Abstract
Microbiome-wide association studies have established that numerous diseases are associated with changes in the microbiota1,2. These studies typically generate a long list of commensals implicated as biomarkers of disease, with no clear relevance to disease pathogenesis1–5. If the field is to move beyond correlations and begin to address causation, an effective system is needed for refining this catalog of differentially abundant microbes and allow for subsequent mechanistic studies1,4. Herein, we demonstrate that triangulation of microbe–phenotype relationships is an effective method for reducing the noise inherent in microbiota studies and enabling identification of causal microbes. We found that gnotobiotic mice harboring different microbial communities exhibited differential survival in a colitis model. Co-housing of these mice generated animals that had hybrid microbiotas and displayed intermediate susceptibility to colitis. Mapping of microbe–phenotype relationships in parental mouse strains and in mice with hybrid microbiotas identified the bacterial family Lachnospiraceae as a correlate for protection from disease. Using directed microbial culture techniques, we discovered Clostridium immunis, a previously unknown bacterial species from this family, that—when administered to colitis-prone mice—protected them against colitis-associated death. To demonstrate the generalizability of our approach, we used it to identify several commensal organisms that induce intestinal expression of an antimicrobial peptide. Thus, we have used microbe–phenotype triangulation to move beyond the standard correlative microbiome study and identify causal microbes for two completely distinct phenotypes. Identification of disease-modulating commensals by microbe–phenotype triangulation may be more broadly applicable to human microbiome studies.
Main Text
The microbiota regulates various facets of host physiology, including immune responses, metabolic functions, and behavior2. Numerous microbiome-wide association studies (MWAS) have linked the microbiome to a panoply of diseases, offering hope that rational microbiome alteration is a feasible treatment modality for many ailments1. However, MWAS have produced long lists of implicated microbes without clearly elucidating their causal role, correlations have not always held up in subsequent studies, and notable differences have been observed between human and animal studies1–3. Although many correlations may simply reflect biomarkers of disease, causal links between the microbiome and disease susceptibility have been sporadically defined in studies that have initially identified immunomodulatory bacteria, functionally categorized the microbiota on the basis of immune recognition, or used complex bioinformatic heuristics to identify disease-modulating bacteria6–11. Ultimately, a generalizable pathway that refines the catalog of differentially abundant microbes identified by MWAS to include only those most likely causally related to the phenotype is lacking.
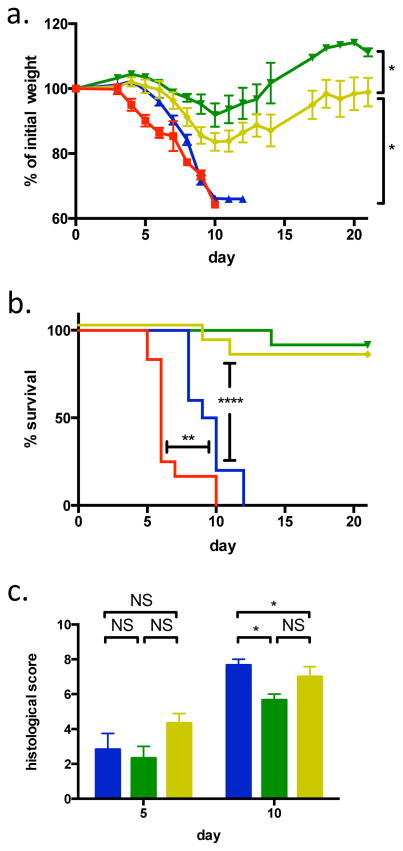
Previously, we generated and characterized gnotobiotic mice colonized with a mouse microbiota (MMb) or a human microbiota (HMb)12. We have now explored how these mice differ in susceptibility to colitis, using a chemically induced model of inflammatory bowel disease (IBD). We found that—similar to germ-free (GF) mice—MMb mice are exquisitely sensitive to dextran sodium sulfate (DSS)–induced colitis, with severe weight loss and 100% mortality (Figs 1A and 1B). In contrast, HMb and specific pathogen–free (SPF) mice lose significantly less weight and rarely die (Figs 1A and 1B). This surprising dichotomy in survival does not reflect lack of colonic inflammation in HMb or SPF mice: the degree of inflammation does not differ at day 5. However, MMb mice have slightly more severe inflammation than either SPF or HMb mice at day 10 (Fig 1C).
Figure 1. MMb mice have more severe colitis than HMb mice.
a and b. Weight change (a) and survival (b) of GF (red; n=12 mice), MMb (blue; n=10 mice), HMb (green; n=12 mice), and SPF (yellow; n=12 mice) mice subjected to DSS-induced colitis. In panel a, mean ± SEM values are depicted. c. Histologic assessment of disease severity in the colon at days 5 (n=6 mice per group) and 10 (n=3 mice per group) after induction of colitis in MMb (blue), HMb (green), and SPF (yellow) mice. Bars depict mean ± sem values. NS, not significant; *, p<0.05; **, p<0.01; ****, p<0.0001 by two-tailed t test (a, c) or log-rank test (b). Panels a and b include data pooled from 3 experiments, and panel c includes data pooled from 2 experiments.
We compared the fecal microbiotas of HMb and MMb mice in an attempt to identify the microbe(s) responsible for modulating disease severity. Given the different underlying microbial sources for these mice, we were not surprised to identify more than 150 bacterial taxa that were differentially abundant between the two groups (Extended Data Fig 1A). Comparison of the fecal microbiotas of MMb mice and SPF mice demonstrated ~100 differentially abundant bacterial taxa (Extended Data Fig 1B). In both cases, the sheer number of implicated microbes makes prioritizing and pursuing potential leads extremely challenging. Focusing on some of the most differentially abundant organisms detected in MWAS has identified some of the few microbes that have been causally related to a phenotype8. Using this approach in HMb and MMb mice, we identified 26 taxa present in one group and absent in the other. We chose four of these organisms, orally administered them to HMb or MMb mice that lacked them, and challenged the mice with DSS. No organism significantly augmented colitis severity (Extended Data Fig 2). This result highlights the challenges of relying solely on organisms that can be dichotomized as present or absent.
The general inability to move beyond correlations and address causation has been the Achilles heel of microbiome research. In considering how to render microbiome analyses more specific, we noted that typical MWAS often compare microbial populations with little similarity to one another1,2—a feature that our study of MMb and HMb mice exemplifies. Comparison of animals with more similar microbiotas should result in a shorter list of phenotype-associated microbial taxa. We reasoned that co-housing of mice with different microbiotas would generate hybrid-microbiota animals microbially related to their parental strains. If the microbial impact on disease were dominant, mapping of microbe–phenotype relationships in parental mouse strains and in hybrid-microbiota mice would enable us to triangulate disease-modulating organisms.
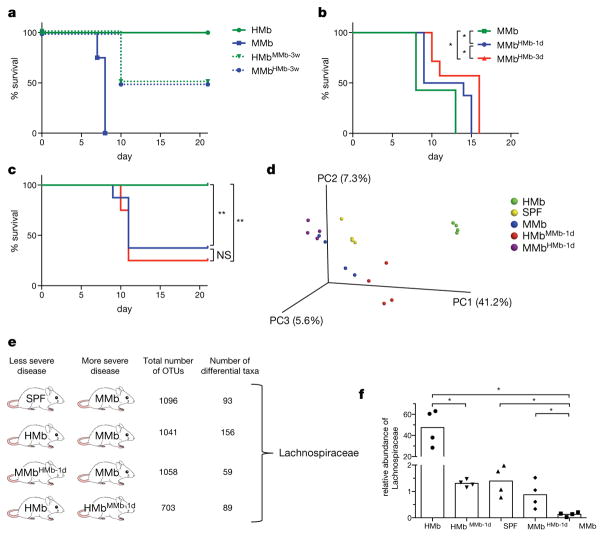
To determine whether the microbiota’s effect is dominant in susceptibility to DSS-induced colitis, we co-housed HMb and MMb mice for 3 weeks. Both HMb mice co-housed with MMb mice (HMbMMb-3w) and MMb mice co-housed with HMb mice (MMbHMb-3w) had intermediate phenotypes (Fig 2A), a result suggesting bi-directional microbe transfer. To limit the degree of microbial change, we defined the shortest period of co-housing producing a phenotypic difference, co-housing HMb and MMb mice for 1 or 3 days and then separating the two groups. We challenged mice with DSS on day 14 after initial co-housing to allow time for physiological changes. Surprisingly, 1 day of co-housing was sufficient to induce significant survival differences, and co-housing duration had a dose-dependent effect (Figs 2B and 2C).
Figure 2. Microbe–phenotype triangulation reveals that the bacterial family Lachnospiraceae is associated with survival from colitis.
a–c. Survival of mice following DSS-induced colitis. a. MMb (blue; n=4 mice) and HMb (green; n=4 mice) mice were co-housed for 3 weeks before induction of colitis. Dashed and solid lines depict mice that were and were not co-housed, respectively. Data are representative of 2 independent experiments. b. MMb mice (n=7 mice) were co-housed with HMb mice for 1 (n=8 mice) or 3 days (n=7 mice). c. HMb mice were co-housed with MMb mice for 1 day (blue) or 3 days (red). Non-co-housed HMb mice are depicted in green (n=8 mice per group). Panels b and c include data pooled from 2 independent experiments. d. A principal-components analysis depicting the fecal microbiota of MMb, SPF, HMb, MMbHMb-1d, and HMbMMb-1d mice. e. The different pairwise comparisons used for triangulation, with 4 mice included per group; the total number of operational taxonomic units (OTUs) in each pair; the number of differentially abundant taxa; and the only taxon identified in all 4 comparisons. f. Relative abundance of Lachnospiraceae in each of the indicated microbiotas (n=4 mice per group); individual (dots) and mean (bars) values are shown. NS, not significant; *, p<0.05; **, p<0.01 by log-rank test (b–c) or two-tailed Mann-Whitney (f).
Along with differences in DSS-induced disease severity, we found distinctive fecal microbiotas in mice co-housed for 1 day (HMbMMb-1d, MMbHMb-1d), parental mouse strains (HMb, MMb), and SPF mice (Fig 2D). We identified differentially abundant bacteria in four pairwise comparisons, including microbially related mouse pairs and pairs in which the two mice varied in disease severity. Each pairwise analysis (representing a MWAS) yielded ~60–160 differentially abundant bacterial taxa (Fig 2E and Extended Data Figs 1A–1D). We reasoned that if any taxa were truly relevant to disease pathogenesis, they would be present in all four comparisons. The only such taxon was Lachnospiraceae. This family of Gram-positive, anaerobic, non-spore-forming bacteria was associated with survival from DSS-induced colitis; its abundance was negligible in MMb mice, intermediate in SPF and co-housed mice, and high in HMb mice (Fig 2F). Interestingly, the Lachnospiraceae have been identified in multiple human studies as inversely correlated with IBD5,13–15, although the significance of this association remains unclear.
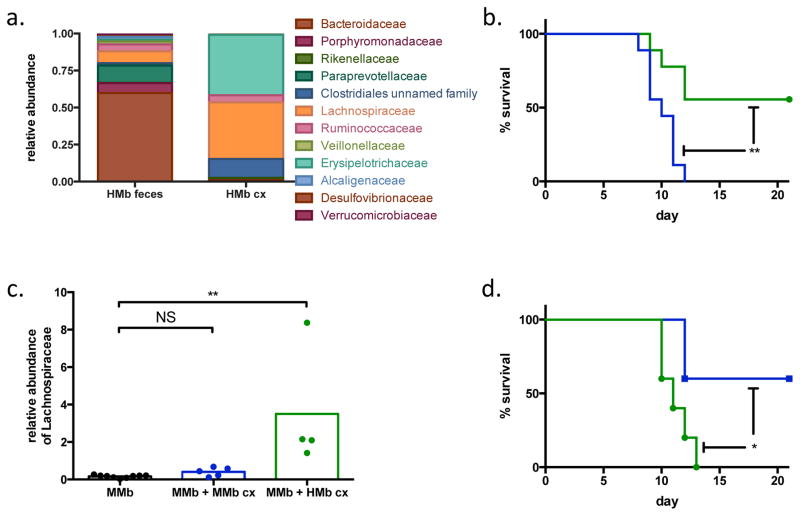
To address whether this taxon is causally linked to improved survival, we cultured feces from HMb mice (HMb cx), obtaining ~5-fold enrichment on semi-selective medium; the cultured bacterial consortium included ~40% Lachnospiraceae (Fig 3A). As a control, we cultured feces from MMb mice (MMb cx) on the same semi-selective medium and found no enrichment of Lachnospiraceae—a finding consistent with the insignificant numbers of this family in MMb mice (Extended Data Fig 3). We orally administered these bacterial cultures to colitis-prone MMb mice to determine whether Lachnospiraceae can protect against colitis-associated death. Although all MMb mice given MMb cx succumbed, MMb mice given HMb cx were remarkably protected (Fig 3B). The microbiotas of these two groups differed (Extended Data Fig 4); MMb mice receiving bacteria from HMb mice had more abundant Lachnospiraceae (Fig 3C). To ensure that increased survival was specifically due to the administered bacteria rather than to an unrelated change in the pre-existing MMb microbiota, we colonized GF mice with HMb cx; they were similarly protected from colitis-associated death (Extended Data Fig 5).
Figure 3. Clostridium immunis protects MMb mice from colitis.
a. Relative abundance of bacterial families present in HMb feces before (left) and after (right) culture. b. Survival of MMb mice orally receiving MMb cx (blue; n=9 mice) or HMb cx (green; n=9 mice) and subjected to DSS-induced colitis. Data are pooled from 2 independent experiments. c. Relative abundance of Lachnospiraceae in a subset of the mice depicted in panel b. The fecal microbiota was assessed before gavage with any material (MMb) or 1 week after gavage with MMb cx or HMb cx (n=9, 5, and 4 mice, respectively). The post-gavage samples represent day 0 of the DSS colitis experiment. Individual (dots) and mean (bars) values are shown. d. Survival of MMb mice orally receiving Clostridium innocuum (green; n=5 mice) or C. immunis (blue; n=5 mice) and subjected to DSS-induced colitis. Data are representative of 2 independent experiments. NS, not significant; *, p<0.05; **, p<0.01 by log-rank (b, d) or a two-tailed Mann-Whitney test (c).
To directly test whether Lachnospiraceae protected mice against colitis, we recovered a single Lachnospiraceae isolate from HMb feces that accounted for ~24% of bacterial abundance in the HMb cx bacterial consortium. The 16S rDNA gene sequence of this isolate shared 98% identity with Clostridium symbiosum; MALDI-TOF analysis identified it as Clostridium clostridioforme. However, its biochemical properties did not fit either species (see Methods). Indeed, this isolate represents a novel bacterial species that we are tentatively designating Clostridium immunis. We orally administered this isolate to MMb mice to assess protection against colitis; as a control, we used a Clostridium innocuum isolate (family Erysipelotrichaceae) that accounts for ~9% of bacterial abundance in HMb cx and is also absent in MMb mice. Remarkably, only MMb mice treated with C. immunis were protected from colitis-associated death (Fig 3D). Clearly, this isolate is causally related to protection from colitis. This outcome provides insight into the clinical observations that the abundance of Lachnospiraceae is inversely correlated to risk of IBD and suggests a causal relationship. Moreover, our findings offer critical preclinical support for use of C. immunis as a probiotic in patients with IBD and lower abundances of Lachnospiraceae5,13–15.
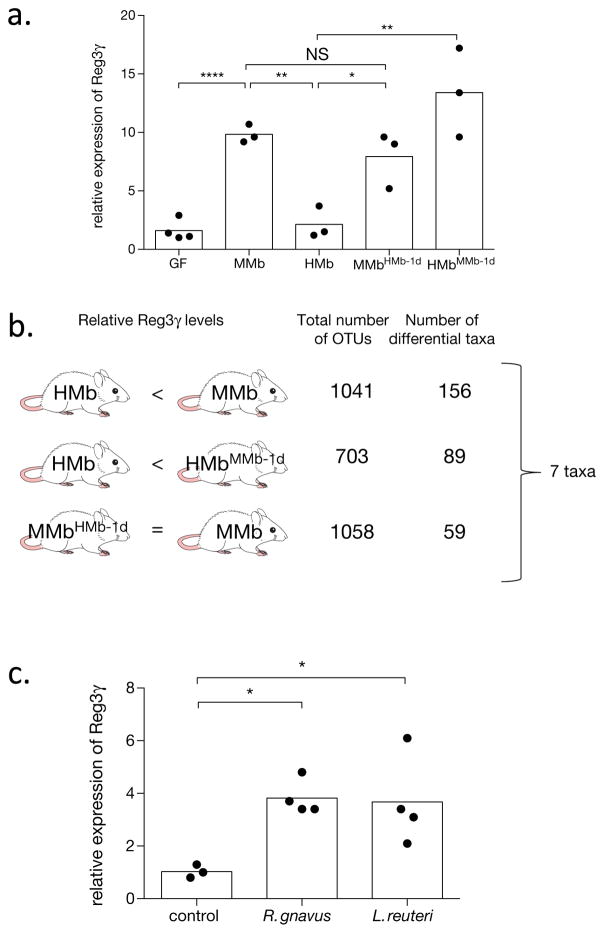
To demonstrate its generalizability, we used microbe–phenotype triangulation to identify commensal bacteria that induce intestinal expression of Reg3γ, a critical antimicrobial peptide that maintains spatial segregation between the epithelial layer and the microbiota16. Although commensal microbes are known to be required for induction of Reg3γ expression17, few specific organisms have been defined. In our recently published comprehensive analysis of the immunomodulatory capacity of taxonomically diverse commensal microbes, none of 28 bacteria assessed for small-intestinal expression of Reg3γ caused significant induction18. We previously showed that HMb mice have ileal levels of Reg3γ expression similar to those in GF mice and lower than those in MMb mice12. We now found that Reg3γ expression in HMbMMb-1d mice is restored to levels comparable to those in MMb mice, with no change observed in MMbHMb-1d mice (Fig 4A). Using microbe–phenotype triangulation, we identified taxa that were differentially abundant in two pairwise comparisons but were unchanged between MMb and MMbHMb-1d mice (Fig 4B). Thus we refined the list of associated microbes to seven taxa, three of which [Ruminococcus gnavus, Lactobacillus reuteri, and segmented filamentous bacteria (SFB)] exhibit bioinformatic resolution to the species level (Fig 4B and Extended Data Table 1).
Figure 4. Ruminococcus gnavus and Lactobacillus reuteri induce intestinal expression of Reg3γ.
a. qPCR analysis of ileal Reg3γ expression in mice with varying microbiotas (n=4 mice for GF, n=3 mice for all other groups). Reg3γ expression is normalized to GF mice. b. The different pairwise comparisons used for triangulation, with 4 mice included per group; the total number of operational taxonomic units (OTUs) in each pair; and the number of differentially abundant taxa. The 7 taxa identified in this analysis are detailed in Extended Data Table S1. c. qPCR analysis of ileal Reg3γ expression in HMb mice after oral administration of control bacteria (n=3 mice), R. gnavus (n=4 mice), or L. reuteri (n=4 mice). Reg3γ expression is normalized to HMb mice + control. Individual (dots) and mean (bars) values are shown in panels a and c. NS, not significant; *, p<0.05; **, p<0.01; ****, p<0.0001 by two-tailed t test (a, c).
HMb mice treated with R. gnavus or L. reuteri had greater ileal Reg3γ expression than a control (Fig 4C). The previous demonstration that SFB induces Reg3γ expression in GF and SPF mice confirms the more general applicability of results obtained using our specific gnotobiotic mice8. Our requirement that organisms be unchanged between MMb and MMbHMb-1d mice correctly excluded a relation between Reg3γ expression and Lactobacillus vaginalis, a close relative of L. reuteri that is also absent in HMb mice (Extended Data Fig 6). Although focusing on taxa that are highly divergent between groups may have suggested that L. reuteri and SFB (present in MMb mice at only 0.2% and 0.3% abundance, respectively, and absent in HMb mice) are relevant to Reg3γ induction, two other taxa present only in MMb mice did not affect Reg3γ expression (Extended Data Fig 6). Additionally, our approach provides phenotype-directed results. Although we used the same datasets for both phenotypes, the taxa identified for Reg3γ induction had no effect in the colitis model (Extended Data Fig 2). Taken together, microbe–phenotype triangulation facilitated identification of taxonomically diverse microbes causally related to two disparate outcomes.
Our results delineate a bioinformatically straightforward approach that can be used to triangulate specific microbiota members that are likely to influence disease pathogenesis. Combination of microbiota analyses from multiple sets of microbially related mice allowed us to trade sensitivity for increased specificity. In both the colitis and Reg3γ examples, we bioinformatically pinpointed a limited number of taxa associated with our phenotype and substantiated these correlations by add-back experiments to fulfill Koch’s postulates. Although Koch’s postulates and their modern revisions traditionally apply only to pathogen identification19–21, our findings with a disease-protective commensal conform to all their major tenets. As microbiome research matures, increasing numbers of microbes will probably be causally linked to protection from disease. We propose that Koch’s postulates be expanded to apply to identification of beneficial organisms, thus ensuring that burgeoning research on probiotics is subject to the same degree of scientific rigor required for research on microbial pathogenesis.
Our microbe–phenotype triangulation approach may be more generally applicable to human microbiome studies. Thoughtful selection of participants and controls may obviate the trend toward increasingly large-scale, complicated study designs while allowing meaningful comparative analyses. Furthermore, the results from other “-omics” datasets (e.g., transcriptomics, metagenomics, metabolomics) can be analyzed by this approach to refine result lists and subsequently determine which are causally related to the phenotype. Microbe–phenotype triangulation may not identify all disease-related microbes, particularly those with small effects, but we envision it as a useful tool facilitating the discovery of key disease-modulating components of the microbiota.
Materials and Methods
Mice
GF Swiss-Webster (SW) mice were bred and maintained in vinyl isolators in the animal facility at Harvard University. MMb and HMb mice have been bred and maintained at this facility in separate vinyl isolators since their initial characterization12. Experimental manipulation of gnotobiotic mice was performed in sterile cages (Innovive; San Diego, CA) in which animals received autoclaved food and water. SPF SW mice obtained from Taconic Biosciences were fed an autoclaved diet similar to that given to gnotobiotic mice for 1 week before the start of the study and for the duration of all experiments. Mice used in experiments were sex- and age-matched (typically 5–10 weeks old) and drawn randomly from the same litter, when feasible. All procedures were approved by the Harvard Medical Area Standing Committee on Animals and were conducted in accordance with NIH guidelines.
DSS colitis
DSS experiments were performed as previously described22. In brief, mice were given 4% DSS (molecular weight, 36,000–50,000 Da; MP Biomedicals) ad libitum in their drinking water for 7 days, with the DSS solution changed every 2–3 days. From day 7 until the end of the experiment, the mice were given autoclaved water with no DSS. Animals were weighed every 1–2 days, and any mouse that appeared moribund was sacrificed. A pathologist blinded to treatment groups conducted a histologic assessment of colons. The histologic score represented the combined scores for inflammation and ulceration; both elements were scored 0–4, with 0 being normal.
Co-housing experiments
For co-housing of MMb and HMb mice prior to DSS experiments, we placed two mice per group together in a cage (total, 4 mice) for the indicated period. For a co-housing period of 1 day or 3 days, the MMb and HMb mice were separated at the relevant time point, placed into a new sterile cage until day 14 (to allow physiological changes to occur), and then challenged with DSS. For co-housing of MMb and HMb mice for the purpose of fecal microbiota analysis, we placed one mouse per group in a cage (total, 2 mice) for 1 day, after which the mice were separated and individually housed until day 14. Fecal pellets were collected on days 0 and 14 and were frozen at −80°C until further processed.
16S rDNA sequencing and analysis
Fecal samples were added to a tube containing 400 μl of zirconia/silica beads (0.1 mm in diameter; Biospec), 250 μl of 20% sodium dodecyl sulfate, 500 μl of buffer PBI (Qiagen), and 550 μl of phenol–chloroform–isoamyl alcohol (25:24:1; pH 7.9; Ambion) and then homogenized by bead beating for 2.5 min. After centrifugation (10,000 × g for 5 min), the DNA in the aqueous phase was purified with a QIAquick PCR purification kit (Qiagen) according to the manufacturer’s instructions except that the PE wash step was performed twice. DNA was eluted with 50 μl of EB buffer (Qiagen).
Detailed protocols used for 16S rDNA amplification and sequencing have been described23. In brief, the V4 region of the 16S rDNA gene was PCR-amplified (35 cycles, primers 515F and 806R) in triplicate; 1 μl of purified DNA and 5′ Hot Master Mix (Five Prime) were used. Amplicons were quantified by Quant-It (Invitrogen), pooled in equimolar concentration, and size-selected (375–425 bp) on the Pippen Prep (Sage Sciences; Beverly, MA) to reduce nonspecific amplification products. Sequencing was performed on a MiSeq sequencer (Illumina; 2×250 bp paired-end reads with V2 chemistry).
Microbial diversity was analyzed with Quantitative Insights into Microbial Ecology (QIIME versions 1.8 and 1.9)24. The closed-reference operational taxonomic unit (OTU) workflow in QIIME and the Greengenes reference database (May 2013) were used to cluster reads into OTUs with 97% identity and to assign taxonomy to representative OTUs24,25. OTU tables were rarified to a depth of 40,000 (Figures 2 and 4) and 13,000 sequences per sample (Figure 3). Principal-coordinates beta-diversity visualizations were created with Emperor as packaged in QIIME26. The Linear Discriminant Analysis Effect Size (LEfSe) Galaxy module (http://huttenhower.sph.harvard.edu/galaxy/) was used for additional statistical analyses27.
Bacterial culture
Serial dilutions of feces from MMb and HMb mice were spread on brain–heart infusion (BHI) agar supplemented with colistin (10 μg/ml), gentamicin (6 μg/ml), and aztreonam (5 μg/ml), and cultures were incubated for 7 days in an anaerobic chamber (Coy Industries; Grass Lake, MI). MMb cx and HMb cx samples were harvested from plates that contained ~500–1000 colonies, resuspended in pre-reduced BHI agar, and frozen at −80°C until needed. To recover specific isolates, we cultured HMb feces on the medium described above for 5–14 days and picked ~60 individual colonies. These colonies were inoculated into pre-reduced chopped meat medium with glucose (Anaerobe Systems; Morgan Hill, CA), and genomic DNA was isolated with the DNeasy Blood and Tissue Kit (Qiagen). We compared the 16S rDNA gene sequence from each of these isolates to Genbank and the Greengenes reference database to identify bacterial taxonomy.
Probiotic administration to mice
MMb, HMb, and GF mice received orally administered Paraprevotella clara, Bacteroides uniformis, SFB, Lactobacillus reuteri (BEI HM-102), Ruminococcus gnavus (ATCC 29149), MMb cx, HMb cx, Clostridium innocuum, or Clostridium immunis (100–150 μl; ~108–109 colony-forming units). P. clara, B. uniformis, C. innocuum, and C. immunis were isolated from the feces of HMb mice; SFB was previously obtained from Y. Umesaki (Yakult; Tokyo, Japan) and propagated in SFB-monocolonized mice at Harvard Medical School. Seven days later, the mice were challenged with DSS. In some experiments, fecal samples were collected before and 7 days after probiotic administration for microbiota analysis. For Reg3γ experiments, HMb mice received orally administered (150–200 μl; ~108–109 colony-forming units) Parabacteroides distasonis (ATCC 8503; control bacteria), R. gnavus (ATCC 29149), L. reuteri (BEI HM-102), Allobaculum stercoricanis (DSM 13633), Muribaculum intestinale (DSM 28989), or Lactobacillus vaginalis (DSM 5837). Mice were sacrificed 7 days later.
Characterization of C. immunis
Our Lachnospiraceae isolate stains gram-negative, is resistant to colistin differential disks (Anaerobe Systems), and is sensitive to vancomycin and kanamycin differential disks (Anaerobe Systems). Taken together, these findings are consistent with the isolate’s being a Gram-positive organism. The closest match for the 16S rDNA gene sequence of this isolate is Clostridium symbiosum, with which it shares 98% identity. MALDI-TOF analysis by VITEK MS (BioMerieux; Durham, NC) revealed that the closest match for this isolate is Clostridium clostridioforme, and MALDI-TOF analysis with the MALDI Biotyper (Bruker; Billerica, MA) was unable to provide a species- or genus-level identification. Biochemical testing demonstrated that our isolate—unlike C. symbiosum28—is resistant to 20% bile and does not produce acid from mannose. Moreover, our isolate—unlike C. clostridioforme28—produces abundant amounts of butyrate from peptone–yeast extract–Fildes solution–glucose broth. Given these genetic, proteomic, and biochemical differences from its closest relatives, we propose that this isolate represents the type strain of Clostridium immunis, a novel species within the family Lachnospiraceae.
We purified genomic DNA, using a Genomic Tip G/100 (Qiagen), and sequenced it on a PacBio RS II at the Yale Center for Genome Analysis (New Haven, CT), following the manufacturer’s instructions for library preparation. With use of the sequence data from 1 SMRT cell, the Hierarchical Genome Assembly Process (HGAP) was able to assemble the genome into 7 contigs, with a N50 contig length of 3.4 Mbp and a total genome size of 5.4 Mbp. We annotated the genome with the RAST server (http://rast.nmpdr.org), which indicated that there are 5905 coding sequences and 75 RNAs. Tetracycline resistance genes were the only antibiotic resistance genes identified by ResFinder29. No virulence genes were identified by VirulenceFinder30, and PathogenFinder predicted that C. immunis is not a human pathogen (probability of being a human pathogen = 0.25)31.
Reg3γ expression analysis
qPCR for Reg3γ was performed as previously described12. The distal 1.5 cm of small intestine was harvested from the indicated mice, frozen immediately in liquid nitrogen, and stored at −80°C until needed. Tissues were homogenized in Trizol (Invitrogen), and RNA was purified according to the manufacturer’s instructions, with a subsequent additional cleaning step (RNeasy Mini Kit; Qiagen). cDNA was prepared with the SuperScript III First-Strand Synthesis System (ThermoFisher Scientific), and qPCR was performed on a LightCycler (Roche) with iQ SYBR Green Supermix (Biorad).
Statistics
Sample-size estimates for each experiment were based on prior lab experience. Prism 6 (GraphPad Software; La Jolla, CA) was used for all statistical analyses.
Data availability
The 16S rDNA sequences from the microbiota analyses have been deposited at ENA under study accession number PRJEB23029. The genome sequence for C. immunis has been deposited at DDBJ/ENA/GenBank under the accession PDDG00000000; the version described in this paper is PDDG01000000. Source data for all figures have been provided with the paper. Any other data that support the findings of this study are available from the corresponding authors upon reasonable request.
Extended Data
Extended Data Figure S1. Individual microbiome-wide association studies reveal a large number of differentially abundant taxa.
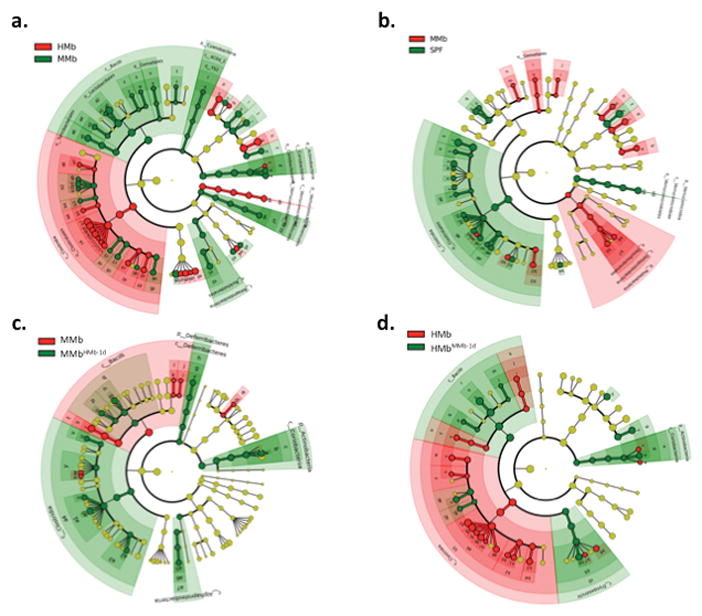
LEfSe was used to identify differentially abundant taxa in the fecal microbiota of various mice. Taxa colored red and green were more abundant in that particular group of mice. Taxa colored yellow did not statistically differ in abundance between groups. Each ring of the cladogram represents a different taxonomic level, starting with kingdom in the center and ending with genus in the outer ring. a. Comparison of HMb and MMb. b. Comparison of MMb and SPF. c. Comparison of MMb and MMbHMb-1d. d. Comparison of HMb and HMbMMb-1d. The family Lachnospiraceae is indicated by the symbols c4 (a), a6 (b), a1 (c), and a9 (d).
Extended Data Figure S2. Several taxa that are differentially present in HMb and MMb mice do not augment colitis severity.
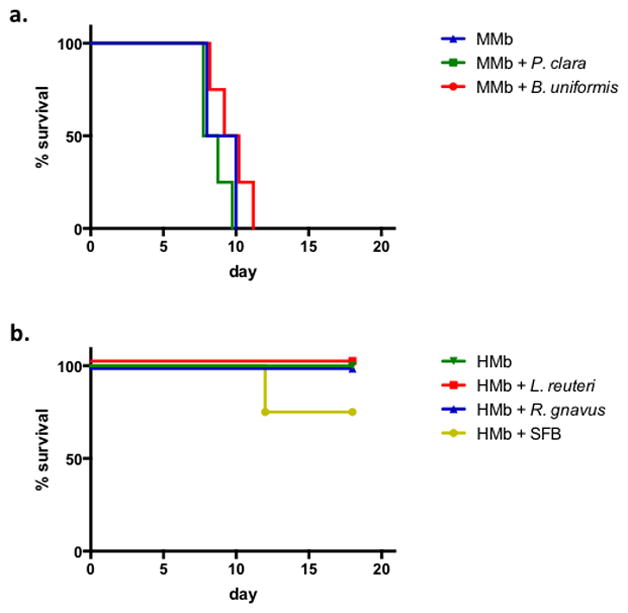
a. Survival of MMb mice (n=2 mice) and MMb mice orally receiving Paraprevotella clara (n=4 mice) or Bacteroides uniformis (n=4 mice) and subjected to DSS-induced colitis. b. Survival of HMb mice (n=2 mice) and HMb mice orally receiving Lactobacillus reuteri (n=4), Ruminococcus gnavus (n=4 mice), or SFB (n=4 mice) and subjected to DSS-induced colitis.
Extended Data Figure S3. Culture of MMb feces on semi-selective medium does not enrich for Lachnospiraceae.
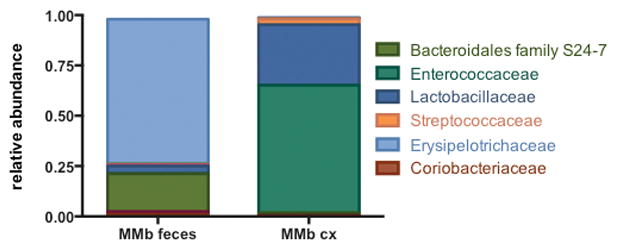
The relative abundance of bacterial families present in MMb feces before and after culture is shown.
Extended Data Figure S4. MMb mice given MMb cx and MMb mice given HMb cx have distinct microbiotas.
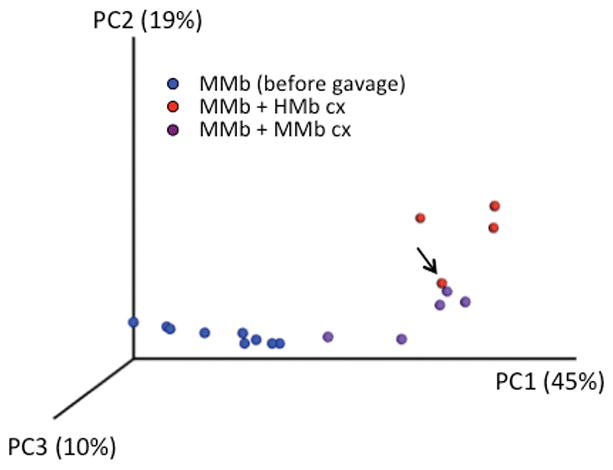
Weighted principal-components analysis of the fecal microbiota of MMb mice before and after gavage with MMb cx or HMb cx is shown. The arrow indicates an MMb mouse that received HMb cx but died after being challenged with DSS.
Extended Data Figure S5. The HMb cx bacterial consortium is sufficient to protect mice from colitis-associated death.
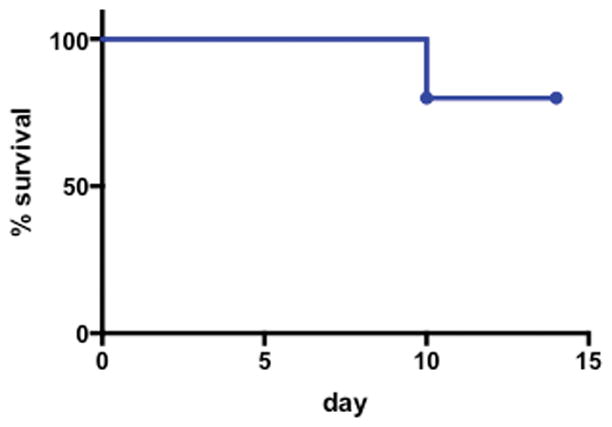
The survival of GF mice orally receiving HMb cx (n=10 mice) and subjected to DSS-induced colitis is shown.
Extended Data Figure S6. Several taxa that are present in MMb mice and absent in HMb mice do not induce Reg3γ expression.
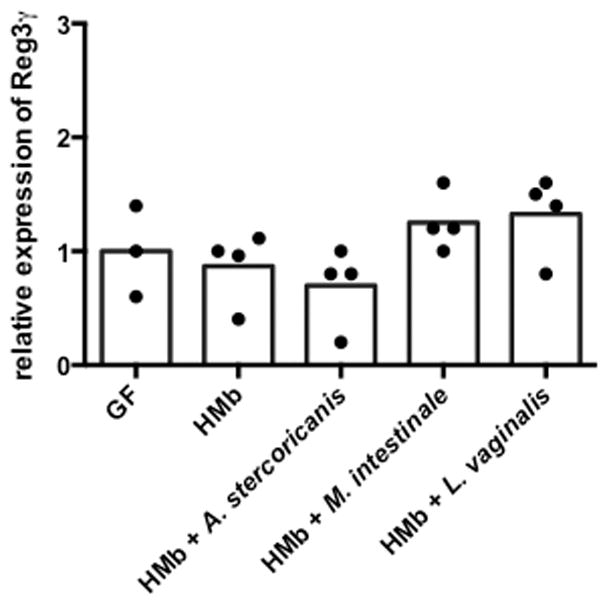
qPCR analysis of ileal Reg3γ expression in HMb mice receiving no organisms (n=4 mice) and in HMb mice receiving orally administered Allobaculum stercoricanis (n=4 mice), Muribaculum intestinale (n=4 mice), or Lactobacillus vaginalis (n=4 mice). Reg3γ expression was normalized to GF mice (n=3 mice). Individual (dots) and mean (bars) values are shown.
Extended Data Table 1.
List of bacterial taxa associated with Reg3γ induction.
| p__Bacteroidetes.c__Bacteroidia.o__Bacteroidales.f__.g__.s |
| p__Bacteroidetes.c__Bacteroidia.o__Bacteroidales.f__.Rikenellaceae.g__.s__ |
| p__Firmicutes.c__Bacilli.o__Lactobacillales.f__.g__.s__ |
| p__Firmicutes.c__Bacilli.o__Lactobacillales.f__Lactobacillaceae.g__Lactobacillus.s__reuteri |
| p__Firmicutes.c__Bacilli.o__Lactobacillales.f__Streptococcaceae.g__AStreptococcus.s__ |
| p__Firmicutes.c__Clostridia.o__Clostridiales.f__Clostridiaceae.g__CandidatusArthromitus.s__ |
| p__Firmicutes.c__Clostridia.o__Clostridiales.f__Lachnospiraceae.g__Ruminococcus.s__gnavus |
p, phylum; c, class; o, order; f, family; g, genus; s, species. Taxonomic levels that lack information (e.g., f__.g__.s__) did not match named taxa present in the Greengenes database.
Acknowledgments
We thank Cheryn Couter for technical assistance; Shakir Edwards, Jaime Ramos, and Tsering Sherpa for assistance with gnotobiotic mice; Roderick Bronson for review of histology; Julie McCoy for editorial assistance; and members of the Kasper lab for helpful discussions. Support for this work was provided by a Career Development Award from Boston Children’s Hospital (N.K.S.) and National Institutes of Health grants K08 AI108690 (N.K.S.) and U19 AI109764 (N.K.S. and D.L.K.).
Footnotes
Author contributions:
N.K.S. conceived the study, designed and performed experiments, and analyzed all data. D.L.K. supervised all aspects of the project. N.K.S. and D.L.K. wrote the paper.
Competing financial interests:
N.K.S. and D.L.K. are inventors on patent application numbers 17/38680 and 62/523330 submitted by Harvard University that cover the therapeutic use of C. immunis.
References
- 1.Lynch SV, Pedersen O. The Human Intestinal Microbiome in Health and Disease. N Engl J Med. 2016;375:2369–2379. doi: 10.1056/NEJMra1600266. [DOI] [PubMed] [Google Scholar]
- 2.Surana NK, Kasper DL. Deciphering the tete-a-tete between the microbiota and the immune system. J Clin Invest. 2014;124:4197–4203. doi: 10.1172/JCI72332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sze MA, Schloss PD. Looking for a Signal in the Noise: Revisiting Obesity and the Microbiome. mBio. 2016;7 doi: 10.1128/mBio.01018-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gilbert JA, et al. Microbiome-wide association studies link dynamic microbial consortia to disease. Nature. 2016;535:94–103. doi: 10.1038/nature18850. [DOI] [PubMed] [Google Scholar]
- 5.Gevers D, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Atarashi K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 7.Blanton LV, et al. Gut bacteria that prevent growth impairments transmitted by microbiota from malnourished children. Science. 2016:351. doi: 10.1126/science.aad3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ivanov, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kau AL, et al. Functional characterization of IgA-targeted bacterial taxa from undernourished Malawian children that produce diet-dependent enteropathy. Sci Transl Med. 2015;7:276ra224. doi: 10.1126/scitranslmed.aaa4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 11.Palm NW, et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell. 2014;158:1000–1010. doi: 10.1016/j.cell.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chung H, et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149:1578–1593. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baumgart M, et al. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn’s disease involving the ileum. The ISME journal. 2007;1:403–418. doi: 10.1038/ismej.2007.52. [DOI] [PubMed] [Google Scholar]
- 14.Frank DN, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kang S, et al. Dysbiosis of fecal microbiota in Crohn’s disease patients as revealed by a custom phylogenetic microarray. Inflamm Bowel Dis. 2010;16:2034–2042. doi: 10.1002/ibd.21319. [DOI] [PubMed] [Google Scholar]
- 16.Vaishnava S, et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geva-Zatorsky N, et al. Mining the Human Gut Microbiota for Immunomodulatory Organisms. Cell. 2017;168:928–943 e911. doi: 10.1016/j.cell.2017.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Falkow S. Molecular Koch’s postulates applied to microbial pathogenicity. Rev Infect Dis. 1988;10(Suppl 2):S274–276. doi: 10.1093/cid/10.supplement_2.s274. [DOI] [PubMed] [Google Scholar]
- 20.Fredricks DN, Relman DA. Sequence-based identification of microbial pathogens: a reconsideration of Koch’s postulates. Clin Microbiol Rev. 1996;9:18–33. doi: 10.1128/cmr.9.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koch R. Untersuchungen über Bakterien: V. Die Ätiologie der Milzbrand-Krankheit, begründet auf die Entwicklungsgeschicte des Bacillus anthracis. Cohns Beitrage zur Biologie der Pflanzen. 1876;2:277–310. [Google Scholar]
- 22.Wirtz S, Neufert C, Weigmann B, Neurath MF. Chemically induced mouse models of intestinal inflammation. Nature protocols. 2007;2:541–546. doi: 10.1038/nprot.2007.41. [DOI] [PubMed] [Google Scholar]
- 23.Caporaso JG, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. The ISME journal. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nature methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McDonald D, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. The ISME journal. 2012;6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vazquez-Baeza Y, Pirrung M, Gonzalez A, Knight R. EMPeror: a tool for visualizing high-throughput microbial community data. GigaScience. 2013;2:16. doi: 10.1186/2047-217X-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Segata N, et al. Metagenomic biomarker discovery and explanation. Genome biology. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaneuchi C, Watanabe K, Terada A, Benno Y, Mitsuoka T. Taxonomic Study of Bacteroides clostridiiformis subsp. clostridiiformis (Burri and Ankersmit) Holdeman and Moore and of Related Organisms: Proposal of Clostridium clostridiiformis (Burri and Ankersmit) comb. nov. and Clostridium symbiosum (Stevens) comb. nov. International Journal of Systematic and Evolutionary Microbiology. 1976;26:195–204. doi: 10.1099/00207713-26-2-195. [DOI] [Google Scholar]
- 29.Zankari E, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joensen KG, et al. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J Clin Microbiol. 2014;52:1501–1510. doi: 10.1128/JCM.03617-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cosentino S, Voldby Larsen M, Moller Aarestrup F, Lund O. PathogenFinder--distinguishing friend from foe using bacterial whole genome sequence data. PLoS One. 2013;8:e77302. doi: 10.1371/journal.pone.0077302. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The 16S rDNA sequences from the microbiota analyses have been deposited at ENA under study accession number PRJEB23029. The genome sequence for C. immunis has been deposited at DDBJ/ENA/GenBank under the accession PDDG00000000; the version described in this paper is PDDG01000000. Source data for all figures have been provided with the paper. Any other data that support the findings of this study are available from the corresponding authors upon reasonable request.