Abstract
Synergistic advances in fluorescent protein (FP) engineering and live-cell imaging techniques in recent years have fueled the concurrent development and application of genetically encoded fluorescent reporters that are tailored for tracking signaling dynamics in living systems over multiple length and time scales. These biosensors are uniquely suited for this challenging task, owing to their specificity, sensitivity, and versatility, as well as to the non-invasive and non-destructive nature of fluorescence and the power of genetic encoding. Over the past 10 years, a growing number of fluorescent reporters have been developed for tracking a wide range of biological signals in living cells and animals, including second messenger and metabolite dynamics, enzyme activation and activity, and cell cycle progression and neuronal activity. Many of these biosensors are gaining wide use and are proving to be indispensable for unraveling the complex biological functions of individual signaling molecules in their native environment, the living cell, shedding new light on the structural and molecular underpinnings of cell signaling. In this review, we highlight recent advances in protein engineering that are likely to help expand and improve the design and application of these valuable tools. We then turn our focus to specific examples of live-cell imaging using genetically encoded fluorescent reporters as an important platform for advancing our understanding of G protein-coupled receptor (GPCR) signaling and neuronal activity.
Keywords: biosensor, fluorescent protein, FRET, GPCR signaling, neuronal activity
Graphical abstract
Genetically encoded fluorescent reporters comprise an ever-expanding array of molecular tools that allow researchers to directly visualize different aspects of intracellular signaling within living cells. This review explores several recent innovations and advances in the design and application of genetically encoded fluorescent reporters to illuminate the mechanics of intracellular signaling pathways with increasing clarity.
Introduction
Cell signaling is the communications network that determines the responses of a cell at multiple levels, from molecular and cellular, to tissue and whole organism. At the molecular level, this network consists of signaling molecules that dictate information flow within the complex cellular environment through often transient changes and interactions. The choreographed interplay of signaling molecules in cellular space and time results in exquisite spatial organization and temporal regulation [1]. As such, it is imperative to study these signaling molecules and activities in their native context, the living cell, in order to elucidate the general working principles and underlying molecular mechanisms of cell signaling [2].
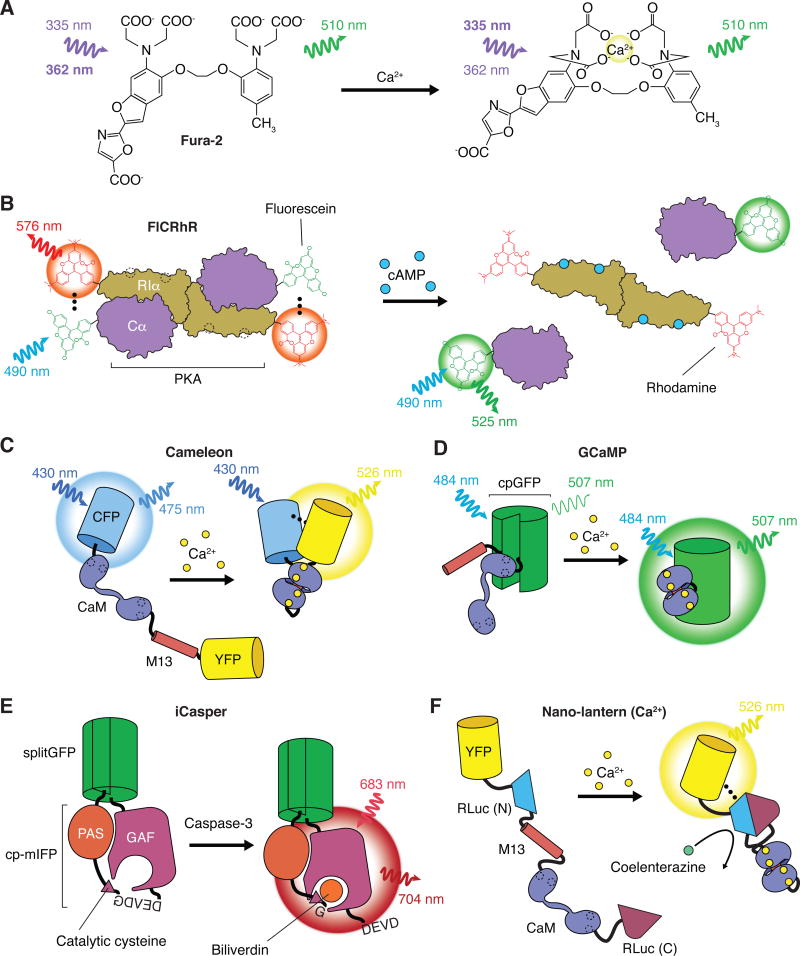
With a typical lifetime of nanoseconds, fluorescence is fast and capable of achieving single-molecule sensitivity and nanometer spatial resolution in living cells when combined with suitable imaging modalities [3]. Together with its desirable non-invasive and non-destructive nature, fluorescence-based reporters, such as synthetic fluorescent indicators [4], have long been recognized as valuable tools for visualizing signaling dynamics with high spatial and temporal resolution in living systems. Conceptually, a fluorescent reporter comprises a fluorescent reporting element that is coupled to a specific signal-sensing element such that the fluorescence readout of the reporting element is modulated by the distinct molecular states of the sensing element, which correspond to the ON and OFF states of the target signal [5]. For example, Fura-2 [4], a widely-used fluorescent indicator for measuring Ca2+ dynamics in living cells, incorporates a fluorescent moiety, the reporting element, into a molecular scaffold derived from a Ca2+ chelator, EGTA, the sensing element. In this case, Ca2+ binding shifts the excitation peak of Fura-2 from 362 nm to 335 nm, thereby providing a dual-excitation ratiometric readout suitable for quantitative measurement of intracellular Ca2+ concentrations (Fig. 1A). The general success of synthetic fluorescent indicators, however, has been limited by the lack of suitable synthetic sensing elements for many of the known signaling molecules [6]. On the other hand, proteins are the workhorses of cellular signal processing in that nearly all signaling molecules and activities are mediated through protein molecules, be it ligand binding, chemical modification, or non-covalent interaction. Thus, in principle, a protein-based sensing element with suitable sensitivity and specificity for a given cellular signal of interest can often be derived from or engineered based on naturally occurring proteins. As an early example, FlCRhR [7], a fluorescent cyclic AMP (cAMP) reporter, was developed utilizing the cAMP-dependent protein kinase (PKA) holoenzyme as the sensing element. FlCRhR was constructed by labeling the purified catalytic and regulatory subunits of PKA holoenzyme with two small molecule fluorophores, fluorescein and rhodamine, respectively. FlCRhR works by a mechanism known as fluorescence resonance energy transfer (FRET), as binding of cAMP to the regulatory subunits leads to the dissociation of the catalytic subunits from the regulatory subunits and therefore the loss of FRET (Fig. 1B), which is only effective when the two fluorophores are in close proximity [8]. The development of FlCRhR not only demonstrated the usefulness of FRET as a general working mechanism for reporter design, but its modular design employing two functionally distinct physical elements also suggests that a fully genetically encoded fluorescent reporter could be made possible with the availability of a genetically encodable fluorophore [9]. To this end, the discovery that the green fluorescent protein (GFP) from the jellyfish Aequorea victoria [10] is an intrinsically fluorescent protein truly revolutionized the field of live-cell imaging. FP-based genetically encoded fluorescent reporters have since been developed for dynamic visualization of a wide range of signaling molecules and activities in their native cellular environment and are still growing at a rapid pace [11–14].
Fig. 1.
An evolving toolkit for live-cell imaging of signaling events. (A) Synthetic fluorescent dyes have long enabled researchers to track the dynamics of various ions in living cells. For instance, Fura-2, which is related to the Ca2+ chelators EGTA and BAPTA, emits light at ~510 nm but undergoes a shift in its preferred excitation wavelength from 362 nm in the Ca2+-free state (left) to 335 nm in the Ca2+-bound state (right). The fluorescence intensity at each excitation wavelength is directly proportional to the concentration of Ca2+, and calculating the emission intensity at each excitation wavelength (i.e., the excitation ratio) offers a quantitative readout of live-cell Ca2+ dynamics. Such dyes can be readily introduced into cells through the use of acetoxymethyl ester derivatives. (B) In the absence of chelators for larger cellular analytes, researcher subsequently turned to chemical labeling to generate fluorescent biosensor from purified proteins. As shown here, FlCRhR utilizes the cAMP-dependent dissociation of the PKA holoenzyme to provide a FRET-based readout of cAMP levels in live cells by chemically linking the PKA Cα (catalytic) and RIα (regulatory) subunits with fluorescein and rhodamine dye, respectively. In the absence of cAMP (left), assembly of the PKA holoenzyme results in FRET between the fluorescein and rhodamine moieties. The complex dissociates when cAMP binds the regulatory subunit (right), leading to a decrease in FRET, which can be monitored by calculating the relative emission from rhodamine or fluorescein upon fluorescein excitation (i.e., the emission ratio). However, such biosensors must be microinjected into cells, potentially limiting their accessibility. (C and D) The advent of genetically encoded fluorescent biosensors introduced nearly endless possibilities for the live-cell detection of intracellular signaling processes. These modular tools couple a conformationally dynamic molecular switch, typically derived from endogenous cellular components, with a fluorescent protein (FP)-based optical readout. For example, cameleon (C) and GCaMP (D) both utilize a Ca2+-dependent molecular switch composed of calmodulin (CaM) and the CaM-binding M13 peptide, whereupon the Ca2+-induced binding of CaM to the M13 peptide produces a conformational change that (C) increases FRET between a flanking FP pair or (D) increases the intensity of a circularly permuted GFP (cpGFP). (E) Unlike GFP and related FPs, infrared fluorescent proteins (IFPs) derived from bacterial phytochromes require an exogenous cofactor, such as biliverdin, that covalently binds a catalytic cysteine residue to form the final chromophore. Researchers have taken advantage of this fact to develop fluorogenic protease sensors such as iCasper, in which the catalytic cysteine of a circularly permuted mIFP (cp-mIFP) is tethered by a short linker and prevented from interacting with the chromophore. Only when the target protease (e.g., caspase-3) cleaves its substrate sequence (e.g., DEVDG) is the chromophore formed. Importantly, whereas FRET-based protease sensors typically dissociate upon cleavage, iCasper is held together by split GFP, allowing it to highlight regions of protease activity during in vivo imaging. (F) Luminescence-based biosensors also require an external cofactor, but obviate the need for external illumination of the sample, which can increase contrast (i.e., reduce background) and limit phototoxicity. Furthermore, BRET-based probes such as the Ca2+ sensor Nano-lantern (Ca2+) incorporate a bright FP (e.g., YFP) to increase light output. Nano-lantern (Ca2+) utilizes a Ca2+-dependent molecular switch, like that found in cameleon, to control the complementation of a split form of Renilla luciferase (RLuc). In the presence of Ca2+, reconstituted RLuc is able to catalyze the oxidation of its substrate, coelenterazine, and subsequent energy transfer results in YFP fluorescence.
Wild-type GFP is a 238-amino-acid, 27-kDa protein that folds into a unique, 11-stranded β-barrel structure of 2.4 nm diameter and 4.2 nm height [15], with an α-helix running through the central axis of the barrel cylinder, which proves to be a common structural feature shared by all currently known GFP-like FPs with intrinsic chromophores. The GFP chromophore – derived from the Ser65-Tyr66-Gly67 tripeptide – is located in the middle of the highly stable β-barrel structure and is effectively shielded from external influence, and its fluorescence is therefore largely insensitive to environmental changes [10]. Over the past 20 years, extensive protein engineering efforts have resulted in a dazzling color palette of FPs, encompassing the whole visible spectrum from ultraviolet (UV) to far-red [16,17]. The latest FPs have overcome many issues of early generations of FPs concerning expression, folding, maturation, brightness, excitation/emission spectra, photostability, oligomerization, and environmental sensitivity, thus becoming increasingly useful as the reporting element for building genetically encoded fluorescent reporters [18]. Based on their emission maxima, these color variants can be conveniently divided into the following spectral classes: UV (401–440 nm), blue (441–470 nm), cyan (471–500 nm), green (501–520 nm), yellow (521–550 nm), orange (551–575 nm), red (576–630 nm) and far-red (631–660 nm).
The burgeoning and expanding toolbox of FP-based biosensors is due, in large part, to continuous protein engineering efforts, as well as the availability of several modular and highly effective design strategies that exploit the unique spectral and structural features of FPs [19,20]. Here, we provide a brief overview of two such designs, namely FRET-based and single FP-based biosensors, that typically employ a naturally occurring or engineered conformational switch specific for the target signal as the sensing element, to highlight several important design elements before turning our attention to more recent developments in protein engineering that are providing exciting new opportunities for the development and application of genetically encoded fluorescent reporters.
FRET is an electrodynamic phenomenon of radiationless energy transfer from an excited-state donor fluorophore to a ground-state acceptor chromophore due to dipole-dipole interaction between the pair [8,21]. FRET efficiency is dependent on the photophysical properties of both the donor and the acceptor, including quantum yield (QY) of the donor, extinction coefficient of the acceptor, and the spectral overlap between the donor emission and the acceptor absorption, as well as the relative distance between the donor and acceptor, which takes both the distance and the relative orientation between the pair into account. FRET is only effective when the donor and acceptor are in molecular proximity (i.e., <10 nm apart), and its efficiency is highly sensitive to the relative distance between the donor and acceptor; therefore, it can provide a sensitive measurement of conformational changes or protein-protein interactions that correspond to specific signaling activities [22,23]. The robust fluorescence properties of FPs due to their stable β-barrel structure make them ideal FRET pairs in FRET-based biosensors. In fact, finding FP pairs optimal for FRET was and still remains one of the major motivations for the continuous engineering and development of FP color variants [10,24]. Consequently, early FP-based biosensors, including the cameleon family of Ca2+ indicators [25], were mostly FRET-based. One notable feature of a cameleon-type biosensor is that its sensing element employs an engineered two-module molecular switch comprising calmodulin (CaM) as the primary Ca2+-sensing module and M13, a CaM-binding peptide, as the auxiliary module to ensure a reliable conformational change that can be readily detected by FRET measurement (Fig. 1C). Importantly, this modular design serves as a general blueprint for engineering conformationally responsive molecular switches for sensing a variety of signaling molecules and activities [26], even in cases where the biochemical signal of interest is not naturally associated with a significant conformational change [27,28]. Furthermore, the two functional elements, namely the CaM/M13 switch and the FRET pair, can be independently engineered and evolved for improved performance or additional purposes. For instance, fundamental properties of the sensing element, such as sensitivity and response kinetics, can be readily tuned or modified by re-engineering the CaM/M13 or an analogous two-module interface, independent of the FRET pair used.
On the other hand, single FP-based biosensors can also be engineered by taking advantage of either the intrinsic sensitivities, such as pH [29] and halide [30], of some FPs, or the distinct localization pattern of the sensing element to which an FP is attached [31]. Alternatively, using a single FP as a reporting element can also be based on fluorescence complementation [32] or fragment complementation [33] of FPs, although the largely irreversible nature of this process limits its use in monitoring signaling dynamics. However, it was not until the development of circularly permuted FPs (cpFPs) [34] that it became practical to modulate the fluorescence properties of an FP with a conformational switch. In a cpFP, the original C- and N-termini of the FP are joined together through a short peptide linker, and new C- and N-termini are created at specific sites within the original amino acid sequence. Therefore, it is conceivable that when the newly-created termini are coupled to a conformational switch and placed close to the chromophore, the fluorescence properties of the cpFP can be modulated by the conformational switch. This design was first proposed in the paper describing the development of cpFPs [34] and was subsequently realized in two series of single FP-based Ca2+ indicators, namely GCaMP [35] (Fig. 1D) and Pericam [36], by incorporating the CaM/M13 switch into cpGFP and cpYFP, respectively.
Recent Advances in Biosensor Development
Next, we highlight recent advances in the development of new and improved genetically encoded fluorescent reporters. We use select examples to illustrate the capabilities as well as the limitations of current biosensors and briefly discuss some future directions.
Dimerization-dependent FPs (ddFPs)
A ddFP [37,38] is a fluorescent heterodimer consisting of a monomeric non-fluorescent FP and a monomeric fluorogenic FP whose fluorescence, as the name suggests, depends on the heterodimerization of the two FPs. Typically, the binding affinity between a ddFP pair is engineered to be relatively weak such that heterodimerization is not significant at normal expression levels for fluorescence imaging (i.e., in the range of one to tens of micromolar). As a result, ddFP fluorogenesis requires additional interaction, which makes ddFPs suitable to serve as a reporting element by combining reversible fluorogenesis with single-color convenience. In addition, spectrally distinct pairs of ddFPs, green, yellow and red, can share the same non-fluorescent FP partner, making them versatile tools for constructing new types of fluorescent reporters [39]. The utility of these ddFP pairs has been demonstrated in biosensors for a variety of biochemical activities, including Ca2+ and cAMP dynamics, protease activity, and more [38,39]. For example, ddGFP was applied to image the membrane contact sites between the endoplasmic reticulum (ER) and mitochondria, known as mitochondria-associated membranes (MAMs), which have emerged as major players in Ca2+ signaling and lipid metabolism between the two organelles in recent years [40]. Using a reporter for MAMs created by fusing the two distinct monomers of a ddGFP pair with the ER protein calnexin and the mitochondrial protein Tom20, respectively, MAMs were found to be clearly visible in both HeLa and wild-type MEF cells but not in knockout MEF cells lacking mitofusin-2, a MAM protein whose functional role at the ER/mitochondria interface was controversial [38]. This study thus provides important evidence that supports the notion that mitofusin-2 functions as a molecular tether at the interface between the ER and mitochondria.
Although current ddFP pairs employ two FPs, the fact that the non-fluorescent FP monomer only serves as a fluorescence dequencher to modulate its interacting partner suggests that different protein modules [41] that can play essentially the same role could be developed and utilized to further extend the utility of ddFP-like protein tools. Additionally, because of their distinct working mechanism, ddFP-based biosensors can also be used to orthogonally validate results from using fluorescent reporters based on other designs such as FRET and fluorescence complementation.
Photochromic FPs for super-resolution imaging and reporting
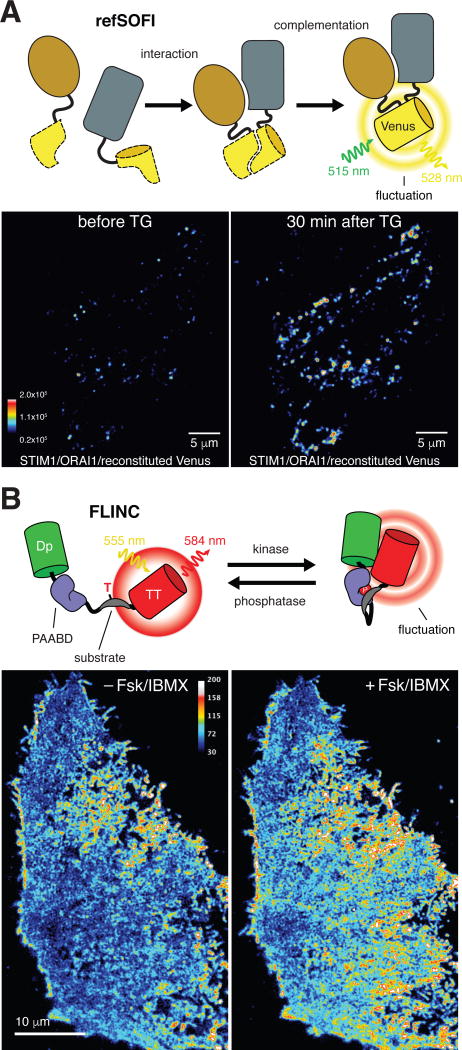
It was recognized early on that some FPs can exhibit complex photochromic behaviors such as blinking under certain illumination conditions due to the chromophore switching between different molecular states [25,42]. Subsequent demonstration that FPs with controlled photochromism can be engineered and harnessed for novel live-cell imaging applications [43] catalyzed the rapid development of a variety of new imaging methods achieving subdiffraction spatial resolution and/or enhanced contrast [44], including photoactivated localization microscopy (PALM) [45] and many others [46]. As many fluorescent biosensors for live-cell imaging are FP-based [47], there has been a growing interest in developing photochromic FP-based biosensors to visualize signaling molecules and activities with improved spatial resolution and enhanced contrast. To date, several strategies based on the reconstitution of FPs that are suitable for super-resolution imaging have been reported for nanoscale imaging of protein-protein interactions in living cells [48–51]. One such strategy, termed reconstituted fluorescence-based stochastic optical fluctuation imaging (refSOFI) [51], utilizes fragment complementation and the reconstitution of photoswitchable FPs induced by specific protein-protein interactions and subsequent fluctuation-based super-resolution imaging to obtain super-resolved maps of protein complexes in living cells (Fig. 2A). refSOFI was applied to probe the interaction between the ER Ca2+ sensor STIM1 and the pore-forming protein ORAI1 at ER-plasma membrane junctions and revealed that stimulation of store-operated Ca2+ entry results in an increase in the number of STIM1/ORAI1 interaction puncta rather than an increase in the size of existing puncta, two plausible mechanisms that are difficult to distinguish otherwise [51]. Despite improved spatial resolution, however, these fluorescence complementation-based methods are irreversible and therefore not suitable for tracking dynamic activities in super-resolution. To overcome this limitation, a new class of fluorescent reporters capable of visualizing dynamic kinase activity and protein-protein interactions in super-resolution has been recently developed [52]. These biosensors were developed based on the serendipitous discovery that fluorescence fluctuation of a red FP, TagRFP-T [53], significantly increases when it is in close proximity with Dronpa [54], a photochromic FP. This phenomenon, termed FLINC, is dependent on inter-FP distance and results from perturbation of the molecular environment of the TagRFP-T fluorophore by Dronpa. FLINC therefore forms the basis for a new class of biosensors in which the molecular proximity between Dronpa and TagRFP-T, and thus TagRFP-T fluctuation characteristics, could be modulated by engineered molecular switches in a similar manner as in FRET-based biosensors. When combined with pcSOFI [55], a super-resolution imaging method based on fluorescence fluctuation, a FLINC-based PKA activity reporter allowed mapping of PKA activity at super-resolution in living cells and uncovered a new functional role of clustered A-kinase anchoring proteins (AKAPs) in maintaining PKA signaling microdomains [52] (Fig. 2B). Importantly, this work further extends the realm of spatiotemporal regulation of cell signaling now that dynamically-structured biochemical activity can be studied with unprecedented detail in cellular space and time.
Fig. 2.
Novel methods for super-resolution imaging of signaling activities. (A) Top – Reconstituted fluorescence-based stochastic optical fluctuation imaging (refSOFI) merges a standard bimolecular fluorescence complementation (BiFC) approach with spontaneous intensity fluctuations produced by reconstituted fluorescent protein such as Dronpa or, under certain illumination conditions, Venus (top). Subsequent analysis of the resulting intensity fluctuations over a period of time enables the generation of super-resolution maps of protein-protein interactions. Bottom – Representative refSOFI images of HeLa cells expressing ORAI1 tagged with an N-terminal Venus fragment and STIM1 tagged with a C-terminal Venus fragment before and after treatment with thapsigargin (TG) to induce dimer formation and Venus reconstitution. (B) Top – Fluorescence fluctuation induced by contact (FLINC) utilizes a novel phenomenon in which the red fluorescent protein TagRFP-T (TT) undergoes spontaneous intensity fluctuations when brought into close proximity with Dronpa (Dp). Much like FRET, FLINC can be coupled to a conformational switch to yield genetically encoded biosensors for super-resolution imaging of enzyme activities. For example, FLINC-AKAR contains a molecular switch composed of a PKA substrate fused to the phosphoamino acid binding domain FHA1, which are flanked by Dp and TT. Phosphorylation of the substrate by PKA promotes binding by the FHA1 domain, leading to a conformational change and an increase in TT intensity fluctuations. SOFI analysis of the TT intensity fluctuations at different time points yields a live-cell super-resolution map of PKA activity dynamics. Bottom – Representative images of HeLa cells expressing FLINC-AKAR before and after treatment with forskolin (Fsk) and 3-isobutyl-1-methyxanthine (IBMX) to activate PKA.
Alternative FPs using endogenous cofactors
The tremendous success of GFP-like FPs has also fueled further discovery and development of new FPs with alternative chromophores, in particular those with long-wavelength excitation/emission maxima (ex/em) between 650 and 900 nm, as these are missing from the existing toolbox of GFP-like FPs. Longer-wavelength FPs are preferable for cellular imaging, especially for in vivo imaging applications, owing to minimal autofluorescence, absorption, and light scattering from endogenous cellular components, and consequently improved contrast and deeper tissue penetration in mammalian systems [56]. The first demonstration of IFP1.4 (ex/em, 684/708 nm; QY, 0.07) [57], an infrared FP (IFP) derived from a bacterial phytochrome (BphP) [58], as a useful fluorescent tag for in vivo mammalian imaging has sparked the rapid development of a series of alternative FPs, including improved infrared IFPs [59,60] and iRFPs [61–63] based on BphPs, far-red smURFP (ex/em, 642/670 nm; QY, 0.18) [64] evolved from a cyanobacterial phycobiliprotein, and green UnaG (ex/em, 498/527 nm; QY, 0.51) [65] derived from a fatty acid binding protein in the skeletal muscle of Japanese eel. These FPs, unlike GFP, utilize endogenous cofactors, such as bilirubin (e.g., UnaG) and biliverdin (e.g., IFPs, iRFPs, and smURFP), as their chromophores, and have distinct structural domains, thus providing new scaffolds for engineering genetically encoded fluorescent reporters. Indeed, a number of biosensors have already been developed based on the unique features of these alternative FPs [66–68]. For example, a fluorogenic caspase activity reporter named iCasper [66] was developed by incorporating a caspase substrate to constrain the physical proximity between two conserved domains, known as the PAS and GAF domains, in an IFP, which is believed to be critical for its chromophore formation and therefore fluorescence. Cleavage of the substrate by the target caspase releases this constraint and allows fluorescence to develop (Fig. 1E). In contrast to FRET-based reporters, this type of caspase activity reporter in principle provides a more sensitive signal because its fluorogenic nature results in low background fluorescence signal. When applied to image apoptosis during fly embryonic development using spinning-disk confocal microscopy, iCasper revealed an interesting spatiotemporal correlation between apoptosis and morphogenesis [66]. In addition, iCasper has also been successfully used to study the dynamics of apoptosis during tumorigenesis in a Drosophila brain tumor model [66].
It is expected that further discovery and development of alternative FPs will continue to create exciting opportunities and new applications for live-cell imaging that are either challenging or not yet feasible with current GFP-like FPs.
Nano-lantern and luminescent proteins
The requirement for excitation light in the case of conventional fluorescence microscopy can limit the use of fluorescent reporters in certain applications, in vivo imaging applications in particular, due to poor light penetration, phototoxicity, photobleaching, and high background autofluorescence from the sample. With luminescence, however, light is instead generated by a chemical reaction, thereby eliminating the need for an external light source. However, most luminescent proteins suitable for live-cell imaging, such as firefly luciferase (FLuc) and Renilla luciferase (RLuc), are dim compared with FPs and, as a result, provide relatively poor spatial and temporal resolution [69].
Nano-lantern [70], a bright luminescent protein, is a chimeric protein consisting of an enhanced RLuc variant fused to a bright YFP, Venus, which serves as a bioluminescence resonance energy transfer (BRET) acceptor of the energy generated by RLuc using its coelenterazine substrate. The resulting improved brightness, about 10 times brighter than wild-type RLuc (QY, ~0.2 for Nano-lantern vs ~0.02 for wild-type RLuc), enables real-time imaging of subcellular structural features such as microfilaments and microtubules in living cells and allows video-rate imaging of small subcutaneous tumors in freely moving unshaved mice [70]. Moreover, cyan and orange color variants of Nano-lantern have been engineered to enable high-speed multicolor luminescence imaging in living cells [71]. More recently, five bright new color variants, namely, cyan, green, yellow, orange and red, of enhanced Nano-lantern (eNL) [72], which is based on NanoLuc [73], the brightest luciferase currently available, have been developed to enable luminescence co-imaging of five distinct subcellular structures in living cells. Notably, Nano-lantern based biosensors for tracking Ca2+ (Fig. 1F), cAMP and ATP dynamics have been successfully created by inserting the corresponding sensing domains into the RLuc region of Nano-lantern, achieving up to 300%, 130%, and 200% dynamic ranges, respectively, in living cells [70]. Similarly, an eNL-based Ca2+ biosensor has also been created with an impressive 500% dynamic range to allow fast and long-term Ca2+ imaging in cardiomyocytes to show drug effects on cardiomyocyte function [72]. These results suggest that new and improved biosensors may be developed by exploiting the structural features of different luciferases, such as the FLuc, NanoLuc, or RLuc.
In another recent development of luminescent proteins, CyOFP1 [74], a bright cyan-excitable orange FP, was used as a BRET acceptor for NanoLuc to create Antares [74], a highly bright red-emitting luminescent protein consisting of NanoLuc sandwiched between two copies of CyOFP1. Compared with orange Nano-lantern, Antares is 20 times brighter overall and 28 times brighter at wavelengths longer than 600 nm under similar test conditions. Moreover, Antares is capable of producing much higher levels of detectable photon emission when expressed in deep tissues of living mice compared with the most commonly used luminescent protein, FLuc. Therefore, it is of great interest to see whether high-performance biosensors can be developed for in vivo imaging of cellular signaling based on Antares or similarly designed luminescent proteins.
Luminescent proteins such as Nano-lantern and Antares hold great promise in enabling new applications of genetically encoded fluorescent reporters by eliminating the need for external illumination and are particularly suitable for applications such as live-cell drug screens and in vivo imaging, for which illumination is undesirable or difficult under certain conditions. In addition, luminescent protein-based biosensors are readily compatible with perturbation techniques that involve the use of light, such as optogenetics, making them valuable tools for all-optical analysis of cell signaling in living systems [75]. Moreover, eliminating the light source could also facilitate the development of compact or miniaturized optical microscopes [76] to enable more convenient and efficient high-content and in vivo imaging applications. However, significant improvements to current luminescent protein-based biosensors, both in terms of brightness and dynamic range, are still required in order to achieve spatial and temporal resolution sufficient for a wide range of live-cell imaging applications.
Nanobodies as a unique sensing element
Owing to their high affinity and exquisite specificity, antibodies, typically consisting of two heavy chains and two light chains, are valuable tools for cell signaling research and are routinely used to visualize signaling molecules in fixed cells and tissues. Their application in living cells, however, is hindered by their large size (~150 kDa), poor solubility and low stability in living cells. On the other hand, some animals, including llamas and sharks, also produce antibodies devoid of light chains [77]. Importantly, the variable domain of these heavy chain-only antibodies, known as nanobodies [78], retains the full capability of specific antigen recognition. Nanobodies can be readily generated following established protocols and screened for desired affinity and specificity [79]. With their small size (~15 kDa), high solubility and stability, nanobodies are well suited for tracking specific targets in living cells [80]. Moreover, many nanobodies recognize conformational epitopes (i.e., a discontinuous sequence of amino acids of a native protein that signifies its conformation) in functional proteins, making them ideal sensing elements for detecting distinct conformational states of a target protein directly in its cellular context [81]. The potential of nanobodies as a valuable tool for live-cell imaging has been demonstrated in a range of applications, including the dynamic tracking of endogenous proteins [82–84] and the development of conformation-sensitive fluorescent reporters for real-time visualization of transient conformational changes in signaling molecules in living cells [85,86].
As a key member of the poly(ADP-ribose) polymerase (PARP) protein family [87], PARP1 plays an important role in DNA repair and other important cellular processes. Upon detecting DNA damage, PARAP1 binding to the damaged DNA initiates PAR chain synthesis on specific target substrates, including itself. These PAR chains in turn serve as a signal for the further recruitment of other DNA repair enzymes. Being at the starting point of the DNA repair process, PARP1 is also an attractive drug target for the development of anti-cancer agents. To address the pressing need to better understand PARP1 dynamics in living cells, a fluorescent reporter for tracking endogenous PARP1, named PARP1 chromobody, was generated by fusing an FP to a well-characterized PARP1 nanobody [84]. Consistent with results from other studies, PARP1 chromobody was shown to be capable of tracking the dynamic localization patterns of endogenous PARP1 under various conditions, suggesting its potential as a useful tool for understanding the multiple functions of PARP1 in its native cellular environment.
Current nanobody-based biosensors typically employ conventional FPs as the reporting element and may suffer from high background signals even in the absence of endogenous targets. One way to overcome this limitation is to match the expression level of the biosensor to that of the target protein [88,89]. Alternatively, nanobody-based sensing elements may be combined with other types of reporting elements, for example ddFPs [39] or split UnaG [90], to generate biosensors whose fluorescence properties change upon recognition of the target to improve signal sensitivity. Moreover, given the growing appreciation for the role of dynamic signaling complexes as critical regulators of cell signaling, nanobodies may also serve as a much-needed specific targeting motif [88] for targeting existing biosensors to specific signaling nanodomains to help elucidate their unique functional roles in more biologically relevant contexts.
Biosensors at Work
GPCR signaling revisited
Results from many live-cell imaging studies using fluorescent reporters have demonstrated unequivocally that spatiotemporal regulation is involved in practically all aspects of cell signaling, and therefore, it is critically important to understand the functional roles of signaling molecules in biologically relevant cellular context, even for those that have been well characterized both biochemically and biophysically. Here, we highlight some important contributions of this approach to the current understanding of GPCR signaling, including GPCR signaling from endosomes and β-arrestin signaling.
With an estimated 800 or more members, G protein-coupled receptors (GPCRs), also known as seven-transmembrane (7TM) receptors, constitute the largest family of cell-surface receptors in the human genome. Functionally, GPCRs mediate distinct physiological responses by specifically transducing a vast array of extracellular stimuli into various intracellular signaling pathways, including the cAMP and phosphatidylinositol signaling pathways, via coupling to heterotrimeric G proteins. Traditionally, GPCRs are thought to signal to second messengers such as cAMP from the cell surface and become desensitized or switch to distinct signaling pathways upon agonist-induced internalization. This desensitization or switching process involves the action of GPCR kinases (GRKs) and subsequent interaction between the phosphorylated GPCR and β-arrestin. Depending on the nature of this interaction, GPCRs are commonly divided into two classes, namely, class A (weak interaction) and class B (strong interaction). However, recent studies using FRET-based cAMP biosensors have shown persistent cAMP production from internalized GPCRs in living cells [91,92], indicating active intracellular G protein signaling from internalized compartments. These results have led to a series of studies to further investigate the underlying molecular mechanisms of such phenomena.
To determine whether internalized GPCRs could remain active upon internalization, a GPCR conformation-specific nanobody-based biosensor named Nb80-GFP was generated to directly probe the activation process of the β2 adrenergic receptor (β2AR), a prototypical class A GPCR, in living cells [85]. Nb80-GFP was created by fusing GFP to a nanobody, Nb80, which specifically recognizes the activated form of β2AR. Live-cell imaging results showed two distinct phases of Nb80-GFP recruitment upon agonist stimulation: an early one to the plasma membrane and a later one to β2AR-containing early endosomes. This observation of a second recruitment to endosomes suggests that internalized β2AR can indeed exist in an active conformation. To test whether internalized β2ARs could actually engage G protein coupling, another G protein conformation-specific nanobody-based biosensor was created by fusing GFP to a different nanobody, Nb37, which specifically binds the G protein in a transitional state, devoid of guanine-nucleotide, during activation. Consistent with the observations using Nb80-GFP, Nb37-GFP also showed a later recruitment to β2AR-containing endosomes. Together with results from a luminescence-based live-cell cAMP assay, these findings support the notion that G protein signaling from internalized receptors might represent a general mechanism of GPCR signaling. Furthermore, using an optogenetic approach, it was also shown that cAMP production from different compartments, e.g., plasma membrane and endosomes, can lead to distinct cellular responses [93].
On the other hand, cAMP production triggered by internalized receptors has also been reported for a number of class B GPCRs, including the thyroid-stimulating hormone receptor (THSR) [91], parathyroid hormone receptor (PTHR) [92], and vasopressin type 2 receptor (V2R) [94], which have strong receptor-arrestin interactions. However, this is even more surprising given the prevailing paradigm that GPCR-arrestin and GPCR-G protein interactions are mutually exclusive. To look into this rather intriguing question, a recent study systematically tested the hypothesis that β-arrestin is actually capable of inducing receptor internalization without disrupting G protein coupling by forming a GPCR-G protein-β-arrestin super-complex [95]. First, real-time cAMP measurements using a FRET-based biosensor [96,97] in living cells established that class B GPCRs can indeed continue to stimulate cAMP production after agonist-induced internalization. Second, cAMP production as a result of G protein signaling from internalized receptors was confirmed using a panel of BRET biosensors [98] in combination with an agonist-washout protocol. Third, fluorescently labeled receptor, β-arrestin, and G protein were shown to co-localize in endosomes after agonist stimulation using confocal microscopy. Furthermore, interactions between key components of the proposed super-complex at the internalized compartments were also validated with BRET measurements. Finally, in vitro preparation and isolation of the hypothesized super-complex was carried out using an engineered β2V2R receptor (a chimeric receptor in which the C-terminal tail of β2AR is replaced with the C-terminal tail of V2R), antibody-stabilized G protein and β-arrestin1; single-particle electron microscopy (EM) analysis revealed a novel structural architecture in which the chimeric receptor simultaneously binds to G protein and β-arrestin via its transmembrane core and C-terminal tail, respectively. Taken together, these results establish a new mode of GPCR signaling that supports sustained G protein signaling, arising from the simultaneous interaction of both G protein and β-arrestin with the activated GPCR within internalized cellular compartments. Interestingly, several lines of evidence in this study also suggest that β2AR, a class A GPCR, does not seem to contribute significantly to this β-arrestin-mediated GPCR signaling from internalized compartments under these specific conditions. A follow-up study using a β-arrestin mutant lacking the ability to interact with the transmembrane core of GPCRs further showed that β-arrestin interaction with the C-terminal tail of GPCRs alone is sufficient to cause receptor internalization but not desensitization [99], consistent with results from another recent study using a mutant GPCR that is deficient in its core interaction [100].
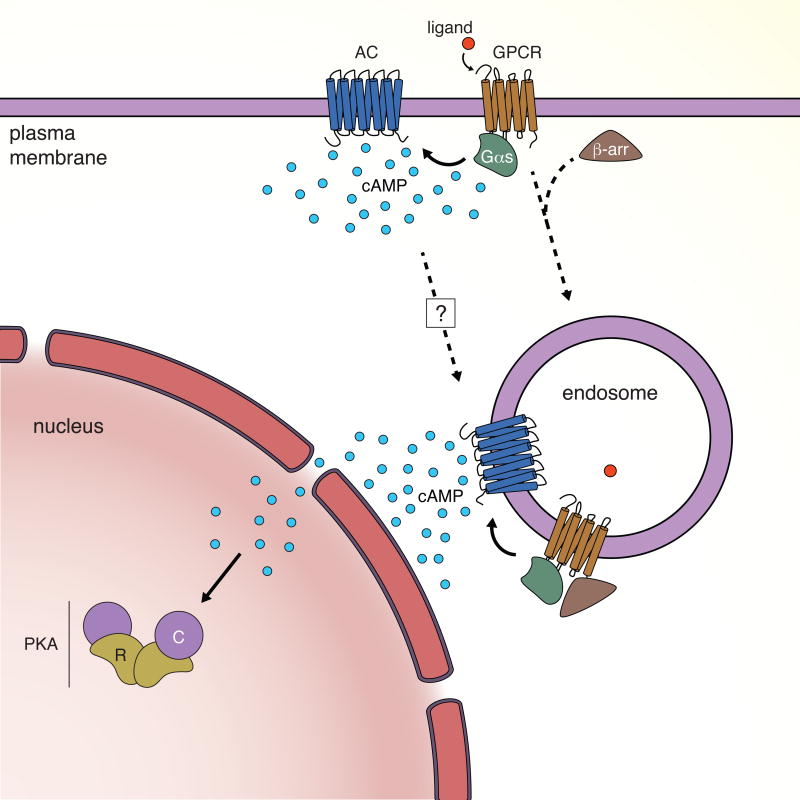
It is increasingly clear from many studies, including those highlighted above, that spatiotemporal regulation is an integral part of GPCR signaling, and therefore, our understanding of the complexity and distinct physiological consequences of GPCR signaling is very much dependent on our ability to visualize various aspects of GPCR signaling in real time in relevant biological contexts, including important downstream signaling events such as cAMP production at different membrane compartments. For instance, a recent study uncovered yet another unexpected mechanism of crosstalk between two distinct GPCRs, namely PTHR and β2AR, at the site of PTHR-containing endosomes [101], which promotes endosomal cAMP production, suggesting the true complexity of GPCR signaling is likely to be much greater than currently appreciated (Fig. 3). Ultimately, live-cell imaging of GPCR signaling with high spatial and temporal resolution in living cells, tissues, and intact organisms will allow researchers to directly establish the causal links between the diverse molecular and mechanistic underpinnings and the distinct physiological effects of GPCR signaling.
Fig. 3.
Updated model of prolonged intracellular GPCR signaling based on live-cell imaging. In the traditional model of G protein-coupled receptor (GPCR) signaling, agonist-induced GPCR activation leads to second messenger production at the plasma membrane, such as Gαs-stimulated production of cAMP by adenylyl cyclase (AC), followed by activity-dependent GPCR internalization and signaling desensitization through β-arrestin (β-arr) binding. However, several recent live-cell imaging studies using genetically encoded fluorescent biosensors have indicated that internalized GPCRs remain active and continue to signal from endosomal compartments, in some cases despite continued association with β-arr. Indirect evidence also suggests the presence of endosomal AC activity, although further studies are needed to clarify how ACs reach the endosome (indicated by a question mark), and such persistent GPCR signaling may therefore be responsible for sustained cAMP production within the cytosol, in contrast to transient cAMP production from the plasma membrane, leading to the activation of PKA signaling within the nucleus.
Illuminating neuronal activity in vivo
The ability to measure neuronal activity in large populations of individual neurons in freely behaving animals is critical to advancing our understanding of how the nervous system works. Membrane potential changes such as action potentials (APs) are considered the common currency for neuronal activity, whereas Ca2+ signals serve as a proxy since APs often trigger intracellular Ca2+ changes. Over the past decade, both genetically encoded calcium indicators (GECIs) and genetically encoded voltage indicators (GEVIs) have been under active development for the imaging of neuronal activity in the intact brains of living animals [102,103].
The first reported GECIs were FRET-based [25], using FP color variants such as the BFP/GFP or CFP/YFP pair. One such reporter, yellow cameleon-2 (YC2) is a fusion protein comprising enhanced CFP (ECFP), CaM, M13, and enhanced YFP (EYFP), from N- to C-terminus (Fig. 1C). As described above, the CaM-M13 fusion functions as an engineered molecular switch, which alters its conformation upon Ca2+ binding to CaM and subsequent CaM/M13 complex formation, to modulate the FRET efficiency between the flanking FP pair. Many FRET-based GECIs have since been developed with improved performance and/or different characteristics for an increasingly broader range of applications in different living systems. For example, two independent approaches have been taken to minimize the perturbation of the YC signal due to endogenous CaM and CaM-binding proteins. In the TN series [104,105], a muscle-specific Ca2+ binding protein, troponin C, was used to replace the CaM/M13 switch; while in the D series of cameleon-type reporters such as D3cpV [106], the CaM/M13 binding interface was computationally redesigned using a bump-and-hole strategy so that they are no longer perturbed by endogenous CaM.
As the first example of single FP-based GECIs, EYFP-calmodulin (also known as camgaroo-1, with CaM inserted into EYFP at position Tyr145) was the result of a rather fortuitous discovery that an ECFP mutant with an insertion of a six-residue peptide between residues 144 and 145 remained fluorescent [34]. This discovery also led to the creation of a family of cpFPs that proved to be useful for developing and improving single FP-based as well as FRET-based GECIs [107].
Early GECIs were not sensitive enough, in terms of signal-to-noise ratio (SNR) and response kinetics, to reliably detect the fast Ca2+ dynamics often associated with neuronal activity in vivo. Over the past 10 years, significant protein engineering efforts, together with large-library screening, have led to the development of many high-performing GECIs, with the most recent ones showing in vivo sensitivities matching or even surpassing the benchmark performance set by synthetic Ca2+ indicators [108]. For example, GCaMP6 indicators [109], the current generation of GCaMP-type GECIs, are suitable for general in vivo imaging of neuronal activity in a variety of model organisms. In addition, a broad selection of GECI viral vectors and transgenic mice are now available, paving the way for widespread use of multi-scale in vivo brain activity imaging in freely behaving animals. In an effort to investigate the long-term place-coding dynamics of CA1 hippocampal place cells, the environment-specific neuronal activities of thousands of neurons in freely moving mice were monitored repeatedly across many sessions over a period of multiple weeks [110]. This was made possible by head-mounting a miniaturized fluorescence microscope [76] on the target animal to allow Ca2+ dynamics in neurons expressing a GECI, in this case GCaMP3 [111], to be tracked repeatedly while the mouse was freely exploring a familiar environment over many sessions. It was found that, at the cellular level, cells with place fields varied greatly from session to session, with only about 15–25% overlap between any two sessions ranging from 5 to 30 days apart; yet, at the ensemble level, this modest overlap was sufficient to preserve the spatial representation of the explored environment over the entire period, given the stable place fields of the cells. Furthermore, there was no discernable correlation between the overlapping cells and their activity patterns as revealed by Ca2+ imaging, suggesting that long-term CA1 place-coding is a dynamic network-level property. In another study, Ca2+ imaging of GCaMP6 transgenic mice was used to generate retinotopic maps of the visual cortex while mice were exposed to visual stimuli [112]. In this case, the superior SNR of GCaMP6 not only enabled rapid mapping but also helped to reveal 4 new regions that were not identified via previous methods, resulting in more complete coverage of the functionally distinct visual regions in the mouse cortex.
Changes in membrane potential are spatially confined yet highly dynamic, and can be very fast in the case of APs. These intrinsic properties of membrane voltage signals proved to be a significant challenge for GEVI development. Early GEVIs utilizing voltage-sensing protein domains derived from various voltage-gated ion channels simply failed to properly localize to the plasma membrane in mammalian cells and are therefore not very useful [113]. More recently, the discovery of voltage-sensitive phosphatases [114] and the engineering of voltage-sensing microbial rhodopsins [115] with improved membrane localization and fast kinetics have provided more effective protein scaffolds for GEVI design and optimization, leading to the development of a series of increasingly better-performing GEVIs and many successful applications for reporting neuronal voltage dynamics in various animal models [116,117]. For example, Ace2N-4AA-mNeon [118], a fast and bright FRET-opsin type GEVI [119], created by coupling a fast opsin-based voltage-sensing domain to a bright FP, mNeonGreen [120], was shown to be capable of high-fidelity recording of single action potentials and subthreshold membrane voltage dynamics in cortical neurons of awake mice. Despite the impressive progress in recent years, however, more breakthroughs in the development of GEVIs, as well as major advances in related imaging modalities, are still needed in order to achieve the long-standing goal of long-term tracking of membrane potential changes in large populations of individual neurons in freely behaving animals. More comprehensive comparisons and discussions of current GEVIs can be found in several recent review articles [103,116,117].
Whole-brain imaging of neuronal activity using a snapshot reporter, CaMPARI
It has been well established that different behavioral tasks involve distinct populations of neurons in the brain. In this regard, whole-brain imaging of neural activity with single-cell resolution in living animals is probably the most straightforward approach to identify specific behavior-associated neural circuits in animal brains [121,122]. However, while genetically encoded fluorescent reporters for neuronal activity, such as GECIs, can be readily expressed in neurons of interest in a whole brain, current in vivo imaging modalities lack the capability to label specific neurons of interest for post hoc analysis and are also limited in their capacity to capture all the targeted neurons in an intact brain with sufficient spatial and temporal resolution. To address this challenge, a snapshot reporter concept was proposed [123] and recently materialized with the development of CaMPARI [124], a calcium-modulated photoactivatable ratiometric integrator.
Briefly, CaMPARI is a single FP-based reporter that undergoes a Ca2+-dependent green-to-red photoconversion upon violet-light illumination. CaMPARI is based on the GCaMP design but uses a photoconvertible FP, mEos2 [125], instead of cpGFP. Expressing this reporter in the brains of living organisms effectively turns the whole brain itself into an optical recording medium for brain activity, and upon temporally controlled illumination, active neurons in animals experiencing specific stimuli or performing certain behavioral tasks change color from green to red, while neurons with low activity at the same moment remain green due to differential photoconversion efficiency of the reporter in different Ca2+-binding states. This color switching is non-perturbing, effectively permanent and remains stable after fixation, making the reporter compatible with both in vivo and ex vivo analysis of the whole brain or selected regions [121,122,126,127]. Therefore, whole-brain imaging of neural activity with single-cell resolution can be performed retrospectively, and additional characterization and analysis using various techniques, including many single-cell analysis techniques, can be performed to obtain cellular activity-dependent neural circuit information at the single-cell, as well as whole-brain, level. The performance and utility of CaMPARI as a light-controlled snapshot reporter for neural activity in vivo have been validated and demonstrated in several model systems, including fish, flies, and mice [124]. For example, upon application of a photoconversion light that coincides with an odor stimulation, CaMPARI was used to label active neurons in response to specific odorant stimuli in the brains of adult files. The results are consistent with previous studies that used other methods such as real-time Ca2+ imaging with GECIs, but more importantly, it demonstrates the utility of CaMPARI as a permanent activity marker in scenarios where real-time monitoring of Ca2+ signal might not be feasible, for example, when imaging a large field of view. In another example, whole-brain imaging of transgenic zebrafish expressing CaMPARI in all neurons under various conditions, such as freely swimming, anesthesia, heat, and cold, showed consistent activity patterns between fish that experienced the same condition but distinct patterns among fish that were exposed to different conditions, with cellular-level resolution.
To properly utilize CaMPARI as a tool for labeling active neurons in vivo during behavior or stimulus, it is important to ensure that the photoconversion light used during the experiment does not alter the behaviors of the living organisms under study. In addition to having been validated in multiple model organisms under various established experimental conditions, CaMPARI can also serve as an internal control since it also functions as a regular GECI in either the original green state or the photoconverted red state. Consequently, Ca2+ signals measured before and after photoconversion light illumination can be compared with each other as well as its normal red/green ratio signal, which is indicative of integrated Ca2+ activity during the illumination period, to determine the potential impact of the illumination light on animal behavior.
Conclusion
In just 20 years, the toolbox of genetically encoded fluorescent reporters has grown from virtually non-existent to being a major resource for studying many aspects of cell signaling. Given the important role of genetically encoded fluorescent reporters in advancing our understanding of the spatiotemporal regulation of signal transduction in relevant biological contexts, current efforts are devoted to growing the diversity of the toolbox as well as improving the performance of the existing tools to be able to visualize essentially all signaling molecules in living cells. With synergistic advances in chemical biology, advanced optical imaging, computational modeling, and genomic editing, the future of illuminating cell signaling is bright.
Acknowledgments
This work was supported by the NIH (R35 CA197622, R01 DK073368, R01 GM111665, and R01 MH111516). We thank Fabian Hertel and Gary Mo for contributing the images used in Fig. 2.
Abbreviations
- AKAP
A-kinase anchoring protein
- AP
action potential
- BFP
blue fluorescent protein
- BphP
bacterial phytochrome
- BRET
bioluminescence resonance energy transfer
- CaM
calmodulin
- cAMP
cyclic AMP
- CFP
cyan fluorescent protein
- cpFP
circularly permuted fluorescent protein
- ddFP
dimerization-dependent fluorescent protein
- ECFP
enhanced cyan fluorescent protein
- EM
electron microscopy
- eNL
enhanced Nano-lantern
- ER
endoplasmic reticulum
- ex/em
excitation/emission maxima
- EYFP
enhanced yellow fluorescent protein
- FLuc
firefly luciferase
- FP
fluorescent protein
- FRET
fluorescence resonance energy transfer
- GECI
genetically encoded calcium indicator
- GEVI
genetically encoded voltage indicator
- GFP
green fluorescent protein
- GPCR
G protein-coupled receptor
- GRK
G protein-coupled receptor kinase
- IFP
infrared fluorescent protein
- iRFP
infrared fluorescent protein
- MAM
mitochondria-associated membrane
- PALM
photoactivated localization microscopy
- PAR
poly(ADP-ribose)
- PARP
poly(ADP-ribose) polymerase
- PKA
cAMP-dependent protein kinase
- PTHR
parathyroid hormone receptor
- QY
quantum yield
- refSOFI
reconstituted fluorescence-based stochastic optical fluctuation imaging
- RFP
red fluorescent protein
- RLuc
Renilla luciferase
- SNR
signal-to-noise ratio
- THSR
thyroid-stimulating hormone receptor
- UV
ultraviolet
- V2R
vasopressin type 2 receptor
- YC
yellow cameleon
- YFP
yellow fluorescent protein
- β2AR
β2 adrenergic receptor
References
- 1.Kholodenko BN, Hancock JF, Kolch W. Signalling ballet in space and time. Nat. Rev. Mol. Cell Biol. 2010;11:414–26. doi: 10.1038/nrm2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mehta S, Zhang J. Reporting from the field: genetically encoded fluorescent reporters uncover signaling dynamics in living biological systems. Annu. Rev. Biochem. 2011;80:375–401. doi: 10.1146/annurev-biochem-060409-093259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xia T, Li N, Fang X. Single-molecule fluorescence imaging in living cells. Annu. Rev. Phys. Chem. 2013;64:459–80. doi: 10.1146/annurev-physchem-040412-110127. [DOI] [PubMed] [Google Scholar]
- 4.Grynkiewicz G, Poenie M, Tsien RY. A new generation of Ca2+ indicators with greatly improved fluorescence properties. J. Biol. Chem. 1985;260:3440–50. [PubMed] [Google Scholar]
- 5.Sample V, Mehta S, Zhang J. Genetically encoded molecular probes to visualize and perturb signaling dynamics in living biological systems. J. Cell Sci. 2014;127:1151–60. doi: 10.1242/jcs.099994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lavis LD, Raines RT. Bright building blocks for chemical biology. ACS Chem. Biol. 2014;9:855–66. doi: 10.1021/cb500078u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adams SR, Harootunian AT, Buechler YJ, Taylor SS, Tsien RY. Fluorescence ratio imaging of cyclic AMP in single cells. Nature. 1991;349:694–7. doi: 10.1038/349694a0. [DOI] [PubMed] [Google Scholar]
- 8.Stryer L. Fluorescence energy transfer as a spectroscopic ruler. Annu. Rev. Biochem. 1978;47:819–46. doi: 10.1146/annurev.bi.47.070178.004131. [DOI] [PubMed] [Google Scholar]
- 9.Tsien RY. Constructing and exploiting the fluorescent protein paintbox (Nobel Lecture) Angew. Chem. Int. Ed. Engl. 2009;48:5612–26. doi: 10.1002/anie.200901916. [DOI] [PubMed] [Google Scholar]
- 10.Tsien RY. The green fluorescent protein. Annu. Rev. Biochem. 1998;67:509–44. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 11.Zhang J, Campbell RE, Ting AY, Tsien RY. Creating new fluorescent probes for cell biology. Nat. Rev. Mol. Cell Biol. 2002;3:906–18. doi: 10.1038/nrm976. [DOI] [PubMed] [Google Scholar]
- 12.Chudakov DM, Matz MV, Lukyanov S, Lukyanov KA. Fluorescent proteins and their applications in imaging living cells and tissues. Physiol. Rev. 2010;90:1103–63. doi: 10.1152/physrev.00038.2009. [DOI] [PubMed] [Google Scholar]
- 13.Newman RH, Fosbrink MD, Zhang J. Genetically encodable fluorescent biosensors for tracking signaling dynamics in living cells. Chem. Rev. 2011;111:3614–66. doi: 10.1021/cr100002u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miyawaki A, Niino Y. Molecular spies for bioimaging--fluorescent protein-based probes. Mol. Cell. 2015;58:632–43. doi: 10.1016/j.molcel.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 15.Ormö M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 1996;273:1392–5. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 16.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat. Methods. 2005;2:905–9. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 17.Rodriguez EA, Campbell RE, Lin JY, Lin MZ, Miyawaki A, Palmer AE, Shu X, Zhang J, Tsien RY. The Growing and Glowing Toolbox of Fluorescent and Photoactive Proteins. Trends Biochem. Sci. 2017;42:111–129. doi: 10.1016/j.tibs.2016.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cranfill PJ, Sell BR, Baird MA, Allen JR, Lavagnino Z, de Gruiter HM, Kremers G-J, Davidson MW, Ustione A, Piston DW. Quantitative assessment of fluorescent proteins. Nat. Methods. 2016;13:557–62. doi: 10.1038/nmeth.3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frommer WB, Davidson MW, Campbell RE. Genetically encoded biosensors based on engineered fluorescent proteins. Chem. Soc. Rev. 2009;38:2833–41. doi: 10.1039/b907749a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palmer AE, Qin Y, Park JG, McCombs JE. Design and application of genetically encoded biosensors. Trends Biotechnol. 2011;29:144–52. doi: 10.1016/j.tibtech.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jares-Erijman EA, Jovin TM. FRET imaging. Nat. Biotechnol. 2003;21:1387–95. doi: 10.1038/nbt896. [DOI] [PubMed] [Google Scholar]
- 22.Sekar RB, Periasamy A. Fluorescence resonance energy transfer (FRET) microscopy imaging of live cell protein localizations. J. Cell Biol. 2003;160:629–33. doi: 10.1083/jcb.200210140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miyawaki A. Development of probes for cellular functions using fluorescent proteins and fluorescence resonance energy transfer. Annu. Rev. Biochem. 2011;80:357–73. doi: 10.1146/annurev-biochem-072909-094736. [DOI] [PubMed] [Google Scholar]
- 24.Bajar BT, Wang ES, Zhang S, Lin MZ, Chu J. A Guide to Fluorescent Protein FRET Pairs. Sensors (Basel) 2016;16:1488. doi: 10.3390/s16091488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, Ikura M, Tsien RY. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature. 1997;388:882–7. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- 26.Hochreiter B, Garcia AP, Schmid JA. Fluorescent proteins as genetically encoded FRET biosensors in life sciences. Sensors (Basel) 2015;15:26281–314. doi: 10.3390/s151026281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ananthanarayanan B, Ni Q, Zhang J. Signal propagation from membrane messengers to nuclear effectors revealed by reporters of phosphoinositide dynamics and Akt activity. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15081–6. doi: 10.1073/pnas.0502889102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang J, Allen MD. FRET-based biosensors for protein kinases: illuminating the kinome. Mol. Biosyst. 2007;3:759–65. doi: 10.1039/b706628g. [DOI] [PubMed] [Google Scholar]
- 29.Miesenböck G, De Angelis DA, Rothman JE. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature. 1998;394:192–5. doi: 10.1038/28190. [DOI] [PubMed] [Google Scholar]
- 30.Jayaraman S, Haggie P, Wachter RM, Remington SJ, Verkman AS. Mechanism and cellular applications of a green fluorescent protein-based halide sensor. J. Biol. Chem. 2000;275:6047–50. doi: 10.1074/jbc.275.9.6047. [DOI] [PubMed] [Google Scholar]
- 31.Várnai P, Balla T. Live cell imaging of phosphoinositide dynamics with fluorescent protein domains. Biochim. Biophys. Acta. 2006;1761:957–67. doi: 10.1016/j.bbalip.2006.03.019. [DOI] [PubMed] [Google Scholar]
- 32.Kerppola TK. Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annu. Rev. Biophys. 2008;37:465–87. doi: 10.1146/annurev.biophys.37.032807.125842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Magliery TJ, Wilson CGM, Pan W, Mishler D, Ghosh I, Hamilton AD, Regan L. Detecting protein-protein interactions with a green fluorescent protein fragment reassembly trap: scope and mechanism. J. Am. Chem. Soc. 2005;127:146–57. doi: 10.1021/ja046699g. [DOI] [PubMed] [Google Scholar]
- 34.Baird GS, Zacharias DA, Tsien RY. Circular permutation and receptor insertion within green fluorescent proteins. Proc. Natl. Acad. Sci. U. S. A. 1999;96:11241–6. doi: 10.1073/pnas.96.20.11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakai J, Ohkura M, Imoto K. A high signal-to-noise Ca(2+) probe composed of a single green fluorescent protein. Nat. Biotechnol. 2001;19:137–41. doi: 10.1038/84397. [DOI] [PubMed] [Google Scholar]
- 36.Nagai T, Sawano A, Park ES, Miyawaki A. Circularly permuted green fluorescent proteins engineered to sense Ca2+ Proc. Natl. Acad. Sci. U. S. A. 2001;98:3197–202. doi: 10.1073/pnas.051636098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alford SC, Abdelfattah AS, Ding Y, Campbell RE. A fluorogenic red fluorescent protein heterodimer. Chem. Biol. 2012;19:353–60. doi: 10.1016/j.chembiol.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alford SC, Ding Y, Simmen T, Campbell RE. Dimerization-dependent green and yellow fluorescent proteins. ACS Synth. Biol. 2012;1:569–75. doi: 10.1021/sb300050j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ding Y, Li J, Enterina JR, Shen Y, Zhang I, Tewson PH, Mo GCH, Zhang J, Quinn AM, Hughes TE, Maysinger D, Alford SC, Zhang Y, Campbell RE. Ratiometric biosensors based on dimerization-dependent fluorescent protein exchange. Nat. Methods. 2015;12:195–8. doi: 10.1038/nmeth.3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Naon D, Scorrano L. At the right distance: ER-mitochondria juxtaposition in cell life and death. Biochim. Biophys. Acta. 2014;1843:2184–94. doi: 10.1016/j.bbamcr.2014.05.011. [DOI] [PubMed] [Google Scholar]
- 41.Kirchhofer A, Helma J, Schmidthals K, Frauer C, Cui S, Karcher A, Pellis M, Muyldermans S, Casas-Delucchi CS, Cardoso MC, Leonhardt H, Hopfner K-P, Rothbauer U. Modulation of protein properties in living cells using nanobodies. Nat. Struct. Mol. Biol. 2010;17:133–8. doi: 10.1038/nsmb.1727. [DOI] [PubMed] [Google Scholar]
- 42.Dickson RM, Cubitt AB, Tsien RY, Moerner WE. On/off blinking and switching behaviour of single molecules of green fluorescent protein. Nature. 1997;388:355–8. doi: 10.1038/41048. [DOI] [PubMed] [Google Scholar]
- 43.Patterson GH, Lippincott-Schwartz J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 2002;297:1873–7. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- 44.Huang B, Babcock H, Zhuang X. Breaking the diffraction barrier: super-resolution imaging of cells. Cell. 2010;143:1047–58. doi: 10.1016/j.cell.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–5. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 46.Nienhaus K, Nienhaus GU. Fluorescent proteins for live-cell imaging with super-resolution. Chem. Soc. Rev. 2014;43:1088–106. doi: 10.1039/c3cs60171d. [DOI] [PubMed] [Google Scholar]
- 47.Zhang J, Ni Q, Newman RH, editors. Fluorescent Protein-Based Biosensors. Humana Press; Totowa, NJ: 2014. [Google Scholar]
- 48.Liu Z, Xing D, Su QP, Zhu Y, Zhang J, Kong X, Xue B, Wang S, Sun H, Tao Y, Sun Y. Super-resolution imaging and tracking of protein-protein interactions in sub-diffraction cellular space. Nat. Commun. 2014;5:4443. doi: 10.1038/ncomms5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nickerson A, Huang T, Lin L-J, Nan X. Photoactivated localization microscopy with bimolecular fluorescence complementation (BiFC-PALM) for nanoscale imaging of protein-protein interactions in cells. PLoS One. 2014;9:e100589. doi: 10.1371/journal.pone.0100589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xia P, Liu X, Wu B, Zhang S, Song X, Yao PY, Lippincott-Schwartz J, Yao X. Superresolution imaging reveals structural features of EB1 in microtubule plus-end tracking. Mol. Biol. Cell. 2014;25:4166–73. doi: 10.1091/mbc.E14-06-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hertel F, Mo GCH, Duwé S, Dedecker P, Zhang J. RefSOFI for Mapping Nanoscale Organization of Protein-Protein Interactions in Living Cells. Cell Rep. 2016;14:390–400. doi: 10.1016/j.celrep.2015.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mo GCH, Ross B, Hertel F, Manna P, Yang X, Greenwald E, Booth C, Plummer AM, Tenner B, Chen Z, Wang Y, Kennedy EJ, Cole PA, Fleming KG, Palmer A, Jimenez R, Xiao J, Dedecker P, Zhang J. Genetically encoded biosensors for visualizing live-cell biochemical activity at super-resolution. Nat. Methods. 2017;14:427–434. doi: 10.1038/nmeth.4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shaner NC, Lin MZ, McKeown MR, Steinbach PA, Hazelwood KL, Davidson MW, Tsien RY. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat. Methods. 2008;5:545–51. doi: 10.1038/nmeth.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ando R, Mizuno H, Miyawaki A. Regulated fast nucleocytoplasmic shuttling observed by reversible protein highlighting. Science. 2004;306:1370–3. doi: 10.1126/science.1102506. [DOI] [PubMed] [Google Scholar]
- 55.Dedecker P, Mo GCH, Dertinger T, Zhang J. Widely accessible method for superresolution fluorescence imaging of living systems. Proc. Natl. Acad. Sci. U. S. A. 2012;109:10909–14. doi: 10.1073/pnas.1204917109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weissleder R. A clearer vision for in vivo imaging. Nat. Biotechnol. 2001;19:316–7. doi: 10.1038/86684. [DOI] [PubMed] [Google Scholar]
- 57.Shu X, Royant A, Lin MZ, Aguilera TA, Lev-Ram V, Steinbach PA, Tsien RY. Mammalian expression of infrared fluorescent proteins engineered from a bacterial phytochrome. Science. 2009;324:804–7. doi: 10.1126/science.1168683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Auldridge ME, Forest KT. Bacterial phytochromes: more than meets the light. Crit. Rev. Biochem. Mol. Biol. 2011;46:67–88. doi: 10.3109/10409238.2010.546389. [DOI] [PubMed] [Google Scholar]
- 59.Yu D, Gustafson WC, Han C, Lafaye C, Noirclerc-Savoye M, Ge W-P, Thayer DA, Huang H, Kornberg TB, Royant A, Jan LY, Jan YN, Weiss WA, Shu X. An improved monomeric infrared fluorescent protein for neuronal and tumour brain imaging. Nat. Commun. 2014;5:3626. doi: 10.1038/ncomms4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu D, Baird MA, Allen JR, Howe ES, Klassen MP, Reade A, Makhijani K, Song Y, Liu S, Murthy Z, Zhang S-Q, Weiner OD, Kornberg TB, Jan Y-N, Davidson MW, Shu X. A naturally monomeric infrared fluorescent protein for protein labeling in vivo. Nat. Methods. 2015;12:763–5. doi: 10.1038/nmeth.3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Filonov GS, Piatkevich KD, Ting L-M, Zhang J, Kim K, Verkhusha VV. Bright and stable near-infrared fluorescent protein for in vivo imaging. Nat. Biotechnol. 2011;29:757–61. doi: 10.1038/nbt.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shcherbakova DM, Verkhusha VV. Near-infrared fluorescent proteins for multicolor in vivo imaging. Nat. Methods. 2013;10:751–4. doi: 10.1038/nmeth.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shcherbakova DM, Baloban M, Emelyanov AV, Brenowitz M, Guo P, Verkhusha VV. Bright monomeric near-infrared fluorescent proteins as tags and biosensors for multiscale imaging. Nat. Commun. 2016;7:12405. doi: 10.1038/ncomms12405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rodriguez EA, Tran GN, Gross LA, Crisp JL, Shu X, Lin JY, Tsien RY. A far-red fluorescent protein evolved from a cyanobacterial phycobiliprotein. Nat. Methods. 2016;13:763–9. doi: 10.1038/nmeth.3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kumagai A, Ando R, Miyatake H, Greimel P, Kobayashi T, Hirabayashi Y, Shimogori T, Miyawaki A. A bilirubin-inducible fluorescent protein from eel muscle. Cell. 2013;153:1602–11. doi: 10.1016/j.cell.2013.05.038. [DOI] [PubMed] [Google Scholar]
- 66.To T-L, Piggott BJ, Makhijani K, Yu D, Jan YN, Shu X. Rationally designed fluorogenic protease reporter visualizes spatiotemporal dynamics of apoptosis in vivo. Proc. Natl. Acad. Sci. U. S. A. 2015;112:3338–43. doi: 10.1073/pnas.1502857112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Erapaneedi R, Belousov VV, Schäfers M, Kiefer F. A novel family of fluorescent hypoxia sensors reveal strong heterogeneity in tumor hypoxia at the cellular level. EMBO J. 2016;35:102–13. doi: 10.15252/embj.201592775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Navarro R, Chen L-C, Rakhit R, Wandless TJ. A Novel Destabilizing Domain Based on a Small-Molecule Dependent Fluorophore. ACS Chem. Biol. 2016;11:2101–4. doi: 10.1021/acschembio.6b00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Herbst KJ, Allen MD, Zhang J. Luminescent kinase activity biosensors based on a versatile bimolecular switch. J. Am. Chem. Soc. 2011;133:5676–9. doi: 10.1021/ja1117396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Saito K, Chang Y-F, Horikawa K, Hatsugai N, Higuchi Y, Hashida M, Yoshida Y, Matsuda T, Arai Y, Nagai T. Luminescent proteins for high-speed single-cell and whole-body imaging. Nat. Commun. 2012;3:1262. doi: 10.1038/ncomms2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Takai A, Nakano M, Saito K, Haruno R, Watanabe TM, Ohyanagi T, Jin T, Okada Y, Nagai T. Expanded palette of Nano-lanterns for real-time multicolor luminescence imaging. Proc. Natl. Acad. Sci. U. S. A. 2015;112:4352–6. doi: 10.1073/pnas.1418468112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Suzuki K, Kimura T, Shinoda H, Bai G, Daniels MJ, Arai Y, Nakano M, Nagai T. Five colour variants of bright luminescent protein for real-time multicolour bioimaging. Nat. Commun. 2016;7:13718. doi: 10.1038/ncomms13718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hall MP, Unch J, Binkowski BF, Valley MP, Butler BL, Wood MG, Otto P, Zimmerman K, Vidugiris G, Machleidt T, Robers MB, Benink HA, Eggers CT, Slater MR, Meisenheimer PL, Klaubert DH, Fan F, Encell LP, Wood KV. Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chem. Biol. 2012;7:1848–57. doi: 10.1021/cb3002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chu J, Oh Y, Sens A, Ataie N, Dana H, Macklin JJ, Laviv T, Welf ES, Dean KM, Zhang F, Kim BB, Tang CT, Hu M, Baird MA, Davidson MW, Kay MA, Fiolka R, Yasuda R, Kim DS, Ng H-L, Lin MZ. A bright cyan-excitable orange fluorescent protein facilitates dual-emission microscopy and enhances bioluminescence imaging in vivo. Nat. Biotechnol. 2016;34:760–7. doi: 10.1038/nbt.3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang J, Cumberbatch D, Centanni S, Shi S-Q, Winder D, Webb D, Johnson CH. Coupling optogenetic stimulation with NanoLuc-based luminescence (BRET) Ca(++) sensing. Nat. Commun. 2016;7:13268. doi: 10.1038/ncomms13268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ghosh KK, Burns LD, Cocker ED, Nimmerjahn A, Ziv Y, Gamal AEl, Schnitzer MJ. Miniaturized integration of a fluorescence microscope. Nat. Methods. 2011;8:871–8. doi: 10.1038/nmeth.1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hamers-Casterman C, Atarhouch T, Muyldermans S, Robinson G, Hamers C, Songa EB, Bendahman N, Hamers R. Naturally occurring antibodies devoid of light chains. Nature. 1993;363:446–8. doi: 10.1038/363446a0. [DOI] [PubMed] [Google Scholar]
- 78.Muyldermans S. Nanobodies: natural single-domain antibodies. Annu. Rev. Biochem. 2013;82:775–97. doi: 10.1146/annurev-biochem-063011-092449. [DOI] [PubMed] [Google Scholar]
- 79.Pardon E, Laeremans T, Triest S, Rasmussen SGF, Wohlkönig A, Ruf A, Muyldermans S, Hol WGJ, Kobilka BK, Steyaert J. A general protocol for the generation of Nanobodies for structural biology. Nat. Protoc. 2014;9:674–93. doi: 10.1038/nprot.2014.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rothbauer U, Zolghadr K, Tillib S, Nowak D, Schermelleh L, Gahl A, Backmann N, Conrath K, Muyldermans S, Cardoso MC, Leonhardt H. Targeting and tracing antigens in live cells with fluorescent nanobodies. Nat. Methods. 2006;3:887–9. doi: 10.1038/nmeth953. [DOI] [PubMed] [Google Scholar]
- 81.Dmitriev OY, Lutsenko S, Muyldermans S. Nanobodies as Probes for Protein Dynamics in Vitro and in Cells. J. Biol. Chem. 2016;291:3767–75. doi: 10.1074/jbc.R115.679811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Traenkle B, Emele F, Anton R, Poetz O, Haeussler RS, Maier J, Kaiser PD, Scholz AM, Nueske S, Buchfellner A, Romer T, Rothbauer U. Monitoring interactions and dynamics of endogenous beta-catenin with intracellular nanobodies in living cells. Mol. Cell. Proteomics. 2015;14:707–23. doi: 10.1074/mcp.M114.044016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Panza P, Maier J, Schmees C, Rothbauer U, Söllner C. Live imaging of endogenous protein dynamics in zebrafish using chromobodies. Development. 2015;142:1879–84. doi: 10.1242/dev.118943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Buchfellner A, Yurlova L, Nüske S, Scholz AM, Bogner J, Ruf B, Zolghadr K, Drexler SE, Drexler GA, Girst S, Greubel C, Reindl J, Siebenwirth C, Romer T, Friedl AA, Rothbauer U. A New Nanobody-Based Biosensor to Study Endogenous PARP1 In Vitro and in Live Human Cells. PLoS One. 2016;11:e0151041. doi: 10.1371/journal.pone.0151041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Irannejad R, Tomshine JC, Tomshine JR, Chevalier M, Mahoney JP, Steyaert J, Rasmussen SGF, Sunahara RK, El-Samad H, Huang B, von Zastrow M. Conformational biosensors reveal GPCR signalling from endosomes. Nature. 2013;495:534–8. doi: 10.1038/nature12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ismail S, Gherardi M-J, Froese A, Zanoun M, Gigoux V, Clerc P, Gaits-Iacovoni F, Steyaert J, Nikolaev VO, Fourmy D. Internalized Receptor for Glucose-dependent Insulinotropic Peptide stimulates adenylyl cyclase on early endosomes. Biochem. Pharmacol. 2016;120:33–45. doi: 10.1016/j.bcp.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 87.Schreiber V, Dantzer F, Ame J-C, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat. Rev. Mol. Cell Biol. 2006;7:517–28. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 88.Gross GG, Junge JA, Mora RJ, Kwon H-B, Olson CA, Takahashi TT, Liman ER, Ellis-Davies GCR, McGee AW, Sabatini BL, Roberts RW, Arnold DB. Recombinant probes for visualizing endogenous synaptic proteins in living neurons. Neuron. 2013;78:971–85. doi: 10.1016/j.neuron.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tang JC, Drokhlyansky E, Etemad B, Rudolph S, Guo B, Wang S, Ellis EG, Li JZ, Cepko CL. Detection and manipulation of live antigen-expressing cells using conditionally stable nanobodies. Elife. 2016;5:e15312. doi: 10.7554/eLife.15312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.To T-L, Schepis A, Ruiz-González R, Zhang Q, Yu D, Dong Z, Coughlin SR, Shu X. Rational Design of a GFP-Based Fluorogenic Caspase Reporter for Imaging Apoptosis In Vivo. Cell Chem. Biol. 2016;23:875–82. doi: 10.1016/j.chembiol.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Calebiro D, Nikolaev VO, Gagliani MC, de Filippis T, Dees C, Tacchetti C, Persani L, Lohse MJ. Persistent cAMP-signals triggered by internalized G-protein-coupled receptors. PLoS Biol. 2009;7:e1000172. doi: 10.1371/journal.pbio.1000172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ferrandon S, Feinstein TN, Castro M, Wang B, Bouley R, Potts JT, Gardella TJ, Vilardaga J-P. Sustained cyclic AMP production by parathyroid hormone receptor endocytosis. Nat. Chem. Biol. 2009;5:734–42. doi: 10.1038/nchembio.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tsvetanova NG, von Zastrow M. Spatial encoding of cyclic AMP signaling specificity by GPCR endocytosis. Nat. Chem. Biol. 2014;10:1061–5. doi: 10.1038/nchembio.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Feinstein TN, Yui N, Webber MJ, Wehbi VL, Stevenson HP, King JD, Hallows KR, Brown D, Bouley R, Vilardaga J-P. Noncanonical control of vasopressin receptor type 2 signaling by retromer and arrestin. J. Biol. Chem. 2013;288:27849–60. doi: 10.1074/jbc.M112.445098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Thomsen ARB, Plouffe B, Cahill TJ, Shukla AK, Tarrasch JT, Dosey AM, Kahsai AW, Strachan RT, Pani B, Mahoney JP, Huang L, Breton B, Heydenreich FM, Sunahara RK, Skiniotis G, Bouvier M, Lefkowitz RJ. GPCR-G Protein-β-Arrestin Super-Complex Mediates Sustained G Protein Signaling. Cell. 2016;166:907–19. doi: 10.1016/j.cell.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.DiPilato LM, Cheng X, Zhang J. Fluorescent indicators of cAMP and Epac activation reveal differential dynamics of cAMP signaling within discrete subcellular compartments. Proc. Natl. Acad. Sci. U. S. A. 2004;101:16513–8. doi: 10.1073/pnas.0405973101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Violin JD, DiPilato LM, Yildirim N, Elston TC, Zhang J, Lefkowitz RJ. beta2-adrenergic receptor signaling and desensitization elucidated by quantitative modeling of real time cAMP dynamics. J. Biol. Chem. 2008;283:2949–61. doi: 10.1074/jbc.M707009200. [DOI] [PubMed] [Google Scholar]
- 98.Galés C, Van Durm JJJ, Schaak S, Pontier S, Percherancier Y, Audet M, Paris H, Bouvier M. Probing the activation-promoted structural rearrangements in preassembled receptor-G protein complexes. Nat. Struct. Mol. Biol. 2006;13:778–86. doi: 10.1038/nsmb1134. [DOI] [PubMed] [Google Scholar]
- 99.Cahill TJ, Thomsen ARB, Tarrasch JT, Plouffe B, Nguyen AH, Yang F, Huang L-Y, Kahsai AW, Bassoni DL, Gavino BJ, Lamerdin JE, Triest S, Shukla AK, Berger B, Little J, Antar A, Blanc A, Qu C-X, Chen X, Kawakami K, Inoue A, Aoki J, Steyaert J, Sun J-P, Bouvier M, Skiniotis G, Lefkowitz RJ. Distinct conformations of GPCR-β-arrestin complexes mediate desensitization, signaling, and endocytosis. Proc. Natl. Acad. Sci. U. S. A. 2017;114:2562–2567. doi: 10.1073/pnas.1701529114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kumari P, Srivastava A, Banerjee R, Ghosh E, Gupta P, Ranjan R, Chen X, Gupta B, Gupta C, Jaiman D, Shukla AK. Functional competence of a partially engaged GPCR-β-arrestin complex. Nat. Commun. 2016;7:13416. doi: 10.1038/ncomms13416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jean-Alphonse FG, Wehbi VL, Chen J, Noda M, Taboas JM, Xiao K, Vilardaga J-P. β2-adrenergic receptor control of endosomal PTH receptor signaling via Gβγ. Nat. Chem. Biol. 2017;13:259–261. doi: 10.1038/nchembio.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Broussard GJ, Liang R, Tian L. Monitoring activity in neural circuits with genetically encoded indicators. Front. Mol. Neurosci. 2014;7:97. doi: 10.3389/fnmol.2014.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lin MZ, Schnitzer MJ. Genetically encoded indicators of neuronal activity. Nat. Neurosci. 2016;19:1142–53. doi: 10.1038/nn.4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Heim N, Griesbeck O. Genetically encoded indicators of cellular calcium dynamics based on troponin C and green fluorescent protein. J. Biol. Chem. 2004;279:14280–6. doi: 10.1074/jbc.M312751200. [DOI] [PubMed] [Google Scholar]
- 105.Mank M, Santos AF, Direnberger S, Mrsic-Flogel TD, Hofer SB, Stein V, Hendel T, Reiff DF, Levelt C, Borst A, Bonhoeffer T, Hübener M, Griesbeck O. A genetically encoded calcium indicator for chronic in vivo two-photon imaging. Nat. Methods. 2008;5:805–11. doi: 10.1038/nmeth.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Palmer AE, Giacomello M, Kortemme T, Hires SA, Lev-Ram V, Baker D, Tsien RY. Ca2+ indicators based on computationally redesigned calmodulin-peptide pairs. Chem. Biol. 2006;13:521–30. doi: 10.1016/j.chembiol.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 107.Mank M, Griesbeck O. Genetically encoded calcium indicators. Chem. Rev. 2008;108:1550–64. doi: 10.1021/cr078213v. [DOI] [PubMed] [Google Scholar]
- 108.Rose T, Goltstein PM, Portugues R, Griesbeck O. Putting a finishing touch on GECIs. Front. Mol. Neurosci. 2014;7:88. doi: 10.3389/fnmol.2014.00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chen T-W, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, Looger LL, Svoboda K, Kim DS. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ziv Y, Burns LD, Cocker ED, Hamel EO, Ghosh KK, Kitch LJ, El Gamal A, Schnitzer MJ. Long-term dynamics of CA1 hippocampal place codes. Nat. Neurosci. 2013;16:264–6. doi: 10.1038/nn.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tian L, Hires SA, Mao T, Huber D, Chiappe ME, Chalasani SH, Petreanu L, Akerboom J, McKinney SA, Schreiter ER, Bargmann CI, Jayaraman V, Svoboda K, Looger LL. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat. Methods. 2009;6:875–81. doi: 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhuang J, Ng L, Williams D, Valley M, Li Y, Garrett M, Waters J. An extended retinotopic map of mouse cortex. Elife. 2017;6:e18372. doi: 10.7554/eLife.18372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Baker BJ, Lee H, Pieribone VA, Cohen LB, Isacoff EY, Knopfel T, Kosmidis EK. Three fluorescent protein voltage sensors exhibit low plasma membrane expression in mammalian cells. J. Neurosci. Methods. 2007;161:32–8. doi: 10.1016/j.jneumeth.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 114.Murata Y, Iwasaki H, Sasaki M, Inaba K, Okamura Y. Phosphoinositide phosphatase activity coupled to an intrinsic voltage sensor. Nature. 2005;435:1239–43. doi: 10.1038/nature03650. [DOI] [PubMed] [Google Scholar]
- 115.Kralj JM, Hochbaum DR, Douglass AD, Cohen AE. Electrical spiking in Escherichia coli probed with a fluorescent voltage-indicating protein. Science. 2011;333:345–8. doi: 10.1126/science.1204763. [DOI] [PubMed] [Google Scholar]
- 116.Storace D, Sepehri Rad M, Kang B, Cohen LB, Hughes T, Baker BJ. Toward Better Genetically Encoded Sensors of Membrane Potential. Trends Neurosci. 2016;39:277–89. doi: 10.1016/j.tins.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yang HH, St-Pierre F. Genetically Encoded Voltage Indicators: Opportunities and Challenges. J. Neurosci. 2016;36:9977–89. doi: 10.1523/JNEUROSCI.1095-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gong Y, Huang C, Li JZ, Grewe BF, Zhang Y, Eismann S, Schnitzer MJ. High-speed recording of neural spikes in awake mice and flies with a fluorescent voltage sensor. Science. 2015;350:1361–6. doi: 10.1126/science.aab0810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bayraktar H, Fields AP, Kralj JM, Spudich JL, Rothschild KJ, Cohen AE. Ultrasensitive measurements of microbial rhodopsin photocycles using photochromic FRET. Photochem. Photobiol. 2012;88:90–7. doi: 10.1111/j.1751-1097.2011.01011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Shaner NC, Lambert GG, Chammas A, Ni Y, Cranfill PJ, Baird MA, Sell BR, Allen JR, Day RN, Israelsson M, Davidson MW, Wang J. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat. Methods. 2013;10:407–9. doi: 10.1038/nmeth.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ahrens MB, Orger MB, Robson DN, Li JM, Keller PJ. Whole-brain functional imaging at cellular resolution using light-sheet microscopy. Nat. Methods. 2013;10:413–20. doi: 10.1038/nmeth.2434. [DOI] [PubMed] [Google Scholar]
- 122.Prevedel R, Yoon Y-G, Hoffmann M, Pak N, Wetzstein G, Kato S, Schrödel T, Raskar R, Zimmer M, Boyden ES, Vaziri A. Simultaneous whole-animal 3D imaging of neuronal activity using light-field microscopy. Nat. Methods. 2014;11:727–30. doi: 10.1038/nmeth.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tsien RY. Very long-term memories may be stored in the pattern of holes in the perineuronal net. Proc. Natl. Acad. Sci. U. S. A. 2013;110:12456–61. doi: 10.1073/pnas.1310158110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Fosque BF, Sun Y, Dana H, Yang C-T, Ohyama T, Tadross MR, Patel R, Zlatic M, Kim DS, Ahrens MB, Jayaraman V, Looger LL, Schreiter ER. Neural circuits. Labeling of active neural circuits in vivo with designed calcium integrators. Science. 2015;347:755–60. doi: 10.1126/science.1260922. [DOI] [PubMed] [Google Scholar]
- 125.McKinney SA, Murphy CS, Hazelwood KL, Davidson MW, Looger LL. A bright and photostable photoconvertible fluorescent protein. Nat. Methods. 2009;6:131–3. doi: 10.1038/nmeth.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ragan T, Kadiri LR, Venkataraju KU, Bahlmann K, Sutin J, Taranda J, Arganda-Carreras I, Kim Y, Seung HS, Osten P. Serial two-photon tomography for automated ex vivo mouse brain imaging. Nat. Methods. 2012;9:255–8. doi: 10.1038/nmeth.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tomer R, Ye L, Hsueh B, Deisseroth K. Advanced CLARITY for rapid and high-resolution imaging of intact tissues. Nat. Protoc. 2014;9:1682–97. doi: 10.1038/nprot.2014.123. [DOI] [PMC free article] [PubMed] [Google Scholar]