each of these residues, the top predicted kinases were identified by GPS 3.0 software43 (Table S-VII). Of these, casein kinase I (CKI) was predicted to phosphorylate multiple residues. CKI is particularly interesting as it both positively and negatively regulates WNT signaling56. If verified as a modifier of DEK, CK1 may help to identify the mechanisms by which DEK supports the WNT-β-catenin signaling loop28. Secondly, phosphorylation of DEK by other kinases has been demonstrated to weaken the protein’s affinity for DNA44.
Fig. 4. Four novel phosphorylation sites were identified by TAP-MS.

The full DEK protein sequence is shown with TAP-MS identified regions bolded. All identified phosphorylation sites are in blue font, and the four novel phosphorylated residues are annotated with the top predicted kinases as calculated by GPS 3.043. This image is a composite of the HF-DEK and untreated samples, and no novel phosphorylated sites were identified in the HU data set. Individual data sets are represented in Supplementary Data 1 and GPS predictions in Table S-VII.
In conclusion, this work has utilized TAP-MS methodology to develop a DEK interactome map, to predict DEK binding partners, and to point to new cellular processes that may be regulated by DEK. While the literature certainly suggests that DEK has multiple functions across different pathways, the extent and variety of its operations has not been mapped or explored in detail. DEK interacting partners identified in the study align with classical DEK functions described in previous reports and potentially expand the number of regulated pathways to include rRNA maturation and nucleotide synthesis. Taken together, these findings support a model whereby the nuclear DEK protein interfaces with numerous cellular pathways via protein-protein interactions.
Supplementary Material
Acknowledgments
We would like to thank the Taplin Mass Spectrometry Facility for conducting the mass spectrometry analysis.
FUNDING
This work was supported by the National Institutes of Health (R01-CA-102357 to S.W., R01-CA-116316 to S.W.) and the University of Cincinnati Department of Cancer and Cell Biology (Cardell Fellowship for Excellence in Graduate Research to E.S.).
Footnotes
The authors have no conflicts of interest to disclose.
References
- 1.von Lindern M, Fornerod M, van Baal S, Jaegle M, de Wit T, Buijs A, Grosveld G. The translocation (6;9), associated with a specific subtype of acute myeloid leukemia, results in the fusion of two genes, dek and can, and the expression of a chimeric, leukemia-specific dek-can mRNA. Mol Cell Biol. 1992;12(4):1687–97. doi: 10.1128/mcb.12.4.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Privette Vinnedge LM, Kappes F, Nassar N, Wells SI. Stacking the DEK: from chromatin topology to cancer stem cells. Cell Cycle. 2013;12(1):51–66. doi: 10.4161/cc.23121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khodadoust MS, Verhaegen M, Kappes F, Riveiro-Falkenbach E, Cigudosa JC, Kim DS, Chinnaiyan AM, Markovitz DM, Soengas MS. Melanoma proliferation and chemoresistance controlled by the DEK oncogene. Cancer Res. 2009;69(16):6405–13. doi: 10.1158/0008-5472.CAN-09-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riveiro-Falkenbach E, Ruano Y, Garcia-Martin RM, Lora D, Cifdaloz M, Acquadro F, Ballestin C, Ortiz-Romero PL, Soengas MS, Rodriguez-Peralto JL. DEK oncogene is overexpressed during melanoma progression. Pigment Cell Melanoma Res. 2016 doi: 10.1111/pcmr.12563. [DOI] [PubMed] [Google Scholar]
- 5.Liu X, Qi D, Qi J, Mao Z, Li X, Zhang J, Li J, Gao W. Significance of DEK overexpression for the prognostic evaluation of non-small cell lung carcinoma. Oncol Rep. 2016;35(1):155–62. doi: 10.3892/or.2015.4365. [DOI] [PubMed] [Google Scholar]
- 6.Ou Y, Xia R, Kong F, Zhang X, Yu S, Jiang L, Zheng L, Lin L. Overexpression of DEK is an indicator of poor prognosis in patients with gastric adenocarcinoma. Oncol Lett. 2016;11(3):1823–1828. doi: 10.3892/ol.2016.4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith EA, Kumar B, Komurov K, Smith SM, Brown NV, Zhao S, Kumar P, Teknos TN, Wells SI. DEK associates with tumor stage and outcome in HPV16 positive oropharyngeal squamous cell carcinoma. Oncotarget. 2017;8(14):23414–23426. doi: 10.18632/oncotarget.15582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martinez-Useros J, Rodriguez-Remirez M, Borrero-Palacios A, Moreno I, Cebrian A, Gomez del Pulgar T, del Puerto-Nevado L, Vega-Bravo R, Puime-Otin A, Perez N, et al. DEK is a potential marker for aggressive phenotype and irinotecan-based therapy response in metastatic colorectal cancer. BMC Cancer. 2014;14:965. doi: 10.1186/1471-2407-14-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ying G, Wu Y. DEK: A novel early screening and prognostic marker for breast cancer. Mol Med Rep. 2015;12(5):7491–5. doi: 10.3892/mmr.2015.4380. [DOI] [PubMed] [Google Scholar]
- 10.Alexiadis V, Waldmann T, Andersen J, Mann M, Knippers R, Gruss C. The protein encoded by the proto-oncogene DEK changes the topology of chromatin and reduces the efficiency of DNA replication in a chromatin-specific manner. Genes Dev. 2000;14(11):1308–12. [PMC free article] [PubMed] [Google Scholar]
- 11.Waldmann T, Eckerich C, Baack M, Gruss C. The ubiquitous chromatin protein DEK alters the structure of DNA by introducing positive supercoils. J Biol Chem. 2002;277(28):24988–94. doi: 10.1074/jbc.M204045200. [DOI] [PubMed] [Google Scholar]
- 12.Bohm F, Kappes F, Scholten I, Richter N, Matsuo H, Knippers R, Waldmann T. The SAF-box domain of chromatin protein DEK. Nucleic Acids Res. 2005;33(3):1101–10. doi: 10.1093/nar/gki258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kappes F, Waldmann T, Mathew V, Yu J, Zhang L, Khodadoust MS, Chinnaiyan AM, Luger K, Erhardt S, Schneider R, et al. The DEK oncoprotein is a Su(var) that is essential to heterochromatin integrity. Genes Dev. 2011;25(7):673–8. doi: 10.1101/gad.2036411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ivanauskiene K, Delbarre E, McGhie JD, Kuntziger T, Wong LH, Collas P. The PML-associated protein DEK regulates the balance of H3.3 loading on chromatin and is important for telomere integrity. Genome Res. 2014;24(10):1584–94. doi: 10.1101/gr.173831.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sawatsubashi S, Murata T, Lim J, Fujiki R, Ito S, Suzuki E, Tanabe M, Zhao Y, Kimura S, Fujiyama S, et al. A histone chaperone, DEK, transcriptionally coactivates a nuclear receptor. Genes Dev. 2010;24(2):159–70. doi: 10.1101/gad.1857410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gamble MJ, Fisher RP. SET and PARP1 remove DEK from chromatin to permit access by the transcription machinery. Nat Struct Mol Biol. 2007;14(6):548–55. doi: 10.1038/nsmb1248. [DOI] [PubMed] [Google Scholar]
- 17.Hollenbach AD, McPherson CJ, Mientjes EJ, Iyengar R, Grosveld G. Daxx and histone deacetylase II associate with chromatin through an interaction with core histones and the chromatin-associated protein Dek. J Cell Sci. 2002;115(Pt 16):3319–30. doi: 10.1242/jcs.115.16.3319. [DOI] [PubMed] [Google Scholar]
- 18.Ko SI, Lee IS, Kim JY, Kim SM, Kim DW, Lee KS, Woo KM, Baek JH, Choo JK, Seo SB. Regulation of histone acetyltransferase activity of p300 and PCAF by proto-oncogene protein DEK. FEBS Lett. 2006;580(13):3217–22. doi: 10.1016/j.febslet.2006.04.081. [DOI] [PubMed] [Google Scholar]
- 19.Lee KS, Kim DW, Kim JY, Choo JK, Yu K, Seo SB. Caspase-dependent apoptosis induction by targeted expression of DEK in Drosophila involves histone acetylation inhibition. J Cell Biochem. 2008;103(4):1283–93. doi: 10.1002/jcb.21511. [DOI] [PubMed] [Google Scholar]
- 20.Delaunay S, Rapino F, Tharun L, Zhou Z, Heukamp L, Termathe M, Shostak K, Klevernic I, Florin A, Desmecht H, et al. Elp3 links tRNA modification to IRES-dependent translation of LEF1 to sustain metastasis in breast cancer. J Exp Med. 2016;213(11):2503–2523. doi: 10.1084/jem.20160397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soares LM, Zanier K, Mackereth C, Sattler M, Valcarcel J. Intron removal requires proofreading of U2AF/3′ splice site recognition by DEK. Science. 2006;312(5782):1961–5. doi: 10.1126/science.1128659. [DOI] [PubMed] [Google Scholar]
- 22.Smith EA, Gole B, Willis NA, Soria R, Starnes LM, Krumpelbeck EF, Jegga AG, Ali AM, Guo H, Meetei AR, et al. DEK is required for homologous recombination repair of DNA breaks. Sci Rep. 2017;7:44662. doi: 10.1038/srep44662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kavanaugh GM, Wise-Draper TM, Morreale RJ, Morrison MA, Gole B, Schwemberger S, Tichy ED, Lu L, Babcock GF, Wells JM, et al. The human DEK oncogene regulates DNA damage response signaling and repair. Nucleic Acids Res. 2011;39(17):7465–76. doi: 10.1093/nar/gkr454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deutzmann A, Ganz M, Schonenberger F, Vervoorts J, Kappes F, Ferrando-May E. The human oncoprotein and chromatin architectural factor DEK counteracts DNA replication stress. Oncogene. 2015;34(32):4270–7. doi: 10.1038/onc.2014.346. [DOI] [PubMed] [Google Scholar]
- 25.Matrka MC, Hennigan RF, Kappes F, DeLay ML, Lambert PF, Aronow BJ, Wells SI. DEK over-expression promotes mitotic defects and micronucleus formation. Cell Cycle. 2015;0 doi: 10.1080/15384101.2015.1044177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wise-Draper TM, Allen HV, Thobe MN, Jones EE, Habash KB, Munger K, Wells SI. The human DEK proto-oncogene is a senescence inhibitor and an upregulated target of high-risk human papillomavirus E7. J Virol. 2005;79(22):14309–17. doi: 10.1128/JVI.79.22.14309-14317.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wise-Draper TM, Allen HV, Jones EE, Habash KB, Matsuo H, Wells SI. Apoptosis inhibition by the human DEK oncoprotein involves interference with p53 functions. Mol Cell Biol. 2006;26(20):7506–19. doi: 10.1128/MCB.00430-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Privette Vinnedge LM, Benight NM, Wagh PK, Pease NA, Nashu MA, Serrano-Lopez J, Adams AK, Cancelas JA, Waltz SE, Wells SI. The DEK oncogene promotes cellular proliferation through paracrine Wnt signaling in Ron receptor-positive breast cancers. Oncogene. 2015;34(18):2325–36. doi: 10.1038/onc.2014.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wise-Draper TM, Morreale RJ, Morris TA, Mintz-Cole RA, Hoskins EE, Balsitis SJ, Husseinzadeh N, Witte DP, Wikenheiser-Brokamp KA, Lambert PF, et al. DEK proto-oncogene expression interferes with the normal epithelial differentiation program. Am J Pathol. 2009;174(1):71–81. doi: 10.2353/ajpath.2009.080330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adams AK, Hallenbeck GE, Casper KA, Patil YJ, Wilson KM, Kimple RJ, Lambert PF, Witte DP, Xiao W, Gillison ML, et al. DEK promotes HPV-positive and -negative head and neck cancer cell proliferation. Oncogene. 2015;34(7):868–77. doi: 10.1038/onc.2014.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Privette Vinnedge LM, McClaine R, Wagh PK, Wikenheiser-Brokamp KA, Waltz SE, Wells SI. The human DEK oncogene stimulates beta-catenin signaling, invasion and mammosphere formation in breast cancer. Oncogene. 2011;30(24):2741–52. doi: 10.1038/onc.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adams AK, Bolanos LC, Dexheimer PJ, Karns RA, Aronow BJ, Komurov K, Jegga AG, Casper KA, Patil YJ, Wilson KM, et al. IRAK1 is a novel DEK transcriptional target and is essential for head and neck cancer cell survival. Oncotarget. 2015;6(41):43395–407. doi: 10.18632/oncotarget.6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pease NA, Wise-Draper T, Privette Vinnedge L. Dissecting the Potential Interplay of DEK Functions in Inflammation and Cancer. J Oncol. 2015;2015:106517. doi: 10.1155/2015/106517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matrka MC, Watanabe M, Muraleedharan R, Lambert PF, Lane AN, Romick-Rosendale LE, Wells SI. Overexpression of the human DEK oncogene reprograms cellular metabolism and promotes glycolysis. PLoS One. 2017;12(5):e0177952. doi: 10.1371/journal.pone.0177952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim DW, Chae JI, Kim JY, Pak JH, Koo DB, Bahk YY, Seo SB. Proteomic analysis of apoptosis related proteins regulated by proto-oncogene protein DEK. J Cell Biochem. 2009;106(6):1048–59. doi: 10.1002/jcb.22083. [DOI] [PubMed] [Google Scholar]
- 36.Choi S, Park SY, Kwak D, Phark S, Lee M, Lim JY, Jung WW, Sul D. Proteomic analysis of proteins secreted by HepG2 cells treated with butyl benzyl phthalate. J Toxicol Environ Health A. 2010;73(21–22):1570–85. doi: 10.1080/15287394.2010.511583. [DOI] [PubMed] [Google Scholar]
- 37.Takata H, Nishijima H, Ogura S, Sakaguchi T, Bubulya PA, Mochizuki T, Shibahara K. Proteome analysis of human nuclear insoluble fractions. Genes Cells. 2009;14(8):975–90. doi: 10.1111/j.1365-2443.2009.01324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tabbert A, Kappes F, Knippers R, Kellermann J, Lottspeich F, Ferrando-May E. Hypophosphorylation of the architectural chromatin protein DEK in death-receptor-induced apoptosis revealed by the isotope coded protein label proteomic platform. Proteomics. 2006;6(21):5758–72. doi: 10.1002/pmic.200600197. [DOI] [PubMed] [Google Scholar]
- 39.Matrka MC, Hennigan RF, Kappes F, DeLay ML, Lambert PF, Aronow BJ, Wells SI. DEK over-expression promotes mitotic defects and micronucleus formation. Cell Cycle. 2015;14(24):3939–53. doi: 10.1080/15384101.2015.1044177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ling C, Ishiai M, Ali AM, Medhurst AL, Neveling K, Kalb R, Yan Z, Xue Y, Oostra AB, Auerbach AD, et al. FAAP100 is essential for activation of the Fanconi anemia-associated DNA damage response pathway. EMBO J. 2007;26(8):2104–14. doi: 10.1038/sj.emboj.7601666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37:W305–11. doi: 10.1093/nar/gkp427. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xue Y, Zhou F, Zhu M, Ahmed K, Chen G, Yao X. GPS: a comprehensive www server for phosphorylation sites prediction. Nucleic Acids Res. 2005;33:W184–7. doi: 10.1093/nar/gki393. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kappes F, Damoc C, Knippers R, Przybylski M, Pinna LA, Gruss C. Phosphorylation by protein kinase CK2 changes the DNA binding properties of the human chromatin protein DEK. Mol Cell Biol. 2004;24(13):6011–20. doi: 10.1128/MCB.24.13.6011-6020.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Calo E, Flynn RA, Martin L, Spitale RC, Chang HY, Wysocka J. RNA helicase DDX21 coordinates transcription and ribosomal RNA processing. Nature. 2015;518(7538):249–53. doi: 10.1038/nature13923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crabtree GW, Henderson JF. Rate-limiting steps in the interconversion of purine ribonucleotides in Ehrlich ascites tumor cells in vitro. Cancer Res. 1971;31(7):985–91. [PubMed] [Google Scholar]
- 47.Sintchak MD, Fleming MA, Futer O, Raybuck SA, Chambers SP, Caron PR, Murcko MA, Wilson KP. Structure and mechanism of inosine monophosphate dehydrogenase in complex with the immunosuppressant mycophenolic acid. Cell. 1996;85(6):921–30. doi: 10.1016/s0092-8674(00)81275-1. [DOI] [PubMed] [Google Scholar]
- 48.Li HX, Meng QP, Liu W, Li YG, Zhang HM, Bao FC, Song LL, Li HJ. IMPDH2 mediate radioresistance and chemoresistance in osteosarcoma cells. Eur Rev Med Pharmacol Sci. 2014;18(20):3038–44. [PubMed] [Google Scholar]
- 49.Zhou L, Xia D, Zhu J, Chen Y, Chen G, Mo R, Zeng Y, Dai Q, He H, Liang Y, et al. Enhanced expression of IMPDH2 promotes metastasis and advanced tumor progression in patients with prostate cancer. Clin Transl Oncol. 2014;16(10):906–13. doi: 10.1007/s12094-014-1167-9. [DOI] [PubMed] [Google Scholar]
- 50.Kato A, Komatsu K. RNF20-SNF2H Pathway of Chromatin Relaxation in DNA Double-Strand Break Repair. Genes (Basel) 2015;6(3):592–606. doi: 10.3390/genes6030592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mottis A, Mouchiroud L, Auwerx J. Emerging roles of the corepressors NCoR1 and SMRT in homeostasis. Genes Dev. 2013;27(8):819–35. doi: 10.1101/gad.214023.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Skalka AM, Katz RA. Retroviral DNA integration and the DNA damage response. Cell Death Differ. 2005;12(Suppl 1):971–8. doi: 10.1038/sj.cdd.4401573. [DOI] [PubMed] [Google Scholar]
- 53.Oakley GG, Patrick SM. Replication protein A: directing traffic at the intersection of replication and repair. Front Biosci (Landmark Ed) 2010;15:883–900. doi: 10.2741/3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol Cell. 2010;37(4):492–502. doi: 10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Santivasi WL, Xia F. Ionizing radiation-induced DNA damage, response, and repair. Antioxid Redox Signal. 2014;21(2):251–9. doi: 10.1089/ars.2013.5668. [DOI] [PubMed] [Google Scholar]
- 56.Cruciat CM. Casein kinase 1 and Wnt/beta-catenin signaling. Curr Opin Cell Biol. 2014;31:46–55. doi: 10.1016/j.ceb.2014.08.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


