SUMMARY
Bactericidal antibiotics alter microbial metabolism as part of their lethality and can damage mitochondria in mammalian cells. In addition, antibiotic susceptibility is sensitive to extracellular metabolites, but it remains unknown if metabolites present at an infection site can affect either treatment efficacy or immune function. Here, we quantify local metabolic changes in the host microenvironment following antibiotic treatment for a peritoneal Escherichia coli infection. Antibiotic treatment elicits microbiome-independent changes in local metabolites but not those distal to the infection site by acting directly on host cells. The metabolites induced during treatment, such as AMP, reduce antibiotic efficacy and enhance phagocytic killing. Moreover, antibiotic treatment impairs immune function by inhibiting respiratory activity in immune cells. Collectively, these results highlight the immunomodulatory potential of antibiotics and reveal the local metabolic microenvironment to be an important determinant of infection resolution.
Keywords: antibiotic, metabolic environment, metabolomics, LC-MS/MS, immunomodulation, respiration, phagocytosis, germ-free, systems biology
eTOC BLURB
Antibiotic susceptibility is sensitive to metabolites but how this impacts in vivo treatment efficacy remains unexplored. Yang and Bhargava et al. characterize antibiotic-induced changes to the metabolic environment during infection and find that direct actions of antibiotics on host cells induce metabolites that alter drug efficacy and impair phagocytic activity.
INTRODUCTION
Although the mechanisms of action for most conventional antibiotics have been well studied (Kohanski et al., 2010), the effects of antibiotic treatment on human physiology and host-microbe interactions are only beginning to become understood (Willing et al., 2011). Antibiotics have been observed to alter immune responses (Anuforom et al., 2015) and there is growing appreciation that non-specific actions by antibiotics may promote disease by creating favorable niches for opportunistic pathogens (Theriot et al., 2014). In light of the pressing challenges of antibiotic resistance and the diminishing drug discovery pipeline (Brown and Wright, 2016), there is an urgent need to better understand the complex consequences of antibiotic treatment during infection.
We have previously shown that bacterial metabolism participates in the efficacy of bactericidal antibiotics (Belenky et al., 2015; Dwyer et al., 2014; Lobritz et al., 2015) and that antibiotic susceptibility is sensitive to extracellular metabolites (Allison et al., 2011; Meylan et al., 2017). During infection, bacterial pathogens dynamically remodel their metabolic environment by inducing host catabolism, disrupting metabolic balance and altering the abundance of energy metabolites, amino acids and lipids (Beisel, 1975; Dong et al., 2012). However, it remains unknown how local changes to the metabolic microenvironment might also alter antibiotic efficacy during treatment.
Here, we sought to determine if antibiotic treatment alters the host metabolic microenvironment and if such changes alter antibiotic susceptibility or immune function. We performed targeted metabolomics on samples from mice receiving oral antibiotics for a peritoneal infection and found that antibiotics treatment systemically alters metabolites in the host, depleting central metabolism intermediates in the peritoneum. We show that these changes are microbiome-independent as they also occur in germ-free mice. Additionally, we demonstrate that metabolites altered by antibiotic treatment during infection may inhibit antibiotic efficacy and potentiate the phagocytic activity of immune cells. Moreover, we show that antibiotics directly inhibit respiratory activity in immune cells and can consequently impair their phagocytic activity. Together, these results indicate that antibiotic-induced changes to host metabolites and metabolic processes can significantly impact both treatment efficacy and immune function.
RESULTS
Antibiotic treatment depletes central metabolism intermediates in the peritoneum
In order to determine if antibiotics alter metabolites in the host environment, we quantified metabolites in samples from mice receiving antibiotic treatment, bacterial infection or their combination. We subjected a cohort of 8-week old C57BL/6J mice to a set of perturbations including antibiotic treatment with 100 μg/mL ciprofloxacin (cipro) delivered in the drinking water (ABX), intraperitoneal infection by Escherichia coli ATCC25922 (INF), or their combination (COMB) (Figure 1A). After 24 h, mice were sacrificed and samples from three tissues were collected for metabolomic profiling: (a) the peritoneum, to characterize changes in metabolites local to infection; (b) plasma, to characterize global changes in circulating metabolites; and (c) the lung, to characterize changes in metabolites distal to infection. We performed targeted LC-MS/MS on these samples (McCloskey et al., 2015), enabling absolute quantification for nearly 80 metabolites supporting bacterial growth, including amino acids, nucleotides and central metabolism intermediates (Table S1).
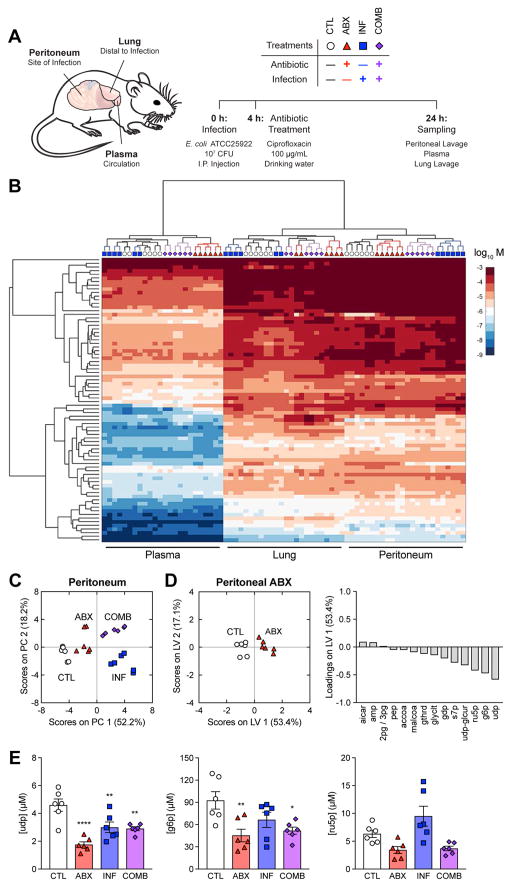
Figure 1. Antibiotic treatment depletes central metabolism intermediates in the peritoneum.
(A) Experimental design for metabolomic profiling. C57BL/6J mice were subjected to control conditions (CTL), antibiotic treatment with 100 μg/mL cipro (ABX), intraperitoneal infection with 107 CFU E. coli (INF), or their combination (COMB). Peritoneal lavage, plasma and lung lavage samples were collected 24 h after infection.
(B) Hierarchically clustered heatmap of metabolite concentrations from CTL, ABX, INF and COMB mice.
(C) PCA projection of metabolomic profiles from peritoneal samples of all four treatment groups.
(D) PLS-DA of peritoneal samples from ABX mice. Metabolites selected by elastic net regularization were depleted for central metabolism intermediates.
(E) Concentrations for metabolites with large LV1-loadings in peritoneal samples from the ABX metabolite signature. Antibiotic treatment depleted uridine diphosphate (udp), glucose-6-phosphate (g6p) and ribulose-5-phosphate (r5p).
Data are represented as mean ± SEM from n = 3 independent biological replicates. Significance reported as FDR-corrected p-values in comparison with corresponding CTL conditions: *: p ≤ 0.05, **: p ≤ 0.01, ****: p ≤ 0.0001.
Hierarchical clustering of these measurements revealed that the metabolomic profiles clustered first by tissue, then by perturbation (Figure 1B), indicating that the metabolic changes elicited by antibiotic treatment or infection were local and tissue specific. Unsupervised principal component analysis (PCA) revealed that antibiotic treatment systemically altered metabolites in the host environment as samples from ABX mice clustered away from control (CTL) samples in all three tissues (Figure 1C, S1A, S1B). In contrast, infection only exerted local changes, eliciting significant changes in metabolites in the peritoneum (Figure 1C), but not in the plasma (Figure S1A) or lung (Figure S1B).
In order to better understand the antibiotic-induced changes in the infection microenvironment, we performed multivariate analysis on the peritoneal samples to identify a metabolite signature corresponding to antibiotic treatment. We applied elastic net regularization and partial least squares-discriminant analysis (PLS-DA) and identified a feature set of metabolites that could discriminate the ABX samples from the union of CTL and INF samples (Ballabio and Consonni, 2013). This yielded 14 metabolites sufficient for explaining 71% of the variance in metabolites and 40% of the variance in treatments with 100% calibration accuracy and 100% cross-validation accuracy (Figure 1D). PLS-DA predicted that antibiotic-associated metabolic changes were most strongly characterized by depletion in uridine diphosphate (udp), glucose-6-phosphate (g6p) and ribulose-5-phosphate (ru5p) (Figure 1E). By inspection, many of these metabolites are intermediates in the pentose phosphate pathway, glycolysis and fatty acid biosynthesis. We performed metabolite set enrichment analysis (MSEA) on these metabolites and found enrichment for “carbohydrate biosynthesis” (p = 6.47e–4) (Table S2).
Performing a similar analysis on the plasma and lung samples, we found different metabolite signatures associated with each tissue (Figure S1C, S1D). While MSEA identified lung metabolites as enriched for “generation of precursor metabolites and energy” (p = 3.46e–3) (Table S3), plasma metabolites were not specifically enriched for any metabolic pathways with FDR-corrected p-values below significance (p = 0.05). We metabolomically profiled additional mice treated with a higher concentration of cipro (Figure S2A) and found that 400 μg/mL cipro mostly amplified the antibiotic-induced changes to host metabolites that we had observed at 100 μg/mL cipro in the peritoneum (Figure S2B, S2C) and plasma (Figure S2B, S2D), indicated by a further projection along the first principal component. Together, these data indicate that antibiotic treatment exerts tissue-specific changes in metabolites in the host environment.
Antibiotic treatment elicits microbiome-independent changes in host metabolites
Antibiotics are thought to systemically alter metabolites in the host environment by acting on the gut microbiome (Reijnders et al., 2016), but the tissue-specific changes we observed suggested that antibiotics may instead act locally and directly on host cells. To test this hypothesis, we profiled metabolites from germ-free mice treated with 100 μg/mL cipro delivered in the drinking water (Figure 2A). Surprisingly, we found that many of the tissue-specific changes in metabolites from the conventional (CONV) mice also occurred in the germ-free (GF) mice lacking a microbiome (Figure 2B).
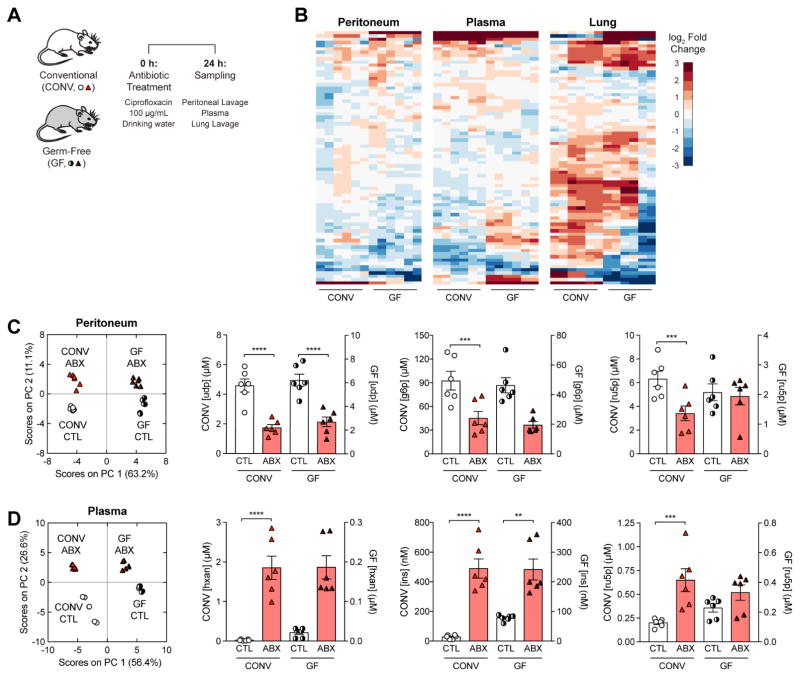
Figure 2. Antibiotic treatment elicits microbiome-independent changes in host metabolites.
(A) Experimental design for germ-free (GF) metabolomic profiling. GF mice were subjected to antibiotic treatment with 100 μg/mL cipro (ABX) and sampled 24 h later.
(B) Hierarchically clustered heatmaps for changes in metabolite concentrations between ABX and control (CTL) mice, by tissue sampled.
(C) PCA projection of metabolomic profiles from CTL and ABX conventional (CONV) and GF mice in the peritoneum (left). Concentrations for peritoneal metabolites with large peritoneal ABX LV1-loadings in CONV and GF mice (right).
(D) PCA projection of metabolomic profiles from CTL and ABX conventional (CONV) and GF mice in the plasma (left). Concentrations for plasma metabolites with large plasma ABX LV1-loadings in CONV and GF mice (right).
Data are represented as mean ± SEM from n = 3 independent biological replicates. Significance reported as FDR-corrected p-values in comparison with corresponding CTL conditions: **: p ≤ 0.01, ***: p ≤ 0.001, ****: p ≤ 0.0001.
In order to better understand if the antibiotic-induced changes to local metabolites in the CONV mice were due to direct effects on host cells versus effects on the microbiome, we performed PCA on CTL and ABX samples from both the CONV and GF mice for each tissue. PCA orthogonally clustered the peritoneal samples into four distinct quadrants along two principal components that captured 74% of the variance and appeared to directly correspond to either presence of a microbiome (PC 1) or of antibiotic treatment (PC 2) (Figure 2C). Moreover, inspection of metabolites with the greatest PLS-DA loadings in the peritoneal ABX signature revealed similar fold-changes in both CONV and GF mice, despite differences in the untreated CTL concentrations. PCA on the plasma samples similarly revealed that 83% of the variance in plasma metabolites could be explained by direct effects on host cells instead of effects on the microbiome (Figure 2D). In contrast, the lung samples did not separate as cleanly along principal components corresponding to a microbiome and antibiotic treatment (Figure S3). Together, these data indicate that most of the antibiotic-induced changes in metabolites are microbiome-independent and likely due to direct actions of antibiotics on local host cells.
Antibiotic treatment elicits unique metabolic changes in the presence of infection
Because antibiotics are administered to treat infection, we sought to characterize metabolic changes that occur in the presence of infection. We first identified an infection-specific metabolic signature by multivariate analysis and then tested if the complex changes observed under combination treatment could be explained by the ABX and INF signatures. We performed elastic net regularization and used PLS-DA to discriminate the INF samples from the union of CTL and ABX samples, identifying a feature set of nine metabolites sufficient for explaining 90% of the variance in metabolites and 44% of the variance in treatments with 100% calibration accuracy and 100% cross-validation accuracy (Figure 3A). These were most strongly characterized by enrichment in guanosine monophosphate (gmp) and depletion in adenosine (adn) and adenosine monophosphate (amp) (Figure 3B). MSEA revealed that these were specifically enriched for purine metabolic pathways, including terms such as “purine nucleotide degradation” (p=1.42e–6) and “purine nucleotide biosynthesis” (p=1.96e–6) (Table S4). Because purine metabolites can function as important immune signaling molecules (Cekic and Linden, 2016), these data suggest that the INF metabolite signature indicated a local induction of host immunity.
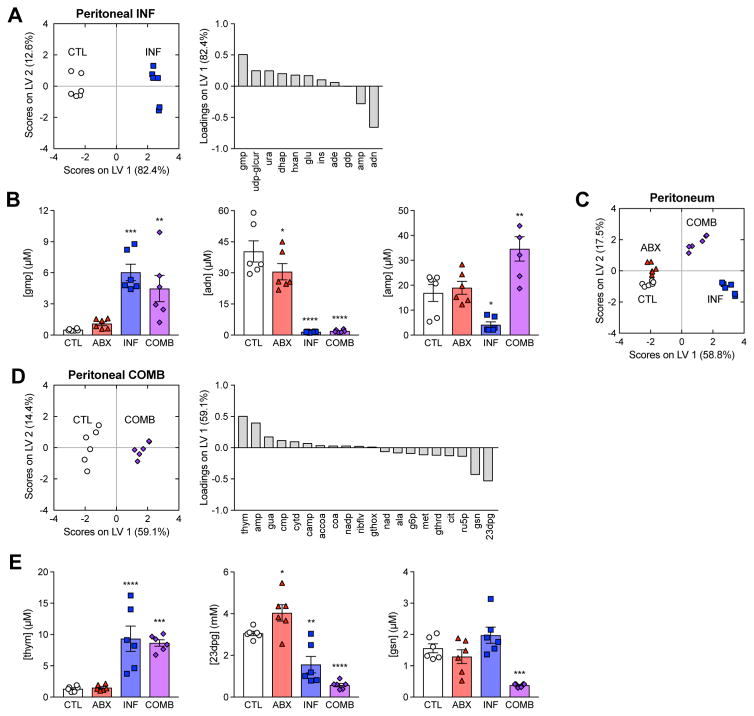
Figure 3. Antibiotic treatment elicits unique metabolic changes in the presence of infection.
(A) PLS-DA of peritoneal samples from INF mice. Metabolites selected by elastic net regularization were enriched for purine metabolites.
(B) Concentrations for metabolites with large LV1-loadings in peritoneal samples from the INF metabolite signature. Peritoneal infection increased the abundance of guanine monophosphate (gmp) and depleted adenosine (adn) and adenosine monophosphate (amp).
(C) PCA projection of peritoneal samples from all four treatment groups using metabolites from the ABX and INF metabolite signatures.
(D) PLS-DA of peritoneal samples from COMB mice. Metabolites selected by elastic net regularization were enriched across diverse pathways.
(E) Concentrations for metabolites with large LV1-loadings in peritoneal samples from the COMB metabolite signature. The combination treatment increased the abundance of thymine (thym) and depleted 3-phospho-D-glyceroyl phosphate (23dpg) and guanosine (gsn).
Data are represented as mean ± SEM from n = 3 independent biological replicates. Significance reported as FDR-corrected p-values in comparison with corresponding CTL conditions: *: p ≤ 0.05, **: p ≤ 0.01, ***: p ≤ 0.001, ****: p ≤ 0.0001.
PLS-DA of the peritoneal samples from all four treatment groups onto the union of metabolites from the ABX and INF signatures revealed that antibiotic treatment and infection formed two orthogonal dimensions describing the infection microenvironment with 88% calibration accuracy and 83% cross-validation accuracy (Figure 3C), in close agreement to the unsupervised PCA of these samples (Figure 1C). This was supported by the observation that udp, g6p and ru5p under combination treatment trended similarly to antibiotic treatment and gmp and adn trended similarly to infection (Figure 3B). Collectively, these results suggest that most of the local changes in host metabolites associated with antibiotic treatment of infection could be explained by the independent actions of either antibiotic treatment or infection.
Interestingly, not all of the metabolite changes in the COMB samples could be explained by the independent actions of antibiotics or infection. For instance, amp was significantly enriched in the COMB samples despite being depleted in the INF samples and unchanged in the ABX samples (Figure 3B). We subjected additional mice to both infection and antibiotic treatment and sampled the peritoneum at earlier time points. We found that amp concentrations peaked at ~3-fold six hours after infection (Figure S4). COMB samples also scored higher on the antibiotic axis (LV 2) than the ABX samples, suggesting infection may potentiate the metabolic changes elicited by antibiotic treatment (Figure 3C). To test this, we performed elastic net regularization and used PLS-DA to identify a COMB-specific metabolite signature discriminating the COMB samples from the union of CTL, ABX and INF samples. This was comprised of 20 metabolites sufficient for explaining 55% of the variance in metabolites and 35% of the variance in treatments with 100% calibration accuracy and 96% cross-validation accuracy (Figure 3D). PLS-DA predicted that this signature was enriched for thymine (thym) and amp and depleted in 3-phospho-D-glyceroyl phosphate (23dpg) and guanosine (gsn) (Figure 3E, 3B). MSEA revealed these to be enriched in diverse metabolic processes, including various nucleotide, energy and amino acid metabolism pathways (Table S5).
Metabolites altered by antibiotic treatment during infection inhibit drug efficacy
Antibiotic efficacy is sensitive to the abundance of extracellular metabolites (Allison et al., 2011; Meylan et al., 2017). We hypothesized that host metabolites induced by antibiotic treatment of infection might feedback on drug susceptibility in the pathogen. To test this hypothesis, we quantified the minimum inhibitory concentration (MIC) of cipro in E. coli cells following 10 mM supplementation with thym or amp, which was ~13 ng/mL without supplementation. While thym supplementation did not appear to alter the MIC (~13.5 ng/mL), amp supplementation significantly decreased cipro susceptibility, increasing the MIC to ~100 ng/mL (Figure 4A). Time-kill experiments with 25 ng/mL cipro revealed that amp supplementation elicited dose-dependent protection (Figure 4B).
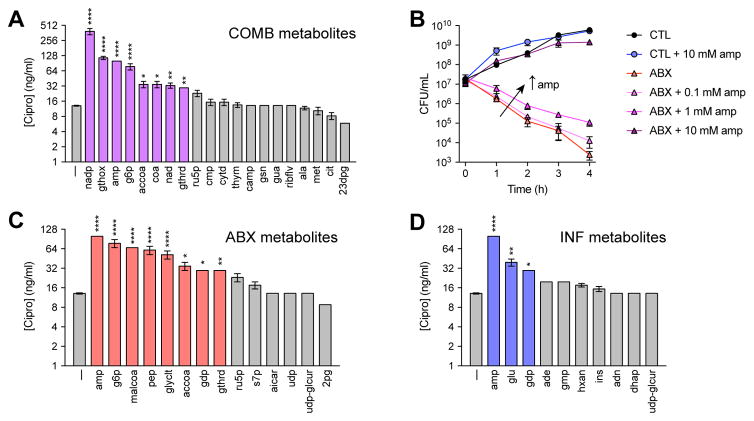
Figure 4. Metabolites altered by antibiotic treatment during infection inhibit drug efficacy.
(A) Cipro MICs following supplementation with 10 mM of each metabolite from the COMB signature.
(B) Dose-dependent reduction in cipro susceptibility by amp. E. coli were treated with 25 ng/mL ciprofloxacin, supplemented with increasing concentrations of amp.
(C) Cipro MICs following supplementation with 10 mM of each metabolite from the ABX signature.
(D) Cipro MICs following supplementation with 10 mM of each metabolite from the INF signature.
Data are represented as mean ± SEM from n ≥ 3 independent biological replicates. Significance reported as FDR-corrected p-values in comparison with corresponding CTL conditions: *: p ≤ 0.05, **: p ≤ 0.01, ***: p ≤ 0.001, ****: p ≤ 0.0001.
We further characterized changes in MIC promoted by supplementation with metabolites from the COMB, ABX or INF signatures and found that many of these metabolites also inhibited drug susceptibility (Figure 4A, 4C, 4D). MSEA on these metabolites identified “amines and polyamines biosynthesis” as the metabolic pathway most enriched by these metabolites (p=3.99e–4) (Table S6). Together, these results demonstrate that host metabolites induced by antibiotics during treatment of infection can feedback and impact antibiotic efficacy.
Direct actions of antibiotic treatment on immune cells inhibit phagocytic killing
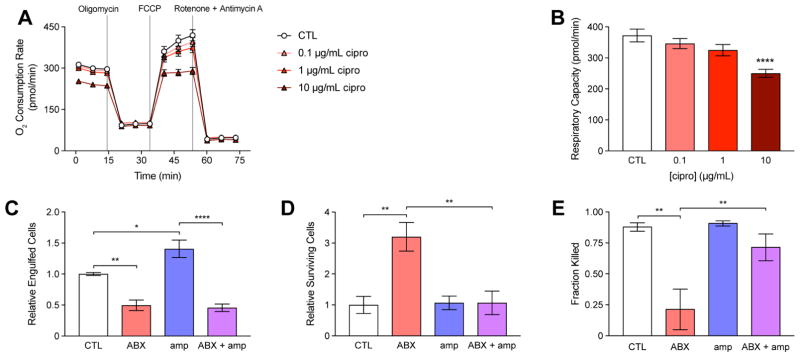
Bactericidal antibiotics can impair mitochondrial function in epithelial cells and inhibit their respiratory activity (Kalghatgi et al., 2013). In addition, metabolites present in the host microenvironment may initiate signaling cascades in immune cells that lead to metabolic reprogramming and functional changes (O’Neill and Hardie, 2013). Because efficient phagocytosis involves induction of a highly energy-dependent respiratory burst (Murphy and Weaver, 2016), we sought to determine if antibiotic treatment might also interfere with the respiratory activity of immune cells. We pre-treated mouse macrophages with cipro and measured changes in oxygen consumption following electron transport chain uncoupling (Figure 5A). These data revealed a dose-dependent inhibition of respiratory capacity (Figure 5B).
Figure 5. Direct actions of antibiotic treatment on immune cells inhibit phagocytic killing.
(A) Changes in macrophage oxygen consumption rate in control (CTL) and cells pre-treated for 3 h with cipro, following electron transport chain uncoupling by 2 μM oligomycin, 1 μM FCCP and 0.5 μM rotenone + antimycin A.
(B) Changes in respiratory capacity following cipro pre-treatment.
(C) Pathogen engulfment by control (CTL) or macrophages treated with 20 μg/mL cipro (ABX), 10 mM amp (amp) or their combination (ABX + amp).
(D) Pathogen survival in CTL, ABX, amp or ABX + amp macrophages.
(E) Phagocytic killing by CTL, ABX, amp or ABX + amp macrophages.
Data are represented as mean ± SEM from n ≥ 3 independent biological replicates. Significance reported as FDR-corrected p-values within the indicated comparisons: *: p ≤ 0.05, **: p ≤ 0.01, ****: p ≤ 0.0001.
We hypothesized that antibiotic-induced impairments in respiration and/or the induction of local metabolites might physiologically alter the phagocytic activity of immune cells recruited to a site of infection. To test this hypothesis, we enumerated E. coli cells attached or killed by macrophages following treatment with cipro and/or amp. Compared with untreated cells, macrophages pretreated with 20 μg/mL cipro for 3 h (ABX) engulfed fewer E. coli cells (Figure 5C, S5A) and possessed more surviving cells following lysis of the macrophages (Figure 5D, S5B), indicating a significant decrease in the killing of engulfed cells (Figure 5E). In contrast, we found that 10 mM amp significantly increased pathogen engulfment (Figure 5C, S5C) and decreased pathogen survival (Figure 5D, S5D), indicating that metabolites induced at the site of infection by antibiotics may feed-forward and potentiate immune function. Interestingly, antibiotic treatment exerted a dominant negative effect over the metabolic potentiation in macrophages subjected to both cipro and amp (ABX+amp), most prominently in the ability to engulf pathogens. Collectively, these results indicate that antibiotic treatment can directly act on immune cells and metabolically impair their function.
DISCUSSION
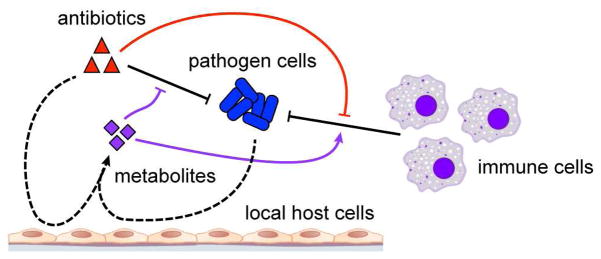
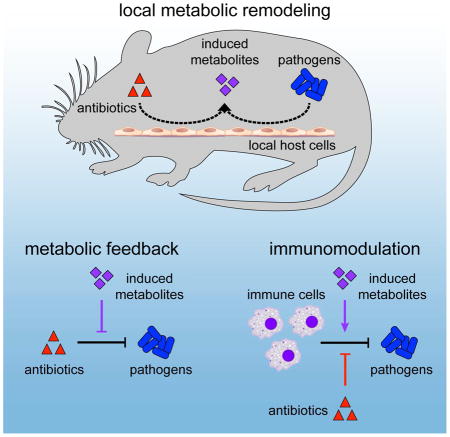
Infections are complex host-microbe interfaces comprised of multiple species, cell types and biochemical species. While local metabolism is understood to be important for pathogens to establish their niche, the contribution of antibiotics to this environment is poorly understood. Here, we investigated the effects of antibiotic treatment on host metabolism in a commonly used and well-defined model of in vivo infection. While antibiotics normally act in concert with host immune cells to remove pathogens at a site of infection, we report that antibiotics also act synergistically with pathogen cells to remodel the local metabolic environment (Figure 6). We demonstrate that metabolites induced in this environment have the capacity to affect both antibiotic efficacy and immune function and that the direct consequences of antibiotics on the metabolism of immune cells can inhibit their phagocytic activity. These data highlight the complex interactions elicited by antibiotics at the site of infection and support prior in vitro studies demonstrating that the host metabolic environment is critical for the function of both antibiotics (Yang et al., 2017) and immune cells (Buck et al., 2017).
Figure 6. Metabolic effects of antibiotic treatment on host cells inhibit drug efficacy and impair immune function.
During infection, antibiotics work in concert with immune cells to clear microbial pathogens (black). Meanwhile, antibiotics and pathogen cells metabolically remodel the local infectious microenvironment by acting on local host cells (dashed). Induced metabolites can inhibit drug efficacy and potentiate immune function (purple). Direct actions by antibiotics on immune cell metabolism can also impair immune cell phagocytic activity (red).
Antibiotics are thought to systemically alter metabolites in the host by manipulating gut microbiome taxonomy (Reijnders et al., 2016). We report that orally administered antibiotics also induce dose-dependent and microbiome-independent changes in metabolites in the host environment. Of significance, we find that most of the metabolic variation observed in conventionally raised mice also appeared in germ-free mice in both the plasma and peritoneum. These results are consistent with our previous observations that bactericidal antibiotics directly induce mitochondrial dysfunction in mammalian cells (Kalghatgi et al., 2013) and our present observations that antibiotics can inhibit the phagocytic activity of immune cells, which require a functioning respiratory chain for their bactericidal oxidative burst. In our in vitro experiments, these effects appeared to occur at concentrations ≥ 1 μg/mL, well within the pharmacokinetic range of 1–5 μg/mL achieved in human serum following oral delivery and ≥ 10 μg/mL in other tissues following intravenous delivery (Vance-Bryan et al., 1990).
Nutrient availability at a site of infection is an important regulator of antibiotic susceptibility (Amato et al., 2014) and previous studies have demonstrated that antibiotic tolerance may be overcome metabolically by stimulating bacterial central metabolism (Allison et al., 2011; Meylan et al., 2017). We report that diverse metabolites induced by antibiotics at the site of infection also have potential to alter both antibiotic efficacy and immune function. An example of this is in the synergistic and local induction of amp, an energy currency precursor and a participant in purine metabolism that exerts immunomodulation through AMP-activated protein kinase (O’Neill and Hardie, 2013). We find that amp alters E. coli sensitivity to cipro and enhances phagocytic killing by macrophages. Although the amp concentrations that we measured from our bulk lavage of the peritoneum (~10–50 μM) did not exceed the ≥ 100 μM required to elicit an effect on either antibiotic efficacy or immune function, it is possible that amp concentrations were higher in regional peritoneal microdomains due to the large surface area and complex geometry of the peritoneum. Moreover, amp concentrations are generally ~200 μM in mammalian tissues (Traut, 1994) and can increase > 100-fold in human tissues following metabolically demanding activities such as exercise (Harkness et al., 1983). Amp therefore may have more significant effects in other tissues or other in vivo infection models.
While here we use amp to exemplify a single form of complex metabolic cross-talk between host and microbe physiology in the context of infection, other metabolites altered at a site of infection likely also significantly impact antibiotic efficacy or immune function through undiscovered mechanisms. MSEA identified polyamine biosynthesis as a metabolic process enriched by the protective metabolites from our metabolite screens. Interestingly, several studies demonstrate that polyamines can protect pathogens against antibiotic treatment (Kwon and Lu, 2006), by inhibiting drug uptake (Sarathy et al., 2013), inducing protective stress responses (Tkachenko et al., 2006), and reducing antibiotic-induced reactive oxygen species (Tkachenko et al., 2012). Future studies will need to clarify the mechanisms by which diverse metabolites may alter antibiotic efficacy (Yang et al., 2017).
Finally, our work identifies the local metabolic microenvironment as an important determinant in the resolution of infection, due to its actions on antibiotic susceptibility and immune function. It is likely that interpersonal differences in the combination of such metabolites may contribute to variable treatment outcomes across patients suffering from similar infections (Lee and Collins, 2011). Additionally, dynamic changes to these environments, for instance by prophylactics, may be useful for maximizing both antibiotic efficacy and immune function. As recognition of metabolic context-dependence for antibiotic efficacy is growing for important human pathogens (Black et al., 2014), our findings support the use of metabolic adjuvants to enhance our existing antibiotic arsenal (Wright, 2016).
STAR METHODS
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, James J. Collins (jimjc@mit.edu).
Experimental Model and Subject Details
Bacterial Strains, Media, Growth Conditions
Escherichia coli strain ATCC25922 was purchased from the American Type Culture Collection (ATCC) and used for all experiments in this study. Cells were cultured in tryptic soy broth (TSB) and grown at 37°C on a rotating shaker at 300 rpm in flasks or 14 mL test tubes or at 900 rpm in 96-well plates.
Vertebrate Animals
Conventional and germ-free 8-week old male C57BL/6J mice were acquired from Jackson Labs (Bar Harbor, ME) and used for all experiments in this study. Once received, conventional mice were housed at the Harvard Institute of Medicine Vivarium and germ-free mice housed at the Gnotobiotic and Microbiology Core Facility at Brigham and Women’s Hospital. All mice were socially housed and monitored daily for ~7 days before experiments to permit acclimatization. Following intra-peritoneal infection and/or antibiotic treatment, mice were monitored twice for signs of dehydration and water intake and observed for signs of potential pain or distress associated with any intestinal discomfort. Animal protocols for the experiments performed in this study were approved by the Harvard Medical School Institutional Animal Care and Use Committee and all experiments conform to relevant regulatory standards.
Cell Lines
Immortalized mouse SV129 macrophages were derived from a bone marrow cell line from male SV129 mice and authenticated by qPCR (Lee et al., 2006). Macrophages were maintained in 1 g/L glucose DMEM (Cell-Gro; Manassas, VA) supplemented with 10% FBS and 1% penicillin/streptomycin. SV129 macrophages were loaded onto 24-well tissue culture treated plates at 5·105 macrophages/well the day before experiment.
Method Details
Materials and Reagents
Ciprofloxacin (cipro) and all metabolites used as metabolic perturbations or metabolomic profiling standards were purchased from Sigma-Aldrich (St. Louis, MO). BD Difco TSB and tryptic soy agar were purchased from Fisher Scientific (Hampton, NH). Uniformly labeled 13C glucose was purchased from Cambridge Isotope Laboratories, Inc. (Tewksbury, MA). LC-MS reagents were purchased from Honeywell Burdick & Jackson® (Muskegon, MI) and Sigma-Aldrich.
Animal Experiments
8-week old male C57BL/6J mice were intraperitoneally injected with 107 CFU E. coli cells in sterile saline. Control water or drinking water containing 100 or 400 μg/mL cipro was introduced 4 h post infection. These concentrations were selected based on an estimate that a typical 25 g mouse drinks 4 mL of water each day, yielding a daily dose of 16 or 64 mg/kg/day cipro, which is in the 10–250 mg/kg/day range typically used to treat humans. Mice were sacrificed 20 h later; blood was harvested by cardiac puncture and plasma was isolated using lithium heparin tubes (Greiner Bio-One; Kremsmünster, Austria), according to the manufacturer’s instructions. The peritoneum was lavaged with 5 mL sterile PBS in 2.5 mL increments. The lung was lavaged through the trachea with 5 mL sterile PBS in 0.7 mL increments. All samples were kept on ice following collection.
Metabolomic Profiling
Metabolites were acquired and quantified on an AB SCIEX Qtrap® 5500 mass spectrometer (AB SCIEX; Framingham, MA) and processed using MultiQuant® 3.0.1, as described previously (McCloskey et al., 2015). Metabolite extractions were performed using a 40:40:20 mixture of acetonitrile, methanol and LC-MS grade water in Phree Phospholipid Removal tubes (Phenomenex; Torrance, CA). Uniformly labeled 13C-standards were generated by growing E. coli in uniformly labeled Glucose M9 minimal media in aerated shake flasks, as previously described (McCloskey et al., 2014). Calibration mixes of standards were split across several mixes, aliquoted, and lyophilized to dryness. All samples and calibrators were equally spiked with the same internal standards. Samples were quantified using isotope-dependent mass spectrometry. Calibration curves were run before and after all biological and analytical replicates. Consistency of quantification between calibration curves was checked by running a Quality Control sample composed of all biological replicates. Values reported are derived from the average of the biological triplicates, analyzed in duplicate (n = 6).
Minimum Inhibitory Concentrations
Minimum Inhibitory Concentrations (MICs) were determined by microbroth dilution in 96-well microtiter plates with 1.5-fold step sizes of cipro dissolved in TSB, as previously described (Andrews, 2001). Approximately 104 CFU E. coli suspended in TSB supplemented with 2–20 mM of each metabolite were added to each well of the 96-well plates to achieve a total volume of 200 μL. Plates were sealed with a breathable membrane and then incubated at 37°C with 900 rpm shaking for 20 h. Following incubation, OD600 was quantified on a SpectraMax M3 plate reader. MIC experiments were performed in at least triplicate from independent overnight cultures with values reported as the mean ± SEM.
Time-Kill Experiments
Time-kill experiments were performed as previously described (Dwyer et al., 2014). Briefly, an overnight culture of E. coli cells in TSB was diluted 1:1,000 and grown to OD600 ~0.1 at 37°C with 300 rpm shaking prior to experiment. 1 mL cultures were then dispensed to 14 mL test tubes and treated with cipro and/or amp. Hourly samples were collected and serially diluted in PBS for colony enumeration 24 h later.
Macrophage Assays
In vitro phagocytosis assays were adapted from previously published protocols (Sokolovska et al., 2012) and performed using SV129 macrophages. On the day of experiment, culture media was replaced with 1 g/L DMEM supplemented with 1% FBS. E. coli cells grown to OD600 ~0.3 in TSB and antibiotics were added to macrophages on ice for 30 min to synchronize phagocytosis. Plates were then incubated at 37°C for 30 min with a control plate maintained on ice to quantify E. coli cells engulfed, but not killed. Cells were washed thrice with PBS + 0.5 mM EDTA. Cells were lysed with pH 11.5 H2O for 4 min and serially diluted for colony enumeration 24 h later.
Oxygen Consumption Rate Quantification
Macrophage respiratory activity was quantified using the Seahorse XF Cell Mito Stress Test Kit and XFe96 Extracellular Flux Analyzer (Seahorse Bioscience; North Billerica, MA), according to the manufacturer’s instructions. Briefly, SV129 macrophages were plated at a density of 125,000 cells per well and cultured overnight before the start of the experiment. Cells were PBS washed and then pre-treated with cipro for 3 h in the XF Base Medium (Seahorse Bioscience; North Billerica, MA). The Mito Stress Test Kit assay was then performed using 2 μM oligomycin, 1 μM FCCP and 0.5 μM rotenone/antimycin A. Respiratory capacity was computed as the difference in oxygen consumption rate following FCCP treatment and following rotenone/antimycin A.
Quantification and Statistical Analysis
Experimental Replicates
All in vivo metabolomic quantification experiments were performed using n = 3 mice in each treatment condition, with samples prepared in technical duplicate, yielding n = 6 samples for each condition. In vitro MIC estimations following metabolite supplementations were performed using n = 3–6 biological replicates of E. coli cells on different days. Time-kill experiments involving E. coli cells were performed in biological triplicate on different days. Oxygen consumption experiments were performed in biological triplicate with n = 5 technical replicates in different wells of the 96-well Seahorse cartridges on different days. In vitro measurements of immune function using SV129 mouse macrophages were performed with n = 3–4 biological replicates on different days, with n = 4 technical replicates on 24-well tissue-culture treated plates.
Metabolite Quantification
Metabolite concentrations were estimated from LC-MS/MS peak heights using previously generated calibration curves. Metabolites found to have a quantifiable variability (RSD ≥ 50%) in the Quality Control samples or possessing individual components with a RSD ≥ 80% were excluded from analysis. Metabolites in blanks with a concentration greater than 80% of that found in the biological samples were similarly excluded. Missing values were imputed by bootstrapping using the R package Amelia II (v. 1.7.4, 1,000 imputations) (Honaker et al., 2011). Remaining missing values were approximated as ½ the lower limit of quantification for the metabolite normalized to the biomass of the sample. Metabolite concentrations from the peritoneum and lung were scaled 300- and 125-fold, respectively, to account for the dilution effect by a 5 mL lavage based on estimated peritoneal fluid volume of ~16 μL (Hartveit and Thunold, 1966) and estimated lung alveolar fluid volume of ~40 μL (Korfhagen et al., 1996).
Multivariate Analysis
Metabolite concentrations were first pre-processed with the generalized log transformation using the R package LMGene (v. 3.3) (Lu et al., 2008) and then centered to achieve an approximately normal distribution. Hierarchical clustering, principal components analysis and elastic net regularization were performed in MATLAB 2016b (Mathworks; Natick, MA). Partial least squares discriminant analysis was performed using the MATLAB package Classification Toolbox (v. 4.0) (Ballabio and Consonni, 2013).
Metabolite Set Enrichment Analysis
Metabolite Set Enrichment Analysis (MSEA) was performed in Ecocyc (v. 20.1) (Keseler et al., 2017). SmartTables were created comprised of metabolites from each metabolite signature and pathways were identified using the “Enrichment and Depletion” analysis type. The Fisher Exact test was performed for each enrichment analysis with FDR correction by the Benjamini-Hochberg method.
Statistical Analysis
Statistical significance testing was performed in Prism v7.0b (Graphpad; San Diego, CA). One-way ANOVA was performed on data from all experiments. Reported p-values reflect false-discovery correction by the Holm-Šídák multiple comparisons test, with comparisons against either only the relevant control condition, or only against specific other conditions as indicated. Although ANOVA is generally robust against lack of normality in the data, statistical tests were not specifically performed to determine if all of the assumptions of ANOVA had been met.
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| Escherichia coli | ATCC | ATCC 25922 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Ciprofloxacin | Sigma-Aldrich | Cat# 17850-25G-F; CAS: 85721-33-1 |
| Adenosine 5′-monophosphate | Acros Organics (Fisher Scientific) | Cat# AC102790050; CAS: 61-19-8 |
| Critical Commercial Assays | ||
| Seahorse XF Cell Mito Stress Test Kit | Seahorse Bioscience | Cat# 103015-100 |
| Experimental Models: Cell Lines | ||
| Mus musculus (SV129) macrophages | (Lee et al., 2006) | N/A |
| Experimental Models: Organisms/Strains | ||
| Mus musculus (C57BL/6J) | Jackson Labs | 000664 |
| Software and Algorithms | ||
| AB SCIEX MultiQuant v. 3.0 | SCIEX | N/A |
| Amelia II v. 1.7.43 | (Honaker et al., 2011) | N/A |
| LMGene v. 3.3 | (Lu et al., 2008) | N/A |
| MATLAB 2016b | Mathworks | N/A |
| Classification Toolbox v. 4.0 | (Ballabio and Consonni, 2013) | N/A |
| Ecocyc v. 20.1 | (Keseler et al., 2017) | N/A |
| Prism v. 7.0b3 | GraphPad | N/A |
Supplementary Material
Metabolites quantified by the targeted LC-MS/MS protocol used in this study; related to Figure 1.
Metabolic pathways enriched from ABX signature metabolites in the peritoneal samples, identified by Metabolite Set Enrichment Analysis in Ecocyc; related to Figure 1.
Metabolic pathways enriched from ABX signature metabolites in the lung samples, identified by Metabolite Set Enrichment Analysis in Ecocyc; related to Figure S1.
Metabolic pathways enriched from INF signature metabolites in the peritoneal samples, identified by Metabolite Set Enrichment Analysis in Ecocyc; related to Figure 3.
Metabolic pathways enriched from COMB signature metabolites in the peritoneal lavage samples, identified by Metabolite Set Enrichment Analysis in Ecocyc; related to Figure 3.
Metabolic pathways enriched from peritoneal ABX, INF and COMB signature metabolites that decreased antibiotic susceptibility, identified by Metabolite Set Enrichment Analysis in Ecocyc; related to Figures 3 and S3.
HIGHLIGHTS.
Antibiotic treatment depletes central metabolism intermediates in the peritoneum.
Antibiotic treatment elicits microbiome-independent changes in host metabolites.
Metabolites altered by antibiotic treatment during infection inhibit drug efficacy.
Antibiotic treatment impairs phagocytic killing by inhibiting respiratory activity.
Acknowledgments
This work was supported by grant HDTRA1-15-1-0051 from the Defense Threat Reduction Agency, grants K99GM118907 and U01AI124316 from the National Institute of Health, grant NNF16CC0021858 from the Novo Nordisk Foundation, the Paul G. Allen Frontiers Group, and the Wyss Institute for Biologically Inspired Engineering. The authors thank Dr. Lynn Bry and the Gnotobiotic and Microbiology Core Facility of the Massachusetts Host-Microbiome Center at Brigham and Women’s Hospital for their assistance in performing experiments on the germ-free mice. Immortalized SV129 mouse macrophages were generously gifted by Dr. Chih-Hao Lee (Harvard Chan School of Public Health). J.J.C. is scientific co-founder and scientific advisory board chair of Enbiotix, an antibiotics startup company.
Footnotes
AUTHOR CONTRIBUTIONS
Conceptualization, J.H.Y. and P.B.; Methodology, P.B., J.H.Y., and D.M.; Investigation, J.H.Y., P.B., D.M., and N.M.; Formal Analysis, J.H.Y., P.B., and D.M.; Visualization, J.H.Y. and P.B.; Writing, J.H.Y., P.B., D.M., B.O.P. and J.J.C.; Resources, J.J.C., B.O.P. and J.H.Y.; Funding Acquisition, J.J.C., B.O.P. and J.H.Y.; Supervision, J.J.C. and B.O.P.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allison KR, Brynildsen MP, Collins JJ. Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature. 2011;473:216–220. doi: 10.1038/nature10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amato SM, Fazen CH, Henry TC, Mok WW, Orman MA, Sandvik EL, Volzing KG, Brynildsen MP. The role of metabolism in bacterial persistence. Front Microbiol. 2014;5:70. doi: 10.3389/fmicb.2014.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews JM. Determination of minimum inhibitory concentrations. J Antimicrob Chemother. 2001;48(Suppl 1):5–16. doi: 10.1093/jac/48.suppl_1.5. [DOI] [PubMed] [Google Scholar]
- Anuforom O, Wallace GR, Piddock LV. The immune response and antibacterial therapy. Med Microbiol Immunol. 2015;204:151–159. doi: 10.1007/s00430-014-0355-0. [DOI] [PubMed] [Google Scholar]
- Ballabio D, Consonni V. Classification tools in chemistry. Part 1: linear models. PLS-DA. Anal Methods-Uk. 2013;5:3790–3798. [Google Scholar]
- Beisel WR. Metabolic response to infection. Annu Rev Med. 1975;26:9–20. doi: 10.1146/annurev.me.26.020175.000301. [DOI] [PubMed] [Google Scholar]
- Belenky P, Ye JD, Porter CB, Cohen NR, Lobritz MA, Ferrante T, Jain S, Korry BJ, Schwarz EG, Walker GC, et al. Bactericidal Antibiotics Induce Toxic Metabolic Perturbations that Lead to Cellular Damage. Cell Rep. 2015;13:968–980. doi: 10.1016/j.celrep.2015.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black PA, Warren RM, Louw GE, van Helden PD, Victor TC, Kana BD. Energy metabolism and drug efflux in Mycobacterium tuberculosis. Antimicrobial agents and chemotherapy. 2014;58:2491–2503. doi: 10.1128/AAC.02293-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown ED, Wright GD. Antibacterial drug discovery in the resistance era. Nature. 2016;529:336–343. doi: 10.1038/nature17042. [DOI] [PubMed] [Google Scholar]
- Buck MD, Sowell RT, Kaech SM, Pearce EL. Metabolic Instruction of Immunity. Cell. 2017;169:570–586. doi: 10.1016/j.cell.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cekic C, Linden J. Purinergic regulation of the immune system. Nat Rev Immunol. 2016;16:177–192. doi: 10.1038/nri.2016.4. [DOI] [PubMed] [Google Scholar]
- Dong F, Wang B, Zhang L, Tang H, Li J, Wang Y. Metabolic response to Klebsiella pneumoniae infection in an experimental rat model. PLoS One. 2012;7:e51060. doi: 10.1371/journal.pone.0051060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer DJ, Belenky PA, Yang JH, MacDonald IC, Martell JD, Takahashi N, Chan CT, Lobritz MA, Braff D, Schwarz EG, et al. Antibiotics induce redox-related physiological alterations as part of their lethality. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E2100–2109. doi: 10.1073/pnas.1401876111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkness RA, Simmonds RJ, Coade SB. Purine transport and metabolism in man: the effect of exercise on concentrations of purine bases, nucleosides and nucleotides in plasma, urine, leucocytes and erythrocytes. Clin Sci (Lond) 1983;64:333–340. doi: 10.1042/cs0640333. [DOI] [PubMed] [Google Scholar]
- Hartveit F, Thunold S. Peritoneal fluid volume and the oestrus cycle in mice. Nature. 1966;210:1123–1125. doi: 10.1038/2101123a0. [DOI] [PubMed] [Google Scholar]
- Honaker J, King G, Blackwell M. Amelia II: A Program for Missing Data. J Stat Softw. 2011;45:1–47. [Google Scholar]
- Kalghatgi S, Spina CS, Costello JC, Liesa M, Morones-Ramirez JR, Slomovic S, Molina A, Shirihai OS, Collins JJ. Bactericidal antibiotics induce mitochondrial dysfunction and oxidative damage in Mammalian cells. Sci Transl Med. 2013;5:192ra185. doi: 10.1126/scitranslmed.3006055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keseler IM, Mackie A, Santos-Zavaleta A, Billington R, Bonavides-Martinez C, Caspi R, Fulcher C, Gama-Castro S, Kothari A, Krummenacker M, et al. The EcoCyc database: reflecting new knowledge about Escherichia coli K-12. Nucleic Acids Res. 2017;45:D543–D550. doi: 10.1093/nar/gkw1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohanski MA, Dwyer DJ, Collins JJ. How antibiotics kill bacteria: from targets to networks. Nat Rev Microbiol. 2010;8:423–435. doi: 10.1038/nrmicro2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korfhagen TR, Bruno MD, Ross GF, Huelsman KM, Ikegami M, Jobe AH, Wert SE, Stripp BR, Morris RE, Glasser SW, et al. Altered surfactant function and structure in SP-A gene targeted mice. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:9594–9599. doi: 10.1073/pnas.93.18.9594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon DH, Lu CD. Polyamines induce resistance to cationic peptide, aminoglycoside, and quinolone antibiotics in Pseudomonas aeruginosa PAO1. Antimicrobial agents and chemotherapy. 2006;50:1615–1622. doi: 10.1128/AAC.50.5.1615-1622.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CH, Kang K, Mehl IR, Nofsinger R, Alaynick WA, Chong LW, Rosenfeld JM, Evans RM. Peroxisome proliferator-activated receptor delta promotes very low-density lipoprotein-derived fatty acid catabolism in the macrophage. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:2434–2439. doi: 10.1073/pnas.0510815103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HH, Collins JJ. Microbial environments confound antibiotic efficacy. Nat Chem Biol. 2011;8:6–9. doi: 10.1038/nchembio.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobritz MA, Belenky P, Porter CB, Gutierrez A, Yang JH, Schwarz EG, Dwyer DJ, Khalil AS, Collins JJ. Antibiotic efficacy is linked to bacterial cellular respiration. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:8173–8180. doi: 10.1073/pnas.1509743112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Lee GC, Shultz M, Dardick C, Jung K, Phetsom J, Jia Y, Rice RH, Goldberg Z, Schnable PS, et al. Assessing probe-specific dye and slide biases in two-color microarray data. BMC Bioinformatics. 2008;9:314. doi: 10.1186/1471-2105-9-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCloskey D, Gangoiti JA, King ZA, Naviaux RK, Barshop BA, Palsson BO, Feist AM. A model-driven quantitative metabolomics analysis of aerobic and anaerobic metabolism in E. coli K-12 MG1655 that is biochemically and thermodynamically consistent. Biotechnol Bioeng. 2014;111:803–815. doi: 10.1002/bit.25133. [DOI] [PubMed] [Google Scholar]
- McCloskey D, Gangoiti JA, Palsson BO, Feist AM. A pH and solvent optimized reverse-phase ion-paring-LC-MS/MS method that leverages multiple scan-types for targeted absolute quantification of intracellular metabolites. Metabolomics. 2015;11:1338–1350. [Google Scholar]
- Meylan S, Porter CB, Yang JH, Belenky P, Gutierrez A, Lobritz MA, Park J, Kim SH, Moskowitz SM, Collins JJ. Carbon Sources Tune Antibiotic Susceptibility in Pseudomonas aeruginosa via Tricarboxylic Acid Cycle Control. Cell Chem Biol. 2017;24:195–206. doi: 10.1016/j.chembiol.2016.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K, Weaver C. Janeway’s immunobiology. 9. New York, NY: Garland Science/Taylor & Francis Group, LLC; 2016. [Google Scholar]
- O’Neill LA, Hardie DG. Metabolism of inflammation limited by AMPK and pseudo-starvation. Nature. 2013;493:346–355. doi: 10.1038/nature11862. [DOI] [PubMed] [Google Scholar]
- Reijnders D, Goossens GH, Hermes GD, Neis EP, van der Beek CM, Most J, Holst JJ, Lenaerts K, Kootte RS, Nieuwdorp M, et al. Effects of Gut Microbiota Manipulation by Antibiotics on Host Metabolism in Obese Humans: A Randomized Double-Blind Placebo-Controlled Trial. Cell Metab. 2016;24:63–74. doi: 10.1016/j.cmet.2016.06.016. [DOI] [PubMed] [Google Scholar]
- Sarathy JP, Lee E, Dartois V. Polyamines inhibit porin-mediated fluoroquinolone uptake in mycobacteria. PLoS One. 2013;8:e65806. doi: 10.1371/journal.pone.0065806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolovska A, Becker CE, Stuart LM. Measurement of phagocytosis, phagosome acidification, and intracellular killing of Staphylococcus aureus. Curr Protoc Immunol. 2012;Chapter 14(Unit14):30. doi: 10.1002/0471142735.im1430s99. [DOI] [PubMed] [Google Scholar]
- Theriot CM, Koenigsknecht MJ, Carlson PE, Jr, Hatton GE, Nelson AM, Li B, Huffnagle GB, JZL, Young VB. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nature communications. 2014;5:3114. doi: 10.1038/ncomms4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkachenko AG, Akhova AV, Shumkov MS, Nesterova LY. Polyamines reduce oxidative stress in Escherichia coli cells exposed to bactericidal antibiotics. Res Microbiol. 2012;163:83–91. doi: 10.1016/j.resmic.2011.10.009. [DOI] [PubMed] [Google Scholar]
- Tkachenko AG, Pozhidaeva ON, Shumkov MS. Role of polyamines in formation of multiple antibiotic resistance of Escherichia coli under stress conditions. Biochemistry (Mosc) 2006;71:1042–1049. doi: 10.1134/s0006297906090148. [DOI] [PubMed] [Google Scholar]
- Traut TW. Physiological concentrations of purines and pyrimidines. Mol Cell Biochem. 1994;140:1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- Vance-Bryan K, Guay DR, Rotschafer JC. Clinical pharmacokinetics of ciprofloxacin. Clin Pharmacokinet. 1990;19:434–461. doi: 10.2165/00003088-199019060-00003. [DOI] [PubMed] [Google Scholar]
- Willing BP, Russell SL, Finlay BB. Shifting the balance: antibiotic effects on host-microbiota mutualism. Nat Rev Microbiol. 2011;9:233–243. doi: 10.1038/nrmicro2536. [DOI] [PubMed] [Google Scholar]
- Wright GD. Antibiotic Adjuvants: Rescuing Antibiotics from Resistance: (Trends in Microbiology 24, 862–871; October 17, 2016) Trends Microbiol. 2016;24:928. doi: 10.1016/j.tim.2016.06.009. [DOI] [PubMed] [Google Scholar]
- Yang JH, Bening SC, Collins JJ. Antibiotic efficacy - context matters. Curr Opin Microbiol. 2017;39:73–80. doi: 10.1016/j.mib.2017.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Metabolites quantified by the targeted LC-MS/MS protocol used in this study; related to Figure 1.
Metabolic pathways enriched from ABX signature metabolites in the peritoneal samples, identified by Metabolite Set Enrichment Analysis in Ecocyc; related to Figure 1.
Metabolic pathways enriched from ABX signature metabolites in the lung samples, identified by Metabolite Set Enrichment Analysis in Ecocyc; related to Figure S1.
Metabolic pathways enriched from INF signature metabolites in the peritoneal samples, identified by Metabolite Set Enrichment Analysis in Ecocyc; related to Figure 3.
Metabolic pathways enriched from COMB signature metabolites in the peritoneal lavage samples, identified by Metabolite Set Enrichment Analysis in Ecocyc; related to Figure 3.
Metabolic pathways enriched from peritoneal ABX, INF and COMB signature metabolites that decreased antibiotic susceptibility, identified by Metabolite Set Enrichment Analysis in Ecocyc; related to Figures 3 and S3.