Abstract
The vascular barrier that separates blood from tissues is actively regulated by the endothelium and is essential for transport, inflammation, and hemostasis1. Hemodynamic shear stress plays a critical role in maintaining endothelial barrier function2, but how this occurs remains unknown. Here, using an engineered organotypic model of perfused microvessels and confirming in mouse models, we identify that activation of the Notch1 transmembrane receptor directly regulates vascular barrier function through a non-canonical, transcription independent signaling mechanism that drives adherens junction assembly. Shear stress triggers Dll4-dependent proteolytic activation of Notch1 to reveal the Notch1 transmembrane domain – the key domain that mediates barrier establishment. Expression of the Notch1 transmembrane domain is sufficient to rescue Notch1 knockout-induced defects in barrier function, and does so by catalyzing the formation of a novel receptor complex in the plasma membrane consisting of VE-cadherin, the transmembrane protein tyrosine phosphatase LAR, and the Rac1 GEF Trio. This complex activates Rac1 to drive adherens junction assembly and establish barrier function. Canonical Notch transcriptional signaling is highly conserved throughout metazoans and is required for many processes in vascular development, including arterial-venous differentiation3, angiogenesis4, and remodeling5; here, we establish the existence of a previously unappreciated non-canonical cortical signaling pathway for Notch1 that regulates vascular barrier function, and thus provide a mechanism by which a single receptor might link transcriptional programs with adhesive and cytoskeletal remodeling.
Keywords: endothelial cells, shear stress, Notch1, VE-cadherin, permeability, mechanotransduction, Rac1, actin cytoskeleton
The selective barrier formed by the vascular endothelial cell monolayer that lines blood vessels prevents tissue edema and maintains plasma volume, while enabling transendothelial exchange of immune cells and nutrients with surrounding tissue, in part through the dynamic regulation of endothelial cell-cell junctions. Dysfunction in barrier maintenance is a hallmark and often the underlying cause of many cardiovascular pathologies. While a host of soluble factors including growth factors, circulating lipids, and reactive oxygen species chemically signal to endothelial cells, hemodynamic forces locally and acutely regulate vascular permeability by remodeling endothelial cell-cell junctions and the cytoskeleton6. The shear stress of steady, laminar flow improves barrier integrity through stabilization of cell-cell junctions, while perfusion defects increase vascular permeability and contribute to pathogenesis of cardiovascular diseases including atherosclerosis7, stroke8, and myocardial infarction9. While in vitro studies have identified putative shear stress-responsive signaling pathways that mediate endothelial junction and cytoskeletal remodeling7, the mechanisms that link hemodynamic shear stress to the regulation of endothelial cell-cell junctions and vascular permeability remain unknown.
Mechanistic studies into the influence of shear stress on the vascular barrier have been challenging in vivo, where control over pressures and flows is limited, and the effects of shear stress cannot be decoupled from changes in nutrient exchange. In vitro approaches have provided better control of these forces, but the use of stiff, planar substrates for cell culture artificially influences cell-extracellular matrix (ECM) and cell-cell adhesion signaling. We therefore developed human engineered microvessels (hEMVs), a biomimetic microvasculature model consisting of a lumen surrounded by ECM, lined by human dermal microvascular endothelial cells (ECs, Fig. 1a), and connected to a microfluidic device to enable precise control of perfusion and hemodynamic stresses. This simplified model lacks other cell types such as pericytes or tissue-specific parenchyma and therefore focuses on biology autonomous to the endothelium. Mimicking in vivo observations2, the endothelial lining of hEMVs in the absence of flow was leaky, allowing 70 kDa fluorescently-labeled dextran to diffuse from the lumen into the interstitial matrix, whereas steady perfusion at flow rates that impart physiologic shear stress to the ECs (>3 Dyne/cm2) promoted the establishment of a functional vascular barrier resistant to transmural leakage and sensitive to known permeability factors (Fig. 1b–c, Extended Data Fig. 1a–e).
Figure 1. Notch1 regulates shear stress-induced vascular barrier function.
a, Organotypic microfluidic devices of human engineered microvessels (hEMVs) consisting of human ECs (red) in physiologic ECM (green), enabling vessel perfusion at a defined luminal shear stress (inlet and outlet in blue). Inset: 3D reconstruction of hEMVs (red - 70kDa dextran, white – VE-cadherin, green – collagen I). b, Real-time assessment of vascular barrier function in hEMVs cultured statically or under flow (heatmap of fluorescent intensity of 70 kDa dextran, imaging plane indicated by dashed line in a). c, Quantification of the diffusive permeability (PD) of 70 kDa dextran across EC barrier as a function of endothelial wall shear stress. d, Relative gene expression for ECs cultured statically or under flow was quantified with qPCR, and Notch target genes regulated by flow are indicated (each column representative of an independent experiment, full gene panel in Extended Data Fig. 2). e, ICD cleavage in static and flow EC lysates as measured by western blot with an antibody specific to cleaved ICD (N1 V1754). f, PD measured in hEMVs under static or flow conditions in the presence of Notch inhibitor (DAPT) or rDll4-coated collagen (rDll4). g, Fluorescent micrographs of hEMVs immunostained for VE-cadherin (magenta) and labeled with phalloidin (actin - green) and DAPI (nucleus – blue). h, Quantification of junctional area measured from VE-cadherin immunostained micrographs. i, PD for hEMVs cultured statically, under flow, or in the presence of rDll4-coated collagen with ECs expressing dnMAML, GFP infection control, or with N1-KO, Dll4-KO, or scramble control ECs. j, Fluorescent micrographs of actin and VE-cadherin for hEMVs under flow with ECs expressing dnMAML or with N1-KO ECs. k, Quantification of junctional area measured from VE-cadherin immunostained micrographs (micrographs of GFP and scramble controls in Extended Data Fig. 3). For (a-k), n≥3 independent hEMVs, mean ± s.e.m. l, Fluorescence intensity heatmaps of Evan’s blue (EB) dye in the mouse dermal vasculature 30min and 60min after IV co-injection of EB and DAPT or DMSO vehicle control. m, Quantification of PD for EB diffusion into the dermal interstitial space (n = 15 vessels across 3 mice/condition). n, Color image of lungs harvested from mice sacrificed 30 min after IV injection of EB. o, Vascular permeability was quantified by eluting and measuring the concentration of EB in lungs relative to the blood EB concentration, (n=4 age, sex-matched littermates, color coded, two-way ANOVA). p, Whole-mount lung vasculature immunostains showing leaked EB and DAPI. *p<0.05, **p<0.01; exact p and n values available in Figure Source Data, all images representative of at least three independent experiments.
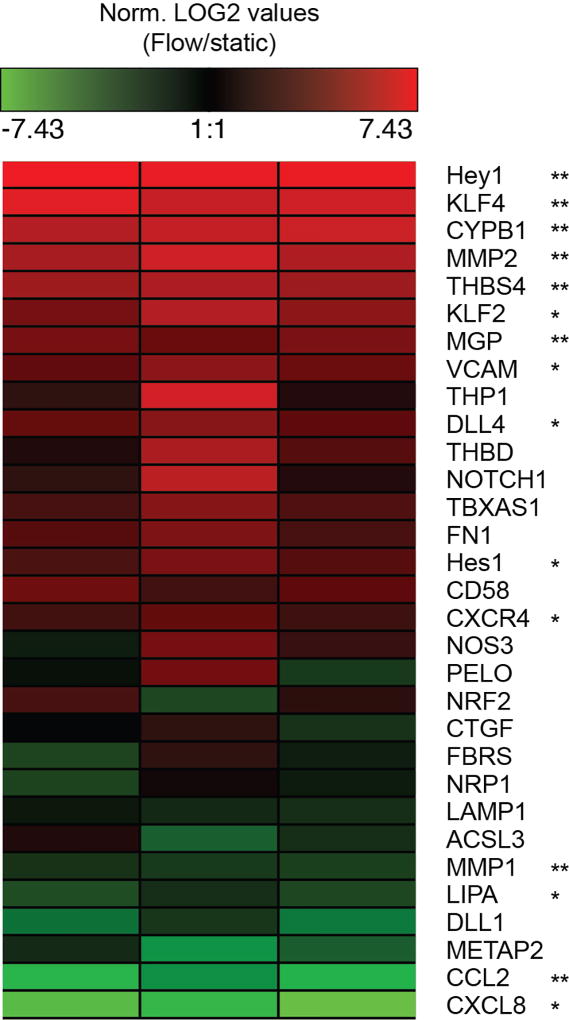
Shear stress regulates numerous mechanosensitive pathways whose signatures can be inferred from upregulation of various genetic networks reported both in vivo and in vitro10. Here, we observed increased expression of many such genes, but noted numerous genes associated with Notch signaling, including the Notch1 ligand DLL4 and the downstream targets of Notch1 activation, HES1 and HEY1 (Fig. 1d, Extended Data Fig. 2). Activation of Notch signaling by shear has recently been shown in zebrafish11, so to investigate whether Notch signaling was involved here in shear-induced barrier function, we treated hEMVs with DAPT (N-[N-(3,5-difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butylester), an inhibitor of γ-Secretase4, which cleaves Notch to release the transcriptionally active Notch intracellular domain (ICD, Extended Data Fig. 3a)12. Notch1 ICD cleavage increases with flow (Fig. 1e), and DAPT treatment in the presence of shear increased permeability (Fig. 1f). Conversely, directly activating Notch by coating the microvessel ablumenal surface with recombinant Dll4 (rDll4, Extended Data Fig. 3a–b) prior to cell seeding reduced permeability even in static (no flow) conditions (Fig. 1f). Examination of the endothelium confirmed that shear stress reduced permeability by inducing the assembly of vascular endothelial (VE-) cadherin-containing adherens junctions (AJs) and the redistribution of F-actin to the cortical membrane (Fig. 1g–h). The integrity of this organization was disrupted by DAPT (Fig. 1g–h), and Notch activation via rDll4 phenocopied the shear-induced phenotype (Extended Data Fig. 3c).
Canonical Notch signaling occurs through ICD-dependent transcription of downstream Notch target genes13. To determine the role of Notch transcriptional signaling in regulating barrier function, we seeded hEMVs with cells expressing the dominant negative Notch transcriptional co-factor Mastermind (dnMAML)14. Unexpectedly, dnMAML expression did not affect permeability (Fig. 1i) or AJ formation (Fig. 1j–k, Extended Data Fig. 3d), despite inhibiting Notch transcriptional signaling (Extended Data Fig. 3e). To investigate potential non-canonical mechanisms by which Notch activity regulates barrier function, we generated hEMVs with cells harboring CRISPR/Cas9-mediated knockout of Notch1 and Dll4 (Notch1-KO, Extended Data Fig. 3f; Dll4-KO, Extended Data Fig. 4a). Notch1-KO cells phenocopied DAPT treatment, resulting in increased permeability (Fig. 1i) and impaired AJ integrity (Fig. 1j–k, Extended Data Fig. 3h–j) without affecting total VE-cadherin levels (Fig. Extended Data Fig. 3j) or cell proliferation (Extended Data Fig. 3k–l). The Notch1 ligand Dll4 was also required for flow-induced barrier formation (Fig. 1i), and needed for cleavage of Notch1 ICD (Fig. Extended Data Fig. 4a) and AJ formation (Fig. Extended Data Fig. 4b–c). Shear stress has been reported to elevate endocytosis in ECs15, and Dll4 internalization appears necessary for ligand-mediated Notch1 activation16. Here, we demonstrate that flow induced dynamin-dependent internalization of Dll4 (Extended Data Fig. 4d–f), and chemically inhibiting dynamin reduced Dll4 internalization and abrogated barrier function (Extended Data Fig. 4g). To validate the finding that Notch activation regulates barrier function, we examined the role of Notch signaling in barrier maintenance in vivo. DAPT was co-injected with Evans blue (EB) dye intravenously in 6–8-week-old BALB/c nude mice, and dermal vascular permeability was quantified in real-time using intravital microscopy (Fig. 1l–m). Inhibition of Notch signaling with DAPT significantly increased vascular permeability (Fig. 1l–m Extended Data Fig. 5a). The implication that Notch might regulate barrier function via a transcriptionally independent pathway was consistent with the observation that DAPT increased vascular permeability within 15–60 minutes in vivo (Fig. 1l) and in vitro (Extended Data Fig. 6a), likely too rapid for a transcription-dependent response. In vivo, knockout of endothelial Notch1 resulted in loss of barrier function, increasing vascular permeability in the lung vasculature, quantified by EB extravasation17 (Fig. 1n–p, Extended Data Fig. 5b). Together, these results suggested an important role for shear stress in maintaining endothelial integrity, and that the Notch1 receptor potentially regulates this effect through a non-transcriptional mechanism.
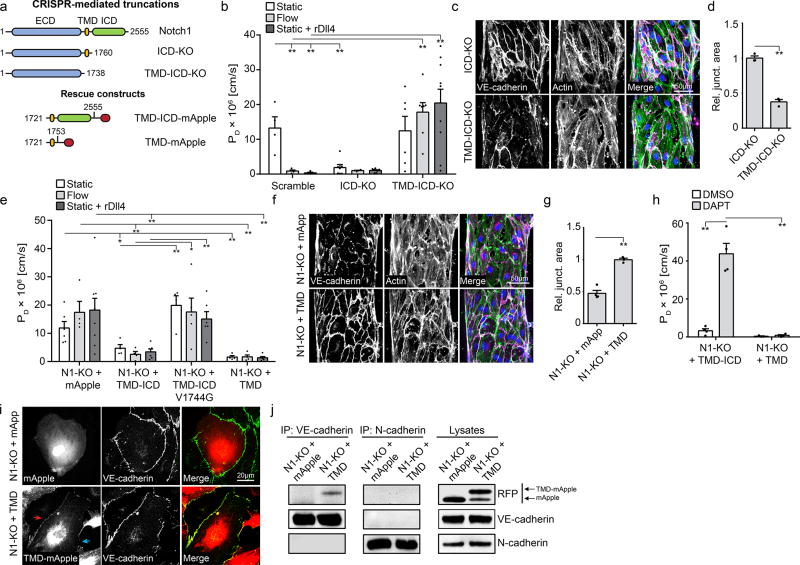
Upon Notch1 activation, the extracellular domain (ECD) of Notch1 is cleaved, which allows γ-Secretase-mediated cleavage of the ICD to leave behind the transmembrane domain (TMD) in the plasma membrane18. Given the observed increase in ICD cleavage with flow, we generated a library of CRISPR/Cas9-mediated Notch1 truncation mutants and recombinant rescue constructs (Fig. 2a, Extended Data Fig. 6e) to determine whether these subdomains of Notch1 contribute to regulating vascular permeability. Truncation of ICD (ICD-KO) resulted in constitutively low permeability and elaborated AJs (Fig. 2b–d) in static conditions, while truncation of both ICD and TMD (ICD-TMD-KO) increased permeability under flow (Fig. 2b). These data suggested that ICD was not critical for Notch1-induced barrier function, while TMD was necessary. Indeed, expression of TMD alone, as well as TMD-ICD, in Notch1-KO cells rescued barrier function and AJ assembly (Fig. 2e–g). Interestingly, Notch1-KO cells expressing TMD-ICD harboring a point mutation that prevents cleavage of the ICD (V1754G19, Extended Data Fig. 6b) failed to rescue barrier function (Fig. 2e), and TMD-ICD failed to rescue in the presence of DAPT, while cells expressing TMD alone maintained barrier function irrespective of DAPT exposure (Fig. 2h). Together, these findings are consistent with a model wherein TMD is the key component of Notch1 for regulating barrier function, and the barrier forming activity of TMD requires cleavage of ICD.
Figure 2. The Notch1 transmembrane domain mediates barrier function through interaction with VE-cadherin.
a, A library of endogenous Notch1 truncation mutants and over-expression rescue constructs were generated to examine the key functional domains of Notch1 that regulate barrier function. b, PD for ECs with CRISPR/Cas9-mediated endogenous truncation of Notch1 ICD (ICD-KO) or truncation of the TMD and ICD (TMD-ICD-KO) cultured statically, under flow, or in the presence of rDll4-coated collagen. c, Fluorescent micrographs of VE-cadherin and actin for ICD-KO and TMD-ICD-KO ECs under flow conditions. d, Quantification of junctional area measured from VE-cadherin immunostained micrographs. e, PD for N1-KO ECs expressing TMD-ICD-mApple, TMD-ICD V1754G-mApple, TMD-mApple, or mApple infection control cultured statically, under flow, or in the presence of rDll4-coated collagen. f, Fluorescent micrographs of VE-cadherin (magenta), actin (green), and DAPI (blue) in static N1-KO cells expressing TMD-mApple or mApple infection control. g, Quantification of junctional area measured from VE-cadherin immunostained micrographs. h, PD for static N1-KO cells expressing TMD-ICD-mApple or ICD-mApple exposed to DAPT or DMSO load control. i, Immunofluorescent images of Notch1-KO cells expressing either mApple or TMD-mApple, co-stained for VE-cadherin. Co-localization of Notch1 TMD and VE-cadherin (red arrow) is lost at free edges (blue arrow). j, Immunoprecipitation of VE-cadherin and N-Cadherin from Notch1-KO cells expressing either mApple or TMD-mApple. Immunoblotting with a RFP antibody identified co-immunoprecipiting TMD-mApple. For all plots, mean ± s.e.m., n≥3 independent hEMVs, *p<0.05, **p<0.01. Exact p and n values available in Figure Source Data, all images representative of at least three independent experiments.
Biological activity of the Notch1 TMD has not been previously demonstrated. To investigate the mechanism by which Notch1 TMD contributed to AJ assembly, we expressed mApple-tagged TMD and observed distinct localization to VE-cadherin-containing cell-cell junctions that was lost in VE-cadherin knockout cells (Fig. 2i; Extended Data Fig. 6c–d). Biochemically, we isolated endogenous VE-cadherin via immunoprecipitation and identified an interaction with Notch1 (Extended Data Fig. 6f). Furthermore, we identified that the TMD specifically co-immunoprecipitated with VE-cadherin, but not with neural (N-) cadherin (Fig. 2j), and this interaction was mediated, in part, by a 6 amino acid stretch in the N-terminal ectodomain of TMD (Extended Data Fig. 6g–h). These data suggest a specific interaction between Notch1 and VE-cadherin that is mediated by the TMD.
A possible link for how Notch1 TMD regulated cell-cell junctions was suggested by the reduced cortical actin and increased actin stress fibers in Notch1-KO cells (Fig. 3a–b). The Rho-family GTPases are powerful regulators of actin redistribution and barrier function20, where Rac1 can stabilize AJs by promoting the formation of cortical actin bundles21. Indeed, Notch1-KO cells and ECs treated with DAPT exhibited reduced Rac1 activity (Fig. 3c–d, Extended Data Fig. 7a–b), while expressing Notch1 TMD increased Rac1 activity (Fig. 3e–f). Rac1 activity increased with flow (Extended Data Fig. 7c), and this increase in Rac1 activity was abrogated with genetic deletion of Notch1 or Dll4 (Extended Data Fig. 7d–e). This Rac1 activity was required for barrier function as Notch1-KO cells expressing Notch1 TMD treated with the Rac1 inhibitor NSC 23766 demonstrated increased permeability (Fig. 3g).
Figure 3. Notch1 assembles a mechanosensory junctional complex involving LAR, Trio, and Rac.
a, Fluorescent micrographs of phalloidin stained ECs. b, Intensity of cortical actin at cell-cell junctions and the number of stress fibers per micron quantified from phalloidin stained ECs (n=3 hEMVs). c, Active Rac1 was isolated from scramble and Notch1-KO cell lysates using a recombinant p21-binding domain of Pak1 (GST-PBD). d, Quantification of western blot band intensity revealed a decrease in Rac1 (~50%) after knockout of Notch1 (n=3 independent lysates). e, Active Rac1 was isolated using GST-PBD from Notch1-KO cell lysates expressing mApple or TMD-mApple. f, Quantification of western blot band intensity revealed an increase in Rac1 (~45%) with expression of TMD-mApple (n=3 independent lysates). g, PD of Notch1-KO ECs expressing TMD-mApple or mApple control in the presence of 50 µM NSC 23766, a Rac1 inhibitor, or vehicle control. h, Immunoprecipitation of VE-cadherin and the Rac1 GEF Trio from ECs cultured statically or under flow. Co-immunoprecipitation of mechanosensory complex proteins was assessed by immunoblotting for Notch1-ECD, Notch1-ICD, LAR (85 kDa P-subunit), and VE-cadherin. i, Immunoprecipitation of VE-cadherin or Trio from scramble and Notch1-KO ECS cultured under flow. Co-immunoprecipitation of mechanosensory complex constituents was assessed by immunoblotting for VE-cadherin, LAR, and Trio. j, PD of LAR-KO and Trio-KO ECs cultured under static or flow conditions or in the presence of rDll4-coated collagen. k, Micrographs of Trio-KO and LAR-KO cells under flow conditions (VE-cadherin – magenta, actin – green, DAPI – blue). l, Quantification of junctional area measured from VE-cadherin immunostained micrographs. m, Immunoprecipitation of VE-cadherin and Trio from Notch1-KO cells expressing TMD-mApple or mApple. Immunoblotting with LAR, VE-cadherin, and RFP antibodies was used to assess co-immunoprecipitation of mechanosensory complex upon expression of TMD. n. PD for scramble, N1KO, LAR-KO, and Trio-KO cells cultured under static conditions expressing TMD-mApple or mApple infection control. For (g,j,n), n≥3 hEMVs. All plots mean ± s.e.m., *p<0.05, **p<0.01. Exact p and n values available in Figure Source Data, all images representative of at least three independent experiments.
To investigate the molecular intermediaries by which Notch1 regulated Rac1, we identified via immunoprecipitation several VE-cadherin interaction partners. Only one—the transmembrane protein tyrosine phosphatase LAR—dissociated from VE-cadherin in Notch1-KO cells (Extended Data Fig. 8a). Interestingly, LAR has been reported to bind the Rac1 guanine exchange factor (GEF) Trio22, which has been shown to play a role in Notch-mediated axon guidance in Drosophila23. Here, we demonstrate that LAR mediates a Notch1-dependent interaction between Trio and VE-cadherin (Extended Data Fig. 8b). Under static conditions, full length Notch1 was associated with VE-cadherin (Fig. 3h). Flow-induced activation of Notch1 released ECD and ICD from VE-cadherin and stimulated the interaction of VE-cadherin with LAR and Trio (Fig. 3h), and the formation of this complex was Notch1-dependent in vitro (Fig. 3i) and in vivo (Extended Data Fig. 8c–d). Using a VE-cadherin biotin ligase proximity labeling construct24, we confirmed that the association of ICD with VE-cadherin decreases, while the interaction of Trio and VE-cadherin increases with stimulation of Notch1 (Extended Data Fig. 8e–f). Furthermore, LAR and Trio are required for the establishment of barrier function as LAR-KO and Trio-KO hEMVs demonstrated reduced Rac1 activity (Extended Data Fig. 7e), high permeability (Fig. 3j), and did not form AJs (Fig. 3k–l). Expression of TMD rescued the interaction of LAR and Trio with VE-cadherin (Fig. 3m), and LAR and Trio were required for Notch1 TMD-mediated rescue of barrier defects (Fig. 3n). Thus, cleavage of Notch1 ICD releases Notch1 TMD to assemble a complex between VE-cadherin, LAR, and Trio, thereby inducing Rac1 activity to assemble junctions and establish barrier function (Extended Data Fig. 9).
Notch receptor signaling is a central regulator of many developmental and homeostatic processes, but how it exerts so many effects has been difficult to explain through the only known downstream signal—ICD-dependent transcription of Notch target genes13. Here, we demonstrate that receptor cleavage, induced by Notch1 receptor activation, also reveals the previously unappreciated TMD, which promotes association among transmembrane receptors and signaling molecules to act as a central pathway for regulating junction assembly and vascular barrier function. Recent work has demonstrated that transmembrane domain interactions among other surface receptors drive signaling in various and diverse biological contexts25, suggesting a more general mechanism for controlling complex formation at the cell surface.
In the context of Notch, this cortical signaling pathway may explain why human mutations in the Notch receptor family have demonstrated vascular defects and barrier dysfunction in human diseases such as CADASIL, Adams-Oliver Syndrome, and Alagille syndrome26–28, and why inhibition of Notch signaling to treat tumors appears to trigger edema29. Identifying ways to isolate the cortical from transcriptional pathway of Notch may provide new opportunities to control these devastating complications. Furthermore, quantitative differences in expression of Notch and Dll4 in different vascular beds, described as a key mechanism for tip/stalk cell specification in angiogenesis30, may underlie differences in shear sensitivity and baseline permeability in organ-specific vascular beds. More broadly, the combination of a non-canonical pathway that regulates adhesive and cytoskeletal processes together with a transcriptional regulator of cell fate programs, channeled through a single receptor, provides a new framework for understanding coordinated developmental programs such as stem cell differentiation and tissue morphogenesis.
METHODS
Cell culture
Human dermal microvascular endothelial cells (hMVEC-Ds, Lonza) were maintained in EGM2-MV growth media (Lonza) and used at passages 2–10. Human HEK293T (Clonetech) were cultured in DMEM (Hyclone) supplemented with 10% fetal bovine serum (Hyclone), 100 U ml−1 penicillin, 100 µg ml−1 streptomycin (Life Technologies) and 2 mM L-glutamine (Life Technologies). Both cell types were maintained at 37 °C in 5% CO2 in a humidified incubator. Cell line authentication (performance, differentiation, and STR profiling) was provided by Lonza. was Mycoplasma testing was performed on all lines using PlasmoTest mycoplasma detection kit from InvivoGen.
Microfluidics
Microfluidic devices were fabricated using soft lithography. Polydimethylsiloxane (PDMS, Sylgard 184, Dow-Corning) was mixed at 10:1 base:curing agent and cured overnight at 60°C on a silicon master. The PDMS was cut from the silicon master, trimmed, and surface activated by plasma treatment for 1 min. Devices were then bonded to glass and treated with 0.01% poly-L-lysine and 1% glutaraldehyde, washed in sterile water overnight, and sterilized with 70% ethanol for 30min. Steel acupuncture needles (160µm diameter, Seirin) were introduced into the device, and devices were UV sterilized for 20min. Collagen type I (Thermo Fisher scientific) solution was buffered with 10x DMEM, titrated to a pH of 8.0 with NaOH, and brought to a final concentration of 2.5mg/mL collagen I in total solution. Collagen solution was injected into microfluidic devices and polymerized for 20min at 37°C. Growth medium was then added to the devices overnight and needles were removed to create 160 µm diameter channels in the collagen gel. hMVECs were harvested with 0.05% Trypsin/EDTA and centrifuged at 200 × g for 5 min. Cells were resuspended at 0.5 × 106 cells/mL in EGM2-MV, and 70µL of cell suspension was introduced into the devices to allow cells to adhere to collagen for 15min before washing with growth medium. To coat collagen with rDll4, 100µL of 1µg/mL recombinant human Dll4 (R&D Systems) in PBS was introduced into devices and incubated on a rocker at 37°C overnight. Devices were washed 2x with PBS prior to cell seeding.
To quantify vascular permeability in the microfluidic device, fluorescent dextran (70 kDa Texas Red or 70 kDa Oregon Green, Thermo Fisher) was introduced into perfusion media at a concentration of 12.5 µg/mL, and diffusion of the dextran was imaged in real time with a Yokogawa CSU-21/Zeiss Axiovert 200M inverted spinning disk microscope with a 10 × air objective and an Evolve EMCCD camera (Photometrics). Time-lapse microscopy was used to measure the flux of dextran into the collagen gel, and the resulting diffusion profile was fitted to a dynamic mass conservation equation as previously described31, with the diffusive permeability coefficient (PD) defined by the following relation:
where J is the mass flux of dextran, cvessel is the concentration of dextran in the vessel, and cECM is the concentration of dextran in the perivascular ECM.
To measure the effects of known permeability inducing factors, flow imparting 5 Dyn/cm2 wall shear stress was applied overnight and 40 ng/mL VEGF (R&D Systems) was added 1hr prior to permeability assay or 1 U/mL thrombin (Sigma) was added 15min prior to permeability assay. For barrier enhancing factors, devices were left under static conditions overnight in growth media supplemented with 10 µM SU-5416 (Cayman Chemical) or 500 nM S1P (Cayman Chemical) added 1hr prior to permeability assay. To inhibit endocytosis in the presence of flow, 60 µM Dynasore hydrate (Sigma) was added 30min prior to applying flow at 5 Dyn/cm2 in media supplemented with 60 µM Dynasore hydrate overnight.
Measurement of vascular permeability in vivo
All in vivo animal studies were reviewed and approved by the Institutional Animal Care and Use Committees of Boston University and Yale University. Mice were bred and maintained under specific-pathogen-free conditions at the AAALAC (Association for Assessment and Accreditation of Laboratory Animal Care International)-accredited Boston University and Yale University, and housed in accordance with the procedures outlined in the Guide for the Care and Use of Laboratory Animals. For intravital measurements of vascular permeability, mice (6–8 weeks old, BALB/c nude mice CrTac:NCr-Foxn1nu strain, female) were anesthetized using an isoflurane nebulizer, retro-orbitally injected with Evans blue dye (10 mg/ml in saline, 100 µl) containing either DMSO or DAPT (20 µM), and immobilized on a microscope stage in an imaging chamber equilibrated to 37°C (schematic, Extended Data Fig. 10a). Vascular permeability images were acquired with a Leica HC FLUOTAR L 25x/0.95 W immersion objective on an upright Leica TCS SP8 multiphoton microscope equipped with a Spectra-Physics Insight DS+ laser tuned to 950 nm excitation and non-descanned emission detection at 680 nm. Vascular permeability was assessed beginning 20 minutes post injection and continued every five minutes up to one hour. The diffusive permeability coefficient was measured for 5 vessels per field of view as described above.
For bulk tissue vascular permeability measurements, inducible Notch1 endothelial cell knockout mice {CDH5-CreERT2(+):NOTCH1fl/fl} were used and their littermates {CDH5-CreERT2(−);NOTCH1fl/fl} as controls. At 4-weeks old, mice received tamoxifen (0.2 mg/day) intraperitoneally for 5 consecutive days. Two weeks after the start of Cre induction by tamoxifen, Evans blue dye (10 mg/ml in saline, 100 µl) was injected retro-orbitally. Thirty minutes post injection animals were euthanized by cervical dislocation under anesthesia and blood and tissue samples including the lung were harvested. Blood samples were clarified at 14,000 rpm for 5 min at 4°C; freeze-dried lung samples were weighed and incubated with 500 µl formamide at 56°C for 48 hours and clarified by centrifugation. Optical density readings at 620 nm (EB) and 740 nm (heme) were collected using a SpectraMax M5 spectrophotometer (Molecular Devices) and the concentrations of Evans blue were calculated based on the standard curve of Evans blue dye solution serial-diluted in formamide. Tissue Evans blue levels were normalized by tissue weight and blood Evans blue concentration and corrected for heme pigment contamination.
Antibodies and reagents
The anti-Trio (H-120, 1:500), anti-Notch1 ICD (C-20-R, 1:1000), anti-Notch1 ECD (H-131, 1:500), anti-VE-cadherin (F-8, 1:200), and anti-VE-PTP (H-300, 1:250) antibodies were from Santa Cruz Biotechnology. Anti-VE-cadherin (ab33168, 1:2,000), anti-RFP (ab124754, 1:1000), and anti-β tubulin (ab6046, 1:5,000) antibodies were from Abcam. Anti-VEGFR2 antibody (55B11, 1:1000), anti-Notch1 V1754G (D3B8, 1:1000 WB), anti-HA (6E2, 1:1000 WB, AF647 conjugate IF) was from Cell Signaling Technologies. Anti-Rac1 antibody (102, 1:1,000) was from BD Biosciences. Rhodamine and Alexa Fluor 488-labelled phalloidin, Alexa Fluor-488, 568 and 647 goat anti-mouse and anti-rabbit IgG secondary antibodies were from Life Technologies. HRP-conjugated donkey anti-mouse, anti-rabbit, and anti-goat IgG secondary antibodies were from Fitzgerald. Anti-PTPRF antibody (SAB4200321, 1:1000), DAPI, DAPT, Evans Blue dye, dynasore hydrate, and formamide were from Sigma. Rac1 inhibitor NSC 23766 was from Santa Cruz Biotechnology.
Cloning and qPCR
Human VE-Cadherin (gift from Michael Davidson) and human Notch1 (gift from Michael Elowitz) cDNAs were used to make C-terminal mApple-tagged constructs in a modified pRRL lentiviral expression vector. PCR amplification of the following amino acid sites was used to make C-terminal mApple tagged Notch1 truncation constructs: Notch1 intracellular domain and transmembrane domain (S2 cleavage site to end of ICD, Val1721 – Lys2555) and Notch1 transmembrane domain (S2 to S3 cleavage site, Val1721 – Gly1753). The dnMAML construct (MSCV-IRES-GFP MAML1 13–74) was from Jon Aster. Human Dll4 cDNA (Sino Biological) was modified with an N-terminal HA tag via PCR and cloned into a modified pRRL lentiviral expression vector. All constructs were expressed via lentiviral transduction.
ECs were lysed with Trizol™ (LifeTechnologies) followed by chloroform phase separation. The aqueous phase was diluted with 70% ethanol at 1:1 ratio, loaded on a RNA micro column (RNA microkit, Qiagen Sciences, Germantown, MD, USA) and RNA extraction proceeded according to the manufacturer’s protocol. Subsequently, 1 µg of total RNA was converted to complementary DNA (cDNA) with qScript™ cDNA Supermix (Quanta Sciences, Gaithersburg, MD, USA). Realtime PCR was performed in 20µl reactions using the Sybr Green Mastermix I (Roche) and a LightCycler480 (Roche), cycling at 95°C for 3 seconds, 60°C for 10 seconds, 72°C for 20 seconds for 40 cycles. Specific gene targets were detected with primers indicated in Table S1. Primers for NOTCH1 and DLL1 were acquired from Qiagen (Quantitec Hs_NOTCH1_2_SG, Quanititec Hs_DLL1_1_SG).
Relative gene expression is expressed as –ddCT in which dCT equals the difference in CT value between the gene of interest (GOI) and a housekeeping gene [Proteasome subunit beta type-2 (PSMB2) or Glyceraldehyde 3-phosphate dehydrogenase (GAPDH)] and ddCT equals the difference between the dCT of the GOI in an experimental condition and the dCT of the same GOI in a control condition. Heatmaps showing –ddCT of GOI under flow normalized to static controls were generated using Genesis v1.8.132. Primer sequences for qPCR are summarized in Supplementary Table 1.
Lentiviral-mediated CRISPR genome editing
Stable CRISPR knockout cell lines in primary hMVEC-Ds were generated using the lentiCRISPRv2 system (gift from Feng Zhang, Addgene plasmid #52961). Specific gRNAs were generated using the Optimized CRISPR Design tool (Feng Zhang) and cloned into the BsmBI site of plentiCRISPRv2. gRNA guides used for both silencing and truncation of genomic targets are summarized in Supplementary Table 2. Individual gRNA-containing plentiCRISPRv2 plasmids were co-transfected with pVSVG, pRSV-REV, and pMDLg/pRRE packaging plasmids into HEK-293T cells using calcium phosphate transfection. After 48 hours, viral supernatants were collected, concentrated using PEG-IT viral precipitator (SBI), and re-suspended in PBS. hMVEC-Ds were transduced in growth media overnight and media was replaced the following morning. 48 hours post infection cells were plated in 6-well plates at 75,000 cells per well and selected with 2 µg/ml puromycin for 3–4 days. All CRISPR modifications were verified by western blot.
GTPase activity assays
pGEX4T-1 construct containing the p21-binding domain of Pak1 (PBD) was from Addgene (Plasmid 12217). Briefly, expression of the GST fusion proteins in BL21 Escherichia coli was induced with 200 µM isopropyl-β-D-thiogalactoside (IPTG) for 12–16 h at room temperature. Bacterial cells were lysed in buffer containing 25 mM HEPES pH 7.6, 1% Triton X-100, 150 mM NaCl, 5 mM MgCl2, 1 mM dithiothreitol, protease and phosphatase inhibitor cocktails (Roche), and the proteins were purified by incubation with glutathione–Sepharose 4B beads (GE Healthcare) at 4 °C. For active Rac1 pulldowns, confluent hMVEC-Ds plated on 50 ug/ml collagen I in reduced serum growth medium (0.5% FBS) were rinsed with cold PBS containing 1 mM Ca2+ and 0.5 mM Mg2+ (PBS+), then lysed with cold 50 mM Tris pH 7.6, 200 mM NaCl, 1% Triton X-100, 0.5% deoxycholate, 5 mM MgCl2, and protease and phosphatase inhibitor cocktails, sonicated at 3 W on ice for 5 s, and clarified at 14,000xg for 5 min. Lysates were equalized for protein content and volume and rotated at 4 °C for 1 h with 20 µg GST–PBD. Bead pellets were washed three times with lysis buffer, extracted in 2X NuPAGE® LDS Sample Buffer (Life Technologies) containing 100 mM DTT, and analyzed by SDS-PAGE and immunoblotting with chemiluminescent HRP detection. Western blots were adjusted for brightness and contrast using ImageJ. Active Rac1 quantifications were normalized to total Rac1 for each condition.
Immunoprecipitation
Confluent hMVEC-Ds plated on 50 ug/ml collagen I and cultured in growth medium were rinsed with cold PBS+, then lysed with cold 25 mM HEPES, pH 7.5, 200 mM NaCl, 1% TX-100, 5 mM MgCl2, 1.5x protease and phosphatase inhibitors (Roche). Lysates were drawn through a 21G syringe 15 times and incubated on ice for 30 minutes before clarification at 14,000xg for 15 minutes at 4°C. Clarified lysates were equalized for protein content and volume and incubated with 2 µg of anti-VE-cadherin (Abcam, ab33168), 2 µg of anti-Trio (Santa Cruz, H-120), or 2 µg anti-Notch1 (Santa Cruz, C-20-R) at 4°C with end-over-end rotation. Antibody complexes were purified with Protein A/G beads (Life Technologies) at 4°C with end-over-end rotation. Bead pellets were washed three times with lysis buffer. Protein complexes were extracted in 2X NuPAGE® LDS Sample Buffer (Life Technologies) containing 100 mM DTT and analyzed by SDS-PAGE and immunoblotting with chemiluminescent HRP detection. Western blots were adjusted for brightness and contrast using ImageJ.
For immunoprecipitation of VE-cadherin and Trio from mouse lung lysates, age and sex-matched CDH5-CreERT2(-):NOTCH1fl/fl and CDH5-CreERT2(+):NOTCH1fl/fllitter mates were euthanized by cervical dislocation under anesthesia and lungs were harvested and immediately snap frozen in liquid nitrogen. Mouse lungs were homogenized in a Potter-Elvehjem tissue grinder in lysis buffer (25mM HEPES pH 7.5, 1mM EDTA, 150mM NaCl, 0.5% Triton X-100, 0.5% NP-40, 1.5x protease and phosphatase inhibitor cocktail (ThermoFisher)), and clarified by centrifugation at 20,000g for 30min at 4C. The soluble supernatants were precleared with protein G sepharose beads and then incubated with protein G sepharose beads with VE-cadherin or Trio antibody (Santa Cruz, 1 mg)for 3 hours at 4C, washed three times with lysis buffer and then processed in SDS–PAGE sample buffer.
Immunofluorescence
For morphological analysis of adherens junctions and actin cytoskeleton, cells cultured in complete medium were perm/fixed with 1% paraformaldehyde/0.03% Triton X-100 in PBS+ at 37°C for 90 seconds. Cells were immediately post-fixed in 4% paraformaldehyde in PBS+ at 37°C for 15 minutes. Cells were rinsed three times with PBS+ and permeabilized with 0.5% Triton X-100 in PBS+ for 10 minutes. Cells were blocked with 2% BSA in PBS+. Primary and secondary antibodies were applied in 2% BSA in PBS+ and rinsed three times over 30 min with PBS+ between each treatment. For examining the localization of endogenous Notch1 and N1 TMD-mApple, cells cultured in complete medium were fixed with 4% paraformaldehyde in PBS+, permeabilized with 0.1% Triton X-100 in PBS+, and blocked with 2% BSA in PBS+. Primary and secondary antibodies were applied in 2% BSA in PBS+ and rinsed three times over 30 min with PBS+ between each treatment. For live cell or immunofluorescent imaging, images were acquired with a Yokogawa CSU-21/Zeiss Axiovert 200M inverted spinning disk microscope with either a Zeiss LD C-Apochromat 40 × , 1.1 N.A or 63X, 1.15 N.A. water-immersion objective and an Evolve EMCCD camera (Photometrics). Fluorescence images were adjusted for brightness and contrast using ImageJ and MATLAB software.
To quantify junctional organization, greyscale micrographs of cells immunostained for VE-cadherin were converted to black and white based on a threshold determined by Otsu’s method, and junctional area was defined as the total number of pixels above the threshold. To quantify actin stress fibers and cortical actin, the intensity of Alexa Fluor 488 phalloidin-labeled cells was plotted along lines drawn between the nucleus centroids of neighboring cells. Local intensity peaks were measured to determine stress fiber locations Extended Data Fig. 10b . Cortical actin was defined as the area under the peak at cell-cell junctions normalized to total area under the curve, and stress fibers were quantified as the number of peaks normalized by the total length of the line between nucleus centroids.
To measure Dll4 endocytosis, ECs expressing Dll4-HA were seeded into flow chambers (Ibidi) and cultured for 6hrs under static conditions. Dynasore hydrate or DMSO load control was added for 30min, followed by AlexaFluor-488 conjugated mouse anti-HA antibody (1:250) in Dynasore or DMSO containing medium for 30min before washing with Dynasore or DMSO containing medium. Devices were then cultured statically for under flow imparting 5 Dyn/cm2 shear stress for 1hr in Dynasore or DMSO containing medium prior to fixation with 4% PFA in PBS+ at 37°C for 15 minutes. Cells were permeabilized with 0.025% saponin (Sigma) and 2% BSA in PBS+ for 30min at RT, and all subsequent antibody incubations and washes were performed in this buffer. Cells were incubated with anti-Notch1 ECD rabbit antibody (1:200) overnight at RT prior to adding AlexaFluor-488 anti-mouse (1:200) and AlexaFluor-568 anti-rabbit (1:200) antibody for 2hrs at 4°C. The fraction of cells with internalized Dll4 were determined by counting the number of cells with >1 HA-positive puncta and the total number of cells labeled by DAPI staining.
Statistical analysis
Sample sizes and p values are reported in each figure legend and statistical analyses were performed using GraphPad Prism 6.0 or Microsoft Excel. Unless otherwise noted in the Figure Legend, statistical differences between sets of data were analyzed with a two-tailed, unpaired Student t-test, and exact p values are provided in each Figure Source Data. Sample sizes of sufficient power were chosen on the basis of similar published research and were confirmed statistically by appropriate tests. Experiments were not randomized.
Data availability
Graphical source data is provided in each figure legend. The source data for the western blots in the figures are available in the Supplementary Information. Additional data that support the findings of this study are available from the corresponding author on reasonable request.
Extended Data
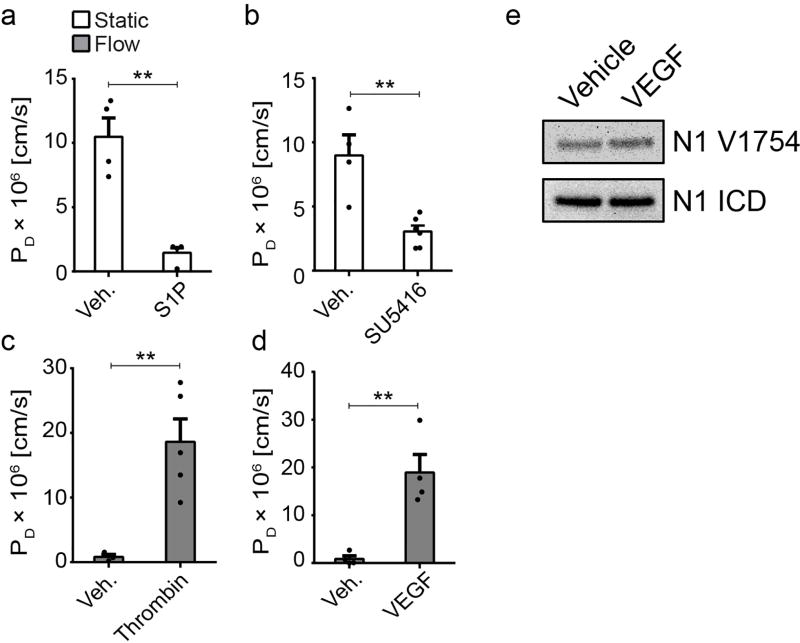
Extended Data Figure 1. Known permeability modulating agents regulate barrier function in hEMVs.
PD measured in hEMVs under static or flow conditions treated with a, 500nM S1P for 1hr, b, 10 µM SU415 (VEGFR2 inhibitor) overnight, c, 1 U/mL thrombin for 15min, or d, 40ng/mL VEGF for 1hr. e, western blots of cleaved Notch1 ICD (V1754) from lysates of ECs treated with vehicle or 40ng/mL VEGF for 1hr. For all plots, mean ± s.e.m., n≥3 hEMVs, **p<0.01. Exact p and n values available in Figure Source Data, western blot representative of two independent experiments.
Extended Data Figure 2. Flow activates expression of mechanotransduction associated gene networks.
Relative gene expression measured by qPCR. –ddCT of GOI under flow normalized to static control is plotted as a heatmap. n=3 flow/static qPCR analyses from distinct hEMV sets, each column representative of an independent experiment. *p<0.05, **p<0.01. Exact p and n values available in Figure Source Data.
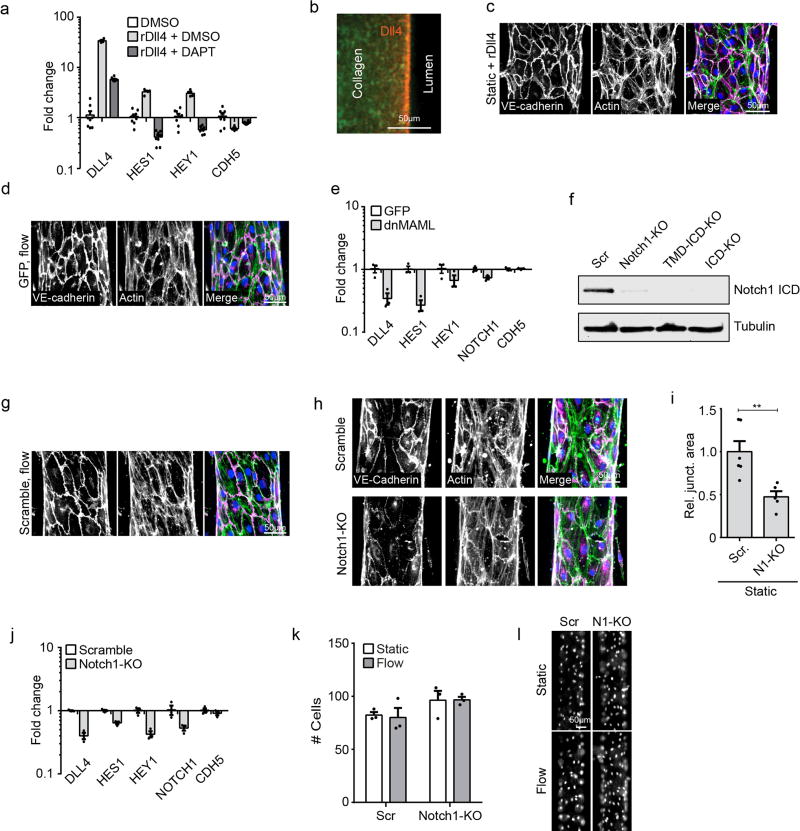
Extended Data Figure 3. Non-transcriptional Notch1 signaling regulates vascular barrier function.
a, Gene expression of Notch1 target genes HES1 and HEY1, the Notch1 ligand DLL4, and VE-cadherin (CDH5) measured via qPCR in ECs treated with DAPT or DMSO load control on Dll4-coated and control tissue culture plastic substrates. b, Fluorescent micrograph of rDll4-coated device prior to cell seeding (green – Alexa Fluor 488 Collagen I, red – immunostain of Dll4). c, Fluorescent micrograph of ECs in hEMVs coated with rDll4 prior to cell seeding. d, Micrographs of GFP-infection control cells under flow. e, Gene expression of HES1, HEY1, DLL4, NOTCH1, and VE-cadherin (CDH5) measured via qPCR in ECs expressing dnMAML or infection control (GFP). f, Western blot validation of Notch1 CRISPR lines: Scramble, Notch1-KO, TMD+ICD-KO, and ICD-KO. g, Fluorescent micrographs of CRISPR/Cas9 scramble control cells under flow. h, Fluorescent micrographs of Scramble and Notch1-KO hEMVs under static conditions immunostained for VE-cadherin and labeled with phalloidin (actin). i, Quantification of junctional area measured from VE-cadherin immunostained micrographs. j, Gene expression measured via qPCR in Notch1-KO cells and scramble control cells. k, Quantification of cell number in f.o.v at 10x magnification of Scramble or Notch1-KO hEMV under static and flow conditions. l, Micrographs of nuclei as visualized by DAPI in Scramble or Notch1-KO hEMVs. For all plots, mean ± s.e.m., n≥3 hEMVs, **p<0.01. Exact p and n values available in Figure Source Data, images representative of at least three independent experiments.
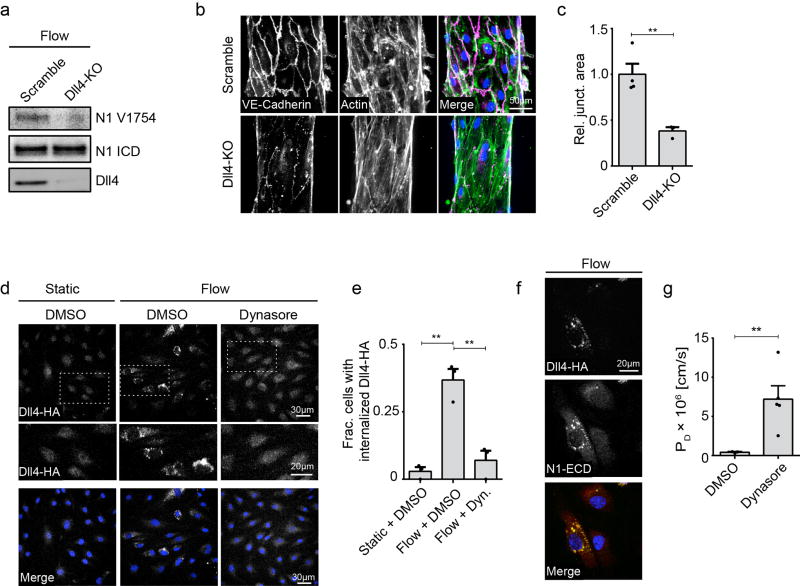
Extended Data Figure 4. Notch1 is activated in response to shear stress by endocytosis of Dll4.
a. ICD cleavage as measured by western blot with an antibody specific to cleaved ICD (N1 V1754) in Scramble and Dll4-KO ECs under flow. b, Fluorescent micrographs of Scramble and Dll4-KO hEMVs under flow conditions immunostained for VE-cadherin and labeled with phalloidin (actin). c, Quantification of junctional area measured from VE-cadherin immunostained micrographs. d, Immunofluorescent micrographs of recombinant Dll4-HA expressing ECs under static + DMSO, flow + DMSO, and flow + Dynasore conditions stained for HA (Dll4-HA) and DAPI. e, Quantification of internalized Dll4-HA in ECs under static + DMSO, flow + DMSO, and flow + Dynasore conditions. Cells with internalized Dll4-HA counted as those with >1 AlexaFluor-488 positive puncta. f, Immunofluorescent micrograph of a Dll4-HA expressing EC under flow stained for HA (Dll4-HA), Notch1 ECD, and DAPI. g, Diffusive permeability of 70kDa dextran measured in cells treated with Dynasore hydrate or DMSO load control and exposed to flow overnight. For all plots, mean ± s.e.m., n≥3 hEMVs, **p<0.01. Exact p and n values available in Figure Source Data, images representative of at least two independent experiments.
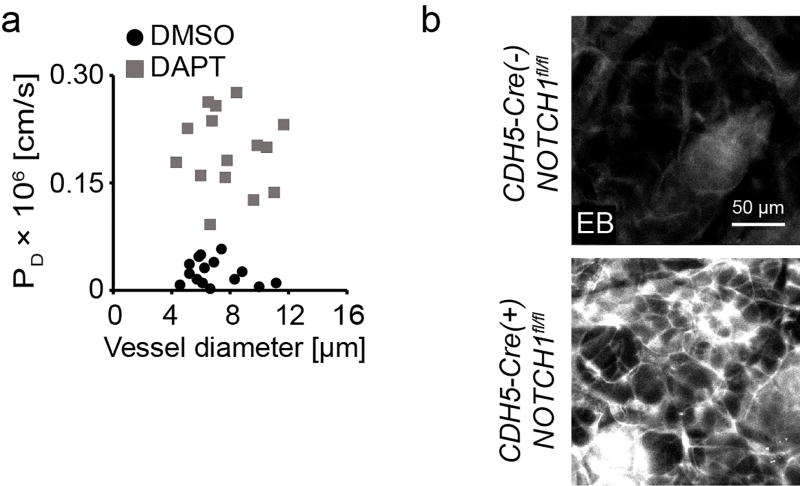
Extended Data Figure 5. DAPT and Notch1 depletion modulate vascular permeability in vivo.
a, Diffusive permeability of dermal vasculature as a function of vessel diameter in the mouse dermal vasculature in mice after 1hr of IV-administered DMSO or DAPT (n = 15 vessels across 3 mice per condition). b, High magnification whole-mount micrographs of Evans Blue in the mouse dermal vasculature. Fluorescent images representative of three independent experiments.
Extended Data Figure 6. Notch1 regulates junctional stability through association with VE-cadherin.
a, Timelapse images of cells expressing VE-cadherin-mApple in the presence of DAPT or DMSO load control demonstrate that AJs disassemble after 30min of exposure to DAPT, leading to macroscopic intercellular gaps (red arrows). b, Fluorescent micrographs of Notch1 KO cells expressing TMD+ICD-mApple or TMD+ICD V1754G-mApple immunostained for cleaved Notch1 ICD V1754 and DAPI. c, Fluorescent micrographs of TMD-mApple expressed in VE-cadherin knockout or scramble control ECs and immunostained for VE-cadherin. d, Western blot for Notch1 ICD and VE-cadherin in Notch1-KO and VE-cadherin KO ECs. e, Western blot validation of Notch1 rescue constructs: mApple, TMD-mApple, ICD+TMD-mApple, and ICD+TMD V1754G-mApple. f, Immunoprecipitation of VE-cadherin from hMVEC-D cells treated with DMSO or DAPT. Co-immunoprecipitation of mechanosensory complex components was assessed by immunoblotting for Notch1 ICD, Trio, and LAR. g. Western blot of VE-cadherin immunoprecipitations from Notch1 KO cells expressing Notch1-TMD truncation constructs (6, 8, 12 amino acids from the N-terminus) fused to mApple. h, Western blot of VE-cadherin immunoprecipitations from Notch1 KO cells expressing single and dual point mutation Notch1-TMD constructs (within the transmembrane segment of Notch1 TMD) fused to mApple. All images representative of at least three independent experiments.
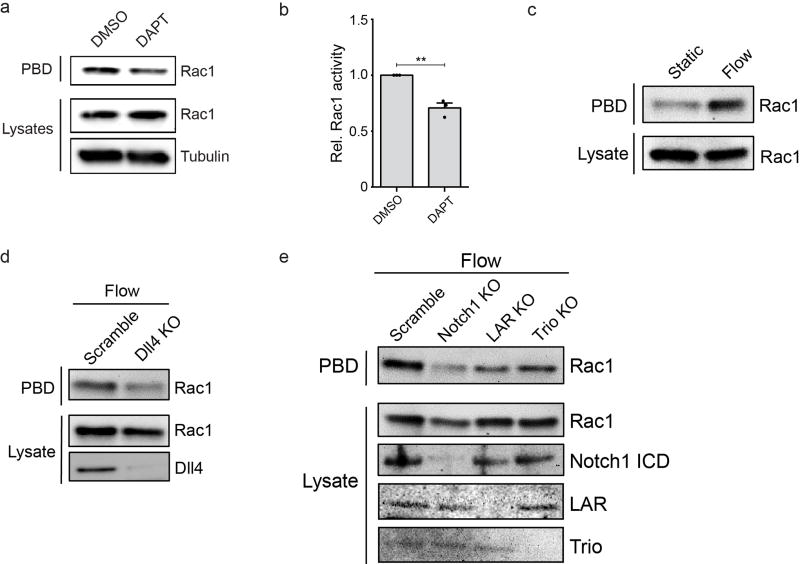
Extended Data Figure 7. Dll4 and the Notch1 mechanosensory complex are critical for increased Rac1 activity in response to shear stress.
a, Active Rac1 was isolated with GST-PBD from hMVEC-D cell lysates treated with DMSO and DAPT (20 µM). b, Quantification of western blot band intensity demonstrates a decrease (~30%) in Rac1 activity with DAPT treatment. c, Active Rac1 was isolated using GST-PBD from hMVEC-D cell lysates from static or shear flow conditions. d, Active Rac1 was isolated using GST-PBD from Dll4-KO cell lysates under flow conditions. e, Active Rac1 was isolated using GST-PBD from Notch1 KO, LAR KO, and Trio KO cell lysates under flow conditions. Mean ± s.e.m., n=3 independent lysates, **p<0.01. Exact p and n values available in Figure Source Data, all images representative of at least three independent experiments.
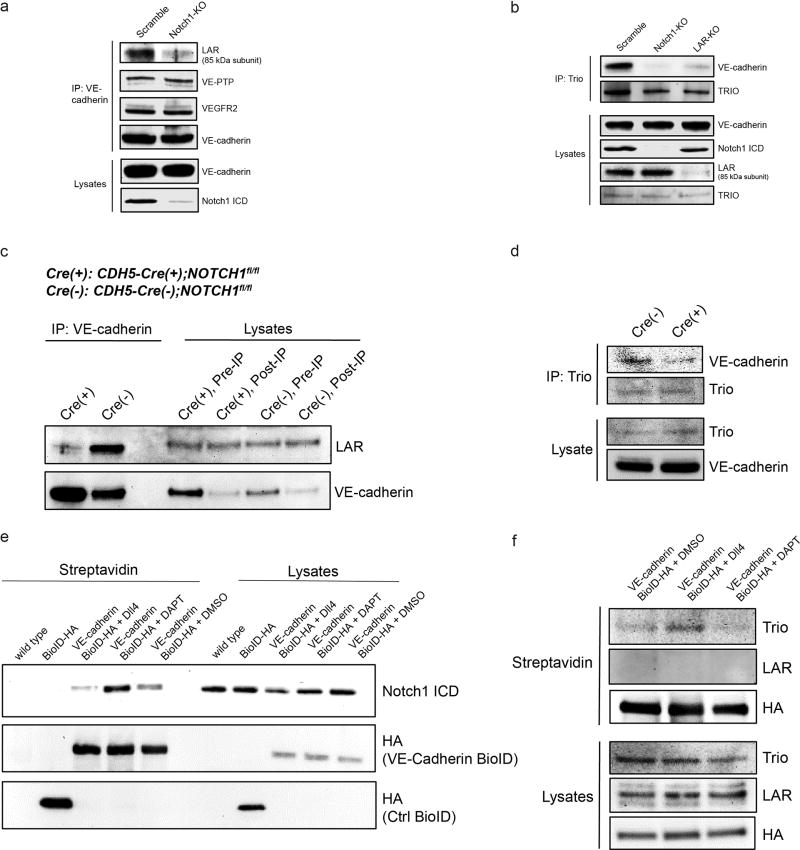
Extended Data Figure 8. Notch1 regulates VE-cadherin interacting proteins to form the Notch1 mechanosensory complex.
a, Immunoprecipitation of VE-cadherin from scramble and Notch1-KO cells. Co-immunoprecipitation of candidate Notch1-dependent, VE-cadherin effectors was assessed by immunoblotting for VE-PTP, VEGFR2, and LAR (85 kDa P-subunit). b, Immunoprecipitation of the Rac1 GEF Trio from scramble, Notch1-KO, and LAR-KO cells. Immunoblotting for VE-cadherin was used to assess impaired Trio-VE-cadherin co-immunoprecipitation upon depletion of Notch1 or LAR. c, Western blots of VE-cadherin immunoprecipitated from the lysates of lungs from CDH5-Cre(+);NOTCH1fl/fl and control CDH5-Cre(+);NOTCH1fl/fl mice and immunoblotted for LAR. d, Western blots of Trio immunoprecipitated from the lysates of lungs from CDH5-Cre(+);NOTCH1fl/fl and control CDH5-Cre(+);NOTCH1fl/fl mice and immunoblotted for VE-cadherin. e, Western blot of proximal interacting proteins extracted with streptavidin from hMVEC-D cells expressing BirA-HA (BioID) or VE-cadherin-BirA-HA (VE-BioID) that were treated with DMSO, Dll4, or DAPT, immunoblotted for HA and Notch1 ICD. f, Western blot of proximal interacting proteins extracted with streptavidin in hMVEC-D cells expressing VE-cadherin-BirA-HA (VE-BioID) that were treated with DMSO, Dll4, or DAPT, immunoblotted for Trio and LAR. All images representative of at least two independent experiments.
Extended Data Figure 9. The Notch1 mechanosensory complex stabilizes cell-cell junctions through activation of Rac1.
Flow induces endocytosis of Dll4, triggering the activation and cleavage of Notch1 ICD and ECD, which allows the N1-TMD domain to scaffold the adaptor protein LAR with VE-cadherin and recruit the Rac1 GEF Trio to AJs. The resulting complex activates Rac1, elaborates cortical actin, and stabilizes cell-cell junctions to establish barrier function.
Extended Data Figure 10. Supplemental methods.
a, Schematic and summary of methods for quantifying vascular permeability in vivo with intravital microscopy. b, Intensity as a function of distance along lines connecting the nucleus centroids of neighboring cells. Blue circles are local maxima used to count the number of stress fibers per unit length, and the shaded region is the area under the peak corresponding to cortical actin and is normalized by the total area under the curve for quantification. Graph representative of three independent experiments.
Supplementary Material
Acknowledgments
This work was supported in part by grants from the National Institutes of Health (EB00262, EB08396, UH3EB017103, HL115553) and the National Science Foundation Center for Engineering MechanoBiology (CMMI15-48571). W.J.P. acknowledges support from a Ruth L. Kirchstein National Research Service Award (F32 HL129733) and from the NIH through the Organ Design and Engineering Training program (T32 EB16652), and M.L.K acknowledges support from The Hartwell Foundation and from the NIH through the Translational Research in Regenerative Medicine Training program (T32 EB005583). We would like to thank Patrick A. Murphy and Rong Wang for helpful discussions of preliminary data and Martin Schwartz for materials and helpful discussions.
Footnotes
AUTHOR CONTRIBUTIONS
W.J.P., M.L.K., and C.S.C. conceived the study and designed experiments. W.J.P. and M.L.K. performed all experiments and data analysis with assistance from J.E. (qPCR) and J.Y. (Notch1 knockout mice). Y.W., H.V., and K.K.H. provided Notch1 knockout mice. W.J.P., M.L.K., and C.S.C. wrote the manuscript with input from all authors.
The authors declare no competing financial interests.
References
- 1.Mehta D, Malik AB. Signaling mechanisms regulating endothelial permeability. Physiol. Rev. 2006;86:279–367. doi: 10.1152/physrev.00012.2005. [DOI] [PubMed] [Google Scholar]
- 2.Chiu J-J, Chien S. Effects of dsisturbed flow on vascular endothelium : pathophysiological basis and clinical perspectives. Physiol Rev. 2011;91:327–387. doi: 10.1152/physrev.00047.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lawson ND, et al. Notch signaling is required for arterial-venous differentiation during embryonic vascular development. 2001;3683:3675–3683. doi: 10.1242/dev.128.19.3675. [DOI] [PubMed] [Google Scholar]
- 4.Hellström M, et al. Dll4 signalling through Notch1 regulates formation of tip cells during angiogenesis. Nature. 2007;445:776–780. doi: 10.1038/nature05571. [DOI] [PubMed] [Google Scholar]
- 5.Krebs LT, et al. Notch signaling is essential for vascular morphogenesis in mice. Genes Dev. 2000;14:1343–1352. [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y-SJ, Haga JH, Chien S. Molecular basis of the effects of shear stress on vascular endothelial cells. J. Biomech. 2005;38:1949–1971. doi: 10.1016/j.jbiomech.2004.09.030. [DOI] [PubMed] [Google Scholar]
- 7.Hahn C, Schwartz MA. Mechanotransduction in vascular physiology and atherogenesis. Nat Rev Mol Cell Biol. 2009;10:53–62. doi: 10.1038/nrm2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ayata C, Ropper AH. Ischaemic brain oedema. J. Clin. Neurosci. 2002;9:113–124. doi: 10.1054/jocn.2001.1031. [DOI] [PubMed] [Google Scholar]
- 9.Dongaonkar RM, Stewart RH, Geissler HJ, Laine GA. Myocardial microvascular permeability, interstitial oedema, and compromised cardiac function. Cardiovasc. Res. 2010;87:331–339. doi: 10.1093/cvr/cvq145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia-Cardeña G, Comander J, Anderson KR, Blackman BR, Gimbrone Ma. Biomechanical activation of vascular endothelium as a determinant of its functional phenotype. Proc. Natl. Acad. Sci. U.S.A. 2001;98:4478–4485. doi: 10.1073/pnas.071052598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen X, et al. Cilia Control Vascular Mural Cell Recruitment in Article Cilia Control Vascular Mural Cell Recruitment in Vertebrates. CellReports. 18:1033–1047. doi: 10.1016/j.celrep.2016.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Artavanis-Tsakonas S, MD R, Lake RJ. Notch Signaling: Cell Fate Control and Signal Integration in Development. Science (80-.) 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 13.Bray SJ. Notch signalling in context. Nat. Rev. Mol. Cell Biol. 2016;9:722–735. doi: 10.1038/nrm.2016.94. [DOI] [PubMed] [Google Scholar]
- 14.Weng AP, et al. Growth Suppression of Pre-T Acute Lymphoblastic Leukemia Cells by Inhibition of Notch Signaling Growth Suppression of Pre-T Acute Lymphoblastic Leukemia Cells by Inhibition of Notch Signaling. Mol. Cell. Biol. 2003;23:655–664. doi: 10.1128/MCB.23.2.655-664.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rizzo V, Morton C, DePaola N, Schnitzer JE, Davies PF. Recruitment of endothelial caveolae into mechanotransduction pathways by flow conditioning in vitro. Am. J. Physiol. - Hear. Circ. Physiol. 2003;285:H1720–H1729. doi: 10.1152/ajpheart.00344.2002. [DOI] [PubMed] [Google Scholar]
- 16.Gordon WR, et al. Mechanical Allostery: Evidence for a Force Requirement in the Proteolytic Activation of Notch. Dev. Cell. 2015;33:729–736. doi: 10.1016/j.devcel.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mammoto T, et al. Angiopoietin-1 requires p190 RhoGAP to protect against vascular leakage in vivo. J. Biol. Chem. 2007;282:23910–23918. doi: 10.1074/jbc.M702169200. [DOI] [PubMed] [Google Scholar]
- 18.Mumm JS, et al. A Ligand-Induced Extracellular Cleavage Regulates -Secretase-like Proteolytic Activation of Notch1. Mol. Cell. 2000;5:197–206. doi: 10.1016/s1097-2765(00)80416-5. [DOI] [PubMed] [Google Scholar]
- 19.Huppert SS, et al. Embryonic lethality in mice homozygous for a processing-deficient allele of Notch1. Nature. 2000;405:966–970. doi: 10.1038/35016111. [DOI] [PubMed] [Google Scholar]
- 20.Spindler V, Schlegel N, Waschke J. Role of GTPases in control of microvascular permeability. Cardiovasc. Res. 2010;87:243–53. doi: 10.1093/cvr/cvq086. [DOI] [PubMed] [Google Scholar]
- 21.Timmerman I, et al. A local VE-cadherin and Trio-based signaling complex stabilizes endothelial junctions through Rac1. J. Cell Sci. 2015;128:3041–54. doi: 10.1242/jcs.168674. [DOI] [PubMed] [Google Scholar]
- 22.Debant A, et al. The multidomain protein Trio binds the LAR transmembrane tyrosine phosphatase, contains a protein kinase domain, and has separate rac-specific and rho-specific guanine nucleotide exchange factor domains. Proc. Natl. Acad. Sci. U.S.A. 1996;93:5466–5471. doi: 10.1073/pnas.93.11.5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le Gall M, De Mattei C, Giniger E. Molecular separation of two signaling pathways for the receptor, Notch. Dev. Biol. 2008;313:556–567. doi: 10.1016/j.ydbio.2007.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roux KJ, Kim DI, Raida M, Burke B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 2012;196:801–810. doi: 10.1083/jcb.201112098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coon BG, et al. Intramembrane binding of VE-cadherin to VEGFR2 and VEGFR3 assembles the endothelial mechanosensory complex. J. Cell Biol. 2015;208:975–986. doi: 10.1083/jcb.201408103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stittrich AB, et al. Mutations in NOTCH1 cause Adams-Oliver syndrome. Am. J. Hum. Genet. 2014;95:275–284. doi: 10.1016/j.ajhg.2014.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McDaniell R, et al. NOTCH2 Mutations Cause Alagille Syndrome, a Heterogeneous Disorder of the Notch Signaling Pathway. Am. J. Hum. Genet. 2006;79:169–173. doi: 10.1086/505332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joutel A, et al. Notch3 mutations in CADASIL, a hereditary adult-onset condition causing stroke and dementia. Nature. 1996;383:707–710. doi: 10.1038/383707a0. [DOI] [PubMed] [Google Scholar]
- 29.Smith DC, et al. A Phase I Dose Escalation and Expansion Study of the Anticancer Stem Cell Agent Demcizumab (Anti-DLL4) in Patients with Previously Treated Solid Tumors. Clin. Cancer Res. 2014;20:6295–303. doi: 10.1158/1078-0432.CCR-14-1373. [DOI] [PubMed] [Google Scholar]
- 30.Bentley K, et al. The role of differential VE-cadherin dynamics in cell rearrangement during angiogenesis. Nat. Cell Biol. 2014;16:309–21. doi: 10.1038/ncb2926. [DOI] [PubMed] [Google Scholar]
- 31.Adamson RH, Lenz JF, Curry FE. Quantitative laser scanning confocal microscopy on single capillaries: permeability measurement. Microcirculation. 1994;1:251–65. doi: 10.3109/10739689409146752. [DOI] [PubMed] [Google Scholar]
- 32.Sturn A, Quackenbush J, Trajanoski Z. Genesis: cluster analysis of microarray data. Bioinformatics. 2002;18:207–8. doi: 10.1093/bioinformatics/18.1.207. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Graphical source data is provided in each figure legend. The source data for the western blots in the figures are available in the Supplementary Information. Additional data that support the findings of this study are available from the corresponding author on reasonable request.